Abstract
The effect of different substrates and various developmental stages (mycelium growth, primordium appearance, and fruiting-body formation) on laccase production in the edible mushroom Lentinula edodes was studied. The cap of the mature mushroom showed the highest laccase activity, and laccase activity was not stimulated by some well-known laccase inducers or sawdust. For our molecular studies, two genomic DNA sequences, representing allelic variants of the L. edodes lac1 gene, were isolated, and DNA sequence analysis demonstrated that lac1 encodes a putative polypeptide of 526 amino acids which is interrupted by 13 introns. The two allelic genes differ at 95 nucleotides, which results in seven amino acid differences in the encoded protein. The copper-binding domains found in other laccase enzymes are conserved in the L. edodes Lac1 proteins. A fragment of a second laccase gene (lac2) was also isolated, and competitive PCR showed that expression of lac1 and lac2 genes was different under various conditions. Our results suggest that laccases may play a role in the morphogenesis of the mushroom. To our knowledge, this is the first report on the cloning of genes involved in lignocellulose degradation in this economically important edible fungus.
Lentinula edodes (Berk.) Pegler. is the second most widely cultivated mushroom in the world, with a worldwide production in 1995 of 1.7 million tons (9). The cultivation of this mushroom uses a significant amount of lignocellulosic substrates and is the largest wood-utilizing bioconversion process (33). Utilization of lignocellulose by L. edodes is dependent on its ability to synthesize hydrolytic and oxidative enzymes which convert the individual components of lignocellulose into low-molecular-weight compounds that can be absorbed and assimilated for nutrition (32, 33). Laccases are one group of enzymes that are believed to be important in the degradation of lignocellulose by the fungus (14, 21, 26, 29).
Laccases are members of the blue copper oxidase enzyme family characterized by having four cupric ions coordinated such that each of the known magnetic species is associated with a single polypeptide chain (26, 46). The copper-binding domains are highly conserved in the blue copper oxidases (39, 46). The crystal structure of the Cu-depleted laccase from Coprinus cinereus has provided a useful model for the structure of the laccase active site (18). In contrast to our understanding of the electron transfer reactions that occur in laccases, few studies have addressed the physiological functions of these enzymes (19, 46). Laccases have been implicated in pigmentation (11, 33), fruiting-body formation (16, 51), pathogenicity (31, 49, 50), and lignin degradation (1, 4, 8, 26). However, only a few of these functions have been experimentally proven (20).
There have been a few studies on laccase production in L. edodes (7, 32, 33). Leatham and Stahmann (32) found that the highest laccase activity is in the pigmented rind of the pileus and in the stipe. Increased laccase activity was found to be associated with rapid growth of nonpigmented aerial mycelium and formation of pigmented primodia and fruiting bodies (32, 33). Although these studies have provided valuable information on mushroom laccase physiology, molecular studies on this enzyme have not been widely investigated, and only a single study on the isolation and characterization of two laccase gene fragments has been reported (17). As such, we aimed to address this issue by cloning and characterizing two allelic variants of the L. edodes lac1 gene. Also, we aimed to compare the kinetics of laccase activity and gene expression (lac1 and lac2) in L. edodes L54 when grown on different substrates (glucose, crystalline cellulose, potato extracts [PE], sawdust, guaiacol, tannic acid, etc.) and in various developmental stages (mycelium growth, primordium appearance, and fruiting-body formation).
MATERIALS AND METHODS
Organisms and culture conditions.
L. edodes L54-A (monokaryon), L54-B (monokaryon), and their mated product L54 (dikaryon) were used in the study, and a defined medium with a high nitrogen level (HN medium) was used to produce laccases (56, 57). The concentrations of nitrogen and copper (CuSO4) in HN medium were 26 and 40 mM, respectively. For solid medium, agar was added to a final concentration of 15 g/liter, and 20 ml of sterilized solid medium was added per 90-mm petri dish. Culture conditions were described previously (55, 57). In liquid medium, stationary cultures were incubated in 500-ml flasks containing 50 ml of liquid medium. To study the effect of various carbon sources, 1% (wt/vol) glucose was replaced by different substrates, i.e., were 1% (wt/vol) crystal cellulose (Sigma), 1% (wt/vol) PE (Difco), and 5% (wt/vol) sawdust. Four well-known laccase inducers, guaiacol, tannic acid, 2,4-xylidine, and veratryl alcohol (all purchased from Sigma), were added at various concentrations to the medium to determine whether they could induce the production of laccases (11, 13, 46). Fruiting-body primodia and mature mushrooms were obtained during a 6-week inoculation. Mycelium block, pregrown for 14 days as colonies on solid HN-PE agar medium supplemented with 0.1% (wt/vol) yeast extract, was used as inoculum for cultures to grow on the fruiting medium. The fruiting process was carried out in HN medium supplemented with 1% (wt/vol) PE and 5% (wt/vol) sawdust. Cultures were incubated at 25°C and 80% relative humidity on a 12-h light and dark cycle. Primordia and mushrooms were harvested from the first appearance of fruiting bodies (5 to 7 days after fruiting-body initiation) to the harvest stage (10 to 12 days after fruiting-body initiation).
Enzyme assays.
