Abstract
The very similar appearance of pollen of the New Zealand Myrtaceous taxa Leptospermum scoparium s.l. (mānuka) and Kunzea spp. (kānuka) has led palynologists to combine them in paleoecological and melissopalynological studies. This is unfortunate, as differentiation of these taxa would improve understanding of past ecological change and has potential to add value to the New Zealand honey industry, where mānuka honey attracts a premium price. Here, we examine in detail the pollen morphology of the 10 Kunzea species and a number of Leptospermum scoparium morphotypes collected from around New Zealand, using light microscopy, SEM, and Classifynder (an automated palynology system). Our results suggest that at a generic level the New Zealand Leptospermum and Kunzea pollen can be readily differentiated, but the differences between pollen from the morphotypes of Leptospermum or between the species of Kunzea are less discernible. While size is a determinant factor–equatorial diameter of Leptospermum scoparium pollen is 19.08 ± 1.28 μm, compared to 16.30 ± 0.95 μm for Kunzea spp.–other criteria such as surface texture and shape characteristics are also diagnostic. A support vector machine set up to differentiate Leptospermum from Kunzea pollen using images captured by the Classifynder system had a prediction accuracy of ~95%. This study is a step towards future melissopalynological differentiation of mānuka honey using automated pollen image capture and classification approaches.
Introduction
Honey from New Zealand mānuka (Myrtaceae: Leptospermum scoparium J.R.Forst. et G.Forst. agg.) attracts a premium value [1–3], arising from the medical benefits of its non-peroxide antibacterial activity [4]. This has created a demand from industry and regulators for accurate and cost-effective authentication testing of the product [5].
Melissopalynology–the pollen analysis of honey–is widely applied internationally, especially in Europe and America, in combination with chemical, physical, and sensory characters to indicate the approximate contributions of nectar from various plants to the honey [6–9]. However, using melissopalynology techniques to determine the nectar contribution of Leptospermum scoparium in New Zealand honeys has been hampered by the very similar appearance of Leptospermum scoparium pollen to pollen from New Zealand kānuka (Kunzea spp.). Kunzea Rchb., the closest relative to Leptospermum J.R.Forst. et G.Forst. in New Zealand [10], is widely distributed throughout the two main islands of New Zealand [11] and is often found growing with Leptospermum J.R.Forst. et G. Forst. It is also frequently visited by honeybees (Apis mellifera Linnaeus, 1758).
Until the 1980s three species of Leptospermum were regarded as endemic to New Zealand, L. scoparium J.R.Forst. et G.Forst. (“mānuka”), L. ericoides A.Rich. (“kānuka”) and L. sinclairii Kirk [12]. Thompson transferred L. ericoides to Kunzea, treating L. sinclairii as a synonym [13]. After a full taxonomic revision of the Kunzea ericoides complex including morphological, cytological and molecular variation as well as hybridisation experiments, de Lange [11] recognised ten Kunzea species, all endemic to New Zealand. Variation in Leptospermum scoparium is also well known, for example in flower colour, growth habit and yield of the antibacterial precursor chemical DHA (dihydroxyacetone) in nectar [12, 14–17]. Nevertheless, until recently only one species L. scoparium with two varieties, L. scoparium var. incanum Cockayne and L. scoparium var. scoparium have been accepted for New Zealand [12, 16, 18]. In her Australasian revision of the genus Thompson [13] only accepted the one species, L. scoparium for New Zealand but she did not critically examine New Zealand plants. At the time of writing, a New Zealand Department of Conservation funded revision of that nation’s Leptospermum scoparium is underway. Initial results confirm the findings of others, notably Buys et al. [19] that New Zealand populations of L. scoparium are distinct from those populations in Australia and Tasmania attributed to L. scoparium, and so are endemic. Buys et al. [19] also suggested that New Zealand plants are worthy of taxonomic segregation (see [16] and references therein). While the findings from the taxonomic revision of New Zealand L. scoparium are still pending, the morphotype analysed in this paper as L. aff. scoparium (c) “Waikato Peat Bog” has already been established as the new species L. repo de Lange et L.M.H.Schmid, and others are pending [16].
Because of their wide geographical distribution, and wide ecological envelope, pollen records of Leptospermum and Kunzea have received little focus in New Zealand palaeovegetation studies. As a result, limited effort has been assigned to pollen morphological study of the two forms. For example, in his seminal work on melissopalynology of New Zealand honeys Moar [20] referred to both forms simply as Leptospermum (although this was prior to Thompson’s 1989 transfer of L. ericoides to Kunzea), and in vegetation history studies he later classed them together as Leptospermum type [21]. However, differences in pollen morphology (at least on a statistical basis) had previously been observed by Pike [22], McIntyre [23], and Harris et al. [24], and later by Moar himself [25].
The increasing economic importance of mānuka honey has led to new focus on Leptospermum and Kunzea pollen [2]. In 2014, the Ministry for Primary Industries initiated preliminary studies to set up a standard for manuka honey. As part of this, we carried out a pilot study to investigate the possibility of differences between pollen of male and hermaphrodite (bisexual) flowers of New Zealand Leptospermum and Kunzea, both of which are at least in part andromonoecious [11, 26] (pers. obs. Newstrom-Lloyd). It was thought possible that the pollen of male and bisexual flowers may differ slightly in morphology, for instance in size, as has been noted in other andromonoecious plants (e.g., Sagittaria guayanensis Kunth [27]). Based on a small number of plant specimens of Kunzea robusta de Lange et Toelken and Leptospermum scoparium s.l. from the East Cape region we found no discernible difference in pollen of the two flower types within each genus, but a significant difference in average size between pollen of the two genera [28]. This conclusion agreed with the previous results of McIntyre [23] and Harris et al. [24] and was further confirmed by measurements of pollen from a small number of plant samples by Holt & Bebbington [29]. The results of our pilot study are summarised in S1 File.
As a result of these findings, in 2016 we set out to examine in detail the pollen morphology of the 10 New Zealand Kunzea species recently established by de Lange [11], and pollen from Leptospermum scoparium and putative segregates from that species, covering a wide geographic spread of populations throughout the New Zealand range, to determine:
if Leptospermum scoparium s.l. and Kunzea pollen can be differentiated, and
if subpopulations of L. scoparium pollen or interspecific variation in Kunzea exist.
A clear differentiation between New Zealand Leptospermum and Kunzea pollen could be used to establish a pollen-based mānuka honey standard and form the basis for mānuka honey melissopalynological testing. The results of our detailed study are reported here. We have not studied specimens of Australian Leptospermum species which occur in New Zealand as a few small, locally naturalised populations unlikely to be significant sources of honey, except possibly L. laevigatum on Matakana Island [30, 31].
In 2017 the New Zealand Government Ministry for Primary Industries (MPI) established an export standard for mānuka honey based on assay of characteristic chemical marker compounds and the presence of DNA of manuka pollen [1, 32], partly because of the then perceived difficulty in discrimination of Leptospermum and Kunzea pollen. At the present time, all New Zealand export honey passing the MPI tests is accepted as “mānuka” honey, without distinction as to geographic (and thus possibly specific) origin, or whether other nectar sources could be predominant. We contend that melissopalynological analysis remains a useful test because a single analysis can routinely quantify major pollen components (as well as non-pollen entities such as honey-dew fungal spores) and thereby provide a more comprehensive view of the nectar sources of a putative mānuka or kānuka honey. Pollen analysis also requires less specialised equipment and is thus more readily available to individual producers as well as overseas laboratories conversant with applying it as part of Codex Alimentarius requirements for certification of monofloral honeys. It may also be less expensive than the MPI test, especially if automated image recognition is routinely achieved.
Materials and methods
Sample collection
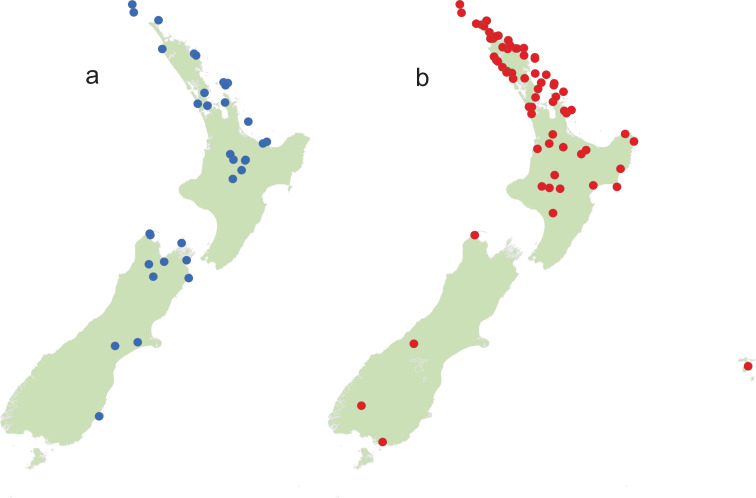
Fifteen geographic/genetic populations from Leptospermum scoparium were selected (see below) across New Zealand, representing 15 or fewer potential species segregates based on growth habit, leaf shape, flower size and colour, capsule shape, size and colour, and chemistry. Additionally, the 10 New Zealand species of Kunzea were selected, along with 10 specimens of putative interspecific Kunzea hybrids. In total, we sampled flowers from 135 herbarium specimens (at least 2 specimens for each Kunzea species and Leptospermum scoparium segregate were taken from both ends of their geographical range) from the Auckland War Memorial Museum herbarium (S1 and S2 Tables). The locations from which the plant specimens were collected are illustrated in Fig 1. Anthers from several fully opened flowers from each herbarium specimen were collected into pre-labelled 15 ml centrifuge tubes. Following the results of our pilot study that demonstrated no difference between the pollen from male and hermaphrodite flowers of Leptospermum scoparium s.l. and Kunzea robusta (S1 File), no attempt was made to separately select male and bisexual flowers in this study.
