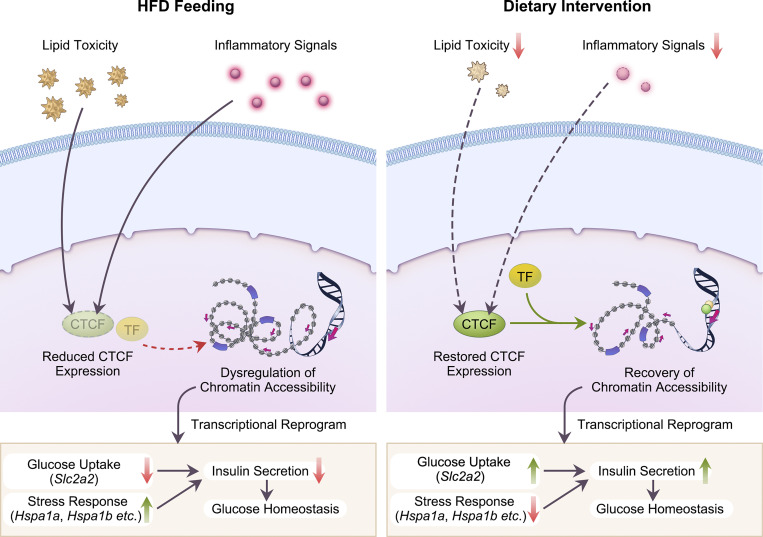
Wang et al. systematically characterize the dynamic nature of β cell functional and transcriptomic adaptation along the progression of diet-induced obesity and T2D. They also identify CTCF as a key mediator of dietary intervention–induced preservation β cell function via transcriptional reprogramming.
Abstract
Pancreatic β cell plasticity is the primary determinant of disease progression and remission of type 2 diabetes (T2D). However, the dynamic nature of β cell adaptation remains elusive. Here, we establish a mouse model exhibiting the compensation-to-decompensation adaptation of β cell function in response to increasing duration of high-fat diet (HFD) feeding. Comprehensive islet functional and transcriptome analyses reveal a dynamic orchestration of transcriptional networks featuring temporal alteration of chromatin remodeling. Interestingly, prediabetic dietary intervention completely rescues β cell dysfunction, accompanied by a remarkable reversal of HFD-induced reprogramming of islet chromatin accessibility and transcriptome. Mechanistically, ATAC-based motif analysis identifies CTCF as the top candidate driving dietary intervention–induced preservation of β cell function. CTCF expression is markedly decreased in β cells from obese and diabetic mice and humans. Both dietary intervention and AAV-mediated restoration of CTCF expression ameliorate β cell dysfunction ex vivo and in vivo, through transducing the lipid toxicity and inflammatory signals to transcriptional reprogramming of genes critical for β cell glucose metabolism and stress response.
Graphical Abstract
Introduction
The modern lifestyle characterized by excessive nutrition and poverty of exercise contributes to the global epidemic of obesity and type 2 diabetes (T2D). Long-term caloric excess results in insulin resistance and β cell dysfunction, the two major causes of T2D (DeFronzo et al., 2015; Taylor et al., 2019). Accumulating evidence suggests that pancreatic islet β cells exhibit significant plasticity during the development of T2D. It has been proposed that during the development of T2D, from obesity and prediabetes to overt diabetes, pancreatic β cells undergo a dynamic adaptation from compensation to eventual decompensation (Weir and Bonner-Weir, 2004). Moreover, recent studies suggest that these adaptive responses could be even more complex since β cells undergo a dynamic alteration in cell identity in response to intra-islet microenvironmental changes during disease progression (Talchai et al., 2012). A better understanding of the dynamic features and underlying molecular bases of β cell adaptation during disease progression is crucial for developing new effective approaches to restoring β cell function in T2D.
It has been reported that long-term exposure of β cells to elevated levels of glucose, fatty acids, and inflammatory cytokines could induce ultrastructural cellular structure damage (Brereton et al., 2014; Pinnick et al., 2010) and ER stress (Huang et al., 2007; Laybutt et al., 2007; Pinnick et al., 2008) and alter gene expression profiles related to insulin secretion (Kelpe et al., 2003; Meng et al., 2006; Zhou and Grill, 1994) and lipid metabolism (Bikopoulos et al., 2008; Pinnick et al., 2008), which would eventually lead to β cell dysfunction in T2D. Removal of these stimuli could reverse most of the above-mentioned cellular responses, leading to the restoration of β cell function (Boucher et al., 2004; Moffitt et al., 2005; Pinnick et al., 2010). However, the mechanisms underlying such reversal are yet to be further understood.
Recently, clinical trials have demonstrated the efficacy of lifestyle interventions in delaying or preventing the development of diabetes in people with prediabetes or even at the early stage of T2D (Dagogo-Jack et al., 2020; Eriksson and Lindgärde, 1991; Knowler et al., 2002; Perreault et al., 2009). A recent Diabetes Remission Clinical Trial also reported that weight loss by dietary intervention achieved remission of T2D in some participants. The extent of remission depends upon the functional recovery of β cells, especially upon the recovery of the first phase of insulin secretion (Al-Mrabeh et al., 2020; Taylor et al., 2018). These findings highlight the importance of elucidating the functional alterations and underlying molecular mechanisms of dietary intervention’s effects on pancreatic β cells. However, due to the specific anatomical properties of pancreatic islets, it is nearly impossible to perform these studies in humans.
As such, it is critical to establish a rodent model of obesity and T2D recapitulating the dynamic features of β cell adaptation during disease progression, enabling the investigation of the effects and molecular mechanisms of dietary intervention on β cell dysfunction during the development of T2D. In this study, by feeding wild-type mice of C57BL/6J background with high-fat diet (HFD) for increasing periods, we established a murine model with similar dynamic adaptation processes in islet β cells as speculated in human studies, and provided a systematic functional and multiomics description of the effects of dietary intervention on pancreatic β cells during disease progression of obesity and T2D. Mechanistically, using assay for transposase-accessible chromatin using sequencing (ATAC-Seq) and motif analysis, we identified the DNA binding protein CCCTC-binding factor (CTCF) as the key driving factor of dietary intervention–mediated recovery of β cell function, through chromatin remodeling and transcriptional regulation of selective genes critical for glucose metabolism, stress response, and β cell identity. CTCF expression was significantly downregulated in pancreatic islet β cells from obese and diabetic mice and humans. More importantly, adeno-associated virus (AAV)–mediated rescue of CTCF expression preserved β cell function in ex vivo cultured human and mouse islets, and in mice.
Results
Pancreatic β cells undergo a compensation-to-decompensation switch during the progression of T2D in mice
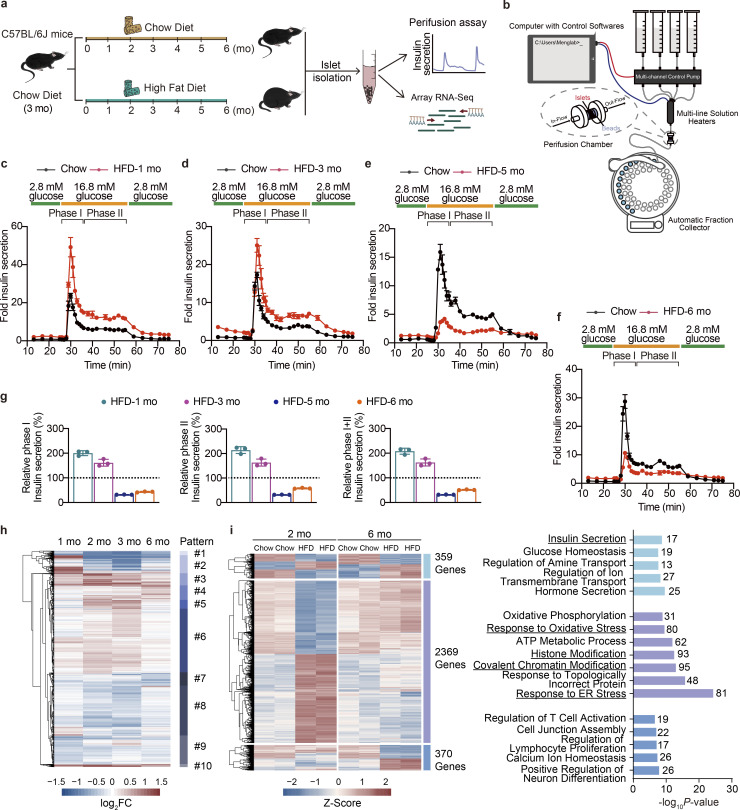
Previous human studies indicated that the development of T2D entails a compensation-to-decompensation process of pancreatic β cell function accompanied by a phase-delayed onset of hyperglycemia (Hudish et al., 2019). However, the dynamic alterations of phase I and phase II insulin secretion from pancreatic islets, and the accompanying molecular basis such as transcriptional networks underlying this adaptation process have not been well established. To solve this issue, we subjected wild-type male mice to increasing durations (1–6 mo) of HFD feeding, and analyzed islet β cell function and transcriptome changes at each time point (Fig. 1 a). Consistent with previous reports (Gupta et al., 2017), HFD feeding induced a continuous weight gain in mice (Fig. S1 a), accompanied by a gradual elevation of fasting blood glucose levels (Fig. S1 b). We next isolated islets from mice fed with HFD and chow diet at each time point and performed islet perifusion studies (Fig. 1 b) to evaluate the dynamic responses of glucose-stimulated insulin secretion (GSIS) from β cells. Intriguingly, 1- and 3-mo HFD feeding significantly elevated both glucose-stimulated phase I (1-mo HFD, 2.0-fold, P = 0.0001; 3-mo HFD, 1.6-fold, P = 0.0002) and phase II (1-mo HFD, 2.1-fold, P = 0.0025; 3-mo HFD, 1.6-fold, P = 0.0025) insulin secretion compared to the chow diet-fed controls (Fig. 1, c and d), while a 21.92% decrease of total insulin secretion (phase I + II) could be observed in mice fed with HFD for 3 mo compared to animals fed with HFD for 1 mo. Moreover, islets isolated from mice fed with HFD for longer durations (5 and 6 mo) exhibited significant decreases in total insulin secretion (68.03% and 48.29%, respectively, P < 0.0001) compared to their respective chow diet-fed control groups (Fig. 1, e and f). This time-series data demonstrates a compensation-to-decompensation course in islet function in response to HFD feeding. The crash of islet function seems to occur between 3 and 5 mo of HFD feeding (Fig. 1 g).
Figure 1.
Dynamic functional and transcriptional profiles of pancreatic islet adaptation in response to continuous HFD feeding. (a) Schematic representation of the experimental setup. (b) Schematic of the islet perifusion system. (c–f) Perifusion analyses of dynamic glucose-stimulated phase I and phage II insulin secretion in islets from mice fed with HFD for 1, 3, 5, and 6 mo and their corresponding controls. Each islet sample was pooled from at least three animals. Data represent mean ± SD; n = 3 technical replicates. (g) Relative phase I and phase II and total (I + II) insulin secretion in islets from mice fed with HFD for 1, 3, 5, and 6 mo shown in c–f determined by calculating the AUC of each HFD group against their respective control group set as 100% (shown in black dashed lines). Each islet sample was pooled from at least three animals. Data represent mean ± SD; n = 3 technical replicates. (h) Unsupervised clustering of differentially expressed genes in timescale HFD feeding. log2FC values against their respective chow diet–fed control group in each time point are shown (scale bar, −1.5 to 1.5). Dendrogram was manually cut to obtain 10 clusters representing 10 change patterns. (i) Differential expression analysis of genes with significant changes in primary islets isolated from mice underwent either 2 or 6 mo of HFD feeding. Heatmap showing scaled fragments per kilobase of exon model per million mapped fragments (as Z-Score) of genes with significant changes in both time points (in light blue), exclusively in the 2 mo (in purple), and exclusively in the 6 mo (in sky blue; left, scale bar indicats −2 to 2), and GO enrichment analysis of biological processes featuring these patterns (right), most significant and nonredundant biological processes with respective gene numbers and P values are shown. Data in c–g are representative of at least two independent experiments.
Figure S1.
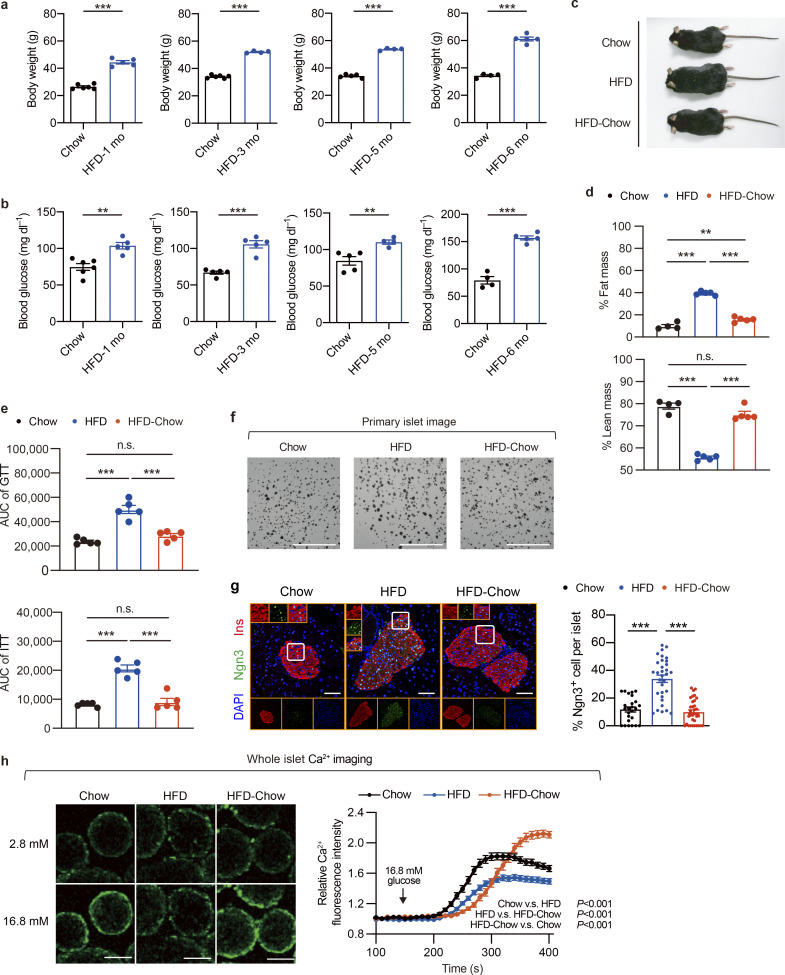
Additional characterization of β cell compensation-to-decompensation model and dietary intervention model. (a) Body weight of mice fed with HFD for 1, 3, 5, and 6 mo. Littermates fed with chow diet were used as controls. Data represent mean ± SEM (n = 4–6 mice per group); ***, P < 0.001; two-tailed unpaired Student’s t test. (b) Overnight fasting blood glucose levels in mice as treated in a. Data represent mean ± SEM (n = 4–6 mice per group); **, P < 0.01; ***, P < 0.001; two-tailed unpaired Student’s t test. (c) Representative images of mice in Chow, HFD, and HFD-Chow groups. (d) Body composition of mice in indicated groups. Data represent mean ± SEM (n = 4–5 mice per group); **, P < 0.01; ***, P < 0.001; one-way ANOVA. (e) AUC quantification of GTT and ITT results shown in Fig. 2 d. Data represent mean ± SEM (n = 4–5 mice per group); ***, P < 0.001; one-way ANOVA. (f) Representative images of mouse islets isolated from indicated groups. Scale bar, 5 mm. (g) Representative immunofluorescent images (left panel) and quantification (right panel) of Ngn3+ cells in islets from indicated groups. Scale bar, 100 μm. Data represent mean ± SEM (n = 25–33 islets from 4 to 5 mice per group); ***, P < 0.001; one-way ANOVA. (h) Glucose-stimulated Ca2+ influx in mouse islets isolated from Chow (n = 53 islets), HFD (n = 52 islets), and HFD-Chow (n = 63 islets) groups. Each islet sample was pooled from at least three animals. Representative fluorescence images of calcium signal in islets treated with high glucose (16.8 mM) for 400 s (left), and dynamic Ca2+ fluorescence intensity in response to high-glucose treatment (right). Scale bar, 100 μm. Data represent mean ± SEM two-way ANOVA with multiple comparisons. Data are representative of at least two independent experiments for all panels.
To further characterize the dynamic alterations of gene expression profiles in mouse islets upon HFD feeding for different periods, we performed bulk RNA sequencing analysis on islets isolated from mice fed with HFD for 1, 2, 3, and 6 mo. We identified 6,038 genes with significant changes (P < 0.05, |log2FC| > 0.3) in at least one time point. With unsupervised clustering, these genes could be further designated into 10 clusters representing diverse change patterns (Fig. 1 h). It was worth noticing that among the highly dynamic transcriptional changes along with the HFD duration, robust signatures could be observed in either the compensation stage (2–3 mo of HFD) or the decompensation stage (6 mo of HFD). To further understand the functional significance of these signatures, we then exclusively compared gene expression changes in islets between 2 and 6 mo of HFD feeding (Fig. 1 i). Among the 3,157 differentially expressed genes (P < 0.05, |log2FC| > 0.3 in either group), we identified 359 genes that were actively altered in both 2- and 6-mo HFD feeding (Fig. 1 i, left, light blue). Gene ontology (GO) analysis showed significant enrichment of hormone transport and insulin secretion pathways related genes (Fig. 1 i, right, light blue). Also, we identified 2,369 genes that were differentially expressed exclusively in mouse islets upon HFD feeding for 2 mo (Fig. 1 i, left, purple) as the signature of the functional compensation stage. Biological processes, including response to ER stress, response to oxidative stress, ATP metabolic processes, and intriguingly, covalent chromatin modification and histone modification, were found to feature such stage (Fig. 1 i, right, purple). Besides, 370 genes showed expression changes exclusively in islets from mice fed with HFD for 6 mo (Fig. 1 i left, sky blue), enriched in biological processes including regulation of neuron differentiation, calcium ion homeostasis, regulation of lymphocyte proliferation, and T cell activation (Fig. 1 i right, sky blue). These results identified distinct transcriptional signatures featuring the compensation to decompensation switch, which may also serve as the key drivers for the impairment of islet function in animals with long-term HFD feeding.
