Abstract
Recent advances have revived interest in the concept of osteocyte perilacunar/canalicular remodeling (PLR) and have motivated efforts to identify the mechanisms regulating this process in bone in the context of normal physiology and pathological conditions. Here, we describe several methods that are evaluating morphological changes associated with PLR function of osteocytes.
Keywords: Osteocytes, perilacunar/canalicular remodeling, lacuno-canalicular network, osteocyte techniques
1. INTRODUCTION
Osteocytes are one of the most abundant cell types in bone and, although previously thought to be quiescent, are now known to be a metabolically active and dynamic cell type.(1,2). These morphologically distinct cells originate from terminally differentiated osteoblasts that are entrapped within the bone matrix(1,2). Osteocytes appear as stellate-shaped that reside in small cavities within the bone matrix termed lacunae and extend their long cytoplasmic projections through narrow canals called canaliculi(1–3). The intricate lacuno-canalicular network (LCN) of osteocytes is estimated to span 215m2 of total bone area, which is much larger than the combined surface areas of the lung, gastrointestinal tract, and skin(4). This unique LCN facilitates osteocyte communication with neighboring bone cells and vasculature and mediates flow of biochemical signals and nutrients among osteocytes(1,2,5–7). The mechanosensitive behavior of osteocytes relies on fluid flow through these channels(8,9). Thus, an impairment in the integrity of the LCN can affect the metabolic and signaling functions of osteocytes, as well as their mechanosensitivity, which can compromise bone health and increase fracture susceptibility(10,11).
Until recently, the multifaceted role of osteocytes in bone mechanotransduction, bone remodeling, and mineral homeostasis was largely thought to occur indirectly through osteocyte regulation of osteoclast and osteoblast function. Apart from these indirect mechanisms, osteocytes also maintain mineral homeostasis directly by remodeling the bone matrix surrounding their lacunar and canalicular spaces through the process known as perilacunar/canalicular remodeling (PLR)(12)(13). Originally termed as ‘osteocyte osteolysis’, the concept of PLR emerged from the hypothesis that the surface area of the LCN is much larger than the surface area available for remodeling by osteoblasts and osteoclasts(12,14,15). Thus, in response to a systemic imbalance in mineral metabolism, engaging osteocytes may be more efficient than recruiting osteoclasts and their progenitors since they can liberate small amounts of mineral across the enormous LCN area(16). With this theory, research during the 1900s demonstrated that osteocytic PLR is elicited during metabolic stress to release calcium from the bone surface to increase its bioavailability in order to restore systemic mineral homeostasis(14,17,18). Several models of induction or suppression of PLR in the context of mineral homeostasis have been previously discussed in other reviews(11,12,19,20).
The osteocyte resorption machinery is not as clearly defined as in osteoclasts. Except for some functional aspects, the structural features of osteocytes that mediate this process remain elusive. Current paradigms in the field suggests that acidifying factors (such as vacuolar H+ ATPase and carbonic anhydrases) and proteolytic enzymes (such as cathepsin K, tartrate resistant acid phosphatases and matrix metalloproteinases) are secreted within the perilacunar and pericanalicular spaces to resorb both the inorganic (crystalline hydroxyapatite) and the organic (mainly collagen) components of bone matrix(18,21–25) In healthy bone, the catabolic activity of osteocytes in resorbing LCN bone matrix is balanced by their anabolic activities. Following a phase of resorption, osteocytes replace the bone matrix by upregulating their expression of genes implicated in mineralization, namely Phex, Ank, Dmp1 and Phospho1(26).
The shift between the catabolic and anabolic phases of PLR appears to be highly dynamic and governed by external stimuli ranging from mechanical to metabolic fluctuations. In lactation, which is the most profound example of PLR in response to metabolic demand, osteocytes exhibit a dramatic increase in lacunar size and canalicular diameter to increase LCN resorptive capacity and meet calcium demand. Upon weaning the osteocyte LCN features return to normal(18,27). However, in less metabolically demanding situations, PLR appears to be crucial in actively ‘rejuvenating’ the LCN and mineralized collagen bone matrix, as well as in maintaining bone quality. Indeed, suppression of enzymes required for PLR can disrupt normal LCN and collagen organization and cause bone fragility, even without a loss of bone mass, implicating PLR as a critical cellular mechanism controlling bone quality(28,29). In this way, osteoblasts and osteoclasts remodel the external bone surfaces to control bone mass; whereas, osteocytes remodel the LCN surface to modulate bone quality to achieve mechanical homeostasis. Thus, it is not surprising that the key regulators of osteoblast and osteoclast function, including vitamin D, parathyroid hormone (PTH) and parathyroid hormone-related peptide (PTHrP), Sclerostin (SOST), and transforming growth factor (TGFβ), also regulate PLR(18,23,30–32). Given the importance of PLR in supporting mineral metabolism and mechanical homeostasis of bone, researchers need more tools to better understand the mechanisms controlling PLR and how it is disrupted in disease.
PLR can be assessed using morphological and molecular approaches. Since changes in the organization of the osteocyte LCN are frequently associated with altered PLR, morphological analysis of osteocyte dendrites or the LCN has been used as a surrogate to assess PLR. Earlier studies documenting PLR used histological stains such as the Periodic Acid-Schiff (PAS) stain to detect unmineralized bone matrix surrounding osteocyte lacunae(33). Similarly, metachromatic toluidine blue that binds to acidified surface was used to demonstrate resorption around the osteocyte lacunae (12,34,35). Histomorphometric analyses with tetracycline labeled bone has been used to demonstrate bone formation around osteocyte lacunae(36). Using back-scatter SEM, changes in LCN features with PLR can be visualized and quantified(27). Morphological changes associated with PLR have been also captured in 3-dimensions using confocal visualization of fluorescently labeled osteocytes or the LCN and using synchrotron radiation tomography (SRμCT) of the LCN(11,20,37). These techniques have elegantly associated changes in osteocyte lacunar sizes, canalicular diameter, and mineralization around these features with altered PLR in normal physiology and in pathological conditions.
Molecular analysis of PLR augments what can be learned from these morphological assessments. Several enzymes, already implicated in bone remodeling by osteoclasts and osteoblasts, are expressed by osteocytes at the time and site of PLR. Studies in genetically modified mouse models have shown a functional role for several of these in PLR, including matrix metalloproteinases (Mmp2, Mmp13, Mmp14), Cathepsin K (Ctsk), carbonic anhydrases (Ca2), vacuolar H+ ATPases (Atp6v1g1, Atp6v0d2, and Atp6v0b), and tartrate resistant acid phosphatases (Acp5) in PLR(18,24,32,38). Analysis of gene expression using qPCR and RNA-Seq shows that many of these genes are regulated in a coordinated manner by agents that modulate PLR, such as PTH, PTHrP, TGFβ, and glucocorticoids (18,23,32,38). Therefore, analysis of changes in mRNA and protein expression of these PLR enzymes provides a good surrogate to quantify dynamic changes in PLR at the molecular level. Use of spatial techniques such as in situ hybridization and immunohistochemistry are needed to complement biochemical analysis of gene or protein expression, since these enzymes are expressed by and regulated in osteoblasts and osteoclasts, as well as in osteocytes.
