Abstract
Emerging bottom-up fabrication methods have enabled the assembly of synthetic colloids, microrobots, living cells, and organoids to create intricate structures with unique properties that transcend their individual components. This review provides an access point to the latest developments in externally driven assembly of synthetic and biological components. In particular, we emphasize reversibility, which enables the fabrication of multiscale systems that would not be possible under traditional techniques. Magnetic, acoustic, optical, and electric fields are the most promising methods for controlling the reversible assembly of biological and synthetic subunits since they can reprogram their assembly by switching on/off the external field or shaping these fields. We feature capabilities to dynamically actuate the assembly configuration by modulating the properties of the external stimuli, including frequency and amplitude. We describe the design principles which enable the assembly of reconfigurable structures. Finally, we foresee that the high degree of control capabilities offered by externally driven assembly will enable broad access to increasingly robust design principles towards building advanced dynamic intelligent systems.
Keywords: Externally driven, reversible assembly, active matter, bioassembly, mesoscale assembly
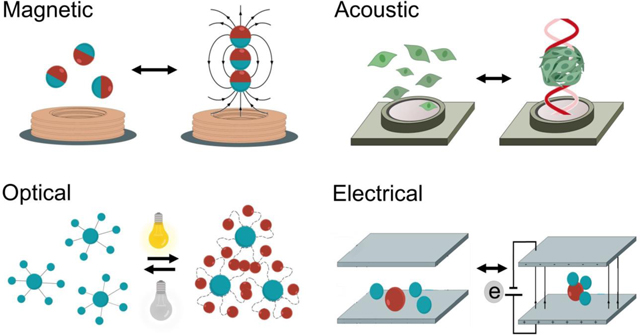
Graphical Abstract
This review provides an access point to the latest developments in externally driven assembly of synthetic and biological components by magnetic, acoustic, optical, and electric fields. In particular, we address how reversibility has the potential to enable the fabrication of multiscale systems that would not be possible under traditional techniques.
Introduction
Translating the fundamentals of small scale assembly into tunable engineering design principles has the potential to revolutionize the development of intelligent nanoscale to mesoscale systems.[1–3] The assembly of synthetic micro/nanoparticles into multiscale structures, ranging from nano, micro and mesoscale, has enabled the translation of their unique properties into large scale materials.[4,5] In a similar fashion, living organisms can be considered as living assemblies, composed of several hierarchical layers with cells as the smallest intact single units.[6–8] In both synthetic and biological assemblies, the complex interplay of these subunits creates robust systems in which slight variations in the unit properties at the small scale can modify the whole system’s functionality at a larger scale.[9] The most common approaches of micro-assembly consist of bottom-up methods based on the assembly of microscale subunits into larger structures.[10–12] The forces driving this “self-organization” consist of changes that are either produced locally[13] (e.g., chemical gradient,[14,15] pH,[16][17] temperature changes [18,19]) or driven by external power sources.[20,21]
Although recent reviews have covered the topic of self-assembly and the use of externally driven forces to drive micro/nanoscale assembly, these works do not fully address the concept of reversible design.[22,23] In this direction, herein, we present the latest developments in reversible assembly at small scales. We define reversible design as a guided process capable of reconfiguring assemblies into new structures and the subsequent ability to restore their building blocks to their initial state. Governing the interface between the subunits, the external field functions as a vector for programable control over the assembly. Although self-assembly methods present unique advantages including simple protocols and scalability, their major drawback is that they produce static structures that cannot be modulated. In the other hand, reversible assembly offers complementary advantages to self-assembly methods, including a high degree of design flexibility, low material consumption, high packing density, when compared to self-assembly methods. Moreover, reversibility in design offers an alternative to fabricate multiscale assemblies not possible by other methods, including the ability to reconfigure the assembly in real-time through the of the applied frequency and amplitude guide assembly formation into complex patterns without the need for scaffolds, providing the homogenous subunits with distinct functionalities based on their assembly and disassembly states and parallel manipulation of swarms of active particles that maintain individual self-locomotive capabilities. Furthermore, the rapid disassembly of the structure into their initial subunits can be typically induced simply by removing the applied stimuli.
Here we will describe the most commonly used external energy sources, including magnetic, acoustic, optical, and electrical fields, which drive and control the assembly of micro and nanoscale programmable mesoscale structures (Figure 1). The use of external fields triggers different processes that lead to the formation of assemblies.[24] They can (i) create pressure nodes or tweezers to concentrate and trap micro-objects in a specific location,[25,26] (ii) modulate particle-particle interactions that can lead to attractive or repulsive behavior between the subunits that compose the assembly[27,28] or (iii) use active matter (micromotors or living organisms) as a seed unit to form assemblies with larger particles, or induce swarming behavior. [29–34] We thoroughly discuss each of the external fields, their mechanisms of action, and the latest applications in the synthetic and biological assembly in the following sections. Finally, we outline the challenges and opportunities for the use of external fields to control reversible assembly at small scales.
Figure 1.
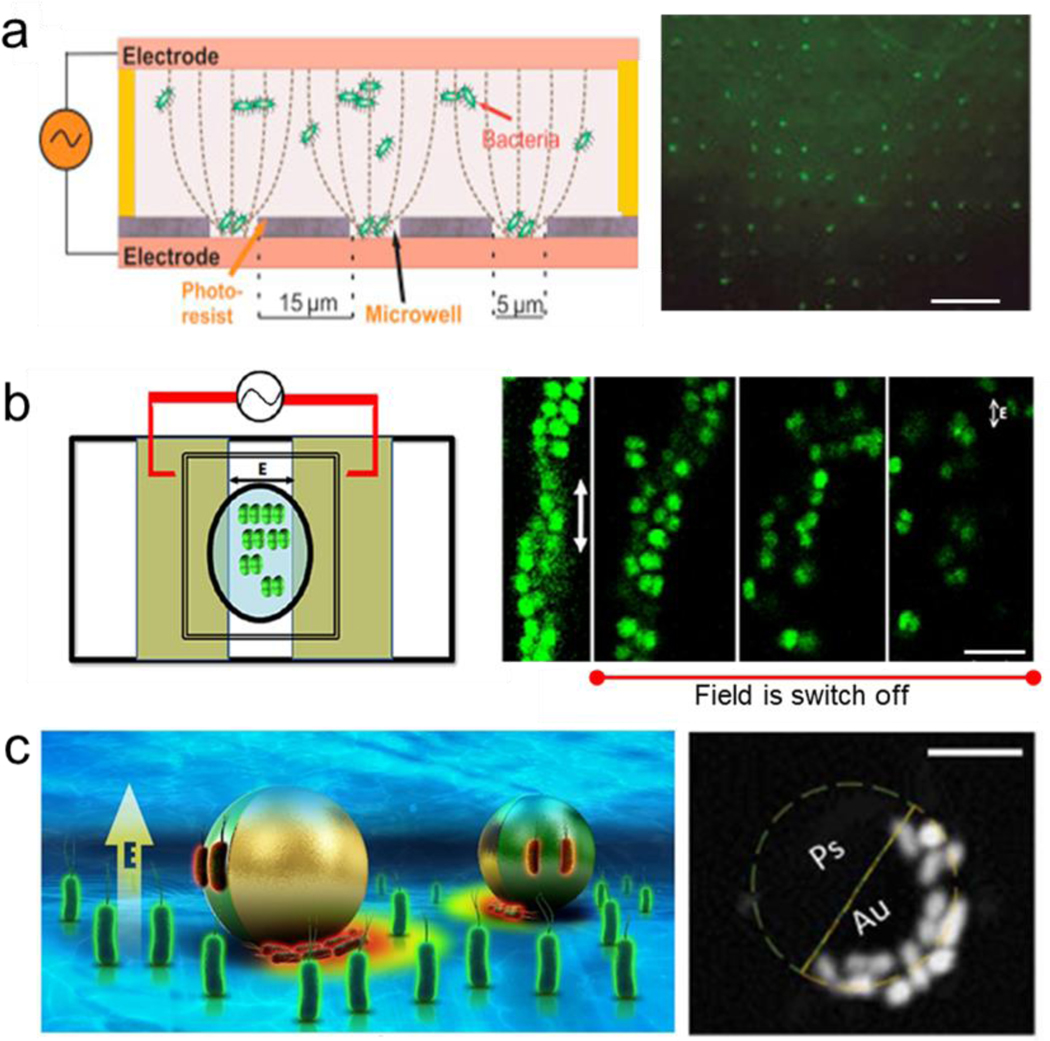
Externally driven reversible assembly at small scales. a) Schematic of the different external fields (magnetic, acoustic, optical and electrical) used for assembling synthetic and biological structures.
Magnetic Reversible Dynamic Assembly
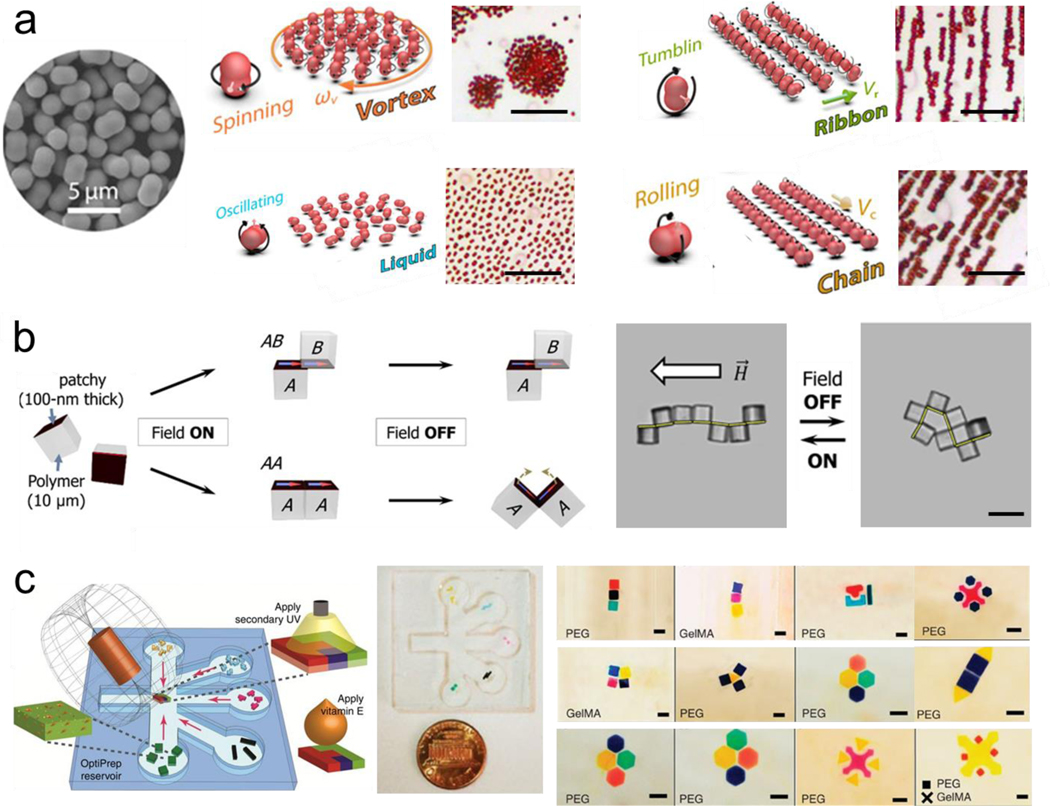
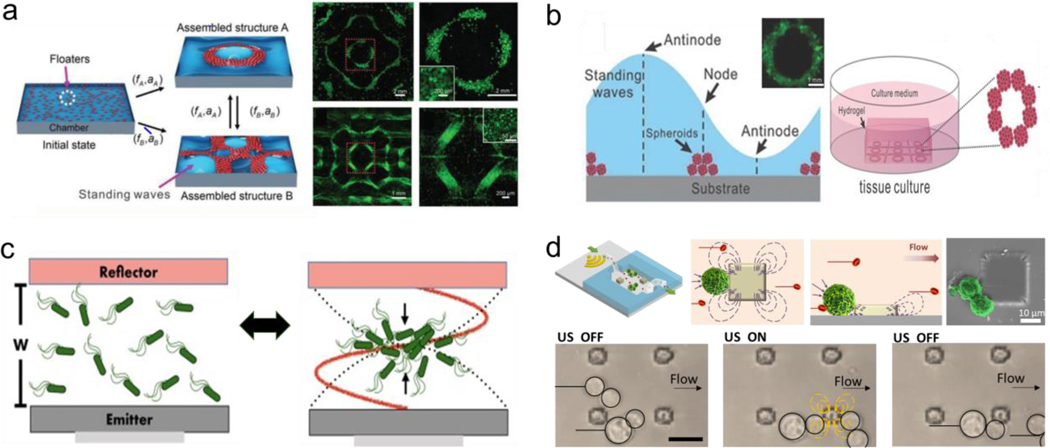
Magnetic assembly has been investigated and successfully applied in guiding the reversible assembly of small scale structures, typically by a simple change in the magnetic field or removal of it, enabling dynamic maneuvering and reconfiguration by guiding the migration of subunits, inducing magnetic levitation or exploiting dipole-dipole interactions. The individual subunits can be magnetized or be inherently magnetic, such as magnetic nanoparticles or paramagnetic iron-containing objects. Moreover, microrobots have been widely used for directing the assembly at a small scale.[35–38] To this end, researchers have worked on developing techniques to code three-dimensional materials with various programmable features, by shape, composition, and surface properties.[39] Reconfigurable swarms of microrobots are one example of assembly involving dynamic group behavior. For instance, alternating magnetic fields have been used to program fast and reversible transformations between different swarm organizations of hematite colloidal particles with permanent magnetic moments, including liquids, chains, vortices, and ribbons. (Figure 2a).[40] Another study explored the latter swarm configuration, using oscillating magnetic fields to program and restructure assemblies of paramagnetic nanoparticles enabling complex behavior, including locomotion, splitting and merging, and passing through channel networks.[41]
Figure 2.