Dietary intervention prevents HFD-induced pancreatic β cell dysfunction
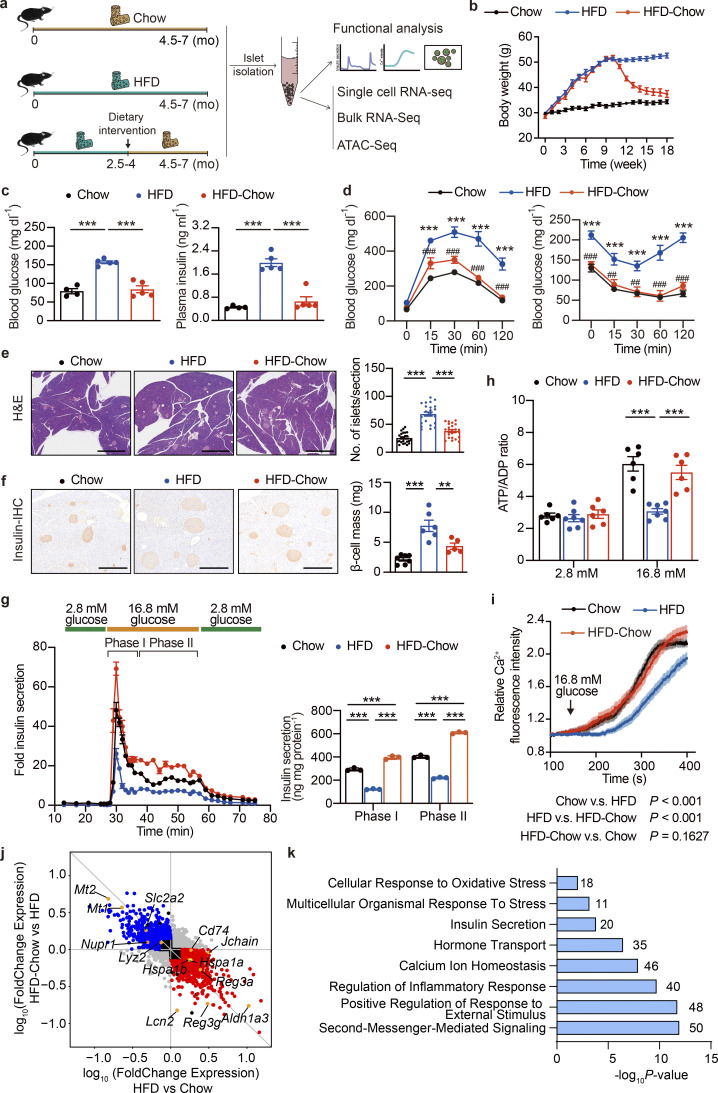
Previous human studies have demonstrated the efficacy of the prediabetic dietary intervention in delaying or even preventing the development of T2D (Al-Mrabeh et al., 2020; Eriksson and Lindgärde, 1991; Knowler et al., 2002; Perreault et al., 2009; Taylor et al., 2018). The establishment of a mouse model closely recapitulating the β cell functional compensation-to-decompensation process in human T2D provided the opportunity to elucidate the effect and underlying mechanism of dietary intervention on β cell GSIS function. Therefore, we established a murine dietary intervention model by switching HFD to a regular chow diet at the 10–16th wk of HFD feeding (the HFD-Chow group), a time during the progression of β cell functional compensation to decompensation and massive transcriptomic changes, to mimic the dietary intervention at prediabetic to early T2D stages in the previous published clinical studies. Mice continuously fed with a regular chow diet (Chow group) or high-fat diet (HFD group) to mimic the healthy diet or nonintervened conditions were used as control groups (Fig. 2 a). As expected, mice under continuous HFD feeding showed sustained body weight gain compared to the Chow group. In contrast, the HFD-Chow group exhibited a significant body weight loss after dietary intervention, and reached similar body weight and body composition as the Chow group (Fig. 2 b; and Fig. S1, c and d). Also, mice in the HFD group developed hyperglycemia, hyperinsulinemia, and significantly impaired glucose tolerance and insulin sensitivity compared to mice in the Chow group (Fig. 2, c and d; andFig. S1 e). In contrast, mice in the HFD-Chow group showed normal glycemic and blood insulin levels compared to those in the HFD group (Fig. 2 c), improved glucose tolerance and insulin sensitivity could be observed (Fig. 2 d and Fig. S1 e). These results demonstrated that prediabetic dietary intervention could almost completely prevent the development and onset of diabetic syndrome and preserve β cell GSIS function in mice.
Figure 2.
Dietary intervention prevents HFD-induced β cell dysfunction and systemic metabolic defects. (a) Schematic representation of the experimental setup. (b) Body weight changes in mice with indicated treatments. Data represents mean ± SEM (n = 7–9 mice per group). (c) Overnight fasting blood glucose levels (left) and plasma insulin levels (right) of Chow, HFD, and HFD-Chow mice. Data represent mean ± SEM (n = 4–5 mice per group); ***, P < 0.001; one-way ANOVA. (d) GTT (left) and ITT (right) of Chow, HFD, and HFD-Chow mice. Data represent mean ± SEM (n = 5 mice per group), ***, P < 0.001, HFD vs. Chow; ##, P < 0.01, ###, P < 0.001, HFD-Chow vs. HFD; two-way ANOVA with multiple comparisons. (e) Representative H&E staining of pancreas sections from Chow, HFD, and HFD-Chow mice (left panel), and islet number per pancreas section (right). Scale bar, 1 mm. Data represent mean ± SEM (n = 24 pancreas sections from six mice/group; at least 4 sections/mouse).***, P < 0.001; one-way ANOVA. (f) Representative IHC images of mouse pancreas sections from Chow, HFD, and HFD-Chow mice immune-stained for insulin (brown) and counterstained with hematoxylin (blue; left). Quantitation of β cell mass in the pancreas of Chow, HFD, and HFD-Chow mice (right). Scale bar, 500 μm. Data represent mean ± SEM (n = 5–7 mice per group). **, P < 0.01; ***, P < 0.001; one-way ANOVA. (g) Perifusion analyses of dynamic glucose-stimulated insulin secretion in islets from Chow, HFD, and HFD-Chow mice (left) and biphasic insulin release levels (right). Each islet sample was pooled from at least three animals. Data represent mean ± SD (n = 3 technical replicates). ***, P < 0.001; one-way ANOVA. (h) ATP/ADP ratios of islets from Chow, HFD, and HFD-Chow mice under low-glucose (2.8 mM) or high-glucose (16.8 mM) treatments, islet samples were pooled from at least three animals. Data represent mean ± SEM (n = 6–7 replicates of islet samples, each sample represent 20 IEQ islets); ***, P < 0.001; one-way ANOVA. (i) Glucose (16.8 mM) stimulated Ca2+ influx in single islet β cells in mice from Chow (n = 128 cells), HFD (n = 178 cells), and HFD-Chow (n = 214 cells) groups. Each islet sample was pooled from at least three animals. Data represent mean ± SEM; two-way ANOVA. (j) Differential gene expression analysis of RNA-Seq of mouse islets from Chow, HFD, and HFD-Chow groups (RNA isolation from islets pooled from three mice). Log10 transferred fold change expression levels of all highly expressed genes (fragments per kilobase of exon model per million mapped fragments >1) in indicated comparisons are shown in the scatter plot. Reversible Genes with |log2FC| > 0.5, P < 0.05 were additionally marked in red (upregulated) and blue (downregulated). (k) GO analysis of Reversible Genes in i. Most significant and nonredundant biological processes with respective gene numbers and P values are shown. All panels report data verified in at least two independent experiments.
To assess the effect of dietary intervention on islet morphology, we performed histochemical and morphometric studies on pancreas sections. H&E staining and quantification analysis of islet number of pancreas sections from three groups demonstrated that the islet number was markedly increased (∼2.7-fold) in the HFD group compared to the Chow group (Fig. 2 e). Further insulin immunohistochemical (IHC) staining on consecutive pancreas sections revealed an ∼3.6-fold increase in β cell mass in the HFD group compared to the Chow group (Fig. 2 f). Notably, a significant decrease in islet number (44.32% decrease, P < 0.001) and β cell mass (43.86% decrease, P = 0.0044) could be observed in HFD-Chow animals compared to those in the HFD group (Fig. 2, e and f). Similarly, the size of primary islets isolated from the HFD group was much bigger than that from the Chow group, and was largely restored by dietary intervention (Fig. S1 f). Consistent with previous studies suggesting that cell de-differentiation and reprogramming play an important role in β cell adaptation and dysfunction in T2D (Talchai et al., 2012), immunofluorescent staining of Neurogin3 (Ngn3), a progenitor cell marker, showed that HFD feeding led to a marked increase in Ngn3+ cell number in islets compared to the Chow group, which was largely reversed by dietary intervention (Fig. S1 g).
To examine the effect of dietary intervention on β cell function, primary islets were isolated for functional analyses. Consistent with the impaired glucose tolerance in the HFD group, the islet perifusion assay revealed a significant decrease in phase I and phase II insulin secretion in islets from the HFD group compared to the Chow group (Fig. 2 g). Intriguingly, HFD-Chow treatment elicited a significant improvement of both phase I and phase II insulin secretion over the HFD group (∼3.3-fold and ∼2.8-fold for phase I and phase II, respectively, P < 0.001; Fig. 2 g). To further dissect the underlying mechanisms responsible for the alterations of GSIS function in primary islets isolated from Chow, HFD, and HFD-Chow groups, we first examined the ATP/ADP ratio in primary islets since the elevation of ATP/ADP ratio in β cells is known to trigger the closure ATP-sensitive potassium (KATP) channel, membrane depolarization, and calcium influx, which is a crucial signal for the following insulin secretion. As shown in Fig. 2 h, upon high-glucose (16.8 mM) treatment, islets from both Chow and HFD-Chow mice exhibited a significant increase in ATP/ADP ratio compared to the basal levels under low-glucose condition. However, the glucose-stimulated elevation of ATP/ADP ratio was dismissed in islets from the HFD group. Consistently, calcium imaging at both islet single cell and whole islet levels demonstrated that dietary intervention almost completely ameliorated the impairment of glucose-stimulated calcium response by HFD feeding (Fig. 2 i and Fig. S1 h).
Dietary intervention improves β cell function through transcriptional reprogramming
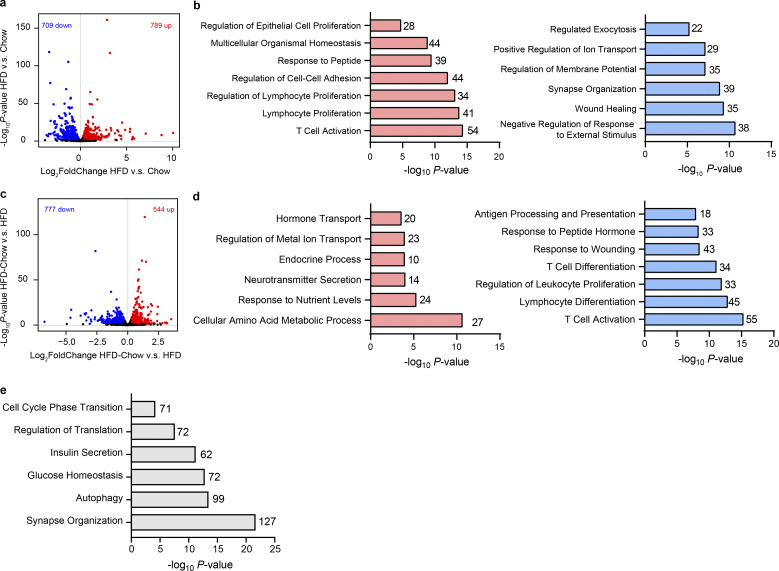
To systematically assess the effects of dietary intervention on the transcriptome in islets, we then performed RNA sequencing (RNA-Seq) analysis on primary islets isolated from the Chow, HFD, and HFD-Chow groups. Pairwise differential expression analysis identified 789 upregulated and 709 downregulated genes in islets from the HFD group compared to the Chow group (Fig. S2 a). Among the upregulated genes, biological processes including epithelial cell proliferation and immune cell proliferation and activation were significantly enriched, whereas biological processes such as regulated exocytosis, regulation of membrane potential, and regulation of ion transport were enriched in the downregulated genes (Fig. S2 b). These results were consistent with the morphological and functional data showing the expanded islet size and impaired β cell insulin secretion triggered by HFD feeding. Moreover, by comparing the HFD-Chow group with the HFD group, we identified 544 upregulated genes and 777 downregulated genes (Fig. S2 c). Notably, biological processes enriched from these upregulated genes were similar to the terms enriched from the downregulated genes in the HFD vs. Chow comparison. Such similarity could also be observed in the terms enriched from downregulated genes in the HFD-Chow vs. HFD comparison and terms enriched from upregulated genes in the HFD vs. Chow comparison (Fig. S2 d). We went on to assess the fold change difference of each gene in these comparisons. We obtained 783 genes (Reversible Genes) that were significantly altered in each comparison and showed an opposite trend of transcriptional changes between the two comparisons (Fig. 2 j). GO analysis revealed that genes associated with cellular response to oxidative stress, insulin secretion, and inflammatory response, such as solute carrier family 2 member 2 (Slc2a2), aldehyde dehydrogenase 1 family member A3 (Aldh1a3), cluster of differentiation 74 (Cd74), regenerating family member 3 α (Reg3a), etc., were enriched in these Reversible Genes (Fig. 2, j and k). These data implied that dietary intervention during the development of T2D preserved β cell function through transcriptional reprogramming. Moreover, to identify the genes that were not reversed by dietary intervention, we performed differential analysis on HFD-Chow islets against Chow islets. Reversible genes shown in Fig. 2 j were excluded from these differential genes to ensure the obtained gene sets were truly not reversible. With this approach, we identified 2,903 genes that remained unaltered by the dietary intervention. GO analysis showed that these genes were mainly related to biological processes such as synapse organization, autophagy, glucose homeostasis, insulin secretion, regulation of translation, and cell cycle phase transition (Fig. S2 e). These results revealed the pathways that are not altered by dietary intervention, which could be critical for the pathogenesis of ultimate β cell failure.
Figure S2.
Pairwise gene expression comparison of bulk islet RNA-Seq data among Chow, HFD, and HFD-Chow groups. (a) Volcano plot showing the gene expression difference in islets between Chow and HFD groups. Genes that are significantly upregulated (log2FC > 0.4, P < 0.05, P values by Wald test) or downregulated (log2FC < −0.4, P < 0.05, P values by Wald test) were marked in red and blue, respectively. (b) GO analysis of significantly upregulated (left) and downregulated (right) genes by HFD compared to Chow group. (c) Volcano plot showing the gene expression difference in islets between HFD and HFD-Chow. Genes that are significantly upregulated (log2FC > 0.4, P < 0.05, P values by Wald test) or downregulated (log2FC < −0.4, P < 0.05, P values by Wald test) were marked in red and blue, respectively. (d) GO analysis of significantly upregulated (left) and downregulated (right) genes by HFD-Chow compared to HFD group. (e) GO analysis of genes unaltered by dietary intervention (|log2FC (HFD-Chow vs. Chow)| > 0.4, P < 0.05 AND NOT Reversible Genes). In a–e, islet samples were pooled from at least three animals. In b, d, and e, most significant and nonredundant biological processes with respective gene number and P value are shown.
