Abstract
Stargardt disease (also known as juvenile macular degeneration or Stargardt macular degeneration) is an inherited disorder of the retina, which can occur in the eyes of children and young adults. It is the most prevalent form of juvenile-onset macular dystrophy, causing progressive (and often severe) vision loss. Images with Stargardt disease are characterized by the appearance of flecks in early and intermediate stages, and the appearance of atrophy, due to cells wasting away and dying, in the advanced stage. The primary measure of late-stage Stargardt disease is the appearance of atrophy. Fundus autofluorescence is a widely available two-dimensional imaging technique, which can aid in the diagnosis of the disease. Spectral-domain optical coherence tomography, in contrast, provides three-dimensional visualization of the retinal microstructure, thereby allowing the status of the individual retinal layers. Stargardt disease may cause various levels of disruption to the photoreceptor segments as well as other outer retinal layers. In recent years, there has been an exponential growth in the number of applications utilizing artificial intelligence for help with processing such diseases, heavily fueled by the amazing successes in image recognition using deep learning. This review regarding artificial intelligence deep learning approaches for the Stargardt atrophy screening and segmentation on fundus autofluorescence images is first provided, followed by a review of the automated retinal layer segmentation with atrophic-appearing lesions and fleck features using artificial intelligence deep learning construct. The paper concludes with a perspective about using artificial intelligence to potentially find early risk factors or biomarkers that can aid in the prediction of Stargardt disease progression.
Key Words: artificial intelligence, assessment, deep learning, fundus autofluorescence, screening, segmentation, spectral-domain optical coherence tomography, Stargardt atrophy, Stargardt disease, Stargardt flecks
Introduction
Stargardt disease (also called Stargardt macular degeneration or juvenile macular degeneration) is a rare inherited disease of the retina (a tissue at the back of the eye that sense light) that occurs in the eyes of children and young adults (Kong et al., 2008; Ma et al., 2011; Binley et al., 2013; Mukherjee and Schuman., 2014; Strauss et al., 2016; Schönbach et al., 2017; Strauss et al., 2017a, b; Cicinelli et al., 2019). Stargardt disease may severely damage the center of the retina (i.e., the macula) and cause vision loss and legal blindness. Due to the disease, visual acuity may decrease gradually, causing patients to end up with 20/200 vision or possibly worse. Since Stargardt disease occurs in children and young adults, it has a significant correlation to lifetime negative economic, psychological, and emotional problems. Currently, there is no treatment for Stargardt disease; however, vision loss can be slowed by taking certain measures. Early detection of Stargardt disease and the understanding of its pathogenesis and progression will be helpful for the early intervention of vision loss and identifying potential treatment solutions.
Color fundus photography (CFP) has been the gold standard method for documenting and assessing Stargardt disease. On CFP images, the disease is typically phenotypically classified by appearance into four stages (Fujinami et al., 2015; Cicinelli et al., 2019). Stage 1 has normal fundus. In stage 2, the macular and/or peripheral regions have flecks, but have no central atrophy. In stage 3, atrophic changes occur in macular central or paracentral regions. In stage 4, atrophic changes occur extensively across the entire macula beyond stage 3. Figure 1 illustrates the 4-stage Stargardt disease classification on CFP images.
Figure 1.
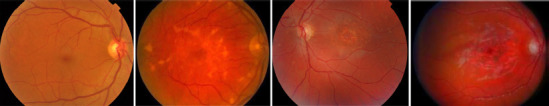
Illustration of 4-stage Stargardt disease classification on color fundus photography images.
Left to right columns: stage 1 to 4. Unpublished data.
In recent years, blue light fundus autofluorescence (FAF) has emerged as a new and useful retinal imaging technique for the assessment of Stargardt disease, particularly because of its high image contrast (Schmitz-Valckenberg, 2008). A FAF image with Stargardt atrophic lesion is shown in Figure 2. Nevertheless, in FAF imaging, the hypofluorescence due to absorption of blue light by luteal pigments may hamper the determination of the involvement of the fovea. Moreover, CFP and FAF are both two-dimensional imaging techniques, which obtain a two-dimensional projection of a three-dimensional (3D) retina. Spectral-domain optical coherence tomography (SD-OCT) has become a frequently used tool in clinics for imaging Stargardt disease, because it provides three-dimensional visualization of the retinal microstructure, thereby allowing the status of the individual retinal layers to be assessed directly and individually (Huang et al., 1991; Fujimoto et al., 1998). Stargardt disease in SD-OCT imaging may show various levels of disruption of the inner and outer photoreceptor segments as well as other outer retinal layers. In Figure 2, the SD-OCT B-scan shows the atrophy, a result of cells wasting away and dying, of outer retinal layers. The light can penetrate the choroid due to atrophy. OCT has an additional advantage over FAF in that it is comfortable and can be performed repeatedly without significant safety concerns in patients with a retinal degenerative disease.
