Abstract
The diversity of the methanotrophic community in mildly acidic landfill cover soil was assessed by three methods: two culture-independent molecular approaches and a traditional culture-based approach. For the first of the molecular studies, two primer pairs specific for the 16S rRNA gene of validly published type I (including the former type X) and type II methanotrophs were identified and tested. These primers were used to amplify directly extracted soil DNA, and the products were used to construct type I and type II clone libraries. The second molecular approach, based on denaturing gradient gel electrophoresis (DGGE), provided profiles of the methanotrophic community members as distinguished by sequence differences in variable region 3 of the 16S ribosomal DNA. For the culturing studies, an extinction-dilution technique was employed to isolate slow-growing but numerically dominant strains. The key variables of the series of enrichment conditions were initial pH (4.8 versus 6.8), air/CH4/CO2 headspace ratio (50:45:5 versus 90:9:1), and concentration of the medium (1× nitrate minimal salts [NMS] versus 0.2× NMS). Screening of the isolates showed that the nutrient-rich 1× NMS selected for type I methanotrophs, while the nutrient-poor 0.2× NMS tended to enrich for type II methanotrophs. Partial sequencing of the 16S rRNA gene from selected clones and isolates revealed some of the same novel sequence types. Phylogenetic analysis of the type I clone library suggested the presence of a new phylotype related to the Methylobacter-Methylomicrobium group, and this was confirmed by isolating two members of this cluster. The type II clone library also suggested the existence of a novel group of related species distinct from the validly published Methylosinus and Methylocystis genera, and two members of this cluster were also successfully cultured. Partial sequencing of the pmoA gene, which codes for the 27-kDa polypeptide of the particulate methane monooxygenase, reaffirmed the phylogenetic placement of the four isolates. Finally, not all of the bands separated by DGGE could be accounted for by the clones and isolates. This polyphasic assessment of community structure demonstrates that much diversity among the obligate methane oxidizers has yet to be formally described.
Methanotrophic bacteria are a physiologically unique group of microorganisms distinguished by their ability to use methane as sole source of carbon and energy (for a review, see reference 24). Microbial ecologists have long been interested in the methanotrophs due to their key role in the global methane cycle, oxidizing CH4 to CO2 (33). The first step in this process is catalyzed by the enzyme methane monooxygenase, of which two forms are known: the particulate methane monooxygenase, a membrane-bound enzyme present in all methanotrophs examined to date (50), and the soluble methane monooxygenase (sMMO), a cytoplasmic enzyme present in only select species (37). Both forms fortuitously oxidize a number of compounds, like the common environmental contaminant trichloroethylene (15, 56). Accordingly, this group of bacteria has attracted interest in regard to their potential for bioremediation (17, 26).
Landfills produce large amounts of CH4 due to methanogenic activity under anaerobic conditions (32). However, very high rates of CH4 oxidation and large methanotrophic populations have been reported in the oxic portion of landfill cover soils (30, 60). CH4 is a potent greenhouse gas, and methanotrophs play an important role in reducing the amount of CH4 released into the atmosphere (54). Thus, understanding the community structure of methanotrophic bacteria in landfill soil may help one better manage the attenuation of CH4 emission by these microorganisms.
One of the most fundamental discoveries made by microbiologists during the last decade has been the realization that the vast majority of bacteria in the environment have not yet been cultured (1). Using molecular biological tools and employing the 16S rRNA gene as a marker, microbiologists have been able to identify the presence of novel, uncultured organisms in situ. These 16S rRNA-based approaches have led to estimates of prokaryotic biodiversity and surveys of the microbial community structure of many environments (3, 9, 36, 57, 59, 61). At present, one of the most burning questions is the relationship between the microbial communities as described by these culture-independent methods and community structure assessment based on culturing. Investigators exploring the wide range of microbial diversity with universal- or domain-specific 16S rRNA probes or primers have only rarely reported the isolation of novel organisms whose presence was suggested by sequence analysis (29, 31, 46, 53). Often the reason is that genetic and metabolic traits cannot be predicted by the phylogenetic position of a clone sequence. Thus, the development of specific enrichment media remains troublesome.
For some bacterial groups, however, phylogenetic placement has been shown to be a reliable indicator of physiology. Traditionally, methane-oxidizing bacteria have been classified into three groups, based primarily on their biochemistry and morphological features (25). The type I methanotrophs employ the ribulose monophosphate pathway for formaldehyde assimilation and have disc-shaped bundles of intracytoplasmic membranes. The type II methanotrophs also assimilate carbon at the oxidation level of formaldehyde but use the serine pathway and show paired membrane structures at the periphery of the cell. The type X methanotrophs have disc-shaped bundles of intracytoplasmic membranes, use the ribulose monophosphate pathway (although they show a low level of activity for some of the enzymes in the serine cycle), and have a functional Calvin cycle. 16S ribosomal DNA (rDNA) sequence analysis has confirmed these three groupings. The type I methanotrophs form a phylogenetically coherent cluster within the gamma subdivision of the Proteobacteria (gamma-Proteobacteria), as do the type II methanotrophs within the alpha-Proteobacteria (8, 10). The type X species also fall within the gamma-Proteobacteria, but in a grouping distinct from type I. Recently, methanotroph taxonomy has been revised by a polyphasic approach, and the type I and type X methanotrophs have been included together in the family Methylococcaceae (6, 7). With one notable exception (14), it seems that within the alpha- and gamma-Proteobacteria no group of methanotrophs has any close relatives that are not methane utilizers (28).
We exploited the restricted phylogeny of the obligate methane oxidizers to design degenerate methanotroph-specific 16S rRNA PCR primers and used them to construct two clone libraries from DNA extracted directly from landfill soil, in an effort to describe the methanotrophic community structure in a culture-independent manner. Simultaneously, a culture-based diversity assessment was undertaken by using a serial dilution enrichment culture technique (11, 49) to isolate numerically dominant but potentially slow-growing species. Recently, novel methanotrophs have been isolated by employing medium of very low ionic strength and low pH (13, 14). Also, there is indication that the amount of CH4 available influences competition between the type I and type II methanotrophs (2, 22). Type I methanotrophs outcompete type II species under conditions of low CH4 and high O2, whereas type II species tend to dominate under the inverse conditions. These factors were varied in our enrichment scheme in an effort to isolate as wide a range of methanotrophs as possible.
MATERIALS AND METHODS
16S rDNA primer design and testing.
New degenerate type I and type II methanotroph primers were designed to amplify part of the 16S rRNA genes of all validly published methanotrophic bacteria sequenced to date. Potential primers were identified by aligning representative 16S rDNA sequences from all major radiations of the domain Bacteria to the type I and type II methanotroph sequences by using the PILE_UP program that is part of the University of Wisconsin’s Genetics Computer Group (GCG) sequence analysis software package. Selected regions unique to each group were identified by using GCG’s BOX_SHADE program, and these were tested against the GenBank, EMBL, and DDJB databases to check specificity. Putative probes were then analyzed for hairpin structures and the potential for probe duplex formation by using the OLIGO program (National Biosciences). The putative methanotroph-specific primers that emerged from these tests are listed in Table 1.
TABLE 1.
Methanotroph-specific primers and probes
| Name (sequence)a | Target genera | E. coli position | Description |
|---|---|---|---|
| MethT1bR (5′-GATTCYMTGSATGTCAAGG-3′) | Methylomonas Methylobacter Methylomicrobium Methylococcus | 988–1006 | Degenerate version of 1035-RuMP (10) broadened to include M. capsulatus; 100% nucleotide identity to unidentified Vibrionaceae clone (GenBank accession no. U14582), gas vacuolate strain 37 (U73721), gas vacuolate strain 174 (U73722), facultative barophile CNPT3 (U91588), marine psychrophile IC004 (U85849), and barophile WHB 46 clone 2 (X54745) |
| MethT1cR (5′-ATCCAATCGAGTTCCCAGGTTAAGCCC-3′) | Methylomonas | 614–640 | 100% identity to Chromatium purpuratum and Gluconobacter oxydans; one mismatch at 5′ end with many Erwinia spp. |
| Methylobacter | |||
| Methylomicrobium | |||
| Methylococcus | |||
| MethT1dF (5′-CCTTCGGGMGCYGACGAGT-3′) | Methylomonas | 84–102 | 100% identity to some Alcaligenes spp., Ectothiorhodospira spp., and Thiobacillus spp., as well as some uncharacterized bacteria |
| Methylobacter | |||
| Methylomicrobium | |||
| Methylococcus | |||
| MethT2R (5′-CATCTCTGRCSAYCATACCGG-3′) | Methylocystis | 997–1017 | Specific for the two target genera |
| Methylosinus |
Y represents C or T; R represents A or G; M represents A or C; S represents C or G.
