Abstract
Background
Inflammatory bowel disease may arise with inadequate immune response to intestinal bacteria. NOD2 is an established gene in Crohn’s disease pathogenesis, with deleterious variation associated with reduced NFKB signaling. We hypothesized that deleterious variation across the NOD2 signaling pathway impacts on transcription.
Methods
Treatment-naïve pediatric inflammatory bowel disease patients had ileal biopsies for targeted autoimmune RNA-sequencing and blood for whole exome sequencing collected at diagnostic endoscopy. Utilizing GenePy, a per-individual, per-gene score, genes within the NOD signaling pathway were assigned a quantitative score representing total variant burden. Where multiple genes formed complexes, GenePy scores were summed to create a “complex” score. Normalized transcript expression of 95 genes within this pathway was retrieved. Regression analysis was performed to determine the impact of genomic variation on gene transcription.
Results
Thirty-nine patients were included. Limited clustering of patients based on NOD signaling transcripts was related to underlying genomic variation. Patients harboring deleterious variation in NOD2 had reduced NOD2 (β = -0.702, P = 4.3 × 10-5) and increased NFKBIA (β = 0.486, P = .001), reflecting reduced NFKB signal activation. Deleterious variation in the NOD2-RIPK2 complex was associated with increased NLRP3 (β = 0.8, P = 3.1475 × 10-8) and TXN (β = -0.417, P = 8.4 × 10-5) transcription, components of the NLRP3 inflammasome. Deleterious variation in the TAK1-TAB complex resulted in reduced MAPK14 transcription (β = -0.677, P = 1.7 × 10-5), a key signal transduction protein in the NOD2 signaling cascade and increased IFNA1 (β = 0.479, P = .001), indicating reduced transcription of NFKB activators and alternative interferon transcription in these patients.
Conclusions
Data integration identified perturbation of NOD2 signaling transcription correlated with genomic variation. A hypoimmune NFKB signaling transcription response was observed. Alternative inflammatory pathways were activated and may represent therapeutic targets in specific patients.
Keywords: IBD, Crohn’s disease, NOD2, WES, transcriptome
Introduction
Pediatric inflammatory bowel disease (IBD) has a complex pathogenesis, reflecting genetic and environmental influences on disease etiology. Contemporary genomics has pointed to the molecular cause of disease being specific to an individual, or family. Despite this, the resulting immune dysfunction frequently converges on common inflammatory pathways such as tumor necrosis factor α production and interleukin (IL)-17 signaling.1 Identification of the precise cause of disease within an individual patient is challenging, and integration of multiomic data to assess the impact of genetic variation on gene expression and immune function and the relationship with intestinal microbiota remains difficult.2 Typically, functional assessment of deleterious genomic variation in an individual is through quantification of downstream cytokine levels.3 Stimulation allows assessment of the impact of a mutation on downstream immune function.3 Analysis may focus on transfection of cell lines with the mutation of interest, although accurate reflection of in vivo response is difficult to achieve when multiple variants are contributing to a complex disease phenotype. While these techniques are effective in Mendelian disease, in polygenic IBD, in which multiple genetic variants appear to result in the same phenotype, establishing the effect of multiple mutations is challenging.
Expression quantitative trait loci have been identified that impact on NOD2 expression.4 Consolidation of genetic variation across a protein complex or pathway would allow grouping of patients with deleterious variation in several interacting genes. Consolidation can occur through a mathematical model based on a whole gene pathogenicity score, which sums the variation within genes, taking into account the key variation metrics.5 This score reflects a patient’s burden of variant pathogenicity within genes of interest.
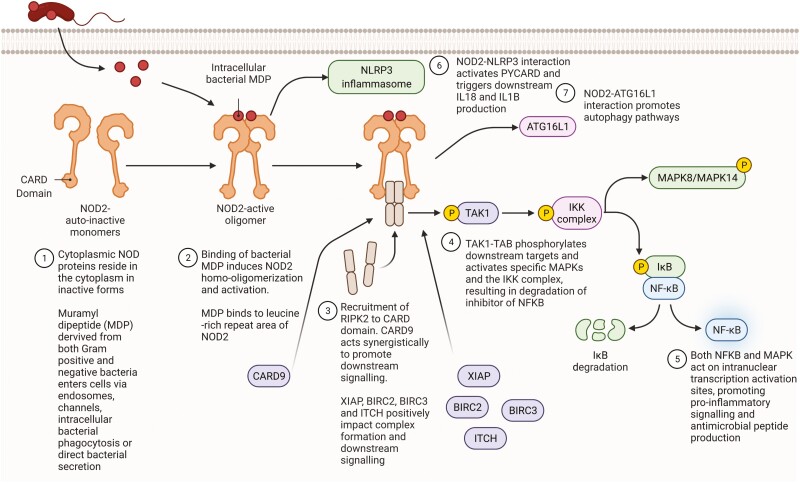
It is recognized that monogenic disease with highly similar phenotypes can result from deleterious variation in different genes, the occurrence of polygenic conditions, such as typical forms of IBD, are also likely to have contribution from variation in a number of related genes within an individual.6 The NOD signaling pathway, an innate immune signaling response, is highly implicated in Crohn’s disease pathogenesis, both through genomic and transcriptomic analysis.7,8NOD2 is the most implicated gene in the Crohn’s disease pathogenesis and is a central player in the pathway, acting as an intracellular pattern recognition receptor for bacterial components, specifically muramyl dipeptide (MDP) (Figure 1).8 Variation within other genes within this pathway, including XIAP, ATG16L1, CARD9, and RIPK2, has been implicated in both monogenic and polygenic forms of IBD.9
Figure 1.
Schematic representation of NOD2 signaling cascade and directly related inflammatory signaling pathways. Adapted from “Detection of Bacterial Peptidoglycan by NOD Receptors,” by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates. IL, interleukin; MAPK, mitogen-activated protein kinase; NFKB, nuclear factor kappa B.