Figure 2.
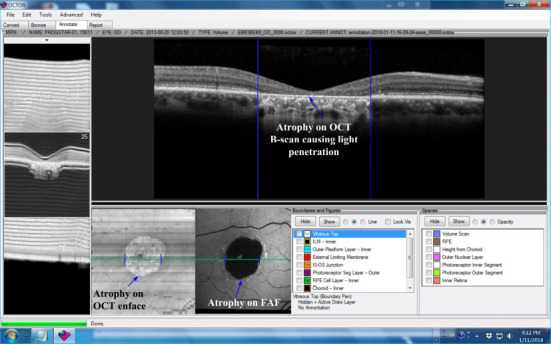
Stargardt atrophy on FAF and OCT images, captured from the grading tool of our lab.
The blue arrows highlight the atrophy on different modality images from a right eye of a patient with Stargardt disease. FAF: Fundus autofluorescence; OCT: optical coherence tomography. Unpublished data.
The Doheny Image Reading Center has conducted studies, which were supported by the Foundation Fighting Blindness to investigate the natural history of Stargardt (i.e., ProgSTAR studies) (Strauss et al., 2015; Schönbach et al., 2017; Strauss et al., 2017a, b). The objective of the ProgSTAR studies was to evaluate possible efficacy measures for potential suitable regulatory endpoints on FAF and SD-OCT images for therapeutic intervention clinical trials for Stargardt disease. More specifically, the studies have assessed the size changes of Stargardt atrophy on FAF images and the disruption to retinal layers on SD-OCT images. The studies have shown that manual delineation of these measures, particularly in 3D OCT images, is extremely tedious, time-consuming, and expensive. Variability between subjective measurements from human graders creates yet another challenge, as the typically slow progression of Stargardt disease over time necessitates a great precision in the measurement for reliable assessment. Automated objective disease assessment techniques would be of significant value for research and in clinical trials of Stargardt disease. Traditional classification methodologies usually require algorithm developers to design hand-crafted filters to extract feasible image features to specific datasets. This causes difficulty in the generalization of developed algorithms for application on large and variable ophthalmic datasets. In contrast, deep learning - artificial intelligence (AI) constructs (Ronneberger et al., 2015; Szegedy et al., 2015, 2016; He et al., 2016) automatically learn relevant image features, which can yield a high level of image processing accuracy and may be more capable of generalization on large variable data.
Recent progress in computer processing power and advances in the design of deep learning algorithms (e.g., deep convolutional neural networks – deep CNNs), have facilitated the state-of-the-art AI approaches to be effectively applied to various imaging data. We have found a limited number of studies conducted using AI for the assessment of Stargardt disease, probably due to Stargardt being a relatively rare disease and as such having a relatively limited amount of accessible data. While there is a deep learning algorithm for the identification of Stargardt hyperautofluorescent flecks on FAF images (Charng et al., 2020), there is only one reported article for the automated assessment (screening and segmentation) of Stargardt macular atrophy on FAF images done using an AI deep learning algorithm (by our team) (Wang et al., 2019). Stargardt atrophy in SD-OCT images may show damage extending from the photoreceptor segment retinal layers to outer retinal layers, including the choroid. For the Stargardt atrophy assessment in SD-OCT images, the quantification of atrophy can be based on the retinal layer segmentation. There are so far only two reported OCT retinal layer segmentation algorithms. Kugelman et al. (2020) developed an approach using graph search along with a deep learning algorithm to segment the retinal layers on SD-OCT images. However, such an approach can only segment two OCT boundaries. We published a graph-based approach with deep learning-derived information to segment twelve SD-OCT retinal layers, which includes all layers associated with Stargardt damage of atrophic-appearing lesions and fleck features (Mishra et al., 2021). This review addressing AI deep learning approaches for the screening and segmentation of Stargardt atrophy on FAF images is first provided, followed by a review on automated retinal layer segmentation with atrophic-appearing lesions and fleck features using AI deep learning constructs. A future perspective regarding further AI application on the assessment of Stargardt disease is provided at the end.
Search Strategy
We conducted a search of PubMed for the past 3 years’ publications with the key words of Stargardt disease, Stargardt atrophy, Stargardt flecks, fundus autofluorescence, FAF, spectral-domain optical coherence tomography, SD-OCT, artificial intelligence, AI, deep learning, automated, screening, segmentation, and assessment.
