Abstract
The extent and nature of tetracycline resistance in bacterial populations of two apple orchards with no or a limited history of oxytetracycline usage were assessed. Tetracycline-resistant (Tcr) bacteria were mostly gram negative and represented from 0 to 47% of the total bacterial population on blossoms and leaves (versus 26 to 84% for streptomycin-resistant bacteria). A total of 87 isolates were screened for the presence of specific Tcr determinants. Tcr was determined to be due to the presence of Tet B in Pantoea agglomerans and other members of the family Enterobacteriacae and Tet A, Tet C, or Tet G in most Pseudomonas isolates. The cause of Tcr was not identified in 16% of the isolates studied. The Tcr genes were almost always found on large plasmids which also carried the streptomycin resistance transposon Tn5393. Transposable elements with Tcr determinants were detected by entrapment following introduction into Escherichia coli. Tet B was found within Tn10. Two of eighteen Tet B-containing isolates had an insertion sequence within Tn10; one had IS911 located within IS10-R and one had Tn1000 located upstream of Tet B. Tet A was found within a novel variant of Tn1721, named Tn1720, which lacks the left-end orfI of Tn1721. Tet C was located within a 19-kb transposon, Tn1404, with transposition genes similar to those of Tn501, streptomycin (aadA2) and sulfonamide (sulI) resistance genes within an integron, Tet C flanked by direct repeats of IS26, and four open reading frames, one of which may encode a sulfate permease. Two variants of Tet G with 92% sequence identity were detected.
Oxytetracycline is currently being used in the United States on a limited basis in apple and pear orchards for the control of fire blight caused by Erwinia amylovora and on a more widespread basis in peach and nectarine orchards for control of bacterial spot caused by Xanthomonas campestris pv. pruni. Although it provides a lower level of disease control than does streptomycin, it is used in apple and pear orchards as a replacement for streptomycin where streptomycin resistance (Strr) in the pathogen has become a serious problem. Repeated use of streptomycin for fire blight control over the past decades has led to the establishment of Strr E. amylovora populations in many pome fruit-growing regions. These bacteria possess either mutations which confer ribosomal insensitivity to streptomycin (12, 42) or the streptomycin-modifying genes strA-strB acquired from other orchard bacteria in association with Tn5393 on conjugative plasmid pEa34 (11, 13) or with RSF1010-type plasmid pEa8.7 (41). Although E. amylovora cannot easily develop tetracycline resistance (Tcr) by chromosomal mutation (27), the use of oxytetracycline may lead to the selection of E. amylovora strains that have acquired tetracycline resistance genes from other orchard bacteria. The time frame of resistance development would depend in part on the availability of tetracycline resistance determinants in orchard bacterial populations and the ability of these determinants to be transferred to E. amylovora.
Tcr in gram-negative bacteria has been found to be conferred by two distinct mechanisms: removal of tetracycline from the cell by an energy-dependent efflux mechanism and protection of the ribosome from the action of tetracycline (for a review, see reference 39). Among enteric genera, only determinants encoding a tetracycline efflux mechanism have been described. Although Tcr can be mediated by chromosomal multidrug resistance genes such as mexA-mexB-oprM in several Pseudomonas species (28, 29) and marRAB in Escherichia coli (19, 20), in most gram-negative species resistance is due to acquisition of a resistance operon of the Tet A family. The resistance operon consists of a structural gene (tetA) and a repressor gene (tetR) that are divergently transcribed from overlapping operator regions. Nine classes of the Tet A family have been described (Tet A to E, G, and H to J). All except Tet I have been sequenced. The structural genes of this family have nucleotide identity ranging from 49 to 75%. Two other examples of Tet A-type determinants have recently been found. A resistance gene, tetY, was found on an IncQ plasmid in E. coli independent of tetR (49), and a dysfunctional Tcr operon was identified on the chromosome of Agrobacterium tumefaciens C58 (30).
The tetracycline resistance genes of the Tet A family have frequently been described in association with conjugative plasmids. The more detailed genetic context of these resistance determinants has been studied in only a few cases. These studies have demonstrated that the Tcr determinants are at least sometimes located in mobile or potentially mobile elements: Tet A in Tn1721 (4), Tet B in Tn10 (15), Tet D flanked by copies of IS26 in a 5.2-kb composite transposon-like structure (3, 26), Tet G within a putative integron (8), and Tet H in Tn5706 (25).
In addition to concerns about antibiotic use leading to the development of resistant plant-pathogenic bacteria, there has recently been some discussion about the impact of agricultural uses of antibiotics on the development of multidrug-resistant human pathogens. The presence of multidrug resistance in bacteria of the phylloplane has not been thoroughly analyzed. It is therefore unclear what the impact of oxytetracycline would be on the establishment of bacterial populations in this environment carrying transferable multidrug plasmids. Furthermore, the extent to which bacteria of the phylloplane can serve as a reservoir of resistance determinants for human pathogens is unknown.
In an effort to address these issues, a study in which reservoirs of Tcr in apple orchards were assessed was initiated. The likelihood that tetracycline resistance would become prevalent in populations of E. amylovora following oxytetracycline application was evaluated by identifying which Tcr determinants were present in orchard bacterial populations and whether the determinants were associated with mobile genetic elements. The role that Tcr bacteria might play in problematic multidrug resistance was also assessed by screening for resistance to other antibiotics.
MATERIALS AND METHODS
Orchard treatments and sample collection.
In 1996 and 1997, blocks of trees three to four rows wide by 12 to 30 trees long were sprayed three times during bloom with 290 g of streptomycin sulfate per ha (orchard 1), 290 g of oxytetracycline hydrochloride per ha (orchard 2), or 290 g of each compound per ha (orchards 1 and 2). Each treatment was replicated three times. Both orchards have long histories of streptomycin usage. Orchard 2 also had a history of approximately two applications of oxytetracycline per season from 1991 to 1995. Orchard 1 had no history of Strr E. amylovora, whereas in orchard 2 Strr E. amylovora had previously been detected.
Samples of 40 to 50 blossoms and 9.6 to 39.2 g of leaves (one per replicate, 1996; three per replicate, 1997) were collected during bloom and 2 weeks postbloom, respectively. Blossom and leaf washes were plated on Luria-Bertani (LB) or King’s B agar amended with 100 μg of cycloheximide per ml plus either 100 μg of streptomycin per ml, 2 μg (1996) or 10 μg (1997) of oxytetracycline per ml, both streptomycin and oxytetracycline, or no antibiotics. Colony counts were made after 3 days at 22°C.
One to eight colonies were subcultured from each sample that yielded Tcr bacteria (347 total). Gram stain tests were performed on each isolate by using the Accustain Gram stain kit (Sigma Chemical Co., St. Louis, Mo.) and a KOH lysis technique (48). In each case, the results from the two methods were in agreement. Seven isolates were gram positive. Gram-negative isolates were used in further studies.
Standard PCR.
Reaction mixtures (20 μl) consisted of 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl), 1.5 mM MgCl2, 0.5 to 1 μM (each) primer, 0.16 mM deoxynucleoside triphosphates (Pharmacia LKB, Piscataway, N.J.), 0.5 U of Taq Polymerase (Gibco BRL, Grand Island, N.Y.), and either 0.1 to 100 ng of template DNA or 1 μl of bacterial culture diluted to an optical density at 600 nm of between 0.1 and 0.5. Reactions were performed in a PTC-100 Thermocycler (MJ Research, Watertown, Mass.). Reaction products were analyzed on 1.5% (wt/vol) agarose gels in 0.5× Tris-borate-EDTA (TBE) buffer run at 10 V/cm followed by ethidium bromide staining.
Fragments of tetracycline resistance genes (293 bp) were amplified with primers TET1-F (5′-GCYRTVGGSATHGGCYTKRTYATGC), TET1-R (5′-ACMGCMCCWGTVGCBCCKGTGAT), TET2-F (5′-GCBATKGGDMTYGGBMTNATYATGC), and TET2-R (5′-ACVGCDCCDGTBGCRCCNGTRAT) with cycling parameters of 94°C for 60 s followed by 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 30 s. HaeIII digestion of PCR products was analyzed by electrophoresis through 10% polyacrylamide gels in 1× TBE followed by ethidium bromide staining.