In this book chapter, we summarize a streamlined approach to assess PLR phenotypes in the bone tissue. While these methods have been most widely applied to rodent models, they also work in other species, including human bone. These approaches entail the detection of morphological changes in osteocytes and the molecular signature associated with PLR. In this chapter we have included the protocols for silver nitrate and Dil stains that were optimized in our labs and been useful to assess morphological changes in osteocytes linked to PLR(20,32,38–40). To assess the molecular changes associated with PLR, we provide protocols for detecting the expression of genes and proteins involved in resorption and formation phases of remodeling using RT-qPCR and immunohistochemistry. Through this chapter we strive to provide a simple, accessible, and robust way to assess a phenotype that might be associated with PLR in your in vivo models. It is important to note that combining these approaches with more sophisticated approaches of synchrotron microCT, back-scatter SEM that can evaluate the mineral deposited or resorbed around the osteocyte lacuna and canaliculi and changes in lacunar volumes will add rigor and power to the assessment of PLR phenotype in bone tissue(41–44). Our recent review more comprehensively discusses the advantages and limitations of these and other approaches currently in use to evaluate PLR(20).
2. MATERIALS
2.1. Morphological Assessment of Osteocytic Changes Associated with PLR Via Silver Nitrate Stain
10% Neutral buffered formalin
1X Phosphate Buffered Saline (PBS)
100% Xylene or Citrisolv
100% ethanol (EtOH)
95% ethanol
70% ethanol
0.5 M Ethylenediaminetetraacetic Acid (EDTA)
Silver Nitrate
Sodium Thiosulfate
Glacial Acetic Acid
Cresyl violet
Gelatin type B, bloom 100 (bloom is the strength of gelatin)
Bone sections
Disposable coplin jars. Holds 20–25ml volume and 4 slides.
Gelatin-formic acid solution: Dissolve 0.2g gelatin type B, bloom 100 in 10ml of warm water (50–55°C). Vortex to dissolve and cool to room temperature (RT). After cooling, add 125μl of 90% formic acid.
50% Silver Nitrate: Dissolve 7.5g of silver nitrate (Spectrum Chemical) in 15ml of water and mix by shaking gently.
Silver Nitrate Working Solution: Mix 50% silver nitrate and gelatin-formic acid solution in a 2:1 ratio. Keep solution in the dark.
5% Sodium thiosulfate: Dissolve 12.5g sodium thiosulfate in 250ml water.
Cresyl violet solution: Dissolve 100.00mg of cresyl violet acetate in 100ml of distilled water.
Cresyl violet Working Solution: Add 300ul of glacial acetic acid with 100ml of cresyl violet solution before use. Adjust pH to ~3.5. Mix well and keep solution in dark.
2.2. Morphological Assessment of Osteocytic Changes Associated with PLR via Dil/Phalloidin Stain
0.5 M Ethylenediaminetetraacetic Acid (EDTA)
Sucrose (10%, 15%, 20% and 30%) solutions prepared in 1X PBS
Normal Donkey Serum (NDS)
Bovine Serum Albumin (BSA)
Dil Stain: 1,1’-Dioctadecyl-3,3,3’,3’-Tetramethylindocarbocyanine Perchlorate (Invitrogen)
DMSO
Alexa Fluor 488 Phalloidin (Invitrogen)
4′−6-Diamidino-2-phenylindole (DAPI)
2,2’-Thiodiethanol (TDE)
Frozen optimum cutting temperature (OCT) embedded sections
48-well plate
Toluene/ formaldehyde-free nail polish
4% PFA: Add 4g of paraformaldehyde (PFA) to solution containing 50 ml of water and 1ml of 1 M NaOH. Dissolve PFA by continuous stirring on a heating block set to 60°C ensuring the solution does not boil. Add 10 mL of 10X PBS and allow the mixture to cool to RT. Adjust the pH to 7.4 with 1 M HCl and bring the volume to 100ml. Store 4% PFA aliquots at −20°C for several months and avoid using solution that is repeatedly frozen and thawed.
Blocking buffer (PBS/1%NDS/1%BSA): Dissolve 2.5μl Bovine Serum Albumin (BSA) and 2.5μl Normal Donkey Serum (NDS) in 200μl PBS.
Dil solution: Dissolve 100μM of Dil stain in 50% DMSO: 50% PBS.
Phalloidin solution: Dissolve 5μl of Alexa Fluor 488 Phalloidin stock solution in blocking buffer.
DAPI solution: Make 4μg/ml DAPI in 1X PBS.
Tissue clearing and mounting media: Prepare 10%, 25%, 50%, 95%, & 97% TDE in 1X PBS using the TDE stock.
2.3. Molecular Assessment of PLR-Associated Genes Via qRT-PCR
Centrifuge Multifuge 3S-R.
Disposable polypropylene round-bottom tubes (15 ml).
Polytron Homogenizer (ThermoSavant or equivalent).
Bone spinner tubes: Make a small hole at the bottom of a 0.2ml and 0.5ml Eppendorf tubes. Place the 0.2 ml tube nested in the 0.5ml tube and insert this 2-tube assembly nested in a 1.5ml Eppendorf tube to make a bone spinner tube.
Polypropylene Eppendorf tubes (1.5 ml).
Petri dish (6 and 10 cm diameter).
Sectioning materials (scissors, forceps, curved forceps).
miRNeasy extraction kit (Qiagen) or equivalent.
iScript™ RT kit (BioRad) or equivalent.
iQ™ SYBR Green Super Mix (BioRad) or equivalent.
Primers for qPCR
MyIQ Single-Color-Real-Time detection system (BioRad) or equivalent.
2.4. Molecular Assessment of Proteolytic Enzymes Associated with PLR Via Immunohistochemistry
Innovex Animal IHC kit
Formalin-fixed, paraffin-embedded (FFPE) sections
Uni-trieve (Innovex)
Primary Antibodies: MMP13 antibody (abcam ab39012); MMP14 antibody (abcam ab38971); Ctsk antibody (abcam ab19027)
2.5. Molecular Assessment of PLR-Associated Enzyme Function via Tartrate Resistant Acid Phosphatase (TRAP) Staining
Acid phosphatase leukocyte kit (Sigma)
Fast Red Violet (Sigma) (CAS# 32348–81-5)
Fast Green (Sigma)(CAS# 2353–45-9)
37% formaldehyde
Formalin-fixed, paraffin-embedded (FFPE) sections
Fast Red Violet Solution: First dissolve 7mg of Fast Red Violet salt in 1ml of 1XPBS.
Diazonium salt solution: Gently mix Fast Red Violet solution and with Sodium Nitrite Solution in a 1:1 ratio by tapping or inverting (depending on the volume of the solution). Allow it to stand at RT for 2minutes.
0.02% Fast Green: Dissolve 0.05g of Fast green dye in 250ml of distilled water.
Fixative Solution: Mix 25ml citrate solution, 65ml acetone, and 8ml 37% formaldehyde. Store solution at 4°C for up to 2 months.
3. METHODS
3.1. Morphological Assessment of Osteocytic Changes Associated with PLR via Silver Nitrate Stain
Silver nitrate staining has been successfully used to visualize the integrity of the osteocyte network, which can be disrupted in conditions in which PLR is suppressed(28,45,46). It is unclear precisely what is being stained by silver nitrate, however, there is some indication that it can bind osteopontin(47).
Dissect the bones and carefully remove the muscle tissue.
Fix the cleaned bones immediately in 10% neutral buffered formalin (NBF) for ~24–48 hours.