Artificial Intelligence for Stargardt Atrophy Screening and Segmentation on Fundus Autofluorescence Images
The so-far only reported Stargardt atrophy screening and segmentation system in FAF images was conducted by our team (Wang et al., 2019). In this system, we first developed a deep learning-based automated screening system using a backbone of residual neural networks (ResNet) (Szegedy et al., 2015, 2016; He et al., 2016), which can differentiate eyes with Stargardt atrophy from normal eyes in FAF images. We further developed another deep learning-based automated system to segment Stargardt atrophic lesions using a fully convolutional neural network – U-Net (Ronneberger et al., 2015). Transfer learning based on a pre-trained model was applied to ResNet in order to facilitate the algorithm training, and excessive data augmentation techniques for both ResNet and U-Net were applied to enhance the algorithm’s generalization ability.
Automated screening of eyes with Stargardt atrophy from normal eyes on FAF images using deep learning
Deep learning algorithms with CNNs are usually made up of a multitude of artificial neurons, which resemble organic neurons as their name suggests, from multiple network layers. Network depth has been shown to be of extreme importance for the accuracy of CNNs. Using more layers allows for more features to be recognized by the network and as such, often higher accuracy. However, one major problem with CNNs is that as the network depth increases, accuracy gets saturated and then degrades rapidly. Deep residual learning frameworks attempt to fix this by fitting stacked nonlinear layers to a residual mapping instead of assuming that these layers will be able to match the desired mapping. Take, for example, a desired underlying mapping H(x). Let the stacked nonlinear layers fit another mapping of F(x)= H(x)–x. Then the original mapping takes on the form F(x)+x. This residual mapping has been shown to be easier to optimize, allowing for improved accuracy. Matching to the residual mapping is done by using shortcut connections. Shortcut connections skip over one or more layers, and only perform identity mapping, allowing the network to still be easily implementable into common libraries. These shortcut connections have no extra parameters. As such, ResNet allows deeper networks to achieve considerably higher accuracy when compared to normal CNNs and is ideal for transfer learning. ResNet has also been shown to vastly outperform its plain counterparts, in large part due to its ability to address the prevalent degradation problem.
Our Stargardt screening system utilized ResNet as the backbone. This structure acted as the starting point for our neural network. Preferably, ResNet should be trained on a larger data set. During the implementation of the screening algorithm, our Stargardt dataset was somewhat small with only 100 FAF images (Spectralis HRA+OCT, Heidelberg Engineering) with Stargardt atrophy and 320 FAF images from normal subjects. Note that the FAF data were from a subset of ProgSTAR studies, which was collected from multiple imaging centers. To overcome the problem of smaller data set, instead of training a new ResNet model from scratch, we utilized a ResNet model (weights) that was already pre-trained on the ImageNet database – a large-scale image set with more than ten million of images. This technique is called transfer learning, which utilizes the cumulative knowledge trained from other datasets to create a new neural network. Training a completely new neural network often requires an extremely large training dataset, which usually has millions of weights to learn. We re-trained our ResNet by taking advantage of the ImageNet model pre-trained from millions of general images and applied the general features learned from the ImageNet data to the ResNet in this project. This allowed our ResNet model to be highly accurate even with a relatively small training dataset. Additionally, as we had a limited amount of FAF images, this project also highly relied on strong data augmentation to increase the number of training samples in both the ResNet and U-Net training. We augmented the training data via several random transformations, containing rotation, vertical and horizontal translation, shearing transformations, zooming, and horizontal flipping of the FAF images. This further helped prevent overfitting and helped the model generalize better. The algorithm-defined screening results were compared to manual gradings by certified reading center graders. With these strategies, we could achieve a good Stargardt atrophy screening result with an accuracy of 0.95, a sensitivity of 0.91, and a specificity of 0.98. Figure 3 provides several example illustrations of the screening system result.
Figure 3.
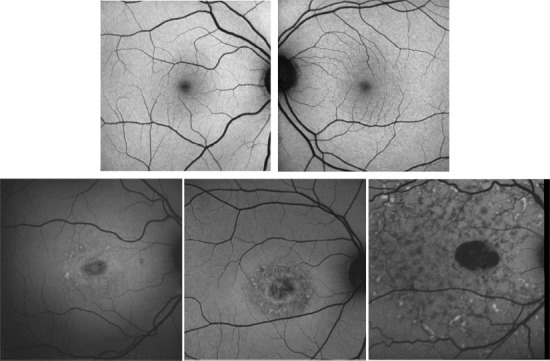
Example illustration of the screening system results on fundus autofluorescence images with 100% accuracy.
Top row shows example images from normal eyes and the bottom row shows example images from eyes with Stargardt atrophy. Note that the Stargardt image in the bottom left has similar intensity distribution as normal images in the top row but still can be differentiated in the accuracy of 100%. Reprinted with permission from Wang et al. (2019).
