Extended Data Fig. 9. TCR profiling and scRNA-seq of p15E-specific T cells isolated from control and Setdb1 KO LLC tumors.
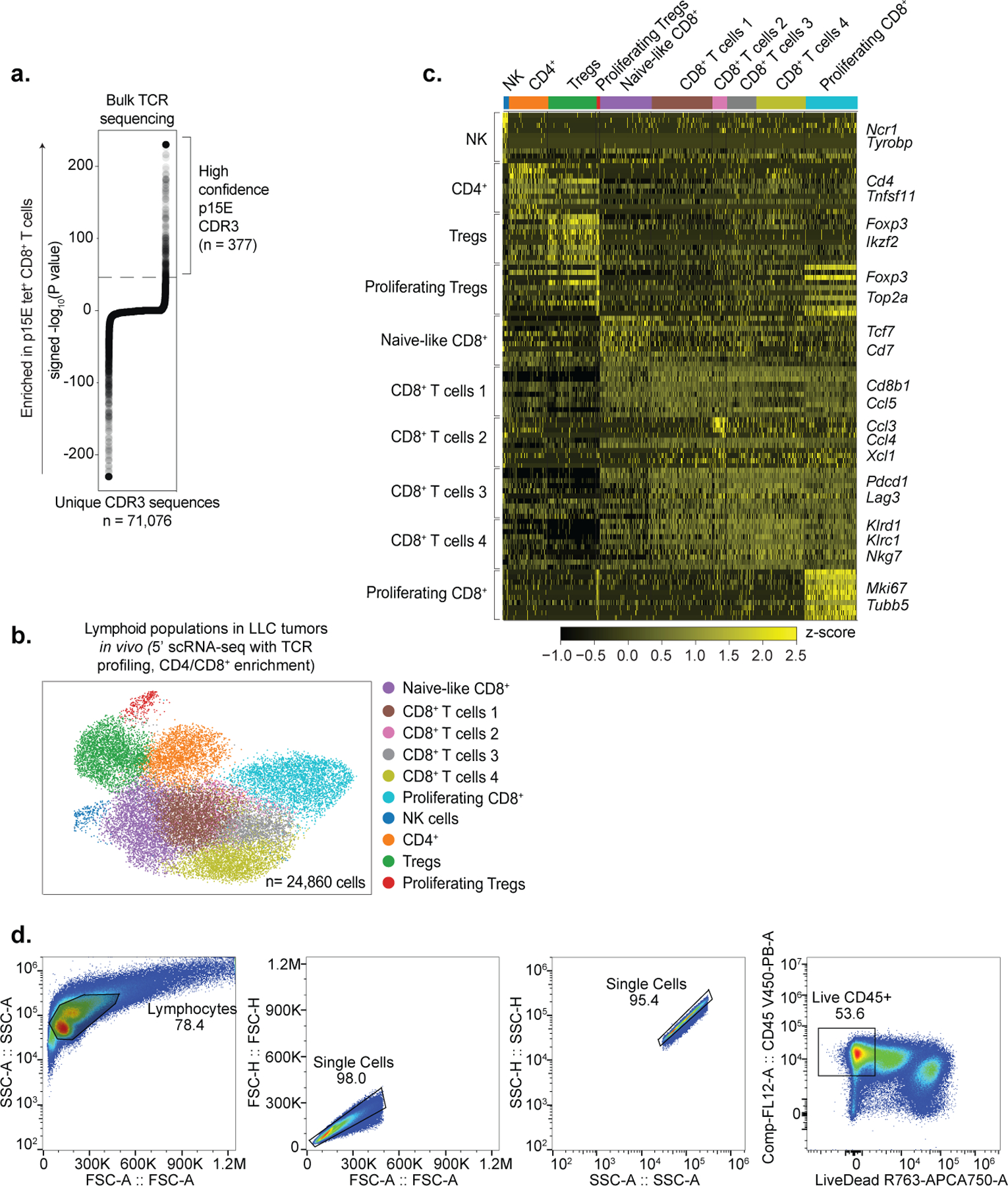
(a) Unique CDR3 sequences (x-axis) identified from TCR sequencing of flow-sorted p15E-tetramer-positive CD8+ T-cells isolated from control LLC tumors. High-confidence CDR3 sequences (n = 377) are highlighted by brackets and identified based on strong statistical enrichment (−log10(P value) > 46 cut-off indicated by dotted line, see Methods) within the p15E-tetramer-positive fraction. (b-c) scRNA-seq (5’) of 24,860 lymphoid cells (CD4+/CD8+-enrichment) isolated from control (n=4 untreated, n=4 ICB-treated) and Setdb1 KO (n=3 untreated, n=3 ICB-treated) LLC tumors. (b) UMAP plot highlights cell populations identified among CD4+/CD8+-enriched lymphoid cells. (c) Representative marker genes used to identify and annotate cell clusters in (b). (d) Representative flow cytometry gating strategy for p15E-tetramer studies. Corresponds to Fig. 4e. *P < 0.05.