Chromosomal DNA was extracted from all strains by using the Easy-DNA kit (Invitrogen) according to the manufacturer’s instructions. For the type I primer testing, DNAs from Methylococcus capsulatus (Bath), Methylobacter luteus ACM 3304, Methylomicrobium album BG8, and Methylomonas methanica S1 were used, with Pseudomonas putida ATCC 12633 and Escherichia coli B serving as negative controls. The type II primer set was tested on Methylosinus trichosporium OB3b, Methylocystis parvus OBBP, and negative controls Agrobacterium tumefaciens NT1 and Sinorhizobium meliloti 1021. PCR was performed by using Ready-to-Go PCR beads (Pharmacia) according to the manufacturer’s directions, except that the protocol was modified for use in a hot-start procedure with AmpliWax PCR gems (Perkin-Elmer). Briefly, 20 μl of distilled, deionized H2O and an Ampliwax gem was added to the PCR bead and heated at 75°C for 5 min. The wax was allowed to cool at room temperature. Then template DNA (approximately 10 to 20 ng) and primers (1 μl each at 10 μM, for a final concentration of 0.4 μM) in 5 μl of double-distilled H2O were added to the top of the cooled wax. PCR was then carried out in a thermocycler under the following conditions: 2 min 30 s at 94°C, followed by 10 cycles of 94°C for 30 s, 56°C for 45 s, and 72°C for 1 min, and then 10 cycles of 94°C for 30 s, 56°C for 1 min, and 72°C for 1 min 30 s, followed by 13 cycles of 94°C for 30 s, 56°C for 1 min 15 s, and 72°C for 2 min 15 s. The final extension step was at 72°C for 7 min 30 s. The presence and size of amplification products were determined by agarose gel electrophoresis and ethidium bromide staining.
Sampling and soil characteristics.
A soil sample was collected from the upper 10-cm soil layer at the Athens-Clarke County Municipal Landfill, Athens, Georgia, on 5 August 1998. The sampling area was a former refuse disposal area that accepted solid waste from 1987 to 1996 and was covered with approximately 2 feet of surrounding soil in 1996 (47a). Visible inspection revealed the soil sample to be mostly red clay. The soil was analyzed for carbon, hydrogen, and nitrogen content by the University of Georgia chemical analysis lab by using a carbon-hydrogen-nitrogen analyzer (Perkin-Elmer 240C elemental analyzer) and for copper content by using an inductively coupled plasma-mass spectrometer. The sample was found to contain 0.48% N, 3.93% C, and 1.80% H by weight, and the copper content of the soil was 139.22 ppm. The soil pH was 4.93, as determined by adding an equal volume of deionized, distilled water to the soil and measuring with a combination pH probe.
DNA extraction.
DNA was extracted and purified directly from the landfill soil by using the FastDNA SPIN Kit for Soil (Bio101). The procedure was slightly modified from that given by the manufacturer in the following areas. To exclude any extracellular DNA present in the sample (55), 25 ml of sterile sodium phosphate buffer (120 mM, pH 7.6) was added to 10 g of soil and the mixture was placed on a shaker table for 10 min at 150 rpm. The slurry was centrifuged at 6,000 × g for 10 min. One-half gram of the washed soil was added to the MULTIMIX2 tissue matrix tube, and 978 μl of the sodium phosphate buffer and 122 μl of the MT buffer were added. The tube was then secured in a mini-beadbeater (Biospec Products) and processed for two 1-min intervals with an intervening minute on ice. The MULTIMIX2 tubes were centrifuged at 14,000 × g for 30 s, 250 μl of the PPS reagent was added to the supernatant, and the tube was inverted by hand 10 times. Precipitate was pelleted at 14,000 × g for 5 min and divided into two tubes to which 0.5 ml of binding matrix suspension was added. The tubes were placed on a rotator for 2 min and then placed in a rack for 3 min to facilitate settling of the matrix. Approximately 250 μl of the supernatant was discarded from each tube, and the remaining binding matrix was pooled, transferred to a SPIN filter, and centrifuged at 14,000 × g for 1 min. Five hundred microliters of SEWS-M was added to the SPIN filter and centrifuged at 14,000 × g for 1 min. The remaining matrix was then dried by centrifuging at 14,000 × g for 2 min and allowed to air dry for 5 min. DNA was eluted from the binding matrix with 100 μl of double-distilled H2O. The resultant DNA was quantified by using a fluorometer with Hoescht dye 33258 (34). The final yield was approximately 9.5 μg of DNA/g (wet weight) of soil.
PCR and construction of clone libraries.
Purified environmental DNA was amplified by PCR by using Ready-to-Go PCR beads (Pharmacia) according to the manufacturer’s instructions. To amplify type I 16S rDNA sequences present in the environmental DNA, the primers MethT1bR and MethT1dF (Table 1) were used at a final concentration of 0.4 μM each, along with approximately 100 ng of environmental DNA as the template. Type II sequences were amplified with primer MethT2R (Table 1) and the Bacteria-specific primer 27F (5′-GAGTTTGATCMTGGCTCAG-3′) (35), both at a final concentration of 0.4 μM and with approximately 100 ng of the environmental DNA. The specifics of the PCR were as described above in the primer testing section. PCR products were visualized on an agarose gel, the correct size was confirmed, and the bands were excised and purified by using the Prep-a-Gene (Bio-Rad) gel purification protocol. To construct the type I and type II clone libraries, the purified PCR products were cloned into pCR2.1 by using the original TA cloning kit (Invitrogen) following the procedure recommended by the manufacturer.
Enrichment conditions and strain isolation.
An extinction-dilution technique was used to isolate the numerically dominant methanotrophs from the landfill soil (11, 49). Seven dilution series were performed with combinations of three variables. First, the initial pH of the medium was adjusted with concentrated phosphoric acid to 4.8 (from the standard 6.8) for some enrichment series. Second, the enrichments were performed under different air/CH4/CO2 headspace ratios (50:45:5 versus 90:9:1). Lastly, the concentration of the nitrate minimal salts (NMS) medium (25) was diluted fivefold (1× NMS medium versus 0.2× NMS medium). The specific conditions of each enrichment series and its letter designation used for the isolate names are given in Table 2. For each series, 0.2 g (wet weight) of landfill soil was added to 1.8 ml of medium and the tube was vortexed vigorously for 3 min before the mixture was serially diluted to a final dilution of 10−10. After 8 days of incubation at 30°C with moderate shaking at 150 rpm, the highest dilution displaying visual turbidity was plated on the same medium solidified with highly purified agar (Becton Dickinson). Colonies from plates were repeatedly picked and streaked on the same medium and incubated under the same conditions in an effort to achieve purity. Culture purity was determined by incubating putative methanotroph isolates with and without methane, checking for growth on nutrient agar, and phase-contrast microscopy. Despite repeated streaking, we were unable to obtain some type II methanotrophs in pure culture. When such cultures were examined microscopically, small cells with morphological features typical of Hyphomicrobium spp. were observed. Hyphomicrobia and similar bacteria are known to copurify with methanotrophs; they likely utilize methanol or other organic waste products that the methanotrophs excrete (23, 25). This was not always an obligate association, however. Many type II methanotrophs, including the novel type II isolates AML-A3 and AML-A6, could be purified with repeated streaking. We were still able to screen methanotrophs not in pure culture by exploiting the specificity of the type I and type II primer pairs described above on DNA isolated from the mixed methanotrophic cultures.