Magnetic induced reversible assembly of synthetic micro/nanoparticles. a) Fast transitions between different collective formations of hematite colloidal microrobots by alternating magnetic fields. The scale bar is 20 μm. Reprinted with permission of reference. [40] Copyright 2019, AAAS. b) Reconfiguration patterns of microcube chains controlled by dipole-field and dipole-dipole interactions. The scale bar is 20 μm. Reprinted with permission of reference.[43] Copyright 2017, AAAS. c) Directed magnetic assembly of magnetoceptive gels into Tetris-like structures. The scale bar is 1 mm. Reprinted with permission of reference.[47] Copyright 2016, Springer Nature.
In another example, an alternating magnetic field was used to guide the microrobot assembly into aster shapes at an immiscible liquid interface.[42] Patchy microcube clusters were assembled using an external magnetic field, self-folding, and self-reconfiguring into a number of possible structures, while releasing energy in the process. In reconfiguration, the original sequence encoded the subsequent function as a result of equilibrium and nonequilibrium states of the cubes. (Figure 2b).[43] Another example in the microrobot field is the use of robotic micro-grippers made of flexible patterned magnetic material, remotely actuated by a magnetic field, to perform programmed 3D assembly of microstructures.[44] Diamagnetically levitated milli-robots were also used for 2D-assembly.[45] The use of an optical microscope was used for feedback, achieving open-loop stability up to 78 hours in this demonstration of microfabrication and manipulation, also show autonomous micro-assembly. Another example for the assembly of superparamagnetic particles combined magnetic fields to assemble a microrobot swarm and an acoustic field to make it rotate demonstrating a behavior similar to a neutrophil rolling on a vessel wall.[46]
The question of assembly in different dimensions and scales presents unique challenges by adding further complexity, while throughput is another factor in implementation. While assembly in two dimensions is well studied, assembly in three dimensions tends to be more challenging. 3D cultures have been particularly important in complex cell culture models because they more closely resemble biological phenomena than the traditional 2D cultures. Towards this goal, guided assembly has been used on tunable gels by using the inherent paramagnetic properties of free radicals as an alternative to magnetic nanoparticle-embedded hydrogels. This rendered the gels “magnetoceptive” and thus capable of constructing complex 3D structures, by combining different building blocks in a Tetris like fashion. The block interacted via magnetic interaction produced by the embed magnetic nanoparticles. Addition of vitamin D eliminated the magnetic reactive oxygen species, resulting in loose of magnetic proprieties, thus disassembly. In this case, the process is not fully reversible but the erasibility of magnetic properties is another potential method for modulation the assembly process.(Figure 2c).[47] Magnetic fields have also been used to guide superparamagnetic colloidal crystals into tunable chromatic arrays that map to structural colors.[48] The guided assembly of colloidal matter thus encompasses a wide variety of actuation techniques and applications.
The magnetic susceptibility of biological objects has been explored as a means for safe and biocompatible dynamic control over biological assembly processes. For example, magnetotactic bacteria, a rare example of living cells with naturally occurring magnetic dipoles, have been manipulated to direct the assembly of synthetic microstructures. These flagellated bacteria occur in aquatic sediments and achieve directed mobility in a magnetic field by a cytoskeletal organization of their characteristic organelle: iron-containing magnetosomes.[49] Such magnetic properties serve to guide their assembly at low Reynolds environment with controlled magnetotaxis, to create a pyramid shape as an example.[50] Others have harnessed the unique properties of assemblies of magnetotactic bacteria to explore their potential to generate mechanical energy, via confinement into a water-in-oil droplet under a constant magnetic field generated by a Helmholtz coil.[51]
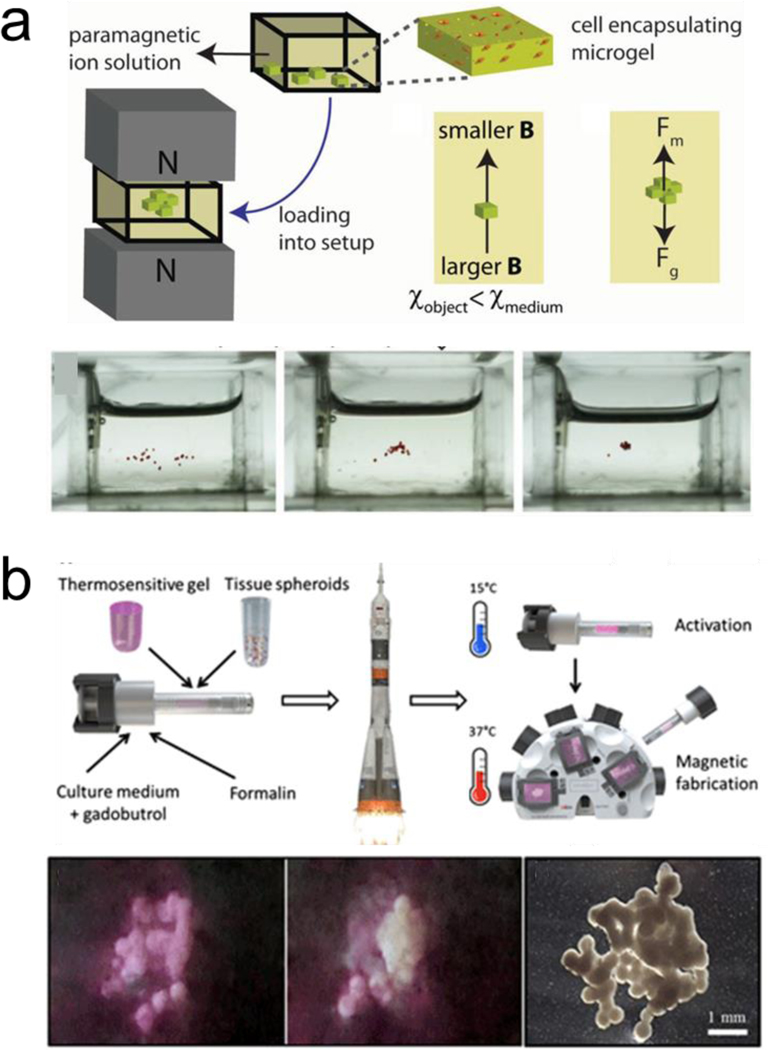
Magnetic levitation is also employed to assemble cells.[52,53] This approach can direct the assembly of subunits with low magnetic susceptibility when they are suspended in a paramagnetic substance within a magnetic gradient. Uniquely, even cells that are diamagnetic can be assembled via this technique without the need of labeling or functionalization, by being suspended in a paramagnetic solution within a magnetic gradient.[54,55] The structures can be disassembled by removing the applied magnetic field.[56] Magnetic levitation has been combined with nanopatterned cell sheets for fast and reversible interactions with their base structure to build thermoresponsive constructs for tissue engineering.[57] Furthermore, cell encapsulating microgels assemblies were generated via control of the magnetic field as well as tuning the paramagnetic medium (Figure3a).[58] 3D scaffolds for bone repair in a clinically-oriented application have also employed magnetic levitation to guide the assembly of the subunits.[59] Moreover, magnetic levitation assembly has been performed at the international space station, to assemble 3D tissue constructs in a setting without actual gravity (Figure 3b).[60]
Figure 3.
Magnetically induced reversible assembly of living cells. a) Magnetic levitation working mechanism based on force equilibrium between magnetic force and gravity for assembling microgels encapsulating living cells. Reprinted with permission of reference.[58] Copyright 2015, Wiley. b) Magnetic levitation of tissue illustrating 3D bioassembly performed in space. The scale bar is 20 μm. Reprinted with permission of reference.[60] Copyright 2020, AAAS.
Magnetic assemblies allow for remote control and manipulation of building blocks that respond to external magnetic fields. Nevertheless, some magnetic assembly systems are complex and dependent on multiple kinds of equipment that can pose a challenge for widespread use. Overall, the magnetic properties of the materials, the applied field, and their magnitudes, are the main parameters governing externally driven magnetic assembly.
Acoustic Reversible Dynamic Assembly
Acoustic actuation technologies have been used as a non-invasive and highly biocompatible power source for rapid and tunable assembly of micro/nanoparticles.[61–65]Acoustic assembly offers non-contact manipulation of objects based on the difference in density between the subunits and their enclosing medium. The acoustic field can drive the migration of dispersed micro/nanoparticles towards a specific location along a pressure node. Different types of acoustic fields result in distinct methods for manipulating micro-objects, including the use of surface traveling waves, standing waves, and acoustic levitation based on projecting focused ultrasound patterns.[66]
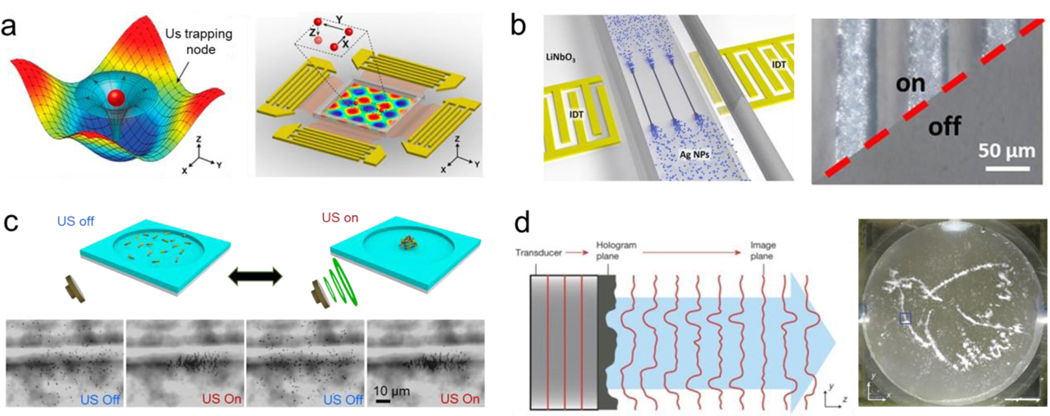
Surface acoustic waves are commonly used for assembling synthetic colloids of different sizes.[67] These types of acoustic fields are generated using an interdigitated microelectrode that converts electrical signals by propagating them as mechanical stress that travels along the surface of a piezoelectric substrate material. These generate pressure nodes that serve as traps to manipulate and assemble micro-objects into predetermined patterns (Figure 4a).[68] One-dimensional (1D) micron-size arrays were constructed by directing the assembly of silver nanoparticles using surface acoustic waves. The nanoparticles were flowed through a microfluidic chip. Upon application of surface acoustic waves, nanoparticles accumulated at pressure nodes forming 1D structures. Turning off the applied field resulted in the dispersion of the 1D assemblies, enabling reversible assembly without any noticeable change even after multiple cycles. Once a final design was selected, the silver nanoparticles were subjected to a sintering process to enable the formation of a stable structure (Figure 4b).[69] Similarly, surface acoustic waves applied in nanoscale pulses enhanced the pattern resolution. This pulsed ultrasound minimized both the noise and acoustic streaming generated by the continuous application of the acoustic field, providing a great degree of control over the formation of the trapping area, leaving nearby areas unaffected.[70] In another case, surface acoustic waves were used to generate two-dimensional acoustic nodes capable of large-scale assembly of nanowires suspended in solution into well-controlled patterns with tunable geometry and spacing. The process enabled real-time dynamic patterning of large-scale assemblies. The acoustic field could be modulated to assemble the nanowires into parallel rows with the same orientation or to generate hybrid patterns of line assemblies with spherical patterns at the junction nodes. The nanowire dispersion was induced by turning off the applied power. The alignment and pattern type were simultaneously switched by changing the direction of propagation of the surface acoustic waves. Apart from this reversible behavior, by evaporating the solution and transferring the pattern into another substrate, an irreversible structure was obtained.[71] Despite the advantages of acoustic fields for induction of reversible assembly, the manipulation of submicron structures with a high spatiotemporal resolution has remained a challenge.[72,73] New methods are starting to show the possibility of driving the assembly of submicron particles down to 200 nm. To this end, a cylindrical chamber was employed, the structure was aligned orthogonally to the wave propagation direction and parallel to the substrate. This approach served to limit formation of competing standing waves or microstreaming forces inside the chamber, and allowed the assembly of smaller nanoparticles.[74]
Figure 4.