Following fixation, rinse the tissue samples in 1X phosphate buffered saline (PBS) solution for 10 minutes and at least three times to remove residual NBF.
Decalcify the fixed bones by keeping them in 0.5M EDTA solution for 2–4 weeks shaking at 60 revolutions/ min at RT. Change EDTA solution every 3–4 days. (see Note 1)
Following decalcification, bones will be rinsed in PBS (three 60 minute washes at 4° C) to remove residual EDTA.
Proceed with paraffin embedding according to standard protocols(48). (see Notes 2 and 3)
Utilizing a microtome, generate 6–8 μm thick sections according to standard protocols or the Demineralized Murine Skeletal Histology chapter of this reference(48).
Deparaffinize and rehydrate tissue samples by immersing the slides in 100% xylene or citrisolv and EtOH gradient as described below. (For frozen sections, see Note 2)
Immerse slides in Citrisolv three times for 5 minutes each.
Immerse slides in 100% EtOH for 5minutes.
Immerse slides in 95% EtOH for 5minutes.
Immerse slides in 70% EtOH for 5minutes.
Immerse slides in distilled water for 5minutes.
Immerse slides in the freshly made silver nitrate working solution for 60 minutes and store the slides in the dark during this incubation. (see Notes 4 and 5)
After silver nitrate incubation, wash the slides three times with distilled water for 5 minutes each.
Incubate slides in 5% sodium thiosulfate for 10 minutes and perform three washes with distilled water for 5 minutes each at RT. (see Note 6)
Following silver nitrate staining, counterstain the slides with cresyl violet working solution for 4–30 minutes. (see Notes 7 and 8)
Rinse the slides in distilled water and proceed to dehydrate the sections by incubating the slides in the EtOH gradient (70%, 95%, 100%, 100%) for 5–10 dips each.
Immerse slides in 100% Isopropyl alcohol for 5–10 dips.
Clear sections by immersing in Citrisolv solution three times each for 5 minutes.
Using a toluene/xylene-based permanent mounting-medium add a coverslip onto the slide and prepare it for microscopy.
3.2. Morphological Assessment of Osteocytic Changes Associated with PLR via Dil/Phalloidin Stain
This method is based on and largely identical to protocols published by Kamel El-Sayed et al, 2015 (40).
Dissect and clean bones of surrounding tissue, remove periosteum from surface of bones.
Fix bone samples by incubating in 4% PFA for 24–48 hours.
Rinse the fixed samples in 1X PBS to remove residual PFA and decalcify the bones in 0.5M EDTA for 2–4 weeks on shaker at 60 revolutions/ minute at RT. Change EDTA solution every 3–4 days.
Following decalcification, bones will be rinsed in PBS (three 60 minute washes at 4° C) to remove residual EDTA.
Cut off the ends of the bone and gently remove the bone marrow by flushing with PBS using a syringe and needle. (see Note 9)
Wash samples in 1XPBS and subject them to a sucrose gradient (10%, 15%, 20%) described in steps 7–9 below.
Immerse samples in 15% sucrose solution for 15 minutes at RT.
Immerse samples in 20% sucrose solution for 15 minutes at RT.
Immerse samples in 30% sucrose solution overnight at 4°C
Following sucrose gradient, gently dab dry samples with tissue, ensuring no liquid remains in the marrow cavity, and proceed to frozen embedding.
Embed samples in OCT media as per standard protocol or as described in the Demineralized Murine Skeletal Histology section of this reference. Cut 40–100 μm thick cryosections of the embedded bones either onto microscope slides or wells of a 48 well plate containing ~250 μl RT 1X PBS. (see Note 10)
Rinse sections three times with 1X PBS for 5 minutes each until all OCT is removed. (see Note 11)
Incubate the sections with the blocking buffer overnight at 4°C (~50 μl per well) in parafilm sealed plate. (for slide mounting see Notes 12 and 13)
Rinse the slides in 1X PBS three times each for 5 minutes.
Incubate the slides with the Dil solution in the dark at 4°C for 1 week. This dye stains hydrophobic structures including cell membranes, which is why long staining time is required for diffusion throughout the LCN. (see Note 14)
Rinse the slides in 1X PBS three times each for 5 minutes.
Incubate the slides in Phalloidin solution overnight at 4°C in parafilm sealed well plate. This dye stains the actin cytoskeleton. (see Note 15)
Rinse the slides in 1X PBS three times each for 5 minutes.
Incubate the slides in DAPI solution at RT for 30 minutes to stain the osteocyte nuclei.
Rinse the slides in 1X PBS five times (~250 μl per well).
For mounting the sections, conduct sequential incubations in TDE gradient solutions namely, 10% TDE, 25% TDE and 50% TDE in PBS. Each of these incubations are 2 hours long and performed at RT. Finally, incubate slides overnight at 4°C in 95% TDE solution in parafilm sealed well plate. (see Note 16)
Place a drop (~10–20 μl) of 97% TDE onto slide (if slide mounted, wick away previous solution with kim wipe and place 97% TDE directly on tissue) and gently place section directly into drop with forceps, curled sections should unroll in solution. Place cover slip and seal edges with toluene/formaldehyde free nail polish.
Image immediately or store slides in −20°C for a few weeks. (see Note 17)
3.3. Molecular Assessment of PLR-Associated Genes Via qRT-PCR
Expression of genes typically associated with the resorptive activity of osteocytes can be monitored to assess induction of PLR. RNA enriched for osteocytic populations can be isolated and quantified as described below.
Dissect the bones and remove the periosteal layer and muscle surrounding the bones as quickly as possible.
Dissect off the epiphyseal ends of the bones and remove the bone marrow by centrifuging the bones in special bone spinner tubes at 9000 rpm for 15 seconds. (see Note 18)
Flash freeze the bones devoid of marrow, periosteum, and epiphyses in liquid nitrogen for 30 seconds. (see Note 19 and 20)
Following flash freezing, immediately immerse the samples in 15 ml polypropylene tubes containing 1.5 ml QIAzol solution for every 20 mg of bone tissue. (see Note 21)
Dissociate the bone tissue in QIAzol using a Polytron tissue homogenizer at full speed in 10 second bursts 3–4 times, icing the samples between bursts for 5–10 seconds. Ensure that the samples are on ice during homogenization step. Homogenized samples can be stored at −80 °C in QIAzol prior to RNA isolation.
For total RNA extraction from homogenized lysates, the miRNeasy Mini Kit (Qiagen), which combines a phenol/guanidine-based lysis and a silica-based on-column purification method for efficient and high-quality RNA extraction, may be used(49). This kit allows collection of both mRNA and miRNA. If only the mRNA is useful for your experiments, the RNeasy Kit (Qiagen) can be used with the same protocol.
Add 300 μl of chloroform (0.2 times the volume of QIAzol) to the QIAzol cell lysate and mix them thoroughly for 15 seconds.
Incubate at room temperature for 2–3 minutes until separation begins to occur.
Centrifuge the samples at 12,000 rcf for 15 minutes at 4°C.
Transfer the upper aqueous phase to new tube being careful not to disturb or contaminate with the lower phase and follow the standard manufacturer’s protocol for isolating RNA. Conduct on-column DNase digestion as recommended in the miRNeasy mini kit protocol.