TABLE 2.
Distribution of type I and type II methanotrophs isolated under various extinction-dilution enrichment series
| Enrichment conditions
|
Methanotroph isolate affiliationc
|
||||
|---|---|---|---|---|---|
| Series letter designation | NMS medium strengtha | Initial pHb | Approx air/CH4/CO2 headspace ratio | Type I | Type II |
| A | 0.2× | 4.8 | 90:9:1 | 0 | 9 |
| C | 1× | 4.8 | 90:9:1 | 7 | 0 |
| D | 1× | 6.8 | 90:9:1 | 3 | 0 |
| E | 0.2× | 4.8 | 50:45:5 | 0 | 6 |
| F | 0.2× | 6.8 | 50:45:5 | 2 | 4 |
| G | 1× | 4.8 | 50:45:5 | 3 | 0 |
| H | 1× | 6.8 | 50:45:5 | 2 | 0 |
For the lower-strength enrichment series, the NMS medium (25) was diluted fivefold (0.2×).
Initial pHs of some media were adjusted with concentrated phosphoric acid.
Affiliation was determined by screening isolate DNA with type I and type II methanotroph-specific primer pairs.
Partial sequencing and phylogenetic analysis.
Ten plasmids with the full-length clone insert (approximately 920 bp for the type I library and 950 bp for the type II library) were sequenced from the type I and type II libraries with primers T1dF and 27F, respectively. Sequencing was done with an automated sequencer (model 373A; Applied Biosystems) at the Molecular Genetics Instrument Facility at the University of Georgia. Sequences were corrected by hand and compared to similar DNA sequences retrieved from the GenBank and EMBL databases by using the FASTA program, which is part of the GCG sequence analysis software package. For the isolates, partial 16S rDNA fragments were amplified with the methanotroph-specific primers described above and the PCR products were sequenced directly. Ten type I and 10 type II isolates of varying colony morphology from different enrichment dilution series were chosen for sequencing. Similar sequences were aligned by using the PILE_UP program of the GCG package. Phylogenetic trees were constructed by using the fastDNAml program (19, 47) run remotely via the World Wide Web at the Pasteur Institute (29a). Bootstrap analyses for 100 resamplings were performed to provide confidence estimates for tree topologies (18).
Sequencing and phylogenetic analysis of the full-length 16S rDNA and partial pmoA genes of selected isolates.
Almost the entire 16S rRNA genes of isolates AML-C10, AML-D4, AML-A3, and AML-A6 were amplified with primers 27F and 1529R (5′-CAKAAAGGAGGTGATCC-3′) (52) under the same conditions as described above, and both strands were sequenced with primers mentioned above (MethT1dF for type I isolates and 27F for type II isolates), in combination with 16S rDNA sequencing primers: 519R, 1392R, 357F, and 926F (35). Phylogenetic trees were constructed in the same manner as described above. Part of the pmoA gene from type II isolates AML-A3 and AML-A6 was amplified with the pmoA-specific primers pmoA189 (5′-GGNGACTGGGACTTCTGG-3′) and pmoA682 (5′-GAASGCNGAGAAGAASGC-3′) (27). Type I isolates AML-C10 and AML-D4 were used with primers pmof1 (5′-GGGGGAACTTCTGGGGITGGAC-3′) and pmor (5′-GGGGGRCIACGTCITTACCGAA-3′) to amplify a slightly smaller section of the pmoA gene (12). The PCR conditions for both sets of primers were the same as those described above. Both strands of the PCR products were sequenced directly with the pmoA-specific PCR primers mentioned above. Phylogenetic analysis of translated gene sequences was performed by using the PROTDIST (20) and neighbor-joining (48) applications that are part of the PHYLIP suite of phylogenetic analysis programs. Bootstrap analyses for 100 resamplings were performed to provide confidence estimates for tree topologies (18).
DGGE.
A nested-PCR approach was used to profile the type I and type II methanotrophic communities by denaturing gradient gel electrophoresis (DGGE). The same amplification products used to construct the clone libraries were used as templates for PCR with the primers GC358F (5′-CGCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCCCCCCTACGGGAGGCAGCAG-3′) and 517R (5′-ATTACCGCGGCTGCTGG-3′), which span variable region 3 (V3 region) of the 16S rRNA gene (44). Primer GC358F has a 40-bp GC clamp added to its 5′ end. Hot-start and touchdown PCRs (16) were done to reduce primer-dimer complexes. The hot-start PCR procedure was as described above; the thermocycling program for touchdown PCR was as follows: initial denaturation was done at 94°C for 2 min 30 s and then at 94°C for 40 s, followed by touchdown primer annealing from 72 to 55°C (the annealing temperature was decreased 1°C for each cycle for the first 17 cycles to touchdown at 55°C), followed by extension at 72°C for 1 min (for each of the 17 cycles). Then 10 cycles of 94°C for 40 s, 55°C for 1 min, and 72°C for 1 min 30 s were done. Then 10 more cycles were performed at 94°C for 40 s, 55°C for 1 min 15 s, and 72°C for 2 min. The final extension step was 72°C for 7 min 30 s. For the clones and isolates, PCR was done directly by employing the primers described above with either plasmid DNA or chromosomal DNA serving as the template, respectively. PCR products were analyzed on agarose gels to confirm the presence of a single amplicand of the expected size. DGGE gels were 6.5% polyacrylamide with a denaturant gradient from 20 to 70% (for type I sequences) and 30 to 60% (for type II sequences). One hundred percent of the denaturant is 7 M urea and 40% deionized formamide. Gels were run in 1× TAE buffer (40 mM Tris base, 20 mM sodium acetate, 1 mM EDTA) for 4 h at 200 V at 60°C and visualized by ethidium bromide staining.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of isolates AML-C10, AML-D4, AML-A3, and AML-A6 have been deposited into GenBank, EMBL, and DDBJ nucleotide sequence databases and assigned accession no. AF177296, AF177297, AF177298, and AF177299, respectively. The partial pmoA gene sequences of these four isolates have been given accession no. AF177325 to AF177328. The partial 16S rDNA sequences of the other unique isolates (AML-A13, AML-A14, AML-E12, AML-E13, and AML-F18) have been assigned accession no. AF177300 to AF177304. The clones have been assigned accession no. AF177305 to AF177324.
RESULTS
Methanotroph-specific primers.
Computer analyses of previously published methanotroph-specific phylogenetic probes indicated that some nondegenerate probes were too specific and would not hybridize to all methane oxidizers of the family Methylococcaceae. For example, probe 1035-RuMP (10), designed to hybridize to the type I methanotrophs, will not detect members of the former type X species, the genus Methylococcus. Since M. capsulatus is an efficient trichloroethylene-degrading species (24), the detection of this group in environmental samples would be beneficial for assessing bioremediation potential. To alleviate this problem, 16S rDNA primers and probes with a broader specificity were designed and tested. According to database searches, no one probe specific for the entire family Methylococcaceae could be identified, as all oligonucleotides chosen showed complete identity to at least one non-methane-utilizing organism or unknown clone sequence (Table 1). However, one oligonucleotide was identified (MethT2R) that was highly specific for all characterized type II methanotrophs (Table 1).