Acoustic induced reversible assembly of synthetic micro/nanoparticles. a) Acoustic surface waves generate acoustic tweezers capable of trapping particles in pressure nodes and assemble them in patterns. Reprinted with permission of reference.[68] Copyright 2016, National Academy of Sciences. b) Surface acoustic waves induce the assembly of silver nanowires into micron scale line patterns. Reprinted with permission of reference.[69] Copyright 2019, ElSevier. c) Standing waves generate the reversible assembly of chemically propelled nanorobots. Reprinted with permission of reference.[83] Copyright 2015, ACS. d) Acoustic hologram assemble three-dimensional microstructures. Reprinted with permission of reference.[91] Copyright 2016, Springer Nature.
Standing waves have also been used to induce migration of particles towards pressure nodes, enabling the formation of small scale assemblies. [75–77] Thus, this principle has enabled ensembles of colloidal particles into “crystal structures” that are reconfigurable in real-time. The fine tuning of the acoustic standing field enabled the lattice geometry modulation.[78] Likewise, the speed of the assembly processes was controlled by modulating the applied amplitude.[79] Another application of standing waves is the assembly of active matter. Self-propelled microswimmers move in random directions, thus making it hard to coordinate their collective behavior. In this direction, tuning the acoustic nodes has permitted to create swarms of microrobots and active particles.[80–82] The application of acoustic standing waves triggered the organization of chemically-powered nanorobots into reversible swarm assemblies. The nanorobot swarm dispersed quickly after the acoustic field was turned off due to their autonomous catalytic locomotion, enabling a fast disassembly process. The swarm assembly generated at the nodal point was moved into different locations by changing the applied frequency. (Figure 4c).[83] The combination of acoustic fields and other external fields, including magnetic[46,84] and optical[85,86] field was capable of generating more complex patterns. The assembly of the active matter was explored using nodes with different strengths. Strong nodes enabled closely packed assemblies, while weak nodes produced less packed assemblies where the subunits suffer constant reconfiguration of the assembly. The ability to tune the strength of the node enabled measurement of the force generated by nanorobots propelled by chemically induced locomotion.[87]
Acoustic levitation has also been used for tunable assembly of microscale units based on particle migration towards projected pressure nodes generated by arrays of acoustic transducers.[88] These create pressure nodes that trap particles, overcoming gravity forces.[89] The scattering of the ultrasound waves on the particles induced attractive interactions that led to controllable assembly. Thus acoustic levitation has been reported to enable the reconfigurable formation of small clusters of microparticles.[90] More recently, the realization of holographic acoustic tweezers was reported. Projections or holograms of acoustic patterns were generated using a mask, where the projected ultrasound wave stored the spatial distribution of the trap particles. These were further reconstructed by the interference of the projected sound waves. The microparticles were enclosed in a water container located above the transducer. Upon turning on the hologram, the particles assembled into the shape of a dove, used as an example of a projected sound pressure image. The ensemble collapsed after the system was turned off, enabling the reversible configuration of the assembly. This holographic approach demonstrated the ability to assemble structures two orders of magnitude larger than their original building blocks. Moreover, phase gradients were also generated using the acoustic hologram, which enabled trapping and transporting particles along predetermined paths. The particles followed the enclosed path indefinitely until the hologram was turned off ( Figure 4d). [91] Further works demonstrated the ability to use the acoustic hologram technique to form a well-defined irreversible assembly of silicon beads by UV induced cross-linking, illustrating the potential of the technique in fabricating permanent assemblies using this bottom-up strategy.[92]
Apart from synthetic particles, acoustic fields have shown great potential in the assembly of living organisms with high cell density and viability towards mimicking real tissue constructs. [93–97] In this direction, acoustic fields have been applied in a scaffold-free manner to drive the assembly of living cells for tissue engineering applications.[98–101] The acoustic method can assemble highly organized cell constructs without the need for conventional scaffold, showing viable promise in building larger 3D tissue-like constructs. Standing waves, were applied for the assembly of microorganisms, which enabled the study of their behavior in closely packed environments.[102] Standing waves were used to assemble microscale biomaterials into ordered structures in a scalable manner at high throughput. These accumulated a large number of cells (∼106) within a few seconds (<5 s). The assembled structures were achieved by modulating the applied frequency, which permitted building structures in the macroscale range (Figure 5a).[103] In another work, 3D cell spheroids were fabricated by a similar method. The cells were first resuspended onto a low-adhesive petri dish surface and incubated for two days to form the cell spheroids. After collection, multiple spheroid assemblies were reversibly patterned by applying acoustic waves for a few seconds of different frequencies and amplitudes. The ability to finely tune the shape of the assembly enabled further crosslinking by thrombin and fibrinogen, enabling long-term cell culture and downstream functional characteristics (Figure 5b).[104]
Figure 5.
Ultrasound induced reversible assembly of living cells. a) Standing waves generate reconfigurable cell assemblies. Reprinted with permission of reference.[103] Copyright 2014, Wiley. b) standing waves fabricate tree dimensional organoids. Reprinted with permission of reference.[102] Copyright 2015, Wiley. c) Standing waves levitation nodes for the assembly and disassembly of bacteria populations. Reprinted with permission of reference.[113] Copyright 2018, Springer Nature. d) Parallel arrays of microstreaming traps capable of assembling cells in a microfluidic chamber. Reprinted with permission of reference.[123] Copyright 2019, Wiley.
Acoustic standing waves were also used to assemble dense myoblast populations in a hydrogel containing type I collagen. The generation of the muscular tissue, and myogenesis behavior, were studied when applying the external field.[105] Cardiomyocytes stem cells were assembled into predefined 3D assemblies using standing waves to mimic cardiac tissue. These methods enabled high-density packing (108–109 cells/mL), which significantly improved cell activity and function, including metabolic activity and cell viability, when compared to randomly generated assemblies.[106] Brain-like construct assemblies have also been built using standing waves. The ability to generate brain models in three dimensions is of great importance as 2D models do not sufficiently recapitulate the neural connectivity of the brain, To build these brain models, acoustic standing waves generated multiple levitation planes which matched the spacing in brain tissues.[107] Using a similar approach, 3D neurospheroids were assemble to model Alzheimer’s disease.[108]
The shape of the assemblies has also been modulated to form ring shape assemblies.[109,110] More recent acoustic methods have utilized ionic crosslinking to rapidly remove hydrogel scaffolds allowing the generation of tissues composed of only cellular assemblies.[111] The use of acoustic forces was also reported towards enhancing cell packing in buckyball microstructures.[112] Moreover, acoustic nodes were also used to induce the swarm formation of Escherichia coli bacterial suspensions. The disassembly was achieved by removal of the acoustic field, although long periods of confinement led to the formation of bacterial seed aggregates (Figure 5c).[113] Different patterns of living cells were generated by modulating the applied ultrasound frequency without affecting cell viability.[114–117] Studies of replication and cell growth under confinement were also performed using cell assemblies exposed to ultrasound fields for more than 12 hours.[118] Other applications consisted of the directional assembly of cells into acoustic wells for localized analysis.[119] Acoustic microstreaming around micro-engineered patterns were also explored using surface acoustic waves. Topographical features of the substrate interact with the traveling wave generating a localized microstreaming flows capable of trapping nearby objects.[120–122] Application of this technique was performed by building arrays of acoustic microstreaming traps that enabled the enrichment of cancer cells from whole blood around the traps. Interestingly, this methodology could also be used to generate organoids, as the cells can be released upon turning off the acoustic field (Figure 5d).[123,124] Acoustic holograms have been utilized for patterning cells enabling rapid 3D tissue fabrication.[125]
In general, acoustic assembly methods offer the ability to concentrate diverse types of subunits into predefined large-scale patterns, controlled by the type of applied acoustic field. These methods rely on the migration of building blocks towards pressure nodes driven by difference in material properties with the medium. A limitation in the use of acoustic fields for small scale assembly relies on restricted capabilities to assemble nanoparticles smaller than 200 nm.
Optical Reversible Dynamic Assembly
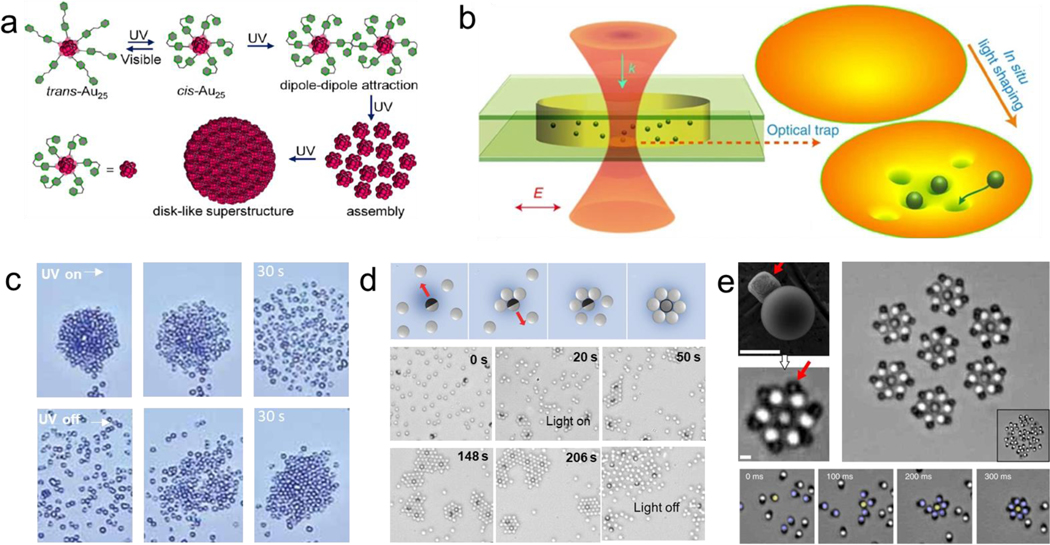
Optical methods have been used to assemble constructs at high speed with high spatiotemporal precision. For example, light can control the reversible assembly of nanoparticles into well-defined colloidal macrostructures.[126] Most methods rely on the use of photoswitchable molecules that can reversibly change between two states in response to specific wavelengths, allowing to selectively trigger the assembly of responsive colloids from a heterogeneous solution. Nanoparticles functionalized with azobenzene groups reported reversible assembly. The exposure to UV light-induced the isomerization of the azobenzene group from trans to cis, which made the nanoparticles lose colloidal stability, thus forming stable assemblies.[127] This process is reversible by removing the UV irradiation.[128] This principle was also used to create “ink” based on nanoparticle assemblies, where the final structure can be tuned to stay stable from a few seconds to days. A photomask was used to create spatial pattern designs that induce the isomerization of azobenzene groups. Exposure to visible light or sunlight resulted in the erasing of the pattern.[129] Au25 nanoclusters functionalized with photoswitchable azobenzene groups reported reversible assembly of large disk-like structures from 100 to 1000 nm under ultraviolet light irradiation. The application of an optical field-induced the assembly and disassembly of the colloidal structure based on dipole-dipole interactions resulting from the trans-cis photoisomerization change of the azobenzene groups (Figure 6a).[130] Nanoparticles functionalized with PMAMC, a photosensitive amphiphilic star-like copolymer, reported reversible self-assembly under optical stimulation. The reversible assembly was based induced by UV light irradiation promoting the PMAMC chain photodimerization (365 nm) and photocleavage (254 nm).[131]
Figure 6-.
Optically induced reversible assembly of synthetic micro/nanoparticles. a) Reversible assembly of nanoparticles based on the photo switchable isomerization of the azobenzene functionalized groups. Reprinted with permission of reference.[130] Copyright 2020, ACS. b) Optical trap generated based on a focused Gaussian beam illustrating manipulation of multiple nanobeads under laser beam irradiation. Reprinted with permission of reference [137] Copyright 2014, Springer Nature. c) Colloidal firework behavior based on the attractive and repulsive forces between the TiO2micromotors and SiO2 passive beads under UV light. Reprinted with permission of reference.[145] Copyright 2010, Wiley. d) Micromotors serving as motile nucleation centers for 2D microcrystal assemblies under UV light. Reprinted with permission of reference [147] Copyright 2017, Wiley. e) Laser-guided assembly of self-spinning microgears. Scale bars e: 10 μm. Reprinted with permission of reference.[153] Copyright 2018, Springer Nature.
