Abstract
The gene loci fcs, encoding feruloyl coenzyme A (feruloyl-CoA) synthetase, ech, encoding enoyl-CoA hydratase/aldolase, and aat, encoding β-ketothiolase, which are involved in the catabolism of ferulic acid and eugenol in Pseudomonas sp. strain HR199 (DSM7063), were localized on a DNA region covered by two EcoRI fragments (E230 and E94), which were recently cloned from a Pseudomonas sp. strain HR199 genomic library in the cosmid pVK100. The nucleotide sequences of parts of fragments E230 and E94 were determined, revealing the arrangement of the aforementioned genes. To confirm the function of the structural genes fcs and ech, they were cloned and expressed in Escherichia coli. Recombinant strains harboring both genes were able to transform ferulic acid to vanillin. The feruloyl-CoA synthetase and enoyl-CoA hydratase/aldolase activities of the fcs and ech gene products, respectively, were confirmed by photometric assays and by high-pressure liquid chromatography analysis. To prove the essential involvement of the fcs, ech, and aat genes in the catabolism of ferulic acid and eugenol in Pseudomonas sp. strain HR199, these genes were inactivated separately by the insertion of omega elements. The corresponding mutants Pseudomonas sp. strain HRfcsΩGm and Pseudomonas sp. strain HRechΩKm were not able to grow on ferulic acid or on eugenol, whereas the mutant Pseudomonas sp. strain HRaatΩKm exhibited a ferulic acid- and eugenol-positive phenotype like the wild type. In conclusion, the degradation pathway of eugenol via ferulic acid and the necessity of the activation of ferulic acid to the corresponding CoA ester was confirmed. The aat gene product was shown not to be involved in this catabolism, thus excluding a β-oxidation analogous degradation pathway for ferulic acid. Moreover, the function of the ech gene product as an enoyl-CoA hydratase/aldolase suggests that ferulic acid degradation in Pseudomonas sp. strain HR199 proceeds via a similar pathway to that recently described for Pseudomonas fluorescens AN103.
Vanillin (4-hydroxy-3-methoxybenzaldehyde) is one of the most important aromatic flavor compounds used in the food- and perfume-producing industries. “Artificial” or “nature-identical” vanillin is currently produced from petrochemicals and from lignin (9), but there is a growing interest in producing “natural” vanillin by biotransformations (17, 21). Phenolic stilbenes, eugenol (4-allyl-2-methoxyphenol), ferulic acid (4-hydroxy-3-methoxycinnamate), and lignin were found to be potential substrates for these biotransformation processes, since vanillin occurs as an intermediate in the corresponding degradation routes (8, 37, 44, 45). Only recently, a biotechnological production of vanillin starting from glucose was proposed (23). A biotransformation process of eugenol to vanillin, based on a new Pseudomonas sp. (strain HR199), which was able to produce methoxyphenol type aroma chemicals from eugenol (32), was developed by Rabenhorst and Hopp (33).
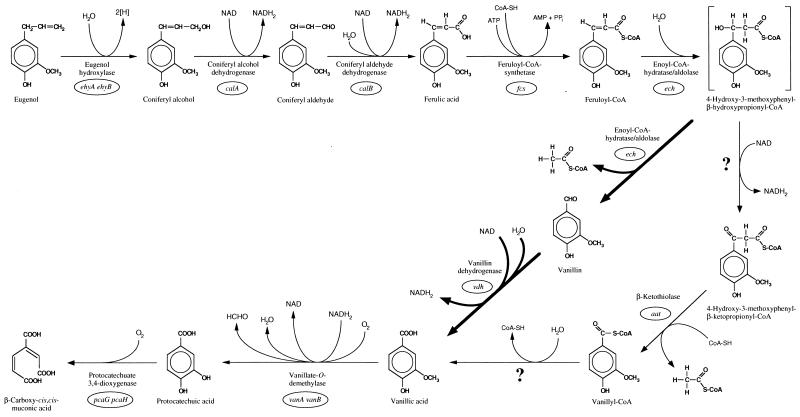
We are currently investigating the physiological and genetic basis for this biotransformation (42). Some of the genes which are essential to the degradation of eugenol by Pseudomonas sp. strain HR199, which proceeds via coniferyl alcohol (4-hydroxy-3-methoxycinnamyl alcohol), coniferyl aldehyde (4-hydroxy-3-methoxycinnamyl aldehyde), ferulic acid, vanillin, vanillic acid (4-hydroxy-3-methoxybenzoate), and protocatechuic acid (3,4-dihydroxybenzoate) (Fig. 1) (32), have already been identified. The genes encoding eugenol hydroxylase (ehyA and ehyB) (30), coniferyl alcohol dehydrogenase (calA) (20), and coniferyl aldehyde dehydrogenase (calB) (3) are responsible for the conversion of eugenol to ferulic acid (Fig. 1). Vanillin dehydrogenase, encoded by vdh, and vanillate-O-demethylase, encoded by vanA and vanB, catalyze the conversion of vanillin to protocatechuic acid (29), which is further metabolized by ortho cleavage (26) (Fig. 1).
FIG. 1.
Proposed route for the catabolism of eugenol in Pseudomonas sp. strain HR199. Fine arrows indicate β-oxidation analogous to that of fatty acid catabolism. Thick arrows indicate ferulic acid degradation via vanillin.
In the present study we identified the genes responsible for the bioconversion of ferulic acid to vanillin in Pseudomonas sp. strain HR199. The essential involvement of these genes in ferulic acid and eugenol catabolism was verified by gene disruption and characterization of the corresponding mutants with respect to the catabolism of ferulic acid and eugenol.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of Pseudomonas and Escherichia coli and the plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Pseudomonas sp. | ||
| HR199 (wild type) | Wild type, eugenol positive, ferulate positive, vanillin positive, vanillate positive, protocatechuate positive, kanamycin and gentamicin sensitive | DSM 7063 32 |
| SK6167 | Eugenol negative, ferulate negative, vanillin positive, vanillate positive, protocatechuate positive | This study |
| SK6202 | Eugenol negative, ferulate negative, vanillin positive, vanillate positive, protocatechuate positive | This study |
| HRaatΩKm | Eugenol positive, ferulate positive, vanillin positive, vanillate positive, protocatechuate positive, kanamycin resistant | This study |
| HRechΩKm | Eugenol negative, ferulate negative, vanillin positive, vanillate positive, protocatechuate positive, kanamycin resistant | This study |
| HRfcsΩGm | Eugenol negative, ferulate negative, vanillin positive, vanillate positive, protocatechuate positive, gentamicin resistant | This study |
| Escherichia coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi hsdR17 (rK mK+) supE44 relA1 λ−lac [F′ proAB lacIqZΔM15 Tn10(Tet)] | 7 |
| S17-1 | recA; harboring the tra genes of plasmid RP4 in the chromosome, proA thi-1 | 38 |
| Plasmids | ||
| pVK100 | Tcr Kmr, cosmid, broad host range | 19 |
| pHP1014 | Tcr Kmr Cmr, broad host range | 31 |
| pE207 | pVK100, harboring fragment E230 with vdh, vanA, and vanB | 29 |
| pE5-1 | pVK100, harboring fragments E12, E18, E30, E58, and E94 with calA and calB | 20 |
| pSUP202 | Tcr Apr Cmr, mob | 39 |
| pBluescript SK− | AprlacPOZ′, T7 and T3 promoter | Stratagene, San Diego, Calif. |
| pSKsym | Derivative of pBluescript SK− with a synthetic symmetrical MCSa: KpnI-SalI-HindIII-EcoRI-SmaI-EcoRI-HindIII-SalI-SacI | 27 |
| pSKsymΩKm | pSKsym harboring ΩKm in the SmaI site of the MCS | 27 |
| pSKsymΩGm | pSKsym harboring ΩGm in the SmaI site of the MCS | 27 |
| pSKaat | pBluescript SK− harboring the PCR product comprising aat as EcoRI fragment | This study |
| pSKechE/H | pBluescript SK− harboring the PCR product comprising ech as EcoRI-HindIII fragment | This study |
| pSKechEE | pBluescript SK− harboring the PCR product comprising ech as EcoRI fragment | This study |
| pSKfcs | pBluescript SK− harboring the PCR product comprising fcs as PstI fragment | This study |
| pSKechE/Hfcs | pBluescript SK− harboring the PCR products comprising ech as EcoRI-HindIII fragment and fcs as PstI fragment downstream from and colinear with the lac promoter | This study |
| pSKaatΩKm | pBluescript SK− harboring the ΩKm-disrupted aat gene | This study |
| pSKechΩKm | pBluescript SK− harboring the ΩKm-disrupted ech gene | This study |
| pSKfcsΩGm | pBluescript SK− harboring the ΩGm-disrupted fcs gene | This study |
| pSUPaatΩKm | pSUP202 harboring the ΩKm-disrupted aat gene in the EcoRI site | This study |
| pSUPechΩKm | pSUP202 harboring the ΩKm-disrupted ech gene in the EcoRI site | This study |
| pSUPfcsΩGm | pSUP202 harboring the ΩGm-disrupted fcs gene in the PstI site | This study |
MCS, multiple cloning site.