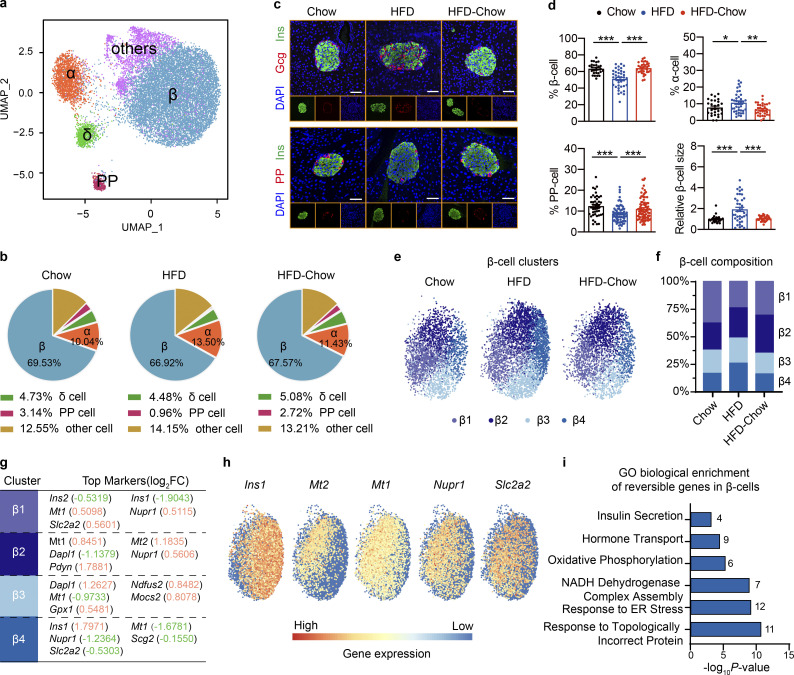
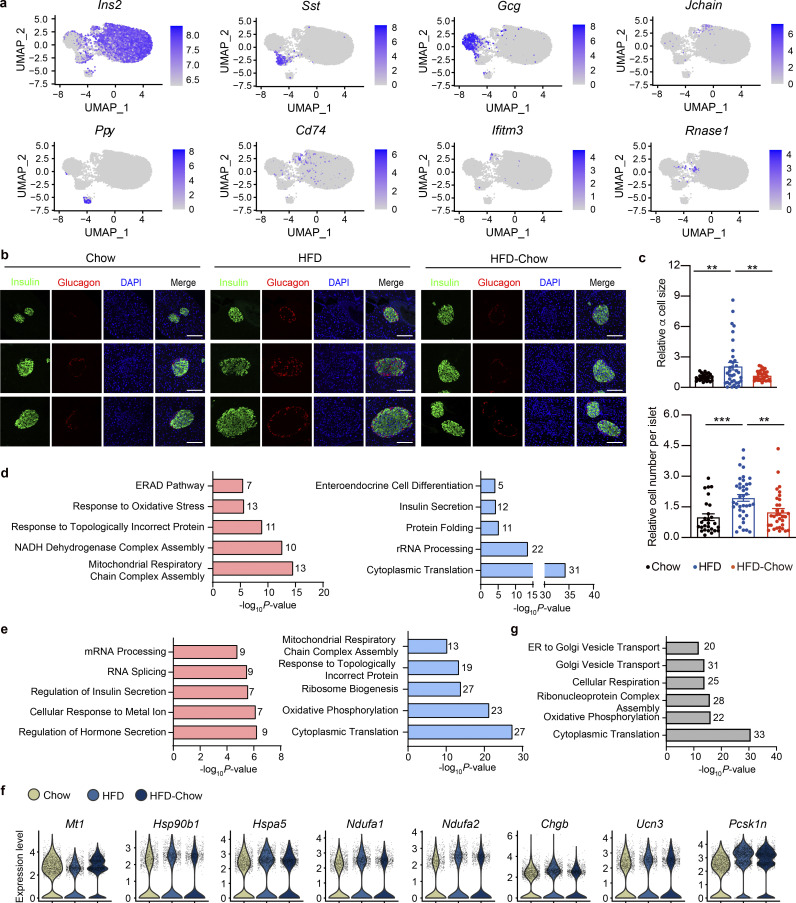
Islet consists of multiple cell types that work in concert to orchestrate their endocrine function. To further reveal how dietary intervention preserves islet β cell function, we then performed microwell-based single cell RNA sequencing (scRNA-Seq; Han et al., 2018) on dissociated primary islet cells from Chow, HFD, and HFD-Chow groups. By Uniform Manifold Approximation and Projection (UMAP), cells were clustered into five major subgroups that were then designated as α cells, β cells, δ cells, pancreatic polypeptide (PP) cells, and other cells mainly based on endocrine hormone gene expression (Fig. 3 a and Fig. S3 a). The “other cells” subgroup mainly consisted of multiple types of immune cells and endothelial cells expressing Rnase1 (Fig. S3 a). Islet cell composition analysis revealed mild differences in the β cell and δ cell proportions among the three groups, while α cell percentage was slightly increased in the HFD group compared to the Chow group. The percentage of PP cells was significantly decreased by ∼70% in islets from the HFD group compared to the Chow group (Fig. 3 b). Interestingly, percentages of both α cells and PP cells in islets from the HFD-Chow group were indistinguishable from those from the Chow group, indicating that dietary intervention could prevent the islet cell composition change toward the track directed by HFD feeding. To further confirm such findings, immunofluorescent staining was performed on pancreas sections. As shown in Fig. 3, c and d; and Fig. S3, b and c, a significant decrease of INS+ β cells, accompanied by the increase of GCG+ α cells and PP+ PP cells could be observed, which were similar to the findings in scRNA-Seq, indicating a possible de-differentiation or trans-differentiation process in islets. Immunofluorescent staining also revealed a significant increase in β cell size in the HFD group compared to the Chow group (∼1.9-fold), which was almost completely reversed by HFD-Chow treatment (Fig. 3, c and d). Among the obtained cell types, the β cell is responsible for GSIS function in islets. Hence, β cell clusters were extracted for further analysis. Previous research has revealed the significant heterogeneity of β cell population in pancreatic islets (Dominguez-Gutierrez et al., 2019; Roscioni et al., 2016). Consistently, with improved resolution in scRNA-Seq, β cells were further designated into four distinct clusters, namely β1 to β4 (Fig. 3 e). Composition analysis revealed a significant decrease of β1 population (from 38.09% in Chow group to 24.23% in HFD group) and an increase of β4 population upon HFD feeding (from 15.75% in Chow group to 25.02% in HFD group), whereas such changes could be largely reversed by dietary intervention. The other two subpopulations showed no significant proportional changes among the three groups (Fig. 3 f). Transcriptome analysis revealed that β1 subcluster was characterized with high expression of metallothionein-1 (Mt1), nuclear protein 1 (Nupr1), and Slc2a2. Interestingly, the β4 subcluster was characterized by significantly reduced levels of Mt1, Nupr1, Secretogranin 2 (Scg2), and Slc2a2. Significantly high level of Ins1 was also detected in the β4 subcluster (Fig. 3, g and h). Previous reports have revealed the role of Mt1 and Nupr1 as antioxidants protecting islet cells against oxidative stress, inflammation, and other cell damage caused by stress reaction (Chen et al., 2001; Li et al., 2004; Päth et al., 2020). Our data, therefore, implied an elevated sensitivity of the β4 subcluster to stress conditions compared to the cells in the β1 subcluster. Also, the significantly elevated Slc2a2 expression in β1 cells implied a higher glucose sensitivity in these cells compared to β4 cells. These findings were in compliance with the functional data showing reduced, but dietary intervention–reversible GSIS function in islets from the HFD group, and our findings in bulk RNA-Seq suggesting dietary intervention might preserve β cell function by reducing cellular stress levels.
Figure 3.
scRNA-Seq analysis of dietary intervention on islet cell composition and β cell specific transcriptome. (a) scRNA-Seq and UMAP visualization of transcriptome similarity-based cell clustering of islet single cells from indicated groups. Islet sample for each treatment was pooled from at least three animals. In total, 4,231 (Chow), 6,454 (HFD), and 4,490 (HFD-Chow) cells were used in the clustering. (b) Cell composition of islets from indicated groups by scRNA-Seq. (c) Representative immunofluorescent images of insulin (Ins), glucagon (Gcg), and PP in islets from Chow, HFD, and HFD-Chow mice. Scale bar, 50 μm. (d) Quantification of β, α, and PP cell percentage and relative β cell size (n = 27–38 islets per group for % β, % α, and relative β cell size; n = 44–82 islets per group for % PP; islets were pooled from 4 to 5 mice; at least 6 islets per mice) in pancreas sections of Chow, HFD, and HFD-Chow mice. Data represent mean ± SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA. Relative β cell size was normalized to the average β cell size in Chow group. (e) UMAP visualization of β cell sub-clusters in indicated groups; cells apart from the major β cell cluster on the UMAP plot were not shown. (f) Composition analysis of β cell sub-clusters in indicated groups. (g) Top markers of indicated β cell clusters. Upregulated genes were marked in red, downregulated genes were marked in green. Log2FC indicates the average log2 transferred fold change of indicated gene expression in given subcluster comparing to the expression level in other β cells. (h) Expression heatmap of indicated genes in all β cells. (i) GO analysis of reversible genes (with P < 0.05 by Wilcoxon Rank Sum test) in β cells. Most significant and nonredundant biological processes with respective gene numbers and P values are shown. Data in c and d are representative of at least two independent experiments.
Figure S3.
Additional details of scRNA-Seq analysis. (a) scRNA-Seq and UMAP visualization of islet single cells from Chow, HFD, and HFD-Chow groups. Expression levels of indicated marker genes are shown. Scale bar indicates gene expression level. (b) Representative immunofluorescent images of insulin (marker for β cell) and glucagon (marker for α cell) staining in pancreas sections from indicated groups. Scale bar, 100 μm. (c) Relative α cell size and islet cell number quantified from immunofluorescent images and normalized to the average α cell size, and the islet cell number, respectively, in the Chow group. Data represent mean ± SEM (n = 4–5 mice per group, dots represent islets); **, P < 0.01; ***, P < 0.001; one-way ANOVA. (d) GO biological process analysis of upregulated (left) and downregulated (right) genes in β cell clusters from HFD group compared to Chow group. (e) GO biological process analysis of upregulated (left) and downregulated (right) genes in β cell clusters from HFD-Chow group compared to HFD group. (f) Violin plot of selected gene expression, expression level in each cell was marked in gray dot. (g) GO biological process analysis of genes unaltered by dietary intervention in β cell clusters (P < 0.05 AND NOT Reversible Genes). In d, e, and g, most significant and nonredundant biological processes with respective gene numbers and P values are shown. Significantly changed genes were identified by P < 0.05 with Wilcoxon Rank Sum test. Data in b and c are representative of two independent experiments.
To further explore the effects of dietary intervention on transcriptional profiles specifically in β cells, differential expression analysis was performed on all the clusters of β cells. GO analysis revealed that, compared to Chow diet, HFD feeding upregulated genes related to ER-associated protein degradation, response to oxidative stress, response to topological incorrect protein and mitochondrial respiratory chain assembly; and downregulated genes related to insulin secretion and cytoplasmic translation processes (Fig. S3 d). In contrast, compared to the HFD group, HFD-Chow treatment significantly elevated the expression of genes involved in the regulation of hormone secretion and cellular response to metal ion, while genes associated with response to topologically incorrect proteins, mitochondrial respiratory chain complex assembly, and oxidative phosphorylation were downregulated by dietary intervention (Fig. S3 e). Among the 502 genes that were either upregulated (158 genes) or downregulated (344 genes) by HFD feeding, 109 genes were significantly reversed by dietary intervention. Among these reversible genes, we also noticed a significant reversion in the expression of Mt1. The gene expression of some members in the heat shock protein family involved in ER response to unfolded proteins, such as Hsp90b1 and Hspa5, was also reversibly expressed. Besides, members of mitochondrial NADH dehydrogenase complex (Ndufa1, Ndufa2, etc.) and genes related to insulin biosynthesis and secretion process, including chromogranin b (Chgb), urocortin 3 (Ucn3), and proprotein convertase 1 inhibitor (Pcsk1n) also showed reversed expression in response to dietary intervention (Fig. S3 f). GO analysis of all reversible genes revealed significant enrichment of biological processes that are potentially important for β cell function, such as response to ER stress, mitochondrial respiratory chain complex I and II assembly, hormone transport, and insulin secretion (Fig. 3 i). Together with the whole-islet RNA-Seq results, these analyses indicated that dietary intervention was likely to preserve islet GSIS function by altering β cell transcriptional programs. Besides, like the RNA-Seq data, we performed differential analysis on mouse islets from the HFD-Chow group against islets from the Chow group. Reversible genes we obtained in β cells were then removed from these differential genes to identify the truly not reversible genes in β cells. We hereby identified 506 genes that were not reversible by dietary intervention. GO analysis showed that biological processes including cytoplasmic translation, ribonucleoprotein complex assembly, cellular respiration, and Golgi vesicle transport were significantly enriched in these genes (Fig. S3 g), suggesting the alterations in these pathways in β cells by HFD feeding could not be reversed by dietary intervention.
CTCF-mediated chromatin remodeling may be responsible for the improvement of β cell function by dietary intervention
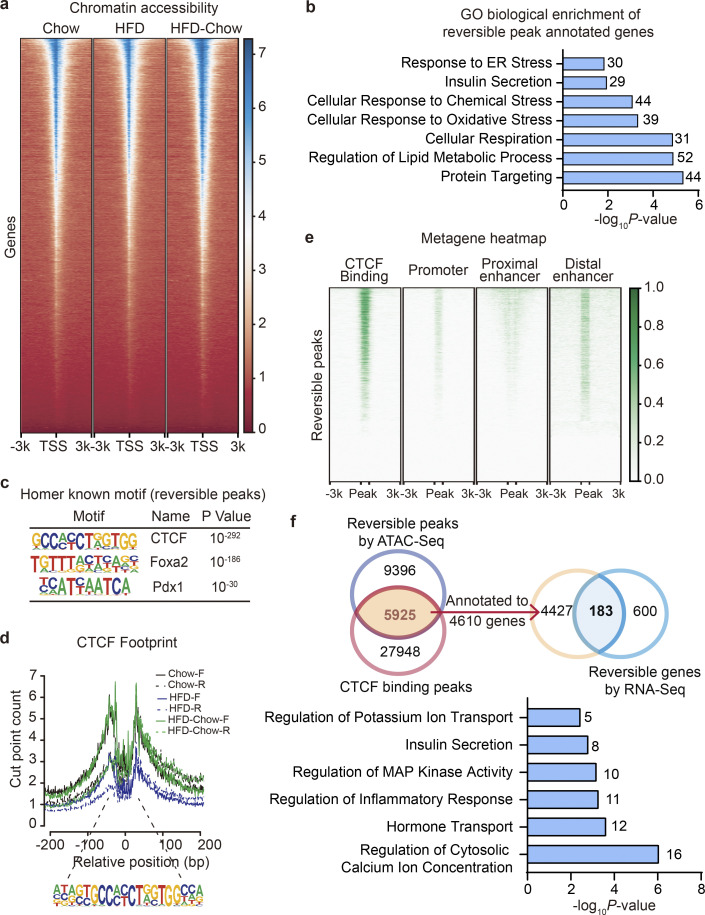
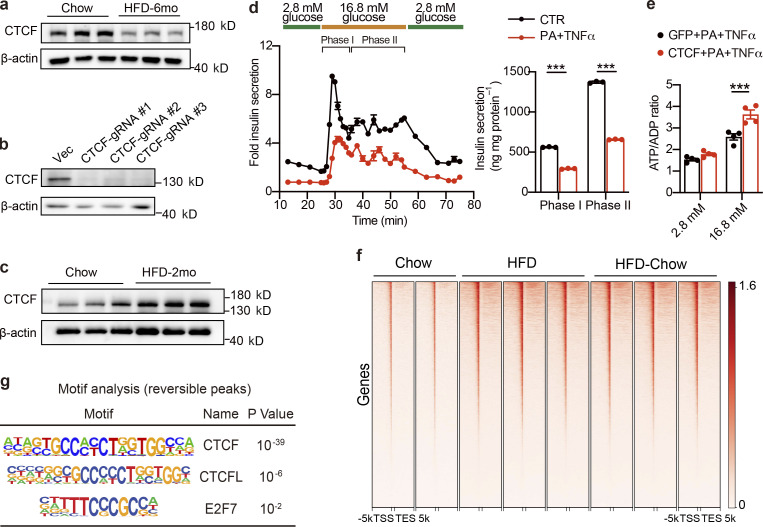
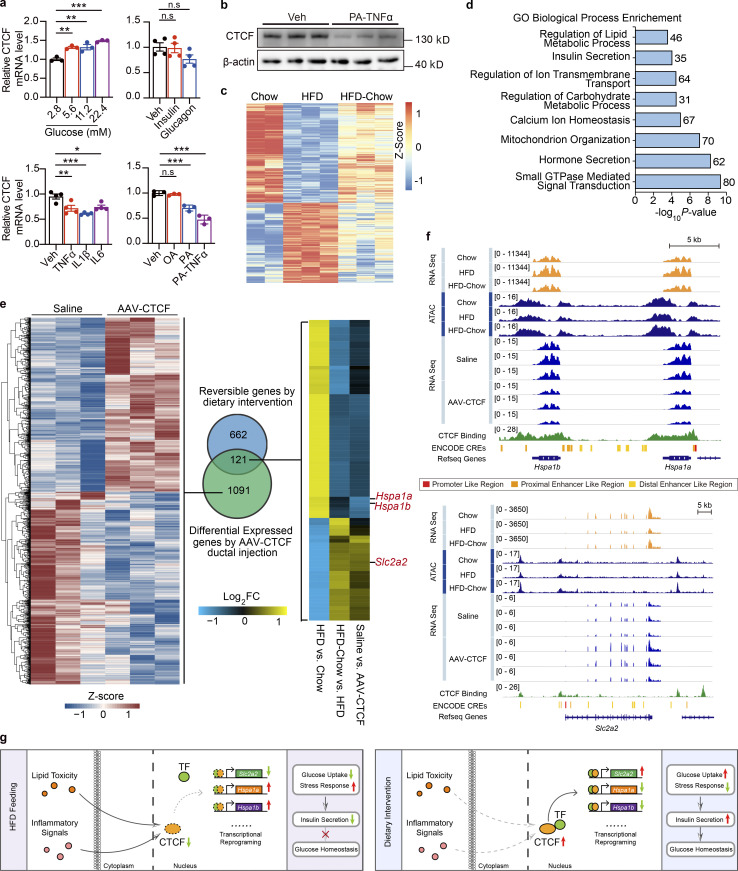
With phenotypical and transcriptional changes systematically described, we then explored the underlying mechanism mediating the preservation of β cell function by dietary intervention. Looking back into the time-series analysis along with HFD duration, we noticed that genes related to chromatin modification and remodeling were significantly altered in the compensation stage (Fig. 1, h and i), implying possible changes in the chromatin landscape and activity during the compensation and decompensation processes of islet β cell function in obesity and T2D. Hence, it is possible that dietary intervention might improve β cell function through chromatin remodeling. To test this possibility, ATAC-Seq analysis was performed on primary islets isolated from the Chow, HFD, and HFD-Chow groups to explore the genome-wide chromatin accessibility changes among these groups. ATAC-Seq totally obtained 41,163 shared open chromatin regions among three groups, which were mainly distributed in introns (43.13%), intergenic regions (36.61%), or at transcriptional start sites (TSSs; 10.37%). Distribution of ATAC signal within the −3 to +3 kb regions flanking TSSs demonstrated a generally less accessible chromatin status in the HFD group compared to the Chow group (Fig. 4 a). Such reduced signal intensity was not observed in the HFD-Chow group, indicating that dietary intervention might prevent the HFD induced remodeling of overall chromatin accessibility. Indeed, we obtained 15,321 chromatin regions that showed opposite accessibility changes in HFD vs. Chow comparison and HFD-Chow vs. HFD comparison (Reversible Peaks), and annotated these regions to their closest genes to identify 2,189 genes that are likely to be epigenetically regulated by dietary intervention. Consistent with the results obtained at the transcriptional level, GO analysis showed significant enrichment of genes involved in cellular response to oxidative stress (39 genes), insulin secretion (29 genes), cell respiration (31 genes), and response to ER stress (30 genes; Fig. 4 b). Meanwhile, we generated differential accessed peaks in HFD-Chow vs. Chow comparison. Reversible peaks we showed in Fig. 4 e were excluded from these differential peaks. The obtained peaks were then annotated to 1,009 genes. GO analysis showed significant enrichment in pathways including cellular translation, cell respiration, and cell cycle alteration (Fig. S4 a), which was similar to the findings we obtained via bulk RNA-Seq (Fig. S2 e).