The optical gradient force of a focused laser has been used to generate light traps that can assemble microparticles into well-defined micropatterns.[132–135] Optical traps were generated by creating an interfering pattern between an annular shaped laser and a reference beam. Nearby objects were trapped inside the spiral interference pattern. The assembly generation was not based on intrinsic material properties, allowing to trap and generate assemblies of diverse particle types, including silica spheres, glass rods and chromosomes.[136] Reversible organization of metal nanoparticle was achieved by Gaussian optical fields to generate assemblies of different geometries. The optical trap generated a focused force point that suppressed the Brownian motion and confined a group of nanoparticles into diverse tridimensional arrangements (Figure 6b).[137] The optical trapping and assembly of microparticles can also take advantage of the propagation of the laser force field to form assemblies outside of the focal point, resulting in the generation of horn-like structures. In another example, an array of photonic-crystal slab could assemble over 100 polystyrene nanoparticles (500 nm) by projecting a laser light from below a silicon patterned with a periodic array of holes. When the laser beam was turned on, the particles were driven to occupy the array lattice. The trapping forces arose from a strong electric-field gradient generated at the holes. Upon removing the laser irradiation, the particles disperse and diffuse away from the patterned holes.[138] An expansion of this work reported the ability to form one dimensional (1D) chains that can be oriented through finely tuning the incident polarization of the light. The chain formation resulted from the competing forces consisting of particle-particle interaction and particle-hole interactions.[139] Holographic optical tweezers have enabled the simultaneous manipulation of multiple microparticles.[140] The assembly process has been automated using closed-loop object recognition.[141,142]
The use of optical fields has been widely used to guide the assembly of active matter.[143,144] For instance, TiO2 micromotors powered by UV light illustrated a firework like behavior based on the diffusiophoresis of charge species gradients generated by chemical propulsion and its interaction with passive SiO2 microbeads. The passive beads aggregated around the TiO2 micromotors as a result of surface charges. On the other hand, exposure to UV light resulted in the passive microbeads rapidly moving away from the TiO2 micromotors. Upon removal of the UV light, the passive bead reaggregated near the TiO2 micromotor surface (Figure 6c_.[145] A similar firework phenomenon has been reported using optoacoustic micromotors, where the interaction of the acoustic nodes and light-induced attractive and repulsive forces resulted in tunable collective behavior.[85,146] In another case, a small number of UV light-powered TiO2–silica Janus micromotors (1.5 μm) were used to create out-of-equilibrium micro assemblies. The motile micromotors served as nucleation centers that condensed silica microbeads into 2D micro-assemblies based on electrostatic interactions. The assembly process was regulated by tuning the applied light intensity and by changing the active–passive particle size ratio, enabling construction of clusters of different geometries, including square, pentagonal, hexagonal, and heptagonal assemblies. The mixture of different cluster types resulted in large scale crystal-like disordered macro/assemblies (>40 μm). The interaction between the micromotor and passive particle assembly remained stable until the UV light source was turned off (Figure 6d).[147] TiO2 micromotors were functionalized with hydroxyl groups, which enabled reversible motor-motor interactions due to electrolyte diffusiophoretic attractions induced by UV irradiation.[148]
SiO2–Pt Janus micromotors functionalized with spiropyran, a photochromic functional group, reported reversible assembly under light irradiation. UV light induced assembly by electrostatic interaction and disassembled under green light.[149] Photoactivated colloidal particles were assembled into 2D living crystals with dynamic behavior including reversible breaking and re-assembly. Such behavior resulted from the interplay between the propulsion of micromotors and particle-particle interactions induced by light generated osmotic and phoretic effects.[150] Au/TiO2 micromotors with rapid on-demand reversible assembly were reported. Application of an optical field resulted in attractive and repulsive forces that drove the assembly into larger structures. The micromotors served as nucleation sites leading to reconfigurable crystal systems that went through fusion and fission transition states. The application of green light led the micromotors to start clustering into small assemblies (~4 particles), which coalesced into medium sized assemblies (~10 particles) in a process similar to fusion or crystallization. The rapid switching to UV illumination led to rapid disassembly where the clusters “explode”, breaking into smaller clusters and individual motors, resembling a fission event.[151] Peanut-shaped hematite micromotors were assembled into twisted colloid ribbons under blue light radiation. The assemblies arose from the force competition between diffusioosmotic propulsion and phoretic attraction, resulting in the formation of large 3D chains. These assemblies diffused without blue light indicating the reversibility of the system.[152] Photoactive micromotors were assembled under light activation, forming rotating gears. The system was composed of microrobots that migrated towards a hematite anchor under exposure to a blue laser focal spot. Once assembled, the laser was turned off, inducing an attractive hydrodynamic force that drove the motile micromotor into a spinning microgear. A collection of seven micromotors were shown to interact in a gear-like fashion illustrating the ability to create engines that communicate with each other (Figure 6e).[153]
AgCl microparticles assembled into swarms under exposure to UV light. Cyclical exposure between visible and UV light resulted in expansion and compression of the swarm area coverage. The assembly was driven by silver ions secretions generated at the AgCl microparticles.[15] The formation of small clusters between polystyrene/Ag/AgCl Janus micromotors and passive polystyrene beads was achieved based on the difference in surface charge produced under blue light illumination. The optical field was used to power the propulsion of the Janus micromotor.[154]
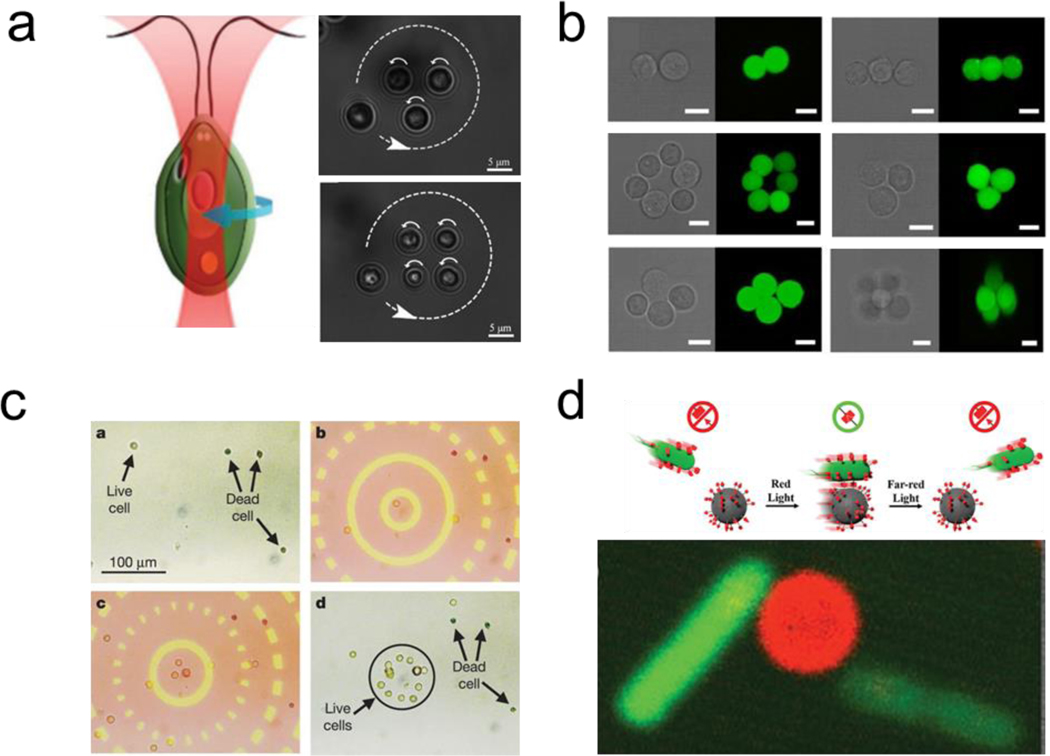
Living organisms can be assembled using optical forces [27,155–157] Optical tweezers generate micro vortices that serve as optical traps. For example, different kinds of cells including yeast (non-motile) and Chlamydomonas reinhardtii (motile), were assembled into micrometer-sized dynamic cellular arrays with well-defined spatiotemporal positioning (Figure 7a).[158] Holographic optical tweezers were used to generate 3D cellular micro architectures. As a proof of concept, a different number of mouse embryonic cells were patterned into 3D assemblies (Figure 7b).[159] An optical image-driven dielectrophoresis mechanism was reported for assembling a large number of microparticles (~15,000) using a light-emitting diode combined with a digital micromirror spatial light modulator array to generate optical patterns of millimeter-sized area. By taking advantage of the dielectric contrast between living and dead cells, this platform reported the ability to separate human cells suspended in solution. The living cells were collected in the assembly regions, while dead cells did not present interaction with the optical field (Figure 7c).[160]
Figure 7-.
Optically induced reversible assembly of living microorganisms. a) use of optical tweezers to induce the assembly of multiple microorganisms into well-defined patterns. Reprinted with permission of reference.[158] Copyright 2020, Wiley. b) Use holographic optical tweezers to build 3d cellular microassemblies. Reprinted with permission of reference.[159] Copyright 2015, Springer Nature. c) Parallel assembly and isolation of live cells from dead ones using optical fields. Reprinted with permission of reference.[160] Copyright 2005, Springer Nature. d) Reversible assembly of bacteria with synthetic micro cargoes under Red/Far-Red Light based on the bonding of and disassembly PhyB and PIF6 proteins. Reprinted with permission of reference.[165] Copyright 2020, Wiley.
Moreover, optical manipulation has been used to manipulate motile living organism by using light patterns to rectify their locomotion. For example, phototaxis capabilities of some microorganisms were used to steer them towards specific regions within a microfluidic chip.[161,162] In this direction, spatially patterned optical fields were used to control the assembly of genetically engineered E. coli that responded to changes in light intensity. A digital mirror device was used to generate light patterns, which induce the bacteria to swim away of the illuminated pattern, accumulating at the edges outside the boundary area of the pattern. The removal of the light pattern led to the dissolution of the bacterial assembly. The optical field was tuned to control the position and sharpness of engineered assemblies, ranging from 10 μm to a few millimeters. [163]
The use of blue light (480 nm) has been used to control engineered bacterial adhesion with functionalized substrates to build reversible biofilm assemblies. The assembly formation was controlled by tuning the photo switchable interaction of motile microorganisms expressing pMag proteins with substrates immobilized with nMag proteins. The interaction between the two proteins is stable in the presence of blue light and unstable in dark environments.[164] Using a similar principle, reversible assembly of bacteria with synthetic micro cargoes under Red/Far-Red Light was reported. The E. coli surface expressed the PhyB protein, which bonded to PIF6 protein functionalized microbead under red light. The interaction was stable upon the removal of the optical stimuli. The dissociation of the protein interaction was further demonstrated under far red light (Figure 7d).[165]
In general, optical assembly methods offer a high degree of tunability and precision. However, they offer smaller throughput fabrication and require a transparent medium to propagate the light source or an inert medium to enable changes in the surface charge required to form the interactions between the colloids, thus limiting the use of this method in heterogeneous opaque microenvironments.
