Abstract
It has been well established that a certain amount of ingested starch can escape digestion in the human small intestine and consequently enters the large intestine, where it may serve as a carbon source for bacterial fermentation. Thirty-eight types of human colonic bacteria were screened for their capacity to utilize soluble starch, gelatinized amylopectin maize starch, and high-amylose maize starch granules by measuring the clear zones on starch agar plates. The six cultures which produced clear zones on amylopectin maize starch- containing plates were selected for further studies for utilization of amylopectin maize starch and high-amylose maize starch granules A (amylose; Sigma) and B (Culture Pro 958N). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to detect bacterial starch-degrading enzymes. It was demonstrated that Bifidobacterium spp., Bacteroides spp., Fusobacterium spp., and strains of Eubacterium, Clostridium, Streptococcus, and Propionibacterium could hydrolyze the gelatinized amylopectin maize starch, while only Bifidobacterium spp. and Clostridium butyricum could efficiently utilize high-amylose maize starch granules. In fact, C. butyricum and Bifidobacterium spp. had higher specific growth rates in the autoclaved medium containing high-amylose maize starch granules and hydrolyzed 80 and 40% of the amylose, respectively. Starch-degrading enzymes were cell bound on Bifidobacterium and Bacteroides cells and were extracellular for C. butyricum. Active staining for starch-degrading enzymes on SDS-PAGE gels showed that the Bifidobacterium cells produced several starch-degrading enzymes with high relative molecular (Mr) weights (>160,000), medium-sized relative molecular weights (>66,000), and low relative molecular weights (<66,000). It was concluded that Bifidobacterium spp. and C. butyricum degraded and utilized granules of amylomaize starch.
The human colon is described as a complex ecosystem, containing over 400 species of microbes (25, 26). The principal nutrients for bacterial growth in the colon are dietary carbohydrates and proteins, which have escaped digestion in the upper gastrointestinal tract, as well as secretions rich in glycoproteins and pancreatic enzymes (11, 20). Although the composition of the colonic microbiota of adults is relatively stable, factors including the diet can alter the composition or metabolism of the gut microbes. A high intake of carbohydrate is likely to elevate the numbers of bifidobacteria in the human colon, while a high-fat diet could lead to an increase in the population of Bacteroides spp. (7, 38). Furthermore, it has been shown that oligosaccharides and, in particular, oligofructose can stimulate the growth of Bifidobacterium spp. in the human colon (24, 31, 40). These findings support the proposal that the balance of the colonic microbes can be manipulated by dietary means.
It is known that starches are one of the major carbohydrates available in the human colon (1, 12, 19). Starch that can escape digestion in the human small intestine can enter the large intestine, where it can be used as a substrate for bacterial fermentation, resulting in the production of gases and volatile fatty acids, of which butyric acid is considered to have specific antiproliferative effects upon colonic epithelial cells (6, 16, 34, 35, 39, 41). These starches that are not degraded in the small intestine are referred to as resistant starch. In an Australian western-style diet, the intake of starch is approximately 132 g per day, which includes approximately 5 g per day of resistant starch (4). Epidemiological studies have shown a correlation between a high intake of dietary starch, e.g., up to 400 g per day for countries with high starch intakes (37), and a lower incidence of colorectal cancer (10). These studies proposed that bacterial fermentation of the large amounts of dietary starch would generate elevated levels of butyric acid in the colon.
Both host pancreatic enzymes and colonic bacterial amylases are involved in the degradation of starches; however, bacterial amylase has been shown to be more active (21). These studies showed that Bifidobacterium, Bacteroides, Fusobacterium, and Butyrivibrio strains were all amylolytic when soluble starch was used; however, up to 58% of amylolytic isolates were Bifidobacterium spp. (21). Other workers (18), using amylose starch, showed that nearly all of the amylolytic bacteria isolated from a South Korean subject were bifidobacteria. Because both the chemical and physical characteristics of starch can vary considerably depending on the species, the granular composition, and the ratio of amylose to amylopectin, the utilization of starches by colonic microbes needs to be studied further, especially starches resistant to pancreatic enzymes.
In this study we examined a wide range of pure cultures of colonic bacteria for their capacity to utilize soluble starch, amylopectin maize starch, and amylomaize starch granules. The fermentation products and starch residues, as well as the activity of the starch-degrading enzymes, were studied in more detail for six bacterial cultures that had the greatest starch-degrading activity.
MATERIALS AND METHODS
Growth substrates.
Soluble starch (S-9765), granular amylopectin maize starch (A-7780), and granular high-amylose maize starch A (A-7043) were purchased from Sigma Chemical Co. High-amylose maize starch granule A contained >70% amylose and 32.5% total dietary fiber, while high-amylose starch granules B (Culture Pro 958N), which were kindly supplied by Starch Australasia, Ltd., contained more than 80% amylose and 33.4% total dietary fiber (7a). The dietary fiber content was measured by using the enzymatic-gravimetric method (AOAC International method no. 991.43) of the Association of Official Analytical Chemists (AOAC) (3). With this method, it was noted that as the amylose content increased, so did its resistance to the action of digestive amylases (9). Both amylomaize granules A and B had comparable high degrees of resistance to amylase-induced degradation (amylolysis) and were all granular (diameter of ca. 10 μm) (8). Unless otherwise specified, all other chemicals used in the experiments were purchased from Sigma.
Bacterial strains.
Bifidobacterium species were obtained from the culture collection of Food Science Australia, Highett, except Bifidobacterium breve, which was kindly donated by The University of Western Sydney. Lactobacillus, Clostridium, Eubacterium, Streptococcus, Staphylococcus, Propionibacterium, and Ruminococcus strains were received from the University of New South Wales Culture Collection in The School of Microbiology and Immunology. Bacteroides spp. were purchased from the Department of Veterinary Science, University of Sydney. All bacteria are listed in Table 1.
TABLE 1.