To determine extracellular enzyme activity in agar medium, plugs containing mycelia from the center of the fungal colony were added to the reaction buffer (at a ratio of 50 mg of plug per ml of reaction buffer). Boiled agar plugs (10 min) served as controls. To detect activity in submerged cultures, culture supernatant was used in the reaction mixtures. Blocks of fresh fruiting bodies from various developmental stages were used for the measurement of enzyme activity. Laccase activity was determined by using 2,2′-azinobis-3-ethylbenthiazoline-6-sulfonate (ABTS) as previously described (57). Oxidation of ABTS was measured by determining the increase in absorbance at 420 nm with an extinction coefficient of 36 mM−1 cm−1. One unit of enzyme activity is defined as the amount of enzyme required to oxidize 1 μmol of ABTS per min. Lignin peroxidase, manganese-dependent peroxidases and tyrosinase activities were determined as previously described (28, 35, 36). All the reactions were performed at 30°C. Enzyme activity was expressed as units per gram of medium or fresh fruiting body. Protein concentrations were estimated as described previously (56).
Preparation of high-molecular-mass DNA and total RNA.
High-molecular-mass genomic DNA (50 to 100 kb) was prepared as previously described (55, 58). Total RNA was prepared by using TRIREAGENT (MEC). Only mycelium or fruiting bodies which showed the highest laccase activity were used in RNA extraction. The RNA concentrations in samples were determined spectrophotometrically (43).
Preparation of genomic libraries.
L. edodes genomic DNA was partially digested with Sau3A and separated on a 0.8% (wt/vol) agarose gel (43). After digestion, the DNA was ligated with DASHII arms (Stratagene) as described by the supplier. The ligation was packaged in vitro with a Gigapack II kit (Stratagene). The titer of the library was determined, and the library was amplified with Escherichia coli K2 cells. The unamplified library was estimated to contain 20,000 independent recombinants, which was calculated to cover at least 99% of the L. edodes (L54) genome (43).
Library screening and product cloning.
Approximately 200 ng of genomic DNA or 50 ng of cDNA was added to a PCR mix containing 2.0 U of Taq polymerase (Promega), 1× buffer (Promega), 2.5 mM MgCl2, 100 μM (each) deoxynucleoside triphosphates, and 1.0 μM concentrations (each) of two primers. Four primers in four combinations were used for library screening: LelacU1 (5′-CACTGGCATGGCCTCTTCCA-3′) (17), LelacL1 (5′-ATGGCTATGGTACCAGAAAGTG-3′) (17), T3 (5′-AATTAACCCTCACTAAAGGG-3′), and T7 (5′-GTAATACGACTCACTATAGGGC-3′). The following PCR cycle parameters were used: 4 min at 94°C for one cycle; 1 min at 94°C, 1 min at 58°C, and 5 min at 72°C for 35 cycles; and 10 min at 72°C for one cycle. PCR products were cloned into PCRscript SK (Stratagene) or sequenced directly (see below).
First-strand cDNA synthesis.
Total RNA (5.0 μg) was used to synthesize cDNA. Reverse transcriptions were carried out in 20-μl reaction mixtures containing 50 U of Moloney murine leukemia virus reverse transcriptase (GIBCO), 15 pmol of oligo(dT)15, and 20 U of RNasin (Promega). Reactions were performed at 25°C for 10 min, at 45°C for 45 min, and at 75°C for 5 min.
Competitive PCR.
Relative transcript levels of L. edodes laccase genes were determined by competitive PCR (5, 6, 14). Included in the PCR mixes was a competitive template in the form of genomic DNA. The competitor was diluted to known concentrations. Introns within the competitive template allowed the target cDNA and genomic product to be size fractionated on agarose gels. PCRs were performed with various dilutions of genomic template. To examine the expression of laccase genes in L54, we designed PCR primers based on the sequence reported here (lac1 sequence) and sequences published in the literature (lac2) (17). Primers Lelac1U1523 (5′-GGTGTAGCATTTGTTTCTCA-3′) and Lelac1L2129 (5′-ATGACCGCGAGAGGAACAGC-3′) were used to amplify lac1. The lengths of the PCR products were 626 and 394 bp for lac1 genomic DNA and cDNA, respectively. Primers Lelac2U1 (5′-CATTGGCATGGTCTCTTCCA-3′) and Lelac2L710 (5′-CGATGATGGTGAGGTTGT-3′) were used to amplify lac2. The lengths of the PCR products were 730 and 450 bp for lac2 genomic DNA and cDNA, respectively. A pair of primers specific for amplification of the constitutively expressed ribosome protein gene was used as the control (55).
The competitive templates consisted of full-length genomic copies of the genes which had been PCR amplified, and the concentrations of templates were estimated by gel electrophoresis. To determine the concentration of cDNA, a serial titration test including 20 to 40 cycles was performed. The optimal competitive PCR conditions were (in a final volume of 20 μl) 0.2 U of Taq polymerase, 1× buffer (Promega), 2.5 mM MgCl2, 100 μM (each) deoxynucleoside triphosphates, and 1.0 μM concentrations (each) of the primers. The following competitive PCR parameters were used: 4 min at 94°C for one cycle; 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C for 30 cycles; and 10 min at 72°C for one cycle. The PCR products were size fractionated in 2% agarose, ethidium bromide stained, and analyzed by using Molecular Analyst Software 1.5 (Bio-Rad).