Growth of bacteria.
Cells of E. coli were grown at 37°C in Luria-Bertani medium (LB) or in M9 mineral salts medium (34). Cells of Pseudomonas strains were grown at 30°C either in a nutrient broth (NB) medium (0.8%, wt/vol; Difco) or in MM (36) or HR-MM (32) mineral salts medium supplemented with carbon sources as indicated below. Ferulic acid, vanillin, vanillic acid, and protocatechuic acid were dissolved in dimethyl sulfoxide and were each added to the medium at final concentrations of 0.1% (wt/vol). Eugenol was directly added to the medium at a final concentration of 0.1% (vol/vol). Tetracycline, kanamycin, and gentamicin were used at final concentrations of 25, 300, and 7.5 μg/ml, respectively, for Pseudomonas strains. Growth of the bacteria was monitored with a Klett-Summerson photometer.
Nitrosoguanidine mutagenesis.
The nitrosoguanidine mutagenesis of Pseudomonas sp. strain HR199 was performed as described previously (29).
Qualitative and quantitative determination of catabolic intermediates.
Culture supernatants were analyzed for excreted intermediates of eugenol catabolism by liquid chromatography without prior extraction, using a high-performance liquid chromatography (HPLC) apparatus (Fa. Knauer, Berlin, Germany). Separation was carried out by reversed-phase chromatography on Nucleosil-100 C18 (5-μm particle size; 250- by 4.0-mm column) with a gradient of 0.1% (vol/vol) formic acid (eluant A) and acetonitrile (eluant B) in a range of 20 to 100% (vol/vol) eluant B and at a flow rate of 1 ml/min. For quantification, all intermediates were calibrated with external standards. The compounds were identified by their retention times and by the corresponding spectra obtained with a diode array detector (WellChrom Diodenarray-Detektor K-2150; Knauer).
Preparation of the soluble fractions of crude extracts.
Cells were disrupted either by a twofold French press passage at 96 MPa or by sonication (1 min/ml of cell suspension with an amplitude of 40 μm) with a Bandelin Sonopuls GM200 ultrasonic disintegrator. Soluble fractions of crude extracts were obtained by centrifugation at 100,000 × g at 4°C for 1 h.
Enzyme assays.
Feruloyl coenzyme A (feruloyl-CoA) synthetase was assayed spectrophotometrically at 30°C by a modified method described by Zenk et al. (49). The reaction mixture (1 ml) contained 100 mM potassium phosphate buffer (pH 7.0), 2.5 mM MgCl2, 0.7 mM ferulic acid, 2 mM ATP, 0.4 mM CoA, and an appropriate amount of extract or enzyme. The assay was started by the addition of ATP, and the initial absorbance increase due to the formation of feruloyl-CoA (ɛ = 10 cm2 μmol−1) was measured at 345 nm.
The activity of the enoyl-CoA hydratase/aldolase was determined by a modified method described by Gasson et al. (16). Extracts were incubated at 30°C in a reaction mixture (0.5 ml) containing 90 mM sodium phosphate buffer (pH 7.0), 3 mM MgCl2, and an appropriate amount of 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA (HMPHP-CoA). HMPHP-CoA was prepared by the method of Gasson et al. (16). The conversion of HMPHP-CoA to vanillin was confirmed by HPLC analysis. Since the concentration of HMPHP-CoA was not determined, only qualitative determination of enoyl-CoA hydratase/aldolase activity was performed.
The amount of soluble protein present was determined as described by Lowry et al. (24).
Electrophoretic methods.
Proteins were separated under nondenaturating conditions in 7.4% (wt/vol) polyacrylamide (PAA) gels as described by Stegemann et al. (41) and under denaturating conditions in 11.5% (wt/vol) PAA gels by the method of Laemmli (22) and stained with Serva Blue R.
Isolation and manipulation of DNA.
Plasmid DNA and DNA restriction fragments were isolated and analyzed by standard methods described in references compiled in a previous study (28).
Transfer of DNA.
Competent cells of E. coli were prepared and transformed by using the CaCl2 procedure as described by Hanahan (18). Conjugations of E. coli S17-1 (donors) harboring hybrid plasmids and of Pseudomonas strains (recipients) were performed on solidified NB medium as described by Friedrich et al. (14), or by a minicomplementation method described previously (29).
Construction of a genomic library of Pseudomonas sp. strain HR199.
Partially EcoRI-digested genomic DNA of Pseudomonas sp. strain HR199 was ligated with EcoRI-linearized cosmid pVK100. The ligation mixture was packaged in λ particles and subsequently transduced into E. coli S17-1. A total of 1,330 transductants were selected on LB-tetracycline agar plates, and the hybrid cosmids of these strains were conjugatively transferred to the ferulic acid-negative mutants SK6167 and SK6202, respectively.
Inactivation of the fcs gene in Pseudomonas sp. strain HR199 by insertion of the omega element ΩGm.