Streptomycin resistance genes and transposon Tn5393 were identified by amplification with primers specific for the strB gene (strB-F [5′-GGAACTGCGTGGGCTACA] and strB-R [5′-GCTAGATCGCGTTGCTCCTCT]; 303 bp) and the tnpA gene of Tn5393 (tnp5393-F [5′-GGCGGGATCTGCTTGTAGAG] and tnp5393-R [5′-CTCCGGAGATGTCTGGCTTACT]; 300 bp). A portion of the 3′ conserved region of integrons was amplified with primers TniB (5′-GCCGGAAATGGAGCAACT) and orf5 (5′-CAGGGGAGCGAATGGACA). A 223-bp fragment of the sulfonamide resistance gene sulI was amplified with primers sulI-F (5′-CCGGGGATGGGATTTTTCT) and sulI-R (5′-GGGTGCGGACGTAGTCAGC). Thermocycling parameters were 94°C for 2 min followed by 30 cycles of 94°C for 30 s; 58°C (strB and Tn5393), 61°C (3′ conserved region), or 59°C (sulI) for 30 s; and 72°C for 30 s or 2 min (3′ conserved region).
Plasmid purification and analysis.
Plasmids were purified by using the Nucleobond Plasmid Midi kit (Clontech, Palo Alto, Calif.), according to the manufacturer’s directions for low-copy-number plasmids, or by an alkaline lysis minipreparation procedure. Plasmid samples were run on 0.8% agarose gels in 0.5× TBE at 6 V/cm and visualized after staining with ethidium bromide.
Probes for tet genes and strB were generated by PCR as described above with substitution of the following nucleotide mixture: 0.2 μM (each) dATP, dCTP, and dGTP; 0.135 μM dTTP; and 0.65 μM digoxigenin-dUTP (Boehringer Mannheim, Indianapolis, Ind.). Hybridization and detection of digoxigenin-labelled probes were performed according to the manufacturer’s instructions (Boehringer Mannheim) with standard hybridization buffer at 68°C and detection with CDP-Star.
DNA sequencing.
Sequencing reactions were performed at the Michigan State University Department of Energy Plant Research Laboratory Sequencing Facility with ABI dye terminator chemistry. The similarity between new sequences and sequence databases was assessed by use of BLAST 2.0 (5), including algorithms blastn and blastx, via the National Center for Biotechnology Information internet server.
Long PCR for detection and analysis of Tn10, Tn1720, and Tn1404.
PCR fragments longer than 3 kb were amplified by using the Expand Long Template PCR system (Boehringer Mannheim Biochemicals). Reaction mixtures (50 μl) consisted of 1× buffer (either no. 1 or no. 3), 0.35 mM deoxynucleoside triphosphates (Gibco BRL), 0.6 μM primer (Tn10) or 0.3 μM (each) primer (all others), 2.6 U of enzyme mix, and 10 to 100 ng of template DNA.
Full-length Tn10 fragments were amplified with a single PCR primer complementary to the most distal sequences of IS10-L and IS10-R (IS10-1: 5′-CTGATGAATCCCCTAATG) with buffer 1. Cycling parameters were 92°C for 2 min; 25 cycles of 92°C for 10 s, 55°C for 30 s, and 68°C for 8 min with an additional 20 s/cycle added to the 68°C extension for cycles 11 through 25; and then 68°C for 7 min.
Tn1720 fragments were amplified with primers IRL-F (5′-GGGGCATAGAGAAAACGG) and TETA-R (5′-GCAGGCAGAGCAAGTAGAG) with buffer 3. Cycling parameters were 95°C for 5 min; 30 cycles of 95°C for 30 s, 63°C for 30 s, and 68°C for 4 min with an additional 20 s/cycle added to the 68°C extension for cycles 11 through 30; and then 68°C for 7 min.
The left and right halves of Tn1404 were amplified with primers specific for the 5′ end of orfA (Tn1404left [5′-AGCGCCCACATCCAAACACTTACT]) and the region downstream of tetR(C) (TETCleft [5′-CCGGCCCGAACTGGAGCGAGGAAC]) and primers specific for the region downstream of tetR(C) (TETCright [5′-GCACCTGCCGCCCCCTGTCATTC]) and the 3′ end of tnpA (Tn1404right [5′-GGCCGAAACTTCCCGTCCTC]). Reactions were performed with buffer 3 with cycling parameters of 94°C for 2 min; 30 cycles of 94°C for 10 s, 62°C for 30 s, and 68°C for 13 min with an additional 20 s/cycle added to the 68°C extension for cycles 11 through 30; and then 68°C for 7 min. The gene cassette region of the Tn1404 integron was amplified with primers intR (5′-CATAAGCCTGTTCGGTTCGTAA) and sulR (5′-GGGTGCGGACGTAGTCAGC) with buffer 1 with cycling parameters of 94°C for 2 min; 30 cycles of 94°C for 10 s, 58°C for 30 s, and 68°C for 4 min with an additional 20 s/cycle added to the 68°C extension for cycles 11 through 30; and then 68°C for 7 min.
Electroporation.
Electroporation-competent bacteria (50 to 100 μl) (43) were mixed with 1 μl of purified plasmid and pulsed at 25 μF, 200 Ω, and 2.5 kV in a Gene Pulser II apparatus (Bio-Rad Laboratories, Richmond, Calif.) with a 0.2-cm electroporation cuvette. SOC (43) (1 ml) was immediately added. After 1 h at 37°C (E. coli) or 22°C (Pseudomonas sp.), aliquots were plated on LB medium amended with 10 μg of oxytetracycline per ml and incubated overnight.
Detection of transposition.
Plasmids carrying Tet A or Tet C genes were introduced into E. coli JM109[pGEM3zf(+)] by electroporation; pACY177 providing kanamycin resistance was introduced into Tet G-carrying Pseudomonas isolate R82. Transformants were selected on LB medium containing 50 μg of ampicillin (E. coli) per ml or 30 μg of kanamycin (R82) per ml and 10 μg of oxytetracycline per ml. Plasmids were purified from transformants and introduced into E. coli JM109 by electroporation. Transformants resistant to both antibiotics were selected on amended LB medium.
Antibiotic susceptibility testing.
Antibiotic susceptibility of E. coli JM109 transformants was tested on Mueller-Hinton agar supplemented with 50 μg of ampicillin per ml, 200 μg of sulfathiazole per ml, or 20 μg of chloramphenicol per ml and by use of antibiotic-impregnated test discs (gentamicin, 10 μg; kanamycin, 30 μg; trimethoprim, 5 μg; cefoxitin, 30 μg; cefotaxime, 30 μg; cephalothin, 30 μg; piperacillin, 100 μg; ciprofloxacin, 5 μg; oxytetracycline, 30 μg; carbenicillin, 100 μg; Becton Dickinson, Cockeysville, Md.) on a seeded bacterial lawn on Mueller-Hinton agar. The susceptibility of transformants was compared to the sensitivity of E. coli JM109.
Nucleotide sequence accession numbers.
The following nucleotide sequences were entered into GenBank: Tn1720 left end (AF157802), IRRI region (AF157803), and right end (AF157804); Tn1404 left end (AF157797), aadA2 region (AF157798), IS26-flanked Tet C element left (AF157799) and right (AF157800) ends, and right end (AF157801); Tet G from pPSTG1 (AF133139); and Tet G from pPSTG2 (AF133140).
RESULTS
Orchard surveys.
To assess reservoirs of antibiotic resistance within apple orchards, bacterial populations in two Michigan orchards were screened for the presence and prevalence of streptomycin and oxytetracycline resistance (Table 1). Both orchards had a long history of streptomycin usage and were treated with streptomycin and/or oxytetracycline during the study period. The application of oxytetracycline regardless of whether it was alone or in combination with streptomycin generally resulted in lower bacterial populations than did the application of streptomycin used alone. Streptomycin resistance was common at all study sites and did not diminish at orchard 2 over the 2 years of the study in the absence of selection pressure. The number of bacteria in the 1997 streptomycin treatment was quite high due to fire blight infection in one section of orchard 1. The majority of these bacteria were Strr E. amylovora isolates carrying pEa34 and Tn5393 (data not shown) detected at this location for the first time. Tetracycline resistance was much less common than was streptomycin resistance. At orchard 1, Tcr bacteria represented less than 5% of the population. At orchard 2, they were more prevalent but their detection was sporadic; in 20% of the samples from orchard 2, no Tcr bacteria were found, and in 20% of the samples, less than 5% of the population was Tcr (data not shown). Interestingly, most Tcr bacteria also appeared to be Strr.
TABLE 1.