DNA sequencing.
The nucleotide sequences of PCR products were determined by using Taq polymerase cycle sequencing and an automated DNA sequencer (ABI 310; Perkin-Elmer Corp.). All DNAs were sequenced on both strands, and the encoded amino acid sequences were predicted by using Gene Jockey (Biosoft). Sequences were aligned by using SeqEd 2.0 software (Applied Biosystems).
Nucleotide sequence accession numbers.
The nucleotide sequences of the L. edodes lac1A and lac1B genes have been deposited in the GenBank database under accession no. AF153610 and AF153611, respectively.
RESULTS
Effect of different media on the production of laccases.
The effect of glucose, cellulose, sawdust, and PE on the production of laccases was studied (Table 1). In strain L54, peak laccase activity (0.081 U/g) in the HN-glucose medium was detected after 20 days. The presence of 1% PE stimulated laccase production 2.8-fold (Table 1). The presence of 5% sawdust in agar medium did not increase laccase activity, and 1% cellulose slightly increased the peak level of laccase for this strain (Table 1). Monokaryotic strains L54-A and L54-B showed a similar pattern of laccase production in agar medium, and L54-A produced more laccase than did L54-B in all the media tested. Both monokaryotic strains showed less laccase activities than did their mated dikaryotic strain L54 (data not shown).
TABLE 1.
Effect of different media and conditions on the production of enzymes and transcript levels of laccase genes
| Medium and conditions | Laccase activity in L54 (U/g)a | cDNA concnb (mol/μg of total RNA) for:
|
Ratio (lac1/lac2) | |
|---|---|---|---|---|
| lac1 | lac2 | |||
| L54 mycelium | ||||
| HN–1% glucose | 0.081 ± 0.005 (20) | (7.2 ± 0.8) × 10−21 | (2.7 ± 0.3) × 10−22 | 26.7 |
| HN–1% cellulose | 0.105 ± 0.004 (24) | (5.8 ± 0.6) × 10−20 | (5.3 ± 0.5) × 10−20 | 1.1 |
| HN–1% glucose–2 mM guaiacol | 0.076 ± 0.005 (24) | (3.0 ± 0.4) × 10−21 | (5.0 ± 0.4) × 10−20 | 0.06 |
| HN–1% glucose–1% PE | 0.307 ± 0.014 (20) | (1.4 ± 0.3) × 10−19 | (5.3 ± 0.8) × 10−20 | 2.6 |
| HN–1% glucose–1% PE–5% sawdust | 0.300 ± 0.018 (24) | (7.2 ± 0.9) × 10−19 | (2.7 ± 0.8) × 10−19 | 2.7 |
| L54 fruiting body | ||||
| Primordia | 0.083 ± 0.005 | (3.6 ± 0.7) × 10−18 | (2.7 ± 0.4) × 10−18 | 1.3 |
| Fruiting body (cap) | 1.113 ± 0.034 | (5.0 ± 0.4) × 10−17 | (3.0 ± 0.3) × 10−17 | 1.7 |
| Fruiting body (stipe) | 0.033 ± 0.002 | (3.6 ± 0.8) × 10−18 | (1.7 ± 0.2) × 10−18 | 2.1 |
Mean ± standard deviation from triplicate cultures. Numbers in parentheses indicate the time required for maximum activity (in days).
Mean ± standard deviation from triplicate cultures.
Addition of 2 mM guaiacol did not stimulate the production of laccase in agar medium significantly, whereas 5 mM tannic acid decreased the peak activity of laccases (data not shown). Both guaiacol and tannic acid decreased the growth of the monokaryotic and dikaryotic strains but had less effect on the latter. Tannic acid (5 mM) fully inhibited the growth of L54-B. Veratryl alcohol and xylidine neither increased the peak activity of laccases nor affected the growth of the strains tested (data not shown).
The production of laccase in various developmental stages was also studied. Interestingly, the laccase activity in the mushroom cap was 1.113 U/g, which was 34-fold higher than that in the stalk (0.033 U/g) and 13-fold higher than that in the primordia (0.083 U/g). No tyrosinase, MnP and LiP activities were detected in various fruiting stages.
Isolation and analysis of the laccase genomic sequence.