We hypothesize that genomic variation across the NOD signaling pathway will have direct impact on transcription of related genes within the pathway. Therefore, we undertook systematic analysis of genes and complexes along the NOD pathway to identify individual contributions to dysregulation of transcriptional programs. We assess whether variation in single genes, and across protein complexes, is associated with altered gene expression within the NOD signaling pathway to identify patterns in immune pathway transcription across a cohort.
Methods
Pediatric IBD patients were recruited through from the Paediatric Gastroenterology service at Southampton Children’s Hospital. Patients were recruited prior to diagnostic endoscopy and are termed treatment-naïve IBD patients. All patients had a terminal ileal biopsy and blood for whole exome sequencing data taken at the time of endoscopy. These analyses included patients with a subsequent diagnosis of Crohn’s disease, ulcerative colitis, or IBD-unclassified. No patients with monogenic IBD were included. While NOD2 variation is associated with Crohn’s disease, ulcerative colitis and IBD-unclassified patients were included to ensure a spectrum of genomic mutation burden.
Whole Exome Sequencing Analysis
Patient DNA was extracted from peripheral venous blood samples collected in EDTA using the salting-out method, or from saliva, as previously described.10 DNA concentration was estimated using the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Approximately 20 µg of each patient’s DNA was extracted for next-generation sequencing.
Raw FASTQ sequencing data from patients with treatment-naïve IBD were processed using our in-house pipeline (https://github.com/UoS-HGIG). VerifyBamID was utilized to check the presence of DNA contamination across the cohort.11 Alignment was performed against the human reference genome (hg38 assembly) using BWA-mem (version 0.7.15).12 Aligned BAM files were sorted, and duplicate reads were marked using Picard Tools (version 2.9.2; Broad Institute of MIT and Harvard, Cambridge, MA, USA). Following GATK v3.813 best practice recommendations,14 variants were called using GATK HaplotypeCaller to produce a gVCF file for each sample and later jointly genotyped. HaplotypeCaller default settings were utilized corresponding to variants with a minimum Phred base quality score of 20 being called.
Annotation of this composite file for variant-based analysis applied Annovar v2018Apr16 using default databases refSeq gene transcripts (refGene), deleteriousness scores databases (dbnsfp35c, CADD v.1.5 and DANN), dbSNP147, and the human genetic mutation database (HGMD Pro 2018) flat file.15 Variant allele frequencies were sourced through the Genome Aggregation Database (gnomAD) v2.1.1.16,17
Application of GenePy
Whole exome sequencing data were transformed in a per gene, per individual GenePy score for integration with transcriptomic analysis.5 Before GenePy scores were calculated, the joint cohort variant call format (VCF) was quality controlled by implementing methods from Carson et al.17 The VCF was then annotated with the databases refGene and gnomAD exome v2.1.117, using Annovar v2018Apr16. The VCF was additionally annotated with deleteriousness metric CADD (v.1.6).18 The GenePy score algorithm was then applied to the annotated cohort VCF (https://github.com/UoS-HGIG/GenePy). Demonstration of the contributions of individual variants to GenePy scores can be seen in Supplementary Data 1.
Normalization and Application of LOEUF Score to GenePy
Some genes can accrue very high GenePy scores (owing to length or mutability), and when summing GenePy scores across a complex, we first needed to normalize the values in order to make each gene comparable. We normalized all GenePy scores between 0 and 1, where 0 represented the lowest GenePy score for that gene across all patients in the analysis and 1 represented the highest score for that gene across all patients in the analysis.
The LOEUF (loss of function observed/expected upper bound fraction) score is a metric developed as part of the gnomAD that assigns a score to each gene based on the gene’s intolerance to inactivation.19 This score can then be applied to determine which genes are able to accrue variation while maintaining activity, compared with those in which variation will be highly damaging. Higher LOEUF scores are associated with increased tolerance to variation. Genes within the NOD pathway that are highly conserved and key to multiple inflammatory processes, such as RIPK2 or TAK1, have very low LOEUF scores, whereas genes in which variation is more commonly seen, such as NOD2, have higher scores.
We integrated the LOEUF score into the GenePy score in order to upweight the importance of variation in genes predicted to be intolerant to inactivation. LOEUF scores for genes in this analysis can be seen in Supplementary Data 2. The normalized GenePy score was divided by the respective LOEUF score for each gene.
Ileal Biopsy RNA Extraction
Terminal ileal biopsies were obtained during routine endoscopy and frozen at -80 in 1 mL of RNAlater (Sigma-Aldrich, St Louis, MO, USA) <30 minutes from collection. RNA was extracted from ileal biopsies using Maxwell processing, as previously described.20 A single biopsy was used for each patient.
Targeted RNA Sequencing
The HTG EdgeSeq Autoimmune Panel (HTG Molecular Diagnostics, Tucson, AZ, USA) was used to measure messenger RNA expression levels in 2002 genes associated with autoimmune disease, including IBD. The targeted sequencing was performed as previously described.20 Briefly, an RNA sample for each patient was thawed, diluted, and loaded onto the HTG EdgeSeq instrument. Following a standardized polymerase chain reaction clean-up procedure, and quantification, each sample underwent a dilution-based normalization process. Individual libraries were pool at equimolar quantities into the final library for sequencing. The library was denatured and prepared for sequencing in line with Illumina (San Diego, CA, USA) and HTG practice guidelines. Sequencing was performed on the Illumina NextSeq platform.