Recovery of bacteria from apple blossoms and leaves sprayed during bloom with oxytetracycline and/or streptomycin
| Yr | Orchard | Treatmenta
|
Bacterial populationb on:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Streptomycin | Oxytetracycline | Blossoms
|
Leaves
|
||||||||
| Total (CFU/blossom) | Strr (%) | Tcr (%) | Strr Tcr (%) | Total (CFU/g) | Strr (%) | Tcr (%) | Strr Tcr (%) | ||||
| 1996 | 1 | + | − | 36,000 a | 28 a | 2 b | NAc | 220,000 a | 43 a | 5 a | NA |
| 1 | + | + | 2,900 b | 56 a | 0 b | NA | 58,000 b | 63 a | <1 a | NA | |
| 2 | − | + | 1,900 b | 56 a | 21 a | NA | 4,500 b | 28 a | 53 a | NA | |
| 2 | + | + | 5,400 b | 26 a | 0 b | NA | 320 b | 22 a | 6 a | NA | |
| 1997 | 1 | + | − | 1,500,000 a | 59 a | 0 b | 0 b | 1,700,000 a | 51 b | <1 b | <1 b |
| 1 | + | + | 150,000 b | 51 a | <1 b | <1 b | 330,000 a | 42 b | <1 b | <1 b | |
| 2 | − | + | 12,000 b | 48 a | 9 b | 7 b | 35,000 a | 46 b | 8 b | 6 b | |
| 2 | + | + | 3,400 b | 68 a | 47 a | 23 a | 110,000 a | 84 a | 21 a | 18 a | |
Antibiotics were applied three times at 290 g/ha. Each treatment pair was replicated three times on three blocks of trees at each orchard. Orchard 1 had not been treated with oxytetracycline prior to the initiation of this experiment, while orchard 2 had been treated approximately twice per season beginning in 1991.
Serial dilutions of washes of 50 blossoms or 100 leaves from three to nine samples per treatment were plated in duplicate on medium with no antibiotics and medium amended as indicated (100 mg of streptomycin sulfate per ml; 2 [1996] or 10 [1997] mg of oxytetracycline per ml). Bacterial colonies were counted after 3 days of incubation at 22°C. Data from each year were tested separately by analysis of variance and Fisher’s range test. Means which were significantly different are indicated by different range indices (a or b). Differences indicated were significant at 0.05.
NA, not assayed.
Tcr colonies (44 from 1996 and 303 from 1997) were subcultured from the orchard survey plates. Gram-negative isolates, representing 98% of the total, were categorized into 87 groups (20 from orchard 1 and 67 from orchard 2) according to source, colony color and morphology, production of fluorescent pigments, and antibiotic resistance levels (data not shown). A representative from each of the 87 groups was tested with Biolog GN Microplates (Table 2). Most isolates were identified as Pantoea (Enterobacter) agglomerans or fluorescent and nonfluorescent Pseudomonas species; several isolates could not be identified by the Biolog system.
TABLE 2.
Tetracycline resistance genes detected in gram-negative bacteria isolated from Michigan apple orchards
| Tcr operon | Bacterial identificationa | 1996 isolate(s)b | 1997 isolate(s) |
|---|---|---|---|
| Tet B | Pantoea (Enterobacter) agglomerans | R79A, R164A, R166A, R168A, R172A | W112, W120, W129, W130, W131, W219, W227, W235, W243, W251, W259, W275, R3, R34, R49, R56, R64, R65, R91, R96, R103, R320, R322, R330, R342, R347, R362, R380, R381, R396, R408, R416, R420, R422, R432, R433 |
| Erwinia chrysanthemi | R35, R107 | ||
| Erwinia stewartii | R392 | ||
| Escherichia vulneris | W260 | ||
| Tet A | Fluorescent Pseudomonas sp. | R1, R88 | |
| Nonfluorescent Pseudomonas sp.—tan | R94A, R171A, R173A | ||
| Nonfluorescent Pseudomonas sp.—yellow | W267, R97, R354, R413, R428 | ||
| Unidentified—fluorescent | R41 | ||
| Unidentified—nonfluorescent | R100, R389 | ||
| Tet C | Fluorescent Pseudomonas sp. | R96A | R9, R17, R25, R33 |
| Nonfluorescent Pseudomonas sp.—tan | R37 | ||
| Nonfluorescent Pseudomonas sp.—yellow | R326, R372 | ||
| Unidentified—tan | R6 | ||
| Unidentified—yellow | R104 | ||
| Tet G | Fluorescent Pseudomonas sp. | R36, R57, R67, R82 | |
| Nonfluorescent Pseudomonas sp.—yellow | R121A | ||
| Unknown | Fluorescent Pseudomonas sp. | W101A, W113A, W205A | |
| Nonfluorescent Pseudomonas sp.—yellow | W199A | W123, W125, R334, R388 | |
| Nonfluorescent Pseudomonas sp.—tan | R370 | ||
| Unidentified—yellow | R72, R338, R407 | ||
| Unidentified—white | R349 | ||
| Unidentified—tan | R424 |
Identification based on MicrologGN plates (Biolog).
Isolates beginning with a W are from orchard 1, and those beginning with an R are from orchard 2. Isolates used in further studies are indicated in bold italics.
Identification of tetracycline resistance determinants by restriction analysis of PCR products.
Alignment of the deduced amino acid sequences of Tet A, B, C, D, E, G, and H (2, 4, 21, 23, 35, 50, 58) revealed several areas of high homology, including regions in putative transmembrane-spanning domains 1 and 4 (16). Two degenerate oligonucleotide primer pairs were designed based on the nucleotide sequences corresponding to these regions. Primers TET1-F and TET1-R were based on the sequences of tetA, -B, and -C, and TET2-F and TET2-R were based on the sequences of tetD, -E, and -H. A PCR product was obtained with at least one pair of primers for 73 of 87 isolates (Fig. 1A and B). TET1-F–TET1-R yielded products for 68 isolates; for several of these isolates TET2-F–TET2-R also gave a product, although it was generally fainter (example in Fig. 1A and B, lane 4) and not always reproducible. For five isolates, a band was obtained only with TET2-F–TET2-R. Isolate R121A gave an additional band of approximately 500 bp which was not observed in other isolates (Fig. 1B, lane 9).
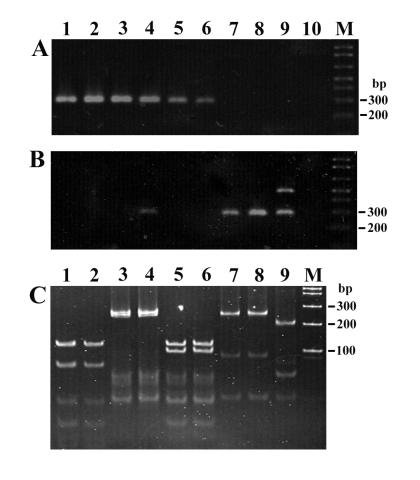
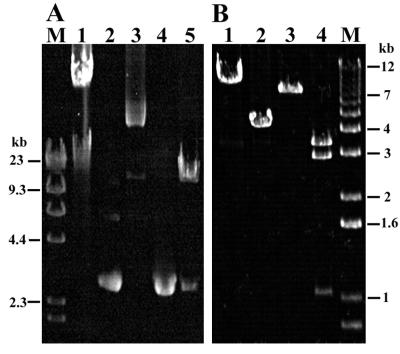
FIG. 1.
Amplification of tetracycline resistance genes from gram-negative bacteria from apple orchards with primers TET1-F–TET1-R (A) and TET2-F–TET2-R (B) and HaeIII restriction analysis of PCR products (C). Lanes 1 to 10 are isolates R171A, R41, R416, W251, R326, R9, R82, R36, R121A, and water control, respectively. Lane M is 1-kb Ladder Plus (Gibco BRL). Amplification results and restriction patterns are consistent with resistance genes tetA (lanes 1 and 2), tetB (lanes 3 and 4), tetC (lanes 5 and 6), and tetG (lanes 7 and 8). Isolate R121A (lane 9) produced a novel result. Similar results were obtained for 64 additional isolates, but no band was amplified from 14 other isolates.
The tet PCR fragments were further classified according to their HaeIII restriction digestion patterns (Fig. 1C). Within the targeted region, tetA to -E, -G, and -H have predicted HaeIII restriction sites which would yield five distinct patterns of major digestion products: 130 and 75 bp (tetA), 240 and/or 254 bp depending on sites within the degenerate primers (tetB and tetG), 123 and 104 bp (tetC), 293 bp (tetE and tetH), and 153 and 140 bp (tetE). Among isolates which yielded products with primers TET1-F–TET1-R, three patterns consistent with the presence of tetA (13 isolates), either tetB or tetG (45 isolates), and tetC (10 isolates) were observed (Table 2). Two patterns were seen among isolates which yielded products only with primers TET2-F–TET2-R: one similar to tetB and tetG (four isolates) and one novel pattern (one isolate). Sequencing of a fragment from each group confirmed the presence of tetA, tetB, and tetC in isolates which yielded products with TET1-F–TET1-R and revealed that the products produced with TET2-F–TET-2R represented distinct variants of tetG which differed at 32 nucleotide positions (data not shown).
