Abstract
Synthesis of the osmolyte glucosylglycerol (GG) in the marine cyanobacterium Synechococcus sp. strain PCC 7002 was characterized. The ggpS gene, which encodes the key enzyme (GG-phosphate synthase [GgpS]) in GG biosynthesis, was cloned by using PCR. A 2,030-bp DNA sequence which contained one open reading frame (ORF) was obtained. The protein deduced from this ORF exhibited 85% similarity to the GgpS of the freshwater cyanobacterium Synechocystis sp. strain PCC 6803. The function of the protein was confirmed by generating a ggpS null mutant, which was not able to synthesize GG and thus exhibited a salt-sensitive phenotype. Expression of the ggpS gene was analyzed in salt-shocked cells by performing Northern blot and immunoblot experiments. While almost no expression was detected in cells grown in low-salt medium, immediately after a salt shock the amounts of ggpS mRNA and GgpS protein increased up to 100-fold. The finding that salt-induced expression occurred was confirmed by measuring enzyme activities, which were negligible in control cells but clearly higher in salt-treated Synechococcus sp. cells. The salt-induced increase in GgpS activity could be inhibited by adding chloramphenicol, while in protein extracts of the freshwater cyanobacterium Synechocystis sp. strain PCC 6803 a constitutive, high level of enzyme activity that was not affected by chloramphenicol was found. A comparison of GG accumulation in the two cyanobacteria revealed that in the marine strain osmolyte synthesis seemed to be regulated mainly by transcriptional control, whereas in the freshwater strain control seemed to be predominantly posttranslational.
Accumulation of compatible solutes is a universal process during cellular acclimation to environments with low water potentials. The importance of this process for successful acclimation to high-salinity conditions has been demonstrated by two kinds of experiments. (i) Mutations that affect genes which encode osmolyte-synthesizing enzymes lead in all cases to significant reductions in both salt tolerance and osmotolerance; therefore, these genes are essential for survival at low water potentials. (ii) Decreases in salt resistance can be phenotypically complemented by feeding mutant cells compatible solutes, since most bacteria express active osmolyte transport systems. Particularly in heterotrophic bacteria, uptake of osmolytes is often preferred over de novo synthesis of these substances; this results in repression of the synthesis of compatible solutes, when such compounds are present in the medium (4). On the basis of their chemical structures, compatible organic solutes can be divided into the following four groups: carbohydrates (e.g., trehalose), heterosides (e.g., glucosylglycerol [GG]), amino acids (e.g., proline) and their derivatives (e.g., glycinebetaine and ectoine), and polyols (e.g., glycerol and mannitol) (5).
The cyanobacteria represent an ancient group of eubacteria in which oxygen-evolving photosynthesis is believed to have arisen. These photoautotrophic bacteria are found in almost all habitats, including waters that contain salt at very low to saturating concentrations. The salt-induced osmolyte spectrum of these organisms has been investigated with more than 100 different strains. This has led to classification of the cyanobacteria into three groups on the basis of their salt tolerance characteristics and dominant compatible solutes; strains that tolerate the lowest salt levels accumulate the disaccharides sucrose and trehalose, moderately halotolerant strains synthesize the heteroside GG, and halophilic strains synthesize the quaternary ammonium compounds glycinebetaine and glutamatebetaine (19). Accumulation of GG has been characterized by using Synechocystis sp. strain PCC 6803 (9), which was initially isolated from a freshwater pond (21). After growth at higher salt concentrations, this strain synthesizes mainly GG and traces of sucrose, which allows it to tolerate up to 1.2 M NaCl (20). The biosynthetic pathway begins with ADP-glucose and glycerol 3-phosphate, which are used by the GG-phosphate synthase (GgpS), and proceeds via the intermediate GG-phosphate, which is dephosphorylated to GG by the GG-phosphate phosphatase (GgpP) (7). These enzyme activities have been found to depend on enhanced salt concentrations in assay mixtures. GG synthesis in vitro can be activated simply by adding salt during extraction of proteins from cells grown in low-salt medium and, conversely, can be inhibited in extracts obtained from cells acclimated to high-salt medium by omitting salt from the homogenization and assay buffers. Besides NaCl, other salts have also been found to effectively promote activation of the GG-synthesizing enzymes in Synechocystis sp. strain PCC 6803 extracts (25). Therefore, synthesis of GG seems to be mainly posttranslationally regulated in Synechocystis sp. strain PCC 6803. The genes that encode the GG biosynthetic enzymes in this cyanobacterial strain have been identified and have been found to exhibit slightly enhanced expression after salt shock (8, 14).
The occurrence of direct activation of preformed GG-synthesizing enzymes in the freshwater organism Synechocystis sp. strain PCC 6803 seems to be favorable from an ecological point of view. In salt-shocked cells of this strain, protein synthesis is nearly completely inhibited, and thus de novo synthesis of enzyme proteins is prevented (10). Direct, salt-dependent activation of the enzyme activities that are involved in synthesis of organic osmolytes also takes place in other organisms. For instance, in Escherichia coli the activity of trehalose-6-phosphate synthase (TPS) is stimulated by high concentrations of K+, as well as other ions, and is further increased by transcriptional induction of de novo synthesis (6). Salt-induced trehalose synthesis has also been found to be salt dependent in the bacterium Ectothiorhodospira halochloris (13). Floridoside phosphate synthase, the key enzyme in osmotically regulated floridoside (galactosylglycerol) synthesis, is stimulated by ammonium sulfate and other salts (15). In addition to osmolyte synthesis, the activity of the osmolyte transporter BetP in Corynebacterium glutamicum is also directly influenced by the external salt concentration (18). However, the molecular mechanisms that lead to these activation processes and are directly dependent upon the salt concentration are not known.