RNA Data Processing
Output files from the NextSeq run were converted from BCL format to FASTQ files. The barcode mismatch filter was set to 0 to ensure that reads were linked to the correct patient. A gene expression count matrix was constructed for each gene and each patient. These were merged to form a single output file containing all genes and all counts. Quality control was performed as previously described, in line with HTG recommendations.20 Gene transcript counts were normalized using quantile normalization for downstream integration with genomic data. Gene expression using the targeted RNA panel has previously been validated utilizing single-cell RNA sequencing.20 We determined if histological inflammation, regardless of underlying genomic variation, was associated with differential gene expression using DEseq2.21
Key Genes and Protein Complexes Within the NOD signaling Pathway
We proposed that summing variation across key protein complexes within the NOD signaling pathway would enable us to discern the impact of underlying genomic variation on gene expression. We utilized a precollated list of 95 genes in the NOD signaling pathway, produced by HTG as part of their autoimmune panel product.22
The NOD signaling pathway genes were cross-referenced with genes known to be implicated in IBD either by genome-wide association study or as a monogenic IBD gene.22,23 Invariant genes or complexes were not assessed. Single genes within the pathway, and implicated in IBD, were included in the analysis. Molecular complexes in the NOD signaling pathway, with 2 or more constituent proteins, containing genes with variation, were included in the analysis. Molecular complexes activated because of multiple inflammatory pathways, including the IKK complex and MAPK complexes, were excluded due to the lack of specificity to NOD signaling.
Summing GenePy Scores for Protein Complexes
We summed GenePy scores across the genes in key protein complexes. For each gene within a molecular complex, the LOEUF-corrected, normalized GenePy scores for each gene were summed to create a GenePy score for the molecular complex (Supplementary Data 2). This provides a quantitative score reflecting the cumulative sum of variation within that complex.
Genomic and Transcriptomic Integration
We utilized a stepwise linear regression model to determine the impact of genomic variation on transcription. The analyses were conducted to determine whether genomic variation within the NOD signaling pathway impacted on gene expression within the same pathway. We utilized the GenePy scores for an individual gene, or summed scores for a complex, as the dependent variable. The quantile normalized (QN) expression values for the 95 NOD signaling genes were entered as independent variables. These data assume an additive model of deleteriousness.
To assess whether patients with higher burden of variation within key NOD2 pathway genes and complexes exhibited similar expression patterns across all NOD signaling genes, we performed principal component analysis (PCA) and hierarchical clustering (HC). Patients within the top 10% of GenePy scores for NOD2, NOD2-RIPK2, and TAK1-TAB were annotated within the analyses, and the top 10% of patients were chosen as per previous evidence.23 These 12 patients were also utilized to determine if high GenePy scores were driving inflammation within the terminal ileum, and therefore altered transcription related to inflammation, rather than altering pathway transcription per se.
Statistical Analysis
Statistical analysis was performed using SPSS (version 25; IBM, Armonk, NY, USA). PCA and HC (Euclidean distance, average clustering) were performed using Reveal (version 3.1; HTG Molecular Diagnostics). Weighted gene coexpression network analysis was performed as previously described using the WGCNA R package.20,24
Ethical Approval
The study has category A ERGO II ethics approval (30630) and a REC approval from Southampton and South West Hampshire Research Ethics Committee (09/H0504/125). All patients and families provided informed consent at recruitment.
Results
Thirty-nine patients were included in the analysis; 27 had a diagnosis of Crohn’s disease, 9 had a diagnosis of ulcerative colitis, and 3 had a diagnosis of IBD-unclassified. The mean age at diagnosis was 13.2 years (range, 2.9-16.8 years). All patients were naïve to any immunomodulating, anti-inflammatory, or monoclonal treatment at the time of biopsy acquisition. The core NOD signaling pathway was interrogated (Figure 1). Three genes and 3 sets of genes encoding protein complexes were selected for analysis based on the criteria described previously (Table 1). The IRAK-TRAF6 complex acts synergistically with the core NOD2 signaling pathway and was included, in addition to the core NOD2-RIPK2 and TAK1-TAB complexes. The IKK complex, consisting of IKKa, IKKb, and NEMO, was excluded, as it is the target of multiple activators including tumor necrosis factor α and IL-1 signaling, and is not specific for NOD-dependent bacterial recognition and response.25 Similarly, MAPK complexes were excluded due to the large number of MAPK genes that may be included and the lack of specificity to NOD signaling (Supplementary Figure 1A). Further data can be seen in the Supplementary Results, including the phenotype-NOD2 genotype correlation (Supplementary Table 2).
Table 1.
Genes and complexes to be entered as dependent variables in regression analysis and the constituent proteins (genes)
| Gene or Complex | Proteins Comprising Complex |
|---|---|
| NOD2 | NOD2 |
| ATG16L1 | ATG16L1 |
| CARD9 | CARD9 |
| NOD2-RIPK2 complex | RIPK2, NOD2, XIAP, BIRC2, BIRC3, ITCH |
| TAK1-TAB complex | TAK1, TAB2, TAB3 |
| IRAK-TRAF6 complex | IRAK1, IRAK2, IRAK4, TRAF6, MYD88 |
All genes’ GenePy scores are scaled to between 0 and 1 and corrected by LOEUF (loss of function observed/expected upper bound fraction) score prior to being summed to form the complex’s GenePy score.
NOD2 GenePy Score Is Not Associated With Specific Gene Expression Modules Across All Autoimmune Genes
Given the established role of NOD2 in Crohn’s disease pathogenesis, we hypothesized that subgroup(s) of patients with accumulation of pathogenic NOD2 variation would be characterized by similar gene expression. To test this hypothesis, we determined gene coexpression modules, identified through WGCNA of all 39 patients, and correlated these modules with NOD2 GenePy score. Patients were clustered by the similarity of gene expression for all 2002 autoimmune gene transcripts (Supplementary Figure 2). NOD2 GenePy scores were entered as a continuous variable. Hierarchical clustering did not demonstrate patients with similar NOD2 GenePy scores in the same clusters.