Tet B associated exclusively with Tn10.
Eighteen tetB-containing isolates were screened for the presence of Tn10 by PCR with a single primer complementary to the ends of the transposon. A 9.3-kb product was amplified from 16 isolates; larger products were amplified from isolates R34 and R380 (Fig. 2A). Restriction analysis of the 9.3-kb product from isolate W251 yielded the expected BglII and BamHI fragments of Tn10 (18). Restriction analysis of the large PCR products from isolates R34 and R380 suggested that isolate R34 possessed an extra 1.2 kb in the right end of the transposon and that isolate R380 contained an extra 5 to 6 kb between the first BamHI and BglII sites (Fig. 2B). The location of the variation in isolate R34 was mapped to within IS10-R, and a partial sequence of the insert (167 bases of the outer end) was obtained with an IS10-specific primer. The insertion site was found to be 814 bp from the right end of IS10-R. The sequence of the insert was 95% identical to the 3′ end of IS911, a 1,250-bp insertion sequence previously described for Shigella dysenteriae (37) (Fig. 3). These findings are consistent with the presence of a copy of an insertion sequence very similar to IS911 within Tn10 in this isolate.
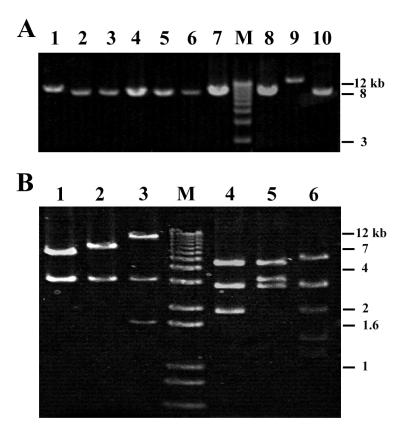
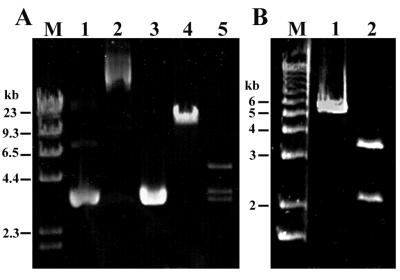
FIG. 2.
Detection and analysis of Tn10 in tetB-containing isolates. (A) PCR amplification of Tn10 with primer IS10-1. Lanes 1 to 10 are isolates R34, R49, R65, W120, W130, W131, W219, W251, R380, and R416, respectively. Reaction products of 9.3 kb were obtained for an additional eight isolates not shown. (B) Variation in BamHI (lanes 1 to 3) and BglII (lanes 4 to 6) restriction digests of Tn10 PCR products from isolates W251 (lanes 1 and 4), R34 (lanes 2 and 5), and R380 (lanes 3 and 6). Lanes M are 1-kb Ladder Plus.
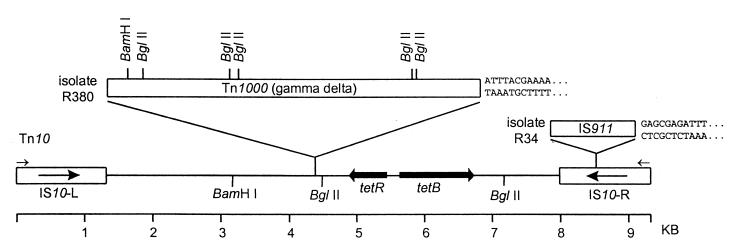
FIG. 3.
Map of Tn10 variants found in isolates of P. agglomerans from Michigan apple orchards. Locations of insertions within Tn10 from isolates R380 (Tn1000) and R34 (IS911) are shown; the sequence flanking the insertion site is given to the right of each element. The positions of BamHI and BglII restriction sites are shown. The priming sites for oligonucleotide IS10-1 used for PCR analysis are indicated by small arrows.
The variation in isolate R380 was mapped to the region downstream of tetR(B). Analysis of the sequence revealed an insertion 332 bp downstream of tetR(B) near the first BglII site of Tn10. The first 435 bases of the insert were sequenced, and they matched the sequence of Tn1000 (9), a 6-kb transposon originally described for E. coli (Fig. 3).
Tet A.
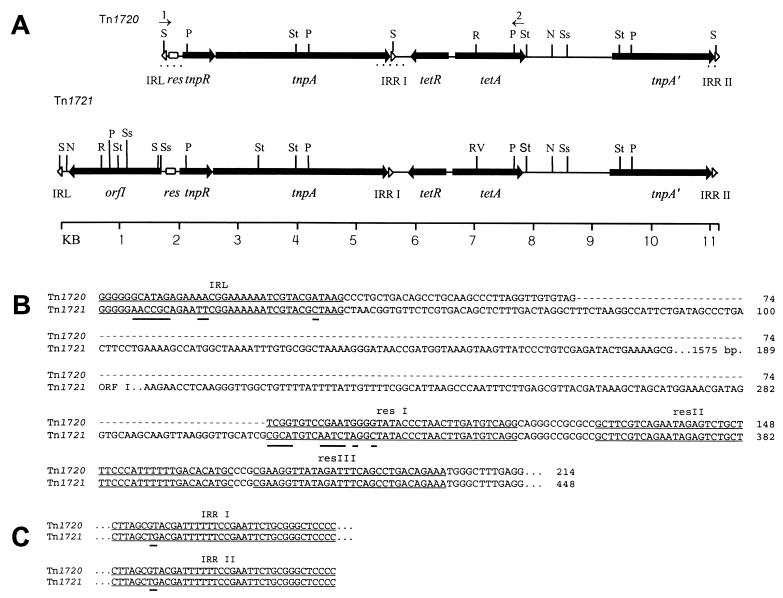
The Tet A operon was located on plasmids in all 10 tetA-containing isolates as determined by Southern blotting of purified plasmids (data not shown). Restriction analysis and partial sequencing of adjacent 11.5- and 12-kb NcoI fragments of the Tcr plasmid from isolate R171A showed that the Tet A operon was located in a 9.3-kb variant of transposon Tn1721, which has been designated Tn1720 (Fig. 4A). Tn1720 and Tn1721 appear to be nearly identical from the resolvase gene (tnpR) region through the final inverted repeat, IRRII. However, at the transposon cointegrate resolution site, located within the first resolvase binding site (resI), the sequences abruptly diverge (Fig. 4B) with Tn1720 lacking the 1,575-bp open reading frame (ORF), orfI, of Tn1721. Minor variation between each of the three inverted repeat sequences was also present (Fig. 4B and C).
FIG. 4.
Comparison of Tn1720, a transposon found on the Tcr plasmid from nonfluorescent Pseudomonas isolate R171A, with Tn1721. (A) Genetic and physical map of Tn1720 and corresponding map of Tn1721. Sequenced regions of Tn1720 are shown as dotted lines beneath the map. Inverted repeat sequences are indicated by open arrowheads and labelled as inverted repeats left (IRL), right I (IRRI), and right II (IRRII). Genes for resolvase (tnpR), transposase (tnpA), tetracycline resistance repressor (tetR), tetracycline resistance protein (tetA), and methyl-accepting chemotaxis-related protein (orfI) and the partial duplication of the transposase gene (tnpA′) are indicated with large filled arrows. Restriction sites are indicated as follows: S, SunI; P, PvuII; St, StuI; R, EcoRV; Ss, SspI; and N, NcoI. The locations of genes indicated in Tn1720 are inferred from similarity of sequence and restriction sites with those of Tn1721. The locations of primer binding sites for IRL-F (1) and TETA-R (2) used for PCR analysis are indicated with numbered small arrows. (B) Alignment of the sequences of Tn1720 and Tn1721 upstream of tnpR. IRL and resolvase binding sites resI, resII, and resIII (40) are underlined. (C) Alignment of the IRRI and IRRII sequences from Tn1720 and Tn1721. Differences within the inverted repeats and res sites are indicated by thick lines under the sequences.