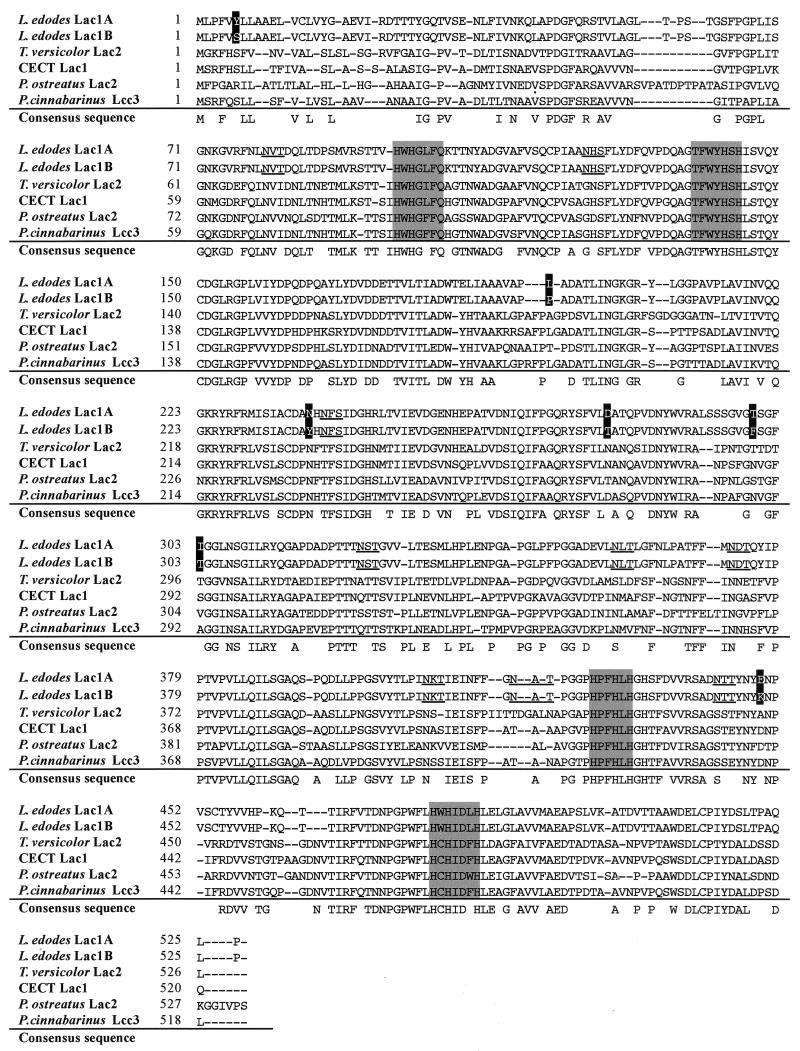
By using a PCR-walking method, which involved gene-specific primers and internal primers (T3 and T7) (55), we obtained four PCR products which showed strong homology to known basidiomycete laccase genes. Based on the sequence overlap analysis, we found that the four fragments formed two laccase genes. By comparing both laccase sequences with the sequences of the corresponding PCR products from monokaryotic parental strains L54-A and L54-B, we were able to demonstrate that the two genes were alleles (Fig. 1); therefore, the laccase genes were designated lac1A and lac1B for strains L54-A and L54-B, respectively.
FIG. 1.
Alignment of the deduced Lac1A and Lac1B amino acid sequences with other basidiomyceteous laccase sequences. The different amino acid sequences between Lac1A and Lac1B are indicated by solid boxes. Four pontential copper-binding domains are shaded. Nine possible N-glycosylation sites are underlined. Accession numbers of sequences are as follows: T. versicolor Lac2, D84235 (13); CECT Lac1, U65400 (37); P. ostreatus Lac2, Q12739 (25); P. cinnabarinus Lcc3, AF025481 (20). Gaps have been introduced for optimal alignment and are indicated by dashes. The consensus sequence is composed of residues shared by at least five of the mature proteins. Alignment was done with SeqEd 2.0 software.
lac1 sequence analysis.
The nucleotide sequence of lac1 was found to contain an open reading frame (ORF) of 1,578 bp capable of coding for a protein of 526 amino acid residues (Fig. 1). It was estimated that Lac1A has a molecular mass of 57,111 Da. The extreme N-terminal portion of this putative protein sequence is positively charged and is separated from the rest of the molecule by a stretch of predominantly hydrophobic residues, features typical of a signal peptide (11, 19, 46). Comparison of the genomic and cDNA sequences of lac1 indicated the presence of 13 introns varying in size from 50 to 65 bp. All of the intron splice junctions correspond to the GT----AG rule. Comparison between lac1A and lac1B revealed that there were 45 nucleotide differences in the ORF and 50 nucleotide differences in all introns except one (data not shown). Seven of the nucleotide changes in the ORF result in amino acid changes, and the remaining 38 nucleotide differences are silent changes. The copper-binding domain structure found in other laccase genes is conserved in the L. edodes Lac1 protein (12, 20). The similarity between the lac1 product and other basidiomycetous laccases is from 65% to 75%, with the region of highest conservation being found in the copper-binding domains (Fig. 1).
Competitive PCR analysis of laccase gene expression.
