Key Words: anterograde, lateral geniculate nucleus, mouse brains, neural circuit, neurons, retina, Sindbis virus, superior colliculus, synapse, tracer
Abstract
Mapping neural circuits is critical for understanding the structure and function of the nervous system. Engineered viruses are a valuable tool for tracing neural circuits. However, current tracers do not fully meet the needs for this approach because of various drawbacks, such as toxicity and characteristics that are difficult to modify. Therefore, there is an urgent need to develop a new tracer with low toxicity and that allows for long-term studies. In this study, we constructed an engineered Sindbis virus (SINV) expressing enhanced green fluorescent protein (EGFP) reporter gene (SINV-EGFP) and found that it had no significant difference in biological characterization compared with the wild-type Sindbis virus in BHK-21 cells and neurons in vitro. We injected the virus into the visual circuit of mouse brain and found that the virus infected neurons in the local injected site and anterogradely spread in the neural circuits. Although the efficiency of transmission was limited, the findings demonstrate that SINV can be used as a new anterograde tracer to map neural circuits in mouse brain and that it spreads exclusively in the anterograde direction. Further, use of SINV in mouse brain research will provide longer time windows for circuit tracing than is possible with herpes simplex virus and vesicular stomatitis virus tracers.
Introduction
Many-branched dendrites and axon fibers form complex networks within the brain. Various neurotropic viruses have been engineered to express reporter genes to trace these neural circuits (Wickersham et al., 2007b; Lo and Anderson, 2011; Zingg et al., 2017). Viral tracers transmit between connected neurons and fluorescent protein is used to visualize the networks. Among these tracers, pseudorabies virus and rabies virus belong to the retrograde tracers. Rabies virus is usually used as a mono-transsynaptic tracer (Kelly and Strick, 2000; Wickersham et al., 2007a) and pseudorabies virus is usually used as a multi-transsynaptic tracer (Jia et al., 2019; Sun et al., 2019). Nearly all adeno-associated viruses (AAVs) are non-transmittable viral tracers that express reporter genes only in the initially injected neurons. AAV vectors label neurons and then transmit to axons, which can provide insight on neural innervation, but are not used for trans-synaptic transmission. Although AAV1 and AAV9 exhibit anterograde trans-synaptic properties with super-high titers, they require cre-dependent transgene expression for amplification (Zingg et al., 2017). The herpes simplex virus type 1 (HSV-1) and vesicular stomatitis virus (VSV) can anterogradely spread within neural circuits (Lo and Anderson, 2011; Harouaka and Wertz, 2012; Lin et al., 2020; Su et al., 2020). HSV-129 is a multi-transsynaptic tracer, and H129-ΔTK-tdT can be used as a mono-transsynaptic tracer with AAVs expressing the thymidine kinase gene. However, a major drawback of HSV-1 is terminal absorption; axon terminals of neurons pick up the virus and retrogradely transport it to the soma, which limits the virus’s use for tracing circuits (Diefenbach et al., 2008). Further, HSV-1 has cytotoxicity and animal toxicity. It normally kills experimental mice in 3–7 days, and brain slices of the animals exhibit severe inflammation and tissue damage in the injected sites. In addition, because of the large genome of HSV-1, it is difficult to engineer and manipulate. VSV also has high cytotoxicity and animal toxicity and can lead to the rapid death of mice, which limits tracing to short periods (Li et al., 2019). These virus-based tracers provide powerful tools and methods for neuroscientists. However, the current drawbacks, such as toxicity, limit their use for labeling neural circuits. Therefore, novel tracers are needed.
Sindbis virus (SINV) is a small, enveloped positive-strand RNA virus that belongs to the alphavirus family (Carrasco et al., 2018). SINV transmits between vertebrates, including birds and mammals, in nature by mosquito bites (Strauss and Strauss, 1994; Ziegler et al., 2019). Previous studies have shown that SINV prefers to infect neurons rather than glial cells (Furuta et al., 2001; Ehrengruber and Lundstrom, 2007). It has been engineered as a vector to express heterogeneous genes with high-level expression in vitro and in vivo (Bredenbeek et al., 1993; Ehrengruber and Lundstrom, 2007; Ghosh et al., 2011; Zhu et al., 2011; Kebschull et al., 2016; Kuramoto, 2019). SINV is approximately 12 kb in length, and encodes four nonstructural proteins (nsP1 to nsP4) and five structural proteins (capsid, E3, E2, 6K, and E1) (Lustig et al., 1988; Zhu et al., 2011). The genome of SINV can be easily engineered, and SINV-based vectors are commonly used in different fields (Huang, 1996; Ghosh et al., 2011; Lundstrom, 2017). Previous studies have shown that SINV infects neurons and spreads within the neural circuits of zebrafish larva with low toxicity (Zhu et al., 2009; Mounce et al., 2016; Passoni et al., 2017), which suggests that SINV might be useful for mapping neural circuits of mouse brain. Therefore, we analyzed the characteristics of SINV and the direction of its spread in mapping the neural circuits of mouse brain.