Reversible Electric Dynamic Assembly
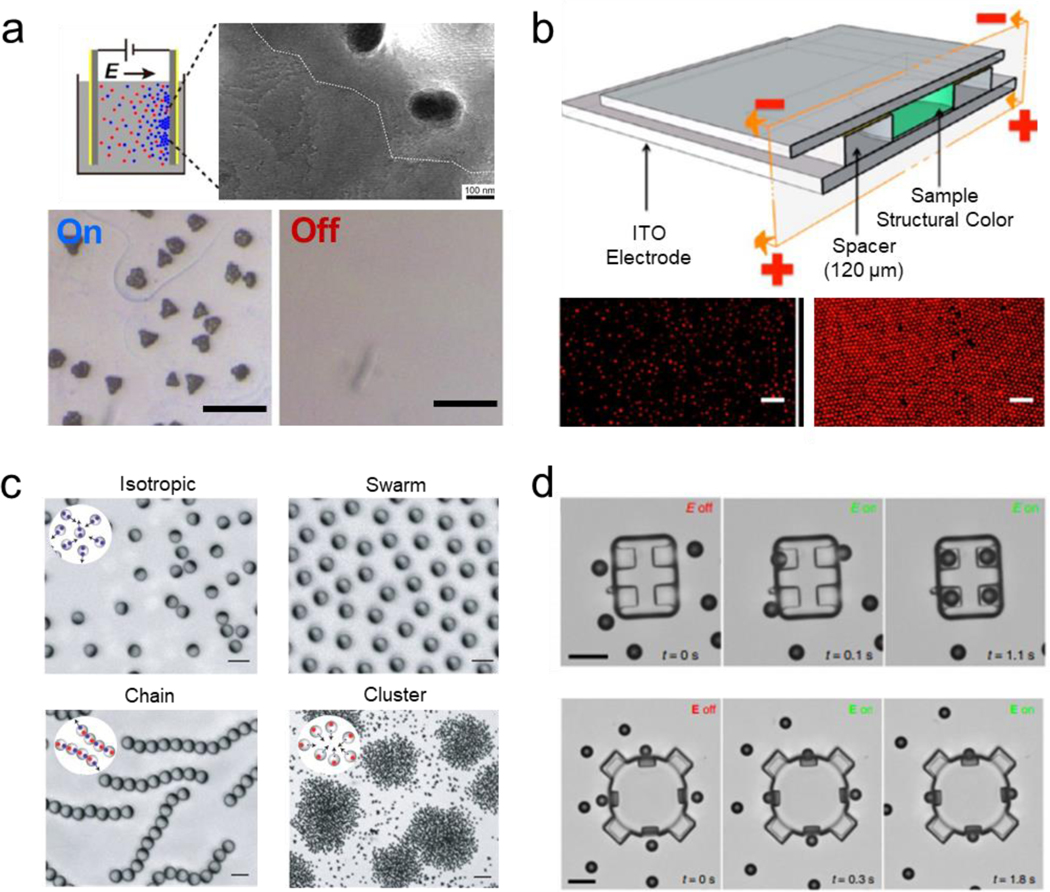
Electric fields have been used as an external stimulus to direct multiscale assemblies of micro/nanoparticles.[166][167] Electrical induced assemblies have demonstrated the ability to generate multifunctional assemblies in which properties arise from their structural design or by tuning the applied field. The use of such electric fields has been shown to induce positive or negative migration into organized structures based on the innate material properties of the subunit particle into assemblies of larger orders of magnitude.[168] For instance, Silver nanoparticles (6 nm) were assembled into micron size superlattice crystals using electric fields. This assembly method was based on the migration of the charged nanoparticles towards a cathode or electrode. Similarly, the addition of tetraoctyl ammonium bromide induced the formation of silver nanocrystals macro-assemblies over the surface of an anode electrode, based on the change in surface charge of the nanoparticles. Removing the electric field, while the electrodes were submerged in the solution, resulted in the silver nanoparticles super lattice to dissolve. This work generated nanocrystal assemblies into large supper lattices without resulting in a change of composition or requiring solvent evaporation (Figure 8a).[169] The large-scale assembly of polymer latex nanoparticles using electrophoretic deposition was exploited to generate reversible assemblies. Upon applying a DC electric field, the positively charged particles assembled over a conductive negatively charged ITO electrode, generating a 20 μm thick crystal lattice with a close-packed structure, capable of maintaining its properties for prolonged periods. The assemblies presented reversibility within 60 s. The reversibility between the ordered and disordered nanoparticles resulted in tunable structural color (Figure 8b).[170] A similar approach relied on the use of predefined patterned scaffolds over an ITO electrode to generate assemblies in the centimeter scale within a few seconds.[171] The electrically driven reversible assembly of plasmonic gold nanoparticles (16 nm) in between two immiscible electrolyte solutions was reported. The nanoparticles were functionalized with hydrophobic chains, which responded to negative polarization, thus accumulating in the liquid-organic interface by induction of positive electric polarization. The assembly resulted in a change in optical response by the colloidal suspension of nanoparticles shifting to red while aggregated and blue when they were dispersed. Such capabilities were used as a proof of concept to generate electric modulated liquid mirrors based on the change in reflectivity, switching from a highly reflective to a transparent material, induced by the reversible assembly of the plasmonic nanoparticles.[172]
Figure 8-.
Electrically induced reversible assembly of synthetic micro/nanoparticles. a) Reversible assembly of silver nanoparticles into large nanocrystal superlattices. Scale bar: 10 μm. Reprinted with permission of reference.[169] Copyright 2017, ACS. b) Reversible structural color based on a reversible assembly of colloids on an ITO electrode induced by application of a direct current electric field. Scale bar: 5 μm. Reprinted with permission of reference.[170] Copyright 2014, ACS. c) Assembly of metal–dielectric Janus colloids into programmable collective states by induced imbalanced polarizable interactions between the colloids. Scale bar: 30 μm. Reprinted with permission of reference.[175] Copyright 2016, Springer Nature. d) Three-dimensional hierarchical assembly of microrobots driven by the dielectrophoretic interactions between a structural frame and multiple microengines. Scale bar: 10 μm. Reprinted with permission of reference.[177] Copyright 2016, Wiley.
Dielectrophoretic methods have been used to perform assembly and sorting of particles with lower electric permeability. For example, the reversible assembly of diverse types of objects, including liquid droplets and cell-loaded hydrogel units, using electric fields was reported. This methodology used a two parallel plate electrode system to induce electrical assembly based on dielectrophoresis and electrowetting. Each building-block could be directed to assemble into three-dimensional (3D) structures.[173] The ability to assemble one dimensional (1D) structures was shown by the electrically-driven assembly of Janus ellipsoids into colloidal fibers. The ellipsoid shape of the microparticles limited the interaction that enabled formation into fibrillary assemblies.[174] The application of a perpendicular applied vertical electrical gradient resulted in the assembly of metal–dielectric Janus colloids into distinct types of swarms by inducing asymmetric interactions between the polarized particles. The mismatched dielectric responses of the metallic and dielectric hemispheres of the Janus colloids to the perpendicular electric field resulted in controlled locomotion and programmable particle-particle interactions. By tuning the amplitude and frequency the Janus colloids assembled into diverse structures, including clusters, chains, swarms and isotropic gas like formations (Figure 8c).[175] The assembly of asymmetric microparticles under electric-fields reported subunits exhibiting chirality, with such behavior generated in part by asymmetric hydrodynamics flows that caused the rotational motion of the assemblies. The use of an alternating current electric field generated dipole-based assemblies between a central microparticles and orbiting dimers.[176]
Moreover, electric fields have been used to power the locomotion of untethered robots, enabling them to interact and assemble into large swarms. Mixing electrically powered micron-sized Janus micromotors with passive silica particles resulted in the ability to create rotating cluster assemblies. For instance, under an applied alternating current electric field, motile micromotors served as seed particles that acted as anchors to passive microbeads, in which the steps of the cluster interaction defined the final structure. The alternating current electrical field induced one dipole at the center of the silica microbead, while the micromotor generated two dipoles at different hemisphere based on the different material properties of the Janus structure, thus generating the repulsion and attraction forces that led to the assembly of different structures with different rotation preferences. These assembled structures lost their stability upon removal of the electric field (Figure 8d).[177] Janus micromotors were also used as carriers of microbeads for targeted delivery using an externally applied electric field to induce the capture or repulsion of cargo carriers without the requirement of previous functionalization or labeling of the micromotor surface. The Janus micromotor presented the ability to perform on-the-fly capture and assembly of different targets ranging from 100 to 720 nm beads, controlled by modulating the applied voltage. The disassembly/release of the beads were achieved by changing the applied frequency.[178] PMMA-Ag Janus micromotors were coordinated to ensemble into assemblies that periodically expand and contract by changing the applied electric field. The synchronization of the micromotors motion created a beating assembly induced by each motor generated chemical field responding to nearby micromotors being attracted or repelled by it.[179] Dielectrophoretic interactions have been used to enable 3D modular assembly of microrobots based on electrically induced attractive interactions generated between a structural body and multiple microengines. Specifically, the application of an electric field induced the polarization of the main body, which generated an asymmetric tailored dielectrophoretic gradient localized at geometrical driven bonding sites that guided the microengines assembly into the desired locations of the microrobot frame. The 3D distribution of the engines resulted in different propulsion modalities and behavior, including axial and rotational motion. Moreover, the motion was used to drive the hierarchical assembly between two microrobots (Figure 8d).[180]
Electric fields can also assemble living microorganisms into assemblies via electrophoresis, where the microorganism migrate to an electrode presenting opposite charge, or by dielectrophoresis by inducing dipoles in the cells via an alternating current electric field. Usually, these methodologies are used for sample sorting and isolation applications. Bacterial assembly is a common process naturally happening in biofilms and plays an important role in many infectious diseases.[181] Despite different bacteria having different shapes and sizes, the clustering can be simplified to spherical colloids as building blocks interacting between each other and with other components in the media. The motion of microorganisms follows random run and tumble or chemotactic behavior towards nutrients. Nevertheless, the use of electrical fields has shown the ability to direct their assembly into bioassemblies based on the polarizability of their surface using an AC electric field.[182] In this directions, the effects of an AC field applied onto E. coli cultures have been studied.[183] AC electric fields applied to bacterial cultures have been shown to produce large-scale aggregates due to the strong attraction between bacteria. This transient phenomenon was reverted with the decrease or the removal of the electric field. The clustering competes with the steric repulsion of bacteria aiming to swim away. Therefore, a critical AC value is required to force the clustering of the living organism and induces the external flow to modify the trajectories of the bacteria. This electric-field controlled fluid flow resulted in long-range interactions between the cells and served to transport cells in the far field.
The induced polarization in bacteria induces the formation of assemblies aligned to the direction of the applied field. Micrococcus luteus bacteria that commonly cluster into tetrad structures (group of four units) under normal incubation conditions, were assembled into one dimensional structures composed of single chains or double helixes. The assemblies disintegrated upon removal of the field. At high frequencies, the dipolar interaction is stronger, creating elongated structures (Figure 9a). [184]
Figure 9-.
Electrically induced reversible assembly of living microorganism. a) Use of electrical induce assembly of multiple microorganisms into microwells. Scale bar: 60 μm. Reprinted with permission of reference.[184] Copyright 2020, ACS. b) Alternative current induced reversible assembly of bacterial tetrads, chain assembly and helix structures. Scale bar: 4 μm. Reprinted with permission of reference.[186] Copyright 2016, ElSevier. c) Reversible assembly of bacteria over the surface of a polarizable dielectric micromotors via dielectrophoresis. Reprinted with permission of reference.[191] Copyright 2020, AAAS.
The assembly of rod-shaped Salmonella Typhimurium in the presence of an AC electric field was also studied. At different field strengths, the bacteria generated different dimensional structures. The use of low fields induced the formation of line assemblies, while at higher fields two dimensional reversible arrays were generated and extended to 3D columnar structures when high bacterial concentration were used.[185] Microarrayed electrodes were used to assemble pathogenic Salmonella and E. coli into predetermined patterns using dielectrophoresis based assembly (Figure 9b).[186] In another case, the aggregation of S. cerevisiae cultures was studied under the presence of an electrical field. Interestingly, close-packed hexagonal 2D crystals were generated over time because of the lateral attraction between the formed chains when using four electrodes for applying an electric field in two perpendicular directions.[187] Interestingly, Yeast cells (S. cerevisiae) formed chains using this format, but did not formed 2D assemblies. Reversible disassembly was induced by turning off the applied electrical field. The use of functionalized microparticles with Concanavalin A, a protein that binds to the polysaccharides at the yeast cell, was used to permanently bond the live cell structures.[188] This concept of dielectrophoresis was exploited for creating hybrid bio-assemblies consisting of yeast bacteria, cells, and inorganic colloidal particles into one and two dimensional assemblies.[189] Application of dielectrophoretic forces were used for assembling microalgae (chlamydomonas reinhardtii) in 2D arrays. The response of the algae assemblies to diverse inorganic pollutants from solutions was used as a probe sensor.[190] E coli were assembled over the surface of a dielectric micromotor, which functions as a motile electrode, under the presence of an electrical field. The disassembly of bacteria was achieved when the electric field was turned off. Moreover, targeted electroporation of the assembled bacterial was achieved with a pulsed signal, which could lead to programable cell assemblies (Figure 9c).[191] In general, electrical assembly methods offer unique opportunities for assembling nanoparticles into large structures. Although their use in biological applications might be hindered by side effects caused by the applied electrical field.