Moderately halotolerant cyanobacteria accumulate GG as their main osmolyte, and these organisms include strains which originate from both freshwater and marine systems (18). One might expect that despite the fact that the organisms use the same kind of osmolyte, there might be differences in the genes and proteins involved in the process and particularly in regulation and that these differences would reflect the adaptations of the organisms to environments in which the salt concentrations are different. The aims of the present work were to analyze GG formation in a cyanobacterial strain isolated from a marine environment and to compare the results obtained with the results obtained for the freshwater organism Synechocystis sp. strain PCC 6803. The marine strain used was Synechococcus sp. strain PCC 7002, which was isolated from coastal water (21) and accumulates GG as its main osmolyte (20). Despite the fact that they use the same osmolyte, these Synechocystis and Synechococcus strains are not as closely related as members of the genera Escherichia and Salmonella are. On the basis of their morphological features, they were classified into two different sections (21), and the distance between the genera Synechocystis and Synechococcus has been confirmed by comparing 16S rRNA sequences (17). The ggpS gene, which encodes GgpS, the key enzyme in GG synthesis, was cloned from the marine strain and sequenced. While the sequences of the GgpS proteins were very similar, differences in the regulation of these proteins were found.
MATERIALS AND METHODS
Strains and culture conditions.
A derivative of Synechocystis sp. strain PCC 6803 with enhanced transforming capacity was used in all of the experiments and was obtained from S. Shestakov (Moscow State University, Moscow, Russia). Synechococcus sp. strain PCC 7002 was obtained from the Pasteur Culture Collection, Paris, France (21). Axenic cells were cultured on plates at 30°C with constant illumination by using mineral medium C (11). Transformants were initially selected on media containing 5 μg of kanamycin (Sigma) per ml, while clones were segregated and mutants were cultivated in the presence of 50 or 100 μg of kanamycin per ml. For physiological characterization, axenic batch cultures of the cyanobacteria were grown photoautotrophically. Cultures were bubbled with CO2-enriched air (5%, vol/vol) at 29°C with constant illumination (175 μmol m−2 s−1) by using a modified mineral medium (1) in which KNO3 and NaCl were replaced by equimolar amounts of NaNO3 and KCl, respectively. Some growth experiments were performed without CO2 gassing in shaken Erlenmeyer flasks at 29°C with constant illumination (65 μmol m−2 s−1) by using medium BG11 (21). All media used for cultivation of Synechococcus sp. strain PCC 7002 were supplemented with vitamin B12 (10 μg ml−1). In order to perform salt shock experiments, NaCl was added to the basal media to obtain the desired concentrations. In some experiments cellular protein synthesis was inhibited by adding chloramphenicol (final concentration, 100 μg ml−1) directly to the medium. E. coli TG1 was used for routine DNA manipulations and was cultivated in Luria-Bertani medium at 37°C (23).
DNA manipulations.
Synechococcus sp. strain 7002 total DNA was isolated from lysozyme-treated cells and was purified by using the detergent cetyltrimethyleammonium bromide as described by Hagemann et al. (8). All of the other methods used, such as the methods used for transformation of E. coli, ligation, and restriction analysis (restriction enzymes were obtained from New England Biolabs), were standard methods (23). Plasmid DNA was isolated by using a QIAprep Spin Miniprep kit (Qiagen). DNA probes were labelled with digoxigenin for Southern hybridization by using a PCR DIG probe synthesis kit (Boehringer, Mannheim, Germany). Sequencing was performed by using the dideoxy chain termination method; a Thermo Sequenase fluorescent labelled primer cycle sequencing kit was used with 7-deaza-dGTP (Amersham Life Science). Universal primers, which were fluorescently labelled with IRD 800 (MWG Biotech), were used for sequencing. So that both strands could be completely sequenced, fragments cloned into pGEMT (Promega) were selectively shortened by using a double-stranded nested deletion kit (Pharmacia). Computer analyses of DNA and protein sequences were performed with the BLAST program (2), Clustal X (version 1.64 b), Align-Plus-V2.0 (Scientific & Educational Software), and Clone-Manager-V4.1 (Scientific & Educational Software) software packages.
Cloning of ggpS.
We used a PCR strategy in which degenerate primers were used. To design primers, the amino acid sequence of GgpS from Synechocystis sp. strain PCC 6803 was compared to the amino acid sequences of TPS from bacteria and yeasts in order to identify well-conserved portions. We selected four boxes (Fig. 1) that yielded two primers which were specific for each end of the gene. For conversion of the genetic code, the codon usage of Synechocystis sp. strain PCC 6803 was used, and this led to four primers (F5A, F5B, F3A, and F3B) which contained only five wobble positions (Table 1). The following primer combinations were used: F5A and F3A, F5A and F3B, F5B and F3A, and F5B and F3B. We predicted that these combinations would produce fragments about 800, 400, 700, and 500 bp long, respectively. The primers were used for PCR performed with the Elongase enzyme mixture (Life Technologies), and chromosomal DNA of Synechococcus sp. strain PCC 7002 was used as the template under relatively low specificity conditions (annealing temperature, 40°C; Mg2+ concentration doubled). PCR fragments were evaluated by performing Southern hybridization with digoxigenin-labelled ggpS probe from Synechocystis sp. strain PCC 6803 and by partial sequencing after the fragments were cloned into pGEM-T (Promega). A 1.4-kb fragment obtained with primers F5B and F3B contained the 5′ end of the ggpS gene from Synechococcus sp. In order to obtain the sequence of the complete gene, we designed additional primers. For the 5′ end, a specific primer (primer G1) (Table 1) was deduced from the sequence already determined. For the unknown 3′ end, we designed primers by using the highly iterated palindrome (22) sequences, which occur frequently in the genomes of several cyanobacterial strains. Using a combination of the anchored primer HIP-A and the specific primer G1, we obtained a 700-bp fragment which was not synthesized when only primer HIP-A was used. This fragment was cloned into pGEM-T (Promega) and completely sequenced on both strands. It contained the complete 3′ part of the putative ggpS gene of Synechococcus sp. strain PCC 7002.
