Abstract
Nitrogen fixation by the microorganisms in the gut of termites is one of the crucial aspects of symbiosis, since termites usually thrive on a nitrogen-poor diet. The phylogenetic diversity of the nitrogen-fixing organisms within the symbiotic community in the guts of various termite species was investigated without culturing the resident microorganisms. A portion of the dinitrogenase reductase gene (nifH) was directly amplified from DNA extracted from the mixed population in the termite gut. Analysis of deduced amino acid sequences of the products of the clonally isolated nifH genes revealed the presence of diverse nifH sequences in most of the individual termite species, and their constituents were considerably different among termite species. A majority of the nifH sequences from six lower termites, which showed significant levels of nitrogen fixation activity, could be assigned to either the anaerobic nif group (consisting of clostridia and sulfur reducers) or the alternative nif methanogen group among the nifH phylogenetic groups. In the case of three higher termites, which showed only low levels of nitrogen fixation activity, a large number of the sequences were assigned to the most divergent nif group, probably functioning in some process other than nitrogen fixation and being derived from methanogenic archaea. The nifH groups detected were similar within each termite family but different among the termite families, suggesting an evolutionary trend reflecting the diazotrophic habitats in the symbiotic community. Within these phylogenetic groups, the sequences from the termites formed lineages distinct from those previously recognized in studies using classical microbiological techniques, and several sequence clusters unique to termites were found. The results indicate the presence of diverse potentially nitrogen-fixing microbial assemblages in the guts of termites, and the majority of them are as yet uncharacterized.
A symbiotic relationship between termites and microorganisms inhabiting their gut enables termites to live exclusively on lignocellulosic materials (4, 6). Nitrogen fixation in termites is one of the crucial aspects of the symbiosis, since the diet of termites is usually low in nitrogen sources (1, 4, 5). The nitrogen fixation activity is associated with the gut microorganisms. Ecologically, termites thrive in great abundance and they play important roles in the turnover of lignocellulose derived from dead plant materials. Considering their great abundance, the ability of termites to fix atmospheric N2 may also play a hitherto unrecognized role in fertilization of ecosystems by replenishing combined nitrogen compounds. For example, it is known that termites are preyed upon by various carnivores as important nitrogen sources (36, 41).
Termites are comprised of a complex assemblage of evolutionarily diverse species, roughly divided into so-called lower and higher termites (17). The lower termites, which comprise six families, harbor a dense and diverse population of both prokaryotes and flagellated protists in their gut. The higher termites comprise only one family but three-quarters of all termite species, and they also harbor a dense and diverse array of prokaryotes. However, the higher termites typically lack flagellated protists, and they have a more elaborate morphology and social organization than do the lower termites. The higher termites, especially, show considerable variation in their feeding behavior, which is not limited to xylophagy. Some feed exclusively on soil, presumably deriving nutrition from the humic compounds therein, and others cultivate and consume cellulolytic fungi. Even in the wood-feeding guilds, which include all lower termites, food preferences range from sound to extensively decayed woods.
A wide variety of nitrogen fixation rates of termite species are known (1, 4, 5). Within the same species, large variations in nitrogen fixation rates have been demonstrated. At least one reason for the variations is the nitrogen content of the termite diet fed prior to the assay (5). Considering the variations in nitrogen-fixing activity and the presence of evolutionarily diverse termite species, differences in microbial populations and differences in the constituents of the resident microorganisms responsible for nitrogen fixation in the gut of termites are of significant interest and need to be elucidated in order to understand the termite symbiotic systems.