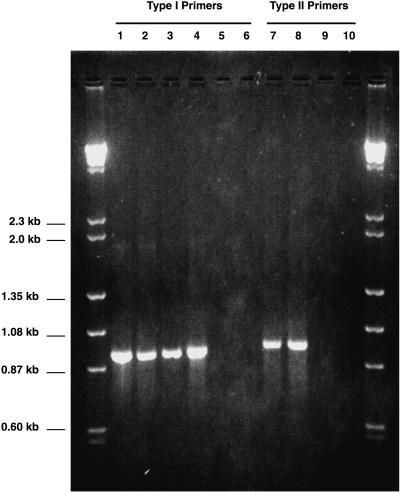
Although one all-inclusive type I methanotroph probe free from the potential for false positives could not be identified, the use of two of these oligonucleotides in combination as PCR primers proved to be methanotroph specific. Figure 1 shows the results of PCR with primers MethT1dF and MethT1bR with template DNA extracted from four methanotrophs from different genera and two close relatives. The expected size of the PCR product with these two primers was approximately 920 bp. Under the PCR conditions used, a product of the correct size was obtained when M. capsulatus (Bath), Methylobacter luteus ACM 3304, Methylomicrobium album BG8, and Methylomonas methanica S1 served as templates for the PCR. Two related gamma-Proteobacteria, P. putida and E. coli, failed to amplify under the PCR conditions employed.
FIG. 1.
Ethidium bromide-stained agarose gel (1.5% [wt/vol]) showing PCR amplification products obtained with methanotroph-specific primers. See the text for details on primer design and reaction conditions. Lanes 1 to 6 show results with the type I primer set, MethT1dF and MethT1bR. Lanes 7 to 10 show results with the type II primer set, 27F and MethT2R. Template DNAs are as follows: lane 1, Methylomicrobium album; lane 2, Methylomonas methanica; lane 3, Methylobacter luteus; lane 4, M. capsulatus; lane 5, P. putida; lane 6, E. coli; lane 7, Methylosinus trichosporium; lane 8, Methylocystis parvus; lane 9, A. tumefaciens; and lane 10, S. meliloti.
The type II specific primer-probe, MethT2R, was used in combination with the Bacteria-specific primer 27F, which hybridizes to all members of the domain Bacteria. Based on computer analysis, the expected product had a size of approximately 950 bp. As shown in Fig. 1, both type species of the two type II methanotroph genera, Methylosinus trichosporium OB3b and Methylocystis parvus OBBP, gave a product of the expected size with the primers. No PCR product was generated by using the DNA from two related alpha-Proteobacteria, A. tumefaciens and S. meliloti.
Type I clone library.
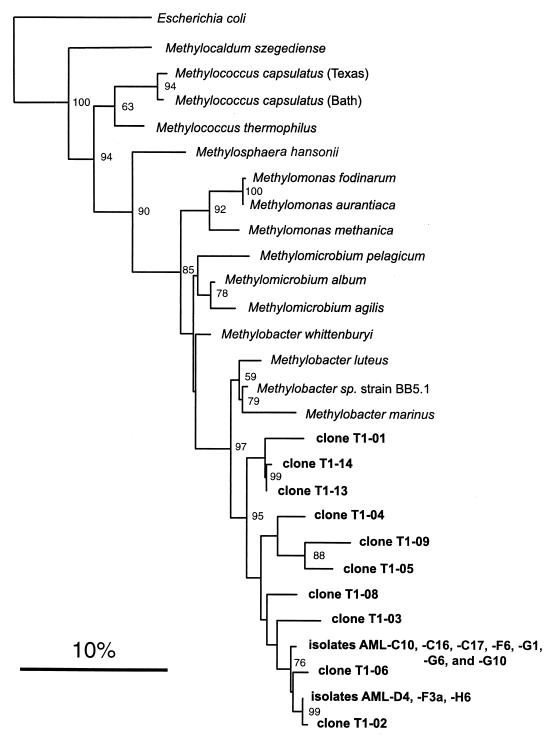
Ten clones that resulted from the use of the type I-specific primer pair on directly extracted environmental DNA from the landfill soil were sequenced with MethT1dF as the primer. This resulted in a range of between 640 and 675 usable bases for each clone. Database searches invariably indicated that the most closely related sequence to each of the 10 clones was the 16S rRNA gene of Methylobacter sp. strain BB5.1 (51). Methylobacter sp. strain BB5.1 is a NaCl-requiring strain recently isolated from estuary sediment. However, the range of sequence identity of the landfill clones with this species was quite low, ranging from 96.1% (clone T1-13) to 94.4% (clone T1-06). To better understand the relationship of clone sequences to the characterized type I methanotrophs, the clones were used in a phylogenetic analysis. Figure 2 shows the results of the phylogenetic analysis of all type I landfill clones and their relationship to some of the characterized type I methanotrophs of the genera Methylococcus, Methylomonas, Methylomicrobium, and Methylobacter and the type strains of two recently described genera, Methylosphaera (5) and Methylocaldum (4). Also included on this tree are some methanotrophic isolates from the landfill soil discussed in detail below. All landfill clones formed a distinct cluster most closely related to members of the genus Methylobacter. However, the high bootstrap value separating the clone sequences from any of the other known methanotrophs suggests that the clones form a monophyletic group distinct from the genus Methylobacter.
FIG. 2.
Phylogenetic tree showing the relationship of landfill soil type I methanotroph 16S rDNA clone sequences and type I AML isolates to some characterized methanotrophs from the family Methylococcaceae. This tree was constructed by using the fastDNAml program (47), which uses a maximum likelihood algorithm (19). A total of 625 aligned bases corresponding to E. coli positions 141 to 766 were used in this analysis. E. coli served as the outgroup. The scale bar represents 0.10 substitutions per base position. The numbers at the nodes of the tree indicate bootstrap values (18) for each node out of 100 bootstrap resamplings (values below 50 are not shown).
Type II clone library.
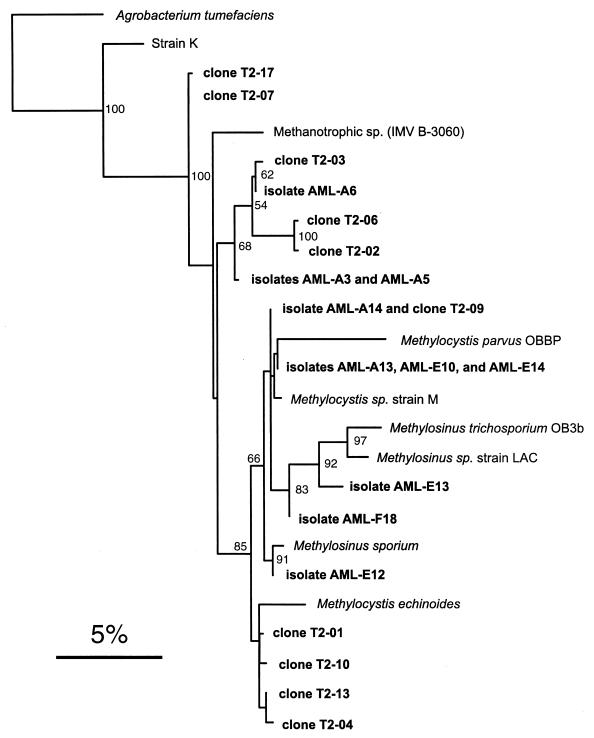
Ten clones were also sequenced from the type II clone library with the 16S rDNA sequencing primer 27F. This resulted in a range of between 638 and 683 usable bases for each clone. Database searches indicated that five clones (T2-01, T2-04, T2-09, T2-10, and T2-13) showed the highest percentages of identity to Methylocystis sp. strain M, an sMMO-containing isolate which can degrade high levels of trichloroethylene (42). The identity of these clones to Methylocystis sp. strain M was high, ranging from 97.9 to 99.1%. Three clones (T2-02, T2-03, and T2-06) were found to be most closely related to an uncharacterized methanotrophic isolate, strain IMV B-3060 (10). These sequence identities were lower, ranging from 95.1 to 96.3%. Clones T2-07 and T2-17 were 96.1% and 96% identical, respectively, to each of three species (Methylocystis sp. strain M, strain IMV B-3060, and Methylosinus sporium). Figure 3 shows the results of the phylogenetic analysis of the type II clone sequences as they relate to some of the validly published type II methanotrophs and some uncharacterized strains. Also included on this tree are some landfill isolates discussed in detail below. According to this analysis, clones T2-01, T2-10, T2-13, and T2-04 group with Methylocystis echinoides, although the bootstrap value supporting this cluster is less than 50. Clone T2-09 is most closely related to a group containing Methylocystis parvus and Methylocystis sp. strain M, although again this cluster is not supported by high bootstrap values. None of the remaining clones clustered closely with any other characterized type II methanotroph.