Outlook
Tremendous progress has been made in engineering reversible assemblies at small scales during the last decade. In this direction, we reviewed the latest advances in guided reversible assembly of diverse building block, ranging from nanoparticles to living cells. The unique material properties of the building blocks govern the response to an external stimulus. For instance, magnetic fields enable assembly by acting on subunits through magnetophoresis or magnetic levitation of subunits. The acoustically driven reversible assembly is achieved by applying diverse types of acoustic waves that generate predefined patterns. Optical reversible assembly methods are based on the migration of particles towards optical traps or the change of surface interaction stimulated by light. Electrically driven assembly relies on tuning the polarizability of the building blocks and their response to the diverse alternating or direct electric fields though dielectrophoretic or electrophoretic mechanism. Reversible assembly provides alternatives for the fabrication of multiscale systems that are not possible by pure chemistry or traditional bottom up manufacturing. For instance, (i) Offering a high degree of design flexibility enabling to reconfigure the assembly in real time. (ii) Localizing and reusing the subunits with a high packing density, thus reducing material consumption. (iii) Giving dual functionality to a single type of building block, as shown in the reversible formation of reflective/transparent surfaces by assembling nanoparticles under an external stimulus. (iv) Tuning the formation rate to increase the resolution of the final structure; a feature that would be hard to control using instantaneous assembly methods. v) Enabling the assembly and guidance of large groups of micromotors as a single swarm, while retaining the individual micromotor ability to propel autonomously after removal of the external field. Nevertheless, despite such great progress there are still many challenges to overcome. Thus, we lay out a set of key challenges to set the foundation of future research:
How to scale up the fabrication towards macroscale assemblies?
How to direct the assembly using heterogeneous materials of different scales?
What applications can these assemblies uniquely enable in medicine and engineering?
How to interface externally-driven assembly and self-assembly to obtain different degrees of reversibility?
In order to advance the use of reversible assembly protocols from laboratory settings to practical applications, new developments will require interdisciplinary convergence between very different backgrounds such as biology, physics, computer science, electronics, and nanotechnology. The combinatorial use of external fields could expand the capabilities to fabricate more complex structures, as each assembly method has unique advantages for specific applications and types of subunits. Combining multiple external fields have enabled to generate more complex and tunable assemblies. For instance, in previous sections we covered how the combination of acoustic fields with magnetic[46,84] and optical fields[85,86] enabled dynamic reorganization of an assembly in real time. This was also shown for combinations of optical and electrical fields.[158]
Moreover, testing the response of novel shapes and materials to diverse external fields could result in an ever-expanding library of potential building blocks. As the field moves towards increasingly complex assemblies at small scales, it will become important to embrace machine learning and artificial intelligence towards advancing parallel object tracking, feedback loops, and real-time remote control. The development of novel methods of assembly could entail a new technological revolution enabling fabrication of materials, where intelligence is embedded in the relationship between the subunits, in a similar fashion to a brain being composed of millions of neurons working together for intelligence and consciousness. The design of reversible assemblies by external fields permits to fabricate constructs that would not otherwise be possible to fabricate. For example, the use of external stimuli to drive the formation of assemblies does not rely on gravity and has low material consumption due to their ability to preconcentrate subunits in close-pack structures. Thus, they would are applicable for constructing assemblies in space.[60] Reprogramming tissue engineering in real time inside a patient is another field of great potential for reversible assembly. Instead of relying on invasive surgery to implant tissue scaffolds or requiring a large quantity of cell to overcome the low targeting efficacy of injections of individual stem cells, externally guided cell assemblies could be reconfigured to fit and secure fixation at a target site.
Considering the great progress made in reversible assembly, we encourage researchers to look for unmet needs and applications by proposing problem orientated engineered structures or methodologies to streamline the adoption of externally driven fabrication methods. The methods may broaden their application areas in a similar way that 3D printing has become ubiquitous in rapid prototyping. We envision that the key challenges we pointed out can be gradually addressed, thus expanding the horizon and applications of guided bottom-up fabrication. The ability to finely tune and modulate the assembly of complex structures could result in diverse practical applications ranging from sensing, 3D biofabrication, tissue engineering, bioprinting, micromanufacturing, patterning and sample manipulation.
Acknowledgements
This work was supported by the CCNE-TD (5U54 CA19907502, NIH), the the International Alliance for Cancer Early Detection (ACED) pilot award, and the Canary Center at Stanford for Cancer Early Detection Seed Award. F.S was supported by Stanford Molecular Imaging Scholars program, 5R25CA118681. (NIH T32 postdoctoral fellowship) S.D. was supported by the Stanford Predictives and Diagnostics Accelerator (SPADA, part of the Stanford Center for Clinical and Translational Research).
Biography

Dr. Fernando Soto is a postdoctoral researcher at Stanford University School of Medicine, Canary Center for Cancer Early Detection, Department of Radiology, where he works on developing micro/nanorobots and bioengineered devices. He received his B.Sc. in Nanotechnology Engineering at Instituto Tecnológico de Tijuana, México and his Ph.D. in NanoEngineering at the University of California, San Diego.

Dr. Jie Wang is a postdoctoral researcher at Stanford University School of Medicine, with a research focus on developing micro/nanomaterial technologies. She has expertise in microfabrication, chemistry, material science for biomedical engineering applications. She has been developing micromotor fabrication and engineering technologies to isolate and detect biotargets from blood, as well as engineered biocompatible micromotors as a drug delivery carrier for tumor diagnostic and therapy.

Shreya Deshmukh is a Bioengineering PhD candidate at Stanford University in the Demirci group, and holds a BSc in Biomedical Engineering from Boston University. She works on developing portable measurement tools for global health applications, such as a microscale levitation tool to analyze malaria-infected blood in resource-limited settings, tested in Uganda. She also works on a portable platform to efficiently process backlogged sexual assault samples.

Dr. Utkan Demirci is a professor with tenure at Stanford University School of Medicine and serves as the interim Division Chief and Director of the Canary Center for Cancer Early Detection in the Department of Radiology. His group focuses on developing innovative microfluidic biomedical technology platforms with broad applications to multiple diseases. Some of his inventions have already been translated into FDA approved products serving patients. He has mentored and trained many successful scientists, entrepreneurs and academicians.
Footnotes
Conflict of Interest
Prof. Utkan Demirci (UD) is a founder of and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic IVF tools and imaging technologies, (ii) Koek Biotech, a company that is developing microfluidic technologies for clinical solutions, (iii) Levitas Inc., a company focusing on developing microfluidic sorters using magnetic levitation, and (iv) Hillel Inc., a company bringing microfluidic cell phone tools to home settings. UD’s interests were viewed and managed in accordance with the conflict of interest policies.
References
- [1].Whitesides GM, Grzybowski B, Science (80-. ). 2002, 295, 2418. [DOI] [PubMed] [Google Scholar]
- [2].Gurkan UA, Tasoglu S, Kavaz D, Demirel MC, Demirci U, Adv. Healthc. Mater. 2012, 1, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang J, Luijten E, Granick S, Annu. Rev. Phys. Chem. 2015, 66, 581. [DOI] [PubMed] [Google Scholar]
- [4].Grzelczak M, Liz-Marzán LM, Klajn R, Chem. Soc. Rev. 2019, 48, 1342. [DOI] [PubMed] [Google Scholar]
- [5].Vogel N, Retsch M, Fustin CA, Del Campo A, Jonas U, Chem. Rev. 2015, 115, 6265. [DOI] [PubMed] [Google Scholar]
- [6].Popkin G, Nature 2016, 529, 16. [DOI] [PubMed] [Google Scholar]
- [7].Mendes AC, Baran ET, Reis RL, Azevedo HS, Wiley Interdiscip. Rev. Nanomedicine Nanobiotechnology 2013, 5, 582. [DOI] [PubMed] [Google Scholar]
- [8].Guven S, Chen P, Inci F, Tasoglu S, Erkmen B, Demirci U, Trends Biotechnol. 2015, 33, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tasoglu S, Gurkan UA, Wang SQ, Demirci U, Chem. Soc. Rev. 2013, 42, 5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM, Nature 2009, 459, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].von Freymann G, Kitaev V, Lotsch BV, Ozin GA, Chem. Soc. Rev. 2013, 42, 2528. [DOI] [PubMed] [Google Scholar]
- [12].Shimomura M, Sawadaishi T, Curr. Opin. Colloid Interface Sci. 2001, 6, 11. [Google Scholar]
- [13].Truby RL, Emelianov SY, Homan KA, Langmuir 2013, 29, 2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kagan D, Balasubramanian S, Wang J, Angew. Chemie Int. Ed. 2011, 50, 503. [DOI] [PubMed] [Google Scholar]
- [15].Ibele M, Mallouk TE, Sen A, Angew. Chemie - Int. Ed 2009, 48, 3308. [DOI] [PubMed] [Google Scholar]
- [16].Askarieh G, Hedhammar M, Nordling K, Saenz A, Casals C, Rising A, Johansson J, Knight SD, Nature 2010, 465, 236. [DOI] [PubMed] [Google Scholar]
- [17].Ghosh A, Haverick M, Stump K, Yang X, Tweedle MF, Goldberger JE, J. Am. Chem. Soc. 2012, 134, 3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gurkan UA, Fan Y, Xu F, Erkmen B, Urkac ES, Parlakgul G, Bernstein J, Xing W, Boyden ES, Demirci U, Adv. Mater. 2013, 25, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li Y, Chen P, Wang Y, Yan S, Feng X, Du W, Koehler SA, Demirci U, Liu B-F, Adv. Mater. 2016, 28, 3543. [DOI] [PubMed] [Google Scholar]