Fig 1.
Collection localities for (a) Kunzea and (b) Leptospermum scoparium specimens.
Leptospermum scoparium putative segregates
At the time this study was initiated in 2016 a taxonomic investigation of New Zealand Leptospermum scoparium populations had not been initiated. However, on examination of herbarium specimens (AK) considering the morphological variation evident it was decided to split those collections into putative segregates or “morphotypes”, to see if any could be differentiated by their pollen. While this investigation was in progress, four of the segregates (representing morphotypes “Auckland”, “Waikato Peat Bog”, “East Cape” and “Three Kings” of this paper respectively) were informally published as L. aff. scoparium (a), (c), (d), and (e) as part of a New Zealand-wide conservation assessment of the indigenous vascular flora [33]. In this paper we also examine a further 10 putative segregates (“Surville Cliffs”, “Central Volcanic Plateau”, “Coromandel Swamp”, “Flat Silver”, “Otaipango”, “Papa”, “South Island Mountain”, “Northern South Island”, “Southern South Island” and “Wellington”) and examples of L. scoparium var. incanum with pink-tinged or uniformly pink flowers. Buys et al. [19] included many of these putative segregates in their molecular analysis, finding evidence to merge some of this variation into subclades within a New Zealand (endemic) L. scoparium clade. For the purposes of this study, whilst a taxonomic revision is in progress, we retain usage of the putative segregates as we sampled them for this study, noting that, and as mentioned above, segregate (c) is now formally recognised as L. repo. We also note that putative segregates recognised as “Wellington”, “South Island Mountain”, “Southern South Island”, based on current thinking (P.J. de Lange unpubl.) may be best treated as L. scoparium s.s.–though this needs further study, and so we retain their usage here.
Pollen preparation
Dried herbarium flower samples were rehydrated overnight in deionized water with several drops of filtered detergent. After being vigorously stirred, each of the sample suspensions was sieved through a 90 μm nylon filter cloth to remove non-pollen flower parts. Pollen was then concentrated by centrifugation.
The residual pollen samples were then prepared using Erdtman’s acetolysis method [34]. Each sample was washed with glacial acetic acid, heated in a 9:1 mixture of acetic anhydride and concentrated sulphuric acid for 5 minutes at 95°C in a fume hood, returned to acetic acid, then twice washed in deionized water. Microscope slides were then made using glycerine jelly with safranin stain as the mounting medium.
Light microscopy and manual measurements
All photographs were taken with Zeiss AxioImager microscopes using 100× oil immersion objectives. Pollen grains in polar and equatorial orientations were measured either using an eyepiece graticule or using the measure function in Zen 3.2, an image-processing and analysis software from ZEISS microscopy. Visual measurements were made under the highest magnification available, using a 100× oil immersion objective, 1.6× tube factor, and a 10× eyepiece with an eyepiece graticule of 100 divisions. At this magnification, each division is equivalent to 0.625 μm. Measurements were made to the nearest half division.
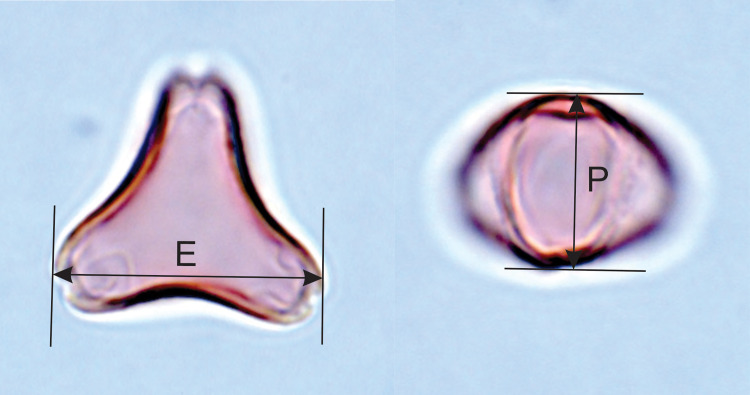
Two dimensions which are usually used to characterise angiosperm pollen grain size were measured: polar diameter (Lg max, longitudo: Iversen and Troels-Smith 1950 [35]; P: Erdtman, Nilsson and Praglowski 1992 [34]) and equatorial diameter (Lt+, longitudo transversa: Iversen and Troels-Smith 1950 [35]; E: Erdtman, Nilsson and Praglowski 1992 [34]), as shown in Fig 2. Polar diameter, P, can be measured only in equatorial views of the pollen grain. Equatorial diameter, E, can be measured in suitable equatorial and oblique views, as well as in polar views. As it is constrained to be the maximum dimension along a line at right angles to a line drawn through the grain centre and the midpoint of one of the apertures, it may or may not be equivalent to the maximum Feret diameter measured by Classifynder, depending on the equatorial contour shape (see below). In the oblate pollen grains of Leptospermum scoparium s.l. and Kunzea spp., which tend to lie in polar and oblique views, it is the dimension most readily measured.
Fig 2.
Acetolysed Leptospermum scoparium pollen in polar view (left) and equatorial view (right) showing equatorial (E) and polar (P) dimensions.
To avoid unconscious bias in the selection of pollen grains for measurement, every grain encountered in suitable orientation in one or two traverses across the middle part of the area under the microscope slide coverslip was measured. This was continued until totals of 30 equatorial diameters and 10 polar diameters were achieved. Folded or damaged pollen grains were ignored. Specimens of pale and thin-walled pollen grains were ignored as suspected immature pollen grains.
Scanning electron microscopy
Specimens from each of the 10 species of Kunzea spp. and 15 morphotypes of Leptospermum scoparium s.l. were selected, and all samples were prepared as described above, including dehydration, filtration and acetolysis treatment. One drop of pollen suspension from each sample was deposited on a conductive carbon tab attached to an aluminium stub and dried in a fume hood overnight. Specimens were then coated with gold, using a vacuum sputter coater, and examined using a JEOL Neoscope JCM-6000e microscope at 15 kV.
Digital image analysis: Automated capture
The 135 pollen samples were further analysed using a Classifynder version 3a. The Classifynder is an automated palynology system [36]. It includes an automated stage and image capture system, where pollen specimens are digitally photographed under monochromatic dark field illumination using a 40× dry objective lens. Fifty parameters are calculated from each pollen image. These include 7 core parameters that are shape- and size-related: elongation, compactness, convex hull, Heywood circularity, hydraulic radius, maximum Feret diameter, and area [29]. The remaining 43 parameters are mainly texture-related but include additional measurements of shape [36, 37]. The parameters measured were selected empirically during development of the system as those best able to allow discrimination between a range of modern pollen types [37]. Here, we used the Classifynder only for image capture and measurement, but used other tools for classification, described below.
A single microscope slide was examined for each sample. The stage configuration of the Classifynder version 3a restricts analysis to a 10×10 mm square within the 20×20 mm glass cover slips used in this study. For each sample, a 10×10 mm area was scanned by the Classifynder, and digital images of all pollen-like objects were automatically collected. These false-colour images were manually sorted to extract two groups of pollen: specimens orientated in polar view, and those orientated in equatorial view. All images properly presented in these two orientations were included, including the immature pollen grains which were ignored during light microscopic manual measurements. Results reported here are based on these two subsets of images. A median number of 158 pollen grains of either equatorial or polar view was captured from 122 samples (minimum = 16, maximum = 1290 grains). Results from the other 13 samples were discarded due to either scarcity or poor presentation of pollen grains. Although 10 Kunzea hybrids were included in the data processing stage, the results were only used as a reference guide but not incorporated in the data analysis.
Initial inspection of parameter plots revealed a sub-population of specimens with optical parameter values far from the main population, resulting in a bimodal distribution on parameter plots. Re-examination of digital images associated with these outlying specimens revealed various imperfections, mainly poor focus. These specimens were excluded from further analysis, discarding 1190 specimens with compactness parameter of <0.55. This resulted in a total dataset of 17835 Leptospermum and 5867 Kunzea pollen grains, of which approximately two-thirds were images of grains in polar view. The number of observations and summary statistics are reported in S3 and S4 Tables.
Parameter data from the pollen data set was processed using packages from the open-source software R (www.r-project.org). Exploratory data analyses to investigate the extent to which the full 50 Classifynder parameters or the 7 core parameters allowed discrimination between the various Leptospermum scoparium morphotypes and Kunzea species included discriminant function analysis (R package MASS) [38], and principal component analysis (PCA) (R package Vegan) [39]. The distribution of eigenvalues in the PCA results was illustrated using a highest posterior density region, calculated using R package emdbook [40].
Digital image analysis: Machine learning
We tested the skill of a Support Vector Machine predictive model at discriminating between Leptospermum and Kunzea in our 23702 pollen grain dataset [41]. This model, which is optimised to discriminate between two classes (here two pollen types), was tested by 99 iterations of a randomised 80:20 split of the data into training and test sets. We first tested the model on the entire pollen grain dataset, using first all 50 available parameters, then the core 7 parameters. Then, we tested the model on the subset of specimens in polar view, again using all 50 available parameters, then the core 7 parameters.