FIG. 1.
Partial amino acid sequence alignment of the GgpS of Synechococcus sp. strain PCC 7002 (line 1), the GgpS of Synechocystis sp. strain PCC 6803 (line 2), and functionally related TPS of Mycobacterium tuberculosis (OtsA; accession no. O06353) (line 3), Aspergillus niger (TpsA; accession no. Q00075) (line 4), Saccharomyces cerevisiae (Tps1; accession no. Q00764) (line 5), E. coli (OtsA; accession no. P31677) (line 6), and Schizosaccharomyces pombe (Tps1; accession no. P40387) (line 7). The boxes indicate conserved sequence elements used to design the oligonucleotide primers which were used to clone the ggpS gene of Synechococcus sp. strain PCC 7002. The lines above the sequences indicate parts of the GgpS proteins that exhibit low levels of similarity (less than 40%, compared to an average level of similarity of 85% for the complete sequence). Amino acid residues identical to the residues of Synechococcus sp. strain PCC 7002 GgpS are shaded, while the dashes indicate breaks in the amino acid sequences introduced to obtain maximal levels of similarity.
TABLE 1.
Primers used to amplify DNA fragments containing the ggpS gene of Synechococcus sp. strain PCC 7002 and to generate gene-specific hybridization probes
| Primer | Sequencea | Enzyme |
|---|---|---|
| F5Ab | 5′-TGGGTGCACGATTACCACYTGATG-3′ | |
| F5Bb | 5′-CACACCCCCTTTCCCTCC-3′ | |
| F3Ab | 5′-GTATTCTTTGGCCACCARGTT-3′ | |
| F3Bb | 5′-CCTTTCACGTAATCCASCCG-3′ | |
| HIP-Ab | 5′-GGCGATCGCCA-3′ | |
| HIP-Cb | 5′-GGCGATCGCCC-3′ | |
| HIP-Gb | 5′-GGCGATCGCCG-3′ | |
| HIP-Tb | 5′-GGCGATCGCCT-3′ | |
| G1b | 5′-CTAGGCAGCAACAAGCTG-3′ | |
| cocPK5_1c | 5′-CGGGATCCGGAGGTCTTAATGAAATC-3′ | BamHI |
| cocPK3_1c | 5′-CGGGATCCGCAAAAAAACGACCTA-3′ | BamHI |
| 16SrRNA27fcd | 5′-AGAGTTTGATCMTGGCTCAG-3′ | |
| 16SrRNA1525cd | 5′-AAGGAGGTGWTCCARCC-3′ |
Generation of insertion mutants.
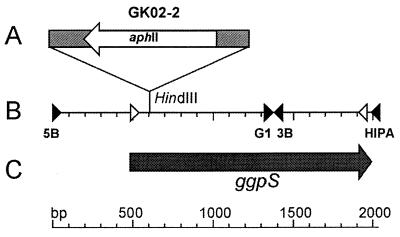
To generate a Synechococcus sp. strain PCC 7002 ggpS mutant, the aphII gene (conferring kanamycin resistance) from pUC4K (26) was inserted into the unique HindIII restriction site of the ggpS after treatment with S1 nuclease (Table 2). Constructs harboring the aphII gene in the same transcription direction or in the opposite transcription direction relative to ggpS were selected. The plasmids obtained, pG16::Km1 and pG16::Km2 (Table 2), were transformed into wild-type cells of Synechococcus sp. strain PCC 7002. Since these constructs do not replicate in Synechococcus sp. strain PCC 7002, continued selection in the presence of kanamycin depends on integration of the aphII gene into the chromosome. After cultivation for several generations with increasing concentrations of the antibiotic, mutants impaired in ggpS were isolated. The lesions in the DNA of these mutants were characterized by performing PCR experiments with primers cocPK5_1 and cocPK3_1 (Table 1), which overlap the start and stop codons of the ggpS gene.
TABLE 2.
Plasmids and Synechococcus sp. strain PCC 7002 mutants used and constructed in this studya
| Plasmid or mutant | Description |
|---|---|
| Plasmids | |
| pG16 | pGEM-T containing a 1.4-kb Synechococcus fragment with 0.9 kb of the 5′ end of the ggpS gene obtained with primers F5B and F3B; final size, 4.4 kb |
| pG2-5 | pGEM-T containing a 0.7-kb Synechococcus fragment representing the 3′ end of the ggpS gene obtained with primers G1 and HIP-A; final size, 3.7 kb |
| pG16::Km1 | pG16 containing an inactivated ggpS gene (the aphII gene is inserted at the unique HindIII site in the same transcription direction as ggpS); final size, 5.65 kb |
| pG16::Km2 | pG16 containing an inactivated ggpS gene (the aphII gene is inserted at the unique HindIII site in the transcription direction opposite that of ggpS); final size, 5.65 kb |
| Mutants | |
| GK02-1 | Salt-sensitive Synechococcus mutant impaired in ggpS obtained after transformation of the wild type with pG16::Km1 |
| GK02-2 | Salt-sensitive Synechococcus mutant impaired in ggpS obtained after transformation of the wild type with pG16::Km2 |
See Fig. 2.
Expression of the ggpS gene.