Materials and Methods
Animals
All procedures were approved by Animal Care and Use Committees at Innovation Academy for Precision Measurement Science and Technology, the Chinese Academy of Sciences (approval No. APM20035A) in 2021. Fetal C57BL/6 mice (for primary cultured neurons) and adult C57BL/6 mice (for virus injection in vivo; n = 28, 8-week-old, 20–25 g) were provided by Hunan SJA Laboratory Animal Co., Ltd., Changsha, China (license No. SCXK (Xiang) 2019-0004).
Construction of plasmids
The SINV backbone was from the hybridTE12 strain, the nonstructural region and capsid were from the Toto1101 strain and the structural region was from the NSV strain, which was isolated after six intracerebral passages of AR339 in mice (Lustig et al., 1988). The wild-type (pSINV-WT) and enhanced green fluorescent protein (pSINV-EGFP) plasmids (both from Institute Pasteur, Paris, France) contained a ubiquitin C promoter to initiate transcription of the virus genome and produce viral particles. The EGFP gene was inserted into the region between ApaI and NotI under the control of a second subgenomic promoter (Hahn et al., 1992). All plasmids were verified by DNA sequence.
Cells and viruses
All experiments were performed in a Biosafety Level 2 laboratory and animal facility. Baby hamster kidney cells (BHK-21; American Type Culture Collection, Manassas, VA, USA, RRID: CVCL_1915) were cultured in Dulbecco’s minimum essential medium (Shanghai BasalMedia Technologies Co., Ltd., Shanghai, China) containing 10% fetal bovine serum (Thermo Fisher, Waltham, MA, USA) and incubated at 37°C in 5% CO2. The plasmid was transfected with lipofectamine 2000 reagent (Thermo Fisher) using opti-MEM (Thermo Fisher). After 6 hours, the supernatant was discarded and replaced with Dulbecco’s minimum essential medium containing 2% fetal bovine serum at 37°C in 5% CO2. Then the supernatant was collected at 24, 36, 48, and 60 hours post-transfection (hpt), and the viral titers were measured by plaque assay (Jia et al., 2016) in BHK-21 cells. The primary culture neurons were selected from fetal C57BL/6 mice brains 3 days before delivery. The pregnant mice were anaesthetized by pentobarbital sodium (50 mg/kg; Sigma-Aldrich, St. Louis, MO, USA), the fetal mice were removed, and then the pregnant mice were sacrificed. The neurons were maintained in neuronal medium (neurobasal medium, penicillin-streptomycin-glutamine, B27 supplement; all from Sigma-Aldrich) on 24-well plates with approximately 3 × 105 cells per well in 5% CO2 at 37°C. After 7 days, the primary cultured neurons were mature, and the neurons and BHK-21 cells were infected with the viruses (SINV-WT and SINV-EGFP) at a multiplicity of infection (MOI) of 1. Two days post-infection, we observed the infection of SINV-EGFP.
Determining the one-step growth curves of SINV-WT and SINV-EGFP
The one-step growth curves of the viruses were detected to compare the replication efficiency of the two viruses. BHK-21 cells were grown in 6-well plates to a density of 90%, and were infected with the virus at a MOI of 0.1 and 1. After 1 hour of incubation at 37°C, we removed the supernatant, washed the cells using phosphate buffered saline, and applied fresh Dulbecco’s minimum essential medium containing 2% fetal bovine serum. Supernatant was harvested at different time points, and the titers were measured by plaque assay.
Virus injection
All animals in the in vivo study are listed in Additional Table 1. Adult C57BL/6 mice (n = 28) were used, and the injection process was performed according to a previous study (Jia et al., 2016). Briefly, 2 μL of SINV-EGFP (3 × 109 plaque forming unit/mL (PFU/mL)) was injected into the vitreous body of the eye, and 150 nL of SINV-EGFP (3 × 109 PFU/mL) was intracerebrally injected into the superior colliculus (SC) and the lateral geniculate nucleus (LGN). The stereotaxic coordinates for the SC were: anterior-posterior, –4.00 mm; medio-lateral, –0.45 mm; and dorso-ventral, –1.80 mm, and for the LGN were: anterior-posterior, –1.80 mm; medio-lateral, –2.00 mm; and dorso-ventral, −2.50 mm from the Bregma (Paxinos and Franklin, 2013). After 24, 48, 72 and 96 hpt, the mice were intraperitoneally injected with pentobarbital sodium (50 mg/kg) and sacrificed. The brains were fixed in 4% paraformaldehyde solution overnight, dehydrated in 30% sucrose solution for 48 to 72 hours, and sectioned in 40-μm-thick slices using a freezing sectioning machine (CryoStar NX50 cryostat, Thermo Scientific, San Jose, CA, USA). The brain slices were collected with antifreeze liquid (50% phosphate buffered saline, 20% glycerine, 30% ethylene glycol) for further staining and imaging.
Additional Table 1.