Bacterial hydrolysis of soluble starch, amylopectin, and granular high-amylose maize starch A (HA A)a
| Bacterial strain | Reference no. | ATCC source | Mean diam of clear zone (mm) ± SD on:
|
||
|---|---|---|---|---|---|
| Soluble starch | HA A | Amylopectin | |||
| Bifidobacterium infantis | FII 510000 | NA | 20.5 ± 3.3 | 7.5 ± 10.6 | 26.3 ± 1.7 |
| Bifidobacterium adolescentis | FII 509400 | 15703 | 18 ± 2.6 | 7.5 ± 8.6 | 21 ± 3.4 |
| Bifidobacterium bifidum | FII 509800 | 11863 | 32.2 ± 2.4 | 22.2 ± 3.1 | 33.6 ± 3.5 |
| Bifidobacterium longum | FII 509700 | NA | 24.6 ± 5.5 | 16.7 ± 1.3 | 26.2 ± 2.9 |
| Bifidobacterium pseudolongum | FII 509500 | 25526 | 31.33 ± 1.5 | 21.7 ± 1.7 | 34.5 ± 3.9 |
| Bifidobacterium breve | CSC 1930 | NA | 30 ± 0 | 16 ± 1.4 | 30.5 ± 0.5 |
| Bacteroides fragilis | UNSW 035100 | 25285 | 19.2 ± 1.0 | 0 | 23 ± 3.6 |
| Bacteroides vulgatus | UNSW 035200 | 8482 | 20.2 ± 1.3 | 0 | 16.3 ± 7.5 |
| Bacteroides thetaiotaomicron | FII 513200 | 29148 | 0 | 0 | 0 |
| Bacteroides distasonis | FII 513300 | 8503 | 0 | 0 | 0 |
| Bacteroides ovatus | FII 513800 | 8483 | 18 ± 1.2 | 0 | 22.8 ± 1.7 |
| Clostridium difficile | UNSW 055800 | NA | 0 | 0 | 0 |
| Clostridium butyricum | UNSW 060600 | NA | 26.3 ± 0.6 | 35.6 ± 5.4 | 37 ± 6.3 |
| Clostridium tyrobutyricum | UNSW 052400 | NA | 0 | 0 | 0 |
| Clostridium sporogenes | UNSW 048400 | 19404 | 0 | 0 | 0 |
| Fusobacterium mortiferum | FII 513500 | 25557 | 0 | 0 | 20.7 ± 1.0 |
| Fusobacterium gonidiaformans | FII 513400 | 25563 | 18.5 ± 0.6 | 0 | 22.7 ± 1.0 |
| Fusobacterium necrogenes | FII 513600 | 25556 | 14.5 ± 0.6 | 0 | 19.7 ± 1.0 |
| Fusobacterium necrophorum | FII 513700 | 25286 | 0 | 0 | 19 ± 0.8 |
| Lactobacillus viridescens | UNSW 023200 | 12706 | 0 | 0 | 0 |
| Lactobacillus fermentum | UNSW 053300 | 9338 | 0 | 0 | 0 |
| Lactobacillus casei | UNSW 017900 | NA | 0 | 0 | 0 |
| Lactobacillus acidophilus | UNSW 060800 | 4356 | 0 | 0 | 0 |
| Lactobacillus plantarum | UNSW 003900 | 8014 | 0 | 0 | 0 |
| Lactobacillus rhamnosus | UNSW 060400 | 7469 | 0 | 0 | 0 |
| Lactobacillus brevis | UNSW 055100 | NA | 0 | 0 | 0 |
| Lactobacillus salivarius | UNSW 056300 | 11741 | 0 | 0 | 0 |
| Streptococcus thermophilus | UNSW 023300 | NA | 0 | 0 | 0 |
| Streptococcus salivarius | UNSW 034800 | 13419 | 19 ± 0 | 0 | 0 |
| Propionibacterium acnes | UNSW 042200 | 6919 | 20 ± 0.2 | 0 | 16 ± 0.1 |
| Propionibacterium freudenreichii | UNSW 035900 | 035900 | 0 | 0 | 0 |
| Eubacterium limosum | UNSW 041602 | 8486 | 26 ± 0.1 | 0 | 25 ± 0.1 |
| Staphylococcus aureus | UNSW 048500 | 9144 | 0 | 0 | 0 |
| Lactococcus lactis | UNSW 018000 | 19435 | 0 | 0 | 0 |
| Peptostreptococcus anaerobius | UNSW 041900 | 27337 | 0 | 0 | 0 |
| Enterococcus faecalis | UNSW 054400 | 19433 | 0 | 0 | 0 |
| Enterococcus hirae | UNSW 032800 | 9790 | 0 | 0 | 0 |
| Escherichia coli | UNSW 048200 | 11775 | 0 | 0 | 0 |
Results are expressed as the size of the cleared zone after growth on agar plates containing the starches (n = 4). NA, not applicable.
Starch hydrolysis on agar plates.
Bacterial strains (n = 38) were subcultured from freeze-dried ampoules into 25 ml of anaerobic PYG broth (14). After incubation at 37°C for 2 to 4 days, these cultures were inoculated (10%) into 10 ml of starch broth in serum tubes and incubated at 37°C in an anaerobic chamber (Mark 3 Workstation; DW Scientific) (N2-CO2-H2 at 80:15:5). Basal medium (BM1) contained (in grams/liter): bacteriological peptone (Oxoid), 5; yeast extract (Oxoid), 5; tryptone (Oxoid), 5; NaCl, 0.1; K2HPO4, 0.04; KH2PO4, 0.04; MgSO4, 0.01; CaCl2, 0.01; NaH2CO3, 2.0; Tween 80, 10 ml; hemin, 0.005; vitamin K1, 0.0002; vitamin B12, 0.00125; and cysteine, 0.5. The pH was adjusted to 6.8, and 5 g of soluble starch (BDH) or glucose was added prior to the medium being boiled and then cooled with nitrogen gas bubbling into it. The medium was then dispensed under a flow of nitrogen gas into serum tubes, which were then sealed and autoclaved at 121°C for 15 min. For an additional control, the basal medium without any carbon source was used. The concentrations of total starch and resistant starch in uninoculated starch media were measured by using the α-amylase amyloglucosidase method (Megazyme, total starch assay kit; Megazyme International, Ltd.). After 24 h of anaerobic incubation in the starch broth or glucose broth, 10 μl of the cultures was transferred to sterile paper discs (6 mm), which were then placed on the surfaces of dried starch agar plates. The starch agar plates were similar to the starch medium, but with 10 g of soluble starch, granular amylopectin maize starch, or granular high-amylose maize starches A and B per liter. The autoclaving caused the disruption and gelatinization of the amylopectin maize starch granules but only partially gelatinized the high-amylose maize starch granules. These later granules do not fully gelatinize until heated to 150 to 170°C. Consequently, the autoclaving left these granules intact, as visualized by light microscopy. Each petri dish contained exactly 30 ml of autoclaved medium. The inoculated plates were incubated in the anaerobic chamber for 5 days at 37°C, after which time the diameters of the clear zones around the paper discs were measured by using a light box and ruler. Less-distinct zones were further enhanced by adding I2-KI solution (0.15% I2 in 1.5% KI).