Northern blot hybridization experiments were performed in order to estimate the steady-state level of ggpS mRNA. Total RNA was isolated from 10-ml portions of cells cultivated for different times in high-salt medium. The cells were taken directly from the cultures, harvested by centrifugation (4,000 × g, 10 min, 2°C), and immediately frozen and stored at −80°C. RNA was extracted with a High Pure RNA isolation kit (Boehringer). The methods used for separating RNA, blotting, and hybridization were described in detail by Hagemann et al. (8). A ggpS-specific DNA hybridization probe was obtained after PCR amplification of its coding sequence (Table 1). The DNA was labelled with [α-32P]dATP (Amersham Buchler) by using a HexaLabel DNA labelling kit (MBI Fermentas). Hybridization signals were recorded and quantified with a phosphoimager (model BAS1000; Fuji). In order to correct the quantitative data for variations in RNA loading, all calculations were made by using the relative intensities of hybridization signals obtained after a radiolabelled 16S ribosomal DNA probe was applied (Table 1 shows the primers used) to the same filters. The amount of GgpS protein was estimated by performing immunoblot experiments with an antibody specific for the Synechocystis sp. strain PCC 6803 GgpS. Rabbit antibody (Eurogentec) was raised against purified protein obtained after overexpression of the Synechocystis sp. strain PCC 6803 ggpS gene in E. coli (14). The protein was isolated and separated by polyacrylamide gel electrophoresis as described by Hagemann et al. (8). Binding of the antibody was detected with an enhanced chemiluminescence kit (Amersham Buchler). The chemiluminescent signals on X-ray films were quantified by videodensitometry with Bioprofil 1D software (Vilbert Lourmat).
Physiological characterization.
The low-molecular-mass carbohydrate content was analyzed by high-performance liquid chromatography (24). The activities of GgpS and GgpP were determined in vitro by using the substrate [14C]glycerol 3-phosphate (Amersham Buchler) and buffers containing no NaCl or an elevated level of NaCl (342 mM). The reaction products were separated by thin-layer chromatography. In vitro determination of these enzyme activities has been described in detail by Hagemann and Erdmann (7). Radioactive spots were quantified by using a phosphoimager (model BAS1000; Fuji). Protein concentrations were measured by the Bradford method (3). Growth and cell density were monitored by determining the optical densities at 750 nm of diluted cyanobacterial suspensions with a spectrophotometer (model U2000; Hitachi).
Nucleotide sequence accession number.
The coding sequence of the Synechococcus ggpS gene has been deposited in the EMBL database under accession no. AJ006298.
RESULTS
Cloning of ggpS.
In order to compare the regulation of GG accumulation in a marine cyanobacterium at the transcriptional and posttranslational levels, the ggpS gene from Synechococcus sp. strain PCC 7002 was cloned by using a PCR strategy. None of the fragment sizes obtained with the degenerate primers (Table 1) matched the size predicted on the basis of the binding of the primer combinations to the ggpS gene of Synechocystis sp. strain PCC 6803. However, after the fragments were analyzed by performing Southern hybridization with a digoxigenin-labelled ggpS probe from Synechocystis sp. strain PCC 6803 and by performing a partial DNA sequence analysis, we obtained a fragment about 1.4 kb long with primers F5B and F3B that was found to contain at least part of the ggpS gene from Synechococcus sp. strain PCC 7002. Only this fragment gave a clear hybridization signal in the Southern blot experiment (data not shown), and the partial DNA sequence of its 3′ end, obtained after cloning into pGEM-T (leading to pG16 [Table 2]), was very similar to the sequences of the ggpS gene from Synechocystis sp. strain PCC 6803 and the otsA genes from other bacteria. After a collection of shorter inserts of pG16 was obtained, the entire sequence of both strands of the 1.4-kb fragment could be obtained. The sequence data showed that the fragment was composed of 488 bp of the upstream noncoding region covering the probable promoter region and 917 bp of the 5′ part of the putative ggpS gene of Synechococcus sp. strain PCC 7002. Therefore, primer F3B bound to the expected sequence of the ggpS gene, while primer F5B did not anneal inside the gene but annealed by chance about 900 bp 5′ to it. Using other primers (G1 and HIP-A [Table 1]), we obtained a 700-bp fragment and cloned it into pGEM-T (leading to pG2-5 [Table 2]), which contained the 3′ end of the ggpS gene of Synechococcus sp. strain PCC 7002. We obtained a 2,030-bp DNA sequence that contained one large open reading frame (ORF) that was 1.5-kb long (Fig. 2). On the basis of this ORF, a protein containing 500 amino acid residues was deduced and this protein was very similar to GgpS from Synechocystis sp. strain PCC 6803 (76% identity, 85% similarity) and TPS from other organisms (Fig. 1).
FIG. 2.
Schematic drawing showing the integration site of the aphII gene in mutant GK02-2 (A), restriction and primer binding sites (B), and the protein-encoding region of the chromosomal site harboring the ggpS gene in Synechococcus sp. strain PCC 7002 (C). Arrow labelled aphII, inserted aphII gene cassette; solid triangles, primer binding sites used to clone the complete ggpS gene; open triangles, primer binding sites used to generate a gene-specific probe.
Mutation of ggpS.
As predicted from the PCR results, the sequence data indicated that the deduced protein represented GGPS. In order to confirm this, the ggpS gene was insertionally inactivated by interposon mutagenesis (Fig. 2). To do this, an aphII resistance gene cassette conferring kanamycin resistance was introduced in the same transcription direction and also in the opposite transcription direction relative to ggpS in a unique restriction site. When the DNA of two mutant of the clones obtained (GK02-1 and GK02-2) (Table 2) were used, PCR analyses revealed fragments that were larger than the fragments obtained with wild-type DNA (data not shown). The size differences corresponded exactly to the increase in size expected if an aphII gene cassette was inserted (1.2 kb). Furthermore, in all of the PCR in which mutant DNA was used, the wild-type fragment was completely absent. The results of the PCR analyses indicated that the plasmid constructs used for transformation of Synechococcus sp. strain PCC 7002 in order to obtain the mutants were correctly integrated by double homologous recombination and that the ggpS::aphII allele completely replaced the wild-type copy of the ggpS gene.