Results
The detailed pollen morphology descriptions that follow are revised from the original descriptions of McIntyre [23] and Moar [25] and are based on LM and SEM observations.
Leptospermum scoparium s.l. visual observation
Pollen grains are isopolar, oblate, triangular in polar view and flattened oval in equatorial view. In polar view, the side of the amb appears to be often concave and the angles to be extended. Pollen is tricolporate and angulaperturate, very rarely di- or tetracolporate. The ectoapertures are usually narrow, and the endoapertures are narrow but lalongate. Pollen grains are normally syncolpate, but very rarely slightly parasyncolpate due to slight widening of the colpi at the poles (however, polar islands are not present). The exine is always slightly patterned: obscurely flecked under LM, but clearly scabrate under SEM (Figs 3–5). The size of the P axis is 12.98 ± 1.66 μm (n = 188) and E axis 19.08 ± 1.28 μm (n = 490).
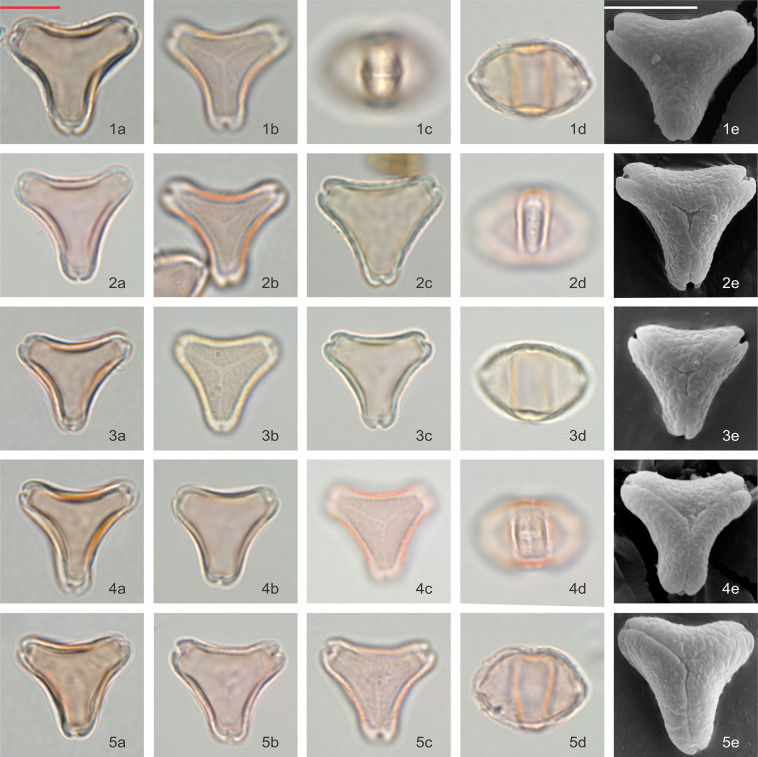
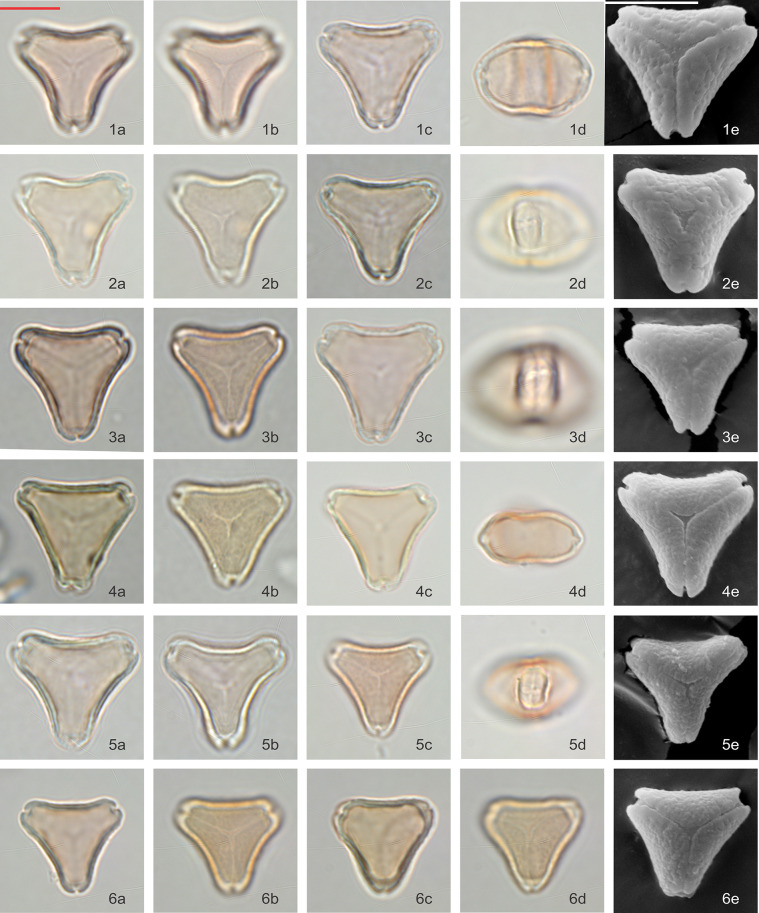
Fig 3. Polar and equatorial views of Leptospermum scoparium, Part 1.
Scale bar = 10 μm. (1a-1e) Surville Cliffs, (2a-2e) Flat Silver, (3a-3e) East Cape, (4a-4e) Waikato Peat Bog, (5a-5e) Coromandel Swamp.
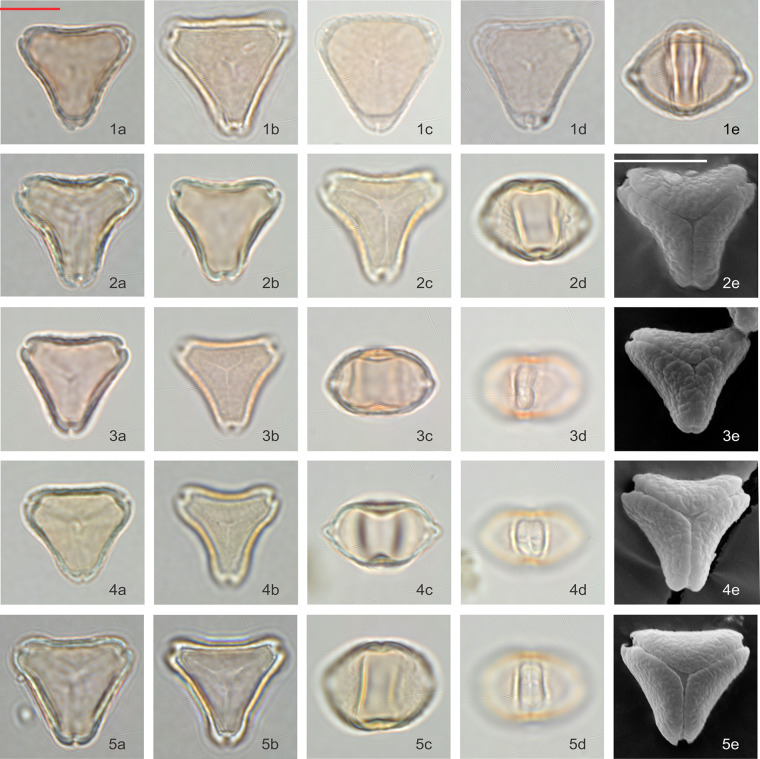
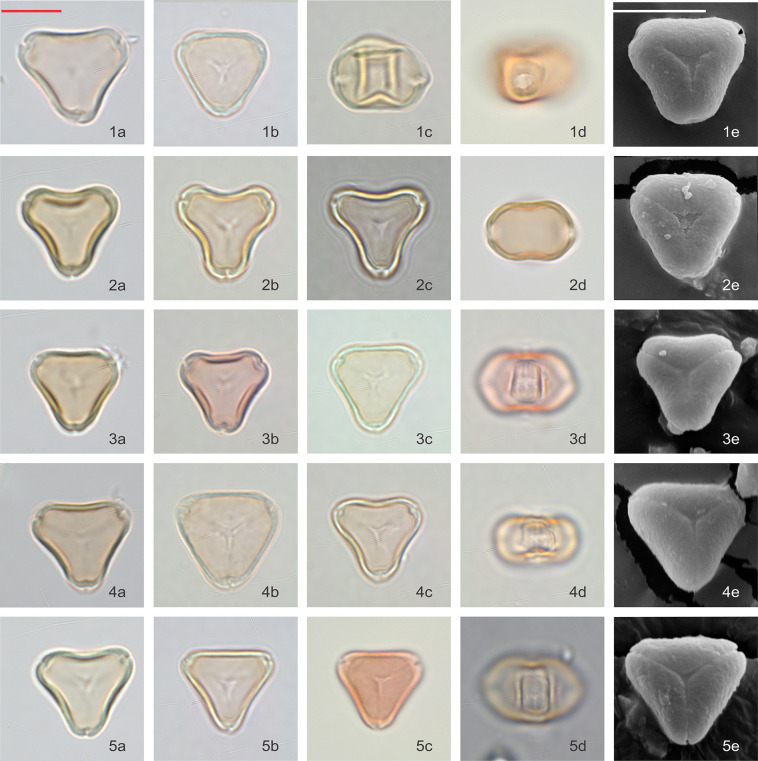
Fig 5. Polar and equatorial views of Leptospermum scoparium, Part 3.