FIG. 3.
Phylogenetic tree showing the relationship of landfill soil type II methanotroph 16S rDNA clone sequences and type II AML isolates to some characterized type II methanotrophs. Also included are some partially characterized methane oxidizers, including a recently isolated acidophilic bacterium, strain K, that diverges from the type II methanotrophs (14). This tree was constructed by using the fastDNAml program (47), which uses a maximum likelihood algorithm (19). A total of 623 aligned bases corresponding to E. coli positions 54 to 731 were used in this analysis. A. tumefaciens served as the outgroup. The scale bar represents 0.05 substitution per base position. The numbers at the nodes of the tree indicate bootstrap values (18) for each node out of 100 bootstrap resamplings (values below 50 are not shown).
Methanotroph isolation via dilution to extinction.
After an 8-day incubation period, the highest dilution that showed growth was the 10−6 dilution for the H and D series (series letter designations are given in Table 2). The A and E series showed growth at the 10−5 dilution. The highest dilution that showed growth for the F and G series was the 10−4 dilution. After being plated on solidified media, several colonies were selected for further purification on the basis of differences in colony morphology. Overall, 64 methane-utilizing cultures were obtained and 36 were screened with the type I- and type II-specific primer sets. The results of the screening are shown in Table 2. Of the three conditions varied, the concentration of the medium was the most important factor in selection for methanotrophic-type affiliation. Type II strains dominated all enrichments in 0.2× NMS medium. Only type I methanotrophs were enriched in full-strength NMS medium. Just one enrichment dilution series, series F, resulted in the isolation of both type I and type II strains.
Ten type I methanotrophs were selected for partial 16S rDNA sequencing, resulting in a range of between 645 and 684 usable bases for each isolate. Seven type I isolates were identical: AML-C10, AML-C16, AML-C17, AML-F6, AML-G1, AML-G6, and AML-G10. Database searches revealed that the most closely related bacterium to these strains was Methylobacter sp. strain BB5.1; AML-C10 showed a 95.6% identity to this species in a 684-bp overlap. The other three type I isolates, AML-D4, AML-F3a, and AML-H6, also had sequences identical to each other. Strain AML-D4 was also most closely related to Methylobacter sp. strain BB5.1, with 94.9% identity in a 668-bp overlap. As seen in Fig. 2, phylogenetic analysis revealed that all isolates fell within the new cluster of type I sequences suggested by the clone library.
Ten type II isolates were also partially sequenced, and the phylogenetic analysis is shown in Fig. 3. For these type II isolates, a range of between 634 and 696 bases was obtained. Strains AML-A13, AML-E10, and AML-E14 were found to be identical. All were most similar to Methylocystis sp. strain M (>99% identity) and clustered with this species and Methylocystis parvus. Three strains grouped with members of the Methylosinus genus: AML-E13, AML-F18, and AML-E12. One isolate, AML-A14, was identical to clone sequence T2-09, with both showing 99.4% identity to Methylocystis sp. strain M. Isolates AML-A3 and AML-A5 were found to have the same sequence, and these strains clustered with isolate AML-A6 in a group containing clones T2-02, T2-03, and T2-06. These strains had the lowest similarity of any cultured type II isolate to the methanotrophs in the databases. All were most similar to methanotrophic isolate IMV B-3060, but only at approximately 96% identity. The phylogenetic analysis indicated that this cluster is distinct from any of the characterized Methylosinus or Methylocystis species, although the bootstrap value supporting this grouping is less than 50.
Novel type I isolates AML-C10 and AML-D4.
Ten-day-old colonies of both AML-C10 and AML-D4 grown on NMS medium were circular and light brown to buff colored and had regular margins. In liquid culture, both strains tended to grow in a flocculent manner, and the flocculent particles settled rapidly when shaking was ceased. The particles resembled irregularly shaped, sarcina-like aggregates when examined by phase-contrast microscopy. Individual AML-C10 cells were motile and coccus to oval shaped and had a diameter of approximately 1 to 1.5 μm. Individual AML-D4 cells were pleiomorphic; they commonly exhibited a fusiform morphology but were also present as long rods and sometimes chains of ovoid cells. Cell length ranged from 1.5 to 6 μm with a diameter of approximately 1 μm. At 30°C, strains AML-C10 and AML-D4 had doubling times of approximately 3.5 and 6.5 h, respectively.
To confirm the phylogenetic uniqueness of these two isolates, both strands of the nearly complete 16S rRNA gene were sequenced. AML-C10 and AML-D4 were 99.0% identical to each other over the length of the full gene. AML-C10 was 95.9% identical to Methylobacter sp. strain BB5.1 in a 1,494-bp overlap. AML-D4 was 95.4% identical to Methylobacter sp. strain BB5.1 in a 1,492-bp overlap. The phylogenetic tree constructed with the full sequence revealed that the placement of these isolates among the Methylococcaceae did not change; both isolates cluster together in a group distinct from the Methylobacter-Methylomicrobium clade with moderately high bootstrap support (data not shown).
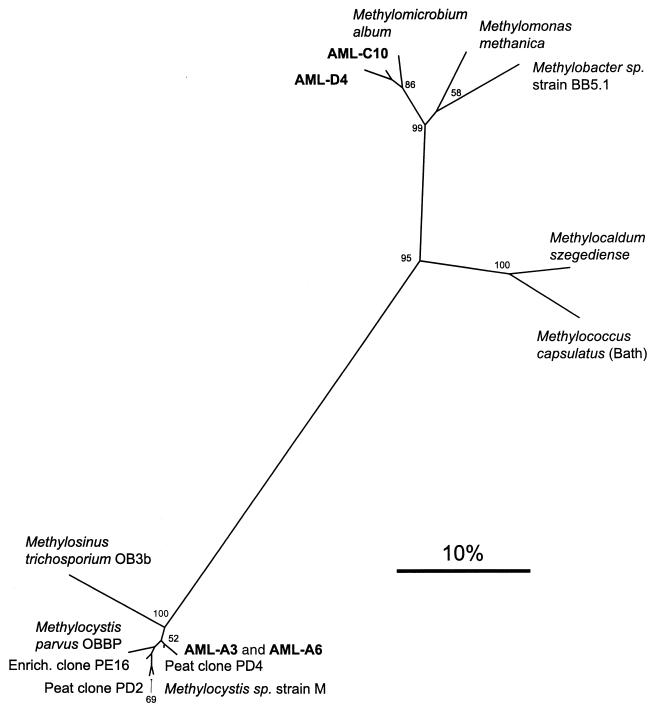
Primer pair A189-A682, shown to successfully amplify the 525-bp pmoA gene in almost all species of methanotrophs (27), did not yield a product of the expected size with isolate AML-D4 as the template. With AML-C10 DNA, a minor band of approximately the correct size was observed, but the reaction lacked specificity, as many other products were obtained despite repeated attempts to optimize the conditions. Therefore, pmoA-specific primers internal to this region were employed. Primer pair pmof1-pmor (12) gave the expected 330-bp product and the derived amino acid sequence used in a phylogenetic analysis (Fig. 4). The partial PmoA protein sequences of AML-C10 and AML-D4 were both most similar to Methylomicrobium album at 97.3 and 94.5% amino acid similarity, respectively. Neither AML-C10 nor AML-D4 yielded any PCR product with primers specific for the alpha subunit of the sMMO hydroxylase component (mmoX gene) (43).
FIG. 4.
Phylogenetic analysis of partial amino acid sequences of the pmoA gene from selected type I and type II methanotrophs, novel isolates and environmental clones (41). This unrooted tree was constructed by using the neighbor-joining method (48) from a matrix of pairwise genetic distances as calculated by the PROTDIST program (20). A total of 110 aligned amino acid positions were used in this analysis. The scale bar represents 10% sequence divergence. Bootstrap analyses (18) for 100 resamplings were performed to provide confidence estimates for tree topologies (values below 50 are not shown).