Transposition of Tn1720 was detected in cultures of E. coli JM109 carrying both pGEM3zf(+) and the Tcr plasmid from isolate R171A. A plasmid of approximately 12 kb conferring Tcr was recovered from these cultures (Fig. 5A). Digestion of the plasmid with SunI yielded the predicted digestion products of Tn1720, a 3.8-kb fragment carrying the tnpR and tnpA genes and a 5.5-kb fragment carrying the Tet A operon, and a fragment the size of pGEM3zf(+). Tn1720 was detected in isolate R171A by PCR with primers specific for the IRL and the 3′ end of tetA. The resulting 6-kb product yielded the expected SunI restriction digestion products (Fig. 5B). All 10 tetA-containing isolates yielded PCR products with identical restriction patterns, indicating that all carry the Tet A operon in Tn1720.
FIG. 5.
(A) Transposition of Tn1720. Shown are results of agarose gel electrophoresis of pGEM3zf(+) undigested (lane 1) and PstI digested (lane 3) and the Ampr Tcr plasmid recovered from E. coli JM109 carrying both pGEM3zf(+) and the Tcr plasmid from Pseudomonas isolate R171A undigested (lane 2), PstI digested (lane 4), and SunI digested (lane 5). Lane M is HindIII-digested lambda DNA. (B) PCR detection of Tn1720. Shown is the PCR product from isolate R171A undigested (lane 1) and Sun I digested (lane 2). Lane M is 1-kb Ladder Plus.
Tet C.
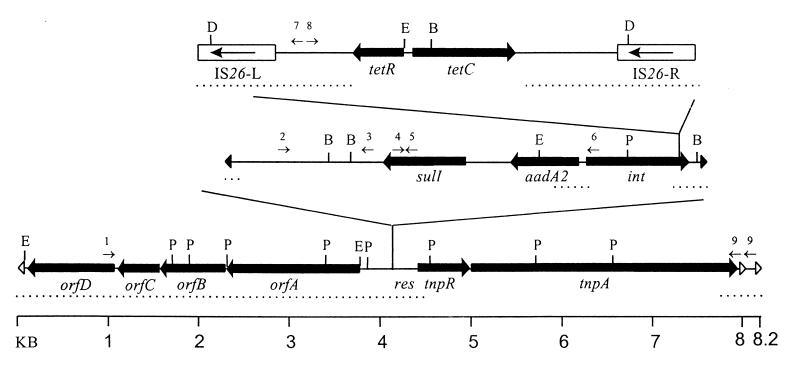
Southern blotting analysis revealed that the Tet C operon was located on plasmids in all nine tetC-containing isolates (data not shown). When the Tcr plasmid from isolate R9 and pGEM3zf(+) were both introduced into E. coli JM109, a plasmid of approximately 20 kb conferring Tcr could be recovered. This plasmid consisted of pGEM3zf(+) with a 19-kb insertion between the SspI sites at positions 2142 and 2580 (Fig. 6A). The insertion was characterized by restriction analysis and sequence analysis of the ends of the element and the regions flanking the Tet C operon (Fig. 7). Inverted repeats of 38 bp similar to those of Tn501 and Tn1721 were found at both ends of the inserted element, and a 5-bp direct repeat of the target sequence flanked the element; the sequence obtained from the right end of the insertion included part of an ORF with high homology (>87%) to the 3′ ends of the tnpA genes of Tn501 and Tn1721. A tandem duplication of the last 216 bp was present, including a portion of the ORF and the 38-bp repeat. The location of PvuII sites matched those predicted for tnpR and tnpA of Tn501, suggesting that both resolvase and transposase genes were present. These data suggest that the mobile element is a transposon related to Tn501, and it has been given the designation Tn1404.
FIG. 6.
(A) Detection of transposition of a Tet C element (Tn1404) from the Tcr plasmid of Pseudomonas sp. isolate R9 to pGEM3zf(+). Shown are the Tcr plasmid from isolate R9 (lane 1), pGEM3zf(+) undigested (lane 2) and SspI digested (lane 4), and the Ampr Tcr plasmid derived from E. coli JM109 carrying both pGEM3zf(+) and the Tcr plasmid from isolate R9 undigested (lane 3) and SspI digested (lane 5). The 0.4-kb SspI fragment of pGEM3zf(+) is not visible in lane 4. Lane M is HindIII-digested lambda DNA. (B) Detection of Tn1404 by PCR with primers pairs Tn1404left-TetCleft and TetCright-Tn1404right. Shown are PCR products from isolate R9 representing the left and right halves of Tn1404 undigested (lanes 1 and 3, respectively) and BamHI digested (lanes 2 and 4, respectively). Lane M is 1-kb Ladder Plus.
FIG. 7.
Physical and genetic map of transposon Tn1404 from Pseudomonas sp. isolate R9. Sequenced regions are indicated with dotted lines beneath the map. Genes for transposase (tnpA), resolvase (tnpR), integrase (int), aminoglycoside adenyltransferase A2 (aadA2), sulfonamide resistance protein (sulI), tetracycline efflux protein class C (tetC), and tetracycline resistance repressor class C (tetR) as well as ORFs designated orfA, orfB, orfC, and orfD are indicated with bold arrows. Insertion sequences are shown as open rectangles (IS26-L and IS26-R) with the direction of the IS26 transposase gene indicated by an arrow. Inverted repeat sequences are indicated by open arrowheads (transposon, 38 bp) and filled arrowheads (integron, 25 bp). Restriction enzyme sites are indicated as follows: D, DraI; E, EcoRI; B, BamHI; and P, PvuII. The locations of primers used for PCR analysis are indicated as numbered small arrows. Primer pairs used for analysis included 1 (Tn1404left) and 7 (TetCleft), 8 (TetCright) and 9 (Tn1404right), 2 (TniB) and 3 (orf5), 4 (sulIR) and 5 (sulIF), and 4 (sulIR) and 6 (intR).
Analysis of the Tet C region demonstrated that copies of IS26 in direct orientation (IS26-L and IS26-R) flank a 3.7-kb sequence whose central portion matches the Tet C region of pSC101 including 622 bases downstream of tetR(C) and 754 bases downstream of tetC. The 145 bases adjacent to IS26-L and the 280 bases adjacent to IS26-R bore no similarity to known sequences.
The sequence surrounding the IS26-flanked Tet C element matched that of the integrase gene (int) of class I integrons and suggested that the Tet C element had been inserted into an int gene 426 bp from the end of an integron. The presence of the sulfonamide resistance gene sulI, found in many class I integrons, was verified by PCR (Fig. 7, primers 4 and 5, and data not shown). A single aadA2 gene cassette (7) was present as determined by analysis of the PCR product from the gene cassette region between sulI and int (Fig. 7, primers 4 and 6). The size of the PCR product from the tni region to downstream of sulI (Fig. 7, primers 2 and 3) was similar to that expected from an integron with truncated tniB but without insertion sequences IS1353 and IS1326, which are found in this region of many class I integrons.
The integron appears to have been inserted into the transposon upstream of the res site. The location of the insertion and the sequence of the res site match exactly the sequence reported for Tn1403 (51), a 19.9-kb transposon from Pseudomonas aeruginosa carrying resistance cassettes PSE-1 (ampicillin), cat (chloramphenicol), and aadA (streptomycin-spectinomycin) within the integron and an additional Strr gene, aphC.
The left end of Tn1404 contained four ORFs of 1,485 (orfA), 849 (orfB), 357 (orfC), and 960 (orfD) bp. The predicted products of orfA and orfB are similar to those from adjacent ORFs in the transposon region of plasmids found in Yersinia enterocolitica (24): a 492-amino-acid (aa) sulfate permease (74% identity; 91% similarity) and a 288-aa protein of unknown function (42% identity; 68% similarity). The predicted orfC product was weakly homologous to DnaK suppressor proteins (DskA) with 43% similarity (22% identity) to the 151-aa product of the E. coli dskA gene. The predicted product of orfD had some similarity (20 to 30% identity; 45 to 48% similarity) to the predicted products of ORFs in a Salmonella typhimurium plasmid (22) (328 aa), the E. coli genome 12 kb upstream of mcrB (10) (323 aa), and an E. coli O157:H7 pathogenicity island (36) (272 aa). Approximately 0.5 kb of sequence from both ends of Tn1403 has been reported; these sequences are identical, suggesting a duplication of the terminal sequences in Tn1403. The sequence from the left end of Tn1404 matched the Tn1403 terminal sequence, and this region of identity included a portion of orfA.