Scale bar = 10 μm. (1a-1e) Otaipango, (2a-2e) Northern South Island, (3a-3e) Wellington, (4a-4e) Papa, (5a-5e) Southern South Island.
Fig 4. Polar and equatorial views of Leptospermum scoparium, Part 2.
Scale bar = 10 μm. (1a-1e) South Island Mountain, (2a-2e) Central Volcanic Plateau, (3a-3e) L. scoparium var. incanum, (4a-4e) Three Kings, (5a-5e, 6a-6e) Auckland.
Pollen of all 15 Leptospermum scoparium s.l. morphotypes were examined under LM and, except for “Otaipango”, under SEM. Acknowledging there is variability within each morphotype, and even within pollen populations collected from individual plants, we make the following observations about pollen morphological characters that assist with discrimination between the morphotypes (Table 1):
Table 1. Pollen morphological characters of Leptospermum scoparium s.l. morphotypes (visual observation).
| Morphotype | Size of Pollen Grains | Characters of Pollen Grains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E (μm) | P (μm) | Sides of Amb | Pattern | Vestibulum | Enlarged Apices | Thickened Arci |
||||
| Average | Range | Average | Range | Concave | Straight | |||||
| Otaipango | 19.2 | 17.0–20.8 | 14.4 | 12.1–16 | + | ++ | ++ | - | - | |
| Papa | 19 | 15.9–20.9 | 11.6 | 10.6–13 | + | ++,+ | ++ | + | + | |
| Wellington | 17.9 | 15.7–20.4 | 11.5 | 10.3–12.6 | + | ++ | +,- | - | - | |
| Northern South Island | 19.1 | 18.0–20.9 | 14.2 | 13.1–15.3 | + | ++ | - | + | + | |
| Southern South Island | 19.1 | 18.4–21.6 | 14.1 | 12.9–16.4 | + | + | - | + | - | |
| Three Kings | 20 | 18.2–21.3 | 10.3 | 9.3–12.1 | + | ++,+ | + | + | - | |
| East Cape | 18.1 | 16–20.7 | 11.6 | 10–13.1 | ++, + | + | - | + | + | |
| Flat Silver | 19 | 16.9–20.3 | 13.4 | 12.7–14.1 | ++, + | ++,+ | +,- | +,- | + | |
| Surville Cliffs | 20.6 | 18.2–22.1 | 13.6 | 13.1–15.1 | ++ | + | + | + | - | |
| Central Volcanic Plateau | 18.5 | 16.6–20.1 | 13 | 11.1–14.3 | + | + | - | + | + | |
| L. scoparium var. incanum | 19.4 | 17.3–21.9 | 16 | 13.5–17.5 | + | ++,+ | + | + | - | |
| South Island Mountain | 19.8 | 17.9–22.2 | 14 | 12.6–15.8 | + | + | + | +,- | + | + |
| Waikato Peat Bog | 18.6 | 16.5–20.1 | 13.4 | 12.7–14.3 | + | + | ++,- | - | +,- | - |
| Auckland | 18.8 | 16–22.4 | 11.7 | 10.7–13.1 | + | + | + | - | + | - |
| Coromandel Swamp | 18.3 | 17.1–20.3 | 14.3 | 12.8–16.1 | + | + | + | - | + | - |
++ denotes feature obviously present
+ denotes feature present
- denotes feature not obviously seen (multiple scores indicate a range of feature states)
Distinctly concave-sided pollen is more prominent in “Surville Cliffs”, “Flat Silver” and “East Cape”, whereas the amb of “Three Kings” and “Otaipango” pollen tends to be relatively straight.
Surface pattern with coarse texture, presented in patches, is especially noticeable in “Otaipango”, “Northern South Island” and “Wellington”, and common in “Papa”, “Three Kings”, L. scoparium var. incanum and “Flat Silver”.
Vestibulum–The ectexine slightly protrudes and is buckled along the sides of the amb and forms a gap with the endexine at the apices of the amb. This feature is usually present in “Otaipango” and “Papa”, and frequently seen in “Three Kings”, “Surville Cliffs” and L. scoparium var. incanum.
Rounded and slightly enlarged apices–this feature is generally distinct in many of the morphotypes except for “Otaipango” and “Wellington”.
Slightly thickened arci, only resolved under SEM, are commonly found in “East Cape”, “Flat Silver”, “Central Volcanic Plateau”, “Papa”, “South Island Mountain” and “Northern South Island” populations.
Both “Waikato Peat Bog” and “Coromandel Swamp” have large variations especially in the shape of their pollen grains, from concave to straighten sides of amb. Tetracolporate pollen grains are frequently observed in these two morphotypes.
Among the specimens examined, “Surville Cliffs” tends to have the largest pollen with average E-axis 20.6 μm, while “Wellington” appears to have the smallest with E-axis 17. 9 μm. A wider range of pollen size is observed within “Auckland” type due to the larger number of specimens examined.
Kunzea visual observations
Pollen grains are isopolar, oblate, triangular in polar view, and flattened oval in equatorial view. In polar view, the side of the amb appears to vary from slightly concave or straight, to slightly convex. Pollen is tricolporate and angulaperturate, very rarely di- or tetracolporate. Ectoapertures are usually narrow, and the endoapertures are narrow but lalongate.
Pollen grains are basically syncolpate, but more often appear to be slightly parasyncolpate, with small apocolpia at the poles. The exine appears uniformly psilate under LM and very obscurely patterned under SEM (Figs 6 and 7). The size of P axis is 11.28 ± 2.33 μm (n = 103), and E axis 16.30 ± 0.95 μm (n = 318).
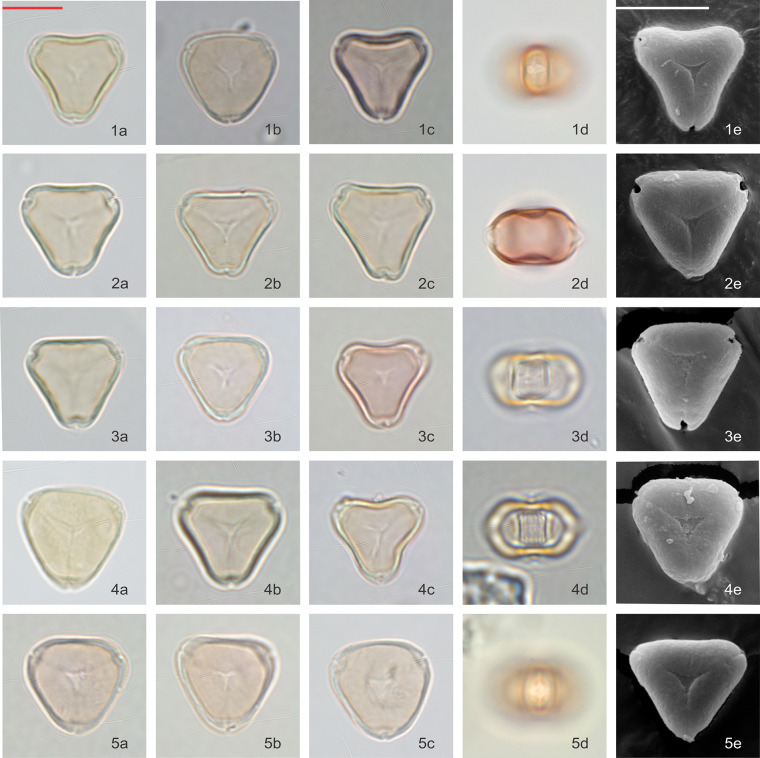
Fig 6. Polar and equatorial views of Kunzea, Part 1.
Scale bar = 10 μm. (1a-1e) K. toelkenii, (2a-2e) K. sinclairii, (3a-3e) K. serotina, (4a-4e) K. amathicola, (5a-5e) K. ericoides.
Fig 7. Polar and equatorial views of Kunzea, Part 2.
Scale bar = 10 μm. (1a-1e) K. linearis, (2a-2e) K. tenuicaulis, (3a-3e) K. salterae, (4a-4e) K. robusta, (5a-5e) K. triregensis.