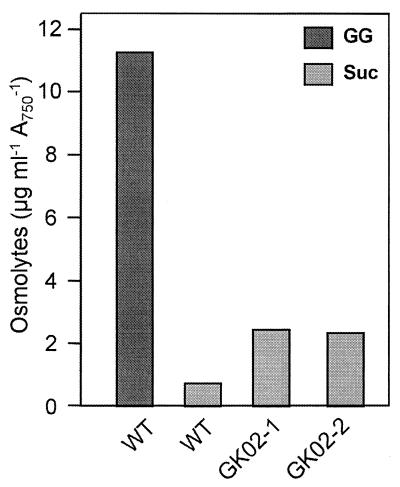
As expected, the mutant clones (50 independent transformants were tested) exhibited decreased salt tolerance when they were cultivated on solid media containing different NaCl concentrations. While wild-type cells were able to grow on media containing more than 684 mM NaCl (4% NaCl), the mutant clones lysed at moderate levels of salinity (513 mM NaCl [3% NaCl]). This was observed with mutant clones in which the aphII gene was introduced in the same transcription direction as the ggpS gene, as well as with clones in which the gene was introduced in the opposite transcription direction. The osmolyte compositions of the mutant cells were compared to the osmolyte composition of the wild type after cultivation in the presence of 324 mM NaCl (Fig. 3). In wild-type cells high levels of GG were detected by high-performance liquid chromatography. In addition, small amounts of sucrose (a second osmolyte) were present. However, GG was not detected in 324 mM NaCl-grown cells of ggpS null mutants GK02-1 and GK02-2. Instead of GG, a larger amount of sucrose accumulated. Compared to wild-type cells, about four times more sucrose was detected; however, this sucrose could not compensate for the smaller amount of total osmolytes resulting from the complete absence of GG. These physiological data are consistent with the expected function of the ggpS gene, which apparently encodes GGPS, the key enzyme in GG synthesis.
FIG. 3.
GG and sucrose (Suc) accumulation in cells of the wild type and two clones of the ggpS null mutant of Synechococcus sp. strain PCC 7002 salt shocked for 12 h with 324 mM NaCl. A750, absorbance at 750 nm; WT, wild type.
Regulation of ggpS expression.
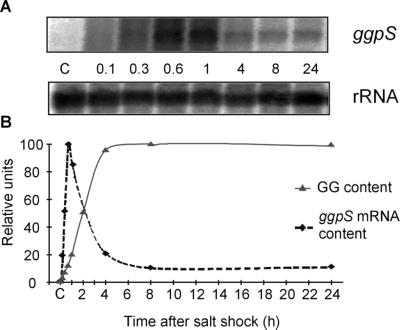
After we cloned the ggpS gene, it was possible to study expression of this gene at the transcriptional level by performing Northern blot hybridization experiments. After hybridization with a gene-specific probe, a major transcript about 2,000 nucleotides long was observed (Fig. 4A). Almost no hybridization signal was obtained with cells grown in the presence of low NaCl concentrations, while cells shocked for 10 min with 684 mM NaCl produced a clear hybridization signal. Hybridization data were quantified on the basis of levels of hybridization to 16S rRNA, which were equivalent for low-salt-grown and salt-shocked cells (Fig. 4A). The steady-state amount of the ggpS transcript increased very quickly in salt-shocked Synechococcus sp. strain PCC 7002 cells and reached a maximum 40 min after the salt was added. The transcript level decreased during the following hours. Nevertheless, a significantly higher ggpS mRNA level was observed in completely salt-acclimated cells, although this level was about 10-fold lower than the peak content. For comparison, Fig. 4B shows the increase in GG content for cells shocked with 684 mM NaCl. After a short lag period, a rapid, linear increase in GG concentration was observed. The saturation level was reached about 5 h after the salt shock.
FIG. 4.
Northern blot hybridization experiments to detect salt stress-induced alterations in the steady-state mRNA levels of the ggpS gene of Synechococcus sp. strain PCC 7002 after 684 mM NaCl was added to control cells for different times. (A) Hybridization signals obtained with a ggpS gene (transcript size, about 2,000 nucleotides) and a 16S rRNA-specific probe, which was used as a control for RNA loading. Probes were generated by PCR (the primers used are shown in Table 1) and were labelled by random priming with [32P-α]dATP. (B) Quantitative estimates of relative transcript levels (ggpS relative to 16S rRNA) as determined with a phosphoimager. The maximum signal was set to a level of 100. For comparison, accumulation of GG in Synechococcus cells shocked with 684 mM NaCl is shown.
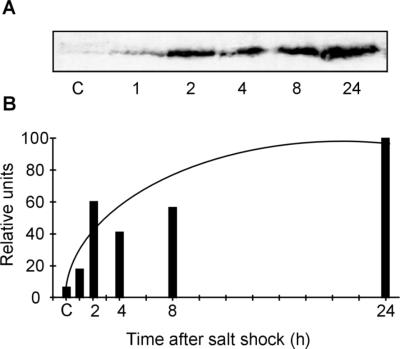
We estimated the amount of GgpS protein in protein extracts obtained from salt-shocked cells of Synechococcus sp. strain PCC 7002 by using an antibody specific for the GgpS protein from Synechocystis sp. strain PCC 6803 in immunoblot experiments (Fig. 5A). This antibody recognized a protein of about 56 kDa, a size which is similar to the predicted size of the protein deduced on the basis of the deduced sequence of the Synechococcus sp. strain PCC 7002 ggpS gene. Only a very faint signal was observed in protein extracts obtained from cells grown in basal medium. The protein level clearly increased in salt-shocked cells. After 24 h the GGPS protein content increased about 20-fold (Fig. 5B). The immunoblot experiments, as well as the Northern blot hybridization experiments, clearly indicated that expression of the ggpS gene is significantly upregulated in salt-shocked Synechococcus sp. strain PCC 7002 cells.
FIG. 5.
Immunoblot experiments to detect salt stress-induced alterations in the steady-state GgpS protein levels in Synechococcus sp. strain PCC 7002 cells after 684 mM NaCl was added for different times. (A) Cross-reactions with protein extracts obtained from Synechococcus sp. strain PCC 7002 with an antibody specific for Synechocystis sp. strain PCC 6803 GgpS. A 100-μg portion of total protein was applied to each lane. Antibody binding was detected with an ECL kit. (B) Quantitative estimates of relative GgpS protein levels as determined by videodensitometry. The maximum signal was set to a level of 100.