Tn1404 was detected in isolate R9 by PCR amplification of the left (Fig. 7, primers 1 and 7) and right (Fig. 7, primers 8 and 9) halves of the transposon. Fragments of 8.5 to 9.5 kb were obtained and yielded the expected BamHI restriction patterns (Fig. 6B). Identical restriction patterns were obtained from nine additional Tet C-containing isolates (Table 3), implying that all carry Tet C within Tn1404.
TABLE 3.
Association of tetracycline resistance with plasmids, transposons, and other resistance genes
| Tcr operon class | No. of isolates | No. of isolates with:
|
Tcr operon on transposon | Other resistance genes detected in transposon | Replication of Tcr plasmid in E. coli JM109d | Phenotype of E. coli transformantse | |
|---|---|---|---|---|---|---|---|
| Tcr operons detected on plasmids | Tn5393 detected on Tcr plasmid | ||||||
| B | 18 | 7a | 7 | Tn10(n = 18) | None | 7/7 | Tcr |
| A | 10 | 10 | 10 | Tn1720(n = 10) | None | 3/10 | Tcr |
| C | 9 | 9 | 8 | Tn1404(n = 9) | aadA2, sulI | 4/9 | Tcr Sulr |
| Gb | 4 | 4 | 4 | Unknown | 0/4 | ||
| Gc | 1 | 1 | 1 | Unknown | 1/1 | Tcr | |
The presence of Tet B on plasmids in the remaining 11 isolates could not be confirmed due to poor plasmid integrity in preparations from these isolates.
Variant found in isolates R36, R57, R67, and R82.
Variant found in isolate R121A.
Number of plasmids which were replicated/total number of plasmids tested.
Transformants were tested for growth on oxytetracycline, streptomycin, sulfathiazole, ampicillin, gentamicin, kanamycin, trimethoprim, cefoxitin, cefotaxime, cephalothin, piperacillin, and ciprofloxacin. Unless otherwise indicated, isolates were sensitive to the antibiotics.
Tet G.
Five isolates carrying Tet G were detected. Isolate R121A, collected in 1996, contained a variant of Tet G which differed from that found in the four 1997 isolates. In all cases, Tet G was associated with a plasmid; Tet G from isolate R121A was located on a plasmid (pPSTG1) that was smaller than the Tet G-containing plasmids from the four isolates from 1997 which showed identical restriction patterns (data not shown). Of the Tet G plasmids, only pPSTG1 was successfully introduced into E. coli JM109. No transposition of either Tet G operon onto recipient plasmids was detected, suggesting either that transposition of these elements is a rare event or that neither variant resides on a functional mobile element.
The complete sequences of the Tet G operons from pPSTG1 and the plasmid from isolate R82, pPSTG2, were obtained. Tet G from pPSTG2 was identical to the Tet G from S. typhimurium previously reported (8). The Tet G sequence from pPSTG1 was 92% identical to that from pPSTG2. The tetG sequences differed at 85 nucleotide positions, resulting in 20 predicted amino acid differences, resulting in overall 93% nucleotide and 95% amino acid identity. The tetR(G) sequences were 89% identical at both the nucleotide and the amino acid levels with 66 and 22 differences, respectively. Ten nucleotide differences were observed in the regulatory region.
Properties of the resistance plasmids.
All but 1 of the 87 representative isolates were able to grow in the presence of 100 μg of streptomycin per ml. The nature of the streptomycin resistance was evaluated by PCR screening for the strA-strB gene pair and Tn5393, the transposon which is a common carrier of the strA-strB resistance genes in plant-associated bacteria (46, 47). The strB and tnpA genes of Tn5393 were present in 42 of 42 selected isolates (data not shown). The location of strB was evaluated by Southern blotting analysis of uncut plasmid DNA; in most isolates, strB was located on the same plasmid as the tet gene (Table 3).
The ability of the Tcr plasmids to be maintained in other species was tested by electroporation of purified plasmids into E. coli JM109 (Table 3). Plasmids from 3 of 10 Tet A-, 7 of 7 Tet B-, 4 of 7 Tet C-, and 1 of 5 Tet G-containing isolates were successfully introduced into and maintained by E. coli. The three Tet A plasmids had nearly identical EcoRV restriction patterns; the restriction fragment patterns of the four Tet C plasmids were also similar to each other. The Tet B plasmids showed more variation in restriction pattern (data not shown), suggesting that Tet B may be located on a wide variety of plasmids. The Tet A, Tet C, and Tet G plasmids which could not be propagated in E. coli were presumably structurally different from their broader-host-range counterparts.
Even though Tn5393 has been shown to confer streptomycin resistance when introduced into E. coli JM109 (13), none of the E. coli strains transformed with the Tcr Strr plasmids were resistant to streptomycin. Even Tet C transformants, which carried both Tn5393 and aadA2, were sensitive to streptomycin.
The Tcr E. coli transformants were screened for the presence of additional resistance genes by testing their sensitivity to sulfathiazole, chloramphenicol, ampicillin, gentamicin, kanamycin, trimethoprim, cefoxitin, cefotaxime, cephalothin, piperacillin, and ciprofloxacin. As expected, the transformant with Tn1404 was resistant to sulfathiazole. However, resistance to the other antibiotics was not detected in any of the transformants.
DISCUSSION
Detectable numbers of Tcr bacteria were found on apple blossoms and leaves collected from both sites included in this study, even in blocks of trees which had never received an oxytetracycline treatment. Although not all samples from either location yielded Tcr bacteria, orchard 2, which received oxytetracycline applications during the 5 years prior to the study, tended to have higher numbers of Tcr bacteria, suggesting that selection pressure may have been responsible for the greater extent of Tcr and that future applications of oxytetracycline may further increase the populations of Tcr orchard epiphytic bacteria. The detection of a pool of Strr bacteria in the orchard environment is consistent with a number of studies on the evaluation of Strr in plant-pathogenic (31, 33, 45) and nonpathogenic (34, 44) bacteria. The long history of streptomycin usage at both sites is probably responsible for the presence of easily detectable populations of Strr bacteria. Consistent with this observation is the finding that in 1996 the site sprayed with streptomycin alone had more surviving bacteria than the sites which received oxytetracycline. Strr appears to be persistent, as evidenced by the significant Strr populations found on trees at orchard 2 which had not received streptomycin treatment for several years.
To identify the genes responsible for Tcr in orchard bacteria, a PCR-based screening assay was developed for the detection of members of the Tet A resistance gene family. By using two sets of primers and HaeIII restriction analysis of the resulting PCR products, tetA, -B, -C, and -G as well as tetD and tetE (data not shown) could be identified. This method allows identification of Tet A family members without gene-specific probes and the differentiation of variants of tet genes, which is not possible with hybridization-based detection methods.
Five distinct Tcr determinants representing four classes of the Tet A family were detected in a group of gram-negative phyllosphere bacteria inhabiting apple orchards in Michigan. The Tcr determinants primarily were associated with bacteria in two genera—Pantoea and Pseudomonas—and most often were located on plasmids. Tet B was found exclusively in P. agglomerans and a few other rare isolates of the family Enterobacteriaceae. Tet B has previously been found in many bacterial genera and, although it has never been found in Pseudomonas sp., appears to have the widest distribution of all the Tet A family members (39). Tet A, C, and G were found primarily in Pseudomonas sp. Tet G had previously been detected in Vibrio anguillarum (58) and S. typhimurium (8); this is the first report of Tet G in Pseudomonas species.
Two variants of Tet G were found among the orchard isolates. The sequences of the variants showed 7% nucleotide sequence divergence. This is a greater degree of divergence than has been reported for other Tet A family members. For example, the sequences of Tet A from Tn1721 (4) and plasmid RK2 (52) show less than 2% nucleotide variation. None of the 20 predicted amino acid differences between the two Tet G proteins correspond to positions identified as essential for the function of other classes of tetracycline efflux pumps (1, 32, 53–57).
Tcr determinants were not identified in some of the isolates. It is unlikely that these isolates harbor variants of Tet A to E or G because DNA from these isolates failed to hybridize with probes for tetA to -E and -G (41a). These isolates may possess Tet H, I, or J or a previously uncharacterized member of the Tet A family, or they may have resistance to tetracycline conferred by multidrug efflux pumps or another mechanism.
Analysis of the genetic context of Tcr in orchard environments revealed that three of the identified Tcr operons were present on transposons. The presence of Tet B in Tn10 was not surprising, because this determinant has been described only in conjunction with this transposon. In two isolates, an inserted element, either Tn1000 or IS911, was found within Tn10. Neither of the inserted elements carries any known resistance genes. It is unclear whether their presence would have an effect on Tn10 transposition or resistance gene expression; Tn1000 has been shown elsewhere to enhance the segregational stability of plasmids (6).