The number of animals used in each experiment
SINV-EGFP (3 × 109 PFU/mL) and VSV-mNeonGreen (2.5 × 108 PFU/mL) were injected intracranially into the SC of randomly selected adult mice (n = 5 per virus) to compare the lethality of the viruses.
Imaging
Brain slices were washed three times with phosphate buffered saline. After staining with 4′,6-diamidino-2-phenylindole (Beyotime, Shanghai, China) for 10 minutes, the brain slices were attached to the microscope slides and sealed with 70% glycerol. All brain slices and retina slices were stained with 4′,6-diamidino-2-phenylindole and imaged with a virtual microscopy slide scanning system (VS120, Olympus, Tokyo, Japan) and a confocal laser scanning system (SP8, Leica, Wetzlar, Germany) for further analysis.
Data analysis
For cell counting, the number of positive cells within each brain area was quantified in every three whole-brain slices by the cell counter plugin in ImageJ software v1.8.0 (National Institutes of Health, Bethesda, MD, USA) (Schneider et al., 2012). Then, the number of positive neurons within a certain brain region was quantified in three mice per group. The percent survival was analysed by Log-rank test. Graphpad Prism8 (Graphpad Software, San Diego, CA, USA; www.graphpad.com) was used for statistical graphs.
Results
Virus rescue and biological characterization in vitro
The replication-competent vector system contained the full length of the SINV genome, which contained the EGFP reporter gene under the control of the 26S promoter (Figure 1A). To assess the ability of the virus production, the 2 μg plasmid was transfected into BHK-21 cells with lipofectamine 2000, and the supernatant was collected at different time points. The fluorescence signals were detected at 12 hpt and increased with time (Figure 1B). After 36–48 hpt, almost all cells were infected and expressed EGFP. In addition, the growth curves of SINV-EGFP and SINV-WT were depicted by titer determination at different time points. SINV-EGFP and SINV-WT had similar characteristics and increased from 12 hpt, peaking at approximately 7 × 108 PFU/mL and 4.7 × 108 PFU/mL, respectively, at 36 hpt. One-step growth curves were determined to compare the replication efficiency of these two viruses. When BHK-21 cells were infected with 0.1 and 1 MOI, the EGFP signals were detected at 12 hours post-infection (hpi), and all cells expressed EGFP at 24 hpi. In vitro, viral titers reached their peaks at 24 hpi, at approximately 4.2 × 108 PFU/mL (SINV-EGFP) and 4.1 × 108 PFU/mL (SINV-WT) when infected with 0.1 MOI, and 7.4 × 108 PFU/mL (SINV-EGFP) and 5.9 × 108 PFU/mL (SINV-WT) with 1 MOI (Figure 1C). In addition, SINV-EGFP and SINV-WT plaques were similar (Figure 1D).
Figure 1.
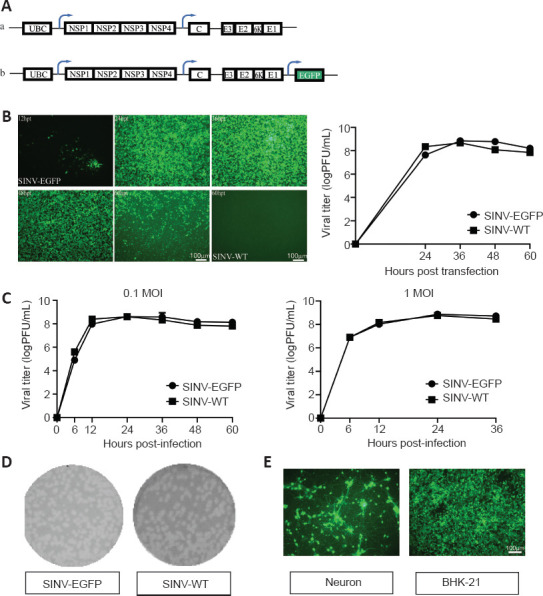
The biological characteristics of the SINV-EGFP in vitro.
(A) Diagram of pSINV-EGFP (a) and pSINV-WT (b) genome structures. (B) The kinetics of virus production. Fluorescent images of BHK-21 cells after transfecting pSINV-EGFP and pSINV-WT at different time points. Fluorescence signals could be detected at 12 hpt and increased with time in the pSINV-EGFP group. No fluorescence was detected in the pSINV-WT group. Virus titers at different time points post-transfection were measured by plaque assay. (C) The single-step growth curves of both viruses. These viruses were collected and titered on BHK-21 cells at different time points post-infection. (D) Plaque sizes of both viruses. The sizes of the viruses were not significantly different. (E) Fluorescent images of cultured primary neurons and BHK-21 cells after infecting with SINV-EGFP. All cells were infected and expressed EGFP. C: Capsid; E3, E2, E1, 6K: structural protein; EGFP: enhanced green fluorescent protein; hpt: hours post-injection; MOI: multiplicity of infection; NSP1–4: nonstructural protein 1–4; UBC: ubiquitin C promoter.