GgpS enzyme activity.
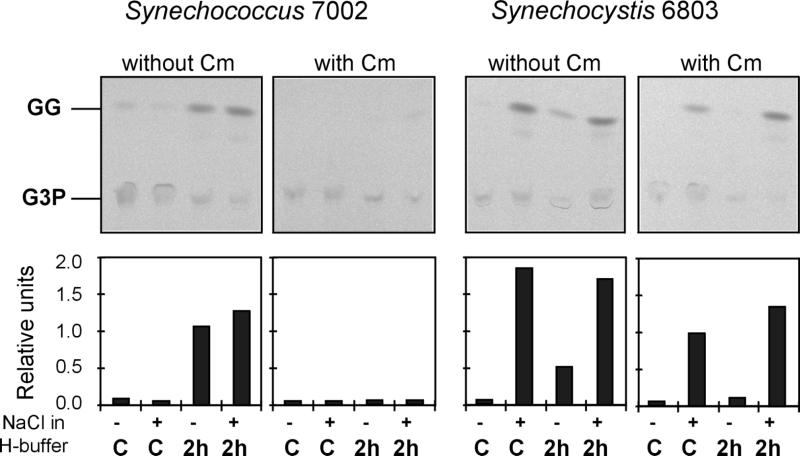
In other experiments the effects of salt in vitro and in vivo on GgpS activity were estimated. To improve the comparison, protein extracts obtained from Synechocystis sp. strain PCC 6803 cells were tested under the same experimental conditions, since it was known that in the freshwater strain the GG-synthesizing enzyme activities could be directly stimulated by salt in protein extracts obtained from low-salt-grown cells (7). This finding was confirmed in the present study, because under salt-free test conditions no GgpS activity was observed in extracts obtained from Synechocystis sp. strain PCC 6803 control cells, while activity was clearly observed in assays in which NaCl was included (Fig. 6). Nearly the same result was obtained with extracts obtained from salt-shocked Synechocystis sp. strain PCC 6803 cells. In contrast, almost no GgpS activity was observed with protein extracts obtained from low-salt-grown Synechococcus sp. strain PCC 7002 cells, whether NaCl was included or not (Fig. 6). However, when protein extracts obtained from Synechococcus sp. strain PCC 7002 cells shocked for 2 h with 684 mM NaCl were used, significant enzyme activities were observed in salt-free assays as well as in salt-containing assays. This finding clearly demonstrates that in Synechococcus sp. strain PCC 7002 the activity of the GG-synthesizing enzymes is present only in salt-treated cells, which use the same biosynthetic pathway for GG as the pathway observed previously for Synechocystis sp. strain PCC 6803. Furthermore, less strict dependence on elevated NaCl concentrations in the assay mixtures was observed for the enzyme obtained from Synechococcus sp. strain PCC 7002, since the enzyme activity was observed in protein extracts obtained with salt-free homogenization buffer; the Synechocystis sp. strain PCC 6803 protein extracts clearly contained diminished GgpS activities (Fig. 6). Corresponding to the data obtained for increased ggpS expression, preformed enzyme activity was completely absent in Synechococcus sp. strain PCC 7002 control cells. This observation was confirmed by measuring the GGPS enzyme activities in protein extracts of cells which had been salt shocked in the presence of the translational inhibitor chloramphenicol. Addition of this antibiotic inhibited the appearance of GgpS activity almost completely in salt-stressed Synechococcus sp. strain PCC 7002 cells, while it did not significantly influence the GgpS activity of control or salt-shocked Synechocystis sp. strain PCC 6803 cells (Fig. 6).
FIG. 6.
Changes in GgpS enzyme activities in control (C) and salt-shocked cells (2 h after 684 mM NaCl was added [2 h]) of the cyanobacteria Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803. Enzyme activities were also determined in cells treated with the protein synthesis inhibitor chloramphenicol (Cm) (final concentration, 100 μg ml−1). Enzyme assays were performed with protein extracts obtained with homogenization buffer (H-buffer) containing 342 mM NaCl (+) or salt-free buffer (−). All assay buffers contained 342 mM NaCl. Autoradiographic images resulting from separation of the reaction products by thin-layer chromatography and quantitative estimation of the radioactivity in GG spots with a phosphoimager are shown. For details of the enzyme assay used see reference 7.
DISCUSSION
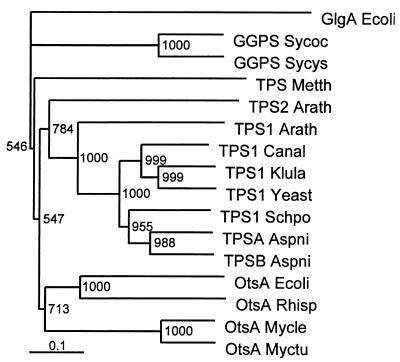
In order to characterize the regulation of GG synthesis in a cyanobacterial strain originating from a marine habitat, we cloned and sequenced the complete ggpS gene of Synechococcus sp. strain PCC 7002 by using a PCR strategy. Only one of the four primers deduced after comparison of the GgpS sequence of Synechocystis sp. strain PCC 6803 with TPS sequences bound to the expected sequence. After the entire sequence of the ggpS gene of Synechococcus sp. strain PCC 7002 was obtained, it became obvious that the other primers exhibited sequence identities that were too low, especially at their 3′ ends. However, the function of the gene could be predicted on the basis of the very high level of sequence similarity between the deduced protein and the previously characterized GgpS protein of the freshwater organism Synechocystis sp. strain PCC 6803 (14). In spite of the overall high level of similarity (85%), two parts of the GgpS from Synechococcus sp. strain PCC 7002 exhibited levels of similarity of less than 40% (Fig. 1). These variable protein sequences might be responsible for the regulatory differences between the two enzymes. The protein sequences were also used to analyze the phylogenetic relationship of cyanobacterial GGPS to functionally related TPS from members of the Archaea, Bacteria, and Eukarya (Fig. 7). Among the TPS sequences, a cluster of eubacterial and eukaryotic sequences was observed, while the archaebacterial TPS sequence was separate. The phylogenetic tree generally reflects the evolutionary relationships obtained when 16S rRNA analyses of the organisms from which the protein sequences originated are performed (16, 27). The structures of the cyanobacterial GgpS proteins are sufficiently different that they form a separate group that branches from all of the TPS sequences regardless of origin. To evaluate the phylogenetic relationships of the enzymes in more detail, it would be interesting to include a TPS sequence of cyanobacterial origin.