In vitro fermentation.
Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bacteroides fragilis, Bacteroides vulgatus, Clostridium butyricum, or Eubacterium limosum was individually grown anaerobically for 24 h in the basal medium (BM1) containing 5 g of soluble starch from BDH per liter anaerobically for 24 h. Aliquots (0.1 ml) from overnight cultures were inoculated into 20 ml of growth medium that consisted of basal medium 2 (BM2) and 10 g of glucose, granular amylopectin maize starch, or granular high-amylose maize starch A or B per liter. BM2, which was modified BM1 with less yeast extract, contained (in grams/liter): bacteriological peptone (Oxoid), 7.5; yeast extract (Oxoid), 2.5; tryptone (Oxoid), 5.0; K2HPO4, 2.0; KH2PO4, 1.0; NaHCO3, 0.2; NaCl, 0.2; MgCl2, 0.2; CaCl2, 0.2; MnCl2, 0.02; CoCl2, 0.02; cysteine, 0.5; Fe2SO4, 0.005; Tween 80, 2 ml; hemin, 0.005; vitamin, B12, 0.00125; and vitamin K1, 0.0002. The tubes were incubated anaerobically at 37°C for 48 h and sampled at 0, 1, 2, 3, 4, 6, 8, 10, 12, and 24 h. The bacterial growth was enumerated by using a modification of the micro-drop method (22). In brief, 0.1-ml aliquots were removed and immediately diluted with 0.9 ml of half-strength Wilkins Chalgren Anaerobic Broth. A Wilkins Chalgrens Anaerobic agar plate was divided into five strips, and 10-μl aliquots from each dilution were dropped onto the strips. The plates were incubated at 37°C for 72 h in the anaerobic chamber. Bacterial populations were quantified as CFU per milliliter. Specific growth rates were calculated according to the equation given by Pirt (32). Samples were collected at 48 h for analysis of the residual amylose and total carbohydrates. In addition, phase-contrast microscopy was used to examine the starch granules before and after autoclaving and growth.
The residual apparent amylose concentrations after 48 h of fermentation were determined by using the Blue Value method (26). This method uses the affinity of iodine for the alpha-helix of the amylose to quantify the amount of amylose, since the resultant polyiodide inclusion complexes have a distinctive Prussian blue color. The intensity of the color is a measure of the apparent amylose content. The concentrations of total carbohydrates in the spent culture fluids were evaluated by using the traditional phenol-sulfuric assay, which produces a pink color (15).
Determination of the activity of starch-degrading enzymes.
The six bacterial strains Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bacteroides fragilis, Bacteroides vulgatus, Clostridium butyricum, and Eubacterium limosum were grown anaerobically for 48 h in 50 ml of BM2 as described previously with 5 g of glucose, granular amylopectin starch, or granular high-amylose maize starch A per liter as the carbon source. The unused amylomaize starch granules were removed from fermentation cultures by centrifugation for 2 min at 1,000 × g, and the bacterial cells were then collected by centrifuge at 5,000 × g for 20 min by using a bench centrifuge. The supernatants were kept on ice for enzyme analyses, and the bacterial pellets were washed twice and resuspended in 0.05 M potassium phosphate buffer (pH 7.0). The cell-bound enzymes were released by sonicating (Branson Sonifier 450) for 5 min the washed cells held on ice (30 1-s pulses/min). The starch-degrading enzymes were quantified by mixing 1 ml of crude cell extracts or culture supernatants with 1 ml of 0.2% soluble starch (BDH) and incubating the mixture in a 37°C water bath for 60 min. The activities of the starch-degrading enzymes were quantified by measuring released glucose by using the reducing sugar method (16). The protein concentration was measured by using the Bio-Rad protein assay kit. One unit of enzyme activity was equivalent to one micromole of glucose released per milligram of protein after 60 min of incubation.
Separation of starch-degrading enzymes by SDS-PAGE.
A modification of the method of Ji et al. (18) was used for the separation of bacterial starch-degrading enzymes by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Briefly, 100 μl of crude cell extracts prepared by sonicating the bacterial cultures was individually mixed with a 50-μl aliquot of loading buffer solution which contained mercaptoethanol (5%), SDS (3.4%), glycerol (15%), and bromophenol blue (0.01%) dissolved in 47 mM Tris-HCl buffer (pH 6.8). The proteins were denatured in a boiling water bath for 3 min and then 50-μl samples were applied to an SDS–10% PAGE gel. After electrophoresis, the gels were washed for 1 h in 2.5% (wt/vol) Triton X-100 at 37°C with gentle mixing to remove the SDS and then soaked overnight in 50 mM acetate buffer (pH 5.0) containing 0.2% soluble starch (BDH). The gel was then rinsed in distilled water and finally stained with iodine solution (0.15% I2, 1.5% KI) for 5 min at room temperature. The translucent bands that were detectable against a dark background indicated the presence of starch-degrading enzymes. The α-amylase (A3306) was used as the positive control, and the molecular weights of the starch-degrading enzymes were determined relative to the molecular weight standard (Bio-Rad SDS-PAGE broad-range standards) included on the same gel and then visualized by separately staining that lane with Coomassie blue and realigning it with the remaining gel after the staining.