Concentration of the extracted RNA is determined using a NanoDrop™ 1000 spectrophotometer (Nanodrop Technologies). RNA with a 260/280 ratio between 1.8–2.0 and a 260/230 ratio of close to 2.0 is considered of good quality. (see Note 22) RNA yields are typically in the range of 7–10 μg from the whole bone (devoid of marrow).
Conduct reverse transcription using the iScript cDNA synthesis kit (VENDOR) as per the manufacturer’s instructions. 1 μg of total RNA is reverse transcribed into cDNA using a final volume of 30 μl reaction mixture.
Prepare reaction master mix as per Table 1 and add RNA samples separately for each reaction.
Mix the RNA and iScript master mix and incubate samples in the thermocycler following the temperature gradients described in Table 2. Store cDNA at −20°C until use for quantitative RT-qPCR. (see Note 23)
Carry out Quantitative RT-qPCR using the iQ-SYBR Green Supermix kit (Bio-Rad) on a MyIQ Single-Color-Real-Time detection system (BioRad). Typically, 50ng of cDNA used in a qPCR reaction mix will obtain Ct readings in the range of 25–30 for the PLR genes in Table 3. While the primers are specific for mouse bones, the protocol for RNA extraction and RT-qPCR is applicable to bone specimens from other species.
Prepare the primer, cDNA and iQ-SYBR green master mix as outlined in Table 4. (see Note 24)
Set up the experiment and program the thermocycler to the quantitative RT-qPCR program outlined in Table 5.
Normalize data to 18s RNA as the internal standard and to the corresponding control group using the 2−ΔΔCT method. For each run, the melting curve and saturation of amplification cycles are controlled by the use of MyIQ software (BioRad). (see Note 25)
Table 1:
Reverse transcription master mix
| Reagents | 1 reaction |
|---|---|
| 5X iScript RT Supermix | 4 μl |
| iScript Reverse Transcriptase | 1 μl |
| RNA template (1 μg) | variable |
| Nuclease free water | variable |
| Total volume | 20 μl |
Table 2:
Thermocycler reverse transcription temperature gradient guide
| Temperature | Time |
|---|---|
| 25°C | 5 minutes |
| 46°C | 20 minutes |
| 95°C | 1 minutes |
| 25°C | Hold |
Table 3:
PLR gene primers for RT-qPCR
| Primers for genes | Sequence |
|---|---|
| 18s RNA (FP) 18s RNA (RP) |
5’-CGAACGTCTGCCCTATCAAC-3’ 5’-GGCCTCGAAAGAGTCCTGTA-3’ |
| Mmp13 (FP) Mmp13 (RP) |
5’-CGGGAATCCTGAAGAAGTCTACA-3’ 5’- CTAAGCCAAAGAAAGATTGCATTTC-3’ |
| Mmp14 (FP) Mmp14 (RP) |
5’-AGGAGACGGAGGTGATCATCATTG-3’ 5’-GTCCCATGGCGTCTGAAGA-3’ |
| Mmp2 (FP) Mmp2 (RP) |
5’-AACGGTCGGGAATACAGCAG-3’ 5’-GTAAACAAGGCTTCATGGGG-3’ |
| Ctsk (FP) Ctsk (RP) |
5’- GAGGGCCAACTCAAGAAGAA-3’ 5’- GCCGTGGCGTTATACATACA-3’ |
| Acp5 (FP) Acp5 (RP) |
5’-CGTCTCTGCACAGATTGCAT-3’ 5’-AAGCGCAAACGGTAGTAAGG-3’ |
| Atp6v0d2 (FP) Atp6v0d2 (RP) |
5’-TCTTGAGTTTGAGGCCGACAG-3’ 5’-GCAACCCCTCTGGATAGAGC-3’ |
| Atp6v1g1 (FP) Atp6v1g1 (RP) |
5’-CCGTTCTCTCAGCCCAAAGT-3’ 5’-CTCCGGTTCTTTCGCTTGC-3’ |
Table 4:
Thermocycler RT-qPCR temperature cycle guide
| Temperature | Time (mins/secs) | No. of cycles |
|---|---|---|
| 95°C | 2–3 min. | 1 |
|
| ||
| 95°C | 10–15 secs | 40 |
| 60°C | 30 secs | |
|
| ||
| 55–95°C (in 0.5°C increments) | 10–30 secs | 1 |
3.4. Molecular Assessment of Proteolytic Enzymes Associated with PLR Via Immunohistochemistry
Dissect the bones and carefully scrape off the surrounding periosteum and muscle tissue.
Fix the cleaned bones immediately in 10% neutral buffered formalin (NBF) for ~24–48 hours.
Following fixation, rinse the tissue samples in the 1X PBS solution to remove residual PFA.
Decalcify the fixed bones using 0.5 EDTA for 2–4 weeks. Following decalcification, rinse bones in PBS (three times each for 60 minutes at 4° C) and then subjected to paraffin embedding.
Utilizing a microtome, generate 6–8 μm thick sections from paraffin blocks according to standard protocols or the Demineralized Murine Skeletal Histology chapter of this book(48).
Deparaffinize and rehydrate tissue sections by immersing the slides in 100% xylene or citrisolv and EtOH gradient as described below.
Immerse slides in Citrisolv three times for 5 minutes each.
Immerse slides in 100% EtOH for 5 minutes
Immerse slides in 95% EtOH for 5 minutes
Immerse slides in 70% EtOH for 5 minutes
Immerse slides in distilled water for 5 minutes.
Incubate slides in Uni-trieve for 30 minutes at 60°C. (see Note 26)
Rinse slides in distilled water two times for 3 minutes each.
Incubate slides with Fc-block (329ANK) for 45 minutes at RT. (see Note 27)
Rinse slides in distilled water two times for 3 minutes each.
Incubate slides with Background Buster for 45 minutes at RT.
Rinse slides in distilled water two times quickly.
Incubate slides with primary antibody in PBS for 1hr at RT or overnight at 4°C. (see Note 28) Recommended dilutions of primary antibodies: MMP13– 1:100 diluted in 1X PBS; MMP14– 1:100 diluted in 1X PBS; CTSK- 1:75 diluted in 1X PBS
The next day, rinse slides in 1X PBS three times for 3 minutes each.
Incubate slides with Secondary Linking Ab (329ANK) for 10 minutes at RT.
Rinse slides in PBS three times for 3 minutes each.
Incubate slides with Peroxidase (HRP) enzyme for 10 minutes at RT.
Rinse slides in 1X PBS three times for 3 minutes each.
Incubate slides with DAB working solution for 5 minutes at RT. (see Note 29)
Rinse slides in 1X PBS thrice for 3 minutes each.
Mount slides with an Innovex advantage permanent mounting media (aqueous-based mounting-medium).
3.5. Molecular Assessment of PLR-Associated Enzyme Function via TRAP Staining
Deparaffinize and rehydrate tissue samples by immersing the slides in 100% xylene or citrisolv and EtOH gradient as described below.
Immerse slides in Citrisolv three times for 5 minutes each.
Immerse slides in 100% EtOH for 5 minutes.
Immerse slides in 95% EtOH for 5 minutes.
Immerse slides in 70% EtOH for 5 minutes.
Immerse slides in distilled water for 5 minutes.
Incubate slides in Fixative Solution for 30 seconds at RT.
Rinse slides in distilled water for 5 minutes.