FIG. 7.
Phylogenetic relationships among cyanobacterial GgpS and functionally related TPS from members of the Archaea, Bacteria, and Eukarya. A cluster analysis was performed by using the Clustal X software package. The glycogen synthase of E. coli (GlgA Ecoli) (accession no. J02616) was used as an outgroup. Sequences were obtained from the EBI sequence database. Bootstrap values based on 1,000 replications are indicated. GGPS Sycys, GgpS of Synechocystis sp. strain PCC 6803 (accession no. D90913); GGPS Sycoc, GgpS of Synechococcus sp. strain PCC 7002 (accession no. AJ006298); TPSA Aspni, TpsA of Aspergillus niger (accession no. Q00075); TPSB Aspni, TpsB of Aspergillus niger (accession no. Q00217); TPS1 Yeast, Tps1 of Saccharomyces cerevisiae (accession no. Q00764); TPS1 Klula, Tps1 of Klyveromyces lactis (accession no. Q07158); TPS1 Schpo, Tps1 of Schizosaccharomyces pompe (accession no. P40387); TPS1 Canal, Tps1 of Candida albicans (accession no. Q92410); TPS Arath1, AtTPPA of Arabidopsis thaliana (accession no. AF007778); TPS Arath2, AtTPPB of Arabidopsis thaliana (accession no. AF007779); OtsA Myctu, OtsA of Mycobacterium tuberculosis (accession no. O06353); OtsA Mycle, OtsA of Mycobacterium leprae (accession no. Q50167); OtsA Rhisp, OtsA of Rhizobium sp. strain NGR234 (accession no. P55612); OtsA Ecoli, OtsA of E. coli (accession no. P31677); TPS Metth, Tps of Methanobacterium thermoautotrophicum (accession no. O27785).
In addition to the prediction concerning the function of the ORF made on the basis of the sequence data, a functional characterization of ggpS null mutants of Synechococcus sp. strain PCC 7002 confirmed the assumed function, since the mutants were not able to synthesize GG. This led in turn to significantly decreased salt tolerance, as was found previously with ggpS null mutants of Synechocystis sp. strain PCC 6803 (14). However, compared with Synechocystis sp. strain PCC 6803, the ggpS null mutants of Synechococcus sp. strain PCC 7002 exhibited a higher level of salt tolerance that reflected the higher level of salt tolerance level of wild-type cells of the marine strain (20). This finding was not based on increased sucrose accumulation, since the reaction, which compensated for the defect in GG synthesis, was also found in the Synechocystis sp. strain PCC 6803 mutants. The marine strain is probably less affected by or sensitive to ion influx, a phenomenon which has been found to occur in Synechocystis sp. strain PCC 6803 to a very great extent and which leads to inhibition of almost all metabolic activities (9). This conclusion is supported by the results of a comparison of the rates of accumulation of GG in both strains after a salt shock. While Synechocystis sp. strain PCC 6803 needs at least 8 h to reach an initial saturating level (10), cells of Synechococcus sp. strain PCC 7002 finished accumulating GG after only 5 h. Furthermore, in Synechococcus sp. strain PCC 7002 gene expression (at least expression of ggpS) was obviously not dramatically inhibited by a salt shock consisting of 684 mM NaCl, as was found to occur in the freshwater organism Synechocystis sp. strain PCC 6803 (9).
After ggpS was cloned from the marine organism Synechococcus sp. strain PCC 7002, salt-regulated expression of this gene was analyzed at the transcriptional and translational levels by using Northern blot and immunoblot techniques. Both methods revealed clearly increased expression of ggpS in salt-shocked cells, while in cells grown in basal medium almost no ggpS transcript or protein was detected. These results indicated that activation of GG synthesis depends on rapid and strong activation of gene expression. The data were confirmed by performing assays to determine GGPS activities. Furthermore, the increase in enzyme activity could be prevented by adding the translational inhibitor chloramphenicol to salt-shocked cells.
In summary, all of the data obtained in order to characterize ggpS expression (mRNA contents, protein contents, enzyme activity, action of an inhibitor) produce a consistent picture which implies that activation of expression of the key enzyme in GG synthesis seems to be the most important regulatory process in the marine organism Synechococcus sp. strain PCC 7002. It has been shown that in the freshwater organism Synechocystis sp. strain PCC 6803 posttranslational activation of a preformed GGPS seems to be the dominant activation mechanism for GG synthesis (9). Significant enzyme activities are present in cells cultivated at low salinities. After acclimation of Synechocystis cells to 684 mM NaCl, the GgpS activity was increased only two- to threefold. The same order of magnitude was observed for accumulation of GgpS protein (7a). The same mechanism seems to regulate GgpP, the second enzyme in GG synthesis, which is transcribed from the separate gene stpA in Synechocystis sp. strain PCC 6803 (8). Constitutive basal expression of this gene has also been revealed by Northern blot and immunoblot experiments. In addition, protein preparations obtained from salt-acclimated cells of Synechocystis sp. strain PCC 6803 by using salt-free homogenization buffers exhibited significantly decreased GG-synthesizing enzyme activities (7), while the Synechococcus sp. strain PCC 7002 enzymes were not affected and exhibited at high levels of activity after this treatment. These results also indicate that the salt dependence characteristics of the two enzymes seem to be different.