RESULTS
Comparison of amylolytic activities of 38 bacterial strains on starch agar plates.
All of the strains used in these experiments are typical of those found in the human colon. They were all able to grow in the basal medium containing 1% glucose. The data in Table 1 only show the sizes of the clear zones formed by bacterial strains previously grown in glucose medium, since 22 of the 38 strains were not able to grow in the medium containing soluble starch. Larger clear zones were observed on the starch agar plates for the bacterial cultures that could be precultured in growth medium containing soluble starch, implying that the starch-degrading enzymes were induced in the presence of starch in the preculture. As seen in Table 1, when amylopectin and soluble starch were included in the agar, large clear zones were formed by all of the Bifidobacterium strains and several of the Bacteroides and Fusobacterium species, while only one strain each of Propionibacterium, Clostridium, Streptococcus, and Eubacterium spp. produced clear zones. In contrast, only Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bifidobacterium longum, Bifidobacterium breve, and Clostridium butyricum formed distinct clear zones on granular amylomaize starch agar plates. There were no detectable zones around Bacteroides vulgatus and Bacteroides fragilis on these plates. It is interesting to note that none of the Lactobacillus strains that were tested showed any starch-degrading activity for any of the starches used.
Analysis of uninoculated starch media.
As presented in Table 2, the concentration of resistant starch in the uninoculated media after autoclaving at 121°C for 15 min was relatively low, with only 6.8 and 5.0% in high-amylose maize starch granules A and B, respectively, and 0.2 and 0.4 in the nongranular amylopectin maize starch and soluble-starch-containing media, respectively.
TABLE 2.
Concentration of total starch and resistant starch in the uninoculated medium prepared by the addition of soluble starch, amylopectin granules, or high-amylose maize granules A or B to basal medium prior to autoclaving at 121°C for 15 mina
| Medium | Total starch (mg/ml) | Resistant starch (%) |
|---|---|---|
| Soluble starch | 9.4 | 0.4 |
| Amylopectin | 9.3 | 0.2 |
| High-amylose maize granules A | 9.7 | 6.8 |
| High-amylose maize granules B | 9.7 | 5.0 |
Only amylomaize granules A and B were granular after autoclaving. The initial total starch concentration was 10 mg · ml−1. Results for resistant starch are expressed as the percentage of total starch.
Fermentation of several starches by selected colonic bacteria.
When examined microscopically, it was noted that amylopectin maize starch and amylomaize starch consisted of granules with an average diameter of 10 μm before autoclaving; however, after the autoclaving complete gelatinization and granule disruption was noted for amylopectin maize starch. The structures of the granules from amylomaize starch granules A and B were maintained, and only a small percentage of the granules were disrupted. Based on the data in Table 1, Bifidobacterium bifidum, Bifidobacterium pseudolongum, Clostridium butyricum, Bacteroides fragilis, Bacteroides vulgatus and Eubacterium limosum were selected for further study by using amylopectin maize starch granules, high-amylose maize starch granules A, and high-amylose maize starch granules B, with glucose as the control. The estimated specific growth rates that were calculated from the viable counts during the log phase are shown in Table 3. Clostridium butyricum had the highest specific growth rates with no demonstrable difference between the glucose- and starch-containing media. Comparable growth rates were also noted for Bifidobacterium spp. on the various starch media. Lower growth rates were noted for Bacteroides fragilis, Bacteroides vulgatus, and Eubacterium limosum in the high-amylose maize starch media.
TABLE 3.
Specific growth rates of selected bacterial strains in autoclaved media containing 1% (wt/vol) glucose, amylopectin, or high-amylose maize granules A (HA A) or B (HA B)a
| Bacterium | Mean specific growth rate/h ± SD in:
|
|||
|---|---|---|---|---|
| Glucose | Amylopectin | HA A | HA B | |
| Clostridium butyricum | 1.33 ± 0.09 | 1.09 ± 0.01 | 1.35 ± 0.07 | 1.07 ± 0.13 |
| Bifidobacterium bifidum | 0.84 ± 0.06 | 0.72 ± 0.14 | 0.51 ± 0.11 | 0.75 ± 0.15 |
| Bifidobacterium pseudolongum | 0.81 ± 0.07 | 0.71 ± 0.09 | 0.58 ± 0.14 | 0.70 ± 0.12 |
| Bacteroides vulgatus | 0.83 ± 0.30 | 0.68 ± 0.20 | 0.33 ± 0.12 | 0.50 ± 0.05 |
| Bacteroides fragilis | 0.64 ± 0.19 | 0.49 ± 0.14 | 0.36 ± 0.11 | 0.40 ± 0.01 |
| Eubacterium limosum | 0.57 ± 0.11 | 0.63 ± 0.14 | 0.34 ± 0.05 | 0.42 ± 0.15 |
Only the HA A and HA B remained granular after autoclaving. The results are from triplicate experiments.
The extent to which the selected strains could degrade the various starches was monitored by measuring residual carbohydrate. As can be seen in Table 4, glucose was metabolized to a greater extent than was noted for amylopectin maize starch, high-amylose maize starch granules A, or high-amylose maize starch granules B. The concentrations of residual carbohydrate were similar for media containing high-amylose maize granules A and B. For all bacterial strains tested, there was a trend toward slightly higher levels of residual carbohydrate in high-amylose maize starch granule media than in glucose broth after 48 h of fermentation. The degradation of high-amylose maize starch granules varied with the different bacterial strains tested, and this was noted for both A and B granules. For example, in the medium containing high-amylose maize granules, only 4 mg of carbohydrate per ml was detected after 48 h of growth of Clostridium butyricum and 4.92 mg/ml after growth of Bifidobacterium bifidum; however, Bacteroides fragilis growth resulted in 8.86 mg of residual carbohydrate per ml. Furthermore, Bacteroides vulgatus and Eubacterium limosum degraded granular amylomaize poorly. Similar observations were also noted in amylopectin maize starch-containing medium. The concentration of total carbohydrates in the uninoculated medium was about 12 mg/ml, of which 10 mg/ml represented the added starch or glucose.
TABLE 4.