In a coplin jar, make TRAP staining solution by mixing Napthol AS-BI Phosphate solution (Sigma), Tartrate solution (Sigma), Diazonium salt solution, Acetate solution (Sigma), and deionized water in a ratio of 1:2:2:4:90 respectively. (see Note 30–32)
Immediately proceed to incubating slides in TRAP working solution for 1hr at 37°C in the dark. (see Note 33)
Rinse slides in distilled water for 3 minutes.
Counterstain by incubating slides in 0.02% Fast Green for 1 min.
Rinse slides in distilled water with 2–3 quick dips.
Dry slides before mounting with coverslip and a toluene/xylene-based mounting-medium. (see Note 34)
Importantly, TRAP is not normally found in high amounts in most osteocytes and that prior work has shown that induced expression of TRAP in osteocytes is associated with perilacunar remodeling(18).
Figure 1:
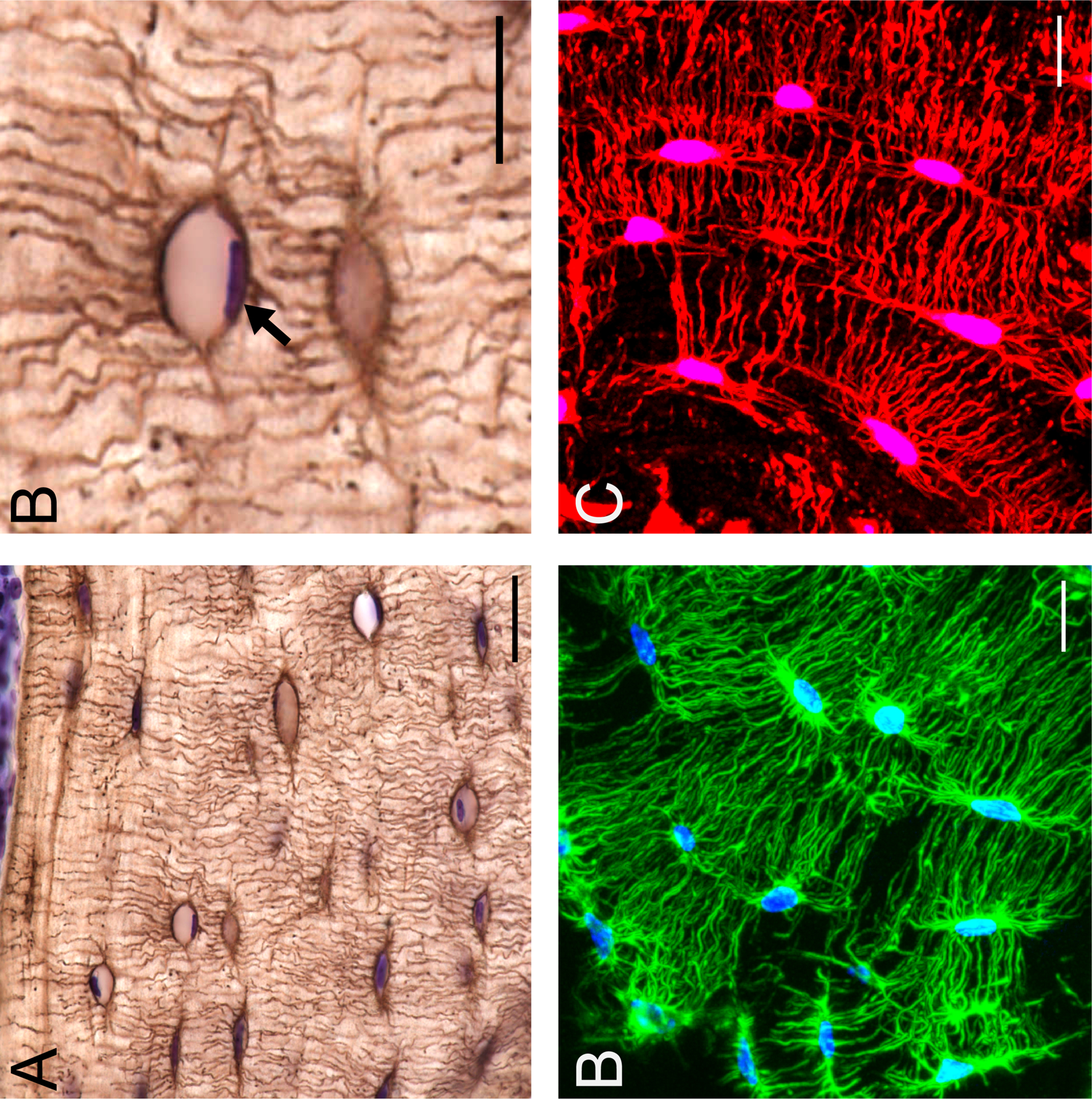
Stains highlighting morphological changes in osteocytes associated with PLR. Silver nitrate stain on C57/BL6 mouse bone demonstrates the osteocyte cell network in the paraffin embedded cortical bone (A, scale bar = 20 μm) with cresyl violet counter stain for nuclei (purple). Enlarged region of section from A (B, scale bar = 10 μm) depicting an osteocyte containing a positively stained cresyl violet nuceli (black arrow). Fluorescent phalloidin (C, scale bar = 10 μm) staining of f-actin (green) and DAPI (blue), can be used to visualize the osteocyte cell network in 3D with frozen,OCT sectioning and confocal microscopy. Dil stain (red) (D, scale bar = 10 μm) intercalates into the hydrophobic cell membrane, further illuminating osteocyte cell bodies and dendritic projections in 3D.