Figure 4.
Identification of CTCF as the top candidate mediating the preservation of β cell function by dietary intervention. (a) Metagene heatmap of the genome-wide ATAC peaks’ occupation in the −3 to +3 kb region flanking the TSS. Scale bar indicates peak density. In each treatment, ATAC-Seq libraries were prepared using islets pooled from three mice. (b) GO analysis of genes annotated to the Reversible Peaks (within −5,000 to +100 bp relative to TSS). Most significant and nonredundant biological processes with respective gene number and P value are shown. (c) Known motif analysis within Reversible Peaks. Consensus islet related motifs (Motif), transcriptional factor names (Name), and P values are shown. (d) Footprint aggregate plots at putative CTCF binding sites. (e) Metagene heatmap of CTCF CUT&Tag peaks and ENCODE annotated CREs around the Reversible Peaks. Scale bar indicates peak density. (f) Upper panel: Venn plot showing the overlap between given datasets and schematic representation of data integration pipeline. Lower panel: GO analysis of light blue shaded genes in the Venn plot. Most significant and nonredundant biological process with respective gene number and P value are shown.
Figure S4.
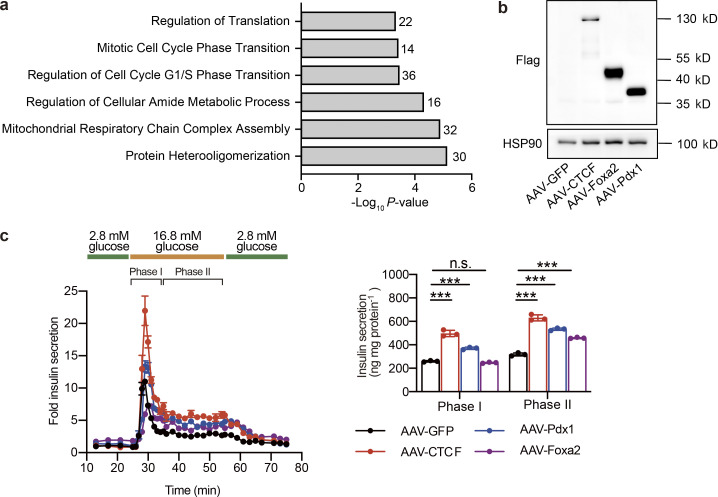
GO analysis of unaltered genes in ATAC-Seq and effects of AAV-mediated overexpression of CTCF, Foxa2, and Pdx1 on islet insulin secretion. (a) GO Biological Process enrichment analysis of genes annotated to peaks with their chromatin accessibility unaltered by dietary intervention (|log2FC (HFD-Chow vs. Chow)| > 0.5 AND NOT Reversible Peaks). (b) Immunoblot of total protein lysates from islets transduced with AAV expressing CTCF, Foxa2, Pdx1, or GFP (as control) for 5 d. (c) Dynamic glucose-stimulated insulin secretion (left) and quantified phase I and phase II insulin levels (right) from islets as treated in b. Islets were isolated from mice fed with HFD for 3 mo. Each islet sample was pooled from at least three animals. Data represent mean ± SD, n = 3 technical replicates; ***, P < 0.001; one-way ANOVA. Data in b and c are representative of two independent experiments. Source data are available for this figure: SourceData FS4.
To explore the possible regulator mediating these chromatin accessibility changes, we then performed known motif enrichment analysis on reversible peaks. Unexpectedly, while binding sites for multiple islet function-related transcription factors such Foxa2 and Pdx1 were significantly enriched in these regions, the CTCF binding site was identified as the most significantly enriched motif in these reversible peaks (P = 10−292; Fig. 4 c). To prioritize these potential factors, primary islets from mice fed with HFD for 1 mo were isolated and subjected to transduction with AAVs carrying either CTCF, Foxa2, Pdx1, or GFP (AAV-CTCF, AAV-Foxa2, AAV-Pdx1, or AAV-GFP) coding sequences. Remarkably, CTCF overexpression showed the most robust improvement in islet insulin secretion. Both phase I (∼1.9-fold, P < 0.001) and phase II (∼2.0-fold, P < 0.0001) insulin secretion was markedly enhanced by CTCF overexpression compared to other groups transduced with AAVs expressing Pdx1, Foxa2, or GFP (Fig. S4, b and c). We next performed footprint analysis to evaluate the ATAC signal distribution within the 200 bp region flanking all CTCF binding sites, and revealed a significant decrease in ATAC signal density in islets from the HFD group compared to the Chow group, suggesting an overall downregulation of chromatin accessibility to CTCF binding sites in response to HFD feeding. Such a decrease in chromatin accessibility was not present in the HFD-Chow group (Fig. 4 d), suggesting that dietary intervention may prevent HFD-induced accessibility loss on CTCF binding sites. With Cleavage Under Targets and Tagmentation (CUT&Tag) analysis (Kaya-Okur et al., 2019), we identified the global CTCF binding sites in primary mouse islets. We then mapped the CTCF binding sites and ENCODE annotated cis-regulatory elements (CREs; Moore et al., 2020) to the Reversible Peaks and their flanking regions (±3 kb). Intriguingly, we observed binding of CTCF on ∼40% of Reversible Peaks. These peaks with CTCF binding were also found to be overlapped with the promoter and distal enhancer signals (Fig. 4 e, lanes 1, 2, and 4). Proximal enhancer signals were also flanking some of these peaks (Fig. 4 e, lane 3). These findings suggested that accessibility changes on CTCF binding sites may directly modulate the transcriptional activation of corresponding genes. As such, we annotated these peaks to their closest genes and found 183 of them overlapped with Reversible Genes as revealed by RNA-Seq analysis (Fig. 4 f). Biological processes, including insulin secretion and regulation of potassium ion transport, were enriched in these genes (Fig. 4 f), suggesting that the CTCF-mediated chromatin accessibility changes might serve as the driving force to trigger transcriptional reprogramming critical for islet β cell function.
CTCF expression is downregulated in islets from obese and diabetic mice and humans
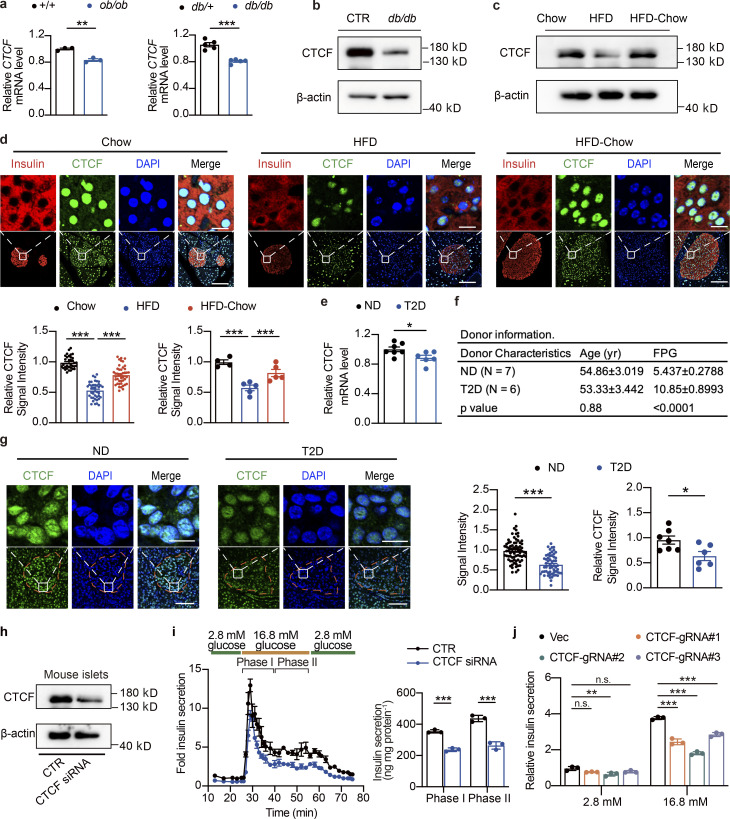
CTCF is a polyfunctional DNA binding protein reported to act as a transcriptional activator, repressor, and insulator (Chen et al., 2013; Gaszner and Felsenfeld, 2006; Gosalia et al., 2014; Klenova et al., 1993). We went on to test whether the expression levels of CTCF are actively regulated in islets during the development of diabetes in mice and humans. Interestingly, CTCF mRNA and protein levels were significantly downregulated in islets from genetically obese and diabetic mouse models, ob/ob and C57BLKS/J (BKS)-db/db mice (Fig. 5, a and b). The ob/ob data were generated by islet RNA-Seq, and the BKS-db/db datasets were obtained from previously published data (Wang et al., 2012). Western blotting analysis also revealed similar downregulation of CTCF protein levels in islets from mice fed with HFD for 6 mo (Fig. S5 a and Fig. 5 c). Intriguingly, HFD-Chow treatment prevented the downregulation of CTCF expression in response to HFD feeding (Fig. 5 c), which is consistent with the pattern of chromatin accessibility change on CTCF binding sites, indicating that CTCF expression level may play an important role in the regulation of chromatin accessibility on genes critical for β cell GSIS function. To further evaluate the protein levels of CTCF in islets and β cells under HFD and dietary intervention conditions, we performed immunofluorescent staining on pancreas sections from the Chow, HFD, and HFD-Chow mice, which revealed a significant decrease of CTCF signal intensity in islet β cells from the HFD group compared to the Chow group (∼46.95% decrease), while a 1.5-fold increase in CTCF signal intensity could be observed in islet β cells from the HFD-Chow group compared to the HFD group (Fig. 5 d). More importantly, significant downregulation of CTCF mRNA expression was also observed in islets from age-matched human T2D (hT2D) patients compared to human nondiabetic (hND) donors (P = 0.0322, Fig. 5 e; data extracted and analyzed from previously published dataset [Dominguez et al., 2011]). Similar to the murine HFD models, CTCF protein levels were markedly decreased in islets from T2D patients compared to age-matched controls (∼35.36%; Fig. 5, f and g). To determine the effects of the reduced CTCF expression on islet β cell function, we knocked down CTCF expression in mouse islets using siRNA transient transfection. Following confirmation of the successful knockdown of CTCF expression by Western blotting (Fig. 5 h), we then performed the islet perifusion study and revealed that CTCF knockdown reduced both phase I (∼35.64%, P < 0.001) and phase II (∼39.83%, P = 0.0018) insulin secretion compared to the control group (Fig. 5 i). To further assess the effects of CTCF ablation on β cell function, we also performed CRISPR-Cas9–mediated CTCF knockout in Min6 cells, a murine pancreatic β cell line (Fig. S5 b). Similar to the findings in primary islets, all CTCF knockout cell lines generated by different single-guide RNAs (sgRNAs) displayed a significantly reduced insulin secretion in response to 16.8 mM glucose treatment, while their basal insulin secretion under 2.8 mM glucose treatment remained merely unchanged (Fig. 5 j).
Figure 5.
CTCF expression is downregulated in islets from obese and diabetic mice and humans. (a) Relative CTCF mRNA expression in islets from 12-wk-old ob/ob mice (n = 3 mice per group; data represent mean ± SEM) or 9-wk-old BKS-db/db mice (n = 5 mice per group; data represent mean ± SEM). The relative gene expression levels of CTCF were calculated using the expression values of CTCF in the RNA-Seq (ob/ob mice) or microarray datasets (BKS-db/db, GEO accession no. GSE31559), normalized to their respective controls. **, P < 0.01; ***, P < 0.001; two-tailed unpaired Student’s t test. (b) Representative immunoblots of total islet protein lysates from control (CTR) and 20-wk-old BKS-db/db mice (pooled islet sample from three mice). (c) Representative immunoblots of total islet protein lysates from Chow, HFD, and HFD-Chow mice (pooled islet sample from three mice). (d) Representative immunofluorescent images (up) and relative signal intensities of CTCF (down) in pancreas sections of indicated mouse groups. Data represent mean ± SEM (dots in left panel represent 34–43 islets from 4 to 5 mice; at least 6 islets/mice; dots in right panel represent 4–5 mice); ***, P < 0.001; one-way ANOVA. CTCF signal intensities were normalized to average signal intensity in Chow group. Scale bar, 10 μm (for zoomed-in view), 50 μm. (e) Relative CTCF mRNA expression in human islets from ND and T2D donors obtained from previously published microarray datasets. Data represent mean ± SEM (n = 6–7 donors per group); *, P < 0.05; two-tailed unpaired Student’s t test. (f) Donor information of ND and T2D donors. (g) Representative immunofluorescent images (left) and relative signal intensity of CTCF (right) in human pancreas sections obtained from ND and T2D donors in f. Data represent mean ± SEM (dots in left panel represent 57–75 islets from 6 to 7 donors; at least 6 islets per donor; dots in right panel represent 6–7 donors); *, P < 0.05; ***, P < 0.001; two-tailed unpaired Student’s t test. CTCF signal intensities were normalized to average signal intensity in control group. Scale bars, 10 μm (for zoomed-in view), 50 μm. (h) Immunoblots (left) of islets transduced with control siRNA (CTR) or CTCF siRNA. Islets were pooled from four wild-type mice. (i) Perifusion analyses of dynamic glucose-stimulated insulin secretion (left) and biphasic insulin release levels (right) of mouse islets as described in h. Islets were pooled from four wild-type mice. Data represent mean ± SD; n = 3 technical replicates; ***, P < 0.001; two-tailed unpaired Student’s t test. (j) GSIS of Min6 cell lines transduced with lentivirus delivering Cas9 and either control (Vec) or CTCF targeting (CTCF-gRNA#1–3) sgRNAs. Data represent mean ± SEM (n = 3); **, P < 0.01; ***, P < 0.001; two-way ANOVA. All panels report data verified in at least two independent experiments. Source data are available for this figure: SourceData F5.
Figure S5.
Regulation of CTCF expression, its effect on islet function, and CUT&Tag analysis of CTCF genome-wide binding profiles. (a) Immunoblot of total islet protein lysates from mice fed with HFD for 6 mo and their corresponding controls (n = 3 biological triplicates). (b) Immunoblot of total protein lysates from Min6 cells transduced with spCas9 and sgRNA expressing lentivirus. Cells transduced with lentivirus expressing only spCas9 were used as the control group (CTR). (c) Immunoblot of total islet protein lysates from mice fed with HFD for 2 mo and their corresponding controls (n = 3 biological triplicates). (d) Dynamic glucose-stimulated insulin secretion (left panel) and biphasic glucose-stimulated insulin release levels (right panel) of mouse islets treated with TNFα (50 ng/ml) and PA (0.5 mM) for 24 h. Each islet sample was pooled from at least three animals. Data represent mean ± SD; n = 3 technical replicates; ***, P < 0.001; two-tailed unpaired Student’s t test. (e) ATP/ADP ratio of CTCF or GFP expressing mouse islets under low-glucose (2.8 mM) or high-glucose (16.8 mM) conditions. Islets were treated with TNFα (50 ng/ml) and PA (0.5 mM) for 24 h prior to glucose stimulation. Each sample was pooled from at least three animals. Data represent mean ± SEM (n = 4 replicates of islet samples, each sample represents 20 IEQ islets); ***, P < 0.001; two-tailed unpaired Student’s t test. (f) Metagene heatmap of the genome-wide CTCF CUT&Tag peaks’ occupation in the −5 to +5 kb regions flanking the TSS in islets from indicated groups. In each treatment, CUT&Tag libraries were prepared using islets pooled from three mice, with 2–3 technical replicates. The scale bar indicates peak density. (g) Known motif analysis within reversible CTCF binding peaks. Consensus non-redundant motifs (Motif), transcriptional factor names (Name), and P values are shown. Data in a–e are representative of two independent experiments. Source data are available for this figure: SourceData FS5.