Concentration of total carbohydrate residues after bacterial growth for 48 h in autoclaved basal medium containing glucose, amylopectin, and high-amylose maize starch granules A (HA A) and B (HA B)a
| Bacterium | Mean concn (mg/ml) ± SD in medium containing:
|
|||
|---|---|---|---|---|
| Glucose | Amylopectin | HA A | HA B | |
| Clostridium butyricum | 2.12 ± 1.77 | 5.36 ± 1.73 | 4.04 ± 0.65 | 4.42 ± 1.00 |
| Bifidobacterium bifidum | 4.05 ± 0.60 | 5.30 ± 0.37 | 4.92 ± 0.21 | 6.96 ± 1.93 |
| Bifidobacterium pseudolongum | 4.64 ± 0.67 | 5.47 ± 0.87 | 7.53 ± 0.46 | 7.58 ± 1.20 |
| Bacteroides vulgatus | 7.08 ± 0.42 | 10.23 ± 1.19 | 9.66 ± 1.34 | 10.31 ± 1.82 |
| Bacteroides fragilis | 7.12 ± 0.45 | 7.86 ± 1.74 | 8.86 ± 0.67 | 9.00 ± 1.49 |
| Eubacterium limosum | 5.39 ± 2.26 | 9.97 ± 1.33 | 10.99 ± 0.73 | 12.45 ± 0.87 |
HA A and HA B remained granular after autoclaving. Total carbohydrates were measured by using the Dubois method and are the means of two determinations from four individual experiments. The initial total carbohydrate concentration was 12 mg/ml.
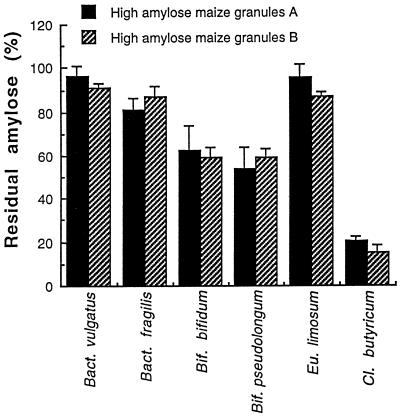
For each of the six cultures tested, the amounts of residual apparent amylose (Fig. 1) after 48 h of growth in the basal medium containing amylomaize starch granules A or B were consistent with the results of the total carbohydrate analyses, as presented in Table 4, when analytical errors are considered. Clostridium butyricum hydrolyzed the high-amylose maize starch granules A and B better than other strains, with only 20% of residual amylose detectable. The degree of amylose degradation varied for the bacterial strains tested, with 60% residual amylose after the growth of Bifidobacterium bifidum and Bifidobacterium pseudolongum and 90% after the growth of Bacteroides fragilis. There was no detectable degradation of the amylose by Bacteroides vulgatus or Eubacterium limosum.
FIG. 1.
Residual apparent amylose from the starch after 48 h of growth of Clostridium butyricum, Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bacteroides vulgatus, Bacteroides fragilis, and Eubacterium limosum in serum tubes containing 1% high-amylose maize granules A or B in the basal medium. Results are expressed as the percent remaining and are expressed as mean values ± the standard deviation of triplicate experiments.
Activity of the starch-degrading enzymes.
The influence of various starches on the activity of starch-degrading enzymes was quantified for the various bacterial strains grown for 48 h in the autoclaved basal medium containing glucose, amylopectin maize starch, or granular high-amylose maize starch A. The enzyme activity was measured by determining the concentration of released reducing sugars after extracts and supernatants were incubated with soluble starch. As shown in Table 5, starch-degrading enzymes were detected in whole-cell extracts of Bifidobacterium and Bacteroides spp. but in very low amounts in the fraction of cell-free supernatant of these bacteria, except for Bacteroides vulgatus grown in glucose-containing medium. For Bacteroides vulgatus, it could be speculated that glucose induces an extracellular enzyme, while high-amylose maize granules A induces a cell-wall-associated enzyme, or that cells grown in glucose do not retain the enzyme on the cell wall. For Clostridium butyricum, a relatively low level of starch-degrading enzyme activity was detected in the crude cell extracts; however, the spent culture supernatant contained up to four times more. In general, the enzymes are induced by high-amylose maize starch or amylopectin maize starch since higher levels were noted for cells grown in the presence of high-amylose maize starch granules or amylopectin maize starch, with maximum enzyme activities obtained for cells grown in high-amylose maize starch granule-containing medium. When the activities of the enzymes from other strains are compared, the highest values were detected in crude cell extracts of Bifidobacterium bifidum and Bifidobacterium pseudolongum. Less enzyme activity was noted in extracts from Bacteroides vulgatus and Bacteroides fragilis than in extracts from Bifidobacterium strains, with the least active enzymes being found in the supernatant and crude cell extracts from Eubacterium limosum.
TABLE 5.
Activities of starch-degrading enzymes in culture supernatants and crude cell extracts from six selected bacterial strains grown in autoclaved anaerobic basal media containing glucose, amylopectin, or granular high-amylose maize starch (HA A)a
| Bacterium | Activity (μmol mg of protein−1 h−1) of starch-degrading enzymes in:
|
|||||
|---|---|---|---|---|---|---|
| Supernatant
|
Crude cell extracts
|
|||||
| Glucose | Amylopectin | HA A | Glucose | Amylopectin | HA A | |
| Bifidobacterium bifidum | 0.104 ± 0.02 | 0.136 ± 0.01 | 0.167 ± 0.03 | 0.188 ± 0.01 | 1.600 ± 0.10 | 1.638 ± 0.09 |
| Bifidobacterium pseudolongum | 0.046 ± 0.01 | 0.091 ± 0.01 | 0.127 ± 0.01 | 0.454 ± 0.16 | 0.515 ± 0.11 | 2.017 ± 1.38 |
| Bacteroides vulgatus | 0.537 ± 0.39 | 0.124 ± 0.01 | 0.125 ± 0.01 | 0.119 ± 0.03 | 0.203 ± 0.00 | 1.194 ± 0.32 |
| Bacteroides fragilis | 0.061 ± 0.00 | 0.302 ± 0.07 | 0.178 ± 0.01 | 0.090 ± 0.01 | 0.550 ± 0.33 | 1.043 ± 0.22 |
| Clostridium butyricum | 0.766 ± 0.15 | 0.913 ± 0.16 | 0.970 ± 0.03 | 0.163 ± 0.02 | 0.306 ± 0.07 | 0.415 ± 0.03 |
| Eubacterium limosum | 0.061 ± 0.00 | 0.148 ± 0.02 | 0.172 ± 0.03 | 0.126 ± 0.04 | 0.291 ± 0.09 | 0.220 ± 0.06 |
The results are expressed as the mean value from triplicate experiments.