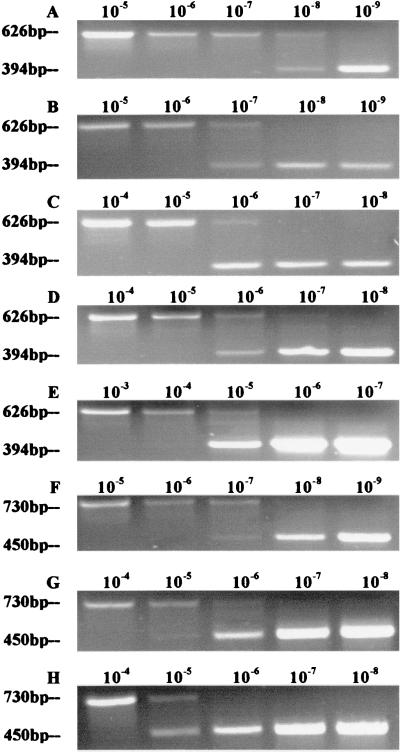
The results of the discriminatory lac1/lac2 competitive PCR from various substrates and different developmental stages are shown in Fig. 2 and Table 1. The highest and lowest levels obtained for lac1 mRNA were 5.0 × 10−17 mol per μg of total RNA in the cap and 3.0 × 10−21 mol per μg of total RNA in HN-glucose-guaiacol medium. Similar results were obtained for lac2. The highest and lowest levels obtained for lac2 mRNA were 3.0 × 10−17 and 2.7 × 10−22 mol, respectively. The use of 1% cellulose as a carbon source increased the levels of lac1 and lac2 mRNA. HN-glucose-PE medium increased the production of both mRNAs, but 5% sawdust did not increase the accumulated mRNA level further (Table 1). The addition of 2 mM guaiacol increased the production of lac2 but not of lac1. The cap of the mushroom showed the highest levels of lac1 and lac2, whereas the stipe and primodia showed lower levels of both mRNAs (Table 1).
FIG. 2.
Competitive PCRs of lac1 (A to E) and lac2 (F to H) transcripts. The amounts of the competitive templates are indicated above the gels in dilution factors. The levels of transcripts in samples were based on estimated equivalence points between competitive product and target cDNAs. The sizes of the PCR products in base pairs are indicated on the left. lac1: HN–1% glucose–guaiacol (A); HN–1% glucose–cellulose (B); HN+1% glucose–PE (C); stipe of fruiting body (D); cap of fruiting body (E). lac2: HN–1% glucose–guaiacol (F); stipe of fruiting body (G); cap of fruiting body (H).
In HN-glucose medium, levels of lac1 mRNA were 27 times higher than those of lac2 mRNA. In the HN-cellulose medium, HN-glucose-PE medium, and HN-glucose-PE-5% sawdust medium, levels of lac1 mRNA were similar to those of lac2 mRNA. Similar levels were also observed in the primordium and fruiting-body stage. However, the levels of lac2 mRNA were 17 times higher than those of lac1 mRNA in the HN-glucose medium supplemented with 2 mM guaiacol (Table 1).
DISCUSSION
Although laccase production in white rot fungi is known to be influenced by a number of factors, little work has been done to study the regulation of laccase gene expression at the molecular level (27, 46). Eggert et al. (19) have shown that laccase activities in culture fluids of Pycnoporus cinnabarinus are dependent on the nitrogen concentration and Collins and Dobson (13) have found that the expression of laccase in Trametes versicolor was regulated at the level of gene transcription by copper and nitrogen. As the concentration of copper or nitrogen in fungal cultures was increased, an increase in laccase activity corresponding to increased laccase gene transcription was observed. Based on these previous reports, we used HN medium supplemented with copper to study the effects of physiological parameters on laccase expression in L. edodes. There have been few reports on the comparison between induced laccases and constitutive laccases (23, 38, 48). Mansur et al. (38) compared transcript levels of three laccase genes from the basidiomycete I-62 under various culture conditions and demonstrated that lcc1 was inducible by veratryl alcohol and lcc3 was not. In our studies, we used a number of substrates to study laccase activity but did not observe any inducing effects, which is consistent with previous reports for L. edodes (32, 33). Laccases in L54 appear to be constitutive, since the enzyme levels did not increase after the use of different inducers or sawdust. However, we cannot conclude from this study that all isozymes of laccase are constitutive. In fact, our results indicated that the level of lac2 mRNA was increased during growth in the medium with 2 mM guaiacol, although the difference was small.
Differential expression of laccase gene families has been reported for a few fungi (38, 40, 47). In the basidiomycete Trametes villosa, lcc1 mRNA was induced approximately 17-fold by the addition of 2,5-xylidine to the culture. The increase in the mRNA level corresponded to a 20-fold increase in enzyme activity. However, lcc2 mRNA is not induced and is present at a constitutive level approximately half that of the uninduced lcc1 mRNA (53, 54). In the basidiomycete Agaricus bisporus, the level of lcc2 mRNA is 300-fold higher than that of lcc1 mRNA in malt extract liquid cultures (41, 44). The transcriptional analysis data from our study agree with the laccase enzyme activity measured with various substrates and under various conditions. However, it must be emphasized that there could be laccase genes besides the ones described in this study or that are posttranslationally modified to become enzymatically active. At present we cannot assign the laccase activity detected to either of these two laccase genes.
An effect of carbon sources on the production of laccases has been demonstrated for the white rot fungus Phanerochaete chrysosporium (1, 17). In addition, malt extract increases the production of laccases in Agaricus bisporus (51, 52). In our study, PE medium increased the peak levels of laccases. However, it should be noted that the increase was not strictly inductive, because it appeared only in the idiophase (46). Interestingly, we found that PE medium was a good substrate for fruiting, which may correlate with the accumulation of laccases in the idiophase. It remains undetermined which compounds in PE medium play important roles in the accumulation of laccases in the fungus.