For the inactivation of the fcs gene by insertion of ΩGm, the hybrid plasmid pSKfcs was digested with BssHII, to delete a 1,284-bp fragment from the fcs gene. After removal of the single-stranded ends by mung bean nuclease treatment, this DNA was ligated with ΩGm, which was recovered from SmaI-digested pSKsymΩGm, whose construction has been described recently (27). E. coli XL1-Blue was transformed with the ligation mixture, and transformants harboring the hybrid plasmid pSKfcsΩGm, which conferred resistance against gentamicin, were obtained. No feruloyl-CoA synthetase activity could be detected in soluble fractions of the crude extracts of the corresponding recombinant strains (see Table 2). For the exchange of the functional fcs gene with the inactivated gene, fcsΩGm had to be cloned in vector pSUP202, which could be transferred from E. coli to Pseudomonas sp. strain HR199 by conjugation. Since pSUP202 cannot be replicated in Pseudomonas strains, it is a suicide plasmid (39), and the integration of the hybrid plasmid by homologous recombination can be forced by selection on media containing antibiotics, the corresponding resistances to which are encoded by genes borne by the plasmid. The disrupted gene fcsΩGm was isolated from pSKfcsΩGm after digestion with PstI and was ligated with PstI-digested pSUP202 DNA. E. coli S17-1 was transformed with the ligation mixture, and transformants harboring the hybrid plasmid pSUPfcsΩGm, which conferred resistance to tetracycline and gentamicin, were obtained. The hybrid plasmid pSUPfcsΩGm was transferred to Pseudomonas sp. strain HR199 by conjugation, and selection was performed on solidified H16-MM containing gentamicin. The obtained transconjugants were tested for tetracycline resistance (encoded by a vector-borne gene) to distinguish between an integration of the whole hybrid plasmid into the chromosome by a single crossover (heterogenotes) or an exchange of the functional fcs gene with the disrupted gene by a double crossover (homogenotes), which resulted in a tetracycline-sensitive phenotype. The exchange of the functional fcs gene with the disrupted gene fcsΩGm in the tetracycline-sensitive mutant Pseudomonas sp. strain HRfcsΩGm was confirmed by amplification of the corresponding gene from genomic DNA of this mutant by PCR with oligonucleotides PCRfcsPU and PCRfcsPD, which resulted in only one PCR product. The sequence of the PCR product was determined, revealing the sequence of fcsΩGm. When genomic DNA of heterogenotic transconjugants were used as template DNA, two PCR products were obtained, corresponding to the functional fcs gene and the disrupted fcs gene, respectively.
TABLE 2.
Feruloyl-CoA synthetase activities in mutant strains of Pseudomonas sp. strain HR199 and in recombinant strains of E. coli XL-1 Bluea
| Strain | Sp act of feruloyl-CoA synthetase (U/mg)b |
|---|---|
| Pseudomonas sp. strain HR199 | 0.08 |
| Pseudomonas sp. strain HRfcsΩGm | <0.01 |
| Pseudomonas sp. strain HRechΩKm | 0.12 |
| Pseudomonas sp. strain HRaatΩKm | 0.07 |
| E. coli XL-1 Blue (pSKfcs) | 0.19 |
| E. coli XL-1 Blue (pSKfcsΩGm) | <0.01 |
| E. coli XL-1 Blue (pBluescript SK−) | <0.01 |
Pseudomonas cells were grown to the late exponential phase at 30°C in HR-MM containing 0.5% (wt/vol) gluconate and 0.1% (vol/vol) eugenol. Recombinant strains of E. coli were grown for 12 h at 37°C in LB in the presence of 1 mM IPTG.
The feruloyl-CoA synthetase activities were determined in soluble fractions of crude extracts at 30°C by a modified method of Zenk et al. (49) as described in Materials and Methods. Specific activities are given as units per milligram of protein.
Inactivation of the ech gene in Pseudomonas sp. strain HR199 by insertion of the omega element ΩKm.
For inactivation of the ech gene by insertion of ΩKm, the ech gene was amplified from PstI-digested genomic DNA of Pseudomonas sp. strain HR199. To obtain a gene which was flanked by EcoRI sites, the aforementioned oligonucleotide PCRechEU and the oligonucleotide PCRechED (5′-AAAGAATTCCCCGCAACATGCCCGCCGCCAGGTAAACG-3′), which was complementary to the nucleotide sequence from bp 364 to 393 downstream of the translational stop codon of ech, were used as primers in the PCR. The isolated PCR product was digested with EcoRI and ligated to EcoRI-digested pBluescript SK−. E. coli XL1-Blue was transformed with the ligation mixture, and transformants harboring the hybrid plasmid pSKechEE were obtained. pSKechEE was digested with NruI to delete a region of 486 bp from the ech gene. The plasmid DNA was ligated with ΩKm, which was recovered from SmaI-digested pSKsymΩKm (27), to obtain the plasmid pSKechΩKm. Subsequently, echΩKm was recovered from EcoRI-digested pSKechΩKm, and the exchange of the functional ech gene with the inactivated gene echΩKm in Pseudomonas sp. strain HR199 was performed as described for the exchange of fcs and fcsΩGm, with the exceptions that the EcoRI site of pSUP202 was used for cloning of echΩKm and gentamicin was replaced by kanamycin in the medium used for selection. The success of the gene replacement in the tetracycline-sensitive mutant Pseudomonas sp. strain HRechΩKm was confirmed by amplification of the corresponding gene from genomic DNA of this mutant by PCR and sequencing of the obtained single PCR product.
Inactivation of the aat gene in Pseudomonas sp. strain HR199 by insertion of the omega element ΩKm.
For the inactivation of the aat gene by insertion of ΩKm, the aat gene was amplified from PstI-digested genomic DNA of Pseudomonas sp. strain HR199. To obtain a gene which was flanked by EcoRI sites, oligonucleotides PCRaatEU (5′-AAAGAATTCGGCGGTCGGCGAAAGTTGATGCG-3′ [the restriction enzyme site is underlined]), which corresponded to the nucleotide sequence from bp 70 to 48 upstream of the translational start codon of aat, and PCRaatED (5′-AAAGAATTCCCACCAACCCTGACAAGGTATGTACAC-3′), which was complementary to the nucleotide sequence from bp 198 to 224 downstream of the translational stop codon of aat, were used as primers in the PCR. The isolated PCR product was digested with EcoRI and ligated to EcoRI-digested pBluescript SK−. E. coli XL1-Blue was transformed with the ligation mixture, and transformants harboring the hybrid plasmid pSKaat were obtained. pSKaat was digested with BssHII, to delete a 59-bp fragment from the aat gene. After removal of the single-stranded ends by mung bean nuclease treatment, this DNA was ligated with ΩKm to obtain plasmid pSKaatΩKm. Subsequently, aatΩKm was recovered from EcoRI-digested pSKaatΩKm and the functional aat gene was exchanged with the inactivated aatΩKm gene in Pseudomonas sp. strain HR199 as described for the exchange of ech and echΩKm. The success of the gene replacement in the tetracycline-sensitive Pseudomonas sp. strain HRaatΩKm mutant was confirmed by amplification of the corresponding gene from genomic DNA of this mutant by PCR and sequencing of the single PCR product thus obtained.
DNA sequence determination and analysis.
DNA sequences were determined by the dideoxy chain termination method (35) with a 4000L DNA sequencer (LI-COR Inc., Biotechnology Division, Lincoln, Neb.). A Thermo Sequenase fluorescence-labelled primer cycle-sequencing kit with 7-deaza-dGTP (Amersham Life Science, Little Chalfont, United Kingdom) was used as specified by the manufacturer, together with synthetic fluorescence-labelled oligonucleotides as primers. The primer-hopping strategy was used (43). Nucleotide and amino acid sequences were analyzed with the Genetics Computer Group sequence analysis software package (GCG package, version 6.2, June 1990) as described by Devereux et al. (11).
Chemicals.
Restriction endonucleases, T4 DNA ligase, lambda DNA, and enzymes or substrates used in the enzyme assays were obtained from C. F. Boehringer & Soehne (Mannheim, Germany) or from GIBCO/BRL-Bethesda Research Laboratories GmbH (Eggenstein, Germany). Agarose type NA was purchased from Pharmacia-LKB (Uppsala, Sweden). Radioisotopes were from Amersham/Buchler (Braunschweig, Germany). Synthetic oligonucleotides were purchased from MWG-Biotech (Ebersberg, Germany). All other chemicals were from Haarmann & Reimer (Holzminden, Germany), E. Merck AG (Darmstadt, Germany), Fluka Chemie (Buchs, Switzerland), Serva Feinbiochemica (Heidelberg, Germany), or Sigma Chemie (Deisenhofen, Germany).
Nucleotide sequence accession number.