Characterization of the starch-degrading enzymes by SDS-PAGE.
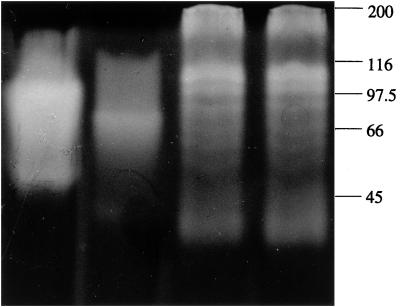
The starch-degrading enzymes in the fraction of crude cell extracts from the six bacterial strains, when grown in amylopectin-containing medium, were separated by SDS-PAGE (Fig. 2). The patterns for Bifidobacterium bifidum and Bifidobacterium pseudolongum were identical, with several active bands with estimated relative molecular (Mr) weights of >160,000, medium-sized molecular weights of >66,000, and low molecular weights of <66,000, suggesting that there is more than one enzyme active in the hydrolysis of starch. The active bands produced from Bacteroides fragilis cells (Mr, <66,000) differed from the one produced by Bacteroides vulgatus (Mr, >66,000), and neither of them showed any similarity to the pattern obtained for the Bifidobacterium spp. No starch-degrading activity was detectable in the lane containing whole-cell extracts of Clostridium butyricum or Eubacterium limosum (data not included in Fig. 2). While the Clostridium butyricum grew well in the amylopectin maize starch broth, the Eubacterium limosum grew poorly.
FIG. 2.
Visualization of starch-degrading enzymes in bacterial cell extracts by SDS-PAGE, followed by an overnight treatment with 0.2% soluble starch and subsequent staining with iodine. Bacteroides vulgatus (lane 1), Bacteroides fragilis (lane 2), Bifidobacterium bifidum (lane 3), Bifidobacterium pseudolongum (lane 4) grown in amylopectin are shown. Values on the right correspond to the relative molecular weights as determined by separately staining with Comassie blue the lane containing the molecular weight markers and then realigning the gels.
DISCUSSION
In this study, the utilization and degradation of amylopectin maize starch and high-amylose maize starch granules by colonic bacteria have been studied. While various factors, including the degree of gelatinization, the biological origin of the starch, the amylose/amylopectin ratio, the starch-protein interaction, amylose-lipid complexes, the percentage of retrograded starch, and the presence of amylase inhibitors, have been shown to affect starch degradation (8), in the present in vitro study it was found that the degradation was most influenced by the granule conformation and the amylose/amylopectin ratio. Starch is composed of two main components, amylose and amylopectin. It has been shown that the higher the amylose content, the more resistant the starch is to degradation (9). Autoclaving completely gelatinized the amylopectin maize starch granules but not the high-amylose maize starch granules A and B, which remained intact when observed by light microscopy. However, the process of autoclaving affected the concentration of resistant starch, with no more than 6.8% of resistant starch detectable in the uninoculated media containing the HA maize starch granules, whereas the amylopectin maize starch granules yielded 0.2% resistant starch after autoclaving (Table 2). Disruption of starch granules during autoclaving rendered the hydrated starch more accessible to degradation by amylases.
Although it was determined that, after autoclaving, there was predominantly digestible starch in both the amylopectin maize and amylomaize (HA) starch broths (Table 2), the colonic bacteria used amylopectin maize starch and amylomaize granules to different extents (Tables 1, 3, and 4). As shown in the screening experiment (Table 1), gelatinized amylopectin maize starch was degraded by a wide range of bacterial strains, including Bifidobacterium, Bacteroides, and Fusobacterium spp. and some species of Clostridium and Eubacterium. In contrast, high-amylose maize starch granules were more resistant to the degradation, with only a few strains of Bifidobacterium and Clostridium capable of utilizing it. Furthermore, the low specific growth rates and a high concentration of residual starch from batch fermentation experiments carried out with six selected bacterial strains confirmed the screening results, indicating that high-amylose maize starch granules A and B were more resistant to degradation than the amylopectin maize starch which gelatinized when the medium was autoclaved and hence lost its granular form. This is consistent with previous findings that high-amylose maize starch does not completely gelatinize until the temperature is in the range 154 to 171°C (14). It was previously noted that the ratio of amylose to amylopectin influenced the degree of digestion both in humans and animals and that high-amylose maize starch granules were resistant to the human pancreatic amylase (4, 28, 29). Consequently, it would be interesting and important to establish the differences in bacterial degradation of amylopectin maize starch and high-amylose maize starch granules.
It has been reported that digestible starch can produce greater amounts of fecal butyrate than less-digestible starch (30). From the data presented here (Tables 1 and 3), it is proposed that the bifidobacteria and Clostridium butyricum would contribute to most of the fermentation of the less-digestible starch. Bifidobacterium spp. do not produce butyrate; however, Clostridium butyricum does have butyrate as an end product (17). Although it has been reported that clostridia are one of the major starch-degrading bacteria in the porcine digestive tract (33), Clostridium spp. are generally less dominant in the human colon and hence the role of Clostridium in the production of butyrate in humans is unclear.