Identification depending on culturing microorganisms may provide limited information on the microbial diversity and the types of organisms that fix nitrogen in termites, because only a few nitrogen-fixing microorganisms have been isolated from termites (12, 18, 31). Moreover, a majority of the members of the symbiotic community in the termite gut have been shown to be as yet uncultivated microorganisms, as demonstrated by culture-independent analyses based on comparisons of PCR-amplified 16S rRNA genes (2, 24–27, 29, 33). However, a similar molecular approach, comparative analysis of a PCR-amplified nitrogen fixation gene, nifH, has provided evidence for a remarkable and previously unexpected diversity of nitrogen-fixing microorganisms in the gut of the lower termite Reticulitermes speratus (28). The gene nifH encoding dinitrogenase reductase is evolutionarily conserved and has often been used as the basis for detecting nitrogen-fixing microorganisms in natural microbial communities (3, 16, 34, 39, 40, 42, 43). Comparative analysis of the nifH gene can provide information about the phylogenetic identity of nitrogen-fixing organisms.
In this report, in an effort to compare the constituents of symbiotic nitrogen-fixing microorganisms in the gut of evolutionarily diverse termites, a portion of the nifH gene was PCR amplified and characterized. The nifH sequences obtained were compared among termite species. Phylogenetic analysis of the cloned nifH sequences revealed that the diazotrophic populations in the termite gut are far more diverse than previously recognized.
MATERIALS AND METHODS
Termites and nitrogen fixation activity.
The termites examined in this study and the time and place of sample collection are shown in Table 1. Nitrogen fixation activity was measured by the acetylene reduction assay (30). Thirty to 200 live workers (or pseudergates) of the termite species were placed in a stoppered 10-ml bottle containing 16% C2H2. After incubation at room temperature for 1 to 3 h, a 0.1-ml gas sample was assayed for C2H4 by using a flame ionization gas chromatograph (Shimazu GC-14B) fitted with packed column J (3 mm by 1 m; GL Science) containing Porapak T (80/100 mesh). Helium was the carrier gas (30 ml/min), and the column temperature was 50°C.
TABLE 1.
Nitrogen fixation (C2H2 reduction) activity of termites examined in this study
| Termite species | Termite family | Collection
|
Nitrogen fixation activitya (nmol of C2H4 formed/ h/g [wet wt]) | |
|---|---|---|---|---|
| Originb | Date | |||
| Lower termites | ||||
| R. speratus | Rhinotermitidae | Ogose | October 1996 | 16 |
| C. formosanus | Rhinotermitidae | Okinawa | April 1997 | 79 |
| N. koshunensis | Kalotermitidae | Okinawa | August 1996 | 210 |
| C. domesticus | Kalotermitidae | Iriomote | April 1997 | 33 |
| G. fuscus | Kalotermitidae | Okinawa | August 1996 | 31 |
| H. sjoestedti | Termopsidae | Yakushima | July 1997 | 34 |
| Higher termites | ||||
| N. takasagoensis | Termitidae | Iriomote | October 1997 | 0.7 |
| O. formosanus | Termitidae | Iriomote | April 1997 | ND |
| P. nitobei | Termitidae | Iriomote | April 1997 | 2.5 |
The activity was measured within 3 days after collection, except for G. fuscus, in which case it was 2 months after collection. Values are the averages of duplicate assays performed separately. ND, not detected.
All collection sites were in the Japan Archipelago. The Ogose district is in the Saitama prefecture at latitude 36°N. Yakushima Island is in the Kagoshima prefecture at latitude 30°N. Okinawa Island and the Iriomote Island are in the Okinawa prefecture at latitudes 26 and 24° N, respectively.
DNA extraction, PCR amplification, and cloning.
DNA was extracted from the mixed population of microorganisms in the whole gut of the termites as described previously (24, 26). The nifH gene was amplified from the extracted DNA by PCR with EX Taq DNA polymerase (Takara) according to the manufacturer’s instructions. The reaction conditions were 30 cycles of 94°C for 30 s, 48°C for 45 s, and 72°C for 2 min. The PCR primers used were IGK and YAA (28), which are specific for a portion of the nifH gene corresponding to amino acid positions 11 to 165 of the Klebsiella pneumoniae nifH sequence. The amino acid sequences of these two primers are the most widely conserved sequences within nifH. PCR products of the expected size (approximately 0.47 kb) were isolated by electrophoresis by using a low-melting-point agarose gel (Seaplaque GTG; FMC Bioproducts) and purified by means of the Wizard PCR prep DNA purification system (Promega). In the case of Neotermes koshunensis, the purified PCR product was cloned in pUC119 as described previously (28). All of the other purified PCR products were cloned in pGEM-T (Promega) according to the manufacturer’s instructions.