Gene Coexpression Modules in Treatment-Naïve Patients
Expression modules were identified within the autoimmune transcripts, based on the 39 treatment-naïve patients. Two large coexpression signatures emerged, the turquoise and blue modules, alongside several smaller coexpression modules (Supplementary Figure 3). In order to determine whether specific sets of coexpressed genes were associated genomic variation in NOD2, we analyzed whether the coexpression modules were correlated with NOD2 GenePy score. None of the 8 modules was significantly correlated with NOD2 (Supplementary Figure 4).
Patients Harboring Deleterious NOD2 Gene Variation Have Reduced NOD2 Gene Expression and Increased Expression of Nuclear Factor Kappa B Inhibitor α
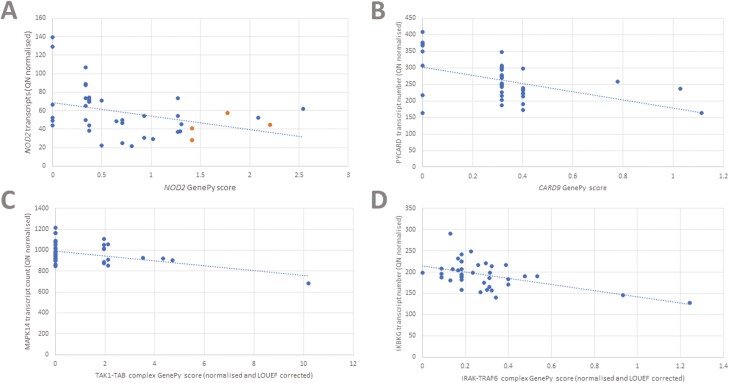
We expected NOD2 (Figure 1.1) gene variation to impact on downstream gene expression, specifically within the signaling pathway. We examined the effect of variation in NOD2 through stepwise linear regression, with NOD2 GenePy score as the dependent variable and all 95 gene transcript levels as the independent variables. Increased NOD2 GenePy scores, reflecting increased deleterious variation, were associated with a decrease in NOD2 transcripts (Figure 2A) and increased expression of NFKBIA, encoding a key inhibitory protein preventing nuclear factor kappa B (NFKB) signaling (Table 2). The effect appears to reflect a decrease in NFKB signaling as a result of deleterious NOD2 variation (Supplementary Figure 1B). CCL5, a T cell chemokine, was also downregulated in patients with high variant deleteriousness in the NOD2 gene.
Figure 2.
A, Relationship between quantile-normalized (QN) NOD2 transcript levels and NOD2 GenePy score. Four patients harboring the 1007fs variant are seen in orange. B, Relationship between QN PYCARD transcript levels and CARD9 complex GenePy score. C, Relationship between QN MAPK14 transcript levels and TAK1-TAB complex GenePy score. D, Relationship between QN IKBKG transcript levels and IRAK-TRAF6 complex GenePy score. LOEUF, loss of function observed/expected upper bound fraction.
Table 2.
Linear regression results of genes and complexes and the impact on NOD signaling transcription
| Dependent Variable: Gene/Complex GenePy Score | Independent Variable: Quantile-Normalized Gene Expression | β Coefficient | R 2 Value for Linear Regression Model | P Value |
|---|---|---|---|---|
| NOD2 | NOD2 | -0.702 | 0.460 | .000043 |
| NFKBIA | 0.486 | .001 | ||
| CCL5 | -0.414 | .008 | ||
| ATG16L1 | IKBKB | 0.504 | 0.403 | .001 |
| IRAK4 | -0.498 | .001 | ||
| CYBA | -0.288 | .041 | ||
| CARD9 | PYCARD | -0.559 | 0.510 | .0007 |
| IFNB1 | 0.333 | .006 | ||
| TRAF2 | 0.371 | .003 | ||
| TANK | -0.294 | .025 | ||
| NOD2-RIPK2 complex (RIPK2, NOD2, XIAP, BIRC2, BIRC3, ITCH) | BIRC2 | 0.800 | 0.687 | 3.1475 × 10-8 |
| TXN | 0.417 | .000084 | ||
| NLRP3 | 0.245 | .014 | ||
| PYCARD | -0.278 | .014 | ||
| IRAK4 | -0.345 | .001 | ||
| UBA52 | -0.224 | .033 | ||
| TAK1-TAB complex (TAK1, TAB2, TAB3) | MAPK14 | -0.677 | 0.438 | .000017 |
| IFNA1 | 0.479 | .001 | ||
| BIRC3 | -0.375 | .008 | ||
| IRAK-TRAF6 complex (IRAK1, IRAK2, IRAK4, TRAF6, MYD88) | IKBKG | -0.508 | 0.258 | .001 |
The 1007fs NOD2 Variant Impacts Transcription But Is Not the Only Driver of Reduced Expression
NOD2 harbors a protein-truncating variant, 1007fs, commonly seen in Crohn’s disease patients. We assessed whether this specific nonsense variant within NOD2 was driving the inverse relationship between the GenePy score and transcript number. Four of the 39 patients were heterozygous for the 1007fs NOD2 variant. A t test demonstrated that there was no significant difference in the NOD2 expression level between those with the 1007fs variant (mean QN transcripts 42.9) and those without (mean QN transcripts 59.5) (P = .12), although only 4 patients with the frameshift variant were seen in the patient group (Figure 2A). No other protein-truncating variants within NOD2 were identified in the 39 patients.
We assessed if NOD2 transcription was impacted by any of the 3 common variants associated with Crohn’s disease: 1007fs (4 patients = heterozygote), R702W (1 patient = heterozygote), and G908R (5 patients = heterozygote). Comparing these 10 patients with the remaining 29 patients, who did not harbor any of these variants, demonstrated a significantly reduced NOD2 transcript count (45.9 vs 61.8; P = .049).