The nucleotide and amino acid sequence data reported in this paper have been submitted to the EMBL, GenBank, and DDBJ nucleotide sequence databases and are listed under accession no. AJ238746.
RESULTS
Cloning of the genes involved in the ferulic acid degradation pathway.
Pseudomonas sp. strain HR199 is able to utilize ferulic acid as the sole carbon source for growth. For identification of the genes, which are involved in the catabolism of ferulic acid, mutants that were unable to grow on ferulic acid but retained the ability to grow on vanillic acid or protocatechuic acid were isolated after nitrosoguanidine mutagenesis. Two of these mutants (SK6167 and SK6202) were chosen as recipients for a genomic library of Pseudomonas sp. strain HR199. Mutant SK6202 was complemented by the hybrid cosmid pE207 harboring a 23-kbp EcoRI fragment (E230), which had recently been identified in a genomic library of Pseudomonas sp. strain HR199 (29). Mutant SK6167 was complemented by the hybrid cosmid pE5-1, which also complemented a coniferyl alcohol dehydrogenase-deficient mutant SK6164, as revealed during our investigations of the eugenol catabolism of Pseudomonas sp. strain HR199 (20). This hybrid cosmid harbored five EcoRI fragments of 1.2, 1.8, 3.0, 5.8, and 9.4 kbp (E12, E18, E30, E58, and E94, respectively).
Subcloning of ech, fcs, and aat.
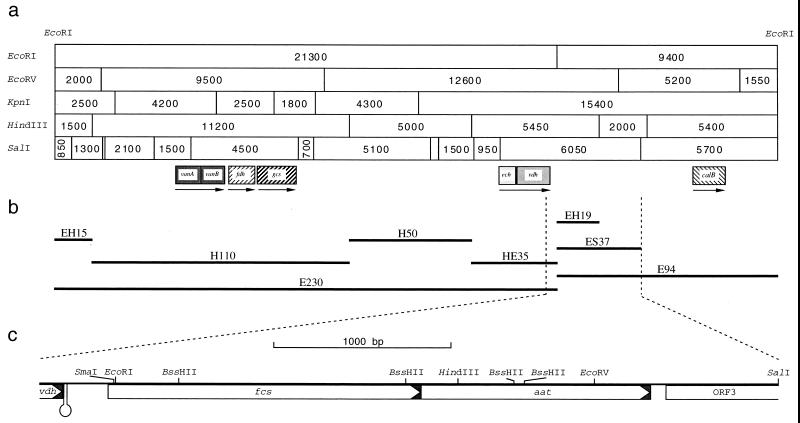
A physical map of E230 was obtained recently (29). This fragment harbored the vanillin catabolism genes vanA, vanB, and vdh, encoding the subunits of vanillate-O-demethylase (vanA and vanB) and vanillin dehydrogenase (vdh) (29) (Fig. 2). The amino acid sequence deduced from an open reading frame (ORF2) identified upstream of vdh showed significant homologies to enoyl-CoA hydratases (29).
FIG. 2.
Localization of fcs, aat, and ORF3. (a) EcoRI restriction fragments E230 and E94 from plasmid pE207 and pE5-1, respectively. (b) Relevant subfragments used in this study. (c) Map of fragment ES37 and part of fragment E230 with the structural genes of the feruloyl-CoA synthetase (fcs), β-ketothiolase (aat), and ORF3. The hairpin-like structure is indicated.
Fragment E230 was isolated from EcoRI-digested plasmid pSKE230 and was digested with HindIII. The resulting 11.2-kbp (H110) and 5.0-kbp (H50) HindIII fragments and also the 1.5-kbp (EH15) and 3.5-kbp (HE35) HindIII-EcoRI fragments were cloned in pHP1014 (Fig. 2). After conjugative transfer of the resulting plasmids from corresponding E. coli S17-1 strains to the ferulic acid-negative mutant SK6202, complementation was achieved only with fragment HE35, harboring the vdh gene and ORF2, now designated ech. Since mutant SK6202 was able to grow on vanillin, like the wild type, this mutant most probably lacked the functional gene product of ech.
The five EcoRI fragments E12, E18, E30, E58, and E94 were cloned separately in vector pHP1014. After conjugative transfer of the resulting plasmids from corresponding E. coli S17-1 strains to the ferulic acid-negative mutant SK6167, complementation was achieved only with fragment E94. Fragment E94 was isolated and cloned in pBluescript SK−, resulting in plasmid pSKE94, and a physical map of this fragment was obtained (Fig. 2). pSKE94 was digested with HindIII and SalI, and the resulting subfragments of E94 were cloned in pHP1014. By conjugative transfer of the resulting plasmids to mutant SK6167, the complementing region was assigned to a 3.7-kbp (ES37) EcoRI-SalI subfragment and an 1.9-kbp (EH19) EcoRI-HindIII subfragment of E94 (Fig. 2).
Nucleotide sequence of fragment ES37.
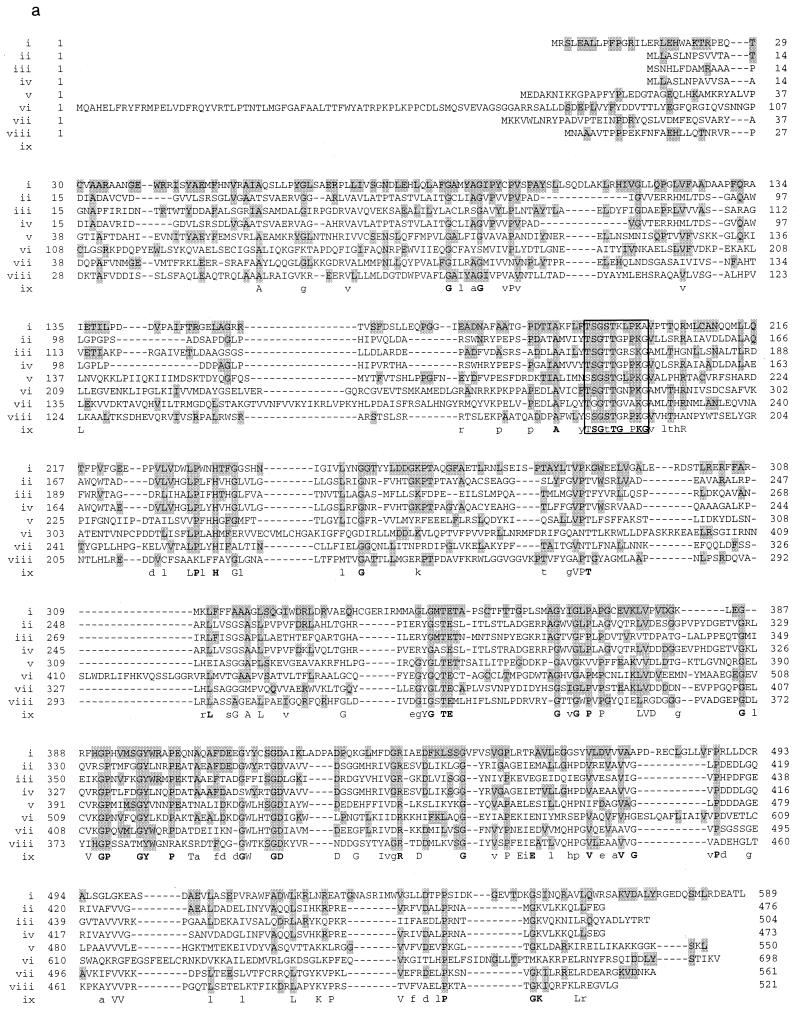
The nucleotide sequence of the entire fragment ES37, which contained the sequence of fragment EH19, was determined (Fig. 2). Fragment EH19 was almost covered by one open reading frame of 1,770 bp (ORF1), whose putative translational product exhibited significant homologies to thiokinases from various sources (Fig. 3). Since the ferulic acid-negative mutant SK6167 was complemented by fragment EH19, it most probably lacked a functional gene product of ORF1. Due to the aforementioned homologies and the proposed degradation mechanism via a β-oxidation analogous mechanism, ORF1 was designated fcs, which most probably encodes a feruloyl-CoA synthetase. The NH2-terminus-encoding part of the fcs gene was not located on fragment EH19 but on the adjacent EcoRI fragment, E230. That both fragments were directly linked, as shown in Fig. 2, was confirmed by using PCR with oligonucleotides PCRfcsPU (5′-AAACTGCAGTCGAGCATCGATTGAGCACTTTACCCAGC-3′) and PCRfcsPD (5′-AAACTGCAGGCCGCGACACACAGCACGTGATCAG-3′), by hybridization to E230 and E94, respectively, and by using genomic DNA of Pseudomonas sp. strain HR199 as template DNA. The translational stop codon of fcs overlapped with the GTG start codon of a second open reading frame of 1,296 bp, which was referred to as aat, since its putative translational product exhibited significant homologies to β-ketothiolases from various sources. Typical Shine-Dalgarno sequences (GGAGGT and GAGG, respectively) preceded the translational start codons of fcs and aat at distances of 9 or 7 nucleotides, respectively.