To determine whether SINV infects neurons, we isolated neurons from fetal mice and cultured them in neuronal medium on 24-well plates in 5% CO2 at 37°C. After the primary neurons were mature, approximately 7 days, the cells were infected with SINV-EGFP. Positive cells were observed in both primary cultured neurons and BHK-21 cells, which demonstrated that SINV-EGFP infected neurons in vitro (Figure 1E).
SINV-EGFP spreads with anterograde direction
To verify whether the virus infects neurons and spreads within neural circuits in vivo, SINV-EGFP was injected into the brains of mice. LGN receives projections directly from retinal ganglion cells, and visual cortex area 1 (V1) receives input from the LGN and transmits to visual area 2 (V2) (Beier et al., 2013) (Figure 2A). The number of the positive cells within each brain area was quantified in every three whole-brain slices (Figure 2B). Following injection with SINV-EGFP in the LGN, positive cells were observed in the LGN (Figure 2C), V1 (Figure 2D), and V2 (Figure 2E) at 96 hpi, which indicated that SINV-EGFP infected the local neurons and spread trans-synaptically in the anterograde direction from the injected sites to primary output V1 and secondary output V2. EGFP was not present in the retinal ganglion cells even at 96 hpi (Figure 2B), which suggested that SINV-EGFP did not spread trans-synaptically in the retrograde direction. The efficiency of its trans-synaptic spread was limited, as not all layers of the brain subregions were labeled in V1 (Figure 2E), and the signals were substantially decreased in secondary neurons (Figure 2B).
Figure 2.
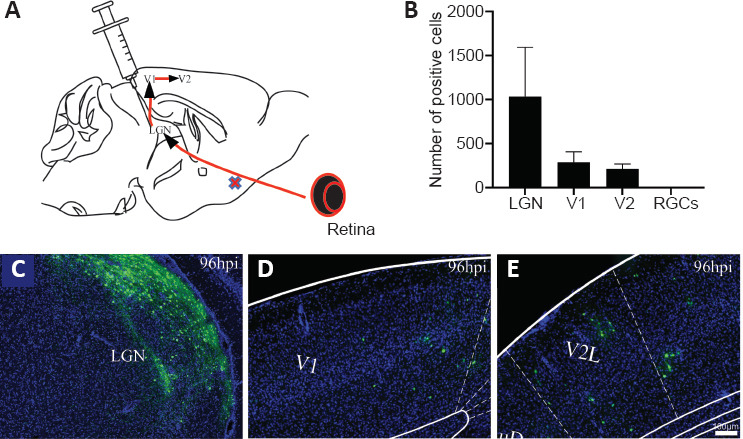
Injection into the LGN of mice to characterize virus trans-synaptic properties.
(A) Diagram of circuits between retinal ganglion cells, LGN, V1 and V2. Data are expressed as mean ± SD. (B) Positive cells of different brain areas at 96 hours post-injection. (C–E) SINV-EGFP spread transsynaptically in the anterograde direction from injected sites to primary output V1 and secondary output V2 by 96 hpi. A large number of neurons were labeled in the LGN (C), and positive cells were also detected in V1 (D) and V2 (E). The experiments were repeated three times. EGFP: Enhanced green fluorescent protein; hpi: hours post-injection; LGN: lateral geniculate nucleus; RGCs: retinal ganglion cells; V1: visual cortex area 1; V2: visual cortex area 2.
Furthermore, we injected the SINV-EGFP into the vitreous body of the eye (Figure 3A), and mice were sacrificed after 96 hpi. Neurons in the retina were infected by the virus (Figure 3B). Positive signals were also present in the LGN and V1 (Figure 3C and D). In addition, we analyzed the anterograde trans-synaptic ability of SINV-EGFP in other pathways. The virus was injected into the SC, and EGFP signals were observed in the lateral posterior thalamic nucleus (LP) at 24 hpi, and in the lateral amygdala (LA) at 96 hpi (Figure 4). Collectively, these results indicate that SINV-EGFP spreads exclusively in the anterograde direction within neural circuits.
Figure 3.
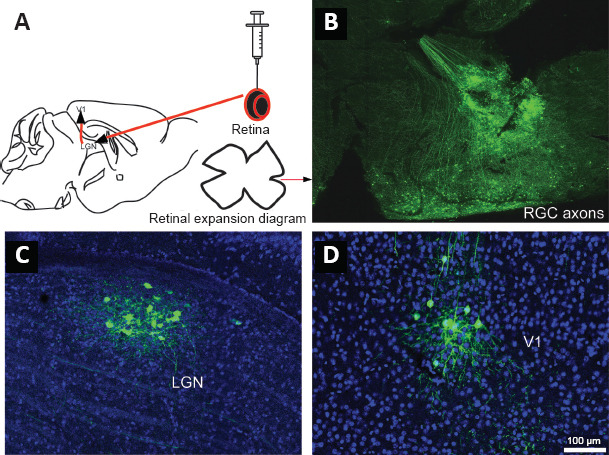
Injection into the retina of mice to characterize the virus.