- [20].Armstrong JPK, Stevens MM, Trends Biotechnol. 2020, 38, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Driscoll M, Delmotte B, Curr. Opin. Colloid Interface Sci. 2019, 40, 42. [Google Scholar]
- [22].Ariga K, Shrestha LK, Adv. Intell. Syst. 2020, 2, 1900157. [Google Scholar]
- [23].Yang D, Zhou C, Gao F, Wang P, Ke Y, Adv. Intell. Syst. 2020, 2, 1900101. [Google Scholar]
- [24].Genix AC, Oberdisse J, Soft Matter 2018, 14, 5161. [DOI] [PubMed] [Google Scholar]
- [25].Brzobohatý O, Karásek V, Šiler M, Chvátal L, Čižmár T, Zemánek P, Nat. Photonics 2013, 7, 123. [Google Scholar]
- [26].Berthelot J, Aćimović SS, Juan ML, Kreuzer MP, Renger J, Quidant R, Nat. Nanotechnol. 2014, 9, 295. [DOI] [PubMed] [Google Scholar]
- [27].Atajanov A, Zhbanov A, Yang S, Micro Nano Syst. Lett. 2018, 6, 1. [Google Scholar]
- [28].Grzelczak M, Vermant J, Furst EM, Liz-Marzán LM, ACS Nano 2010, 4, 3591. [DOI] [PubMed] [Google Scholar]
- [29].Stenhammar J, Wittkowski R, Marenduzzo D, Cates ME, Sci. Adv. 2016, 2, e1501850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu C, Xu T, Xu LP, Zhang X, Micromachines 2017, 9, 10. [Google Scholar]
- [31].Zhang J, Luijten E, Grzybowski BA, Granick S, Chem. Soc. Rev. 2017, 46, 5551. [DOI] [PubMed] [Google Scholar]
- [32].Tsang ACH, Demir E, Ding Y, Pak OS, Adv. Intell. Syst. 2020, 1900137. [Google Scholar]
- [33].Soto F, Wang J, Ahmed R, Demirci U, Adv. Sci. 2020, 2002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ozin GA, Manners I, Fournier-Bidoz S, Arsenault A, Adv. Mater. 2005, 17, 3011. [DOI] [PubMed] [Google Scholar]
- [35].Li J, De Ávila BEF, Gao W, Zhang L, Wang J, Sci. Robot. 2017, 2, DOI 10.1126/scirobotics.aam6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang H, Pumera M, Chem. Rev. 2015, 115, 8704. [DOI] [PubMed] [Google Scholar]
- [37].Mei Y, Solovev AA, Sanchez S, Schmidt OG, Chem. Soc. Rev. 2011, 40, 2109. [DOI] [PubMed] [Google Scholar]
- [38].Soto F, Kupor D, Lopez-Ramirez MA, Wei F, Karshalev E, Tang S, Tehrani F, Wang J, Angew. Chemie Int. Ed. 2020, 59, 3480. [DOI] [PubMed] [Google Scholar]
- [39].Tasoglu S, Diller E, Guven S, Sitti M, Demirci U, Nat. Commun. 2014, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xie H, Sun M, Fan X, Lin Z, Chen W, Wang L, Dong L, He Q, Sci. Robot. 2019, 4, DOI 10.1126/scirobotics.aav8006. [DOI] [PubMed] [Google Scholar]
- [41].Yu J, Wang B, Du X, Wang Q, Zhang L, Nat. Commun. 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Snezhko A, Aranson IS, Nat. Mater. 2011, 10, 698. [DOI] [PubMed] [Google Scholar]
- [43].Han K, Shields CW, Diwakar NM, Bharti B, López GP, Velev OD, Sci. Adv. 2017, 3, e1701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Diller E, Sitti M, Adv. Funct. Mater. 2014, 24, 4397. [Google Scholar]
- [45].Hsu A, Cowan C, Chu W, Mccoy B, Wong-Foy A, Pelrine R, Velez C, Arnold D, Lake J, Ballard J, Randall J, in Int. Conf. Manip. Autom. Robot. Small Scales, MARSS 2017 - Proc., Institute Of Electrical And Electronics Engineers Inc., 2017. [Google Scholar]
- [46].Ahmed D, Baasch T, Blondel N, Läubli N, Dual J, Nelson BJ, Nat. Commun. 2017, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tasoglu S, Yu CH, Gungordu HI, Guven S, Vural T, Demirci U, Nat. Commun. 2014, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].He L, Wang M, Ge J, Yin Y, Acc. Chem. Res. 2012, 45, 1431. [DOI] [PubMed] [Google Scholar]
- [49].Uebe R, Schüler D, Nat. Rev. Microbiol. 2016, 14, 621. [DOI] [PubMed] [Google Scholar]
- [50].Martel S, Mohammadi M, in Proc. - IEEE Int. Conf. Robot. Autom, 2010, pp. 500–505. [Google Scholar]
- [51].Vincenti B, Ramos G, Cordero ML, Douarche C, Soto R, Clement E, Nat. Commun. 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Puluca N, Durmus NG, Lee S, Belbachir N, Galdos FX, Ogut MG, Gupta R, Hirano K, Krane M, Lange R, Wu JC, Wu SM, Demirci U, Adv. Biosyst. 2020, 4, 1900300. [DOI] [PubMed] [Google Scholar]
- [53].Ilievski F, Mirica KA, Ellerbee AK, Whitesides GM, Soft Matter 2011, 7, 9113. [Google Scholar]
- [54].Durmus NG, Tekin HC, Guven S, Sridhar K, Yildiz AA, Calibasi G, Ghiran I, Davis RW, Steinmetz LM, Demirci U, Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tocchio A, Durmus NG, Sridhar K, Mani V, Coskun B, El Assal R, Demirci U, Adv. Mater. 2018, 30, DOI 10.1002/adma.201705034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tasoglu S, Khoory JA, Tekin HC, Thomas C, Karnoub AE, Ghiran IC, Demirci U, Adv. Mater. 2015, 27, 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Penland N, Choi E, Perla M, Park J, Kim D-H, Nanotechnology 2017, 28, 075103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tasoglu S, Yu CH, Liaudanskaya V, Guven S, Migliaresi C, Demirci U, Adv. Healthc. Mater. 2015, 4, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Parfenov VA, Mironov VA, Koudan EV, Nezhurina EK, Karalkin PA, Das Pereira F, Petrov SV, Krokhmal AA, Aydemir T, Vakhrushev IV, Zobkov YV, Smirnov IV, Fedotov AY, Demirci U, Khesuani YD, Komlev VS, Sci. Rep. 2020, 10, 4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Parfenov VA, Khesuani YD, Petrov SV, Karalkin PA, Koudan EV, Nezhurina EK, DAS Pereira F, Krokhmal AA, Gryadunova AA, Bulanova EA, Vakhrushev IV, Babichenko II, Kasyanov V, Petrov OF, Vasiliev MM, Brakke K, Belousov SI, Grigoriev TE, Osidak EO, Rossiyskaya EI, Buravkova LB, Kononenko OD, Demirci U, Mironov VA, Sci. Adv. 2020, 6, eaba4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mulvana H, Cochran S, Hill M, Adv. Drug Deliv. Rev. 2013, 65, 1600. [DOI] [PubMed] [Google Scholar]
- [62].Drinkwater BW, Lab Chip 2016, 16, 2360. [DOI] [PubMed] [Google Scholar]
- [63].Zhu Y, Serpooshan V, Wu S, Demirci U, Chen P, Güven S, in Methods Mol. Biol, Humana Press Inc., 2019, pp. 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xu F, Finley TD, Turkaydin M, Sung Y, Gurkan UA, Yavuz AS, Guldiken RO, Demirci U, Biomaterials 2011, 32, 7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Liu YJ, Ding X, Lin S-CS, Shi J, Chiang I-K, Huang TJ, Adv. Mater. 2011, 23, 1656. [DOI] [PubMed] [Google Scholar]
- [66].Bernassau AL, Glynne-Jones P, Gesellchen F, Riehle M, Hill M, Cumming DRS, Ultrasonics 2014, 54, 268. [DOI] [PubMed] [Google Scholar]
- [67].Ou FS, Shaijumon MM, Ajayan PM, Nano Lett. 2008, 8, 1853. [DOI] [PubMed] [Google Scholar]
- [68].Guo F, Mao Z, Chen Y, Xie Z, Lata JP, Li P, Ren L, Liu J, Yang J, Dao M, Suresh S, Huang TJ, Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sazan H, Piperno S, Layani M, Magdassi S, Shpaisman H, J. Colloid Interface Sci. 2019, 536, 701. [DOI] [PubMed] [Google Scholar]
- [70].Collins DJ, Devendran C, Ma Z, Ng JW, Neild A, Ai Y, Sci. Adv. 2016, 2, e1600089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen Y, Ding X, Steven Lin SC, Yang S, Huang PH, Nama N, Zhao Y, Nawaz AA, Guo F, Wang W, Gu Y, Mallouk TE, Huang TJ, ACS Nano 2013, 7, 3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Connacher W, Zhang N, Huang A, Mei J, Zhang S, Gopesh T, Friend J, Lab Chip 2018, 18, 1952. [DOI] [PubMed] [Google Scholar]
- [73].Zhang N, Manor O, Friend J, J. Acoust. Soc. Am. 2019, 146, 2866. [Google Scholar]
- [74].Akther A, Marqus S, Rezk AR, Yeo LY, Anal. Chem. 2020, DOI 10.1021/acs.analchem.0c01757. [DOI] [PubMed] [Google Scholar]
- [75].Courtney CRP, Ong C-K, Drinkwater BW, Wilcox PD, Demore C, Cochran S, Glynne-Jones P, Hill M, J. Acoust. Soc. Am. 2010, 128, EL195. [DOI] [PubMed] [Google Scholar]
- [76].Courtney CRP, Ong C-K, Drinkwater BW, Bernassau AL, Wilcox PD, Cumming DRS, Proc. R. Soc. A Math. Phys. Eng. Sci. 2012, 468, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Glynne-Jones P, Hill M, Lab Chip 2013, 13, 1003. [DOI] [PubMed] [Google Scholar]
- [78].Caleap M, Drinkwater BW, Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Xu T, Cheng G, Liu C, Li T, Zhang X, Chem. – An Asian J. 2019, 14, 2440. [DOI] [PubMed] [Google Scholar]
- [80].Wang W, Duan W, Zhang Z, Sun M, Sen A, Mallouk TE, Chem. Commun. 2015, 51, 1020. [DOI] [PubMed] [Google Scholar]
- [81].Zhou C, Zhao L, Wei M, Wang W, ACS Nano 2017, 11, 12668. [DOI] [PubMed] [Google Scholar]
- [82].Wang W, Castro LA, Hoyos M, Mallouk TE, ACS Nano 2012, 6, 6122. [DOI] [PubMed] [Google Scholar]
- [83].Xu T, Soto F, Gao W, Dong R, Garcia-Gradilla V, Magaña E, Zhang X, Wang J, J. Am. Chem. Soc. 2015, 137, 2163. [DOI] [PubMed] [Google Scholar]
- [84].Li J, Li T, Xu T, Kiristi M, Liu W, Wu Z, Wang J, Nano Lett. 2015, 15, 4814. [DOI] [PubMed] [Google Scholar]
- [85].Tang S, Zhang F, Zhao J, Talaat W, Soto F, Karshalev E, Chen C, Hu Z, Lu X, Li J, Lin Z, Dong H, Zhang X, Nourhani A, Wang J, Adv. Funct. Mater. 2019, 29, DOI 10.1002/adfm.201809003. [DOI] [Google Scholar]
- [86].Zhou D, Gao Y, Yang J, Li YC, Shao G, Zhang G, Li T, Li L, Adv. Sci. 2018, 5, 1800122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Takatori SC, De Dier R, Vermant J, Brady JF, Nat. Commun. 2016, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Shi Q, Di W, Dong D, Yap LW, Li L, Zang D, Cheng W, ACS Nano 2019, 13, 5243. [DOI] [PubMed] [Google Scholar]
- [89].Marzo A, Drinkwater BW, Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lim MX, Souslov A, Vitelli V, Jaeger HM, Nat. Phys. 2019, 15, 460. [Google Scholar]
- [91].Melde K, Mark AG, Qiu T, Fischer P, Nature 2016, 537, 518. [DOI] [PubMed] [Google Scholar]
- [92].Melde K, Choi E, Wu Z, Palagi S, Qiu T, Fischer P, Adv. Mater. 2018, 30, 1704507. [DOI] [PubMed] [Google Scholar]
- [93].Truby RL, Lewis JA, Nature 2016, 540, 371. [DOI] [PubMed] [Google Scholar]
- [94].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Adv. Mater. 2014, 26, 3124. [DOI] [PubMed] [Google Scholar]
- [95].Bose S, Vahabzadeh S, Bandyopadhyay A, Mater. Today 2013, 16, 496. [Google Scholar]
- [96].Wei G, Ma PX, Biomaterials 2009, 30, 6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gauvin R, Chen YC, Lee JW, Soman P, Zorlutuna P, Nichol JW, Bae H, Chen S, Khademhosseini A, Biomaterials 2012, 33, 3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Haake A, Neild A, Radziwill G, Dual J, Biotechnol. Bioeng. 2005, 92, 8. [DOI] [PubMed] [Google Scholar]
- [99].Neild A, Oberti S, Radziwill G, Dual J, Biotechnol. Bioeng. 2007, 97, 1335. [DOI] [PubMed] [Google Scholar]
- [100].Liu J, Kuznetsova LA, Edwards GO, Xu J, Ma M, Purcell WM, Jackson SK, Coakley WT, J. Cell. Biochem. 2007, 102, 1180. [DOI] [PubMed] [Google Scholar]
- [101].Kurashina Y, Takemura K, Friend J, Lab Chip 2017, 17, 876. [DOI] [PubMed] [Google Scholar]