FLT-RFLP analysis.
The primers used for the fluorescently labeled terminal-restriction fragment length polymorphism (FLT-RFLP) analysis were IGK-Cy5 (5′-TGYGAYCCNAARGCNGA-3′ labeled at the 5′ end with Cy5; synthesized and purified by Pharmacia) and YAA. The reaction conditions were the same as those for the standard PCR described above. The products of the expected size were purified as described above and then digested with HhaI. The lengths of the fluorescently labeled terminal restriction fragments from the PCR products were determined after electrophoresis by means of an automated sequencer, ALFred Express (Pharmacia), and analyzed by using Fragment Manager software (Pharmacia).
Nucleotide sequencing and phylogenetic analysis.
Plasmid DNAs were prepared from randomly picked recombinant clones and used as templates for sequencing performed by using the Dye Primer Cycle Sequencing Kit (Applied Biosystems) with sequencing primers T7 and SP6 and an automatic sequence analyzer (Applied Biosystems model 377). The names assigned to the clones are shown in Table 2. The previously determined nifH sequences included in comparisons in this study were retrieved from the GenBank, EMBL, and DDBJ nucleotide sequence databases. Sequences were aligned by using the CLUSTAL W package (38) and then corrected by manual inspection. Phylogenetic analyses were restricted to unambiguously aligned amino acid residues. The programs used to infer phylogenetic trees were those contained in the PHYLIP package (11). PROTDIST with the Dayhoff PAM matrix option was used to calculate evolutionary distances. Phylogenetic trees were constructed from evolutionary distance data by the neighbor-joining method (32), implemented through the program NEIGHBOR. A total of 100 bootstrapped replicate resampling data sets for PROTDIST were generated with the program SEQBOOT, to provide confidence estimates for tree topologies (10).
TABLE 2.
Assignment of the nifH clones from the symbiotic microbial community in the gut of termites to phylogenetic groups
| Termite species | Clone name taga | Number of clones
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proteo-cyano | Anaerobe | anf-methano | Pseudo nif | Other | Frameshiftc | Total | Different AAd | Different DNAe | ||
| R. speratusb | RSN-TKY | 1 | 18 | 0 | 3 | 0 | 4 | 26 | 13 | 23 |
| C. formosanus | CFN | 0 | 23 | 0 | 1 | 0 | 0 | 24 | 11 | 15 |
| N. koshunensis | NKN | 3 | 5 | 10 | 4 | 0 | 1 | 23 | 12 | 20 |
| C. domesticus | CDN | 0 | 8 | 10 | 4 | 0 | 1 | 23 | 15 | 21 |
| G. fuscus | GFN | 0 | 12 | 2 | 6 | 4 | 0 | 24 | 14 | 14 |
| H. sjoestedti | HSN | 0 | 1 | 22 | 0 | 0 | 1 | 24 | 6 | 15 |
| N. takasagoensis | NTN | 0 | 11 | 0 | 12 | 0 | 1 | 24 | 15 | 17 |
| O. formosanus | OFN | 0 | 0 | 1 | 23 | 0 | 0 | 24 | 7 | 7 |
| P. nitobei | PNN | 0 | 2 | 0 | 20 | 0 | 0 | 22 | 13 | 16 |
Termite origins of the isolated clones were clarified with these clone name tags.
Numbers of clones derived from R. speratus are cited from reference 28 and are restricted to those amplified with the primer pair of IGK and YAA.