Deleterious Variation in CARD9 Is Associated With Decreased Expression of PYCARD and TANK
CARD9 interacts directly with NOD2 (Figure 1.3), resulting in activation of downstream proinflammatory signaling through mitogen-activated protein kinase (MAPK) activation alongside activation of the NLRP3 inflammasome. It also functions independently as a signal transduction complex alongside BCL10 and MALT1, largely in response to fungal infection, which then activates the IKK complex, triggering NFKB activation, or through PYCARD, activating the NLRP3 inflammasome.26 Regression analysis with the CARD9 GenePy score as the dependent variable demonstrated that increased deleterious variation in CARD9 was associated with a decrease in PYCARD, an upstream activator of the NLRP3 inflammasome (Figure 2B, Table 2). Additionally, we observe an increase in increased transcription of TRAF2, and a decrease in TANK, the TRAF family member–associated NFKB activator. TANK directly inhibits TRAF2, and these data reflect a potential reduced ability to inhibit downstream NFKB signaling, through TRAF2 and IKK, within the NOD signaling cascade.27
Deleterious Variation in the NOD2-RIPK2 Complex Is Associated With Increased Expression of BIRC2, TXN, and NLRP3
Following activation by MDP, NOD2 forms a complex with RIPK2 (Figure 1.3). XIAP, BIRC2, BIRC3, and ITCH all positively associate with the complex promoting downstream RIPK2 kinase activity. Regression analysis, using the summed NOD2-RIPK2 complex as the dependent variable, revealed that increased deleterious variation within the complex was related to an increased expression of the NLRP3 and TXN (Table 2). Both of these genes are involved in the proinflammatory NLRP3 inflammasome; however, we also observed a decrease in PYCARD expression, encoding a key protein in the inflammasome activation pathway (Figure 1.6). BIRC2 was highly significantly upregulated in patients harboring deleterious variation in this complex, which includes BIRC2. The impact can be seen in Supplementary Figure 1C.
Genomic Variation Within the TAK1-TAB Complex Leads to Reduced MAPK14 Expression
The TAK1-TAB complex, including TAK1, TAB2, and TAB3, is a key signal transducer, both from the NOD2-RIPK2 complex and Toll-like receptors (TLRs) (Figure 1.4). Summed GenePy scores for the TAK1-TAB complex were used as the dependent variable. Expression of MAPK14 and BIRC3 was reduced in the presence of deleterious variation within the TAK1-TAB complex. There was also increased expression of IFNA1, reflecting activation of a type I interferon response pathway (Figure 2C, Table 2). MAPK14 autophosphorylates in the presence of the TAK1-TAB complex, leading to downstream activation of proinflammatory and antimicrobial gene expression (Supplementary Figure 1D).
Deleterious Variation in ATG16L1 Increases Expression of IKBKB
ATG16L1 encodes for a protein key in autophagy pathways. Activated NOD2 works synergistically with ATG16L1 to promote autophagy (Figure 1.7). Variation within ATG16L1 is an established risk for Crohn’s disease development. Regression analysis utilizing ATG16L1 GenePy score as the dependent variable demonstrated that an increased ATG16L1 GenePy score was associated with an increase in IKBKB, an activator of NFKB signaling (Table 2). Autophagy represents a different pathway to core NOD2 signaling, and variation in ATG16L1 may be proinflammatory via NKFB signaling, while reducing autophagy.
Variation in the IRAK-TRAF6 Complex, Within the TLR Signaling Pathway, Results in Decreased Expression of the NFKB Activating Protein IKBKG (NEMO)
The IRAK-TRAF6 complex (IRAK1, IRAK2, IRAK4, TRAF6, MYD88) plays a key role in signal transduction from TLRs through interaction with MYD88. It acts synergistically with NOD2 activation and one result of IRAK-TRAF6 activation is downstream activation of the TAK1-TAB complex and subsequently activation of NFKB signaling. Variation in the IRAK-TRAF6 complex was associated with a decrease in a single gene transcript, IKBKG, a component of the NFKB activating complex (Figure 2D).
Patients With Deleterious Variants in NOD2, RIPK2, and TAK1-TAB Have Variable NOD Signaling Expression Clustering
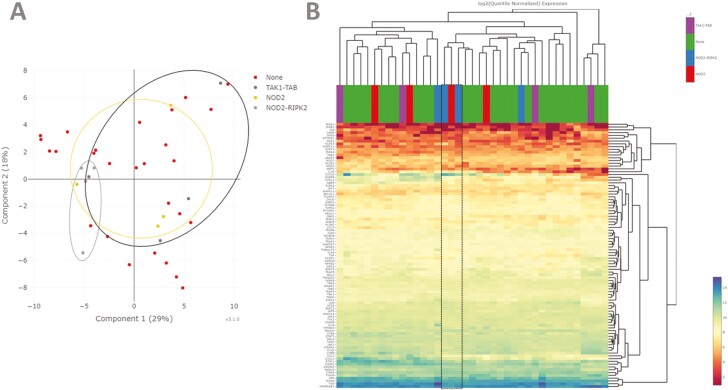
Patients with high burden of variation (top 10% of GenePy scores) in the central NOD2 signaling complexes were assessed for similarity of all NOD signaling gene expression. PCA revealed variable clustering (Figure 3A). The 4 patients with highest burden variation in the NOD2-RIPK2 complex did cluster together, along with a patient with high mutation burden in NOD2 and a patient with high burden in the TAK1-TAB complex.
Figure 3.
A, Principal component analysis using quantile-normalized data from 95 NOD signaling genes demonstrating clustering of patients with the top 10% (4 patients) of deleterious genetic variation within NOD2-RIPK2 but with less defined grouping for NOD2 and TAK1-TAB.B, Heatmap analysis using quantile-normalized data from 95 NOD signaling genes and average linkage clustering demonstrates highly variable clustering of patients. Two patients with top 10% of deleterious genetic variation in NOD2 and 1 with top 10% in NOD2-RIPK2 do cluster (boxed area), with high transcription of CXCL8 (interleukin-8) and low expression of TFNA1.