Automated semantic segmentation of Stargardt atrophic lesions on FAF images using deep learning
The Stargardt atrophic lesion segmentation system was completed by a deep learning CNN, U-Net (Ronneberger et al., 2015). U-Net is a state-of-the-art deep learning algorithm for semantic segmentation, relying on an encoder-decoder type network architecture. It is based on fully convolutional networks instead of fully connected layers and does not require sliding windows. The network consists of a contracting path to capture context and an expansive (upsampling) path to enable precise localization. The U-Net in this project has a similar structure to the one originally reported (Ronneberger et al., 2015), which was composed of downsampling layers of convolutional operation, max pooling, ReLU activation; and upsampling layers of transpose convolutional operation, ReLu, concatenation, and convolutional operation. However, we further enhanced it using a more effective training optimization approach and better loss function.
Note that as mentioned, we only had 100 FAF images with Stargardt atrophy. To overcome the smaller data set barrier for a deep learning algorithm, we used the same U-Net architecture and trained a larger FAF data set with atrophic lesions from a different disease known as age-related macular degeneration for the initialization of the segmentation of Stargardt atrophy. This was because the atrophic lesion profiles of age-related macular degeneration in FAF images have a similar appearance to those caused by Stargardt disease. Compared to the manual gradings, we could achieve a reasonably good Stargardt atrophy segmentation result with the DICE similarity coefficient and the overlapping ratio of 0.87 ± 0.13 and 0.78 ± 0.17 respectively. Figure 4 illustrates the results of our U-Net Stargardt atrophic lesion segmentation system.
Figure 4.
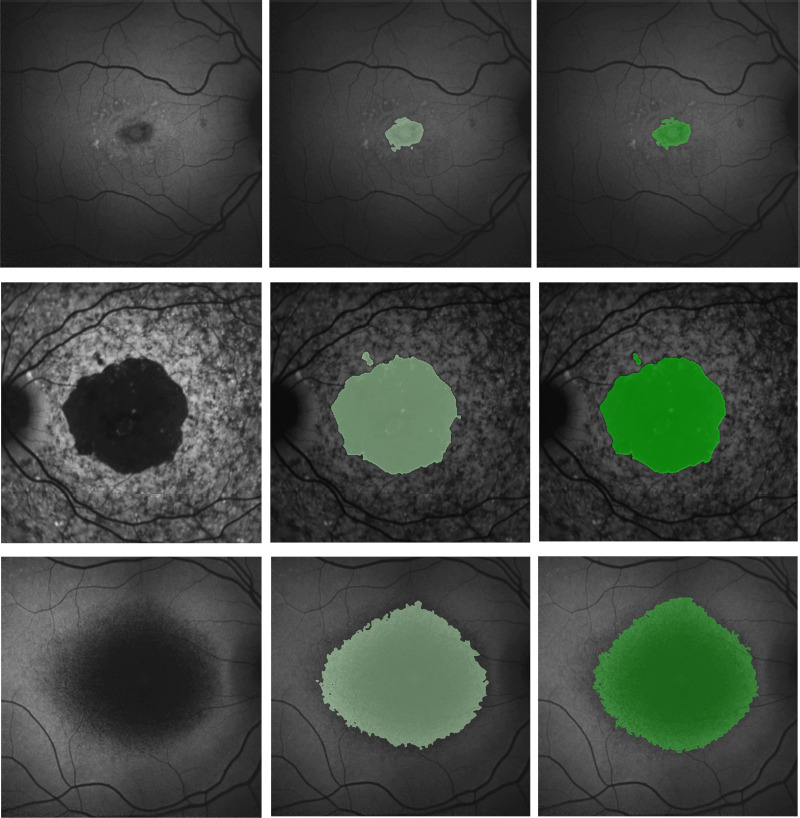
Example illustration of the results of the Stargardt atrophic lesion segmentation on fundus autofluorescence images.
Left column: Fundus autofluorescence images. Middle column: U-Net segmentation results (light green) overlapping on fundus autofluorescence. Right column: manual delineation results (darker green) overlapping on fundus autofluorescence. Reprinted with permission from Wang et al. (2019).
Furthermore, analysis of data from the ProgSTAR study has demonstrated that the areas of decreased autofluorescence (AF) can have variable manifestations with at least three distinct morphologies defined to date: “DDAF” (definite decreased AF), “WDQDAF” (well-defined questionable decreased AF), and “PDQDAF” (poorly defined questionable decreased AF). We recently adopted our deep learning approaches for FAF Stargardt atrophy segmentation using a sub-set of the ProgSTAR data, and all the three distinct morphologies were detected and segmented with reasonably good performance as shown in Figure 5.
Figure 5.