As in the previous report on the analysis of nifH sequences in R. speratus (28), clones having significant similarity to nifH but with frameshift-like mutations, showing no translation of the nifH protein, were isolated from the four termites.
Number of clones with different amino acid (AA) sequences found in a single termite species.
Number of clones with different DNA sequences found in a single termite species.
Nucleotide sequence accession numbers.
The nifH sequences determined in this study will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB011841 to AB011964.
RESULTS
Nitrogen fixation activity in diverse termites.
Nitrogen fixation was measured in six lower termites and three higher termites by the acetylene reduction assay (Table 1). All six lower termites exhibited significant levels of nitrogen fixation activity. Among them, the highest activity was found in N. koshunensis. On the other hand, three higher termites, including a wood feeder (Nasutitermes takasagoensis), a fungus grower (Odontotermes formosanus), and a soil feeder (Pericapritermes nitobei), exhibited only low levels of activity.
FLT-RFLP analysis of the amplified nifH genes.
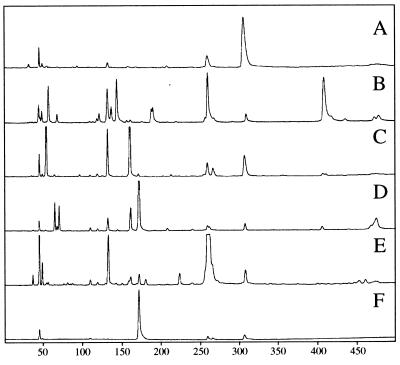
In the six lower termites which exhibited relatively high levels of nitrogen fixation activity, the variation in the amplified nifH sequences was examined and the sequences were compared among the termite species by FLT-RFLP analysis (Fig. 1). This technique is based on RFLP analysis but it differs from conventional RFLP analysis in that a single fluorescent fragment from one terminal side forms the sole focus of the analysis, in contrast to the profile of multiple fragments in RFLP analysis (7, 21). In FLT-RFLP analysis of PCR-amplified DNA from a mixed population, the single fragment length corresponds to a unique sequence or a subclass of sequences. Thus, this technique is expected to be useful for measuring sequence variation and comparing community structures in the ecosystems under investigation. In most of the lower termites, a remarkable diversity of the amplified nifH sequences was detected. Although some terminal restriction fragments (T-RFs) were common among several termites, the profiles of the T-RFs were quite dissimilar among the termite species. The results indicated that the nitrogen fixation genes within the members of the symbiotic microbial community in the termite gut were significantly different among the various termite species. Thus, we decided to further analyze the amplified nifH sequences by cloning and sequencing.
FIG. 1.
Comparison of the diversity of nifH genes in the guts of six termite species by FLT-RFLP analysis. Electropherograms of HhaI-digested nifH sequences amplified with a fluorescence-labeled primer are shown. Base lengths are indicated below the electropherograms. Electropherograms: A, R. speratus; B, C. formosanus; C, N. koshunensis; D, C. domesticus; E, G. fuscus; F, H. sjoestedti.
Only one major and a few minor T-RFs were detected in the FLT-RFLP profiles of nifH sequences from R. speratus and Hodotermopsis sjoestedti, suggesting low levels of heterogeneity of the amplified nifH sequences. In the case of R. speratus, for which nifH sequences have already been reported (28), the FLT-RFLP profile was congruent with that predicted on the basis of the cloned sequences. A majority of the cloned nifH sequences shared identical predicted T-RFs, and they were phylogenetically clustered together in the anaerobe nif group (see below). Nevertheless, a small degree of heterogeneity of around a 10% difference in amino acid residues was observed.
Although the three higher termites exhibited only low levels of nitrogen fixation activity, amplification of nifH genes was successfully attained in each case. We also cloned the amplified nifH sequences in the case of these higher termites. Because the results of both the FLT-RFLP and the cloning analyses were well correlated in the case of R. speratus and the other termites (see below), we did not conduct the FLT-RFLP analysis in the case of these higher termites.