Novel type II isolates AML-A3 and AML-A6.
Since phylogenetic analysis based on a partial 16S rDNA sequence showed AML-A3 and AML-A6 clustering with clones T2-02, T2-03, and T2-06 in a group distinct from the well-described type II methanotrophs, they were chosen for further analysis. Both AML-A3 and AML-A6 formed circular, mucoid colonies on NMS medium that were pink and white pigmented, respectively. In liquid culture, both strains grew in an evenly dispersed manner. Both were nonmotile and exhibited a coccus-shaped morphology. The doubling time at 30°C was approximately 12.75 h for AML-A3 and 16.5 h for AML-A6.
The full 16S rRNA gene sequence revealed that AML-A3 and AML-A6 were 98.9% identical to each other over the length of the gene. AML-A3 was 95.9% identical to strain IMV B-3060 in a 1,307-bp overlap. AML-A6 was 96.3% identical to strain IMV B-3060 in a 1,314-bp overlap. Again, the dendrogram built with the full 16S rDNA sequence was consistent with the one based on the partial sequences; both isolates clustered together in a group distinct from the Methylosinus-Methylocystis clade, although with a bootstrap value of less than 50 (data not shown).
Both isolates AML-A3 and AML-A6 gave a single PCR product of the predicted size when PCR was performed with the primer pair A189-A682. The translated gene sequences showed that there was only one amino acid difference between the two strains out of 165 homologous positions. Interestingly, both are most similar to pmoA clones retrieved directly from environmental DNA extracted from a blanket peat bog (41). AML-A3 and AML-A6 are most closely related to peat clone PD4, both showing similarities of >98% and identities of >95%. This association is evident on the phylogenetic tree shown in Fig. 4. Neither strain proved positive for the mmoX gene, as assayed by PCR (described above).
DGGE.
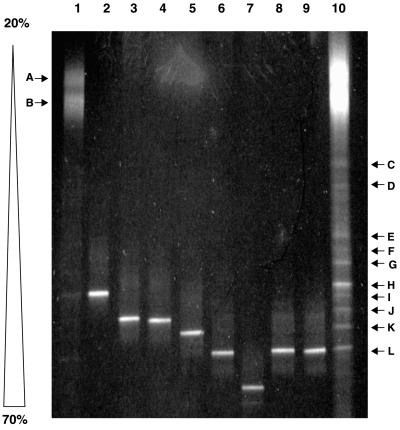
DGGE was employed here to provide genetic fingerprints that could serve as an overview of the diversity of the type I and type II methanotrophic communities in the landfill soil. Figure 5 shows the results of DGGE analysis with the primer pair GC358F-517R (the V3 region) on type I sequences amplified from the landfill environmental DNA and selected clones and isolates. The type I community profile consists of 12 unique bands (Fig. 5, lanes 1 and 10). Two diffuse bands (labeled A and B) that denatured at relatively low denaturant concentrations accounted for the majority of the amplification products. Ten other discernable bands (C through L) denatured at higher denaturant concentrations and were present in a lesser abundance.
FIG. 5.
Type I methanotroph DGGE analysis. Shown is an ethidium bromide-stained 6.5% polyacrylamide denaturing gradient gel (20 to 70%) showing separation patterns based on sequence difference of the V3 region (approximate E. coli positions 341 to 534) of the 16S rDNA. Lanes 1 and 10, the type I landfill methanotrophic community (250 ng and 1.5 μg, respectively); lane 2, clone T1-08; lane 3, clone T1-01; lane 4, clone T1-14; lane 5, clone T1-09; lane 6, clone T1-02; lane 7, clone T1-04; lane 8, isolate AML-C10; lane 9, isolate AML-D4. Bands A through L are explained in the text.
In order to identify some of the bands and compare the DGGE profile to the diversity assessments based on cloning and isolation, the individual clones and isolates were amplified with the same primer pair and separated via DGGE. Of the 10 clones, six distinct mobilities were identified and one example of each is included on the gel in Fig. 5. Running to the same position as clone T1-01 were clones T1-03 and T1-13. Clones T1-05 and T1-06 both ran to the same position as clone T1-02. Both isolates AML-C10 and AML-D4 also migrated to this position on the gel. Of the 12 bands that constitute the type I landfill profile, the clones and isolates can account for only 4 of the bands. Clone T1-08 corresponded to band H; clones T1-01, T1-03, and T1-13 migrated to the same position as band J; clone T1-09 corresponded to band K; clones T1-02, T1-05, and T1-06 and both isolates ran to the same position as band L.
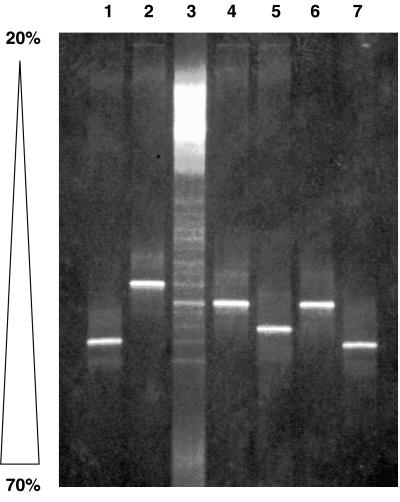
We were curious as to the identity of the large, diffuse bands (A and B) that were the major constituents of the type I profile. We speculated that these bands may be an artifact of the mixed template reaction, since no single methanotroph V3 region 16S rDNA sequence denatured at this relatively low denaturant concentration, including DGGE analysis of the representatives of the Methylococcaceae used in Fig. 1 (data not shown). It is known that heteroduplex molecules that form during PCR from mixed template reactions can complicate community profiles (21, 45). Mismatches in the two similar but nonidentical strands result in a lower denaturing temperature than that of homoduplex molecules; hence, the heteroduplexes melt at lower denaturant concentrations and appear as upper bands in parallel gradient DGGE analysis. Bands A and B were each excised from the gel, purified, and reamplified with the same V3 region primer pair (without the CG clamp). The amplification products were cloned into pCR2.1, and two clones from band A and two clones from band B were sequenced. Clones EB-A1 (for excised band A-1) and EB-B1 were identical to clones T1-09 and T1-08 from the type I clone library, respectively. Clone EB-A2 was approximately 95% identical to clone T1-08, and clone EB-B2 was approximately 99% identical to clone T1-09. Figure 6 shows the mobilities of these four individual clones as they relate to clones T1-08 and T1-09 and the type I methanotroph community profile. The clones from bands A and B that were reamplified with the V3 region GC-clamped primer set were shown to melt at much higher denaturant concentrations than those of the bands from which they were originally excised. The mixed template PCR seems to result in an artifact (possibly heteroduplex molecule formation) that causes the majority of the amplification products to denature at premature denaturant concentrations.
FIG. 6.
DGGE analysis of cloned DNA extracted from type I community bands A and B (Fig. 5). Shown is an ethidium bromide-stained 6.5% polyacrylamide denaturing gradient gel (20 to 70%) showing separation patterns based on sequence differences in the V3 region (approximate E. coli positions 341 to 534) of the 16S rDNA. See Results for experimental details. Lane 1, clone EB-A1; lane 2, clone EB-A2; lane 3, type I landfill methanotrophic community profile; lane 4, clone EB-B1; lane 5, clone EB-B2; lane 6, clone T1-08; lane 7, clone T1-09.