CTCF overexpression mimics the beneficial effect of dietary intervention on islet β cell function ex vivo and in vivo
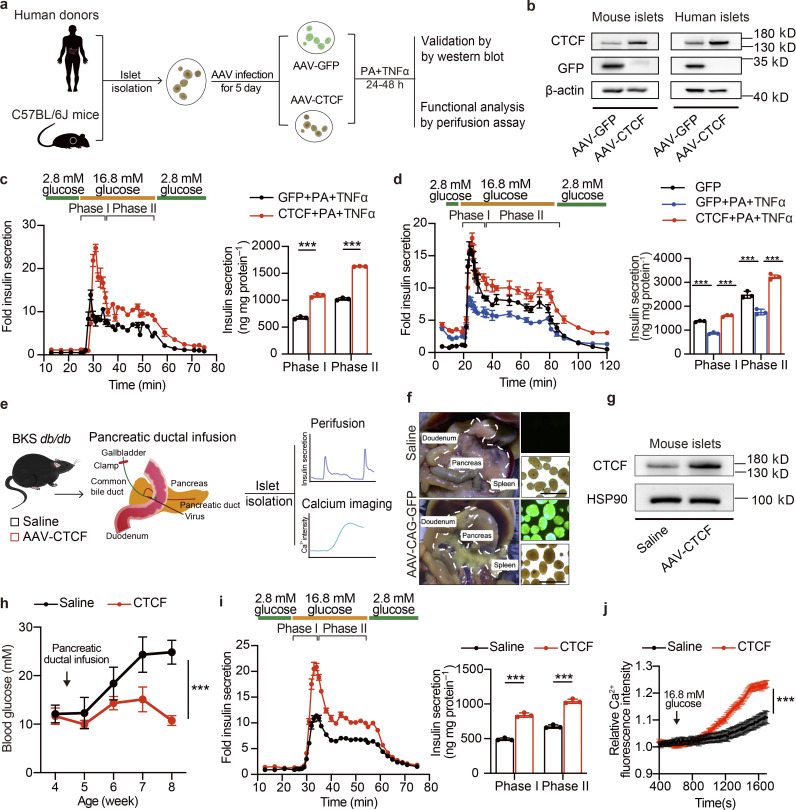
The above findings indicate that dietary intervention during the progression of T2D may preserve β cell function by preventing the downregulation of CTCF expression and its associated dysregulation of chromatin remodeling and transcriptional reprogramming. In addition, we also observed the compensatory enhancement of islet GSIS function in response to HFD feeding for 2 mo is associated with a robust induction of CTCF protein expression (Fig. S5 c). These results suggest that CTCF gain-of-function may promote GSIS function in islets. To test this, two distinct experimental systems were used. Firstly, isolated islets from healthy wild-type mice were treated with palmitic acid (PA) and TNFα to establish β cell dysfunction model ex vivo, and were used to evaluate whether CTCF overexpression could restore β cell dysfunction elicited by PA and TNFα treatment (Fig. 6 a). As expected, PA and TNFα treatment markedly inhibited phase I and phase II insulin secretion by ∼50% in healthy mouse islets (Fig. S5 d). Notably, AAV-mediated overexpression of CTCF (AAV-CTCF), as validated by Western blotting (Fig. 6 b, left panel), significantly protected the mouse islets from PA and TNFα treatment-induced damage of GSIS function (Fig. 6 c). In accordance, AAV-mediated CTCF overexpression significantly enhanced glucose-stimulated increase in ATP/ADP ratio in mouse islets treated with PA and TNFα (Fig. S5 e). Moreover, similar results were also obtained in islets isolated from human donors. While PA and TNFα treatment significantly impaired GSIS function (GFP + PA + TNFα vs. GFP), CTCF overexpression markedly augmented both phase I and phase II insulin secretion in human islets treated with PA and TNFα (83.92% increase for phase I, and 83.75% increase for phase II insulin secretion; CTCF + PA + TNFα vs. GFP + PA + TNFα, P < 0.0001; Fig. 6 b, right panel, and Fig. 6 d). To further validate the protective effect of CTCF overexpression on β cell function in vivo, we delivered AAV expressing CTCF directly into the pancreas of BKS-db/db mice via pancreatic ductal infusion. Islet perifusion and calcium imaging assays were then conducted to evaluate the islet GSIS function (Fig. 6 e). Fluorescent imaging of islets isolated from AAV expressing GFP (AAV-GFP) injected mice showed high transduction efficiency in the pancreatic islets (Fig. 6 f). As shown in Fig. 6 g, the protein level of CTCF in islets isolated from the AAV-CTCF injected mice was successfully elevated compared to those isolated from the saline injected mice. Notably, CTCF overexpression attenuated the progressive increase in blood glucose levels as seen in BKS-db/db mice injected with saline (Fig. 6 h). Further perifusion study of isolated islets showed remarkable improvement of islet GSIS function by AAV-CTCF injection (Fig. 6 i, left panel). Both phase I and phase II insulin secretion were significantly elevated (Fig. 6 i, right panel). In addition, calcium imaging analysis revealed robust improvement of glucose-stimulated calcium influx in islets isolated from AAV-CTCF injected mice compared to controls (Fig. 6 j). Together, these results demonstrate that restoring CTCF expression significantly improves β cell insulin secretion ex vivo in primary mouse and human islets and in vivo in diabetic mice, resembling the therapeutic effect of dietary intervention on β cell dysfunction in T2D.
Figure 6.
AAV-mediated restore of CTCF expression ameliorates β cell dysfunction ex vivo and in vivo. (a) Schematic representation of experimental procedures for b–d. (b) Immunoblots of total cell lysates from mouse or human islets transduced with AAVs expressing CTCF or GFP for 5 d, followed by treatment with TNFα (50 ng/ml) and PA (0.5 mM) for an additional 24 h. (c) Perifusion analyses of dynamic glucose-stimulated insulin secretion (left) and biphasic insulin release levels (right) of mouse islets as described in b. Each islet sample was pooled from at least three animals. Data represent mean ± SD; n = 3 technical replicates; ***, P < 0.001; two-tailed unpaired Student’s t test. (d) Perifusion analyses of dynamic glucose-stimulated insulin secretion (left) and biphasic insulin release levels (right) of human islets as described in b. Data represent mean ± SD; n = 3 technical replicates; ***, P < 0.001; one-way ANOVA. (e) Schematic diagram of pancreatic ductal infusion of AAV and experimental design for f–j. (f) Representative image of pancreas and islets underwent pancreatic ductal infusion of AAV-GFP. Scale bar, 500 μm. (g) Immunoblots of total cell lysates from mouse islets from BKS-db/db mice 5 wk following AAV pancreatic ductal infusion. (h) Random blood glucose levels of BKS-db/db mice injected with saline or AAV-CTCF via pancreatic ductal infusion. Data represent mean ± SEM (n = 6 mice per group). ***, P < 0.001; two-way ANOVA. (i) Perifusion analyses of dynamic glucose-stimulated insulin secretion (left) and biphasic insulin release levels (right) of islets isolated from BKS-db/db mice as described in g. Each islet sample was pooled from at least three animals. Data represent mean ± SD; n = 3 technical replicates; ***, P < 0.001; two-tailed unpaired Student’s t test. (j) Glucose-stimulated Ca2+ influx in islets isolated from db/db mice as described in g. Each islet sample was pooled from at least three animals. Data represent mean ± SEM (n = 53–60 islets per group); ***, P < 0.001; two-way ANOVA. All panels report data verified in at least two independent experiments. Source data are available for this figure: SourceData F6.
CTCF selectively regulates the transcription of genes involved in β cell glucose metabolism and stress response
Seeing the importance of CTCF in regulating β cell function, we then explored the upstream signaling and downstream targets of CTCF in β cells. It is well known that glucose toxicity, lipid toxicity, dysregulated hormone levels, and meta-inflammation play important roles in the development of β cell dysfunction in T2D (Campbell et al., 1988; Meng et al., 2009a; Meng et al., 2009b; Meng et al., 2006; Meng et al., 2012; Zhou and Grill, 1994). To identify the upstream signals responsible for the regulation of CTCF expression by dietary intervention in obesity and T2D, we treated Min6 cells with different concentrations of glucose, hormones, inflammatory cytokines, or lipids, and performed quantitative PCR (qPCR) analyses of CTCF expression. As shown in Fig. 7 a, while glucose, insulin, and glucagon treatments showed mild effects on the mRNA expression of CTCF, treatments with inflammatory cytokines including TNFα, interleukin 1 β (IL1β), or interleukin 6 (IL6) significantly decreased CTCF expression compared to control (decreased by 23.44%, 34.15%, and 20.72%, P = 0.2344, P = 0.0003, and P = 0.0138, respectively). In addition, PA, but not oleic acid (OA) was capable of downregulating CTCF expression in Min6 cells (by 29.68%, P = 0.0007). Moreover, co-treatment with both PA and TNFα elicited a much more robust downregulation of CTCF expression in both mRNA (Fig. 7 a, lower right panel, by 52.64%, P < 0.0001) and protein levels (Fig. 7 b). Indeed, it has been reported that weight-loss treatment could effectively lower plasma fatty acid and inflammation cytokine levels in overweight individuals (Lee et al., 2018; Ryan and Nicklas, 2004), suggesting that dietary intervention may restore the expression of CTCF in islets through alleviating the lipid toxicity and inflammatory signaling in obesity and T2D.
Figure 7.
CTCF regulates glucose metabolism and stress response related gene expression in murine islets. (a) Relative CTCF mRNA expression in Min6 cells treated with indicated reagents for 12 h. Data represent mean ± SEM; n = 3–4 biological replicates; expression levels were normalized to their respective controls; *, P < 0.05; **, P < 0.005; ***, P < 0.001; one-way ANOVA; Veh, Vehicle. (b) Immunoblots of total cell lysates from Min6 cells treated with PA and TNFα. (c and d) Heatmap showing scaled, normalized read-counts (as Z-Score) of significantly reversible CTCF bound peaks in islets between HFD vs. Chow and HFD-Chow vs. HFD comparison and GO analysis of genes annotated to these peaks. Most significant and nonredundant biological processes with respective gene number and P value are shown. (e) Overlapping analysis of significantly altered genes in islets isolated from BKS-db/db mice treatment with AAV-CTCF or saline via pancreatic ductal infusion and reversible genes by dietary intervention. Heatmap representation of significantly altered genes between AAV-CTCF and saline treatments (left, normalized read-counts were shown as Z-Score), Venn plot between two datasets (middle), and heatmap representation of overlapped genes with similar regulatory trends (right) are shown. (f) Representative RNA-Seq and ATAC-Seq browser tracks displaying dietary intervention and CTCF overexpression regulated gene loci including Hspa1a, Hspa1b, and Slc2a2. CTCF CUT&Tag tracks and ENCODE annotated CREs are also displayed. (g) Schematic representation of the working model presented in this paper. Data in a and b are representative of at least two independent experiments. Source data are available for this figure: SourceData F7.
Given that mRNA and protein expression of CTCF was remarkably downregulated in islets from obese and type 2 diabetic mice and humans, we next went on to test whether CTCF binding to DNA is also altered in islets in response to HFD feeding and dietary intervention using CUT&Tag. We did not observe significant global changes in the overall DNA binding landscape by CTCF in islets among three groups (Fig. S5 f). Instead, highly localized changes in binding profiles were observed. Among the total 30,022 peaks shared by the three groups, 2,484 peaks were robustly reversed by the dietary intervention (|log2FC| > 0.3, P < 0.05; Fig. 7 c). Interestingly, GO analysis demonstrated that biological processes, such as lipid metabolic process, insulin secretion, ion transmembrane transport, and hormone secretion, mitochondrion organization, were enriched in the 2,184 genes annotated by the 2,484 reversible peaks by the dietary intervention (Fig. 7 d). These data are consistent with the aforementioned findings in transcriptomic and DNA accessibility levels as revealed by RNA-Seq and ATAC-Seq analyses, respectively (Fig. 2, j and k; and Fig. 4, b and f). These results further confirmed the key role of CTCF in mediating the preservation of β cell function by dietary intervention through direct DNA binding, chromatin remodeling, and selective transcriptional regulation of genes important for β cell function.
To examine the effects of CTCF expression on islet transcriptome in vivo, we performed RNA-Seq analysis on islets isolated from mice treated with AAV-CTCF or saline via pancreatic ductal infusion. Compared to controls, we identified 1,212 genes that were either significantly upregulated or downregulated by CTCF overexpression (Fig. 7 e, left panel, P < 0.05, |log2FC| > 0.3). Potential CTCF downstream target genes were then identified by comparing these 1,212 significantly regulated genes with the 783 reversible genes by dietary intervention (Fig. 7 e, middle panel). Among the 121 overlapped genes, we identified 85 genes with a similar changing pattern between CTCF overexpression and dietary intervention (Fig. 7 e, right panel). Interestingly, ER stress-associated genes heat shock protein family A member 1A (Hspa1a) and 1B (Hspa1b; Skórzyńska-Dziduszko et al., 2018) were downregulated upon either CTCF overexpression or dietary intervention. Moreover, Slc2a2, encoding the key glucose transporter Glut2 in β cell GSIS pathway, was also upregulated by CTCF overexpression and dietary intervention. Representatively, genome browser tracks of our sequencing data on Hspa1a/Hspa1b or Slc2a2 coding regions and flanking loci exhibited reduced ATAC signals on the CTCF bound enhancer and promoter region upon HFD feeding, accompanied by either upregulation (for Hspa1a and Hspa1b) or downregulation (for Slc2a2) of their transcriptional levels as revealed by RNA-Seq analysis. Remarkably, dietary intervention almost completely prevented the alterations of chromatin accessibility, and both CTCF overexpression and dietary intervention largely restored the alterations on mRNA signals of Hspa1a, Hspa1b, and Slc2a2 genes elicited by HFD feeding (Fig. 7 f). In addition, motif analysis of genomic regions with reversible CTCF binding upon dietary intervention revealed significant enrichment of binding motifs for transcriptional factors such as CCCTC-binding factor (zinc finger protein)–like and E2F transcription factor 7 besides CTCF itself (Fig. S5 g), suggesting that CTCF might interact with this transcriptional factor to exert its chromatin remodeling and transcriptional regulatory functions. Altogether, these results reveal that dietary intervention prevents β cell dysfunction in T2D through CTCF-mediated enhancer and promoter chromatin remodeling and transcriptional reprogramming of genes important for β cell glucose uptake and stress response process (Fig. 7 g).
Discussion
Dietary intervention has long been proven as an effective treatment to improve glycemic control, insulin sensitivity, and liver steatosis in murine models of obesity and T2D, and T2D patients (Al-Mrabeh et al., 2020; Eriksson and Lindgärde, 1991; Knowler et al., 2002; Perreault et al., 2009; Taylor et al., 2018). However, the effect and the underlying mechanisms of dietary intervention on β cell dysfunction during the progression of T2D are yet to be determined. In this study, we established a dietary intervention mouse model in the context of HFD-induced obesity and β cell dysfunction. We found that HFD feeding induced a functional compensation-to-decompensation process in mouse islets overtime, closely resembling the development of β cell dysfunction in human T2D. This compensatory process is accompanied by dynamic and robust transcriptional signatures. In addition to the well-demonstrated activation of ER stress and oxidative stress in the development of islet dysfunction (Kajimoto and Kaneto, 2004; Laybutt et al., 2007; Hasnain et al., 2016), which was also observed in our transcriptomic data, it was interesting to notice the significant alteration of genes involved in oxidative phosphorylation and chromatin remodeling at the early phase of HFD feeding (2 mo). While the correlation between oxidative phosphorylation, mitochondrial respiration, and β cell function has been reported elsewhere (Ling et al., 2008; Supale et al., 2012; Yoon et al., 2003), how mitochondrial function is regulated and involved in the islet β cell functional compensation process remains to be determined in future studies. Intriguingly, we also noticed a significant decrease of 18 insulin release–related genes even at the compensatory stage (Fig. 1 i, the top cluster of 359 genes), such as Stx1a, Cacna1c, Cacna1d, Itpr3, and Slc2a2, suggesting that accumulating deterioration of glucose metabolism and calcium-coupled insulin secretion machinery could serve as key driver leading to the ultimate β cell failure. Consistent with the previous published studies showing the reduced glucose transport and calcium signaling in β cells during T2D development (Ohtsubo et al., 2011; Yang and Berggren, 2005), we also observed a significant reduction in glucose uptake and ATP-induced calcium influx machineries starting from the compensatory stage. Our studies reveal the dynamic and temporal regulation of these important processes during the development of obesity and T2D, and suggest that these glucose metabolism and calcium-secretion coupling related processes are affected at the early stage of T2D, and could possibly serve as key drivers leading to the ultimate β cell decompensation and failure in overt T2D.
By administering dietary intervention when the islet β cells are still within the compensatory stage, we established a murine dietary intervention model to mimic prediabetic dietary intervention in human subjects. Using functional assays and multiomics techniques, we systematically characterized the alterations in islets isolated from animals under chow diet, continuous HFD, and prediabetic dietary intervention. Consistent with the changes in systemic glucose homeostasis, we found that dietary intervention could prevent the HFD-induced impairment of glucose-stimulated calcium response and insulin secretion from β cells. Both whole islet and β cell specific transcriptional profiling revealed reversibility in the expression of genes involved in cellular stress response and insulin secretion pathways. Considering the importance of oxidative stress and ER stress responses in islet functional regulation as previously reported (Kajimoto and Kaneto, 2004; Laybutt et al., 2007; Hasnain et al., 2016), these results implied that relief of cellular stress response might serve as the key player in the preservation of β cell function by dietary intervention.
Moreover, at the chromatin accessibility levels, we identified the enrichment of binding sites from multiple islet-function-related transcriptional factors, e.g., Pdx1, Foxa2, Hnf1b, Pax6, and Mafa, in the reversible peaks induced by dietary intervention. Among these transcriptional factors, the overexpression of Pdx1, Pax6, and Mafa has also been reported to preserve β cell function under T2D conditions (Matsuoka et al., 2015; So et al., 2021; Xiao et al., 2018). In these top candidate transcriptional factors, we surprisingly identified CTCF as the most significantly enriched target, which was however consistent with the previous reports showing enriched CTCF binding on distal regulatory elements on T2D susceptibility regions (Parker et al., 2013; Stitzel et al., 2010). Notably, (Fang et al 2014; Tsui et al 2012) have reported the decreased glucagon levels, increased insulin levels, and reduced blood glucose levels in CTCF whole-body transgenic mice (Fang et al., 2014; Tsui et al., 2012). Whole-body transgenic overexpression of CTCF robustly impaired α cell development and population in mice (Tsui et al., 2012). In addition, they also found that CTCF inhibited glucagon-secreting function in α cells likely through suppressing Pax6 expression. These results revealed the potential important role of CTCF in regulating islet α cell development and function. However, the roles and mechanisms of CTCF in the control of β cell biology and function remain unclear. In the dietary intervention model, we illustrated that the peaks with CTCF binding were found in proximity to promoter regions and distal enhancer regions, especially in genes important for β cell insulin secretion like Slc2a2. Consistently, recent studies have shown that beyond its insulator-binding boundary function, CTCF could also regulate long-range chromatin interaction between promoters and distal enhancers in certain genes in a context-dependent manner, thereby regulating target gene expression and specific biological function (Merkenschlager and Nora, 2015; Pugacheva et al., 2020; Ren et al., 2017). Our findings suggest that CTCF likely preserves islet β cell function underpinning dietary intervention through governing the accessibility and activity of enhancers and promoters on key genes important for β cell glucose metabolism and stress response. Future study of genome-wide chromatin interactions with Hi-C and computational based chromatin 3D structure modeling might provide more new insights regarding the nature and consequences of such changes.
While CTCF mRNA and protein levels were dramatically downregulated in islet β cells from multiple murine obesity and diabetic models and human T2D patients, dietary intervention almost completely reversed the decreased CTCF expression elicited by HFD feeding. In addition, our in vitro experiments identified inflammatory cytokines and lipids as the potential upstream regulators of CTCF expression during the development of T2D. As the reduction of these signals, which have been observed in diabetic patients underwent weight-loss treatment (Lee et al., 2018; Ryan and Nicklas, 2004), our findings illustrated an axis connecting dietary intervention to β cell functional preservation by the alleviation of lipid toxicity and inflammatory signaling, and thereby the restoration of CTCF expression and function. Interestingly, Tsui et al. (2014) also demonstrated that CTCF might be involved in the regulation of glucose-stimulated β cell survival. Notably, we observed that the compensatory enhancement of islet GSIS function in mice fed with HFD for 2 mo is accompanied by a robust induction of CTCF expression. We are also aware that long-term overexpression of any transcription factors may lead to deleterious effects on biological functions. This might be an important reason for the impairment of islet function and systemic glucose homeostasis seen in the whole body CTCF transgenic mice (Tsui et al., 2012). Moreover, while our ex vivo and in vivo data showed that transient CTCF overexpression during the decompensatory stage could significantly improve β cell GSIS function, whether CTCF overexpression affects β cell function during the compensatory phase remain unclear. Together, these findings suggest that fine-tuning of CTCF expression at an appropriate level and in a specific temporal window would be critical for achieving the beneficial effects of CTCF overexpression on β cell function and systemic glucose homeostasis.