The different regulatory properties of the GG-synthesizing enzymes of the two strains correlate well with the demands of their natural habitats. In the freshwater organism Synechocystis sp. strain PCC 6803 lethal salt shock is prevented by constitutive expression of the GG-synthesizing enzymes, which are regulated mainly at the activity level by biochemical mechanisms. This strategy ensures that there is a rapid acclimation process when the cells are exposed to high salt concentrations. In the marine organism Synechococcus sp. strain PCC 7002 another strategy seems to be manifested. In cells grown at low salt concentrations, almost no GG-synthesizing enzymes are present, in contrast to Synechocystis sp. strain PCC 6803. However, since the marine strain lives permanently in the presence of high levels of salinity it is never exposed to a salt shock without a preexisting basal level of ggpS expression. At the fluctuating levels of salinity in coastal waters, the level of GG seems to be determined mainly by different levels of expression of the encoding genes. This acclimation mode is probably supported by the low permeability of the membrane of the marine strain to inorganic ions, particularly Na+ and Cl−. Therefore, cyanobacterial strains PCC 6803 and PCC 7002, which were isolated from freshwater and marine water, respectively, represent valuable models for investigating the genetic and biochemical mechanisms that evolved to regulate osmolyte synthesis on the basis of the external salt concentration.
ACKNOWLEDGMENTS
We thank D. A. Bryant, Pennsylvania State University, for critically reading the manuscript. The excellent technical assistance of B. Brzezinka and Ilse Doerr is greatly appreciated.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Allen M B, Arnon D I. Studies on nitrogen-fixing blue-green algae. II. The sodium requirement of Anabaena cylindrica. Physiol Plant. 1955;8:653–660. doi: 10.1104/pp.30.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Csonka L N, Epstein W. Osmoregulation. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1210–1223. [Google Scholar]
- 5.Galinski E A. Compatible solutes of halophilic eubacteria: molecular principles, water-solute interaction, stress protection. Experientia. 1993;49:487–496. [Google Scholar]
- 6.Giaever H M, Styrvold O B, Kaasen I, Strom A R. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J Bacteriol. 1988;170:2841–2849. doi: 10.1128/jb.170.6.2841-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagemann M, Erdmann N. Activation and pathway of glucosylglycerol synthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 1994;140:1427–1431. [Google Scholar]
- 7a.Hagemann, M., et al. Unpublished data.
- 8.Hagemann M, Schoor A, Jeanjean R, Zuther E, Joset F. The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J Bacteriol. 1997;179:1727–1733. doi: 10.1128/jb.179.5.1727-1733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagemann M, Schoor A, Mikkat S, Effmert U, Zuther E, Marin K, Fulda S, Vinnemeier J, Kunert A, Milkowski C, Probst C, Erdmann N. The biochemistry and genetics of the synthesis of osmoprotective compounds in cyanobacteria. In: Oren A, editor. Microbiology and biogeochemistry of hypersaline environments. Boca Raton, Fla: CRC Press LLC; 1999. pp. 177–186. [Google Scholar]
- 10.Hagemann M, Woelfel L, Krueger B. Alterations of protein synthesis in the cyanobacterium Synechocystis sp. PCC 6803 after a salt shock. J Gen Microbiol. 1990;136:1393–1399. [Google Scholar]
- 11.Kratz W A, Myers J. Nutrition and growth of several blue-green algae. J Bot. 1955;42:282–287. [Google Scholar]
- 12.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 13.Lippert K, Galinski E A, Truper H G. Biosynthesis and function of trehalose in Ectothiorhodospira halochloris. Antonie Leeuwenhoek. 1993;63:85–91. doi: 10.1007/BF00871735. [DOI] [PubMed] [Google Scholar]
- 14.Marin K, Zuther E, Kerstan T, Kunert A, Hagemann M. The ggpS gene from Synechocystis sp. strain PCC 6803 encoding glucosyl-glycerol-phosphate synthase is involved in osmolyte synthesis. J Bacteriol. 1998;180:4843–4849. doi: 10.1128/jb.180.18.4843-4849.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng J, Srivastava L M. Extraction, assay and some properties of floridoside phosphate synthase from Porphyra perforata (Rhodophyta) J Phycol. 1990;26:683–688. [Google Scholar]
- 16.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patensky F, Hess W R, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter H, Burkovski A, Kramer R. Osmo-sensing by N- and C-terminal extensions of the glycine betaine uptake system BetP of Corynebacterium glutamicum. J Biol Chem. 1998;273:2567–2574. doi: 10.1074/jbc.273.5.2567. [DOI] [PubMed] [Google Scholar]
- 19.Reed R H, Browitzka L J, Mackay M A, Chudek J A, Foster R, Warr S R C, Moore D J, Stewart W D P. Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol Rev. 1986;39:51–56. [Google Scholar]
- 20.Reed R H, Stewart W D P. Osmotic adjustment and organic solute accumulation in unicellular cyanobacteria from freshwater and marine habitats. Mar Biol. 1985;88:1–9. [Google Scholar]
- 21.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 22.Robinson N J, Robinson P J, Gupta A, Bleasby A J, Whitton B A, Morby A P. Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res. 1995;23:729–735. doi: 10.1093/nar/23.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Schoor A, Erdmann N, Effmert U, Mikkat S. Determination of the cyanobacterial osmolyte glucosylglycerol by high-performance liquid chromatography. J Chromatogr A. 1995;704:89–97. [Google Scholar]
- 25.Schoor A, Hagemann M, Erdmann N. Glucosylglycerol-phosphate synthase: target for ion-mediated regulation of osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. Arch Microbiol. 1999;171:101–106. doi: 10.1007/s002030050684. [DOI] [PubMed] [Google Scholar]
- 26.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 27.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]