FIG. 3.
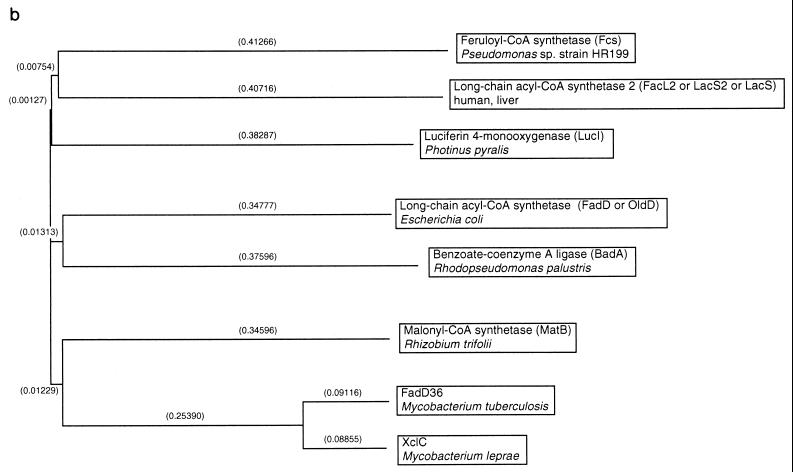
Homologies of Pseudomonas sp. strain HR199 feruloyl-CoA synthetase to thiokinases from various sources. (a) The amino acid sequence of the feruloyl-CoA synthetase from Pseudomonas sp. strain HR199 deduced from the fcs gene (i) was aligned with the amino acid sequence of the xylC gene product from Mycobacterium leprae (U15181) (40) (ii), the malonyl-CoA synthetase (matB gene product) from Rhizobium trifolii (AF022387) (4) (iii), the amino acid sequence of the fadD36 gene product from Mycobacterium tuberculosis (Z93777) (10) (iv), the luciferin 4-monooxygenase (lucI gene product) from Photinus pyralis (P08659) (12) (v), the long-chain acyl-CoA synthetase 2 (facL2 gene product) from human liver (P33121) (2) (vi), the long-chain acyl-CoA synthetase (fadD or oldD gene product) from Escherichia coli (P29212) (15) (vii), and the benzoate-CoA ligase (badA gene product) from Rhodopseudomonas palustris (L42322) (13) (viii). Amino acids are specified by standard one-letter abbreviations. Dashes indicate gaps introduced into the sequences to improve the alignment. Amino acid residues which are identical to the Pseudomonas sp. strain HR199 fcs gene product at one particular sequence position are shaded. The consensus sequence is given (ix). The signature sequence of the AMP-binding domain (5) is boxed. (b) The relationship of the proteins in panel a is displayed as dendrogram, which was constructed by using the program CLUSTAL from pairwise similarity scores generated by the method of Wilbur and Lipman (48) with the following parameters: k-tuple length, 1; gap penalty, 3; number of diagonals, 5; diagonal window size, 5; gap opening penalty, 10; gap extension penalty, 0.10; protein weight matrix, blosum. Relatedness is represented by the branch length (distances are given as 10−2 percent divergence in parentheses).
The G+C contents of fcs and aat were 57.6 and 57.0 mol%, respectively, and the G+C contents for the different codon positions corresponded well to the theoretical values calculated by the method of Bibb et al. (6). In addition, the codon usages of fcs and aat were very similar to the codon usage for other genes of this bacterium (3, 20, 26, 29, 30), indicating that fcs and aat represented coding regions in Pseudomonas sp. strain HR199.
At 85 bp downstream of aat, the start of another open reading frame (ORF3), whose putative translational product exhibited high homologies (up to 37% identity in a 326-amino-acid [aa] overlap) to methyl-accepting chemotaxis proteins from different sources, was identified.
Putative functions of the ech, fcs, and aat gene products.
The amino acid sequences deduced from ech, fcs, and aat were compared with those collected in GenBank. With the ech gene product, the highest homology was found to the enoyl-SCoA hydratase/lyase of P. fluorescens AN103 (88% identity in a 276-aa overlap) (16). With the fcs gene product, the highest homology was found to the long-chain acyl-CoA synthetase 2 of human liver (36% identity in a 114-aa overlap) (2). The relationship of the feruloyl-CoA synthetase from Pseudomonas sp. strain HR199 to enzymes of the thiokinase family is shown in Fig. 3. With the aat gene product, the highest homology was found to the mitochondrial 3-ketoacyl-CoA thiolase of human liver (35% identity in a 399-aa overlap) (1).
Heterologous expression of ech and fcs in E. coli.
The ech gene was amplified in a PCR with PstI-digested genomic DNA of Pseudomonas sp. strain HR199 as the template DNA, together with the primers PCRechEU (5′-AAAGAATTCGCCTGGCGACGAAAGGGCGGCAGGC-3′), which corresponded to the nucleotide sequence from bp 242 to 217 upstream of the translational start codon of ech, and PCRechHD (5′-AA AAAGCTTCCCCGGCGCATTTATCAGCGCTTGTAGGT CTGC-3′), which was complementary to the nucleotide sequence comprising the last 19 bp of the ech gene and an additional 14 bp downstream of the translational stop codon of ech. Since the upstream primer exhibited an EcoRI site (underlined) and the downstream primer exhibited a HindIII site (underlined), the obtained 1.1-kbp PCR product was cloned in pBluescript SK− with the ech gene colinear to and downstream of the lacZ promoter. The resulting hybrid plasmid, pSKechE/H, conferred enoyl-CoA hydratase/aldolase activity to recombinant strains of E. coli XL1-Blue, which was revealed by the enzyme assay described in Materials and Methods.
The fcs gene was amplified in a PCR with PstI-digested genomic DNA of Pseudomonas sp. strain HR199 as template DNA together with primers PCRfcsPU (5′-AAACTGCAGTCGAGCATCGATTGAGCACTTTACCCAGC-3′), which corresponded to the nucleotide sequence from bp 339 to 307 upstream of the translational start codon of fcs, and PCRfcsPD (5′-AAACTGCAGGCCGCGACACACAGCACGTGATCA G-3′), which was complementary to the nucleotide sequence from bp 346 to 370 downstream of the translational stop codon of fcs. The 2.5-kbp PCR product thus obtained was cloned in pBluescript SK−. The resulting hybrid plasmid pSKfcs, with the fcs gene colinear to and downstream of the lacZ promoter, conferred feruloyl-CoA synthetase activity to recombinant strains of E. coli XL1-Blue (Table 2).