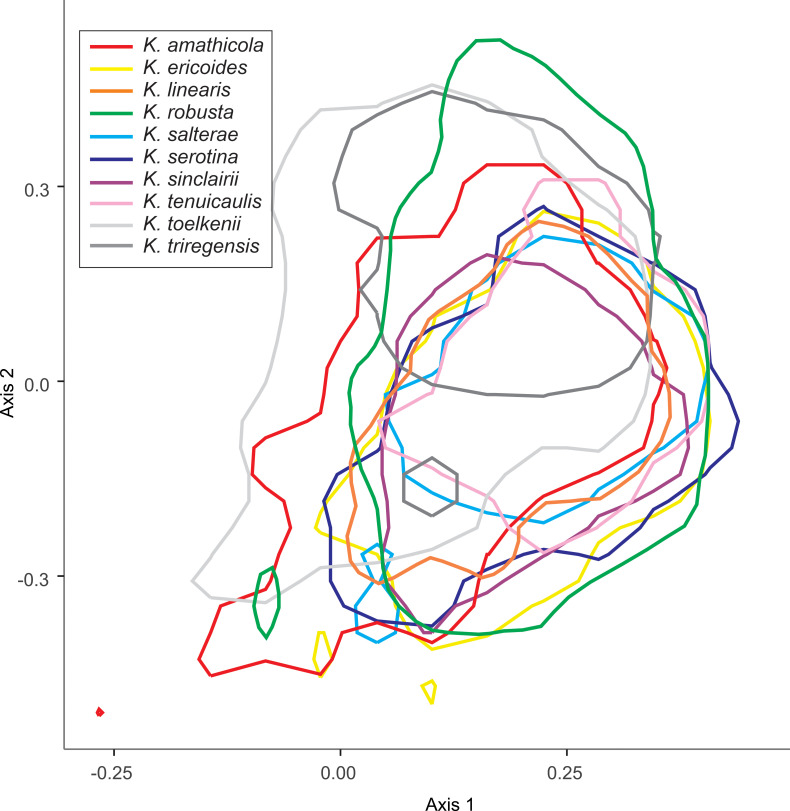
Of the ten species, K. toelkenii is the only species that could be separated from the others–by size–with mean E-axis of 17.8 μm compared with <17 μm for other Kunzea spp., along with slightly enlarged apices (Table 2, Fig 6). Of the remaining species, we could see no apparent difference in the size or surface texture. We do note some range of amb appearance in polar view, described in Table 2, but we observe considerable variability in all populations as well, such that we regard these observations a tendency, rather than a diagnostic feature. Given we do not observe consistent repeatable differences between the other species of Kunzea, we have not tabulated observations from the small number of hybrid combinations sampled.
Table 2. Pollen morphological characters of Kunzea species (visual observation).
| Kunzea species | Size of Pollen Grains | Characters of Pollen Grains | |||||||
|---|---|---|---|---|---|---|---|---|---|
| E (μm) | P (μm) | Sides of Amb | Pattern | Vestibulum | Enlarged Apices | Thickened Arci | |||
| Average | Range | Average | Range | ||||||
| K. toelkenii | 17.8 | 15.2–20 | 10 | 9.4–10.6 | slightly concave | - | - | + | - |
| K. linearis | 16.9 | 16.3–17.5 | 10.5 | 10–10.6 | slightly concave | - | - | - | - |
| K. amathicola | 16.8 | 15.6–18.1 | 9.6 | 8.8–11.3 | slightly concave | - | - | - | - |
| K. triregensis | 16.7 | 15.2–17.6 | 9.1 | 8.8–10 | slightly convex | - | - | - | - |
| K. tenuicaulis | 16.1 | 15.0–16.9 | 9.6 | 8.8–10.6 | straight | - | - | - | - |
| K. robusta | 16 | 15.0–17.5 | 9.9 | 9.4–10.6 | slightly concave | - | - | - | - |
| K. ericoides | 16.6 | 14.4–18.1 | 10.2 | 9.4–11.3 | slightly concave | - | - | - | - |
| K. sinclairii | 16.2 | 15.2–17.6 | 9.7 | 8.8–10 | more concave | - | - | - | - |
| K. salterae | 15.4 | 14.4–16.8 | 9.8 | 9.4–10.6 | straight | - | - | - | - |
| K. serotina | 15 | 13.8–16.3 | 9.4 | 8.8–10.6 | slightly concave | - | - | - | - |
+ denotes feature present
- denotes feature not obviously seen
Our size measurements of Kunzea pollen tend to be larger than those previously reported by de Lange [11]. This is likely due to the acetolysis treatment applied in this study, which tends to enlarge pollen grains [42, 43].
Classifynder measurements of Leptospermum scoparium s.l and Kunzea spp.
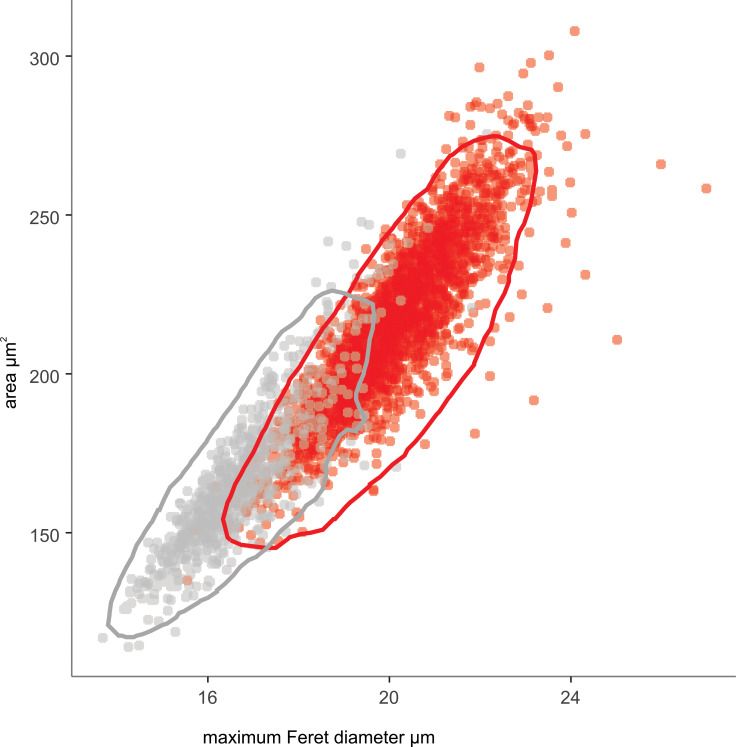
Inspection of distribution histograms (raw scan data is included in S5 Table) revealed maximum Feret diameter and area are the two parameters (of the 50 parameters described above) that most clearly differentiate Leptospermum scoparium s.l. and Kunzea pollen. This is consistent with the observations of Holt and Bebbington [29], who conducted a similar study on four samples of L. scoparium s.l. and Kunzea. An overlap between the two genera was observed for both parameters (Fig 8).
Fig 8. Classifynder measurements of area vs maximum Feret diameter of pollen in polar view.
Leptospermum scoparium (red, n = 11932 specimens) and Kunzea (grey, n = 3678). Polygons show the 95% range of the highest posterior density region for each species, calculated using R package coda [44].
Using the Classifynder, the 2.s.d. equatorial diameter of L. scoparium s.l. pollen is 17.93–22.18 μm (n = 11932), while the 2.s.d. equatorial diameter of Kunzea pollen is 14.32–19.00 μm (n = 3678), calculated from the maximum Feret diameter measurements (MFD) of polar view images. While the relative difference between L. scoparium s.l. and Kunzea was similar for both the visual measurements and the Classifynder, maximum Feret diameter from the Classifynder measurements is consistently 0.5–1 μm larger than light-microscope measurements (S1 and S2 Figs). One reason for this discrepancy is that for highly convex grains, the MFD measurement may not necessary be the equatorial diameter as defined in Fig 2, but a capture of a different axis, which is also observed by Holt and Bebbington [29]. This is supported by the observation that the smallest offsets between MFD and visual equatorial measurements are found in the Leptospermum scoparium morphotypes Otaipango, Flat Silver and Surville Cliffs, which have ambs either straight or slightly concave-sided, while relatively larger deviations are observed within convex-sided Kunzea species. We chose to rely on visual measurements of equatorial diameter for the taxonomic descriptions.
Ordination of Classifynder measurements
Although Linear Discriminant Analysis (LDA) seeks to maximize class separation (i.e., between-class variance), LDA of our pollen data produced a very similar result to the ‘unsupervised’ ordination of Principal Components Analysis. Both these analyses show distinct groups of L. scoparium s.l. and Kunzea pollen in ordination space, but with overlap: we report on and illustrate the PCA results here.
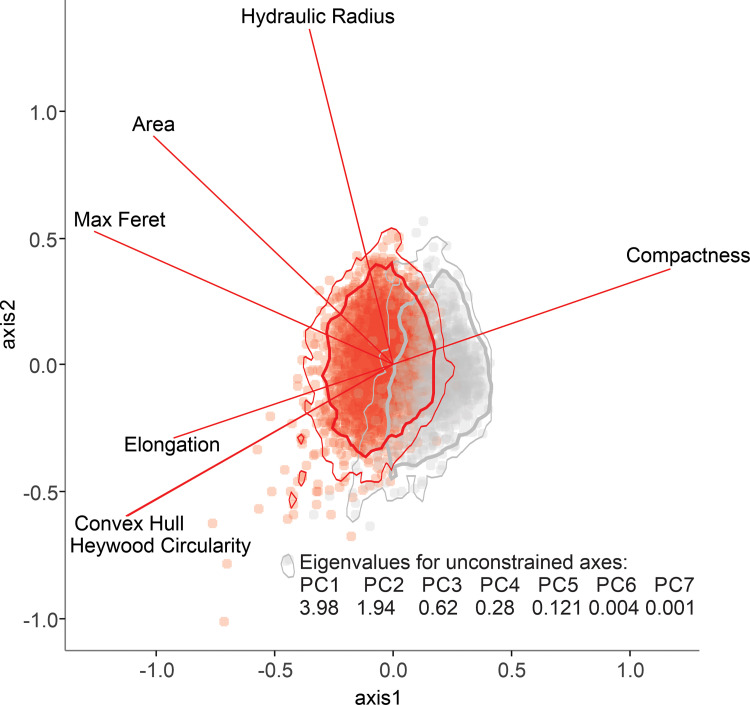
For a PCA constrained by the 7 ‘core’ parameters, 85% of the variance is described by the first two axes (Fig 9), while only 66% variance is described by the first two axes for a PCA constrained by all 50 parameters. Although there is overlap between L. scoparium s.l. and Kunzea pollen in the ordination, we see a clear differentiation between the two populations. In the ordination of the 7 ‘core’ parameters, the most important for discriminating between L. scoparium s.l. and Kunzea are maximum Feret diameter, compactness, convex hull, and Heywood Circularity.