Cloning and analysis of nifH sequences.
The nucleotide sequences of around 24 clones in our libraries of nifH sequences were analyzed for each termite species. We found several completely identical DNA sequences and completely identical amino acid sequences within the library of a single termite species (Table 2). Completely identical amino acid sequences were also encountered four times in comparisons between different termite species: between NKN12 and RSN-TKY19 (in this case, the nucleotide sequence was also identical), NKN9 and OFN35, GFN8 and PNN16, and OFN1 and PNN31. From eight termite species, 125 different nucleotide sequences which encoded 92 different amino acid sequences were newly identified in this study (these numbers do not include the previously reported sequences from R. speratus [28]). These results indicate that notably heterogeneous nitrogenase sequences are present in the symbiotic microbial community in the gut of termites, and most of them are different between termite species.
As shown in Fig. 1, the amplified nifH sequences derived from H. sjoestedti exhibited low heterogeneity in the T-RFs. In fact, a majority of the nifH clones from H. sjoestedti (22 of 24) shared predicted T-RFs of identical length and shared high sequence similarity (less than two amino acids difference). In the case of the other termites, most of the dominant T-RFs detected could be assigned to isolated clones. The sequences corresponding to the T-RFs of 57 and 143 bases in Coptotermes formosanus and that of 65 bases in Cryptotermes domesticus could not be identified, suggesting that further sampling of clones in these termites should give more diversity of nifH sequences.
Phylogenetic locations.
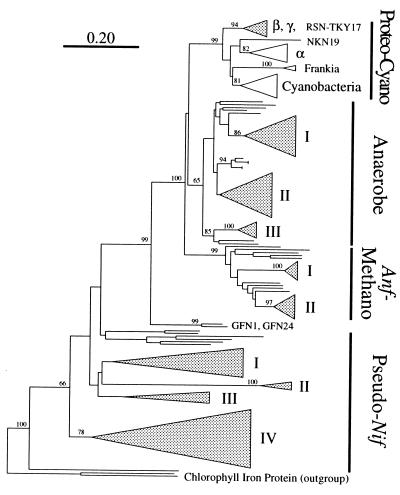
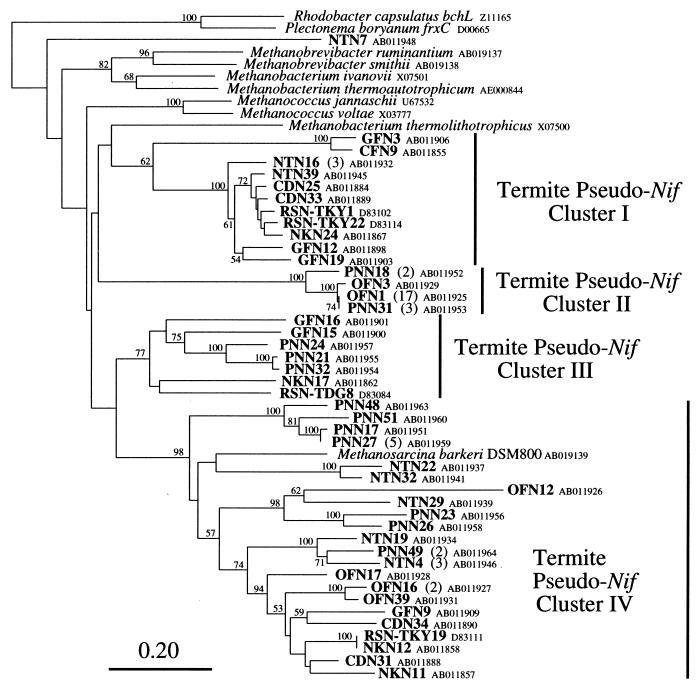
The nifH amino acid sequences from the termites were compared with each other and with sequences in the databases, and their phylogenetic relationships were investigated. Figure 2 shows a large phylogenetic tree representing four major groups of nifH sequences; the proteobacteria-cyanobacteria (proteo-cyano) group, the anaerobe group, the alternative nif methanogen (anf-methano) group, and the pseudo nif group. These four groups corresponded to the previously recognized group of nifH phylogeny (8). As described previously (8) and in this report (see below), the pseudo nif group is the most divergent nif group and is considered to function in some process other than nitrogen fixation. nifH sequences from the termites are present in each of the four groups; however, the majority of the sequences belong to three of the four groups, the anaerobe group, the anf-methano group, and the pseudo nif group. Table 2 summarizes the number of clones in each phylogenetic group detected (see below). The nifH sequences from termites were not dispersed among the nifH sequences described thus far; instead, most of the termite sequences seemed to form several sequence clusters (Fig. 2).