Using HC we were able to discern that several of the patients with high NOD2-RIPK or NOD2 mutation were characterized by increased expression of CXCL8 (encoding for the IL-8 neutrophil chemokine) and low expression of IFNA1 (Figure 3B). Interestingly, while deleterious variation in the NOD2-RIPK complex appears to negatively impact on downstream NFKB signaling, there is still an overall inflammatory response, mediated through alternative inflammatory pathways. Further analysis of the patients with high genomic burden in NOD2, NOD2-RIPK2, and TAK1-TAB revealed that of the 12 patients with the highest burden, 7 had histological inflammation, while 5 were noninflamed. This indicates that transcription differences noted through regression are not solely driven by inflammation status of the biopsy.
Differential Gene Expression Within the NOD Signaling Pathway Is Observed Between Inflamed and Noninflamed Tissue
Within the NOD signaling pathway, there were 17 genes with significant differential expression (corrected P < .05): 8 were upregulated in inflamed biopsies and 9 were downregulated. Further analysis of the patients with the top 10% of genomic burden in NOD2, NOD2-RIPK2, and TAK1-TAB revealed that of the 12 patients with the highest burden, 7 had histological inflammation, while 5 were noninflamed. This indicates that transcription differences noted through regression are not solely driven by inflammation status of the biopsy, but rather by altered transcription related to underlying genomic differences.
Discussion
The impact of genomic variation across key genes, and complexes, within the NOD signaling pathway appears to be associated with a hypoinflammatory response, with reduced activation (or increased inhibition) of NFKB signaling or reduced upstream activation of proinflammatory signaling. Variation in NOD2, and directly related complexes, appears to act synergistically to reduce NOD2 transcription, while simultaneously increasing transcription of alternative inflammatory pathways including the NLRP3 inflammasome and interferon signaling. We identify variation across the TAK1-TAB complex directly correlated with reduced MAPK14 transcription. Variation in NOD2-synergistic activators of autophagy (ATG16L1), or NOD signaling (CARD9), impact on downstream transcription, either increasing or decreasing NFKB signaling, respectively.
Previous data have pointed to a hypoimmune response in both Crohn’s disease patients and murine models harboring deleterious NOD2 variants.3,8NOD2 variants are thought to be loss of function, leading to impaired NOD2 activation.28 Studies detailing the direct effect of deleterious NOD2 variants repeatedly identify a decrease in proinflammatory cytokine response after MDP stimulation in peripheral blood mononuclear cells, specifically reduced NFKB production.8 Overall, the mechanism whereby NOD2 variants increase susceptibility to Crohn’s disease appears to be through impaired bacterial recognition or response leading to reduced bacterial clearance and increased chronic inflammation through non-NOD2 proinflammatory pathways.29 In addition to NOD2, several additional risk susceptibility genes, or monogenic IBD genes, lie within the NOD signaling pathway including XIAP, CARD9, and TAB2.1 Loss-of-function variants within these genes are associated with severe monogenic forms of Crohn’s-like IBD (XIAP, CARD9), or increased risk of “classical” Crohn’s disease (TAB2). Several studies have described the function of these genes in downstream NFKB signaling, including variants in XIAP30 and CARD931 leading to reduced NFKB production. While the mechanism by which these genes leads to disease may not have the evidence base seen with NOD2, it appears that a hypoinflammatory response is implicated, potentially alongside activation of additional aberrant pathways. It was important to include patients with ulcerative colitis is this analysis, as ulcerative colitis patients should have no impact of NOD2 variation on transcription, and it is therefore vital to include these patients to demonstrate a full spectrum of genomic variation and transcription levels. In addition, variation in the wider NOD2 signaling pathway may impact ulcerative colitis patients, and moving IBD toward a precise molecular diagnosis would equate to patients’ being categorized by molecular perturbation, rather than by endoscopic or histological disease location.
Within this study, we hypothesized that variation across the NOD signaling pathway, with a focus on NOD2 signaling, would result in transcription-level defects associated with a hypoinflammatory response. Previous data have inferred that disruption at any step on the NOD2 signaling cascade will result in decreased downstream NFKB or MAPK activation, although direct impact of genetic variation at each step has not been previously identified.32 Through single gene, and whole complex, deleteriousness scoring, we identify a consistent pattern of defects associated with decreased transcription of downstream NFKB or MAPK genes. Importantly, by summing deleteriousness across a complex, we were able to observe cohort-level effects that may be missed if assessing a single variant or a single gene within an individual patient. As individual variant effects on gene transcription are likely to be very mild, or private to an individual, the ability to sum the effects of interacting genes allows a statistical association to emerge across a cohort. It is clear that for most patients with IBD, the effect of multiple genomic variants, rather than a strong effect from a single gene, leads to disease.23
At an individual gene level, we reveal a striking decrease in NOD2 transcripts associated with increased NOD2 deleteriousness. This is driven not only by patients harboring the nonsense 1007fs variant, with only 4 of the 39 patients being heterozygote. No patients were homozygote for this protein-truncating variant. It is not possible to determine whether several of the more common variants harbored by patients with low NOD2 transcript levels are in linkage disequilibrium with noncoding variants in the promotor region of NOD2. ATG16L1 synergistically acts with NOD2 to promote antibacterial autophagy and also has a role in negative regulation of proinflammatory MAPK and NFKB activation cascades.33 Our results demonstrate the direct impact of this additional ATG16L1 role, with ATG16L1 variation leading to an increase in the NFKB activation transcript IKBKB. Where deleterious variation in the NOD2 canonical pathway appears to lead to reduced NFKB or MAPK activation of downstream inflammatory signaling, variation in ATG16L1 may lead to increased inflammation through impaired autophagy, or directly through the inability to negatively regulate NOD2 activity. Interestingly, recent ileal transcriptomic data integrating with NOD2 genotyping of the patients have identified an activated and dysregulated fibroblast cell signature in those with higher mutation burden.34 Within the article the authors identify an apparent STAT3-driven regulation of downstream transcription, specifically related to mutant NOD2 alleles.