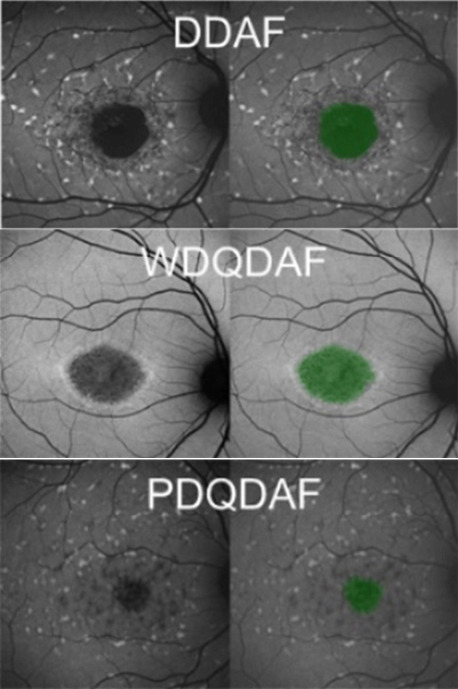
Example illustration of the Stargardt atrophic lesion segmentation on fundus autofluorescence images with three different morphologies.
From top to bottom: definite decreased AF (DDAF), well-defined questionable decreased AF (WDQDAF), and poorly defined questionable decreased AF (PDQDAF). Unpublished data.
Artificial Intelligence for Stargardt Atrophy Segmentation on Spectral-Domain Optical Coherence Tomography Images
Retinal layer segmentation and analysis on SD-OCT images have been very active research topics, as the retinal layer thickness and intensity may be affected locally or globally depending on the specific retinal disease (Carrera Fernandez et al., 2005; Li et al., 2006; Garvin et al., 2008; Yazdanpanah et al., 2009; Chiu et al., 2010; Hu et al., 2013a, b, c). To date, there are three major retinal layer segmentation algorithms: active contour method (Yazdanpanah et al., 2009), a graph-based shortest path with dynamic programming (Chiu et al., 2010), and 3D graph search (Carrera Fernandez et al., 2005; Li et al., 2006; Garvin et al., 2008; Hu et al., 2013a, b, c). Of these approaches, graph-based approaches have generally performed best. We have intensively worked on OCT layer segmentation using graph-based approaches and have supported clinical research and many clinical trials.
The advantages of the 3D graph search frameworks over the shortest path are that it is less dependent on initialization and can detect multiple optimal surfaces simultaneously from a constructed 3D graph based on an OCT image. However, the 3D graph search frameworks are relatively slow compared with shortest-path frameworks, as shortest-path does not request interaction between different retinal surfaces.
The 3D graph search approach we utilized was an evolution of the strategy previously described by Li et al. (2006). It is an unsupervised computer vision approach without manual ground truth for training. The segmentation of multiple surfaces using 3D graph search could be considered as an optimization problem with the goal being to find a set of surfaces with the minimum cost such that the found surface set was feasible. The 3D graph search had two major components: a) the formulation of the cost function, and b) the specification of the layer-based parameters encoding the surface feasibility constraints, which makes it more suitable for accurately identifying retinal layers in various diseases. Having said that, applying a suitable cost function and surface constraints, particularly in regions severely disrupted by retinal disease was not trivial. The cost function in our implementation was a signed edge-based term, favoring a dark-to-bright or bright-to-dark intensity transition based on different surfaces. It was achieved by applying two different 3 by 3 Sobel kernels in the vertical direction convolving with the original SD-OCT image to calculate vertical derivative approximations. Surface feasibility constraints included smoothness constraints within a particular surface and interaction constraints between different surfaces. The smoothness constraints and interaction constraints played an important role in accurately segmenting the multiple OCT layers. In Li’s previous approach, both the smoothness and interaction constraints were of a constant value (Li et al., 2006). In our adaptation, the smoothness constraints for both the single-surface and double graph search were still constant. However, for the interaction constraints of the double-surface graph searches, varying constraints of the estimated morphological shape models were employed. Our first attempt of OCT retinal layer segmentation with late-stage Stargardt atrophy utilized a 3D graph search algorithm to detect eight retinal surfaces. Figure 6 illustrates the eight OCT layer segmentation associated with late-stage Stargardt atrophy.
Figure 6.

Illustration of graph search segmentation of optical coherence tomography retinal surfaces associated with Stargardt atrophic-appearing lesion, as well as retinal deposits corresponding to the characteristic flecks of Stargardt disease.
From top to bottom: reference surface in vitreous layer, internal limiting membrane, outer plexiform and outer nuclear junction, external limiting membrane, inner-outer photoreceptor segmentation junction, inner retinal pigment epithelium, outer retinal pigment epithelium/Bruch’s membrane, choroidal-scleral junction. Unpublished data.