We injected 2 μL SINV-EGFP into the vitreous body of the eye of mice, and mice were sacrificed at 96 hpi. (A) Diagram of neural circuit between the RGCs of retina, the LGN and V1. (B) At 96 hpi of subretinal cells, we isolated and expanded retinal cells. RGC axons were labeled by SINV-EGFP. (C, D) Positive signals were detected in the LGN and V1, which indicates that the virus anterogradely spread in the neural circuit. The experiments were repeated three times. EGFP: Enhanced green fluorescent protein; hpi; hours post-injection; LGN: lateral geniculate nucleus; RGCs: retinal ganglion cells; V1: visual cortex area 1.
Figure 4.
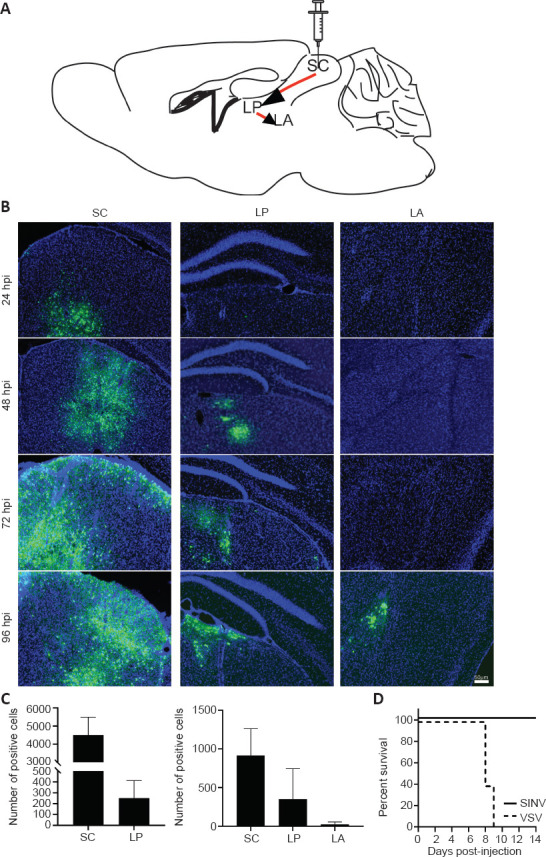
The time course of SINV-EGFP spread within the neural circuit.
The superior colliculus (SC) was chosen as the injection site. (A) Diagram of the SC outputs in the neural circuits. (B) The virus was injected into the SC (150 nL, 3 × 109 PFU/mL) in adult mice. Three adult mice per group were used for SC injection, and were sacrificed at 24, 48, 72, and 96 hpi. Positive cells were first observed at 24 hpi, and the number of labeled neurons in the injection sites increased with time. In the thalamic nucleus (LP), many positive cells were detected at 48 hpi, and in the LA, positive cells were detected at 96 hpi. (C) The positive cells within each brain area were quantified in every three whole-brain slices. Data are expressed as mean ± SD (n = 3 animals for each time point). (D) The lethality after intracranial injection with SINV-EGFP (150 nL, 3 × 109 PFU/mL) and VSV-mNeonGreen (150 nL, 2.5 × 108 PFU/mL). Survival rate of SINV and VSV infected mice. Two viruses were injected intracranially into the SC of adult mice. The percent survival was analysed by Log-rank test (n = 5 in each group). EGFP: Enhanced green fluorescent protein; hpi: hours post-injection; LA: lateral amygdala; LP: lateral posterior thalamic nucleus; SC: superior colliculus.
The time course of SINV anterograde spread
To characterize the time course of SINV spread in mouse brain, we chose the SC as the injection site. Neurons in the SC project to LP, and form a circuit from the SC through the LP to the LA (Wei et al., 2015; Zhou et al., 2019). This projection tract is unidirectional (Figure 4A). Consequently, we injected the virus into the SC (150 nL, 3 × 109 PFU/mL) in adult mice, and the mice were sacrificed at 24, 48, 72, and 96 hours. The positive cells were first observed at 24 hpi, and the number of labeled neurons in the injection sites increased with time (Figure 4B). At 48 hpi, positive cells were detected in LP, which indicated that the virus had transmitted to output neurons from SC. At 96 hpi, many neurons were labeled in the LA, which indicated that the virus had trans-synaptically spread to secondary neurons. We counted the number of positive cells in the SC, LP, and LA. In LP, the number of positive cells was increased at 96 hpi compared with that at 72 hpi, whereas in the LA, positive cells were first detected at 96 hpi (Figure 4C). When we injected SINV-EGFP into the SC, no signals were detected in V1, which confirmed that SINV-EGFP spread with trans-synaptic anterograde properties. The signals in the injection sites were reduced at 96 hpi compared with those at 72 hpi, which indicates that SINV had cytotoxic effects and damaged some neurons.