Tn1720, a variant of Tn1721, was found to be the sole source of Tet A in the two populations surveyed. Tn1720 differs from Tn1721 from the left inverted repeat to the first resolvase binding site, resI. In this region, Tn1721 contains a gene encoding a 56-kDa protein (4) which is not present in Tn1720. The divergence between the two transposons begins precisely at the transposon cointegrate resolution site within resI. Perhaps illegitimate recombination at resI between a Tn1721-type transposon and a related transposon with similar 38-bp inverted repeats and res site led to the formation of Tn1720.
The Tet C operon was carried by a 19-kb transposon designated Tn1404 which appears to consist of a base transposon into which an integron and an IS26-flanked Tcr element have been inserted. Within the base transposon, four ORFs were identified in addition to the transposition genes. One of these is predicted to encode a sulfate permease protein; the function of the remaining three is unknown.
Tn1404 appears to be similar to Tn1403, which was originally identified in P. aeruginosa. Although Tn1403 does not confer Tcr, the transposons have identical terminal inverted repeats, integron insertion sites upstream of res, and sequence of the orfD end extending through at least the first 535 bp. The transposons have different complements of gene cassettes within their integrons: Tn1403 has bla, cat, and aadA cassettes, whereas Tn1404 has a single aadA2 cassette.
Transposition of Tet G was not detected with either Tet G variant. Recent analysis of the ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline resistance gene cluster of S. typhimurium DT104 has revealed the presence of Tet G flanked by integrons in this strain (8). The sequences upstream of the Tet G operons in the Pseudomonas sp. isolates appear to be related to the sequence upstream of Tet G in S. typhimurium DT104 (41a), and further analysis of the Tet G region may provide clues about the divergence and dissemination of these genes.
Given that in Michigan apple orchards, several plasmid-borne Tcr elements, the majority of which are associated with transposable elements, are found in epiphytic bacteria, what is the risk of development of populations of Tcr plant pathogens such as E. amylovora? E. amylovora has been shown to become Tcr upon introduction of pBR322 (tetC; data not shown) or RP1 (tetA) (14, 27), demonstrating the function of these genes in E. amylovora. Because E. amylovora has not been shown to be naturally transformation competent and bacteriophage capable of infecting E. amylovora cannot readily infect P. agglomerans or Pseudomonas sp. (38, 41a), making them unlikely transducing agents, acquisition of resistance genes would most likely occur via conjugation. However, attempts to transfer Tcr plasmids from orchard isolates into E. amylovora or E. coli via conjugation failed. In one Tet A-carrying plasmid, Tn1720 appears to have been inserted within a gene cluster responsible for mating pair formation (data not shown), suggesting that this plasmid would be nonconjugative. Among the plasmids carrying Tn1720 and the plasmids carrying Tn1404 were examples where the restriction pattern of the Tcr plasmid from a fluorescent Pseudomonas sp. matched that from a nonfluorescent Pseudomonas sp., suggesting that the plasmid had been mobilized from one species to another. Furthermore, the plasmids which appear to have been transferred from one Pseudomonas sp. to another could be maintained in E. coli when introduced by electroporation, suggesting that they may be broad host range.
In the case of Strr in Michigan apple orchards, the presence of resistance genes in a bacterial population placed under selection pressure has not lead to rapid dissemination of these genes between all species. The linked aminoglycoside phosphotransferase-encoding genes strA-strB have been shown to be present in variants of Tn5393 on a variety of plasmids in many orchard bacterial species (17, 44). However, only strA-strB in conjunction with a specific variant of Tn5393 on conjugative plasmid pEa34 (11, 31) found in P. agglomerans (Erwinia herbicola) (17, 44) appears to have been transferred to E. amylovora. Furthermore, despite the long history of aggressive oxytetracycline use in peach orchards in the southern United States for control of bacterial spot caused by X. campestris pv. pruni and in pear orchards in the western United States for control of fire blight, there have been no reports of oxytetracycline resistance in these pathogens even though miscellaneous bacteria inhabiting the peach and pear orchards presumably carry Tcr determinants. There have also been no reports of resistance problems in the control of fire blight with oxytetracycline during the 20 years that it has been used in some regions of the United States. These observations taken together suggest that development of Tcr E. amylovora will not necessarily be a short-term consequence of oxytetracycline application in apple orchards.
To better understand the potential impacts of the agricultural use of oxytetracycline on human health, the association of Tcr with resistance to other antibiotics was studied. In the majority of the isolates, Tcr plasmids also carried Strr genes. In the case of Tet C-carrying plasmids, Tcr was also linked to sulfonamide resistance and an additional streptomycin resistance determinant. No other resistance phenotypes were detected in association with the broader-host-range Tcr plasmids (i.e., those capable of being maintained in E. coli in addition to the original isolate) which were screened for resistance to a variety of antibiotics. These results suggest that application of oxytetracycline to apple orchards could lead to increasing numbers of commensal bacteria inhabiting the aerial parts of apple trees which possess resistance to tetracyclines, streptomycin, and sulfonamides on plasmids which may be transferred to other bacterial species but that buildup of resistance to other antibiotics in these populations would not be an immediate consequence.
ACKNOWLEDGMENTS
This research was supported in part by the Michigan Agricultural Experiment Station, the Michigan Apple Research Committee, and USDA/CREEES grant no. 97-34367-3967.
REFERENCES
- 1.Allard J D, Bertrand K P. Membrane topology of the pBR322 tetracycline resistance protein. TetA-PhoA gene fusions and implications for the mechanism of TetA membrane insertion. J Biol Chem. 1992;267:17809–17819. [PubMed] [Google Scholar]
- 2.Allard J D, Bertrand K P. Sequence of a class E tetracycline resistance gene from Escherichia coli and comparison of related tetracycline efflux proteins. J Bacteriol. 1993;175:4554–4560. doi: 10.1128/jb.175.14.4554-4560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard J D, Gibson M L, Vu L H, Nguyen T T, Bertrand K P. Nucleotide sequence of class D tetracycline resistance genes from Salmonella ordonez. Mol Gen Genet. 1993;237:301–305. doi: 10.1007/BF00282811. [DOI] [PubMed] [Google Scholar]
- 4.Allmeier H, Cresnar B, Greck M, Schmitt R. Complete nucleotide sequence of Tn1721—gene organization and a novel gene-product with features of a chemotaxis protein. Gene. 1992;111:11–20. doi: 10.1016/0378-1119(92)90597-i. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellani M, Shlain V, Nudel C, SanchexRivas C. Tn1000 (gamma delta) insertions stabilize pBR322-derived recombinant plasmids in Escherichia coli. Biotechnol Lett. 1997;19:1–5. [Google Scholar]
- 7.Bito A, Susani M. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob Agents Chemother. 1994;38:1172–1175. doi: 10.1128/aac.38.5.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broom J E, Hill D F, Hughes G, Jones W A, McNaughton J C, Stockwell P A, Petersen G B. Sequence of a transposon identified as Tn1000 (gamma delta) DNA Sequence. 1995;5:185–189. doi: 10.3109/10425179509029361. [DOI] [PubMed] [Google Scholar]
- 10.Burland V, Plunkett G R, Sofia H J, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome VI: DNA sequence of the region from 92.8 through 100 minutes. Nucleic Acids Res. 1995;23:2105–2119. doi: 10.1093/nar/23.12.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou C S, Jones A L. The analysis of plasmid-mediated streptomycin resistance in Erwinia amylovora. Phytopathology. 1991;81:710–714. [Google Scholar]
- 12.Chiou C S, Jones A L. Molecular analysis of high-level streptomycin resistance in Erwinia amylovora. Phytopathology. 1995;85:324–328. [Google Scholar]
- 13.Chiou C S, Jones A L. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J Bacteriol. 1993;175:732–740. doi: 10.1128/jb.175.3.732-740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho J J, Panopoulos N J, Schroth M N. Genetic transfer of Pseudomonas aeruginosa R factors to plant pathogenic Erwinia species. J Bacteriol. 1975;122:192–198. doi: 10.1128/jb.122.1.192-198.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman D C, Chopra I, Shales S W, Howe T G, Foster T J. Analysis of tetracycline resistance encoded by transposon Tn10: deletion mapping of tetracycline-sensitive point mutations and identification of two structural genes. J Bacteriol. 1983;153:921–929. doi: 10.1128/jb.153.2.921-929.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckert B, Beck C F. Topology of the transposon Tn10-encoded tetracycline resistance protein within the inner membrane of Escherichia coli. J Biol Chem. 1989;264:11663–11670. [PubMed] [Google Scholar]
- 17.Fernando, G. D. W., and A. L. Jones. 1999. Unpublished observations.