Figure 2:
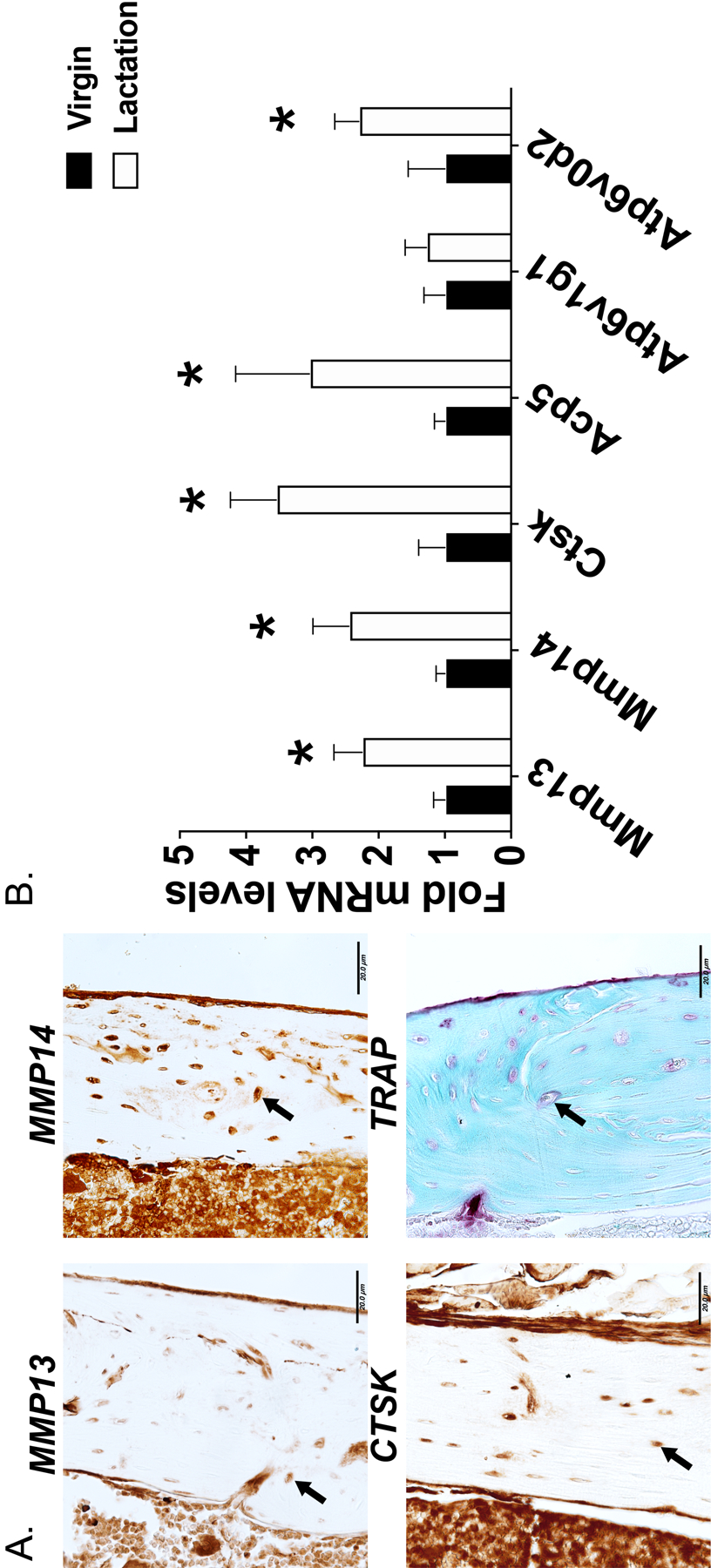
Molecular assessment of PLR. Immunohistochemistry for PLR-associated proteins MMP13, MMP14, CTSK and TRAP on femurs isolated from 8-week old C57BL6 mice. 7 μm thin paraffin embedded bone sections are is shown (A). Arrows indicate the osteocytes positive for the respective stain. Scale bar is 20 μm. RT-qPCR highlights the lactation mediated induction in PLR-related genes (B). All genes were normalized to 18s RNA as an internal control gene and relative changes in gene expression in the lactating C57/BL6 mice were compared against virgin controls. N=10 mice in each group and *p>0.05, different from the virgin mice, as calculated from the Student’s t-test.
ACKNOWLEDGEMENTS:
The authors gratefully acknowledge funding from NIH R01DE019284, P30AR061312, R21AR070403; NSF 1636331 and Center for Disruptive Musculoskeletal Innovation, DOD OR170044, and the Read Research Foundation (TA), NIA-1F31AG063402-01A1 (CS), and NIH P01AG039355 and R21AR054449 (SD). We acknowledge use of the UMKC Confocal Microscopy Core supported by NIH grants S10RR027668 and S10OD021665.
4. NOTES
Bones must be decalcified prior to being paraffin embedded.
This protocol will also work with mineralized frozen embedded sections. If starting with mineralized frozen sections, replace steps 8–12 in 3.1 methods section with a couple of rinses of distilled water to remove OCT. Decalcify sections on the slide prior to staining by immersing in 0.5M EDTA for an hour. You may also decalcify bones prior to frozen embedding following standard protocol (3.1. section step 4 in methods).
This protocol is for paraffin or frozen OCT embedded bones. We have had mixed success with silver nitrate staining of plastic embedded non-decalcified sections and have learned that other laboratories have adapted this protocol for that purpose.
Gelatin-formic acid solution can be prepared ahead of time and stored at RT before incorporation into the working silver nitrate solution.
Silver nitrate mixed with water is highly unstable and should be made fresh and right before use. Silver nitrate is a strong oxidizer and stains/blackens everything, we suggest use of disposable coplin jars to stain our slides. Use care when handling silver solutions. Keep in dark.
Sodium thiosulfate, that serves as a developing solution, is made fresh and used immediately.
Cresyl violet working solution must be at a pH of ~3.5. The solution can be filtered to remove precipitated particles if necessary. This counterstain will also stain cartilage and un-mineralized bone. Adjust staining time accordingly to the species and type of tissue as too long of incubation can remove silver nitrate stain.
Cresyl violet counterstain is optional. If skipping the counterstain, dehydrate sections in the EtOH gradient (70%, 95%, 100%, 100%) and 100% Isopropyl alcohol for 5 minutes each.
The bone marrow will preferentially uptake dye, which will make visualizing osteocytes difficult.
To flatten and adjust sections onto microscope slides, use cryostat chilled parafilm and Q-tips. For longer term storage of thicker sections in 48 well plates, replace 1X PBS with 0.5% Sodium Azide/PBS, wrap with parafilm and store at 4°C.
To aspirate liquid from wells, tilt the well chamber so that the tissue sections lightly adhere to the well then slowly remove as far from the tissue as possible. Gently add liquid on the side of the well and ensure sections do not adhere to the wells.
For sections mounted on slides, use a humid chamber for all the incubation steps. Proceed with all steps after Day 1 in a light proof container or cover with foil. Use only enough liquid to cover tissue section on slide (no more than 50 μl). Use of a barrier pen is suggested.
Blocking buffer can also be with just 1% NDS in 1X PBS.
DiI stain is hydrophobic and may aggregate overtime from the 50/50 DMSO/PBS solution. For slide mounted samples, ensure slides are dry before staining or excess liquid will crash the dye out of solution. Check slides every other day to ensure DiI in droplet hasn’t aggregated while sitting in the humid chamber. After 1 week of staining, place wells or slides in vacuum chamber for 2 hours if dye penetration is incomplete as seen under scope.
Phalloidin stain is very sensitive to ethanol and other polar solvents. Ensure that the tissues are not exposed to ethanol or polar solvents during dissection, fixation and staining and that no tools or machines used around this dye or sample with this dye have any ethanol on them.
2,2’-Thiodiethanol (TDE) used for tissue clearing prior to Dil stain is a restricted compound in the United States and may require additional steps to procure.
Due to possibility of photobleaching and the AlexaFluor signal fading, image as soon as possible. TDE mounting media will begin to degrade AlexaFluor-phalloidin signal in 1–2 days if left at room temperature. If imaging must be delayed, store slides at −20°C. For imaging, remove slides from −20°C and thaw before imaging and work quickly. Minimize freeze-thaw cycles if possible.
Bone marrow should spin out of the bone into the bottom of the 1.5ml Eppendorf tube with bone remaining in the 0.2ml tube. This method works well on appendicular skeleton bones. Make sure the hole at the bottom of 0.2ml tubes is not too large to prevent bones from slipping through.
Harvested and cleaned bones devoid of marrow can be preserved in RNAlater (Invitrogen) if immediate homogenization is not possible. Incubate a single bone in 1 ml of RNAlater for 24 hours at 4°C ensuring no single dimension of the bone is greater than 1 cm, portion bones accordingly. For long-term storage, remove bones from RNAlater solution using RNase AWAY treated forceps, dab with Kimwipes to remove traces of the solution, and moved to −80°C or liquid nitrogen bank. While the manufacturer recommends that tissues can be preserved in RNAlater for months prior to RNA extraction, we have stored bones in RNAlater for up to 1 week.
When ready to extract RNA, thaw the preserved bones that were flash frozen in liquid nitrogen prior to homogenizing using Qiazol as described in the RNA extraction protocol.