Bacteroides is the numerically dominant genus in the colonic ecosystem. It has been considered that Bacteroides species are the primary starch-degrading colonic microbes, since high activities of neopullulanase, α-glucosidase, and amylase have been detected in cell extracts of Bacteroides spp. (22, 36). In the present study, the Bacteroides spp. did not hydrolyze the high-amylose maize starch granules. From the data (Tables 1, 3, and 4), it may be suggested that the genus of Bifidobacterium is probably the principal amylose degrader of the 38 strains examined. This observation is consistent with the finding that all of the amylolytic isolates from a human fecal sample were Bifidobacterium spp. (18). Several genera of amylolytic bacteria were isolated from human feces by Macfarlane and Englyst (21), namely, Bifidobacterium, Bacteroides, Fusobacterium, and Butyrivibrio. These workers used soluble starch as the sole carbon in the selective agar plates, and hence the finding that Bacteroides and Fusobacterium strains tested were amylolytic on the soluble starch agar is in agreement with similar findings presented in Table 1.
Starch-degrading enzymes were detected in the Bifidobacterium, Bacteroides, and Clostridium butyricum cultures grown with amylopectin maize starch and high-amylose maize starch granules, with the highest activities detectable in cells grown in medium containing high-amylose maize starch granules A, suggesting that these enzymes are induced by the amylose. Apart from Clostridium butyricum, as well as glucose-grown cells of Bacteroides vulgatus for which extracellular starch-degrading enzymes were detected (Table 5), the enzymes produced by the bifidobacteria and bacteroides were cell bound, a finding which is in agreement with previous work (2). SDS-PAGE patterns revealed that the starch-degrading bands from Bacteroides fragilis (Mr, <66,000) and Bacteroides vulgatus (Mr, >66,000) differed from one another (Fig. 2), indicating a diversity of starch-degrading enzymes of the Bacteroides species. This observation is supported by the work that the starch-degrading enzyme produced from Bacteroides vulgatus was different from that of Bacteroides ovatus, since the former only produced a single α-glucosidase with amylolytic activities but the latter is able to synthesize several starch-degrading enzymes, including α-glucosidase, α-amylase, and pullulanase (14). Despite the reported starch-degrading enzymes produced by Bacteroides species, the strains tested here had no detectable amylose-hydrolyzing capacity when high-amylose maize granules A were used (Table 1), even though they had detectable soluble-starch-degrading enzymes.
For bifidobacteria, the cell-bound fraction yielded several starch-utilizing bands with different relative molecular weights (Fig. 2), suggesting that there was more than one enzyme present. In the present study, although it is difficult based on the SDS-PAGE pattern to characterize and identify the starch-degrading enzymes from Bifidobacterium bifidum and Bifidobacterium pseudolongum, our results differ from those of Ji et al. (18), who reported that Bifidobacterium spp. of human origin were able to release extracellular amylase that was detectable by SDS-PAGE (Mr, ca. 66,000). In another study, it was suggested that a Bifidobacterium pseudolongum strain produced only two types of α-glucosidase (Mr, ca. 160,000) (13). Those workers failed to detect α-amylase by using the reducing-sugar method; however, in the present study, this was demonstrated for the strains tested using this assay. Since the starch-degrading enzymes produced from the strains of bifidobacteria used here have a range of molecular weights, we proposed that the degrading enzymes produced by the Bifidobacterium pseudolongum FII 509500 and Bifidobacterium bifidum FII 509800 may include both α-amylase and α-glucosidase.
In conclusion, it has been shown that both amylopectin maize starch and high-amylose maize starch granules were fermented by several colonic bacteria and that Bifidobacterium spp. may play an important role in the utilization of starch granules, particularly high-amylose maize starch granules. Consequently, dietary high-amylose maize starch granules may enhance desirable colonic bacteria and thereby induce beneficial effects.
ACKNOWLEDGMENTS
This work was supported by the CRC for Food Industry Innovation.
The assistance of Cherise Ang and Nedhal Elkaid is acknowledged.
REFERENCES
- 1.Anderson I H, Levine A S, Levitt M D. Incomplete absorption of the carbohydrate in all-purpose wheat flour. N Engl J Med. 1981;304:891–892. doi: 10.1056/NEJM198104093041507. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K L, Salyers A A. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Association of Official Analytical Chemists. Official methods of analysis. 16th ed. Arlington, Va: AOAC; 1986. [Google Scholar]
- 4.Ayano Y, Furuhashi T, Watanabe Y, Suzuki T, Takai Y. On the in vitro digestion of raw amylomaize VII starch and on the growth of weaning rats fed the starch as a sole carbohydrate source. J Jpn Soc Food Nutr. 1977;30:123–130. [Google Scholar]
- 5.Baghurst P A, Baghurst K I, Record S J. Dietary fibre, non-starch polysaccharides and resistant starch—a review. Food Aust. 1996;48:1–36. [Google Scholar]
- 6.Bingham S. Meat, starch and non-starch polysaccharides: are epidemiological and experimental findings consistent with acquired genetic alterations in sporadic colorectal cancer? Cancer Lett. 1997;114:25–34. doi: 10.1016/s0304-3835(97)04618-1. [DOI] [PubMed] [Google Scholar]
- 7.Borriello S P. Microbial flora of the gastrointestinal tract. In: Hill M J, editor. Microbial metabolism in the digestive tract. London, England: CRC Press; 1986. pp. 1–20. [Google Scholar]
- 7a.Brown, I. L. Personal communication.