- 18.Foster T J, Davis M A, Roberts D E, Takeshita K, Kleckner N. Genetic organization of transposon Tn10. Cell. 1981;23:201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- 19.George A M, Levy S B. Gene in the major co-transduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol. 1983;155:541–548. doi: 10.1128/jb.155.2.541-548.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hachler H, Cohen S P, Levy S B. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1991;173:5532–5538. doi: 10.1128/jb.173.17.5532-5538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen L M, McMurry L M, Levy S B, Hirsh D C. A new tetracycline resistance determinant, Tet H, from Pasteurella multocida specifying active efflux of tetracycline. Antimicrob Agents Chemother. 1993;37:2699–2705. doi: 10.1128/aac.37.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heffernan E J, Harwood J, Fierer J, Guiney D. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillen W, Schollmeier K. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 1983;11:525–539. doi: 10.1093/nar/11.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann B, Strauch E, Gewinner C, Nattermann H, Appel B. Characterization of plasmid regions of foodborne Yersinia enterocolitica biogroup 1A strains hybridizing to the Yersinia enterocolitica virulence plasmid. Syst Appl Microbiol. 1998;21:201–211. doi: 10.1016/S0723-2020(98)80024-6. [DOI] [PubMed] [Google Scholar]
- 25.Kehrenberg C, Werckenthin C, Schwarz S. Tn5706, a transposon-like element from Pasteurella multocida mediating tetracycline resistance. Antimicrob Agents Chemother. 1998;42:2116–2118. doi: 10.1128/aac.42.8.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E H, Aoki T. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microbiol Immunol. 1994;38:31–38. doi: 10.1111/j.1348-0421.1994.tb01741.x. [DOI] [PubMed] [Google Scholar]
- 27.Lacy G H, Stromberg V K, Cannon N P. Erwinia amylovora mutants and in planta-derived transconjugants resistant to oxytetracycline. Can J Plant Pathol. 1984;6:33–39. [Google Scholar]
- 28.Li X Z, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Z Q, Farrand S K. Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J Bacteriol. 1998;181:618–626. doi: 10.1128/jb.181.2.618-626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McManus P S, Jones A L. Epidemiology and genetic analysis of streptomycin-resistant Erwinia amylovora from Michigan and evaluation of oxytetracycline for control. Phytopathology. 1994;84:627–633. [Google Scholar]
- 32.McMurry L M, Stephan M, Levy S B. Decreased function of the class B tetracycline efflux pump protein Tet with mutations at aspartate 15, a putative intramembrane residue. J Bacteriol. 1992;174:6294–6297. doi: 10.1128/jb.174.19.6294-6297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minsavage G V, Canteros B I, Stall R E. Plasmid-mediated resistance to streptomycin in Xanthomonas campestris pv vesicatoria. Phytopathology. 1990;80:719–723. [Google Scholar]
- 34.Norelli J L, Burr T J, Lo Cicero A M, Gilbert M T, Katz B H. Homologous streptomycin resistance gene present among diverse gram-negative bacteria in New York State apple orchards. Appl Environ Microbiol. 1991;57:486–491. doi: 10.1128/aem.57.2.486-491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peden K W C. Revised sequence of the tetracycline resistance gene of pBR322. Gene. 1983;22:277–280. doi: 10.1016/0378-1119(83)90112-9. [DOI] [PubMed] [Google Scholar]
- 36.Perna N T, Mayhew G F, Posfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prere M F, Chandler M, Fayet O. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J Bacteriol. 1990;172:4090–4099. doi: 10.1128/jb.172.7.4090-4099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie D F, Klos E J. Isolation of Erwinia amylovora bacteriophage from the aerial parts of apple trees. Phytopathology. 1977;67:101–104. [Google Scholar]
- 39.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 40.Rogowsky P, Halford S E, Schmitt R. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 1985;4:2135–2141. doi: 10.1002/j.1460-2075.1985.tb03904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnabel E L, Jones A L. Instability of a pEA29 marker in Erwinia amylovora previously used for strain classification. Plant Dis. 1998;82:1334–1336. doi: 10.1094/PDIS.1998.82.12.1334. [DOI] [PubMed] [Google Scholar]
- 41a.Schnabel, E. L., and A. L. Jones. Unpublished observations.
- 42.Schroth M N, Thompson S V, Moller W J. Streptomycin resistance in Erwinia amylovora. Phytopathology. 1979;69:565–568. [Google Scholar]
- 43.Seidman C E, Struhl K, Sheen J, Jessen T. Introduction of plasmid DNA into cells. In: Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 1.8.1–1.8.10. [DOI] [PubMed] [Google Scholar]
- 44.Sobiczewski P, Chiou C-S, Jones A L. Streptomycin-resistant epiphytic bacteria with homologous DNA for streptomycin resistance in Michigan apple orchards. Plant Dis. 1991;75:1110–1113. [Google Scholar]
- 45.Sundin G W, Bender C L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. Syringae. Appl Environ Microbiol. 1993;59:1018–1024. doi: 10.1128/aem.59.4.1018-1024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundin G W, Demezas D H, Bender C L. Genetic and plasmid diversity within natural populations of Pseudomonas syringae with various exposures to copper and streptomycin bactericides. Appl Environ Microbiol. 1994;60:4421–4431. doi: 10.1128/aem.60.12.4421-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundin G W, Monks D E, Bender C L. Distribution of the streptomycin-resistance transposon Tn5393 among phylloplane and soil bacteria from managed agricultural habitats. Can J Microbiol. 1995;41:792–799. doi: 10.1139/m95-109. [DOI] [PubMed] [Google Scholar]
- 48.Suslow T V, Schroth M N, Isaka M. Application of a rapid method for gram differentiation of plant pathogenic and saprophytic bacteria without staining. Phytopathology. 1982;72:917–918. [Google Scholar]
- 49.Tietze E. Tet Y, a novel tetracycline resistance determinant encoded by the IncQ plasmid pIE1120. Unpublished data. GenBank accession no. AF070999. 1998. [Google Scholar]
- 50.Varela M F, Griffith J K. Nucleotide and deduced protein sequences of the class D tetracycline resistance determinant: relationship to other antimicrobial transport proteins. Antimicrob Agents Chemother. 1993;37:1253–1258. doi: 10.1128/aac.37.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vezina G, Levesque R C. Molecular characterization of the class II multiresistance transposable element Tn1403 from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:313–321. doi: 10.1128/aac.35.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waters S H, Rogowsky P, Grinsted J, Altenbuchner J, Schmitt R. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 1983;11:6089–6105. doi: 10.1093/nar/11.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi A, Adachi K, Akasaka T, Ono N, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon Tn10. Histidine 257 plays an essential role in H+ translocation. J Biol Chem. 1991;266:6045–6051. [PubMed] [Google Scholar]
- 54.Yamaguchi A, Akasaka T, Ono N, Someya Y, Nakatani M, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. Roles of the aspartyl residues located in the putative transmembrane helices. J Biol Chem. 1992;267:7490–7498. [PubMed] [Google Scholar]
- 55.Yamaguchi A, Ono N, Akasaka T, Noumi T, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon, Tn10. The role of the conserved dipeptide, Ser65-Asp66, in tetracycline transport. J Biol Chem. 1990;265:15525–15530. [PubMed] [Google Scholar]
- 56.Yamaguchi A, Ono N, Akasaka T, Sawai T. Serine residues responsible for tetracycline transport are on a vertical stripe including Asp-84 on one side of transmembrane helix 3 in transposon Tn10-encoded tetracycline/H+ antiporter of Escherichia coli. FEBS Lett. 1992;307:229–232. doi: 10.1016/0014-5793(92)80773-a. [DOI] [PubMed] [Google Scholar]
- 57.Yamaguchi A, Someya Y, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by transposon Tn10. The role of a conserved sequence motif, GXXXXRXGRR, in a putative cytoplasmic loop between helices 2 and 3. J Biol Chem. 1992;267:19155–19162. [PubMed] [Google Scholar]
- 58.Zhao J, Aoki T. Nucleotide-sequence analysis of the class-G tetracycline resistance determinant from Vibrio anguillarum. Microbiol Immunol. 1992;36:1051–1060. doi: 10.1111/j.1348-0421.1992.tb02109.x. [DOI] [PubMed] [Google Scholar]