We identify summed variation in the TAK1-TAB complex directly associated with reduced MAPK14 transcription. Alongside this, we observe an increase in gene transcription associated with alternative inflammatory pathways (TXN and NLRP3) seen with NOD2-RIPK2 complex variation. Previous data have indicated a key role for TAK1 in MDP-stimulated NOD2 signaling, with absence of TAK1 completely admonishing downstream NFKB and MAPK signaling.35 Variation in the Toll-like receptor transduction complex, IRAK-TRAF6, was associated with a decrease in IKBKG transcription, a potent activator of NFKB signaling. This membrane receptor–triggered pathway acts in parallel to intracellular NOD2 signaling and also leads to NFKB activation in response to bacterial recognition and response. These data imply that variation across this related complex also impairs antimicrobial response. The activation of the NLRP3 inflammasome has been reported as the key inflammatory pathway leading to colitis in NOD2 knockout murine models.36 Here, we present data suggesting that transcripts in the pathway are upregulated in IBD patients with genomic variation in a number of NOD2 signaling genes. In contrast, we identify variation in CARD9 resulting in a decrease in PYCARD, a key signal transduction protein in the NLRP3 inflammasome, also leading to caspase-1 activation and IL-1B/IL-18 processing. It is possible that reduced NLRP3 activity is the key hypoimmune response for a subset of individuals, as observed in murine models, with concurrent increase in NFKB signaling in these patients.37 Previous data have indicated both a protective and an antagonistic role for NLRP3, with the potential for related molecular variation within related pathways leading to activation or suppression in some patients, with a subsequent hyper- or hypoinflammatory response leading to disease.38
We hypothesize that for many patients, IBD appears to arise due to multiple “hits” across complexes or genes contributing to impairment of inflammatory pathways. These patients then fail to clear bacteria, allowing invasion and chronic inflammation to develop due to perturbation of a number of innate immune responses. The precise immune impairment within an individual is likely to lead to commonality between subgroups of patients, with many Crohn’s disease patients having disease attributable to impaired NOD2 signaling.3 This provides an opportunity to target novel molecules with therapeutics, including the potential for NOD2 activators. Primarily these data provide the framework for molecular profiling of patients. Long-term follow-up data for these individuals will allow integration of treatment response and disease course and may allow prediction of outcomes at the point of diagnosis.
This study has several strengths. We provide a level of validation of genomic findings without the need for time-consuming functional assays, although caution should be exercised in interpretation of these results. Through use of targeted sequencing, we enable identification of lowly expressed transcripts, which are key in many of these analyses. The Supplementary Results indicate that NOD2 does not drive a specific gene expression signature across all genes targeted by the artificial intelligence panel, with a more select analysis revealing transcription differences. Additionally, while inflamed and noninflamed tissue demonstrated a degree of differential gene expression, the main analyses included patients with high variant burden but no ileal inflammation, demonstrating that genetic variation may be driving altered gene transcription independent of inflammation status. Additionally, we have previously determined that ileal transcriptomic profiles are not driven by inflammatory status.20
We acknowledge several limitations of this study. The curated list of NOD signaling genes could include either fewer or more genes. This gene list was specifically designed for use with the targeted autoimmune gene panel, which itself has the potential to limit wider findings related to WGCNA. We chose to not expand the list to retain statistical power to determine the impact of genomic variation on the most closely related transcripts. In addition, the inability of whole exome sequencing to capture promotor or regulatory variation and the dependence of GenePy on in silico deleteriousness metrics and inability to determine compound heterozygosity. Our group has previously studied cytokine induction in the context of NOD2 variation, focused on MDP-induced responses.3 Within this study, we did not study cytokine levels in relation to NOD signaling gene variation. Based on previous data, we hypothesize that these effector cytokines (IL-18, IL-1, tumor necrosis factor) are the common endpoint of numerous immune pathways, and the hypoimmune response seen in specific IBD-related pathways, such as NOD signaling, is overwhelmed by alternative activation leading to high levels of effector cytokines. Finally, while the gene transcript number is frequently related to immune function, there is also posttranslational regulation of signaling molecules, meaning that inference of functional consequences of altered transcription must be cautious.
Conclusions
These data demonstrate a pathway-wide genomic variation in NOD signaling genes, associated with reduced proinflammatory gene transcription within this pathway. Integration of genomic and transcriptomic data allows for statistical association of genomic variation with downstream transcription. We observe variation at each stage of the NOD2 signaling pathway resulting in broadly reduced NFKB signaling, with frequent upregulation of other inflammatory genes including NLRP3 and interferons. Expanding these analyses to additional pathways implicated in IBD may allow for precise “immunotyping” of patients, identifying defects in specific immune pathways and paving the way for personalized therapy.
Supplementary Data
Supplementary data is available at Inflammatory Bowel Diseases online.
Supplementary data 2 can be found at the following link: https://www.dropbox.com/s/mr7xwgow3q5macc/Supplementary%20data%202-%20Genomic%20and%20RNA%20integration%20GenePy%20scores%20QN%20transcripts.xlsx?dl=0
Author Contributions
J.J.A., R.M.B., and S.E. conceived the study. Patients were recruited by J.J.A. and R.H. Patient samples were acquired by J.J.A., R.H., T.A.C., A.B., N.A.A., and R.M.B. Samples were processed and sequenced by J.J.A., K.B., I.S.S., and G.C. Analyses were performed by J.J.A., K.B., J.D., I.S.S., and G.C. under the guidance of M.P., A.W., and S.E. B.V. performed histological analysis for all patients. J.J.A. wrote the manuscript with help from all authors. All authors approved the final manuscript prior to submission.