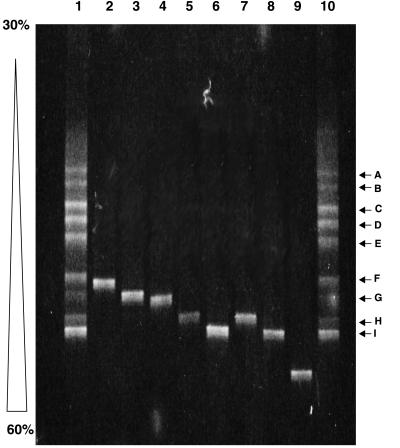
The type II methanotroph DGGE community profile consists of nine distinct bands labeled A through I (Fig. 7). Analysis of the clones and isolates revealed that they could account for three of the bands. Clones T2-07 and T2-17 ran to the same point as band G. Clone T2-02 and isolates AML-A3 and AML-A6 all migrated to the same position as band H. Clones T2-01, T2-04, T2-09, T2-10, and T2-13 and isolates AML-F18, AML-E10, AML-E13, AML-E12, AML-A13, AML-E14, and AML-A14 all corresponded to band I. Clones T2-06 and T2-03 and isolate AML-E13 each ran to a unique position not represented in the DGGE community profile.
FIG. 7.
Type II methanotroph DGGE analysis. Shown is an ethidium bromide-stained 6.5% polyacrylamide denaturing gradient gel (30 to 60%) showing separation patterns based on sequence differences in the V3 region (approximate E. coli positions 341 to 534) of the 16S rDNA. Lanes 1 and 10, the type II landfill methanotrophic community; lane 2, clone T2-06; lane 3, clone T2-07; lane 4, clone T2-03; lane 5, clone T2-02; lane 6, clone T2-01; lane 7, isolate AML-A3; lane 8, isolate AML-F18; lane 9, isolate AML-E13. Bands A through I are explained in the text.
DISCUSSION
In this report we describe the isolation of new methanotrophic bacteria whose presence in the landfill cover soil was predicted by the phylogenetic analysis of clone sequences. The initial detection of microbes via molecular methods is an important first step in community analysis, but detailed investigation into physiology is possible only when organisms are isolated in pure culture. One reason that diversity assessments based on culture-independent analyses have historically been quite different than those based on culturing is the phenomenon of competitive exclusion during enrichment (58). It has been recognized that rare but opportunistic bacteria tend to outcompete the numerically dominant, oligotrophic species under the typical set of enrichment conditions (11). However, in cultures diluted to extinction, the most abundant organisms become favored during enrichment since the opportunistic species may no longer be present in the highest dilutions. Our isolates may represent such species; they may be unfit to compete successfully with some of the more typically isolated methanotroph strains. Equally important is the ability of the enrichment to reproduce as closely as possible the resources and conditions in situ. Dedysh et al. (13, 14) recently had success isolating novel methane oxidizers by engineering the enrichment media and incubation parameters to more accurately simulate the natural conditions of acidic ombrotrophic peat bogs. We adopted a similar approach; methanotrophs living in landfill cover soil enjoy some of the highest levels of methane on the planet, so we used some enrichment series with relatively high (approximately 50%) methane concentrations in the headspace. Also, we adjusted the initial pH of some of the enrichment series to more accurately reflect the actual pH of the landfill soil. Interestingly, it seems the most important variable in determining the isolation of type I versus type II species in our sample was the strength of the medium. Type II species dominated enrichments in which the NMS medium was diluted fivefold (A, E, and F series) regardless of initial pH or methane concentration of the headspace. This suggests a propensity for type II species to better acquire nutrients when they are more scarce. Alternatively, they may be sensitive to the higher salt concentration of the full-strength medium, rendering them at a selective disadvantage.
The results described here add to the growing body of evidence suggesting that the formally described methanotrophs are but a subset of the actual diversity of this physiological type. Previous research using phylogenetic probes and primers directed against the methanotrophs has also detected novel methanotroph sequences. Holmes et al. (28) discovered an unusual 16S rDNA sequence from organisms related to the genus Methylomonas in seawater methane enrichments. Organisms containing this sequence were visualized by in situ hybridization and found to be quite numerous (45% of all cells) in the enrichment, although they could not be obtained in pure culture. McDonald et al. (38) investigated methanotroph diversity in peat cores and found 16S rDNA clone sequences related to but distinct from the characterized type II methanotrophs that may represent novel acidophilic species. Culture-independent diversity assessments have also been performed by exploiting certain conserved functional genes. McDonald et al. (39) used primers specific for the mmoX gene (which codes for the alpha subunit of the hydroxylase component of the sMMO) and presented work that suggests that a large number of unusual methanotrophs, with mmoX sequences different from those of the extant species, are present in the peat samples. Similar results were obtained from peat samples by using the gene for the large subunit of the methanol dehydrogenase, mxaF (40), and the pmoA gene (41). We have some evidence suggesting that our novel type II methanotroph isolates are related to some of the organisms detected in the suite of molecular ecology experiments performed by McDonald and coworkers on acidic peat samples. As seen in Fig. 4, the PmoA amino acid sequences of AML-A3 and AML-A6 are highly similar to clone PD4, a clone sequence retrieved from the peat (41). However, the novel 16S rDNA clones retrieved from the same samples diverge significantly from our isolates: Moorhouse peat core clones MHP14 and MHP17 were each <94% identical to both AML-A3 and AML-A6 (38). It remains to be seen whether the organisms isolated here can account for some of the molecular methanotroph diversity observed in peat. Although both types of environments are acidic, the pH of the peat bog (3.6) is more extreme than that of the landfill soil. Furthermore, one may expect that methanotrophs adapted to the high methane levels and unique soil chemistry prevalent in landfill soil differ from species that thrive in organic-rich peat.
At first glance, the diversity assessment via DGGE analysis seems to be the most encompassing of the three techniques employed here. DGGE profiles of the type I and type II methanotrophic communities resulted in distinct banding patterns that were not too complex, as is often the case when universal- or domain-specific PCR primers are used (45). We found that only some of the bands present in the community profile could be accounted for by the clones and isolates, suggesting that some species were not detected by cloning and sequencing or culturing. However, it is likely that not all bands in the profiles correspond to actual 16S rRNA sequence types. The formation of heteroduplex molecules is expected to be a problem with the community analysis of a highly related phylogenetic cluster, since the mixed PCR products of closely related species may have enough sequence similarity to anneal together. We noted that with the DGGE analysis of both the type I and type II community members, the clones and isolates always accounted for bands that melted at the higher denaturant concentrations. It is possible that bands that denature at the lower concentrations (upper bands on the gel) are due to heteroduplex formation, although migration could also be retarded by secondary structure or other unknown associations. Clearly the diffuse bands A and B in the type I profile are the result of an artifact that occurs with the mixed template DNA, as DNAs reamplified and cloned from these excised bands migrated to a position of much higher denaturant concentration when reanalyzed by DGGE. Also of note, some clones and isolates migrated to positions not represented on the DGGE profiles. For example, isolate AML-E13 melted at a denaturant concentration higher than any band seen in the type II community profile (Fig. 7). Clearly caution should be exercised when making diversity judgments based on DGGE banding patterns.
The two novel type I isolates are part of a monophyletic line of descent and are seemingly just the culturable members of a larger phylotype present in the landfill soil. This phylotype may represent a group of organisms that are especially well adapted to this habitat, since no other type I sequence was retrieved. In contrast, both the clone-based and culturing assessments suggested that the type II methanotroph species were much more diverse, as several distinct clusters of clones and isolates were observed. Evidently the characteristics of the landfill soil support a broad range of type II species. Our future plans include a formal characterization of our unique isolates in order to determine their valid taxonomical status. Whether these groups are different enough to warrant the status of new genera will depend on further physiological, morphological, and genetic tests.
ACKNOWLEDGMENTS
We thank Brad Ricard for allowing access to the Athens-Clarke County Municipal Landfill. We also thank Colin Murrell, Andria Costello, and Robin Brigmon for graciously providing methanotroph strains. Andria Costello generously shared the corrected 16S rDNA sequence of M. trichosporium OB3b. Tim Hoover provided the S. meliloti strain. Finally, we thank Dan Kearns for comments on an earlier version of the manuscript.