Biotransformation of ferulic acid to vanillin by coexpression of the fcs and ech genes in E. coli.
Since the expression of fcs enabled the corresponding E. coli strains to convert ferulic acid to feruloyl-CoA and the expression of ech led to the conversion of 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA to vanillin, coexpression of these genes should enable E. coli to convert ferulic acid to vanillin.
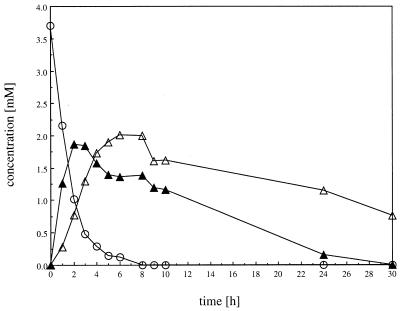
The 2.5-kbp PstI fragment containing fcs was isolated from plasmid pSKfcs after PstI digestion. This fragment was ligated with PstI-digested plasmid pSKechE/H, and the ligation mixture was transformed into E. coli XL1-Blue. Hybrid plasmids of the transformants were analyzed by restriction analysis, with respect to the presence and orientation of the genes ech and fcs. One transformant, which harbored a hybrid plasmid with both genes colinear to and downstream of the lacZ promoter (pSKechE/Hfcs), was grown overnight in 50 ml of LB containing 12.5 μg of tetracycline per ml, 100 μg of ampicillin per ml, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, washed twice in 100 mM potassium phosphate buffer (pH 7.0), and resuspended in 50 ml of HR-MM containing 3.7 mM ferulic acid. With resting cells of E. coli XL1-Blue harboring pSKechE/Hfcs, conversion rates up to 0.037 μmol of ferulic acid to 0.022 μmol of vanillin per min per ml of culture were readily obtained. The course of this biotransformation is summarized in Fig. 4. Beside vanillin, vanillyl alcohol was detected in the medium as a result of a reduction of vanillin by the E. coli cells. This reduction was also observed in a control experiment, when cells of E. coli XL1-Blue harboring only the vector pBluescript SK− were incubated in HR-MM in the presence of 2 mM vanillin (data not shown in detail).
FIG. 4.
Biotransformation of ferulic acid to vanillin by resting cells of E. coli XL1-Blue harboring pSKechE/Hfcs. Cells were grown overnight in 50 ml of LB containing 12.5 μg of tetracycline per ml, 100 μg of ampicillin per ml, and 1 mM IPTG. The cells were harvested, washed, and resuspended in 50 ml of HR-MM containing 3.8 mM ferulic acid. Incubation was performed at 37°C, and samples were taken and analyzed by HPLC. Symbols: ○, ferulic acid; ▴, vanillin; ▵, vanillyl alcohol.
Characterization of the Pseudomonas sp. strain HRfcsΩGm, HRechΩKm, and HRaatΩKm mutants.
The phenotypes of the Pseudomonas sp. strain HRfcsΩGm, HRechΩKm, and HRaatΩKm mutants on solidified MM with eugenol, ferulic acid, vanillin, vanillic acid, and gluconate as the sole carbon source, respectively, were investigated. The HRfcsΩGm and HRechΩKm mutants were not able to grow on eugenol or ferulic acid, but they retained the ability to grow on vanillin or vanillic acid, thus exhibiting the same phenotype as the nitrosoguanidine-induced mutants SK6167 and SK6202 of Pseudomonas sp. strain HR199. In contrast, no difference from the phenotype of the wild-type Pseudomonas sp. strain HR199 was detected with the mutant Pseudomonas sp. strain HRaatΩKm.
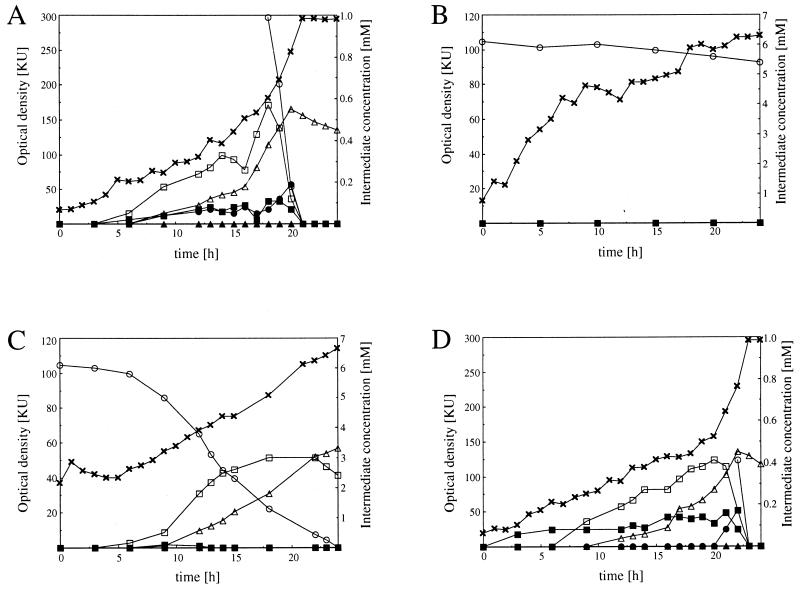
For further physiological characterization, cells of Pseudomonas sp. strain HRfcsΩGm, HRechΩKm, and HRaatΩKm were precultured overnight in HR-MM containing 0.5% (wt/vol) sodium gluconate as the carbon source. The cells were harvested, washed twice with MM, and used for inoculation of 50 ml of HR-MM containing about 6.5 mM eugenol as the carbon source in 250-ml Klett flasks. The cultures were incubated for 24 h at 30°C, and 1-ml samples were taken and analyzed by HPLC for the appearance or disappearance of catabolic intermediates. Cells of Pseudomonas sp. strain HR199 were incubated in the same way and used as a control. In cultures of the wild type, the intermediates coniferyl alcohol, coniferyl aldehyde, ferulic acid, and vanillic acid were obtained (Fig. 5A). Similar results were obtained with Pseudomonas sp. strain HRaatΩKm (Fig. 5D). In cultures of Pseudomonas sp. strain HRfcsΩGm, only a slight decrease in the eugenol concentration from 6.1 to 5.4 mM within 24 h was observed, but no intermediates were detectable (Fig. 5B). Interestingly, Pseudomonas sp. strain HRechΩKm differed significantly from Pseudomonas sp. strain HRfcsΩGm, since eugenol was completely consumed by this mutant. Coniferyl alcohol accumulated to a maximum concentration of 3 mM after about 18 h and was then completely converted to ferulic acid (Fig. 5C). After 40 h, ferulic acid was detected as the only product at a concentration of 5.9 mM, which was not further metabolized even after 72 h (data not shown in detail). Thus, eugenol was converted to ferulic acid by this mutant with a molar yield of about 97%.
FIG. 5.
Characterization of the mutants Pseudomonas sp. strain HRfcsΩGm, HRechΩKm, and HRaatΩKm. Cells were precultured with gluconate as the carbon source and used for inoculation of 50 ml of HR-MM containing about 6.5 mM eugenol as the carbon source in 250-ml Klett flasks. The cultures were incubated at 30°C, and growth was monitored with a Klett-Summerson photometer. Samples were taken from the cultures, and supernatants were analyzed by HPLC as described in Materials and Methods. (A) Pseudomonas sp. strain HR199; (B) Pseudomonas sp. strain HRfcsΩGm; (C) Pseudomonas sp. strain HRechΩKm; (D) Pseudomonas sp. strain HRaatΩKm. x, optical density; ○, eugenol concentration; □, coniferyl alcohol concentration; ■, coniferyl aldehyde concentration; ▵, ferulate concentration; ▴, vanillin concentration; ●, vanillate concentration.