Lethality of SINV-EGFP
To evaluate the lethality of SINV-EGFP, we injected either SINV-EGFP (3 × 109 PFU/mL) or VSV-mNeonGreen (2.5 × 108 PFU/mL) intracranially into the SC of mice. All of the mice injected with SINV-EGFP survived until day 14 and did not die after an extended period (data not shown), whereas mice injected with VSV-mNeonGreen were dead in 9 to 10 days after injection (Figure 4D). This indicated that SINV-EGFP was not lethal to adult mice.
Discussion
Mapping neural circuits is critical for understanding how the brain works and the mechanisms of neurological diseases. Although several virus-based tracers have been developed to map brain networks, they have some limitations, such as toxicity and non-specific entry sites (Lo and Anderson, 2011; Wojaczynski et al., 2015; Li et al., 2019). Therefore, better viral tracers are needed.
The replication-competent vector system of SINV rapidly replicated and produced new progeny virus. After infection in BHK-21 cells, SINV-EGFP viral titer reached the peak at 36 hpi, and its titer was similar to that of the wild-type. Therefore, SINV-EGFP produced a high titer virus suitable for research without requiring additional concentration methods. This will allow for easy preparation of the virus, in contrast to other virus-based tracers that usually need to be concentrated for mapping neural circuits.
Two pathways were selected to determine the direction of SINV-EGFP spread in mouse neural circuits. For the visual circuit, SINV-EGFP was injected into the LGN region, and the signals were observed in V1, but no signals were detected in the retina. Next, SINV-EGFP was injected into the retina, and the signals were first observed in the LGN, then signals were detected in V1. These findings indicate that SINV-EGFP spreads in only the anterograde direction in mouse neural circuits. For the collicular-thalamic-amygdala circuit, SINV-EGFP was injected into the SC, and the signals were first observed in the LP, and then observed in the LA. These findings further support that SINV-EGFP anterogradely spreads in mouse neural circuits.
In an analysis of the lethality of SINV-EGFP, all of the virus-infected animals survived for well beyond our 14-day time point. In contrast, other anterograde tracers, such as VSV and HSV-129, have short time windows for tracing experiments (Lichty et al., 2004; Beier et al., 2011; Su et al., 2020). Thus, SINV-EGFP may provide longer time windows for circuit tracing.
Interestingly, SINV infects and transmits between vertebrates including birds and mammals. SINV was first isolated from febrile patients in 1961 (Adouchief et al., 2016). In addition, seroprevalence surveys showed that humans can be infected by SINV (Kurkela et al., 2008). Current viral tracers are not used for primate research. There are no useful tracers based on HSV-1, and VSV does not transport anterogradely in primates. Therefore, these reports and our results suggest that SINV may be a potential tool to trace trans-synaptic spread in primate neural circuits. We will investigate SINV-EGFP in primate neural circuits in the future.
In the present work, we developed a new method to produce SINV through plasmid transfection, which was easier than traditional methods to produce high titer viruses. Then, we determined the direction of SINV-EGFP transport between neurons in mouse brain, although it is necessary to note that the mechanism of anterograde transmission was not determined and should be investigated in further research. Although SINV was effective for tracing neural circuits, the efficiency of trans-synaptic spread was limited, as not all layers of the brain subregions were labeled. This effect might be related to the characteristics of SINV. Previous research has reported that different trans-synaptic viral tracers, such as rabies virus and pseudorabies virus, exhibit discrepant neurotropism within certain brain regions, even cortical layer preference (Sun et al., 2019; Zhu et al., 2020). Even the same virus with different serotypes targets different regions and can be chosen for specific applications in the nervous system (Burger et al., 2004). Additionally, the signals in the injected sites at 96 hpi were reduced compared with those at 72 hpi, indicating cytotoxicity of SINV. In future research, we will investigate ways to further reduce the cytotoxicity of SINV.
In conclusion, we constructed an SINV-EGFP tracer for mapping neural circuits of mouse brain. We could easily produce a high-titer of the virus expressing a reporter gene in vitro, and it did not influence the biological characterization of SINV. SINV-EGFP labeled neurons and spread exclusively in the anterograde direction within neural circuits. The efficiency of its trans-synapse synaptic spread was limited and should be improved. SINV was less toxic to adult mice than other anterograde viral tracers and has potential as a useful tracer for neuroscience research in the future.
Editor note: FJ is an Editorial Board member of Neural Regeneration Research. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal’s standard procedures, with peer review handled independently of this Editorial Board member and their research groups.
Additional file:
Additional Table 1: The number of animals used in each experiment.
Acknowledgments:
We are grateful to Dr. Jean-Pierre Levraud (Macrophages and Development of Immunity, Institut Pasteur) for his kind gift of SINV vector.
Footnotes
Conflicts of interest: The authors declare no competing financial interests.