- 8.Brown I L. The structure of Australian maize starch. MSc. thesis. Armidale, Australia: University of New England; 1993. [Google Scholar]
- 9.Brown I L, McNaught K, Moloney E. Hi-maize: new directions in starch technology and nutrition. Food Aust. 1995;47:272–275. [Google Scholar]
- 10.Cassidy A, Bingham S A, Cummings J H. Starch intake and colorectal cancer risk: an international comparison. Br J Cancer. 1994;69:937–942. doi: 10.1038/bjc.1994.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings J H, Gibson G R, Macfarlane G T. Quantitative estimate of fermentation in the hindgut of man. Acta Vet Scand. 1989;86:76–82. [PubMed] [Google Scholar]
- 12.Cummings J H, Pomare E W, Branch W J, Naylor C P E, Macfarlane G T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degnan B A, Macfarlane G T. Synthesis and activity of α-glucosidase produced by Bifidobacterium pseudolongum. Curr Microbiol. 1994;29:43–47. [Google Scholar]
- 14.Doublier J L, Choplin L. A rheological description of amylose gelation. Carbohydr Res. 1989;193:215–226. [Google Scholar]
- 15.Dubois M, Giles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Ann Biochem. 1956;28:350–356. [Google Scholar]
- 16.Englyst H N, Macfarlane G T. Breakdown of resistant and readily digestible starch by human gut bacteria. J Sci Food Agric. 1986;37:699–706. [Google Scholar]
- 17.Holdeman L V, Cato E P, Moore W E C. Anaerobic laboratory manual. Blacksburg, Va: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 18.Ji G E, Han H K, Yun S W, Rhim S L. Isolation of amylolytic Bifidobacterium sp. Int-57 and characterization of amylase. J Microbiol Biotechnol. 1992;2:85–91. [Google Scholar]
- 19.Levitt M D, Hirsch P, Fetzer C A, Sheahan M, Levine A. H2 excretion after ingestion of complex carbohydrates. Gastroenterology. 1987;92:383–389. doi: 10.1016/0016-5085(87)90132-6. [DOI] [PubMed] [Google Scholar]
- 20.Macfarlane G T, Cummings J H, Macfarlane S, Gibson G R. Influence of retention time on degradation of pancreatic enzymes by human colonic bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol. 1989;67:521–527. [PubMed] [Google Scholar]
- 21.Macfarlane G T, Englyst H N. Starch utilization by the human large intestinal microflora. J Appl Bacteriol. 1986;60:195–201. doi: 10.1111/j.1365-2672.1986.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy R E, Salyers A A. The effects of dietary fiber utilization on the colonic microflora. In: Rowland I R, editor. Role of the gut flora in toxicity and cancer. London, England: Academic Press, Ltd.; 1988. pp. 295–314. [Google Scholar]
- 23.Meynell G G, Meynell E. Bacterial growth. In: Meynell G G, Meynell E, editors. Theory and practice in experimental bacteriology. Cambridge, England: Cambridge University Press; 1970. pp. 1–34. [Google Scholar]
- 24.Mitsuoka T, Hidaka H, Eida T. Effect of fructooligosaccharides on intestinal microflora. Nahrung. 1987;31:427–436. doi: 10.1002/food.19870310528. [DOI] [PubMed] [Google Scholar]
- 25.Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore W E C, Holdeman L V. Discussion of current bacteriological investigation of the relationship between intestinal flora, diet and colon cancer. Cancer Res. 1975;35:3418. [Google Scholar]
- 27.Morrison W R, Laignelet B. An improved colorimetric procedure for determining apparent and total amylose in cereal and other starches. J Cereal Sci. 1983;1:9–20. [Google Scholar]
- 28.Muir J G, Birkett A, Brown I, Jones G, O’Dea K. Food processing and maize variety affects amounts of starch escaping digestion in the small intestine. Am J Clin Nutr. 1995;61:82–89. doi: 10.1093/ajcn/61.1.82. [DOI] [PubMed] [Google Scholar]
- 29.Muir J G, O’Dea K. Measurement of resistant starch: factors affecting the amount of starch escaping digestion in vitro. Am J Clin Nutr. 1992;56:123–127. doi: 10.1093/ajcn/56.1.123. [DOI] [PubMed] [Google Scholar]
- 30.Nordgaard-Andersen I, Clausen M R, Mortensen P B. Short-chain fatty acids, lactate and ammonia in ileorectal and ileal pouch contents: a model of cecal fermentation. J Parenter Enteral Nutr. 1993;34:324–331. doi: 10.1177/0148607193017004324. [DOI] [PubMed] [Google Scholar]
- 31.Oku T. Metabolism of new sweetener fructooligosaccharides (Neosugar) and its application. Nutr Mag. 1986;44:291–306. [Google Scholar]
- 32.Pirt S J. Parameters of growth and analysis of growth data. In: Pirt S J, editor. Principles of microbe and cell cultivation. Oxford, England: Blackwell Scientific Publication; 1985. pp. 5–14. [Google Scholar]
- 33.Reid C A, Hillman K, Henderson C, Glass H. Fermentation of native and processed starches by the porcine caecal anaerobe Clostridium butyricum (NCIMB 7423) J Appl Bacteriol. 1996;80:191–198. [Google Scholar]
- 34.Rephaeli A, Rabizadeh E, Aviram A, Shaklai M, Ruse M, Nudelman A. Derivatives of butyric acids as potential antineoplastic agents. Int J Cancer. 1991;49:66–72. doi: 10.1002/ijc.2910490113. [DOI] [PubMed] [Google Scholar]
- 35.Sakata T. Stimulatory effect of short chain fatty acids on epithelial cell proliferation in rat intestine. Br J Nutr. 1987;58:95–103. doi: 10.1079/bjn19870073. [DOI] [PubMed] [Google Scholar]
- 36.Smith K, Salyers A A. Purification and characterization of enzymes involved in starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1991;173:2962–2968. doi: 10.1128/jb.173.9.2962-2968.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephen A A, Sieber G M, Gerster Y A, Morgan D R. Intake of carbohydrate and its components—international comparisons, trends over time and effects of changing to low-fat diets. Am J Clin Nutr. 1995;62:851S–867S. doi: 10.1093/ajcn/62.4.851S. [DOI] [PubMed] [Google Scholar]
- 38.Tannock G W. Effect of dietary and environmental stress on the gastrointestinal microbiota. In: Hentges D J, editor. Human intestinal microflora in health and disease. London, England: Academic Press; 1983. pp. 517–534. [Google Scholar]
- 39.van Munster I P, Nagengast F M. The role of carbohydrates fermentation in colon cancer prevention. Scand J Gastroenterol. 1993;28:80–86. doi: 10.3109/00365529309101581. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Gibson G R. Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J Appl Bacteriol. 1993;75:373–380. doi: 10.1111/j.1365-2672.1993.tb02790.x. [DOI] [PubMed] [Google Scholar]
- 41.Whitehead H R, Young G P, Bhatal P S. Effect of short chain fatty acids on a new human colon carcinoma cell line (LM1215) Gut. 1986;27:1457–1463. doi: 10.1136/gut.27.12.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]