In conclusion, our work establishes a murine model of obesity and T2D recapitulating the dynamic nature of β cell adaptation during disease progression in human T2D subjects, demonstrates a comprehensive functional and transcriptional profiling of islet and β cell adaptation in this process, provides a systematic illustration of how dietary intervention during the development of T2D triggers the morphological and functional improvement of islet β cells downstream of chromatin remodeling-mediated transcriptional reprogramming, and finally identifies CTCF, a chromatin architecture regulator, as a key mediator of dietary intervention–induced preservation of β cell function through selectively transcriptional reprogramming.
Materials and methods
Animal models
Male C57BL/6J ob/ob and wild-type mice as well as BKS-db/db mice were obtained from The Jackson Laboratory. All mice were bred in the specific pathogen–free animal facility at the Zhejiang University School of Medicine, and maintained on a 12-h light/dark cycle at an ambient temperature of 23°C. Water and food were provided ad libitum. Age-matched male mice were used for all the animal experiments unless otherwise indicated.
To generate diet-induced obese mice, 12-wk-old C57BL/6J male mice were fed with either a chow diet (10 kcal% fat, 70 kcal% carbohydrate, and 20 kcal% protein; 1010088; Jiangsu Xietong Pharmaceutical Bio-engineering Co., Ltd.) or an HFD (60 kcal% fat, 20 kcal% carbohydrate, and 20 kcal% protein; D12492; Research Diets) for 4–28 wk. For dietary intervention, mice were switched to chow diet feeding for a period of 6–15 wk following HFD feeding for 10–16 wk. Pancreatic ductal infusion of AAVs was performed on BKS-db/db mice at the age of 4.5 wk. The islets of ob/ob mice were isolated and prepared for RNA-Seq at the age of 12 wk.
Human islets
Primary human islets were isolated from healthy male nondiabetic donors (27 and 53 yr old). Islets were cultured in CMRL 1066 (15-110-CVR; Corning), supplemented with 50 μg/ml streptomycin, 50 U/ml penicillin, 10% FBS, 10 U/ml heparin sodium, and 90 U/ml insulin.
Cell culture
Min6 cells originally obtained from Dr. Liangyou Rui’s lab at the University of Michigan were cultured in DMEM (11995065; Gibco) supplemented with 15% ES-FBS (SE200-ES; Vistech), 50 μM 2-β mercaptoethanol (M3148; Sigma-Aldrich), 50 μg/ml streptomycin, and 50 U/ml penicillin in a humidified incubator at 37°C with 5% CO2 as previously described (Meng et al., 2012). CTCF knockout Min6 cells were generated by transducing Min6 cells with spCas9 and sgRNA expressing lentivirus followed up by puromycin selection. For hormone, cytokine, glucose, or lipid treatment, Min6 cells were cultured in DMEM medium supplemented with 0.1% BSA overnight, and then treated with glucose (2.8, 5.6, 11.2 or 22.4 mM), insulin (50 nM), glucagon (50 nM), TNFα (50 ng/ml), IL1β (10 ng/ml), IL6 (10 ng/ml), PA (0.5 mM), or OA (0.5 mM) for 12 h before subsequent analysis.
Plasmid construction
The sgRNAs targeting CTCF were annealed from oligos and inserted into lentiCRISPRv2 puro plasmid (#52961; Addgene). The sgRNA targeting sites (gRNA#1: 5′-CAAGTGCCCAGACTGCGATA-3′, gRNA#2: 5′-GACGATCCAAATTTGAACGC-3′, gRNA#3: 5′-ACTTACCAGAGACGCCGGGA-3′) were designed with Benchling. Shuttle vectors encoding Flag/HA tagged mouse Pdx1 and Foxa2 (pAAV-CAG-FH-mPdx1 and pAAV-CAG-FH-mFoxa2) were generated with the Gateway cloning followed by standard sub-cloning into pAAV-CAG-GFP plasmid vector (28014; Addgene). In brief, coding sequences of Pdx1 and Foxa2 were amplified from mouse islet cDNA sample using FmPDX1-attb: 5′-GGGGACAACTTTGTACAAAAAAGTTGGCTTCACCATGAACAGTGAGGAGCAGTACTACG-3′, RmPDX1-attb: 5′-GGGGACAACTTTGTACAAGAAAGTTGGGTCACCGGGGTTCCTGCGGT-3′, FmFoxa2-attb: 5′-GGGGACAACTTTGTACAAAAAAGTTGGCTTCACCATGCTGGGAGCCGTGAAGAT-3′ and RmFoxa2-attb: 5′-GGGGACAACTTTGTACAAGAAAGTTGGGTTAGGATGAGTTCATAATAGGCCTGG-3′. The amplified fragments were hence cloned into pDONR223 entry vectors using the BP Clonase (11789020; Thermo Fisher Scientific) and subsequently transferred into pcDNA3.0-Flag/HA destination vector with LR Clonase. The Flag/HA tagged sequences were subcloned into pAAV-CAG-GFP by Hind III and Kpn I digestion afterwards.
Shuttle vector encoding mouse CTCF was generated by gene synthesis. Mouse CTCF coding sequence was obtained from CTCF mRNA sequence on GenBank (NCBI reference sequence: NM_181322.3, CCDS22606.1). CTCF coding sequence was synthesized and inserted into the pAAV-CAG-GFP plasmid vector to replace the original GFP coding sequence.
AAV preparation
AAV production and purification were performed by ChuangRui Bio. In brief, AAV293 cells (240073; Agilent) were cultured at 37°C in a humidified incubator with 5% CO2. Cells were cultured in DMEM (11995065; Gibco) with 10% (vol/vol) FBS (SE100-011; Vistech). 1 d before transfection, AAV293 cells were seeded on 20 15-cm culture dishes and grown to ∼90% confluency before transfection. For each 15-cm dish, 7 μg of AAV shuttle vector carrying target genes, 20 μg of Δ F6 helper vector (112867; Addgene), 7 μg of RC2/9 vector (112865; Addgene), and 250 μl of polyethylenimine (23966-2; Polysciences) were added to 1-ml DMEM, incubated at room temperature for 15 min and supplemented into cell cultures. Culture mediums were replaced with DMEM containing 0.5% (vol/vol) FBS at 24 h after transfection. Cells were collected at 72 h after transfection and resuspended in 5 ml cell lysis buffer (150 mM NaCl, 20 mM Tris, pH 8.0) after washing with 20-ml PBS solution. AAVs were purified via discontinuous iodixanol gradient (D1556-250ML; Sigma-Aldrich). In brief, cells in lysis buffer were freeze-thawed for three times, followed by adding 1 M MgCl2 and 25 kU/ml Benzonase (E8263-25k; Sigma-Aldrich) solutions to the final concentrations of 1 mM and 250 U/ml, respectively. Cell lysates were incubated at 37°C for 15 min and centrifuged at 4,000 rpm for 30 min under 4°C. Iodixanol solutions were added into the ultracentrifuge tube (361625; Beckman) in the order of 60%, 40%, 25%, and 17%. Cell lysates were transferred onto the top layer gently, and the remaining volume of the ultracentrifuge tube was filled up with cell lysis buffer. Virus was then isolated from the 40% layer after centrifugation at 53,000 rpm (Beckman Optima XPN-100 Ultracentrifuge with Type 70Ti rotor) at 14°C for 2 h 40 min. Viral titer was determined by qPCR assay with the standard curve generated by serial dilutions of the AAV shuttle vector.
Metabolic measurements
Body fat and lean mass were measured by the Nuclear Magnetic Resonance analyzer (QMR06-090H; Niumag). Fasting blood glucose levels were determined with Bayer Contour blood glucometer by tail-snip blood sampling after overnight fasting (∼16 h). For plasma insulin level measurement, mice were fasted overnight before tail-snip sampling of 150 μl blood. Blood samples were subsequently mixed with EDTA and centrifuged at 8,000 rpm for 8 min to obtain plasma samples from the supernatant. Plasma insulin levels were measured using a commercial ELISA kit (90080; Crystal Chem) according to the manufacturer’s instruction.
Glucose tolerance test (GTT) and insulin tolerance test (ITT) assays
GTT and ITT were performed as previously described (Meng et al., 2017). For GTT, mice were fasted overnight (∼16 h) and i.p. injected with glucose saline solution (1.2 g/kg body weight, glucose concentrations were adjusted accordingly to obtain equal injection volume for each mouse). Blood glucose levels were measured by tail-snip blood sampling pre-injection and 15-, 30-, 60-, and 120-min after injection. For ITT, mice were fasted for 4 h and i.p. injected with insulin saline solution (1 U/kg body weight, concentrations were adjusted accordingly to equalize injection volume for each mouse). Blood glucose levels were measured by tail-snip blood sampling pre-injection and 15-, 30-, 60-, and 120-min after injection.
Murine primary islet isolation
Mouse islets were isolated from the pancreas of 2% pentobarbital sodium (P3761; Sigma-Aldrich) sedated mice by intraductal collagenase (Collagenase type V, 1 mg/ml, C9263; Sigma-Aldrich) digestion, and purified by Histopaque 1077 (10771; Sigma-Aldrich) density gradient centrifugation as previously described (Meng et al., 2009a; Meng et al., 2009b). Purified islets were then hand-picked under stereo microscope. Isolated islets were incubated overnight in RPMI-1640 medium (11875-093; Gibco) containing 50 μg/ml streptomycin, 50 U/ml penicillin, and 10% FBS in the humidified incubator at 37°C with 5% CO2.
RNA-Seq of primary murine islets
Primary murine islets cultured overnight after isolation were used for RNA extraction. 200 ng of RNA extracted by Trizol Reagent (Invitrogen) was sent to library preparation and sequencing in The Beijing Genomics Institute. In brief, mRNAs were enriched from total RNA and fragmentized, followed by reverse transcription and second-strand cDNA synthesis. The cDNAs were then ligated with sequencing adaptors and amplified for paired-end sequencing. Data were processed following the standard Beijing Genomics Institute mRNA analysis pipeline. Statistical analysis was performed with Deseq2 (v.1.20.0) package (Love et al., 2014). Statistical parameters to call differential expressed genes in each analysis were described in the figure legend. Unsupervised clustering and heatmap visualization were performed with pheatmap (V1.0.12, https://cran.r-project.org/web/packages/pheatmap/index.html). GO and pathway grouping and enrichment analysis were performed by clusterProfiler (V3.12.0; Yu et al., 2012). If not mentioned in the figure legends, significantly changed genes were identified by |log2FC| > 0.3, P < 0.05 (P value by Wald test).
scRNA-Seq of primary murine islets
Single-cell library preparation was conducted following the previously reported protocol (Han et al., 2018). In brief, 200 islet equivalents (IEQ) mouse islets were isolated and dissociated into single cells (∼2.5 × 105 cells) by TrypLE (12604013; Thermo Fisher Scientific). Approximately, 1 × 105/sample of resuspended single cells were loaded onto microwell containing agarose plates. Barcoded beads were loaded after cell doublets removal. Cells were lysed using cold lysis buffer (0.1 M Tris-HCl, pH 7.5, 0.5 M LiCl, 1% SDS, 10 mM EDTA, and 5 mM dithiothreitol). The beads were then collected and washed once with 1 ml of 6 × SSC, once with 500 μl of 6 × SSC, and once with 200 μl of 50 mM Tris-HCl, pH 8.0. Samples were then conducted reverse transcription following the Smart-seq2 protocol (Picelli et al., 2013). The obtained cDNAs were treated with exonuclease I (NEB) and amplified using HiFi HotStart Readymix (Kapa Biosystems) with primers used in the previous report (Han et al., 2018). Amplified cDNA libraries were then purified with AMPure XP beads (Beckman Coulter), fragmented, and PCR amplified following the instruction of TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme) replacing the index two primers with the primers reported previously (Han et al., 2018). PCR products were purified with AMPure XP beads (Beckman Coulter) and were subjected to sequencing on the Illumina HiSeq systems by Annoroad Gene Technology.
The scRNA-Seq data processing pipeline was adapted from the previous study (Han et al., 2018). Briefly, raw reads were filtered and extracted for barcode and unique molecular identifier with bbmap (https://sourceforge.net/projects/bbmap/). Filtered reads were then mapped to mm10 genome with STAR (V2.5.2a; Dobin et al., 2012) with default parameters. Unique alignments were then used for unique molecular identifier counting to generate the digital gene expression data matrix. Cells with <200 genes detected were excluded. Seurat (V3.2.1; Stuart et al., 2019) package was used for follow-up dimension reduction and clustering. After initial clustering using principal component analysis with top 500 highly variable genes, the expression level of Ins1, Ins2, Gcg, Sst, and Ppy in immune cell cluster (with H2-Ab1 as the major cluster marker) were considered as background signal to eliminate the possible contamination of highly expressed hormone RNAs. Cells were then reclustered using principal component analysis with top 500 highly variable genes. Follow-up differential gene expression analysis was performed with Seurat and ggplot2 (V3.2.1; Wickham, 2016) was used for plotting results.
ATAC-Seq of primary murine islets
The protocol of ATAC-Seq was adapted from a previous report (Buenrostro et al., 2013). In brief, 50,000 cells were extracted from isolated mouse islets by TrypLE (12604013; Thermo Fisher Scientific). Isolated cells were lysed with lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% [vol/vol] Igepal CA-630) at 4°C for 10 min. Tagmentation and amplification were done following the manufacture’s instruction with TruePrep DNA library Prep Kit V2 (TD501; Vazymes). Paired end sequencing was performed by Annoroad Gene Technology, and clean reads with trimmed adapters were aligned to mm10 reference genome with Bowtie2 (2.3.4.1) package (Langmead and Salzberg, 2012).
Downstream analysis pipeline was adapted from a previous study (Liu et al., 2020). Briefly, broad peaks were called by Macs2 (2.1.1.20160309) package (Zhang et al., 2008) using the parameter of (—nomodel—shift−100—extsize 200 B—broad) and differential accessed peaks were called using Deseq2 (v.1.20.0) package (Love et al., 2014). Reversible accessed peaks were thereby called by |log2FC| > 0.5 with reversible accessibility by dietary intervention. Motif enrichment analysis and peak-associated-gene annotation were performed by HOMER (V4.10; Heinz et al., 2010) using peaks filtered by corresponding criteria mentioned in the figure legends. Browser tracks were visualized by IGV browser (V2.4.14; Robinson et al., 2011) after normalizing the reads from each individual sample to its own library size. To compare ATAC-Seq signal with CUT&Tag data or ENCODE cis regulatory elements (Moore et al., 2020), metagene analysis was performed with Deeptools2 (V3.1.2; Ramirez et al., 2016) using ATAC peaks against the BigWig list of CUT&Tag or ENCODE CREs peaks. For optimal visualization of the CUT&Tag peaks around the ATAC peaks, the ATAC peaks plotted are scaled into 800 bp length, CUT&Tag peak distribution from the −3 to +3 kb region relative to the peak start sites and peak end sites were analyzed. For transcriptional footprint analysis, CTCF binding motifs were scanned by fimo (V5.0.5; Grant et al., 2011) throughout the whole mm10 genome; footprints were then called with CENTIPEDE (V1.2; Pique-Regi et al., 2011) as described previously (Liu et al., 2020). Ataqv (V1.0.0, https://github.com/ParkerLab/ataqv) package developed by Parker’s lab from the University of Michigan was used to perform the ATAC-Seq data quality control analysis.
CUT&Tag and analysis
The library preparation for CUT&Tag was performed as previously reported (Kaya-Okur et al., 2019). In brief, ∼1 × 105 single cells were washed twice in 1 ml PBS. Cells were incubated with 10 μl of activated concanavalin A coated magnetic beads (BP531; Bangs Laboratories) in 500 μl Wash Buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, and protease inhibitor cocktail; Roche) for 10 min under room temperature. Cell-bound beads were collected and resuspended with 50 μl Dig-Wash Buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, protease inhibitor cocktail, 0.05% Digitonin) containing 2 mM EDTA, 0.1% BSA, and a 1:50 dilution of the primary antibody (Rabbit anti-CTCF, Active Motif, 61311; or normal Rabbit IgG, #2729; Cell Signaling Technology), and incubated at 4°C overnight. Secondary antibody (Goat anti-Rabbit IgG, SAB3700883; Sigma-Aldrich) diluted at 1:50 in 100 μl of Dig-Wash buffer was then administered into the beads, and incubated for 60 min at room temperature following primary antibody removal with magnet stand (CM101; Vazyme). The preparation of pG-Tn5 adapter complex was performed according to manufacturer’s instruction with Hyperactive pG-Tn5 Transposase for CUT&Tag (S602; Vazyme). Standard tagmentation and amplification were performed as reported previously (Kaya-Okur et al., 2019). Amplified DNA libraries were purified with VAHTS DNA Clean Beads (N411; Vazyme) and shipped for next-generation sequencing by Annoroad Gene Technology.
Reads were filtered and mapped to mm10 genome, and the pipeline before generating the peak matrix was adapted from the workflow established by Ye Zheng in the website https://yezhengstat.github.io/CUTTag_tutorial/. Peaks were then called to generate peak matrix with a pipeline similar to ATAC-Seq analysis mentioned above. The cell number used for mouse islet CUT&Tag was 100,000 in Chow group and that was 65,000 in HFD/HFD-Chow groups. So, we divided 0.65 into each read of peaks in HFD/HFD-Chow groups to eliminate the difference from cell numbers. Then the new peak matrices of Chow and HFD/HFD-Chow were combined to perform the differential analysis. Differential peaks were identified by |log2FC| > 0.3, P < 0.05 (P value by Wald test).