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: McCollum L, Yu J, Song LP; T-Editor: Jia Y
Funding: This work was supported by the National Natural Science Foundation of China, Nos. 31830035, 91732304, 91632303, 81661148053, and 31771156 (all to FQX); the Key-Area Research and Development Program of Guangdong Province of China, No. 2018B030331001 (to FQX); the SIAT Innovation Program for Excellent Young Researchers of China, No. E1G023 (to FJ); the Guangdong Basic and Applied Basic Research Foundation of China, No. 2021A1515011235 (to FQX); Shenzhen Key Laboratory of Viral Vectors for Biomedicine of China, No. ZDSYS20200811142401005 (to FQX); the National Basic Research Program (973 Program) of China, No. 2015CB755600 (to FQX); and the Strategic Priority Research Program (B) of China, No. XDB32030200 (to FQX).
References
- 1.Adouchief S, Smura T, Sane J, Vapalahti O, Kurkela S. Sindbis virus as a human pathogen-epidemiology, clinical picture and pathogenesis. Rev Med Virol. 2016;26:221–241. doi: 10.1002/rmv.1876. [DOI] [PubMed] [Google Scholar]
- 2.Beier KT, Borghuis BG, El-Danaf RN, Huberman AD, Demb JB, Cepko CL. Transsynaptic tracing with vesicular stomatitis virus reveals novel retinal circuitry. J Neurosci. 2013;33:35–51. doi: 10.1523/JNEUROSCI.0245-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beier KT, Saunders A, Oldenburg IA, Miyamichi K, Akhtar N, Luo L, Whelan SP, Sabatini B, Cepko CL. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci U S A. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. Sindbis virus expression vectors:packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco L, Sanz MA, González-Almela E. The regulation of translation in alphavirus-infected cells. Viruses. 2018;10:70. doi: 10.3390/v10020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diefenbach RJ, Miranda-Saksena M, Douglas MW, Cunningham AL. Transport and egress of herpes simplex virus in neurons. Rev Med Virol. 2008;18:35–51. doi: 10.1002/rmv.560. [DOI] [PubMed] [Google Scholar]
- 8.Ehrengruber MU, Lundstrom K. Alphaviruses:Semliki Forest virus and Sindbis virus vectors for gene transfer into neurons. Curr Protoc Neurosci Chapter. 2007;4:Unit 4.22. doi: 10.1002/0471142301.ns0422s41. [DOI] [PubMed] [Google Scholar]
- 9.Furuta T, Tomioka R, Taki K, Nakamura K, Tamamaki N, Kaneko T. In vivo transduction of central neurons using recombinant Sindbis virus:Golgi-like labeling of dendrites and axons with membrane-targeted fluorescent proteins. J Histochem Cytochem. 2001;49:1497–1508. doi: 10.1177/002215540104901203. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S, Larson SD, Hefzi H, Marnoy Z, Cutforth T, Dokka K, Baldwin KK. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 2011;472:217–220. doi: 10.1038/nature09945. [DOI] [PubMed] [Google Scholar]
- 11.Hahn CS, Hahn YS, Braciale TJ, Rice CM. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992;89:2679–2683. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harouaka D, Wertz GW. Second-site mutations selected in transcriptional regulatory sequences compensate for engineered mutations in the vesicular stomatitis virus nucleocapsid protein. J Virol. 2012;86:11266–11275. doi: 10.1128/JVI.01238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang HV. Sindbis virus vectors for expression in animal cells. Curr Opin Biotechnol. 1996;7:531–535. doi: 10.1016/s0958-1669(96)80057-7. [DOI] [PubMed] [Google Scholar]
- 14.Jia F, Zhu X, Xu F. A single adaptive point mutation in Japanese encephalitis virus capsid is sufficient to render the virus as a stable vector for gene delivery. Virology. 2016;490:109–118. doi: 10.1016/j.virol.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Jia F, Lv P, Miao H, Shi X, Mei H, Li L, Xu X, Tao S, Xu F. Optimization of the fluorescent protein expression level based on pseudorabies virus bartha strain for neural circuit tracing. Front Neuroanat. 2019;13:63. doi: 10.3389/fnana.2019.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kebschull JM, Garcia da Silva P, Zador AM. A new defective helper RNA to produce recombinant sindbis virus that infects neurons but does not propagate. Front Neuroanat. 2016;10:56. doi: 10.3389/fnana.2016.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- 18.Kuramoto E. Method for labeling and reconstruction of single neurons using Sindbis virus vectors. J Chem Neuroanat. 2019;100:101648. doi: 10.1016/j.jchemneu.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Kurkela S, Rätti O, Huhtamo E, Uzcátegui NY, Nuorti JP, Laakkonen J, Manni T, Helle P, Vaheri A, Vapalahti O. Sindbis virus infection in resident birds, migratory birds, and humans, Finland. Emerg Infect Dis. 2008;14:41–47. doi: 10.3201/eid1401.070510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Liu T, Dong Y, Kondoh K, Lu Z. Trans-synaptic neural circuit-tracing with neurotropic viruses. Neurosci Bull. 2019;35:909–920. doi: 10.1007/s12264-019-00374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichty BD, Power AT, Stojdl DF, Bell JC. Vesicular stomatitis virus:re-inventing the bullet. Trends Mol Med. 2004;10:210–216. doi: 10.1016/j.molmed.