The inactivation of the ech gene in mutant Pseudomonas sp. strain HRechΩKm was confirmed by the enzyme assay for enoyl-CoA hydratase/aldolase activity described in Materials and Methods. The mutants Pseudomonas sp. strain HRfcsΩGm, HRechΩKm, and HRaatΩKm were also investigated with respect to feruloyl-CoA synthetase activity. The mutants and the wild-type Pseudomonas sp. strain HR199 were grown in HR-MM containing 0.5% (wt/vol) gluconate and 0.1% (vol/vol) eugenol. Cells were harvested in the late exponential growth phase, and the corresponding soluble fractions of crude extracts were analyzed for feruloyl-CoA synthetase activity (Table 2). No feruloyl-CoA synthetase activity was detectable in extracts of Pseudomonas sp. strain HRfcsΩGm, whereas disruption of the ech and aat genes apparently had no influence on the feruloyl-CoA synthetase activity (Table 2).
DISCUSSION
Pseudomonas sp. strain HR199 is able to utilize eugenol and ferulic acid as the sole carbon source for growth. The genes, which are essential in ferulic acid catabolism, were identified on DNA fragments E230 and E94, which had been cloned recently in studies of the catabolism of eugenol (20, 29). The fcs and aat genes, which encoded proteins with homologies to thiokinases and β-ketothiolases, respectively (this study), were localized in the immediate neighborhood of the ech and vdh genes, encoding an enoyl-CoA hydratase-homologous protein and a vanillin dehydrogenase, respectively (29). Moreover, fcs and aat are most probably constituents of one operon, since the translational start codon of aat overlapped with the translational stop codon of fcs. All these data suggested that ferulic acid degradation in Pseudomonas sp. strain HR199 is initiated by the activation of ferulic acid to feruloyl-CoA, followed by a β-oxidation mechanism analogous to the β-oxidation pathway of fatty acid catabolism (Fig. 1), which had been previously proposed for the degradation of substituted cinnamic acids in plants (47). This pathway would include the thioclastic cleavage of 4-hydroxy-3-methoxyphenyl-β-ketopropionyl-CoA to yield acetyl-CoA and vanillyl-CoA, catalyzed by the aat gene product (β-ketothiolase). However, genes encoding a 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA dehydrogenase or a vanillyl-CoA hydrolase were not identified in Pseudomonas sp. strain HR199.
The identity of the fcs gene product as a feruloyl-CoA synthetase was concluded from a comparison of its amino acid sequence with those of fatty acid-CoA synthetases. FCS showed significant homologies to these CoA ligases and also possessed an amino acid sequence (aa 190 to 199) proposed to be involved in acyl-adenylate formation (5), which is typical for this kind of enzymes. This assumption was confirmed by expression of this gene in E. coli. Feruloyl-CoA synthetase activity was detected by a spectrophotometric assay, and the formation of feruloyl-CoA was directly indicated by HPLC analysis. Thus, this is the first report of the cloning and molecular characterization of a bacterial fcs gene, encoding a feruloyl-CoA synthetase.
During our investigations of the eugenol catabolism of Pseudomonas sp. strain HR199, Gasson et al. reported the isolation and characterization of a gene of the enoyl-SCoA hydratase/isomerase superfamily from Pseudomonas fluorescens AN103, which encoded an enzyme for the hydration and nonoxidative cleavage of feruloyl-SCoA to vanillin and acetyl-SCoA (16). From amino acid sequence comparison, it was obvious that the ech gene product of Pseudomonas sp. strain HR199 had the same function as the enzyme reported by Gasson et al. This assumption was confirmed by expression of the ech gene in E. coli and detection of enoyl-CoA hydratase/aldolase activity for the obtained gene product. Recently, Venturi et al. reported the identification of two proteins designated Fca and Vdh and asserted that these two proteins were responsible for the conversion of ferulic acid to vanillic acid in Pseudomonas putida WCS358 (46). However, they did not provide any evidence that Fca catalyzed the direct cleavage of ferulic acid to acetate and vanillin. Moreover, the high homology of Fca to the enoyl-CoA hydratase/aldolase of Pseudomonas sp. strain HR199 and the similar arrangement of the fca and vdh genes in P. putida WCS358 and ech and vdh in Pseudomonas sp. strain HR199 (29) suggest that Fca also represents an enoyl-CoA hydratase/aldolase rather than a ferulic acid deacetylase. In conclusion, ferulic acid degradation in Pseudomonas sp. strain HR199, as in P. putida WCS358 and Pseudomonas fluorescens AN103, proceeds via feruloyl-CoA, which is hydrated and cleaved by the action of the ech gene product (Fig. 1). This conclusion was also confirmed by the coexpression of the fcs and ech genes from Pseudomonas sp. strain HR199 in E. coli. Corresponding recombinant strains were able to convert ferulic acid to vanillin. The conversion rate obtained was in a biotechnologically interesting range; however, the coformation of vanillyl alcohol, which could be regarded as a detoxification reaction, might cause problems in such a process.
To confirm the essential involvement of fcs and ech in ferulic acid catabolism and to investigate the role of the aat gene, these genes were inactivated by disruption. These experiments clearly demonstrated that the activation of ferulic acid to the corresponding CoA thioester is absolutely necessary for the catabolism of this aromatic compound. Also, mutants with an inactivated ech gene were not able to grow on ferulic acid, confirming the involvement of the enoyl-CoA hydratase/aldolase in this catabolism. Although aat is most probably cotranscribed with fcs, the encoded β-ketothiolase seems not to be involved in the ferulic acid degradation pathway, since the mutant Pseudomonas sp. strain HRaatΩKm, with a defective aat gene, exhibited the same phenotype as the wild type. Thus, the presence of the aat gene and the arrangements of fcs and aat might reflect an evolutionary relic, as a result of gene duplication and diversification.
If this CoA-dependent, non-β-oxidative mechanism of ferulic acid cleavage, which was already proposed by Lute (25) and was first published by Gasson et al. (16), is generally realized in bacteria or if β-oxidation-like mechanisms and direct nonoxidative deacetylation mechanisms can be confirmed at the genetic level, they will be the subject of further studies.
ACKNOWLEDGMENTS
The synthesis of 4-hydroxy-3-methoxyphenyl-β-hydroxypropionate methylester by Dirk Fabritius and his assistance during the synthesis of 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl-CoA are gratefully acknowledged.
H.P. and A.S. are indebted to Haarmann & Reimer GmbH for providing a collaborative research grant.