However, our first attempt with Stargardt atrophy OCT data only could segment eight retinal surfaces and the algorithm did not take advantage of the manual gradings for supervised learning with more accurate layer segmentation performance. Recently, we published another twelve-retinal-layer-segmentation algorithm aimed at more OCT layer segmentation and better segmentation accuracy (Mishra et al., 2021). The ProgSTAR studies have provided accurate manual-annotated retinal layers which can be used as ground truth for algorithm training, allowing us to use a supervised AI deep learning construct for enhanced OCT layer segmentation. Our twelve retinal layer segmentation utilized graph-based shortest path along with deep learning classification derived information. This method achieved a mean border difference that was within the subpixel accuracy range for retinal layers with Stargardt features and alongside associated retinal layers. Figure 7 provides several examples of our twelve-layer OCT segmentation results. To the best of our knowledge, this is the first automated algorithm for twelve retinal layer segmentation on OCT in eyes with Stargardt disease. Furthermore, from this segmentation, various thickness and intensity feature maps characterizing the outer retinal layers were generated, allowing for visualization of the feature differences between eyes diagnosed with Stargardt disease and normal eyes.
Figure 7.
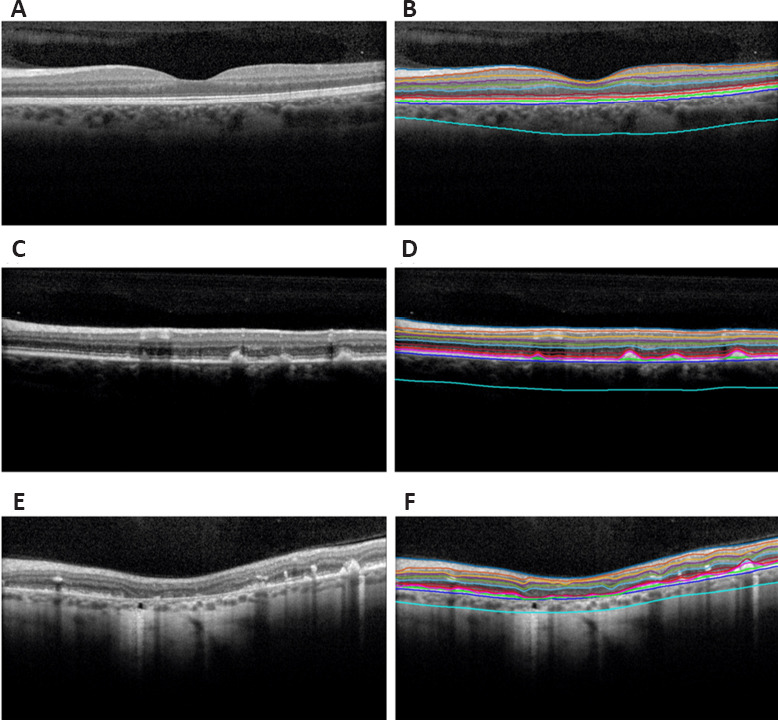
Segmentation of normal eyes and eyes diagnosed with Stargardt disease overlaid on spectral-domain optical coherence tomography B-scans.
(A) and (B) are the original B-scan and segmentation of a normal eye. (C) and (D) are the original B-scan and segmentation of an eye diagnosed with Stargardt disease showcasing mild degeneration and deposits corresponding to the characteristic flecks of Stargardt disease. (E) and (F) are the original B-scan and segmentation of an eye diagnosed with Stargardt disease showcasing severe degeneration and an atrophic-appearing lesion. The layers in order from top to bottom are the internal limiting membrane, nerve fiber-ganglion cell junction, ganglion cell-inner plexiform, inner plexiform-inner nuclear junction, inner nuclear-outer plexiform junction, outer plexiform-outer nuclear junction, external limiting membrane, inner-outer photoreceptor segmentation junction, Stargardt features, inner retinal pigment epithelium, outer retinal pigment epithelium, and choroidal-scleral junction. Reprinted with permission from Mishra et al. (2021).
Conclusion and Future Perspective
We reviewed the major reported articles (Wang et al., 2019; Mishra et al., 2021) using AI for the automated assessment of Stargardt macular atrophy associated features on FAF and SD-OCT images. Since Stargardt disease is rare and image data may be inaccessible or hard to access for most groups conducting AI research on ophthalmology, the publications in this field are limited. Nevertheless, the most representative AI deep learning algorithms from our team (Wang et al., 2019; Mishra et al., 2021) for the automated screening and segmentation of Stargardt atrophy associated features have both presented very promising algorithm performances compared with manually delineated ground truth from the reading center’s certified graders. These robust systems indicate great potential in using AI to facilitate large-scale Stargardt clinic trials and research and eventually transition to regular clinical application.