- [102].Garvin KA, Hocking DC, Dalecki D, Ultrasound Med. Biol. 2010, 36, 1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chen P, Luo Z, Güven S, Tasoglu S, Ganesan AV, Weng A, Demirci U, Adv. Mater. 2014, 26, 5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Chen P, Güven S, Usta OB, Yarmush ML, Demirci U, Adv. Healthc. Mater. 2015, 4, 1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Armstrong JPK, Puetzer JL, Serio A, Guex AG, Kapnisi M, Breant A, Zong Y, Assal V, Skaalure SC, King O, Murty T, Meinert C, Franklin AC, Bassindale PG, Nichols MK, Terracciano CM, Hutmacher DW, Drinkwater BW, Klein TJ, Perriman AW, Stevens MM, Adv. Mater. 2018, 30, 1802649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chen P, Chen P, Wu H, Lee S, Sharma A, Hu DA, Venkatraman S, Ganesan AV, Usta OB, Yarmush M, Yang F, Wu JC, Demirci U, Wu SM, Biomaterials 2017, 131, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bouyer C, Chen P, Güven S, Demirtaş TT, Nieland TJF, Padilla F, Demirci U, Adv. Mater. 2016, 28, 161. [DOI] [PubMed] [Google Scholar]
- [108].Cai H, Ao Z, Hu L, Moon Y, Wu Z, Lu H-C, Kim J, Guo F, bioRxiv Bioeng. 2020, 2020.03.03.972299. [Google Scholar]
- [109].Canadas RF, Ren T, Marques AP, Oliveira JM, Reis RL, Demirci U, Adv. Funct. Mater. 2018, 28, 1804148. [Google Scholar]
- [110].Canadas RF, Ren T, Tocchio A, Marques AP, Oliveira JM, Reis RL, Demirci U, Biomaterials 2018, 181, 402. [DOI] [PubMed] [Google Scholar]
- [111].Ren T, Chen P, Gu L, Ogut MG, Demirci U, Adv. Mater. 2020, 32, 1905713. [DOI] [PubMed] [Google Scholar]
- [112].Ren T, Steiger W, Chen P, Ovsianikov A, Demirci U, Biofabrication 2020, 12, 025033. [DOI] [PubMed] [Google Scholar]
- [113].Gutiérrez-Ramos S, Hoyos M, Ruiz-Suárez JC, Sci. Rep. 2018, 8, 4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ding X, Lin SCS, Kiraly B, Yue H, Li S, Chiang IK, Shi J, Benkovic SJ, Huang TJ, Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Guo F, Li P, French JB, Mao Z, Zhao H, Li S, Nama N, Fick JR, Benkovic SJ, Huang TJ, Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ding X, Li P, Lin SCS, Stratton ZS, Nama N, Guo F, Slotcavage D, Mao X, Shi J, Costanzo F, Huang TJ, Lab Chip 2013, 13, 3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Li S, Guo F, Chen Y, Ding X, Li P, Wang L, Cameron CE, Huang TJ, Anal. Chem. 2014, 86, 9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Vanherberghen B, Manneberg O, Christakou A, Frisk T, Ohlin M, Hertz HM, Önfelt B, Wiklund M, Lab Chip 2010, 10, 2727. [DOI] [PubMed] [Google Scholar]
- [119].Collins DJ, Morahan B, Garcia-Bustos J, Doerig C, Plebanski M, Neild A, Nat. Commun. 2015, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Lu X, Zhao K, Liu W, Yang D, Shen H, Peng H, Guo X, Li J, Wang J, ACS Nano 2019, 13, 11443. [DOI] [PubMed] [Google Scholar]
- [121].Shen H, Zhao K, Wang Z, Xu X, Lu J, Liu W, Lu X, Micromachines 2019, 10, 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Lu X, Soto F, Li J, Li T, Liang Y, Wang J, ACS Appl. Mater. Interfaces 2017, 9, DOI 10.1021/acsami.7b15237. [DOI] [PubMed] [Google Scholar]
- [123].Lu X, Martin A, Soto F, Angsantikul P, Li J, Chen C, Liang Y, Hu J, Zhang L, Wang J, Adv. Mater. Technol. 2019, 4, DOI 10.1002/admt.201800374. [DOI] [Google Scholar]
- [124].Garg N, Westerhof TM, Liu V, Liu R, Nelson EL, Lee AP, Microsystems Nanoeng. 2018, 4, 1. [Google Scholar]
- [125].Ma Z, Holle AW, Melde K, Qiu T, Poeppel K, Kadiri VM, Fischer P, Adv. Mater. 2020, 32, 1904181. [DOI] [PubMed] [Google Scholar]
- [126].Klajn R, Stoddart JF, Grzybowski BA, Chem. Soc. Rev. 2010, 39, 2203. [DOI] [PubMed] [Google Scholar]
- [127].Manna A, Chen PL, Akiyama H, Wei TX, Tamada K, Knoll W, Chem. Mater. 2003, 15, 20. [Google Scholar]
- [128].Balasubramaniam S, Pothayee N, Lin Y, House M, Woodward RC, St. Pierre TG, Davis RM, Riffle JS, Chem. Mater. 2011, 23, 3348. [Google Scholar]
- [129].Klajn R, Wesson PJ, Bishop KJM, Grzybowski BA, Angew. Chemie - Int. Ed. 2009, 48, 7035. [DOI] [PubMed] [Google Scholar]
- [130].Rival JV, Nonappa ES Shibu, ACS Appl. Mater. Interfaces 2020, 12, 14569. [DOI] [PubMed] [Google Scholar]
- [131].Chen Y, Wang Z, He Y, Yoon YJ, Jung J, Zhang G, Lin Z, Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Dienerowitz M, J. Nanophotonics 2008, 2, 021875. [Google Scholar]
- [133].Juan ML, Righini M, Quidant R, Nat. Photonics 2011, 5, 349. [Google Scholar]
- [134].Grier DG, Nature 2003, 424, 810. [DOI] [PubMed] [Google Scholar]
- [135].Parreira R, Özelçi E, Sakar MS, Adv. Intell. Syst. 2020, 2000062. [Google Scholar]
- [136].Paterson L, MacDonald MP, Arlt J, Sibbett W, Bryant PE, Dholakia K, Science (80-. ). 2001, 292, 912. [DOI] [PubMed] [Google Scholar]
- [137].Yan Z, Gray SK, Scherer NF, Nat. Commun. 2014, 5, 1. [DOI] [PubMed] [Google Scholar]
- [138].Jaquay E, Martínez LJ, Mejia CA, Povinelli ML, Nano Lett. 2013, 13, 2290. [DOI] [PubMed] [Google Scholar]
- [139].Jaquay E, Martínez LJ, Huang N, Mejia CA, Sarkar D, Povinelli ML, Nano Lett. 2014, 14, 5184. [DOI] [PubMed] [Google Scholar]
- [140].Bhebhe N, Williams PAC, Rosales-Guzmán C, Rodriguez-Fajardo V, Forbes A, Sci. Rep. 2018, 8, 17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Chapin SC, Germain V, Dufresne ER, Opt. Express 2006, 14, 13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Chizari S, Lim MP, Shaw LA, Austin SP, Hopkins JB, Small 2020, 16, 2000314. [DOI] [PubMed] [Google Scholar]
- [143].Kong L, Mayorga-Martinez CC, Guan J, Pumera M, Small 2019, 1903179. [DOI] [PubMed] [Google Scholar]
- [144].Zheng J, Lam SH, Huang H, Shao L, Adv. Intell. Syst. 2020, 2, 1900160. [Google Scholar]
- [145].Hong Y, Diaz M, Córdova-Fteueroa UM, Sen A, Adv. Funct. Mater. 2010, 20, 1568. [Google Scholar]
- [146].Zhou D, Gao Y, Yang J, Li YC, Shao G, Zhang G, Li T, Li L, Adv. Sci. 2018, 5, 1800122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Singh DP, Choudhury U, Fischer P, Mark AG, Adv. Mater. 2017, 29, DOI 10.1002/adma.201701328. [DOI] [PubMed] [Google Scholar]
- [148].Mou F, Zhang J, Wu Z, Du S, Zhang Z, Xu L, Guan J, iScience 2019, 19, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhang Q, Dong R, Chang X, Ren B, Tong Z, ACS Appl. Mater. Interfaces 2015, 7, 24585. [DOI] [PubMed] [Google Scholar]
- [150].Palacci J, Sacanna S, Steinberg AP, Pine DJ, Chaikin PM, Science (80-. ). 2013, 339, 936. [DOI] [PubMed] [Google Scholar]
- [151].Vutukuri HR, Lisicki M, Lauga E, Vermant J, Nat. Commun. 2020, 11, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Lin Z, Si T, Wu Z, Gao C, Lin X, He Q, Angew. Chemie Int. Ed. 2017, 56, 13517. [DOI] [PubMed] [Google Scholar]
- [153].Aubret A, Youssef M, Sacanna S, Palacci J, Nat. Phys. 2018, 14, 1114. [Google Scholar]
- [154].Wang X, Baraban L, Misko VR, Nori F, Huang T, Cuniberti G, Fassbender J, Makarov D, Small 2018, 14, 1802537. [DOI] [PubMed] [Google Scholar]
- [155].Nahmias Y, Odde DJ, Nat. Protoc. 2006, 1, 2288. [DOI] [PubMed] [Google Scholar]
- [156].Ashkin A, Dziedzic JM, Yamane T, Nature 1987, 330, 769. [DOI] [PubMed] [Google Scholar]
- [157].Norregaard K, Jauffred L, Berg-Sørensen K, Oddershede LB, Phys. Chem. Chem. Phys. 2014, 16, 12614. [DOI] [PubMed] [Google Scholar]
- [158].Zou X, Zheng Q, Wu D, Lei H, Adv. Funct. Mater. 2020, DOI 10.1002/adfm.202002081. [DOI] [Google Scholar]
- [159].Kirkham GR, Britchford E, Upton T, Ware J, Gibson GM, Devaud Y, Ehrbar M, Padgett M, Allen S, Buttery LD, Shakesheff K, Sci. Rep. 2015, 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Chiou PY, Ohta AT, Wu MC, Nature 2005, 436, 370. [DOI] [PubMed] [Google Scholar]
- [161].Akolpoglu MB, Dogan NO, Bozuyuk U, Ceylan H, Kizilel S, Sitti M, Adv. Sci. 2020, 2001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Nagai M, Hirano T, Shibata T, Micromachines 2019, 10, DOI 10.3390/mi10020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Arlt J, Martinez VA, Dawson A, Pilizota T, Poon WCK, Nat. Commun. 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Chen F, Wegner SV, ACS Synth. Biol. 2017, 6, 2170. [DOI] [PubMed] [Google Scholar]
- [165].Sentürk OI, Schauer O, Chen F, Sourjik V, Wegner SV, Adv. Healthc. Mater. 2020, 9, 1900956. [DOI] [PubMed] [Google Scholar]
- [166].Yang X, Johnson S, Wu N, Adv. Intell. Syst. 2019, 1, 1900096. [Google Scholar]
- [167].Huang Y, Liang Z, Alsoraya M, Guo J, (Emma) Fan D, Adv. Intell. Syst. 2020, 1900127. [Google Scholar]
- [168].Fan DL, Zhu FQ, Cammarata RC, Chien CL, Nano Today 2011, 6, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Yu Y, Yu D, Orme CA, Nano Lett. 2017, 17, 3862. [DOI] [PubMed] [Google Scholar]
- [170].Shah AA, Ganesan M, Jocz J, Solomon MJ, ACS Nano 2014, 8, 8095. [DOI] [PubMed] [Google Scholar]
- [171].Zhang H, Cadusch J, Kinnear C, James T, Roberts A, Mulvaney P, ACS Nano 2018, 12, 7529. [DOI] [PubMed] [Google Scholar]
- [172].Montelongo Y, Sikdar D, Ma Y, McIntosh AJS, Velleman L, Kucernak AR, Edel JB, Kornyshev AA, Nat. Mater. 2017, 16, 1127. [DOI] [PubMed] [Google Scholar]
- [173].Chiang MY, Hsu YW, Hsieh HY, Chen SY, Fan SK, Sci. Adv. 2016, 2, e1600964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Shah AA, Schultz B, Zhang W, Glotzer SC, Solomon MJ, Nat. Mater. 2015, 14, 117. [DOI] [PubMed] [Google Scholar]
- [175].Yan J, Han M, Zhang J, Xu C, Luijten E, Granick S, Nat. Mater. 2016, 15, 1095. [DOI] [PubMed] [Google Scholar]
- [176].Ma F, Wang S, Wu DT, Wu N, Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Zhang J, Yan J, Granick S, Angew. Chemie Int. Ed. 2016, 55, 5166. [DOI] [PubMed] [Google Scholar]
- [178].Boymelgreen AM, Balli T, Miloh T, Yossifon G, Nat. Commun. 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhou C, Suematsu NJ, Peng Y, Wang Q, Chen X, Gao Y, Wang W, ACS Nano 2020, 14, 5360. [DOI] [PubMed] [Google Scholar]
- [180].Alapan Y, Yigit B, Beker O, Demirörs AF, Sitti M, Nat. Mater. 2019, 18, 1244. [DOI] [PubMed] [Google Scholar]
- [181].Battin TJ, Besemer K, Bengtsson MM, Romani AM, Packmann AI, Nat. Rev. Microbiol. 2016, 14, 251. [DOI] [PubMed] [Google Scholar]
- [182].Colla T, Mohanty PS, Nöjd S, Bialik E, Riede A, Schurtenberger P, Likos CN, ACS Nano 2018, 12, 4321. [DOI] [PubMed] [Google Scholar]
- [183].Igwe I, Joseph E, FUDMA J. Sci 2020, 4, 722. [Google Scholar]
- [184].Samantaray K, Mishra SR, Purohit G, Mohanty PS, ACS Omega 2020, 5, 5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Samantaray K, Bhol P, Sahoo B, Barik SK, Jathavedan K, Sahu BR, Suar M, Bhat SK, Mohanty PS, ACS Omega 2017, 2, 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Goel M, Verma A, Gupta S, Biosens. Bioelectron. 2018, 111, 159. [DOI] [PubMed] [Google Scholar]
- [187].Lumsdon SO, Kaler EW, Velev OD, Langmuir 2004, 20, 2108. [DOI] [PubMed] [Google Scholar]
- [188].Gupta S, Alargova RG, Kilpatrick PK, Velev OD, Soft Matter 2008, 4, 726. [DOI] [PubMed] [Google Scholar]
- [189].Gupta S, Alargova RG, Kilpatrick PK, Velev OD, Langmuir 2010, 26, 3441. [DOI] [PubMed] [Google Scholar]
- [190].Siebman C, Velev O, Slaveykova V, Biosensors 2015, 5, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Wu Y, Fu A, Yossifon G, Sci. Adv. 2020, 6, eaay4412. [DOI] [PMC free article] [PubMed] [Google Scholar]