To minimize RNA degradation, bones are homogenized in cold Qiazol kept at 4°C (on ice) prior to harvesting bones.
RNA that will be utilized for RNA-Seq should be assessed on a Bioanalyzer (Agilent RNA 6000 Nano Kit) for RNA degradation. Using our RNA extraction protocol, we are able to isolate intact RNA with RIN> 7.0.
Although we have described the protocol for using the iScript RT kit for cDNA synthesis, in the past we have conducted RT using SuperScript II reverse transcriptase.
To minimize the standard deviation among the technical replicates, we recommend preparing the replicates as one sample in the Eppendorf tubes and then distributing the samples (containing cDNA and qPCR master mix) among two wells on the 96-well plate for qPCR. Variation of >1Ct between technical replicates may indicate a problem with RNA quality.
Typically, we perform RT-qPCR with at- least two technical replicates per sample and biological replicates ranging from 4–10 within an experiment. The number of biological replicates is determined by considering the effect size and performing power analysis.
Decalcified bone paraffin sections should be on charged slides to ensure slides adhere during the antigen retrieval step.
All incubations should be done in humid chambers. We recommend using the commercially available histochemistry staining trays with black lids (Simport stain tray™). These trays are designed with a reservoir to hold water beneath the slides.
Primary antibody concentration should be optimized for various samples.
DAB working solution must be made fresh every time, right before adding it onto the sections.
The diazonium salt solution must be made fresh and not incubated longer than 5 minutes.
The Fast green solution can re-used for staining at least five times before its final discard.
Bring the fixative solution to RT before use and discard if evaporation is noted.
It is possible that the sections might stain positive for TRAP within 30 minutes of exposure. So, we recommend monitoring the stain development within the 1 hour. At times incubations longer than an hour might be required. We have conducted TRAP staining for 2 hours for better development of stain color.
Osteocyte TRAP positive cells are visible on sagittal sections of long bones only. For axial sections of long bones, we recommend performing IHC using Cathepsin K antibody.
REFERENCES:
- 1.Bonewald LF. The amazing osteocyte. J. Bone Miner. Res 2011;26(2):229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell … and more. Endocr. Rev 2013;34(5):658–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You LD, Weinbaum S, Cowin SC, Schaffler MB. Ultrastructure of the osteocyte process and its pericellular matrix. Anat. Rec. - Part A Discov. Mol. Cell. Evol. Biol 2004;278(2):505–13. [DOI] [PubMed] [Google Scholar]
- 4.Buenzli PR, Sims NA. Quantifying the osteocyte network in the human skeleton. Bone [Internet]. Elsevier Inc.; 2015;75:144–50. [DOI] [PubMed] [Google Scholar]
- 5.Schaffler MB, Henderson SC, Wang Y, Wang L, Weinbaum S, Majeska RJ, Han Y. In situ measurement of solute transport in the bone lacunar-canalicular system. Proc. Natl. Acad. Sci 2005;102(33):11911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein-Nulend J, Bakker AD, Bacabac RG, Vatsa A, Weinbaum S. Mechanosensation and transduction in osteocytes. Bone 2013;54(2):182–90. [DOI] [PubMed] [Google Scholar]
- 7.Scheiner S, Théoval A, Pivonka P, Smith DW, Bonewald LF. Investigation of nutrient transport mechanisms in the lacunae-canaliculi system. IOP Conf. Ser. Mater. Sci. Eng 2014;10(1). [Google Scholar]
- 8.Fritton SP, Weinbaum S. Fluid and Solute Transport in Bone: Flow-Induced Mechanotransduction. Annu. Rev. Fluid Mech 2009;41:347–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L Solute Transport in the Bone Lacunar-Canalicular System (LCS). Curr. Osteoporos. Rep 2018;16(1):32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Hove RP, Nolte PA, Vatsa A, Semeins CM, Salmon PL, Smit TH, Klein-Nulend J. Osteocyte morphology in human tibiae of different bone pathologies with different bone mineral density — Is there a role for mechanosensing? Bone 2009;45(2):321–9. [DOI] [PubMed] [Google Scholar]
- 11.Tsourdi E, Jähn K, Rauner M, Busse B, Bonewald LF. Physiological and pathological osteocytic osteolysis. J. Musculoskelet. Neuronal Interact 2018. [PMC free article] [PubMed]
- 12.Qing H, Bonewald LF. Osteocyte Remodeling of the Perilacunar and Pericanalicular Matrix. Int. J. Oral Sci 2009;1(2):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wysolmerski JJ. Osteocytic osteolysis: time for a second look? Bonekey Rep 2012;1:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belanger LF. Osteocytic osteolysis. Calcif Tissue Res 1969;4(1):1–12. [DOI] [PubMed] [Google Scholar]
- 15.Jande SS, Bélanger LF. Electron microscopy of osteocytes and the pericellular matrix in rat trabecular bone. Calcif. Tissue Res 1970;6(1):280–9. [DOI] [PubMed] [Google Scholar]
- 16.Bonewald LF. Osteocytes as dynamic multifunctional cells. Ann. N. Y. Acad. Sci 2007;1116:281–90. [DOI] [PubMed] [Google Scholar]
- 17.McGee-Lawrence ME, Carey HV, Donahue SW. Mammalian hibernation as a model of disuse osteoporosis: the effects of physical inactivity on bone metabolism, structure, and strength. Am. J. Physiol. Integr. Comp. Physiol 2008;295(6):R1999–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qing H, Ardeshirpour L, Divieti Pajevic P, Dusevich V, Jähn K, Kato S, Wysolmerski J, Bonewald LF. Demonstration of osteocytic perilacunar/canalicular remodeling in mice during lactation. J. Bone Miner. Res 2012;27(5):1018–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teti A, Zallone A. Do osteocytes contribute to bone mineral homeostasis? Osteocytic osteolysis revisited. Bone 2009;44(1):11–6. [DOI] [PubMed] [Google Scholar]
- 20.Yee CS, Schurman CA, White CR, Alliston T. Investigating Osteocytic Perilacunar/Canalicular Remodeling. Curr. Osteoporos. Rep 2019;17(4):157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue K, Mikuni-Takagaki Y, Oikawa K, Itoh T, Inada M, Noguchi T, Park JS, Onodera T, Krane SM, Noda M, Itohara S. A crucial role for matrix metalloproteinase 2 in osteocytic canalicular formation and bone metabolism. J. Biol. Chem 2006;281(44):33814–24. [DOI] [PubMed] [Google Scholar]