The ProgSTAR study has provided the bulk of the information regarding the natural evolution and progression of Stargardt disease. For example, ProgSTAR confirmed that on average, progression of atrophy in Stargardt is slow but also highly variable. However, in the initial ProgSTAR analysis, only a few risk factors/biomarkers (e.g., heterogeneous background autofluorescence and loss of photoreceptors) were found to predict the speed of progression of Stargardt atrophy overtime. This is a serious limitation for future trials as a slow and highly variable progression rate will require large samples and/or long trials. Therefore, there is a critical need to uncover novel predictive biomarkers, which may be encoded within rich multimodal imaging datasets, and particularly in 3D OCT. The AI techniques using deep learning would be well-suited for addressing the current critical unmet challenges in finding “true” novel predictive Stargardt biomarkers. While traditional AI machine learning approaches use hand-crafted (human designed) filters to extract image features, AI deep learning algorithms are different, automatically learning relevant image features and hence being objective. While we expect that some of the “objectively-learned” OCT biomarkers may overlap with the “subjectively specified” ones, we also anticipate that novel biomarkers with novel deep learning techniques may be identified. Further research would be needed, and we would expect interesting findings in this field.
Additional file: Open peer review report 1 (118.9KB, pdf) .
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
P-Reviewers: Subhi Y, Shah M; C-Editors: Zhao M, Zhao LJ, Qiu Y; T-Editor: Jia Y
Open peer reviewers: Mital Shah, Oxford Eye Hospital, Oxford University Hospitals NHS Foundation Trust, UK; Yousif Subhi, Zealand Univ Hosp, Denmark.
Funding: This project was supported by the National Eye Institute of the National Institutes of Health under Award Number R21EY029839 (to ZJH).
References
- 1.Binley K, Widdowson P, Loader J, Kelleher M, Iqball S, Ferrige G, de Belin J, Carlucci M, Angell-Manning D, Hurst F, Ellis S, Miskin J, Fernandes A, Wong P, Allikmets R, Bergstrom C, Aaberg T, Yan J, Kong J, Gouras P, et al. Transduction of photoreceptors with equine infectious anemia virus lentiviral vectors:safety and biodistribution of StarGen for Stargardt disease. Invest Ophthalmol Vis Sci. 2013;54:4061–4071. doi: 10.1167/iovs.13-11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabrera Fernandez D, Salinas HM, Puliafito CA. Automated detection of retinal layer structures on optical coherence tomography images. Opt Express. 2005;13:10200–10216. doi: 10.1364/opex.13.010200. [DOI] [PubMed] [Google Scholar]
- 3.Charng J, Xiao D, Mehdizadeh M, Attia MS, Arunachalam S, Lamey TM, Thompson JA, McLaren TL, De Roach JN, Mackey DA, Frost S, Chen FK. Deep learning segmentation of hyperautofluorescent fleck lesions in Stargardt disease. Sci Rep. 2020;10:16491. doi: 10.1038/s41598-020-73339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt Express. 2010;18:19413–19428. doi: 10.1364/OE.18.019413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cicinelli MV, Battista M, Starace V, Battaglia Parodi M, Bandello F. Monitoring and management of the patient with Stargardt disease. Clin Optom (Auckl) 2019;11:151–165. doi: 10.2147/OPTO.S226595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimoto JG, Bouma B, Tearney GJ, Boppart SA, Pitris C, Southern JF, Brezinski ME. New technology for high-speed and high-resolution optical coherence tomography. Ann N Y Acad Sci. 1998;838:95–107. doi: 10.1111/j.1749-6632.1998.tb08190.x. [DOI] [PubMed] [Google Scholar]
- 7.Fujinami K, Zernant J, Chana RK, Wright Ga, Tsunoda K, Ozawa Y, Tsubota K, Robson AG, Holder GE, Allikmets R, Michaelides M, Moore AT. Clinical and molecular characteristics of childhood-onset Stargardt disease. Ophthalmology. 2015;122:326–334. doi: 10.1016/j.ophtha.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garvin MK, Abramoff MD, Kardon R, Russell SR, Wu X, Sonka M. Intraretinal layer segmentation of macular optical coherence tomography images using optimal 3-D graph search. IEEE Trans Med Imaging. 2008;27:1495–1505. doi: 10.1109/TMI.2008.923966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He K, Zhang X, Ren S, Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) New York: IEEE; 2016. Deep Residual Learning for Image Recognition; pp. 770–778. [Google Scholar]
- 10.Hu Z, Wu X, Hariri A, Sadda S. Automated multilayer segmentation and characterization in 3D spectral-domain optical coherence tomography images. Proc SPIE. 2013a8567 doi:10.1117/12.2003172. [Google Scholar]
- 11.Hu Z, Wu X, Hariri A, Sadda SR. Multiple layer segmentation and analysis in three-dimensional spectral-domain optical coherence tomography volume scans. J Biomed Opt. 2013b;18:76006. doi: 10.1117/1.JBO.18.7.076006. [DOI] [PubMed] [Google Scholar]