REFERENCES
- 1.Abe H, Ohtake A, Yamamoto S, Satoh Y, Takayanagi M, Amaya Y, Takiguchi M, Sakuraba H, Suzuki Y, Mori M, Niimi H. Cloning and sequence analysis of a full length cDNA encoding human mitochondrial 3-oxoacyl-CoA thiolase. Biochim Biophys Acta. 1993;1216:304–306. doi: 10.1016/0167-4781(93)90160-f. [DOI] [PubMed] [Google Scholar]
- 2.Abe T, Fujino T, Fukuyama R, Minoshima S, Shimizu N, Toh H, Suzuki H, Yamamoto T. Human long-chain acyl-CoA synthetase: structure and chromosomal location. J Biochem. 1992;111:123–128. doi: 10.1093/oxfordjournals.jbchem.a123707. [DOI] [PubMed] [Google Scholar]
- 3.Achterholt S, Priefert H, Steinbüchel A. Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. strain HR199 and molecular characterization of the gene. J Bacteriol. 1998;180:4387–4391. doi: 10.1128/jb.180.17.4387-4391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An J H, Kim Y S. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii: cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem. 1998;257:395–402. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- 5.Babbitt P C, Kenyon G L, Martin B M, Charest H, Slyvestre M. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- 6.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 7.Bullock W O, Fernandez J M, Stuart J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 8.Chen C L, Chang H M, Kirk T. Aromatic acids produced during degradation of lignin in spruce wood by Phanerochaete chrysosporium. Holzforschung. 1982;36:3–9. [Google Scholar]
- 9.Clark G S. Vanillin. Perfum Flavor. 1990;15:45–54. [Google Scholar]
- 10.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Sqares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egland P G, Gibson J, Harwood C S. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J Bacteriol. 1995;177:6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich B, Hogrefe C, Schlegel H G. Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J Bacteriol. 1981;147:198–205. doi: 10.1128/jb.147.1.198-205.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulda M, Heinz E, Wolter F P. The fadD gene of Escherichia coli K12 is located close to rnd at 39.6 min of the chromosomal map and is a new member of the AMP-binding protein family. Mol Gen Genet. 1994;242:241–249. doi: 10.1007/BF00280412. [DOI] [PubMed] [Google Scholar]
- 16.Gasson M J, Kitamura Y, McLauchlan W R, Narbad A, Parr A J, Parsons E L H, Payne J, Rhodes M J C, Walton N J. Metabolism of ferulic acid to vanillin. A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester. J Biol Chem. 1998;273:4163–4170. doi: 10.1074/jbc.273.7.4163. [DOI] [PubMed] [Google Scholar]
- 17.Hagedorn S, Kaphammer B. Microbial biocatalysis in the generation of flavor and fragrance chemicals. Annu Rev Microbiol. 1994;48:773–800. doi: 10.1146/annurev.mi.48.100194.004013. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 20.Kresse, A. U., J. Overhage, H. Priefert, and A. Steinbüchel. Unpublished results.
- 21.Krings U, Berger R G. Biotechnological production of flavours and fragrances. Appl Microbiol Biotechnol. 1998;49:1–8. doi: 10.1007/s002530051129. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Li K, Frost J W. Synthesis of vanillin from glucose. J Am Chem Soc. 1998;120:10545–10546. [Google Scholar]
- 24.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Lute J R. The pathway of phenylpropanoid degradation in Pseudomonas putida MT-2. Ph.D. thesis. The University of Michigan; 1986. [Google Scholar]
- 26.Overhage J, Kresse A U, Priefert H, Sommer H, Krammer G, Rabenhorst J, Steinbüchel A. Molecular characterization of the genes pcaG and pcaH, encoding protocatechuate 3,4-dioxygenase, which are essential for vanillin catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol. 1999;65:951–960. doi: 10.1128/aem.65.3.951-960.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overhage, J., H. Priefert, J. Rabenhorst, and A. Steinbüchel. Biotransformation of eugenol to vanillin by a mutant of Pseudomonas sp. strain HR199 constructed by disruption of the vanillin dehydrogenase (vdh) gene. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 28.Priefert H, Hein S, Krüger N, Zeh K, Schmidt B, Steinbüchel A. Identification and molecular characterization of the Alcaligenes eutrophus H16 aco operon genes involved in acetoin catabolism. J Bacteriol. 1991;173:4056–4071. doi: 10.1128/jb.173.13.4056-4071.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priefert H, Rabenhorst J, Steinbüchel A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J Bacteriol. 1997;179:2595–2607. doi: 10.1128/jb.179.8.2595-2607.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Priefert, H., J. Overhage, and A. Steinbüchel. Identification and molecular characterization of the eugenol hydroxylase genes (ehyA/ehyB) of Pseudomonas sp. strain HR199. Arch. Microbiol., in press. [DOI] [PubMed]
- 31.Pries A, Priefert H, Krüger N, Steinbüchel A. Identification and characterization of two Alcaligenes eutrophus gene loci relevant to the poly(β-hydroxybutyric acid)-leaky phenotype which exhibit homology to ptsH and ptsI of Escherichia coli. J Bacteriol. 1991;173:5843–5853. doi: 10.1128/jb.173.18.5843-5853.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabenhorst J. Production of methoxyphenol type natural aroma chemicals by biotransformation of eugenol with a new Pseudomonas sp. Appl Microbiol Biotechnol. 1996;46:470–474. [Google Scholar]
- 33.Rabenhorst J, Hopp R. Verfahren zur Herstellung von natürlichem Vanillin. European patent 0 405 197 A1. 1990. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: wachstumsphysiologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 37.Shiotsu Y, Samejima M, Habu N, Yoshimoto T. Enzymatic conversion of stilbenes from the inner back of Picea glehnii into aromatic aldehydes. Mukuzai Gakkaishi. 1989;35:826–831. [Google Scholar]
- 38.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulations of Gram negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria-plant interaction. Berlin, Germany: Springer-Verlag KG; 1983. pp. 98–106. [Google Scholar]
- 40.Smith, D. R., and K. Robison. Unpublished results.
- 41.Stegemann H, Francksen H, Macko V. Potato proteins: genetic and physiological changes, evaluated by one or two-dimensional PAA-geltechniques. Z Naturforsch Sect C. 1973;28:722–732. doi: 10.1515/znc-1973-11-1213. [DOI] [PubMed] [Google Scholar]
- 42.Steinbüchel A, Priefert H, Rabenhorst J. Syntheseenzyme für die Herstellung von Coniferylalkohol, Coniferylaldehyd, Ferulasäure, Vanillin und Vanillinsäure und deren Verwendung. European patent 0 845 532 A2. 1998. [Google Scholar]
- 43.Strauss E C, Kobori J A, Siu G, Hood L E. Specific-primer-directed DNA sequencing. Anal Biochem. 1986;154:353–360. doi: 10.1016/0003-2697(86)90536-1. [DOI] [PubMed] [Google Scholar]
- 44.Tadasa K, Kayahara H. Initial steps of eugenol degradation pathway of a microorganism. Agric Biol Chem. 1983;47:2639–2640. [Google Scholar]
- 45.Toms A, Wood J M. The degradation of trans-ferulic acid by Pseudomonas acidovorans. Biochemistry. 1970;9:337–343. doi: 10.1021/bi00804a021. [DOI] [PubMed] [Google Scholar]
- 46.Venturi V, Zennaro F, Degrassi G, Okeke B C, Bruschi C V. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology. 1998;144:965–973. doi: 10.1099/00221287-144-4-965. [DOI] [PubMed] [Google Scholar]
- 47.Vollmer K O, Reisener H J, Grisebach H. The formation of acetic acid from p-hydroxycinnamic acid during its degradation to p-hydroxybenzoic acid in wheat shoots. Biochem Biophys Res Commun. 1965;21:221–225. doi: 10.1016/0006-291x(65)90275-5. [DOI] [PubMed] [Google Scholar]
- 48.Wilbur W J, Lipman D J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci USA. 1983;80:726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenk M H, Ulbrich B, Busse J, Stöckigt J. Procedure for the enzymatic synthesis and isolation of cinnamoyl-CoA thiolesters using a bacterial system. Anal Biochem. 1980;101:182–187. doi: 10.1016/0003-2697(80)90058-5. [DOI] [PubMed] [Google Scholar]