AAV-mediated gene expression in primary islets
Primary islets from wild-type mice or human donors were isolated for AAV treatment. 150 IEQ islets were treated with AAV2/9 viruses expressing CTCF or GFP control at the titer of 1012 viral genomes per ml (vg/ml). Virus was administrated into cultured islets at the final concentration of 20 μl/ml culture medium. Islets were harvested at 5–7 d after transduction, enabling robust exogenous gene expression for Western blot analysis, calcium imaging, and perifusion assay.
siRNA-mediated gene silencing in primary islets
Primary islets were isolated from C57BL6/J wild-type mice for CTCF gene silencing. 300 IEQ islets were transiently transfected with 100 nM of Control siRNA or CTCF siRNA (5′-GGAGGAGTCTGAAACTTTCATTAAA-3′) using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific) according to the manufacturer's instruction. At 48 h after transfection, islets were harvested for protein extraction and Western blotting analysis of CTCF protein levels or perifusion study to evaluate GSIS function.
Pancreatic ductal infusion of AAV
Pancreatic ductal infusion of AAV was performed as previously described (Xiao et al., 2014). In brief, animals were anesthetized using isoflurane. Purified AAV2/9 viruses expressing CTCF were diluted with saline to 4 × 1012 vg/ml with saline. A total volume of 150 μl viral solution was delivered into mouse pancreas through intraductal infusion catheter (CMF31G; World Precision Instruments) at the rate of 10 μl/min. Animals were kept on the heating blanket for recovery after the procedure.
qPCR
TRIzol reagent (Life Technologies) was applied to extract total RNA from islets and cultured cells. For each sample, a total of 1 μg of RNA was reverse-transcribed using the HiScript II Q RT SuperMix for qPCR (R222-01; Vazyme). Gene expression analyses were performed using SYBR Green reagent (S1816; Roche). Relative gene expression levels were calculated and normalized to mouse or human ribosomal protein 36B4 as previously described (Meng et al., 2013; Meng et al., 2017). The following primers were used for the qPCR analysis of CTCF: 5'-AGCCACACTGATGAGAGACCAC-3' (forward) and 5'-ATCGCAGTCTGGGCACTTGTGA-3' (reverse).
Western blot analysis
Primary murine islets were lysed with radioimmunoprecipitation assay buffer, followed by brief sonication (Bioruptor Pico, with 10 of 15:30 s ultrasound:cooling cycles) and centrifugation. Proteins were extracted from the supernatant and then subjected to denaturing and SDS-PAGE electrophoresis. Afterwards, proteins were transferred onto a polyvinylidene difluoride membrane (Millipore), followed by immunoblotting with the following primary antibodies: Flag (1:500–1:1,000; A8592; Sigma-Aldrich), HSP90 (1:1,000; sc-7947; Santa Cruz), GFP (1:10,000; homemade), CTCF (1:1,000; 61311; Active Motif), Foxa2 (1:1,000; ab108422; Abcam), Pdx1 (1:1,000; 5679; CST), β-actin (1:1,000; A4700; Sigma-Aldrich). Secondary antibodies were applied as followed: Anti-Mouse (dilution 1:5,000; A4416; Sigma-Aldrich) and Anti-Rabbit (dilution 1:5,000; A6154; Sigma-Aldrich).
Islet perifusion assay
Primary murine islets were cultured overnight after isolation before perifusion assays. For each assay, perifusion chambers were loaded with 150 IEQ islets. Islets were first perifused with Kreb’s buffer (135 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 1.5 mM CaCl2, 2 mM NaHCO3, 10 mM Hepes, and 0.1% BSA, pH 7.4) containing 2.8 mM glucose for 30 min, followed by perifusion with Kreb’s buffers containing 2.8 mM glucose, 16.8 mM glucose, and 2.8 mM glucose for 25, 30, and 20 min, respectively, under 1 ml/min flow rate. Fractions were collected every minute. Islets were recollected from the perifusion chamber after assay to obtain total protein content via BCA protein assay. Insulin secretion at each time point was quantified using HTRF insulin assay kit (62INSPEC; Cisbio) according to manufacturer’s instructions and normalized to total protein content. To quantify the biphasic insulin secretion, the phase I and phase II of insulin secretion were manually separated based on the plateau formation of the II phase and quantified by calculating the area under curve (AUC). Insulin levels were normalized to the average baseline insulin secretion in buffer containing low glucose (2.8 mM) and presented as fold insulin secretion.
Intracellular calcium imaging of primary islets
Primary murine islets cultured overnight after isolation were washed twice with Kreb’s buffer containing 2 mM glucose, loaded with Kreb’s buffer containing 5.6 µM Fluo-4 AM (F14201; Thermo Fisher Scientific), 0.4‰ Pluronic F127 (P2443; Sigma-Aldrich), and 2 mM glucose for 45 min. Islets were then washed twice with Kreb’s buffer containing 2 mM glucose, seeded on 35-mm glass-bottom dish (MatTek), and cultured for another 15 min. Islets were imaged in a humidified 37°C cell culture chamber with 5% CO2 mounted on a Zeiss LSM 800 inverted confocal microscope. Continuous image acquirement with 10-s interval started 2 min of glucose supplementation to reach 16.8 mM final concentration until fluorescent plateaus was reached. Fluorescent intensity dynamics were calculated via ImageJ (1.52q; NIH).
Measurement of ATP/ADP ratio in primary islets
Primary murine islets cultured overnight were washed twice with Kreb’s buffer. For each sample, 20 IEQ islets were dissociated into single cells (∼1.0 × 104 cells) by TrypLE (12604013; Thermo Fisher Scientific), followed by incubation in Kreb’s buffer containing 2.8 or 16.8 mM glucose for 30 min. ATP/ADP ratios were measured using the ADP/ATP Ratio Assay kit (MAK135; Sigma-Aldrich) according to the manufacturer’s protocol.
H&E staining of murine pancreas tissue and quantification of islet numbers
Freshly dissected murine pancreas tissues were fixed in 4% paraformaldehyde (P6148; Sigma-Aldrich) for 16 h at 4°C, embedded with paraffin and stained with H&E. H&E-stained tissue slides were scanned using Aperio ScanScope XT scanner (Leica). Islet number on each pancreas section was quantified by ImageScope software (Leica v12.3.3.5048). A total of 24 pancreas sections (four sections/mouse with at least 50 μm apart from six mice/group) were included in the quantification of islet numbers.
Immunofluorescent staining of pancreas sections and quantification
Freshly dissected murine pancreas tissues were fixed in 4% paraformaldehyde (P6148; Sigma-Aldrich) for 16 h at 4°C, embedded in paraffin, and sliced into sections with 4-μm thickness. Tissue sections were deparaffinized in xylene and rehydrated through graded concentrations of ethanol (100, 95, 70%), followed by double distlled H2O rinse. Afterwards, sections were socked in sodium citrate (E673001; Sangon Biotech); antigen activity retrieval was then conducted by high-pressure cooker for 10 min. After cooling to room temperature, sections were washed with PBS and blocked with 10% BSA (36106ES25; Yeasen) in PBS for 1 h at room temperature before incubating with primary antibodies in PBS containing 5% BSA overnight at 4°C. Next day, slides were washed with PBS for three times and then incubated with Alexa Fluor conjugated secondary antibodies (dilution 1:200; Invitrogen) at room temperature for 1.5 h. Nuclei were counterstained with DAPI (dilution 1:2,000; P3693l; Invitrogen). Sections were then mounted with Fluoromount-G mounting medium (36307ES08; Yeasen), and sealed with nail polish. Images were acquired using ZEISS LSM 800 and quantified by ImageJ 1.52q software (NIH).
The percentages of α, β, PP, and Ngn3+ cells in each islet were quantified by dividing the insulin and DAPI double positive (insulin+/DAPI+), glucagon and DAPI double-positive (glucagon+/DAPI+), pancreatic polypeptide and DAPI double-positive (PP+/DAPI+), or Neurog3 (Ngn3) and DAPI double-positive (Ngn3+/DAPI+) cell numbers by the total DAPI numbers (total cell number/islet). Average β cell size in each islet was calculated by dividing the area of insulin-positive region (total β cell area/islet) with the insulin and DAPI double-positive cell number (total β cell number/islet). At least six islets/mouse were included in the analysis. First antibodies applied include: Insulin (dilution 1:10,000; homemade), Glucagon (dilution 1:2,000; G2654; Sigma-Aldrich), Pancreatic Polypeptide (dilution 1:200; H-054-02; Phoenixpeptide), Ngn3 (dilution 1:20; F25A1B3-c; DSHB), and CTCF (dilution 1:200; 61311; Active Motif).
β cell mass analysis
Insulin immunohistochemistry staining was performed on pancreas sections to determine β cell mass as previously described (Shrestha et al., 2020; Talchai et al., 2012; Xuan et al., 2010). In brief, pancreata from Chow, HFD, and HFD-Chow groups (5–7 mice/group) were dissected, cleared of fat, weighed, fixed in 4% paraformaldehyde (P6148; Sigma-Aldrich) overnight at 4°C. The tissues were embedded in paraffin, and consecutively sectioned with 3-μm thickness. Five sections at least 160 μm apart were subjected to insulin immunohistochemistry staining and scanned using the Olympus-VS200 scanner for each pancreas. Total insulin positive area and total pancreas area on each section were measured by the cellSens software (Olympus). The β cell mass was obtained through multiplying the pancreas weight by the ratio of insulin-positive area/pancreas area.
GSIS assay of Min6 cells
Min6 cells were preincubated for 30 min in glucose free Kreb’s buffer, followed by incubation in Kreb’s buffer containing 2.8 and 16.8 mM glucose for 1 h. The supernatant fractions were obtained after the static incubation for insulin measurement. Cells were lysed using cold lysis buffer to obtain total protein content via BCA protein assay. Insulin secretion at each condition was quantified using HTRF insulin assay kit (62INSPEC; Cisbio) according to manufacturer's instructions and normalized to total protein content.
Statistics
Statistical analyses were carried out using GraphPad Prism 8. Statistical differences were evaluated using two-tailed unpaired Student’s t test for comparisons between two groups or ANOVA and appropriate post hoc analyses for comparisons of more than two groups. For ITT, GTT, random blood glucose, and calcium imaging studies, two-way ANOVA with multiple comparisons was used for statistical analysis. Furthermore, area under curve was calculated for each mouse, and evaluated by two-tailed unpaired Student’s t test for statistical differences between two groups. P value of <0.05 was considered statistically significant. Statistical parameters and corresponding P values for data shown in each panel were included in the figure legends. Except for the ATAC-Seq accessibility analysis, P values obtained in next-generation sequencing were adjusted by FDR. Key differential expressed genes obtained by RNA-Seq were validated via qPCR analysis.
Study approval
Paraffin embedded pancreas tissue sections used in this study were obtained from patients underwent pancreatectomy due to benign tumors from the First Affiliated Hospital of Zhejiang University (Ethical Approval: No. 2018-753-1) under the informed consent of the patients. The patients’ characteristics have been described in Fig. 5 f.
The isolation of human islets for virus infection and functional analysis was performed in Tianjin First Central Hospital (Ethical Approval: No. 2016N066KY) and the First Affiliated Hospital of Zhejiang University (Ethical Approval: No. 2017-410-1), under the informed consent of deceased organ donors’ families.
All procedures involving animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Zhejiang University and conducted in accordance with the policies of institutional guidelines on the care and use of laboratory animals.
Code availability
All packages and codes used in this study are open source and publicly available.
Online supplemental material
Fig. S1 shows the progressive weight gain and hyperglycemia in mice under HFD feeding and provides additional data on the dietary intervention-induced β cell function preservation and diabetes remission. Fig. S2 illustrates the pairwise gene expression comparisons among islets from Chow, HFD, and HFD-Chow groups. Fig. S3 includes the expression of cell-type specific markers in each cell cluster and provides additional information on cell composition and β cell transcriptome changes among Chow, HFD, and HFD-Chow groups. Fig. S4 shows additional information on ATAC-Seq of islets from Chow, HFD, and HFD-Chow groups, and the effects of AAV-mediated CTCF, Foxa2, or Pdx1 overexpression on islet insulin secretion. Fig. S5 includes the validation of sgRNA-induced CTCF knockout and additional data demonstrating that PA and TNFα treatment impairs GSIS function in primary mouse islets. Further information of CTCF chromatin binding landscapes in islets from Chow, HFD, and HFD-Chow groups and enriched transcriptional binding motifs on the differential CTCF binding peaks are also included.
Supplementary Material
contains original blots for Fig. 5.
contains original blots for Fig. 6.
contains original blots for Fig. 7.
contains original blots for Fig. S4.
contains original blots for Fig. S5.
Acknowledgments
The authors thank Drs. Jiandie Lin and Henry Kuang from the University of Michigan for critical reading of the manuscript, and Dr. Liangyou Rui from the University of Michigan for providing the Min6 cells. We thank the Meng lab members for the helpful discussion and technical support for this study. We thank Dr. Wei Ding for assistance with human pancreas sections. We thank the Core Facilities of Zhejiang University School of Medicine for technical support.
This work was supported by grants from the Training Program of the Major Research Plan of the National Natural Science Foundation of China (91857110), the National Key Research and Development Programme of China (2018YFA0800403 and 2021YFC2701903), the National Natural Science Fund for Excellent Young Scholars of China (81722012), the Zhejiang Provincial Natural Science Foundation of China (LZ21H070001), the National Natural Science Foundation of China (81670740), the Construction Fund of Key Medical Disciplines of Hangzhou (No. OO20200055), the Innovative Institute of Basic Medical Sciences of Zhejiang University, and the Fundamental Research Funds for the Central Universities to Z.-X. Meng. The present study was also supported by grants from the National Key Research and Development Programme of China (2020YFA0803704), the National Natural Science Foundation of China (82070805 and 81870535) to S. Wang, grants from the National Natural Science Foundation of China (31870855) to L. Zhu, and Tianjin Municipal Human Resources and Social Security Bureau (XB202011) to S. Wang. The authors gratefully acknowledge the support of K.C. Wong Education Foundation.
Author contributions: Z.-X. Meng, R.-R. Wang, and X. Qiu, conceived and designed the research. R.-R. Wang, X. Qiu, R. Pan, Q. Wang, H. Fu, Z. Zhang, H. Chen, Q.-Q. Wu, X. Pan, and Y. Zhou performed the experiments. X. Qiu carried out most of the bioinformatics analysis with help from Z. Zhang and Q. Wang. H. Fu and S. Wang helped with the human islet related studies. L. Zhu, M. Zheng, P. Shan, and G. Guo contributed to discussion and data interpretation. Z.-X. Meng and X. Qiu wrote the manuscript with help from other authors. Z.-X. Meng initiated and supervised the project. X. Qiu, R.-R.Wang, R. Pan, and H. Fu are co-first authors; Z. Zhang, Q. Wang and H. Chen contributed equally. The authorship orders of the co-first authors and equally contributed authors were determined by rolling dice.
Data availability
RNA-Seq, scRNA-Seq, ATAC-Seq and CUT&Tag datasets are available for the data presented in Figs. 1, h–i, 2, j–k, 3, a–b, e–i, 4, a–f, and 7, c–f; and Figs. S2, S3, S4 a, and S5, f–g. These data have been deposited in the Mendeley Data (http://dx.doi.org/10.17632/mwgxv7m927.2). All other data and image files are available from the corresponding author upon reasonable request. All packages and codes used in this study are open source and publicly available. Plasmids generated in this study are available from the corresponding author upon signing material transfer agreement due to institutional requirements.