- 12.Hu Z, Wu X, Ouyang Y, Ouyang Y, Sadda SR. Semiautomated segmentation of the choroid in spectral-domain optical coherence tomography volume scans. Invest Ophthalmol Vis Sci. 2013c;54:1722–1729. doi: 10.1167/iovs.12-10578. [DOI] [PubMed] [Google Scholar]
- 13.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Fujimoto JG. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong J, Kim SR, Binley K, Pata I, Doi K, Mannik J, Zernant-Rajang J, Kan O, Iqball S, Naylor S, Sparrow JR, Gouras P, Allikmets R. Correction of the disease phenotype in the mouse model of Stargardt disease by lentiviral gene therapy. Gene Ther. 2008;15:1311–1320. doi: 10.1038/gt.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kugelman J, Alonso-Caneiro D, Chen Y, Arunachalam S, Huang D, Vallis N, Collins MJ, Chen FK. Retinal boundary segmentation in stargardt disease optical coherence tomography images using automated deep learning. Transl Vis Sci Technol. 2020;9:12. doi: 10.1167/tvst.9.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K, Wu X, Chen DZ, Sonka M. Optimal surface segmentation in volumetric images--a graph-theoretic approach. IEEE Trans Pattern Anal Mach Intell. 2006;28:119–134. doi: 10.1109/TPAMI.2006.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L, Kaufman Y, Zhang J, Washington I. C20-D3-vitamin A slows lipofuscin accumulation and electrophysiological retinal degeneration in a mouse model of Stargardt disease. J Biol Chem. 2011;286:7966–7974. doi: 10.1074/jbc.M110.178657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra Z, Wang Z, Sadda SR, Hu Z. Automatic segmentation in multiple OCT layers for Stargardt disease characterization via deep learning. Transl Vis Sci Technol. 2021;10:24. doi: 10.1167/tvst.10.4.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee N, Schuman S. EyeNet Magazine. San Francisco: American Academy of Ophthalmology; 2014. Diagnosis and management of Stargardt disease. [Google Scholar]
- 20.Ronneberger O, Fischer P, Brox T. U-Net:convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. MICCAI 2015. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2015. pp. 234–241. [Google Scholar]
- 21.Schmitz-Valckenberg S, Holz FG, Bird AC, Spaide RF. Fundus autofluorescence imaging:review and perspectives. Retina. 2008;28:385–409. doi: 10.1097/IAE.0b013e318164a907. [DOI] [PubMed] [Google Scholar]
- 22.Schonbach EM, Wolfson Y, Strauss RW, Ibrahim MA, Kong X, Munoz B, Birch DG, Cideciyan AV, Hahn GA, Nittala M, Sunness JS, Sadda SR, West SK, Scholl HPN, ProgStar Study G. Macular sensitivity measured with microperimetry in Stargardt disease in the progression of atrophy secondary to Stargardt disease (ProgStar) Study:report No. 7. JAMA Ophthalmol. 2017;135:696–703. doi: 10.1001/jamaophthalmol.2017.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauss RW, Ho A, Munoz B, Cideciyan AV, Sahel JA, Sunness JS, Birch DG, Bernstein PS, Michaelides M, Traboulsi EI, Zrenner E, Sadda S, Ervin AM, West S, Scholl HP Progression of Stargardt Disease Study G. The natural history of the progression of atrophy secondary to Stargardt disease (ProgStar) studies:design and baseline characteristics:ProgStar Report No. 1. Ophthalmology. 2016;123:817–828. doi: 10.1016/j.ophtha.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Strauss RW, Muñoz B, Ho A, Jha A, Michaelides M, Cideciyan AV, Audo I, Birch DG, Hariri AH, Nittala MG, Sadda S, West S, Scholl HPN. Progression of Stargardt disease as determined by fundus autofluorescence in the retrospective progression of Stargardt disease study (ProgStar Report No. 9) JAMA Ophthalmol. 2017a;135:1232–1241. doi: 10.1001/jamaophthalmol.2017.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss RW, Munoz B, Ho A, Jha A, Michaelides M, Mohand-Said S, Cideciyan AV, Birch D, Hariri AH, Nittala MG, Sadda S, Scholl HPN, ProgStar Study G. Incidence of atrophic lesions in Stargardt disease in the progression of atrophy secondary to Stargardt disease (ProgStar) study:report No. 5. JAMA Ophthalmol. 2017b;135:687–695. doi: 10.1001/jamaophthalmol.2017.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) New York: IEEE; 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 27.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) New York: IEEE; 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 28.Wang Z, Sadda S, Hu Z. Deep learning for automated screening and semantic segmentation of age-related and juvenile atrophic macular degeneration. Proc SPIE. 201910950 doi:10.1117/12.2511538. [Google Scholar]
- 29.Yazdanpanah A, Hamarneh G, Smith B, Sarunic M. Intra-retinal layer segmentation in optical coherence tomography using an active contour approach. Med Image Comput Comput Assist Interv. 2009;12:649–656. doi: 10.1007/978-3-642-04271-3_79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