Supported By
J.J.A. was funded during this work by an Action Medical Research training fellowship. J.J.A. is currently funded by an NIHR clinical lectureship and an European Society for Paediatric Research postdoctoral grant.
Conflicts of Interest
The authors declare no conflicts of interest.
Data Availability
The RNA sequencing data underlying this article are available in Gene Expression Omnibus repository and can be accessed with accession number GSE153974. The exome sequencing data underlying this article cannot be shared publicly due to ethical considerations. The data will be shared on reasonable request to the corresponding author.
References
- 1. Graham DB, Xavier RJ.. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashton JJ, Mossotto E, Ennis S, Beattie RM.. Personalising medicine in inflammatory bowel disease-current and future perspectives. Transl Pediatr. 2019;8:56-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coelho T, Mossotto E, Gao Y, et al. . Immunological profiling of paediatric inflammatory bowel disease using unsupervised machine learning. J Pediatr Gastroenterol Nutr. 2020;70:833-840. [DOI] [PubMed] [Google Scholar]
- 4. Naranbhai V, Fairfax BP, Makino S, et al. . Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mossotto E, Ashton JJ, O’Gorman L, et al. . GenePy—a score for estimating gene pathogenicity in individuals using next-generation sequencing data. BMC Bioinformatics. 2019;20:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uhlig HH, Muise AM.. Clinical genomics in inflammatory bowel disease. Trends Genet. 2017;33:629-641. [DOI] [PubMed] [Google Scholar]
- 7. Andreoletti G, Shakhnovich V, Christenson K, et al. Exome analysis of rare and common variants within the NOD signaling pathway. Sci Rep. 2017;7:46454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caruso R, Warner N, Inohara N, Núñez G.. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crowley E, Warner N, Pan J, et al. Prevalence and clinical features of inflammatory bowel diseases associated with monogenic variants, identified by whole-exome sequencing in 1000 children at a single center. Gastroenterology. 2020;158:2208-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller SA, Dykes DD, Polesky HF.. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jun G, Flickinger M, Hetrick KN, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint. Posted online May 26, 2013. arXiv 1303.3997. doi:arxiv.org/tabs/1303.3997 [Google Scholar]
- 13. McKenna A, Hanna M, Banks E, et al. . The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DePristo MA, Banks E, Poplin R, et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stenson PD, Mort M, Ball EV, et al. . The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136:665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium. . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carson AR, Smith EN, Matsui H, et al. . Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinformatics. 2014;15:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shihab HA, Rogers MF, Gough J, et al. . An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karczewski KJ, Francioli LC, Tiao G, et al. . Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. Preprint. Posted online April 8, 2020. bioRxiv 531210. doi:10.1101/531210 [Google Scholar]
- 20. Ashton JJ, Boukas K, Davies J, et al. . Ileal transcriptomic analysis in paediatric Crohn’s disease reveals IL17- and NOD-signalling expression signatures in treatment-naïve patients and identifies epithelial cells driving differentially expressed genes. J Crohn’s Colitis. 2021;15:774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anders S, Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gene List HTG EdgeSeq Autoimmune Panel. 2020. [Google Scholar]
- 23. Ashton JJ, Mossotto E, Stafford IS, et al. . Genetic sequencing of pediatric patients identifies mutations in monogenic inflammatory bowel disease genes that translate to distinct clinical phenotypes. Clin Transl Gastroenterol. 2020;11:e00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langfelder P, Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Israël A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb Perspect Biol. 2010;2:a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malik A, Sharma D, Malireddi RKS, et al. . SYK-CARD9 signaling axis promotes gut fungi-mediated inflammasome activation to restrict colitis and colon cancer. Immunity. 2018;49:515-530.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bonen DK, Ogura Y, Nicolae DL, et al. . Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124:140-146. [DOI] [PubMed] [Google Scholar]
- 29. Heel DAV, Ghosh S, Butler M, et al. . Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn’s disease. Lancet. 2005;365:1794-1796. [DOI] [PubMed] [Google Scholar]
- 30. Parackova Z, Milota T, Vrabcova P, et al. . Novel XIAP mutation causing enhanced spontaneous apoptosis and disturbed NOD2 signalling in a patient with atypical adult-onset Crohn’s disease. Cell Death Dis. 2020;11:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bruyne MD, Hoste L, Bogaert DJ, et al. . A CARD9 founder mutation disrupts NF-κB signaling by inhibiting BCL10 and MALT1 recruitment and signalosome formation. Front Immunol. 2018;9:2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Warner N, Burberry A, Franchi L, et al. . A genome-wide siRNA screen reveals positive and negative regulators of the NOD2 and NF-κB signaling pathways. Sci Signal. 2013;6:rs3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sorbara MT, Ellison LK, Ramjeet M, et al. . The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity. 2013;39:858-873. [DOI] [PubMed] [Google Scholar]
- 34. Nayar S, Morrison JK, Giri M, et al. . A myeloid-stromal niche and gp130 rescue in NOD2-driven Crohn’s disease. Nature. 2021;593:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim JY, Omori E, Matsumoto K, et al. . TAK1 is a central mediator of NOD2 signaling in epidermal cells. J Biol Chem. 2008;283:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Umiker B, Lee HH, Cope J, et al. . The NLRP3 inflammasome mediates DSS-induced intestinal inflammation in Nod2 knockout mice. Innate Immun. 2019;25:132-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hirota SA, Ng J, Lueng A, et al. . NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17:1359-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhen Y, Zhang H.. NLRP3 inflammasome and inflammatory bowel disease. Front Immunol. 2019;10:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data underlying this article are available in Gene Expression Omnibus repository and can be accessed with accession number GSE153974. The exome sequencing data underlying this article cannot be shared publicly due to ethical considerations. The data will be shared on reasonable request to the corresponding author.