References
- Al-Mrabeh, A., Hollingsworth K.G., Shaw J.A.M., McConnachie A., Sattar N., Lean M.E.J., and Taylor R.. 2020. 2-year remission of type 2 diabetes and pancreas morphology: A post-hoc analysis of the DiRECT open-label, cluster-randomised trial. Lancet Diabetes Endocrinol. 8:939–948. 10.1016/S2213-8587(20)30303-X [DOI] [PubMed] [Google Scholar]
- Bikopoulos, G., Pimenta da Silva A., Lee S.C., Lakey J.R., Der S.D., Chan C.B., Ceddia R.B., Wheeler M.B., and Rozakis-Adcock M.. 2008. Ex vivo transcriptional profiling of human pancreatic islets following chronic exposure to monounsaturated fatty acids. J. Endocrinol. 196:455–464. 10.1677/JOE-07-0174 [DOI] [PubMed] [Google Scholar]
- Boucher, A., Lu D., Burgess S.C., Telemaque-Potts S., Jensen M.V., Mulder H., Wang M.Y., Unger R.H., Sherry A.D., and Newgard C.B.. 2004. Biochemical mechanism of lipid-induced impairment of glucose-stimulated insulin secretion and reversal with a malate analogue. J. Biol. Chem. 279:27263–27271. 10.1074/jbc.M401167200 [DOI] [PubMed] [Google Scholar]
- Brereton, M.F., Iberl M., Shimomura K., Zhang Q., Adriaenssens A.E., Proks P., Spiliotis I.I., Dace W., Mattis K.K., Ramracheya R., et al. 2014. Reversible changes in pancreatic islet structure and function produced by elevated blood glucose. Nat. Commun. 5:4639. 10.1038/ncomms5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J.D., Giresi P.G., Zaba L.C., Chang H.Y., and Greenleaf W.J.. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10:1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, I.L., Iscaro A., and Harrison L.C.. 1988. IFN-gamma and tumor necrosis factor-alpha. Cytotoxicity to murine islets of langerhans. J. Immunol. 141:2325–2329 [PubMed] [Google Scholar]
- Chen, H., Carlson E.C., Pellet L., Moritz J.T., and Epstein P.N.. 2001. Overexpression of metallothionein in pancreatic beta cells reduces streptozotocin-induced DNA damage and diabetes. Diabetes. 50:2040–2046. 10.2337/diabetes.50.9.2040 [DOI] [PubMed] [Google Scholar]
- Chen, X.-F., Zhang Y.-w., Xu H., and Bu G.. 2013. Transcriptional regulation and its misregulation in Alzheimer’s disease. Mol. Brain. 6:44. 10.1186/1756-6606-6-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagogo-Jack, S., Brewer A.A., Owei I., French L., Umekwe N., Rosenthal R., and Wan J.. 2020. Pathobiology and Reversibility of Prediabetes in a Biracial Cohort (PROP-ABC) study: Design of lifestyle intervention. BMJ Open Diabetes Res. Care. 8:e000899. 10.1136/bmjdrc-2019-000899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo, R.A., Ferrannini E., Groop L., Henry R.R., Herman W.H., Holst J.J., Hu F.B., Kahn C.R., Raz I., Shulman G.I., et al. 2015. Type 2 diabetes mellitus. Nat. Rev. Dis. Prim. 1:15019. 10.1038/nrdp.2015.19 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Gutierrez, G., Xin Y., and Gromada J.. 2019. Heterogeneity of human pancreatic β cells. Mol. Metabol. 27S:S7–S14. 10.1016/j.molmet.2019.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, V., Raimondi C., Somanath S., Bugliani M., Loder M.K., Edling C.E., Divecha N., da Silva-Xavier G., Marselli L., Persaud S.J., et al. 2011. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J. Biol. Chem. 286:4216–4225. 10.1074/jbc.M110.200295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, K.F., and Lindgärde F.. 1991. Prevention of Type 2 (non-insulin-dependent) diabetes mellitus by diet and physical exercise the 6-year Malmö feasibility study. Diabetologia. 34:891–898. 10.1007/BF00400196 [DOI] [PubMed] [Google Scholar]
- Fang, Y., Gao J., Qi L., and Li N.. 2014. CTCF-regulating endocrine function of pancreatic islet cells in transgenic mice. Horm. Metab. Res. 46:419–423. 10.1055/s-0034-1372593 [DOI] [PubMed] [Google Scholar]
- Gaszner, M., and Felsenfeld G.. 2006. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7:703–713. 10.1038/nrg1925 [DOI] [PubMed] [Google Scholar]
- Gosalia, N., Neems D., Kerschner J.L., Kosak S.T., and Harris A.. 2014. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 42:9612–9622. 10.1093/nar/gku648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, C.E., Bailey T.L., and Noble W.S.. 2011. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 27:1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, D., Jetton T.L., LaRock K., Monga N., Satish B., Lausier J., Peshavaria M., and Leahy J.L.. 2017. Temporal characterization of β cell-adaptive and -maladaptive mechanisms during chronic high-fat feeding in C57BL/6NTac mice. J. Biol. Chem. 292:12449–12459. 10.1074/jbc.M117.781047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., et al. 2018. Mapping the mouse cell atlas by microwell-Seq. Cell. 173:1091–1107.e17. 10.1016/j.cell.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.J., Lin C.Y., Haataja L., Gurlo T., Butler A.E., Rizza R.A., and Butler P.C.. 2007. High expression rates of human islet amyloid polypeptide induce endoplasmic reticulum stress mediated beta cell apoptosis, a characteristic of humans with type 2 but not type 1 diabetes. Diabetes. 56:2016–2027. 10.2337/db07-0197 [DOI] [PubMed] [Google Scholar]
- Hudish, L.I., Reusch J.E., and Sussel L.. 2019. β Cell dysfunction during progression of metabolic syndrome to type 2 diabetes. J. Clin. Invest. 129:4001–4008. 10.1172/JCI129188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto, Y., and Kaneto H.. 2004. Role of oxidative stress in pancreatic beta cell dysfunction. Ann. NY Acad. Sci. 1011:168–176. 10.1007/978-3-662-41088-2_17 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur, H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., and Henikoff S.. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10:1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelpe, C.L., Moore P.C., Parazzoli S.D., Wicksteed B., Rhodes C.J., and Poitout V.. 2003. Palmitate inhibition of insulin gene expression is mediated at the transcriptional level via ceramide synthesis. J. Biol. Chem. 278:30015–30021. 10.1074/jbc.M302548200 [DOI] [PubMed] [Google Scholar]
- Klenova, E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., and Lobanenkov V.V.. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13:7612–7624. 10.1128/mcb.13.12.7612-7624.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler, W.C., Barrett-Connor E., Fowler S.E., Hamman R.F., Lachin J.M., Walker E.A., Nathan D.M., and Diabetes Prevention Program Research Group . 2002. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 346:393–403. 10.1056/NEJMoa012512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybutt, D.R., Preston A.M., Akerfeldt M.C., Kench J.G., Busch A.K., Biankin A.V., and Biden T.J.. 2007. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 50:752–763. 10.1007/s00125-006-0590-z [DOI] [PubMed] [Google Scholar]
- Lee, Y.J., Lee A., Yoo H.J., Kim M., Kim M., Jee S.H., Shin D.Y., and Lee J.H.. 2018. Effect of weight loss on circulating fatty acid profiles in overweight subjects with high visceral fat area: A 12-wk randomized controlled trial. Nutr. J. 17:28. 10.1186/s12937-018-0323-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Chen H., and Epstein P.N.. 2004. Metallothionein protects islets from hypoxia and extends islet graft survival by scavenging most kinds of reactive oxygen species. J. Biol. Chem. 279:765–771. 10.1074/jbc.M307907200 [DOI] [PubMed] [Google Scholar]
- Ling, C., Del Guerra S., Lupi R., Rönn T., Granhall C., Luthman H., Masiello P., Marchetti P., Groop L., and Del Prato S.. 2008. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 51:615–622. 10.1007/s00125-007-0916-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T., Mi L., Xiong J., Orchard P., Yu Q., Yu L., Zhao X.-Y., Meng Z.-X., Parker S.C.J., Lin J.D., and Li S.. 2020. BAF60a deficiency uncouples chromatin accessibility and cold sensitivity from white fat browning. Nat. Commun. 11:2379. 10.1038/s41467-020-16148-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, T.-a., Kaneto H., Kawashima S., Miyatsuka T., Tochino Y., Yoshikawa A., Imagawa A., Miyazaki J.-i., Gannon M., Stein R., and Shimomura I.. 2015. Preserving Mafa expression in diabetic islet β cells improves glycemic control in vivo. J. Biol. Chem. 290:7647–7657. 10.1074/jbc.M114.595579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z.-X., Li S., Wang L., Ko H.J., Lee Y., Jung D.Y., Okutsu M., Yan Z., Kim J.K., and Lin J.D.. 2013. Baf60c drives glycolytic metabolism in the muscle and improves systemic glucose homeostasis through Deptor-mediated Akt activation. Nat. Med. 19:640–645. 10.1038/nm.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z., Lv J., Luo Y., Lin Y., Zhu Y., Nie J., Yang T., Sun Y., and Han X.. 2009a. Forkhead box O1/pancreatic and duodenal homeobox 1 intracellular translocation is regulated by c-Jun N-terminal kinase and involved in prostaglandin E2-induced pancreatic beta cell dysfunction. Endocrinology. 150:5284–5293. 10.1210/en.2009-0671 [DOI] [PubMed] [Google Scholar]
- Meng, Z.X., Gong J., Chen Z., Sun J., Xiao Y., Wang L., Li Y., Liu J., Xu X.Z.S., and Lin J.D.. 2017. Glucose sensing by skeletal myocytes couples nutrient signaling to systemic homeostasis. Mol. Cell. 66:332–344.e4. 10.1016/j.molcel.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z.X., Nie J., Ling J.J., Sun J.X., Zhu Y.X., Gao L., Lv J.H., Zhu D.Y., Sun Y.J., and Han X.. 2009b. Activation of liver X receptors inhibits pancreatic islet beta cell proliferation through cell cycle arrest. Diabetologia. 52:125–135. 10.1007/s00125-008-1174-x [DOI] [PubMed] [Google Scholar]
- Meng, Z.X., Sun J.X., Ling J.J., Lv J.H., Zhu D.Y., Chen Q., Sun Y.J., and Han X.. 2006. Prostaglandin E2 regulates Foxo activity via the akt pathway: Implications for pancreatic islet beta cell dysfunction. Diabetologia. 49:2959–2968. 10.1007/s00125-006-0447-5 [DOI] [PubMed] [Google Scholar]
- Meng, Z.X., Yin Y., Lv J.H., Sha M., Lin Y., Gao L., Zhu Y.X., Sun Y.J., and Han X.. 2012. Aberrant activation of liver X receptors impairs pancreatic beta cell function through upregulation of sterol regulatory element-binding protein 1c in mouse islets and rodent cell lines. Diabetologia. 55:1733–1744. 10.1007/s00125-012-2516-2 [DOI] [PubMed] [Google Scholar]
- Merkenschlager, M., and Nora E.P.. 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17:17–43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- Moffitt, J.H., Fielding B.A., Evershed R., Berstan R., Currie J.M., and Clark A.. 2005. Adverse physicochemical properties of tripalmitin in beta cells lead to morphological changes and lipotoxicity in vitro. Diabetologia. 48:1819–1829. 10.1007/s00125-005-1861-9 [DOI] [PubMed] [Google Scholar]
- Moore, J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., Kaul R., et al. ENCODE Project Consortium . 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 583:699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo, K., Chen M.Z., Olefsky J.M., and Marth J.D.. 2011. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat. Med. 17:1067–1075. 10.1038/nm.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, S.C.J., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Black B.L., et al. 2013. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA. 110:17921–17926. 10.1073/pnas.1317023110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Päth, G., Mehana A.E., Pilz I.H., Alt M., Baumann J., Sommerer I., Hoffmeister A., and Seufert J.. 2020. NUPR1 preserves insulin secretion of pancreatic β cells during inflammatory stress by multiple low-dose streptozotocin and high-fat diet. Am. J. Physiol. Endocrinol. Metab. 319:E338–E344. 10.1152/ajpendo.00088.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault, L., Kahn S.E., Christophi C.A., Knowler W.C., Hamman R.F., and Diabetes Prevention Program Research G.. 2009. Regression from pre-diabetes to normal glucose regulation in the diabetes prevention program. Diabetes Care. 32:1583–1588. 10.2337/dc09-0523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli, S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., and Sandberg R.. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 10:1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Pinnick, K., Neville M., Clark A., and Fielding B.. 2010. Reversibility of metabolic and morphological changes associated with chronic exposure of pancreatic islet β cells to fatty acids. J. Cell. Biochem. 109:683–692. 10.1002/jcb.22445 [DOI] [PubMed] [Google Scholar]
- Pinnick, K.E., Collins S.C., Londos C., Gauguier D., Clark A., and Fielding B.A.. 2008. Pancreatic ectopic fat is characterized by adipocyte infiltration and altered lipid composition. Obesity. 16:522–530. 10.1038/oby.2007.110 [DOI] [PubMed] [Google Scholar]
- Pique-Regi, R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., and Pritchard J.K.. 2011. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 21:447–455. 10.1101/gr.112623.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva, E.M., Kubo N., Loukinov D., Tajmul M., Kang S., Kovalchuk A.L., Strunnikov A.V., Zentner G.E., Ren B., and Lobanenkov V.V.. 2020. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA. 117:2020–2031. 10.1073/pnas.1911708117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., and Manke T.. 2016. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, G., Jin W., Cui K., Rodrigez J., Hu G., Zhang Z., Larson D.R., and Zhao K.. 2017. CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to cell variation of gene expression. Mol. Cell. 67:1049–1058.e6. 10.1016/j.molcel.2017.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscioni, S.S., Migliorini A., Gegg M., and Lickert H.. 2016. Impact of islet architecture on β cell heterogeneity, plasticity and function. Nat. Rev. Endocrinol. 12:695–709. 10.1038/nrendo.2016.147 [DOI] [PubMed] [Google Scholar]
- Ryan, A.S., and Nicklas B.J.. 2004. Reductions in plasma cytokine levels with weight loss improve insulin sensitivity in overweight and obese postmenopausal women. Diabetes Care. 27:1699–1705. 10.2337/diacare.27.7.1699 [DOI] [PubMed] [Google Scholar]
- Shrestha, N., Liu T., Ji Y., Reinert R.B., Torres M., Li X., Zhang M., Tang C.H.A., Hu C.C.A., Liu C., et al. 2020. Sel1L-Hrd1 ER-associated degradation maintains beta cell identity via TGF-beta signaling. J. Clin. Invest. 130:3499–3510. 10.1172/JCI134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skórzyńska-Dziduszko, K.E., Kimber-Trojnar Ż., Patro-Małysza J., Stenzel-Bembenek A., Oleszczuk J., and Leszczyńska-Gorzelak B.. 2018. Heat shock proteins as a potential therapeutic target in the treatment of gestational diabetes mellitus: What we know so far. Int. J. Mol. Sci. 19:3205. 10.3390/ijms19103205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So, W.Y., Liu W.N., Teo A.K.K., Rutter G.A., and Han W.. 2021. Paired box 6 programs essential exocytotic genes in the regulation of glucose-stimulated insulin secretion and glucose homeostasis. Sci. Transl. Med. 13:eabb1038. 10.1126/scitranslmed.abb1038 [DOI] [PubMed] [Google Scholar]
- Stitzel, M.L., Sethupathy P., Pearson D.S., Chines P.S., Song L., Erdos M.R., Welch R., Parker S.C.J., Boyle A.P., Scott L.J., et al. 2010. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metabol. 12:443–455. 10.1016/j.cmet.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain, S.Z., Prins J.B., and McGuckin M.A.. 2016. Oxidative and endoplasmic reticulum stress in β cell dysfunction in diabetes. J. Mol. Endocrinol. 56:R33–R54. 10.1530/jme-15-0232 [DOI] [PubMed] [Google Scholar]
- Supale, S., Li N., Brun T., and Maechler P.. 2012. Mitochondrial dysfunction in pancreatic β cells. Trends Endocrinol. Metabol. 23:477–487. 10.1016/j.tem.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Talchai, C., Xuan S., Lin H.V., Sussel L., and Accili D.. 2012. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell. 150:1223–1234. 10.1016/j.cell.2012.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R., Al-Mrabeh A., and Sattar N.. 2019. Understanding the mechanisms of reversal of type 2 diabetes. Lancet Diabetes Endocrinol. 7:726–736. 10.1016/S2213-8587(19)30076-2 [DOI] [PubMed] [Google Scholar]
- Taylor, R., Al-Mrabeh A., Zhyzhneuskaya S., Peters C., Barnes A.C., Aribisala B.S., Hollingsworth K.G., Mathers J.C., Sattar N., and Lean M.E.J.. 2018. Remission of human type 2 diabetes requires decrease in liver and pancreas fat content but is dependent upon capacity for beta cell recovery. Cell Metab. 28:547–556.e3. 10.1016/j.cmet.2018.08.010 [DOI] [PubMed] [Google Scholar]
- Tsui, S., Dai W., and Lu L.. 2014. CCCTC‐binding factor mediates effects of glucose on beta cell survival. Cell Prolif. 47:28–37. 10.1111/cpr.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, S., Gao J., Wang C., and Lu L.. 2012. CTCF mediates effect of insulin on glucagon expression. Exp. Cell Res. 318:887–895. 10.1016/j.yexcr.2012.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, I.M., Zhang B., Yang X., Zhu J., Stepaniants S., Zhang C., Meng Q., Peters M., He Y., Ni C., et al. 2012. Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol. Syst. Biol. 8:594. 10.1038/msb.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, G.C., and Bonner-Weir S.. 2004. Five stages of evolving beta cell dysfunction during progression to diabetes. Diabetes. 53:S16–S21. 10.2337/diabetes.53.suppl_3.s16 [DOI] [PubMed] [Google Scholar]
- Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis. Springer, New York. [Google Scholar]
- Xiao, X., Guo P., Prasadan K., Shiota C., Peirish L., Fischbach S., Song Z., Gaffar I., Wiersch J., El-Gohary Y., et al. 2014. Pancreatic cell tracing, lineage tagging and targeted genetic manipulations in multiple cell types using pancreatic ductal infusion of adeno-associated viral vectors and/or cell-tagging dyes. Nat. Protoc. 9:2719–2724. 10.1038/nprot.2014.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X., Guo P., Shiota C., Zhang T., Coudriet G.M., Fischbach S., Prasadan K., Fusco J., Ramachandran S., Witkowski P., et al. 2018. Endogenous reprogramming of alpha cells into beta cells, induced by viral gene therapy, reverses autoimmune diabetes. Cell Stem Cell. 22:78–90.e4. 10.1016/j.stem.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, S., Szabolcs M., Cinti F., Perincheri S., Accili D., and Efstratiadis A.. 2010. Genetic analysis of type-1 insulin-like growth factor receptor signaling through insulin receptor substrate-1 and -2 in pancreatic beta cells. J. Biol. Chem. 285:41044–41050. 10.1074/jbc.M110.144790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.N., and Berggren P.O.. 2005. Beta cell CaV channel regulation in physiology and pathophysiology. Am. J. Physiol. Endocrinol. Metab. 288:E16–E28. 10.1152/ajpendo.00042.2004 [DOI] [PubMed] [Google Scholar]
- Yoon, J.C., Xu G., Deeney J.T., Yang S.N., Rhee J., Puigserver P., Levens A.R., Yang R., Zhang C.Y., Lowell B.B., et al. 2003. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev. Cell. 5:73–83. 10.1016/s1534-5807(03)00170-9 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y.P., and Grill V.E.. 1994. Long-term exposure of rat pancreatic islets to fatty acids inhibits glucose-induced insulin secretion and biosynthesis through a glucose fatty acid cycle. J. Clin. Invest. 93:870–876. 10.1172/JCI117042 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. 5.
contains original blots for Fig. 6.
contains original blots for Fig. 7.
contains original blots for Fig. S4.
contains original blots for Fig. S5.
Data Availability Statement
RNA-Seq, scRNA-Seq, ATAC-Seq and CUT&Tag datasets are available for the data presented in Figs. 1, h–i, 2, j–k, 3, a–b, e–i, 4, a–f, and 7, c–f; and Figs. S2, S3, S4 a, and S5, f–g. These data have been deposited in the Mendeley Data (http://dx.doi.org/10.17632/mwgxv7m927.2). All other data and image files are available from the corresponding author upon reasonable request. All packages and codes used in this study are open source and publicly available. Plasmids generated in this study are available from the corresponding author upon signing material transfer agreement due to institutional requirements.