During recent years, laccase genes have been isolated from several basidiomycetes (3, 25, 30, 42). The sequences of these genes display a common pattern in that they encode polypeptides of approximately 520 to 550 amino acid residues including an N-terminal signal peptide (12, 20, 25). In addition, the single cysteine residue and 10 histidine residues involved in binding the four catalytic cupric ions found in each laccase molecule are conserved, together with a small amount of sequence around the four regions in which the copper ligands are clustered (20, 46). It is in the copper-binding amino acid residues and their general distribution in the polypeptide chain that the laccases are all similar (12, 20, 25). Alignment of the polypeptide sequence derived from lac1 with the sequences derived from other basidiomycete laccase genes shows that the domain structure of Lac1 protein is conserved. Lac1 showed the conserved sequences in the single cysteine residue and 10 histidine residues. The N-terminal lac1 sequence is separated from the C-terminal catalytic domain by a hinge region (46). The latter appears to be duplicated but is typically rich in serine residues. Our data showed that there are seven amino acid differences between two allelic proteins, which was unusual in the filamentous fungi (24, 34). In the white rot fungus P. chrysosporium, although there was 97% similarity between two cellobiose dehydrogenase alleles, their translation products have identical amino acid sequences (34). It remains unknown whether the differences in Lac1B amino acid sequence result in the low level of laccase production in the L54-B strain.
Laccases play a role in the lignification in loblolly pine xylem (2). Ander and Eriksson (1) showed a reduced ability to degrade lignin in laccase-minus mutant Sporotrichum pulverulentum and recovered lignolytic ability in revertants. In an in vitro system involving pure laccases, it was demonstrated that the laccase mediator system can degrade radiolabelled lignin (8). Bourbonnais et al. (4) have shown that the laccase from T. versicolor can degrade kraft lignin in the presence of ABTS. However, other reports have suggested that there is little correlation between laccase activity and ligninolysis. For example, Evans (22) showed that lignin degradation in Coriolus versicolor remained the same after laccase activity was inhibited by a specific antibody. Our results indicated that there were no increases of laccase production in L. edodes strains grown in media supplemented with various phenolic compounds or sawdust. Taken together, these data suggest that laccase can degrade a significant proportion of the components found in lignin, but the role of this enzyme in ligninolysis remains unresolved. All the enzymes possibly involved in lignin degradation produce some highly toxic species from which the fungal mycelium must be protected. It is possible that one of the major functions of L. edodes laccases is to scavenge these compounds by promoting polymerization before they enter the hypha, as previously suggested (10, 32).
In some fungi, laccase has a well-understood function that is unrelated to ligninolysis (46). It was reported that low-laccase-yielding mutants of Pleurotus florida had poor mycelial growth and could not form fruiting bodies whereas the revertants from the same mutants were similar to the parent in mycelial growth and fruiting-body formation (15). In Schizophyllum commune, the dikaryotic strains that are able to form fruiting bodies can secrete high levels of laccases but the monokaryotic strains cannot (16). In A. bisporus, laccase activity is strongly regulated during growth and declines rapidly after fruiting bodies develop (51, 52). Interestingly, like L. edodes, no inducible laccases were found in A. bisporus (51) and S. commune (16). Fruiting-body formation may involve phenol oxidase-catalyzed formation of extracellular pigments coupled to oxidative polymerization of cell wall components strengthening cell-to-cell adhesion (32, 46). Our results demonstrated a strong laccase activity in the fruiting stage and indicated that laccases may catalyze the formation of extracellular pigments by oxidative polymerization and therefore may play an important role in the morphogenesis of the fungus.
ACKNOWLEDGMENTS
J. Zhao was supported by CUHK postdoctoral fellowships. The work described in this paper was partially supported by grants from the Research Grants Council of the Hong Kong Special Administrative Region, China (RGC CUHK189/94M and CUHK364/95M).
We thank Eddie Deane for his critical review.
REFERENCES
- 1.Ander P, Eriksson K E. The importance of phenol oxidase activity in lignin degradation by the white rot fungus Sporotrichum pulverulentum. Arch Microbiol. 1976;109:1–8. [Google Scholar]
- 2.Bao W, O’Malley D M, Whetten R, Sederoff R R. A laccase associated with lignification in loblolly pine xylem. Science. 1993;260:672–674. doi: 10.1126/science.260.5108.672. [DOI] [PubMed] [Google Scholar]