This research was supported by Financial Assistance award DE-FC09-96SR18546 from the U.S. Department of Energy to the University of Georgia Research Foundation.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral J A, Archambault C, Richards S R, Knowles R. Denitrification associated with group I and II methanotrophs in a gradient enrichment system. FEMS Microbiol Ecol. 1995;18:289–298. [Google Scholar]
- 3.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodrossy L, Holmes E M, Holmes A J, Kovacs K L, Murrell J C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol. 1997;168:493–503. doi: 10.1007/s002030050527. [DOI] [PubMed] [Google Scholar]
- 5.Bowman J P, McCammon S A, Skerratt J H. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology. 1997;143:1451–1459. doi: 10.1099/00221287-143-4-1451. [DOI] [PubMed] [Google Scholar]
- 6.Bowman J P, Sly L I, Stackebrandt E. The phylogenetic position of the family Methylococcaceae. Int J Syst Bacteriol. 1995;45:182–185. doi: 10.1099/00207713-45-1-182. [DOI] [PubMed] [Google Scholar]
- 7.Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol. 1993;43:735–753. [Google Scholar]
- 8.Bratina B J, Brusseau G A, Hanson R S. Use of 16S rRNA analysis to investigate phylogeny of methylotrophic bacteria. Int J Syst Bacteriol. 1992;42:645–648. doi: 10.1099/00207713-42-4-645. [DOI] [PubMed] [Google Scholar]
- 9.Britschgi T B, Giovannoni S J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brusseau G A, Bulygina E S, Hanson R S. Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol. 1994;60:626–636. doi: 10.1128/aem.60.2.626-636.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y S, Halsey J L, Fode K A, Remsen C C, Collins M L P. Detection of methanotrophs in groundwater by PCR. Appl Environ Microbiol. 1999;65:648–651. doi: 10.1128/aem.65.2.648-651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dedysh S N, Panikov N S, Tiedje J M. Acidophilic methanotrophic communities from Sphagnum peat bogs. Appl Environ Microbiol. 1998;64:922–929. doi: 10.1128/aem.64.3.922-929.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedysh S N, Panikov N S, Liesack W, Grosskopf R, Zhou J, Tiedje J M. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science. 1998;282:281–284. doi: 10.1126/science.282.5387.281. [DOI] [PubMed] [Google Scholar]
- 15.DiSpirito A A, Gulledge J, Murrell J C, Shiemke A K, Lidstrom M E, Krema C L. Trichloroethylene oxidation by the membrane associated methane monooxygenase in type I, type II, and type X methanotrophs. Biodegradation. 1992;2:151–164. [Google Scholar]
- 16.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ’Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ensley B D. Biochemical diversity of trichloroethylene metabolism. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. Confidence limits of phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 21.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham D W, Chaudhary J A, Hanson R S, Arnold R G. Factors affecting competition between type I and type II methanotrophs in continuous-flow reactors. Microb Ecol. 1993;25:1–17. doi: 10.1007/BF00182126. [DOI] [PubMed] [Google Scholar]
- 23.Hanson R S. Ecology of methylotrophic bacteria. In: Burlage R, et al., editors. Techniques in microbial ecology. New York, N.Y: Oxford University Press; 1998. pp. 137–162. [Google Scholar]
- 24.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson R S, Netrusov A I, Tsuji K. The obligate methanotrophic bacteria Methylococcus, Methylomonas and Methylosinus. In: Balows E, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 2352–2364. [Google Scholar]
- 26.Hazen T C, Lombard K H, Looney B B, Enzien M V, Dougherty J M, Fliermans C B, Wear J, Eddy-Dilek C A. Summary of in-situ bioremediation demonstration (methane biostimulation) via horizontal wells at the Savannah River site integrated demonstration project. In: Gee G W, Wing R, editors. In-situ remediation: scientific basis for current and future technologies. Vol. 1. Richland, Wash: Battelle Press; 1994. pp. 137–150. [Google Scholar]
- 27.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 28.Holmes A J, Owens N J P, Murrell J C. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiology. 1995;141:1947–1955. doi: 10.1099/13500872-141-8-1947. [DOI] [PubMed] [Google Scholar]
- 29.Huber R, Burggraf S, Mayer T, Barns S M, Rossnagel P, Stetter K O. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature. 1995;376:57–58. doi: 10.1038/376057a0. [DOI] [PubMed] [Google Scholar]
- 29a.Institut Pasteur Website. 11 September 1998, revision date. [Online.] http://bioweb.pasteur.fr. [27 September 1999, last date accessed.]
- 30.Jones H A, Nedwell D B. Methane emission and methane oxidation in landfill cover soil. FEMS Microbiol Ecol. 1993;74:309–323. [Google Scholar]
- 31.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kightley D, Nedwell D B, Cooper M. Capacity for methane oxidation in landfill cover soils measured in laboratory-scale soil microcosms. Appl Environ Microbiol. 1995;61:592–601. doi: 10.1128/aem.61.2.592-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King G M. Ecological aspects of methane oxidation, a key determinant of global methane dynamics. Adv Microb Ecol. 1992;12:431–468. [Google Scholar]
- 34.Labarca C, Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980;102:344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- 35.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1991. pp. 115–175. [Google Scholar]
- 36.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipscomb J D. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 38.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microbiol Ecol. 1996;21:197–211. [Google Scholar]
- 39.McDonald I R, Kenna E M, Murrell J C. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl Environ Microbiol. 1995;61:116–121. doi: 10.1128/aem.61.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald I R, Murrell J C. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;63:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald I R, Murrell J C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol Lett. 1997;156:205–210. doi: 10.1111/j.1574-6968.1997.tb12728.x. [DOI] [PubMed] [Google Scholar]
- 42.McDonald I R, Uchiyama H, Kambe S, Yagi O, Murrell J C. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl Environ Microbiol. 1997;63:1898–1904. doi: 10.1128/aem.63.5.1898-1904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miguez C B, Bourque D, Sealy J A, Greer C W, Groleau D. Detection and isolation of methanotrophic bacteria possessing the soluble methane monooxygenase (sMMO) genes using the polymerase chain reaction. Microb Ecol. 1997;33:21–31. doi: 10.1007/s002489900004. [DOI] [PubMed] [Google Scholar]
- 44.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 46.Nold S C, Kopczynski E D, Ward D M. Cultivation of aerobic chemoorganotrophic proteobacteria and gram-positive bacteria from a hot spring microbial mat. Appl Environ Microbiol. 1996;62:3917–3921. doi: 10.1128/aem.62.11.3917-3921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 47a.Ricard, B. Personal communication.
- 48.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 49.Schut F, De Vries E J, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semrau J D, Christoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes A J, Finch R, Murrell J C, Lidstrom M E. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith K S, Costello A M, Lidstrom M E. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl Environ Microbiol. 1997;63:4617–4620. doi: 10.1128/aem.63.11.4617-4620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki M T, Rappe M S, Haimberger Z W, Winfield H, Adair N, Stroebel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Topp E, Hanson R S. Metabolism of radiatively important trace gases by methane-oxidizing bacteria. In: Roger J E, Whitman W B, editors. Microbial production and consumption of greenhouse gases: methane, nitrogen oxides, and halomethanes. Washington, D.C.: American Society for Microbiology; 1991. pp. 71–90. [Google Scholar]
- 55.Tsai Y L, Olson B H. Rapid method for direct extraction from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsien H-C, Brusseau G A, Hanson R S, Wackett L P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989;55:3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. In: Marshall K C, editor. Advances in microbial ecology. Vol. 12. New York, N.Y: Plenum Press; 1992. pp. 219–286. [Google Scholar]
- 58.Ward D M, Ferris M J, Nold S C, Bateson M M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward N, Rainey F A, Goebel B, Stackebrandt E. Identifying and culturing the ‘unculturables’: a challenge for microbiologists. In: Allsopp D, Colwell R R, Hawksworth D L, editors. Microbial diversity and ecosystem function. Wallingford, United Kingdom: CAB International; 1995. pp. 89–109. [Google Scholar]
- 60.Whalen S C, Reeburgh W S, Sandbeck K A. Rapid methane oxidation in a landfill cover soil. Appl Environ Microbiol. 1990;56:3405–3411. doi: 10.1128/aem.56.11.3405-3411.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]