- 22.Tang SY, Herber R-P, Ho SP, Alliston T. Matrix metalloproteinase-13 is required for osteocytic perilacunar remodeling and maintains bone fracture resistance. J. Bone Miner. Res 2012;27(9):1936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jähn K, Kelkar S, Zhao H, Xie Y, Tiede-Lewis LAM, Dusevich V, Dallas SL, Bonewald LF. Osteocytes Acidify Their Microenvironment in Response to PTHrP In Vitro and in Lactating Mice In Vivo. J. Bone Miner. Res 2017;32(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotinun S, Ishihara Y, Nagano K, Kiviranta R, Carpentier VT, Neff L, Parkman V, Ide N, Hu D, Dann P, Brooks D, Bouxsein ML, Wysolmerski J, Gori F, Baron R. Cathepsin K– deficient osteocytes prevent lactation-induced bone loss and parathyroid hormone suppression. J. Clin. Invest 2019;129(8):3058–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada S, Billinghurst RC, Chrysovergis K, Inoue S, Poole AR, Holmbeck K, Pidoux I, Birkedal-Hansen H, Bianco P, Wu W. The metalloproteinase MT1-MMP is required for normal development and maintenance of osteocyte processes in bone. J. Cell Sci 2004;118(1):147–56. [DOI] [PubMed] [Google Scholar]
- 26.Tokarz D, Martins JS, Petit ET, Lin CP, Demay MB, Liu ES. Hormonal Regulation of Osteocyte Perilacunar and Canalicular Remodeling in the Hyp Mouse Model of X-Linked Hypophosphatemia. J. Bone Miner. Res 2018;33(3):499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaya S, Basta-Pljakic J, Seref-Ferlengez Z, Majeska RJ, Cardoso L, Bromage T, Zhang Q, Flach CR, Mendelsohn R, Yakar S, Fritton SP, Schaffler MB. Lactation Induced Changes in the Volume of Osteocyte Lacunar-Canalicular Space Alter Mechanical Properties in Cortical Bone Tissue. J. Bone Miner. Res 2017;32(4):688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dole NS, Mazur CM, Acevedo C, Lopez JP, Monteiro DA, Fowler TW, Gludovatz B, Walsh F, Regan JN, Messina S, Evans DS, Lang TF, Zhang B, Ritchie RO, Mohammad KS, Alliston T. Osteocyte-Intrinsic TGF-β Signaling Regulates Bone Quality through Perilacunar/Canalicular Remodeling. Cell Rep 2017;21(9):2585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang SY, Herber RP, Ho SP, Alliston T. Matrix metalloproteinase-13 is required for osteocytic perilacunar remodeling and maintains bone fracture resistance. J. Bone Miner. Res 2012. Sep;27(9):1936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amling M, Püschel K, Rolvien T, Yorgan T, Jeschke A, Busse B, Krause M, Demay MB, Schinke T. Vitamin D regulates osteocyte survival and perilacunar remodeling in human and murine bone. Bone 2017;103:78–87. [DOI] [PubMed] [Google Scholar]
- 31.Kogawa M, Wijenayaka AR, Ormsby RT, Thomas GP, Anderson PH, Bonewald LF, Findlay DM, Atkins GJ. Sclerostin Regulates Release of Bone Mineral by Osteocytes by Induction of Carbonic Anhydrase 2. J. Bone Miner. Res 2013;28(12):2436–48. [DOI] [PubMed] [Google Scholar]
- 32.Dole NS, Mazur CM, Acevedo C, Lopez JP, Monteiro DA, Fowler TW, Gludovatz B, Walsh F, Regan JN, Messina S, Evans DS, Lang TF, Zhang B, Ritchie RO, Mohammad KS, Alliston T. Osteocyte-Intrinsic TGF-β Signaling Regulates Bone Quality through Perilacunar/Canalicular Remodeling. Cell Rep 2017;21(9):2585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heller-Steinberg M Ground substance, bone salts, and cellular activity in bone formation and destruction. Am. J. Anat 1951;89(3):347–79. [DOI] [PubMed] [Google Scholar]
- 34.Chappard D, Baslé MF, Legrand E, Audran M. New laboratory tools in the assessment of bone quality. Osteoporos. Int 2011;22(8):2225–40. [DOI] [PubMed] [Google Scholar]
- 35.Tazawa K, Hoshi K, Kawamoto S, Tanaka M, Ejiri S, Ozawa H. Osteocytic osteolysis observed in rats to which parathyroid hormone was continuously administered. J. Bone Miner. Metab 2004;22(6):524–9. [DOI] [PubMed] [Google Scholar]
- 36.Baylink DJ, Wergedal JE. Bone formation by osteocytes. Am. J. Physiol 1971;221(3):669–78. [DOI] [PubMed] [Google Scholar]
- 37.Webster DJ, Schneider P, Dallas SL, Müller R. Studying osteocytes within their environment. Bone 2013;54(2):285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fowler TW, Acevedo C, Mazur CM, Hall-Glenn F, Fields AJ, Bale HA, Ritchie RO, Lotz JC, Vail TP, Alliston T. Glucocorticoid suppression of osteocyte perilacunar remodeling is associated with subchondral bone degeneration in osteonecrosis. Sci. Rep 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiede-Lewis LM, Xie Y, Hulbert MA, Campos R, Dallas MR, Dusevich V, Bonewald LF, Dallas SL. Degeneration of the osteocyte network in the C57BL/6 mouse model of aging. Aging 2017. Oct 26;9(10):2190–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamel-ElSayed SA, Tiede-Lewis LM, Lu Y, Veno PA, Dallas SL. Novel approaches for two and three dimensional multiplexed imaging of osteocytes. Bone 2015;76:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider P, Meier M, Wepf R, Müller R. Towards quantitative 3D imaging of the osteocyte lacuno-canalicular network. Bone 2010;47(5):848–58. [DOI] [PubMed] [Google Scholar]
- 42.Dong P, Pacureanu A, Zuluaga MA, Olivier C, Grimal Q, Peyrin F. Quantification of the 3d morphology of the bone cell network from Synchrotron Micro-CT images. Image Anal. Stereol 2014;33(2):157. [Google Scholar]
- 43.Schneider P, Meier M, Wepf R, Müller R. Serial FIB/SEM imaging for quantitative 3D assessment of the osteocyte lacuno-canalicular network. Bone 2011;49(2):304–11. [DOI] [PubMed] [Google Scholar]
- 44.Sano H, Kikuta J, Furuya M, Kondo N, Endo N, Ishii M. Intravital bone imaging by two-photon excitation microscopy to identify osteocytic osteolysis in vivo. Bone 2015;74:134–9. [DOI] [PubMed] [Google Scholar]
- 45.Fowler TW, Acevedo C, Mazur CM, Hall-Glenn F, Fields AJ, Bale HA, Ritchie RO, Lotz JC, Vail TP, Alliston T. Glucocorticoid suppression of osteocyte perilacunar remodeling is associated with subchondral bone degeneration in osteonecrosis. Sci. Rep 2017;7:44618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazur CM, Woo JJ, Yee CS, Fields AJ, Acevedo C, Bailey KN, Kaya S, Fowler TW, Lotz JC, Dang A, Kuo AC, Vail TP, Alliston T. Osteocyte dysfunction promotes osteoarthritis through MMP13-dependent suppression of subchondral bone homeostasis. Bone Res 2019;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaudin-Audrain C, Gallois Y, Pascaretti-Grizon F, Hubert L, Massin P, Baslé M-F, Chappard D. Osteopontin is histochemically detected by the AgNOR acid-silver staining. Histol. Histopathol 2008;23(4):469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skinner RA. Decalcification of Bone Tissue BT - Handbook of Histology Methods for Bone and Cartilage In: An YH, Martin KL, editors. Totowa, NJ: Humana Press; 2003. p. 167–84. [Google Scholar]
- 49.Monleau M, Bonnel S, Gostan T, Blanchard D, Courgnaud V, Lecellier C-H. Comparison of different extraction techniques to profile microRNAs from human sera and peripheral blood mononuclear cells. BMC Genomics 2014;15(1):395. [DOI] [PMC free article] [PubMed] [Google Scholar]