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Lin K, Zhong X, Ying M, Li L, Tao S, Zhu X, He X, Xu F. A mutant vesicular stomatitis virus with reduced cytotoxicity and enhanced anterograde trans-synaptic efficiency. Mol Brain. 2020;13:45. doi: 10.1186/s13041-020-00588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundstrom K. Oncolytic alphaviruses in cancer immunotherapy. Vaccines. 2017;5:9. doi: 10.3390/vaccines5020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lustig S, Jackson AC, Hahn CS, Griffin DE, Strauss EG, Strauss JH. Molecular basis of Sindbis virus neurovirulence in mice. J Virol. 1988;62:2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mounce BC, Poirier EZ, Passoni G, Simon-Loriere E, Cesaro T, Prot M, Stapleford KA, Moratorio G, Sakuntabhai A, Levraud JP, Vignuzzi M. Interferon-induced spermidine-spermine acetyltransferase and polyamine depletion restrict Zika and Chikungunya viruses. Cell Host Microbe. 2016;20:167–177. doi: 10.1016/j.chom.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Passoni G, Langevin C, Palha N, Mounce BC, Briolat V, Affaticati P, De Job E, Joly JS, Vignuzzi M, Saleh MC, Herbomel P, Boudinot P, Levraud JP. Imaging of viral neuroinvasion in the zebrafish reveals that Sindbis and chikungunya viruses favour different entry routes. Dis Model Mech. 2017;10:847–857. doi: 10.1242/dmm.029231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paxinos G, Franklin KB. The mouse brain in stereotaxic coordinates. San Diego:Elsevier 2013 [Google Scholar]
- 29.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ:25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strauss JH, Strauss EG. The alphaviruses:gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su P, Ying M, Han Z, Xia J, Jin S, Li Y, Wang H, Xu F. High-brightness anterograde transneuronal HSV1 H129 tracer modified using a Trojan horse-like strategy. Mol Brain. 2020;13:5. doi: 10.1186/s13041-020-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Tang Y, Yan K, Yu J, Zou Y, Xu W, Xiao K, Zhang Z, Li W, Wu B, Hu Z, Chen K, Fu ZF, Dai J, Cao G. Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol Neurodegener. 2019;14:8. doi: 10.1186/s13024-019-0308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei P, Liu N, Zhang Z, Liu X, Tang Y, He X, Wu B, Zhou Z, Liu Y, Li J, Zhang Y, Zhou X, Xu L, Chen L, Bi G, Hu X, Xu F, Wang L. Processing of visually evoked innate fear by a non-canonical thalamic pathway. Nat Commun. 2015;6:6756. doi: 10.1038/ncomms7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wojaczynski GJ, Engel EA, Steren KE, Enquist LW, Patrick Card J. The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties. Brain Struct Funct. 2015;220:1395–1420. doi: 10.1007/s00429-014-0733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z, Liu X, Chen S, Zhang Z, Liu Y, Montardy Q, Tang Y, Wei P, Liu N, Li L, Song R, Lai J, He X, Chen C, Bi G, Feng G, Xu F, Wang L. A VTA GABAergic Neural Circuit Mediates Visually Evoked Innate Defensive Responses. Neuron. 2019;103:473–488.e6. doi: 10.1016/j.neuron.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 38.Zhu P, Narita Y, Bundschuh ST, Fajardo O, Schärer YP, Chattopadhyaya B, Bouldoires EA, Stepien AE, Deisseroth K, Arber S, Sprengel R, Rijli FM, Friedrich RW. Optogenetic dissection of neuronal circuits in zebrafish using viral gene transfer and the Tet system. Front Neural Circuits. 2009;3:21. doi: 10.3389/neuro.04.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu W, Li J, Tang L, Wang H, Li J, Fu J, Liang G. Glycoprotein is enough for sindbis virus-derived DNA vector to express heterogenous genes. Virol J. 2011;8:344. doi: 10.1186/1743-422X-8-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu X, Lin K, Liu Q, Yue X, Mi H, Huang X, He X, Wu R, Zheng D, Wei D, Jia L, Wang W, Manyande A, Wang J, Zhang Z, Xu F. Rabies virus pseudotyped with CVS-N2C glycoprotein as a powerful tool for retrograde neuronal network tracing. Neurosci Bull. 2020;36:202–216. doi: 10.1007/s12264-019-00423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziegler U, Fischer D, Eiden M, Reuschel M, Rinder M, Müller K, Schwehn R, Schmidt V, Groschup MH, Keller M. Sindbis virus- a wild bird associated zoonotic arbovirus circulates in Germany. Vet Microbiol. 2019;239:108453. doi: 10.1016/j.vetmic.2019.108453. [DOI] [PubMed] [Google Scholar]
- 42.Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, Zhang LI. AAV-mediated anterograde transsynaptic tagging:mapping corticocollicular input-defined neural pathways for defense behaviors. Neuron. 2017;93:33–47. doi: 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]