- 3.Berka R M, Schneider P, Golightly E J, Brown S H, Madden M, Brown K M, Halkier T, Mondorf K, Xu F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl Environ Microbiol. 1997;63:3151–3157. doi: 10.1128/aem.63.8.3151-3157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourbonnais R, Paice M G, Reid I D, Lanthier P, Yaguchi M. Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol. 1995;61:1876–1880. doi: 10.1128/aem.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broda P, Birch P R J, Brooks P R, Sims P F G. PCR-mediated analysis of lignocellulolytic gene transcription by Phanerochaete chrysosporium: substrate-dependent differential expression within gene families. Appl Environ Microbiol. 1995;61:2358–2364. doi: 10.1128/aem.61.6.2358-2364.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broda P, Birch P R J, Brooks P R, Sims P F G. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol Microbiol. 1996;19:923–932. doi: 10.1046/j.1365-2958.1996.474966.x. [DOI] [PubMed] [Google Scholar]
- 7.Buswell J A, Cai Y J, Chang S T. Effect of nutrient nitrogen and manganese on manganese peroxidase and laccase production by Lentinula edodes. FEMS Microbiol Lett. 1995;128:81–88. [Google Scholar]
- 8.Call H P, Mücke I. History, overview and applications of mediated lignolytic systems, especially laccase-mediator-systems (Lignozym®-process) J Biotechnol. 1997;53:163–202. [Google Scholar]
- 9.Chang S T, Buswell J A. Mushroom nutriceuticals. World J Microbiol Biotechnol. 1996;12:473–476. doi: 10.1007/BF00419460. [DOI] [PubMed] [Google Scholar]
- 10.Chefetz B, Chen Y, Hadar Y. Purification and characterization of laccase from Chaetomium thermophilium and its role in humification. Appl Environ Microbiol. 1998;64:3175–3179. doi: 10.1128/aem.64.9.3175-3179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coll P M, Fernandez-Abalos J M, Villanueva J R, Santamaria R, Perez P. Purification and characterization of a phenoloxidase (laccase) from the lignin-degrading basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:2607–2613. doi: 10.1128/aem.59.8.2607-2613.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coll P M, Tabernero C, Santamaria R, Perez P. Characterization and structural analysis of the laccase I gene from the newly isolated lignolytic basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:4129–4135. doi: 10.1128/aem.59.12.4129-4135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins P J, Dobson A D W. Regulation of laccase gene transcription in Trametes versicolor. Appl Environ Microbiol. 1997;63:3444–3450. doi: 10.1128/aem.63.9.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen D. Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 15.Das N, Sengupta S, Mukherjee M. Importance of laccase in vegetative growth of Pleurotus florida. Appl Environ Microbiol. 1997;63:4120–4122. doi: 10.1128/aem.63.10.4120-4122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vries O M H, Kooistra W H C F, Wessels G H. Formation of an extracellular laccase by Schizophyllum commune dikaryon. J Gen Microbiol. 1986;132:2817–2826. [Google Scholar]
- 17.D’Souza T M, Boominathan K, Reddy C A. Isolation of laccase gene-specific sequences from white rot and brown rot fungi by PCR. Appl Environ Microbiol. 1996;62:3739–3744. doi: 10.1128/aem.62.10.3739-3744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducros V, Brzozowski A M, Wilson K S, Brown S H, Ostergaard P, Schneider P, Yaver D S, Pedersen A H, Davies G J. Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nat Struct Biol. 1998;5:310–316. doi: 10.1038/nsb0498-310. [DOI] [PubMed] [Google Scholar]
- 19.Eggert C, Temp U, Eriksson K E. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol. 1996;62:1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggert C, Lafayette P R, Temp U, Eriksson K E L, Dean J F D. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl Environ Microbiol. 1998;64:1766–1772. doi: 10.1128/aem.64.5.1766-1772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eriksson K E L, Blanchette R A, Ander P. Microbial and enzymatic degradation of wood and wood components. Berlin, Germany: Springer-Verlag KG; 1990. [Google Scholar]
- 22.Evans C S. Laccase activity in lignin degradation by Coriolus versicolor in vivo and in vitro studies. FEMS Microbiol Lett. 1985;27:339–343. [Google Scholar]
- 23.Garzillo A M V. Differentially-induced extracellular phenol oxidases from Pleurotus ostreatus. Phytochemistry. 1992;31:3685–3690. [Google Scholar]
- 24.Germann U A, Müller G, Hunziker P E, Lerch K. Characterization of two allelic forms of Neurospora crassa laccase. J Biol Chem. 1988;263:885–896. [PubMed] [Google Scholar]
- 25.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakka A. Lignin-modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbiol Rev. 1994;13:125–135. [Google Scholar]
- 27.Kalisz H K, Wood D A, Moore D. Regulation of extracellular laccase production of Agaricus bisporus by nitrogen sources in the medium. FEMS Microbiol Lett. 1986;34:65–88. [Google Scholar]
- 28.Kanda K, Sato T, Ishii S, Enei H, Ejiri S. Purification and properties of tyrosinase isozymes from the gill of Lentinus edodes fruiting body. Biosci Biotechnol Biochem. 1996;60:1273–1278. doi: 10.1271/bbb.60.1273. [DOI] [PubMed] [Google Scholar]
- 29.Kirk T K, Cullen D. Enzymology and molecular genetics of wood degradation by white-rot fungi. In: Young R A, Akhtar M, editors. Environmentally friendly technologies for the pulp and paper industry. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 273–308. [Google Scholar]
- 30.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 31.Larson T G, Choi G H, Nuss D L. Regulatory pathways governing modulation of fungal gene expression by a virulence-attenuating mycovirus. EMBO J. 1992;11:4539–4548. doi: 10.1002/j.1460-2075.1992.tb05555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leatham G F, Stahmann M A. Studies on the laccase of Lentinus edodes: specificity, localization and association with development of fruiting bodies. J Gen Microbiol. 1981;125:147–157. [Google Scholar]