Fig 9. Principal component analysis of the seven core parameters for Leptospermum scoparium and Kunzea pollen in polar view.
Polygons show the highest posterior density region for each species, thick lines = 95% range, thin lines = 99% range, calculated using R package coda [44].
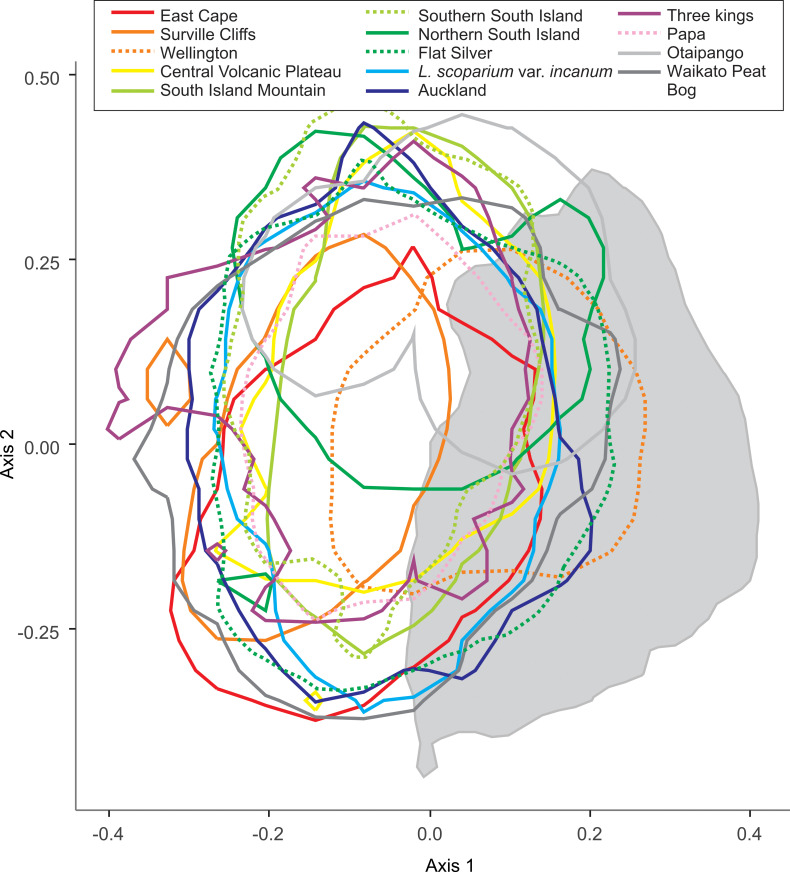
We explored the overlap of the morphotypes of L. scoparium, and the species of Kunzea, in subsequent PCA analyses. In both cases, we do not see strong differentiation between L. scoparium s.l. morphotypes (Fig 10), or Kunzea species (Fig 11).
Fig 10. Principal component analysis of the seven core parameters for Leptospermum scoparium morphotypes pollen in polar view.
Polygons show the 95% range highest posterior density region for each sub species. Underlying grey polygon is 95% range of Kunzea from Fig 9.
Fig 11. Principal component analysis of the seven core parameters for Kunzea species pollen in polar view.
Polygons show the 95% range highest posterior density region for each sub-species.
Differentiation of Leptospermum scoparium s.l. and Kunzea spp. by Support Vector Machine
The Support Vector Machine trial shows acceptable skill at discrimination between L. scoparium s.l. and Kunzea pollen in our dataset (Table 3). The best results are obtained where all 50 parameters are used to discriminate between specimens in polar view, with a mean prediction accuracy of 97.2%. Where only the seven core parameters are used, and the model is required to discriminate between specimens in both polar view and equatorial view, prediction accuracy drops to 94.7%.
Table 3. Cross-validation results of Support Vector Machine to predict pollen type, based on 99 iterations of random 80:20 splits of data into training:test sets.
| Model parameters | precision rate ± s.d. |
|---|---|
| 50 parameters, polar view and equatorial view | 96.1% ± 0.3 |
| 50 parameters, polar view only | 97.2% ± 0.3 |
| 7 parameters, polar view and equatorial view | 94.7% ± 0.3 |
| 7 parameters, polar view only | 95.9% ± 0.3 |
Discussion
Generic discrimination
The results of this study confirm observations from previous work based on smaller sample sizes. The large sample size and comprehensive sampling approach in the present study allows differences between the pollen of the New Zealand representatives of Leptospermum and Kunzea to be identified with more confidence. Firstly, there is a significant difference in average size between pollen of the two genera. Our results suggest that it should be possible to estimate the relative proportions of the two genera in a mixed population from a statistical study of equatorial pollen dimensions. Leptospermum scoparium s.l. pollen (18.88–21.50 μm, n = 11932) could be discriminated from Kunzea spp. pollen (15.49–17.83 μm, n = 3768) using the one standard deviation ranges of equatorial diameter. Only one species of Kunzea, K.toelkenii, has a pollen size comparable to that of the smallest pollen of Leptospermum scoparium morphotypes, e.g “Wellington” and part from “Auckland”. However, specimens of this large Kunzea pollen should be easily separated from small Leptospermum scoparium s.l. specimens on their less concave amb, less patterned surface, and presence of apocolpia.
In addition to the size, our observations indicate shape is another characteristic that allows discrimination of the two pollen types. Nearly all Leptospermum scoparium s.l. pollen is clearly concave-sided in polar view and more angular, while Kunzea pollen is generally nearly straight-sided. Pollen of K. sinclairii is slightly concave-sided, but with angles much less extended compared to Leptospermum scoparium s.l.. McIntyre [23] concluded that 55% of Kunzea pollen (as L. ericoides) have convex sides, compared with 4% of Leptospermum scoparium s.l. grains. It is hard to assess the range of the coverage of McIntyre’s specimens, taking the new taxonomic revision of Kunzea into account [11]. Another observation by McIntyre [23], that the colpi are more frequently parasyncolpate in Kunzea than in Leptospermum scoparium s.l., is confirmed by the present study.
The surface sculpture of Australian Leptospermum and Kunzea pollen was discussed by Thornhill et al. [45]. In contrast to Thornhill et al. [45], who noted both scabrate and psilate exine patterning in Australian Leptospermum and Kunzea pollen, our study of New Zealand grains suggests exine pattern can be used to distinguish between the two. For Leptospermum scoparium s.l., the exine tends to be more coarsely sculptured (i.e., scabrate), while for Kunzea pollen, the exine is less patterned and appears to be more psilate. While exine of L. scoparium s.l. exhibits different levels of coarseness, it is a feature that is consistently present through all specimens examined, and is clearly resolved under LM and prominent under SEM. In contrast, the exine of Kunzea pollen is consistently nearly psilate.
Variability within New Zealand Leptospermum and Kunzea
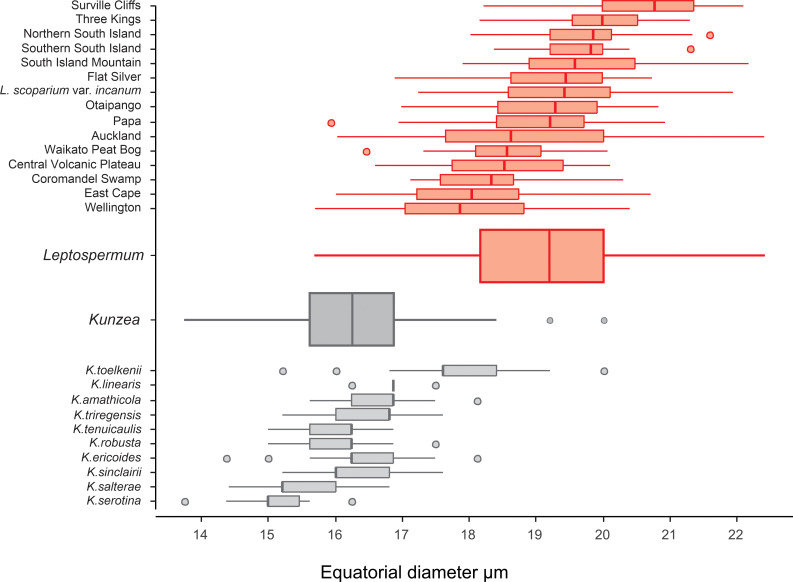
Although we see a clear distinction between pollen of Leptospermum scoparium s.l. and Kunzea spp., differences in pollen between the morphotypes of Leptospermum scoparium s.l. and between species of Kunzea are more subdued. For Kunzea, the average pollen size, combined with concaveness of amb and feature of apices, make it possible to separate K. toelkenii from the others under LM (Figs 6 and 12). Despite these noticeable characteristics, taking intraspecific variation into account, pollen of species within Kunzea spp. are practically indistinguishable from one another. For Leptospermum scoparium s.l., our initial results suggest that some morphological characteristics observed under LM, including size, texture, concaveness of amb, and presence of vestibulum, will be useful to characterise groups of morphotypes. However, Classifynder measurements made in this study do not support differentiation within species of Kunzea or morphotypes of Leptospermum scoparium. This could be partly due to limitations of the apparatus (as discussed below), partly to intraspecific pollen variation within each entity.