- 33.Leatham G F. Extracellular enzymes produced by the cultivated mushroom Lentinus edodes during degradation of a lignocellulosic medium. Appl Environ Microbiol. 1985;60:3447–3449. doi: 10.1128/aem.50.4.859-867.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Nagalla S R, Renganathan V. Cellobiose dehydrogenase from Phanerochaete chrysosporium is encoded by two allelic variants. Appl Environ Microbiol. 1997;63:796–799. doi: 10.1128/aem.63.2.796-799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Alic M, Gold M H. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, Alic M, Brown J A, Gold M H. Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Appl Environ Microbiol. 1995;61:341–345. doi: 10.1128/aem.61.1.341-345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansur M, Suárez T, Fernández-Larrea J B, Brizuela M A, González A E. Identification of a laccase gene family in the new lignin-degrading basiodiomycete CECT 20197. Appl Environ Microbiol. 1997;63:2637–2646. doi: 10.1128/aem.63.7.2637-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansur M, Suárez T, González A E. Differential gene expression in the laccase gene family from basidiomycete I-62 (CECT 20197) Appl Environ Microbiol. 1998;64:771–774. doi: 10.1128/aem.64.2.771-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messerschmidt A, Ladenstein R, Huber R, Bolognesi M, Avigliano L, Petruzzelli R, Rossi A. Refined crystal structure of ascorbate oxidase at 1.9 Å resolution. J Mol Biol. 1992;224:179–205. doi: 10.1016/0022-2836(92)90583-6. [DOI] [PubMed] [Google Scholar]
- 40.Munoz C, Guillen F, Martinez A T, Martinez M J. Laccase isoenzymes of Pleurotus eryngii: characterization, catalytic properties, and participation in activation of molecular oxygen and Mn2+ oxidation. Appl Environ Microbiol. 1997;63:2166–2174. doi: 10.1128/aem.63.6.2166-2174.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry C R, Smith M, Britnell C H, Wood D A, Thurston C F. Identification of two laccase genes in the cultivated mushroom Agaricus bisporus. J Gen Microbiol. 1993;139:1209–1218. doi: 10.1099/00221287-139-6-1209. [DOI] [PubMed] [Google Scholar]
- 42.Saloheimo M, Niku-Paavola M-L, Knowle J K C. Isolation and structural analysis of the laccase gene from the lignin-degrading fungus Phlebia radiata. J Gen Microbiol. 1991;137:1537–1544. doi: 10.1099/00221287-137-7-1537. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Smith M, Shnyreva A, Wood D A, Thurston C F. Tandem organization and highly disparate expression of the two laccase genes lcc1 and lcc2 in the cultivated mushroom Agaricus bisporus. Microbiology. 1998;144:1063–1069. doi: 10.1099/00221287-144-4-1063. [DOI] [PubMed] [Google Scholar]
- 45.Srinivasan C, D’Souza T M, Boominathan K, Reddy C A. Demonstration of laccase in the white rot basidiomycete Phanerochaete BKM-F1767. Appl Environ Microbiol. 1995;61:4274–4277. doi: 10.1128/aem.61.12.4274-4277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurston C F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 47.Wahleithner J A, Xu F, Brown K M, Brown S H, Golightly E J, Halkier T, Kauppinen S, Pederson A, Schneider P. The identification and characterisation of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr Genet. 1996;29:395–403. doi: 10.1007/BF02208621. [DOI] [PubMed] [Google Scholar]
- 48.White N A, Boddy L. Differential extracellular enzyme production in colonies of Coriolus versicolor, Phelbia radiata and Phlebia rufa: effect of gaseous regime. J Gen Microbiol. 1992;138:2589–2598. [Google Scholar]
- 49.Williamson P R. Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front Biosci. 1997;12:99–107. doi: 10.2741/a231. [DOI] [PubMed] [Google Scholar]
- 50.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood D A. Inactivation of extracellular laccase during fruiting of Agaricus bisporus. J Gen Microbiol. 1980;117:339–345. [Google Scholar]
- 52.Wood D A, Leatham G F. Lignocellulose degradation during the life cycle of Agaricus bisporus. FEMS Microbiol Lett. 1983;20:421–424. [Google Scholar]
- 53.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalbøge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yaver D S, Golightly E J. Cloning and characterization of three laccase genes from the white-rot basidiomycete Trametes villosa: genomic organization of the laccase gene family. Gene. 1996;181:95–102. doi: 10.1016/s0378-1119(96)00480-5. [DOI] [PubMed] [Google Scholar]
- 55.Zhang M, Xie W, Leung G S W, Deane E E, Kwan H S. Cloning and characterization of the gene encoding beta subunit of mitochondrial processing peptidase from the basidiomycete Lentinula edodes. Gene. 1998;206:23–27. doi: 10.1016/s0378-1119(97)00576-3. [DOI] [PubMed] [Google Scholar]
- 56.Zhao J, Janse B J H. Comparison of H2O2-producing enzyme in selected white rot fungi. FEMS Microbiol Lett. 1996;139:215–221. [Google Scholar]
- 57.Zhao J, de Koker T H, Janse B J H. Comparative studies of lignin peroxidases and manganese-dependent peroxidases produced by selected white rot fungi in solid media. FEMS Microbiol Lett. 1996;145:393–399. [Google Scholar]
- 58.Zhao J, Chang S T. Intergeneric hybridization between Pleurotus ostreatus and Schizophyllum commune by PEG-induced protoplast fusion. World J Microbiol Biotechnol. 1996;12:573–578. doi: 10.1007/BF00327717. [DOI] [PubMed] [Google Scholar]