Fig 12. Size range of Leptospermum scoparium and Kunzea in equatorial diameter under light microscope.
Leptospermum scoparium–upper, red; Kunzea–lower, grey.
Intraspecific pollen variation is found in many of the Myrtaceae, but separation of ‘true’ variability from confounding issues of experiment design, such as variability in sample processing techniques or environmental morphotypes, remains a major challenge and a matter of debate [46]. Some types of morphological variations, such as the presence of four apertures (especially frequently presented in hybrids), are likely due to natural factors (e.g., genetic variation), as noted in some Australian Myrtaceae species [46]. Although a few aberrations, e.g. where a single flower produces two morphologically different pollen grains, are consistently found in some species of Eucalyptus and occasionally in other Myrtaceae species [46], this is rare or not apparent in morphotypes of Leptospermum scoparium s.l. we have examined. However, other types of morphological variation, for example variability in size, or shape, may reflect a combination of true variability and experimental treatment.
Pollen size can be affected by chemical treatment and mounting medium, maturity of grains, and pollen preservation [43, 47–51]. Although pollen grains shrink under dry and expand under wet conditions, we do not expect freshness of herbarium specimens to be a significant contributor to the intra-specific size variation observed, because shrinkage caused by dehydration is reversible [43]. In addition, because we have applied consistent preparation procedures, chemical treatment and mounting medium are not likely to be the cause of within-group variation in grain size. We do consider it likely that intraspecific size variability could arise from different levels of pollen maturity, along with other factors such as variable sporopollenin content, as discussed by Adeleye et al. [46]. In their study, a small proportion of larger, thinner-walled and convex-sided pollen grains were observed along with “normal” concave-sided grains from the same individual specimen of Leptospermum. This diversity is also observed in some of the samples we analysed and may be partly due to variation in exine development at different levels of maturity of pollen sampled from multiple anthers of the same specimen.
Utility of the Classifynder
The relative ease of collection of a large dataset, and the skill of the Support Vector Machine at discrimination between Leptospermum scoparium s.l. and Kunzea spp. pollen (~95%), shows promise for routine application of the Classifynder or similar device. In some respects we find this level of skill surprising, because some of the characteristics important for a human observer using a 100x oil immersion objective to discriminate between the two types are not available to the lower-resolution Classifynder. In particular, we expect the surface sculptural elements of pollen grains from Leptospermum scoparium s.l. and Kunzea spp., normally less than 1 μm, are of sizes at or below the limit of resolution of the digital imaging used in the Classifynder, which is approximately 1 μm [52]. More generally, we speculate that there may be potential to further improve the skill of a Classifynder-like machine for narrow two-class pollen discrimination problems such as this one. This is because the 50 features captured by the Classifynder were selected during machine design to allow discrimination of a wide range of pollen morphology, with a particular focus on 5 disparate end-member forms [37]. It is possible that a machine optimised for discrimination between a smaller range of pollen forms would be built to capture a different array of features.
Over the past decade, the rapid increase in capability in computational intelligence has resulted in considerable achievements in automatic pollen identification. Deep Learning Convolutional Neural Networks (DLCNN) is one of the most promising applications in the field of image recognition for pollen identification. Compared to a traditional neural network, DLCNN contains many more layers of neurons and is thus appropriate to very large data sets. Based on its capability to process and filter important signals from each layer of complex artificial neurons, DLCNN could extract the features that the network determines to be the most discriminatory aspects of the classes provided in the training sets. This process of feature identification is automatic and free from manual intervention, which obviates the procedure of manually selecting polar and equatorial orientation of pollen grains as practised in this study, while increasing reliability. As a result, over 97% accuracy on a set of 23 different pollen types from Brazil has been achieved by applying Deep Learning Convolutional Neural Networks (DLCNN) to a pollen image dataset composed of 805 microscope images [53]. A further exploration of DLCNN using a larger dataset of over 19,000 pollen images generated using the Classifynder produced even more promising results [54], with a classification success rate up to 98% across 46 different pollen types including New Zealand Leptospermum scoparium and Kunzea ericoides. Further improvements are likely, through increased sample sizes, refined measurement criteria, and improved discrimination networks. The deployment of the Classifynder for image capture and application of DLCNN for pollen classification could be a potential solution.
Conclusion and next steps
We have demonstrated that consistent differentiation between Leptospermum scoparium s.l. and Kunzea spp. pollen collected from New Zealand herbarium specimens is possible, using both visual and Classifynder light microscope measurements. The key characteristics that differentiate L. scoparium and Kunzea pollen are size, shape of amb, and surface texture. A Support Vector Machine trial based on the Classifynder measurements showed acceptable skill at discrimination between L. scoparium and Kunzea pollen. The best results were obtained when all 50 Classifynder parameters were used to discriminate between specimens in polar view, with a mean prediction accuracy of 97.2%.
While there is some indication that differentiation between pollen from some morphotypes of L. scoparium may be possible, further work on a larger sample set (preferably in conjunction with taxonomic studies) would be required to confirm this. An exploration of DLCNN on the larger dataset might have potential to improve the understanding of this issue.
These results could provide a first step to applying melissopalynology techniques to determine the relative contributions of L. scoparium s.l. and Kunzea nectar in manuka honey. Further analysis of honey samples would be required to determine if there are significant correlations between bioactivities and certain types of L. scoparium pollen. Application to paleoecological studies and the geological history of Myrtaceae in New Zealand are other fields yet to be explored.
Supporting information
(DOCX)
Comparison for Leptospermum scoparium (red) and Kunzea (grey) of equatorial diameter measured by palynologist using a light microscope, and maximum Feret diameter measured by Classifynder.
(TIF)
Comparison for Leptospermum scoparium (upper) and Kunzea (lower) of equatorial diameter measured by palynologist using a light microscope (denoted by suffix “_h”), and maximum Feret diameter measured by Classifynder (denoted by suffix “_d”).
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Ewen Cameron of Auckland Museum Herbarium (AK) for permission to destructively sample Kunzea and Leptospermum specimens under his care, and Dallas Mildenhall and Giuseppe Cortese for reviewing the `paper before submission. Andrew Boyes assisted with drafting Fig 1. We also thank two external reviewers for their constructive comments.
Data Availability
All relevant data are within the article and its Supporting Information files.
Funding Statement
An initial study of pollen of male and hermaphrodite flowers by JIR was funded by New Zealand Ministry for Primary Industry. XL received funding [no serial number] from KiwiNet (https://www.mbie.govt.nz/science-and-technology/science-and-innovation/funding-information-and-opportunities/investment-funds/preseed-accelerator-fund/kiwi-innovation-network-limited/) and GNS Science SSIF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McDonald CM, Keeling SE, Brewer MJ, Hathaway SC. Using chemical and DNA marker analysis to authenticate a high-value food, manuka honey. NPJ Science of Food. 2018;2(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ministry for Primary Industries [New Zealand]. 2020. Apiculture monitoring report data. Available from: https://www.mpi.govt.nz/dmsdocument/45082-2020-Apiculture-monitoring-report-data
- 3.Taunton E. Honey exports hit $425m export sweet spot. 2021. Feb 22. Available from: https://www.stuff.co.nz/business/farming/124318150/honey-exports-hit-425m-export-sweet-spot [Google Scholar]
- 4.Carter DA, Blair SE, Cokcetin NN, Bouzo D, Brooks P, Schothauer R, et al. Therapeutic manuka honey: no longer so alternative. Frontiers in Microbiology. 2016; 7. doi: 10.3389/fmicb.2016.00569 WOS:000374374100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quirke S, Ball R. Counterfeiting in the primary industry sector and the threat to New Zealand’s economy. National Security Journal. 201;1(1):61–74. doi: 10.36878/nsj201901.61 [DOI] [Google Scholar]
- 6.Codex Alimentarius Commission. Revised codex standard for honey codex Stan 12–1981, Rev. 1 (1987), Rev. 2 (2001). Codex Standard. 1981;12:1–7. [Google Scholar]
- 7.Sawyer R. Honey Identification. Cardiff, U.K.: Cardiff Academic Press; 1988. [Google Scholar]
- 8.D’Albore GR. Textbook of melissopalynology: Apimondia Publishing House; 1997. [Google Scholar]
- 9.Oddo LP, Piro R, Bruneau É, Guyot-Declerck C, Ivanov T, Piskulová J, et al. Main European unifloral honeys: descriptive sheets. Apidologie. 2004;35 (Suppl. 1):S38–S81. [Google Scholar]
- 10.de Lange PJ, Smissen RD, Wagstaff SJ, Keeling DJ, Murray BG, Toelken HR. A molecular phylogeny and infrageneric classification for Kunzea (Myrtaceae) inferred from rDNA ITS and ETS sequences. Australian Systematic Botany. 2010;23(5):309–19. [Google Scholar]
- 11.de Lange PJ. A revision of the New Zealand Kunzea ericoides (Myrtaceae) complex. PhytoKeys. 2014;(40):1–185. doi: 10.3897/phytokeys.40.7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan HH. Flora of New Zealand. Volume I. Christchurch: Manaaki Whenua Press; 1961. [Google Scholar]
- 13.Redefinitions Thompson J. and nomenclatural changes within the Leptospermum suballiance of Myrtaceae. Telopea. 1983;2(4):379–83. [Google Scholar]
- 14.Perry NB, Brennan NJ, Van Klink JW, Harris W, Douglas MH, McGimpsey JA, et al. Essential oils from mānuka and kānuka: Chemotaxonomy of Leptospermum. Phytochemistry. 1997;44:1485–1495. [Google Scholar]
- 15.Porter NG, Smale PE, Nelson MA, Hay AJ, Van Klink JW, Dean CM. Variability in essential oil and plant morphology within a Leptospermum scoparium population. New Zealand Journal of Botany. 1998;36:125–133. [Google Scholar]
- 16.de Lange P, Schmid LMH. Leptospermum repo (Myrtaceae), a new species from northern Aotearoa/New Zealand peat bog habitats, segregated from Leptospermum scoparium sl. Ukrainian Botanical Journal. 2021;78(4):247–265. doi: 10.15407/ukrbotj78.04.2472021 [DOI] [Google Scholar]
- 17.Adams CJ, Manley-Harris M, Molan PC. The origin of methylglyoxal in New Zealand mānuka (Leptospermum scoparium) honey. Carbohydrate Research. 2009;344:1050–1053. doi: 10.1016/j.carres.2009.03.020 [DOI] [PubMed] [Google Scholar]
- 18.Cockayne L. Notes on New Zealand floristic botany, including descriptions of new species &c. (No. 2). Transactions and Proceedings of the New Zealand Institute. 1917;49:56–65. [Google Scholar]
- 19.Buys MH, Winkworth RC, de Lange PJ, Wilson PG, Mitchell N, Lemmon AR, et al. The phylogenomics of diversification on an island: applying anchored hybrid enrichment to New Zealand Leptospermum scoparium (Myrtaceae). Botanical Journal of the Linnean Society. 2019;191(1):1–17. [Google Scholar]
- 20.Moar N. Pollen analysis of New Zealand honey. New Zealand Journal of Agricultural Research. 1985;28(1):39–70. [Google Scholar]
- 21.Moar N, Wilmshurst J, McGlone M. Standardizing names applied to pollen and spores in New Zealand Quaternary palynology. New Zealand Journal of Botany. 2011;49(2):201–29. [Google Scholar]
- 22.Pike KM. Pollen morphology of Myrtaceae from the south-west Pacific area. Australian Journal of Botany. 1956;4(1):13–53. [Google Scholar]
- 23.MacIntyre DJ. Pollen morphology of New Zealand species of Myrtaceae. Transactions of the Royal Society of New Zealand. 1963;2(7):83–107. [Google Scholar]
- 24.Harris W, Porter N, Dawson M. Observations on biosystematic relationships of Kunzea sinclairii and on an intergeneric hybrid Kunzea sinclairii × Leptospermum scoparium. New Zealand Journal of Botany. 1992;30(3):213–30. [Google Scholar]
- 25.Moar NT. Pollen grains of New Zealand dicotyledonous plants. Christchurch: Manaaki Whenua Press; 1993. [Google Scholar]
- 26.Primack RB, Lloyd DG. Andromonoecy in the New Zealand montane shrub manuka, Leptospermum scoparium (Myrtaceae). American Journal of Botany. 1980;67(3):361–8. [Google Scholar]
- 27.Huang SQ. Flower dimorphism and the maintenance of andromonoecy in Sagittaria guyanensis ssp. lappula (Alismataceae). New Phytologist. 2003;157(2):357–64. doi: 10.1046/j.1469-8137.2003.00676.x [DOI] [PubMed] [Google Scholar]
- 28.Raine JI, Li X. Pollen morphology of mānuka and kānuka. GNS Science Consultancy Report; 2014/118LR. [Google Scholar]
- 29.Holt KA, Bebbington MS. Separating morphologically similar pollen types using basic shape features from digital images: a preliminary study. Applications in Plant Sciences. 2014;2(8). doi: 10.3732/apps.1400032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogle CC, de Lange PJ, Cameron EK, Parris BS, Champion PD. Checklist of dicotyledons, gymnosperms and pteridophytes naturalised or casual in New Zealand: Additional records 2007–2019. Perspectives in Biosecurity. 2020;5:45–116. [Google Scholar]
- 31.Schönberger I, Wilton AD, Boardman KF, Breitwieser I, Cochrane M, de Lange PJ, et al. Checklist of the New Zealand Flora–Seed Plants. Lincoln: Manaaki Whenua Landcare Research; 2022. [Google Scholar]
- 32.Ministry for Primary Industries [New Zealand]. Criteria for identifying mānuka honey: A summary of the mānuka honey science programme, MPI Technical Paper No:2017/28. ISBN No: 978-1-77665-542-7 (online). Available from: https://www.mpi.govt.nz/dmsdocument/17314-criteria-for-identifying-manuka-honey-summary-report
- 33.de Lange PJ, Rolfe JR, Barkla JW, Courtney SP, Champion PD, Perrie LR, et al. Conservation status of New Zealand indigenous vascular plants 2017. New Zealand Threat Classification Series 22. Department of Conservation, Wellington. Available from: https://www.doc.govt.nz/documents/science-and-technical/nztcs22entire.pdf [Google Scholar]
- 34.Erdtman G, Nilsson ST, Praglowski J. Erdtman’s Handbook of Palynology. 2nd ed. Copenhagen: Munksgaard; 1992. [Google Scholar]
- 35.Iversen J, Troels-Smith J. Pollenmorfologiske definitioner og typer. Danmarks Geologiske Undersøgelse IV Række. 1950;3(8):1–54. [Google Scholar]
- 36.Holt K, Allen G, Hodgson R, Marsland S, Flenley J. Progress towards an automated trainable pollen location and classifier system for use in the palynology laboratory. Review of Palaeobotany and Palynology. 2011;167(3–4):175–83. [Google Scholar]
- 37.Zhang Y, Fountain DW, Hodgson RM, Flenley JR, Gunetileke S. Towards automation of palynology 3: Pollen pattern recognition using Gabor transforms and digital moments. Journal of Quaternary Science. 2004;19(8):763–8. [Google Scholar]
- 38.Ripley B, Venables B, Bates DM, Hornik K, Gebhardt A, Firth D, et al. Package ‘mass’. Cran r. 2013;538:113–20. [Google Scholar]
- 39.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, et al. Vegan: community ecology package. R package version 2.2–1. 2015. [Google Scholar]
- 40.Bolker B. Emdbook: support functions and data for “ecological models and data”. R package version. 2016; 1(9). [Google Scholar]
- 41.Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F, Chang C, et al. Misc. functions of the Department of Statistics, Probability Theory Group (formerly: E1071). Package e1071 TU Wien. 2015. [Google Scholar]
- 42.Christensen BB. Measurement as a means of identifying fossil pollen. Danmarks Geologiske Undersøgelse IV Række. 1946;3(2):1–22. [Google Scholar]
- 43.Reitsma T. Size modification of recent pollen grains under different treatments. Review of Palaeobotany and Palynology. 1969;9(3–4):175–202. [Google Scholar]
- 44.Plummer M, Best N, Cowles K, Vines K. CODA: convergence diagnosis and output analysis for MCMC. R News. 2006;6(1):7–11. [Google Scholar]
- 45.Thornhill AH, Wilson PG, Drudge J, Barrett MD, Hope GS, Craven LA, et al. Pollen morphology of the Myrtaceae. Part 3: tribes Chamelaucieae, Leptospermeae and Lindsayomyrteae. Australian Journal of Botany. 2012;60:225–259. [Google Scholar]
- 46.Adeleye MA, Hopf FVL, Haberle SG. Myrtaceae pollen morphology study from Bass Strait islands, Australia, is effective in separating region-specific fossil Myrtaceae pollen types. Review of Palaeobotany and Palynology. 2020;281:104273. [Google Scholar]
- 47.Andersen ST. Silicone oil as a mounting medium for pollen grains. Danmarks Geologiske Undersøgelse IV Række. 1960;4(1):1–24. [Google Scholar]
- 48.Fægri K, Deuse P. Size variations in pollen grains with different treatments. Pollen et Spores. 1960;2(2):293. [Google Scholar]
- 49.Faegri K, Kaland PE, Krzywinski K. Textbook of pollen analysis. John Wiley & Sons Ltd.; 1989. [Google Scholar]
- 50.Mäkelä EM. Size distinctions between Betula pollen types—a review. Grana. 1996;35(4):248–56. [Google Scholar]
- 51.Meltsov V, Poska A, Saar M. Pollen size in Carex: the effect of different chemical treatments and mounting media. Grana. 2008;47(3):220–33. [Google Scholar]
- 52.Allen G. An automated pollen recognition system. M. Sc Thesis, Massey University. 2006. Available from: https://mro.massey.ac.nz/bitstream/handle/10179/613/02whole.pdf
- 53.Sevillano V, Aznarte JL. Improving classification of pollen grain images of the POLEN23E dataset through three different applications of deep learning convolutional neural networks. PLoS ONE. 2018;13(9). doi: 10.1371/journal.pone.0201807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sevillano V, Holt K, Aznarte JL. Precise automatic classification of 46 different pollen types with convolutional neural networks. PLoS ONE. 2020;15(6). doi: 10.1371/journal.pone.0229751 [DOI] [PMC free article] [PubMed] [Google Scholar]