FIG. 2.
A large phylogenetic tree showing the relative positions of the major nifH groups and the major clusters of nifH genes isolated from the microbial communities of termite guts. The tree was constructed by the neighbor-joining method, and bootstrap values above 50 from 100 resamplings are shown for each node. Chlorophyll iron proteins were used as outgroups. The scale bar denotes 0.20 substitutions per site. Shaded wedges indicate the clusters consisting of sequences derived from termites. The depths and widths of the wedges reflect the branching lengths and the numbers of clones within the clusters, respectively. The proteo-cyano group includes conventional nifH sequences from proteobacterial clades (α, β, and γ), Frankia spp., and cyanobacteria. The anf-methano group represents anfH genes of molybdenum- and vanadium-independent nitrogenases and functional molybdenum-dependent nitrogenase genes from methanogenic archaea. The anaerobe group includes sequences from (low G+C gram-positive) clostridia, sulfate reducers (δ-proteobacteria), and M. barkeri (methanogenic Archaea domain). The pseudo nif group includes sequences from divergent genes of methanogens that might not encode active nitrogenases. The roman numbers indicate the clusters consisting of the sequences from termites in each group (Fig. 3 to 5 and see text).
Interestingly, the sequences GFN1 and GFN24 could not be assigned to any of the four nifH phylogenetic groups, indicating that these genes were derived from a novel, as yet uncharacterized group of organisms. They clustered with the proteo-cyano, anaerobe, and anf-methano groups, as supported by a 99% bootstrap value, but are clearly distinct from these three groups and deeply branched, suggesting that they might not involve nitrogen fixation as do members of the pseudo nif group. GFN1 and GFN24 share 90.0% amino acid identity but show less than 70% identity to the other nifH sequences. The nucleotide sequence of each of the other two clones derived from Glyptotermes fuscus was found to be identical to GFN1.
One nifH sequence derived from R. speratus (RSN-TKY17) and one from N. koshunensis (NKN19) were found to belong to the proteo-cyano group. The RSN-TKY17 sequence which was assigned into the β- and γ-proteobacteria clusters is most closely related to the sequences of Azoarcus spp. within the β-proteobacteria cluster (14), especially to that of Azoarcus indigens (94.7% amino acid identity). The NKN19 sequence showed significant similarity (90.9% amino acid identity) with the sequence of nifH derived from zooplankton in the Gulf of Mexico (GM24) (42). They also shared a unique sequence feature, a 12-amino-acid residue insertion (42), suggesting the presence of related nitrogen-fixing organisms in both the termite and the zooplankton. The NKN19 sequence seemed to root with the α-proteobacteria cluster; however, analysis of the zooplankton sequence led to its placement in the γ-proteobacteria cluster (42). Since NKN19 and the zooplankton sequence are deeply branched within the proteo-cyano group, the identity of these organisms is difficult to predict. The nucleotide sequences of each of the other two clones derived from N. koshunensis were found to be identical to NKN19.
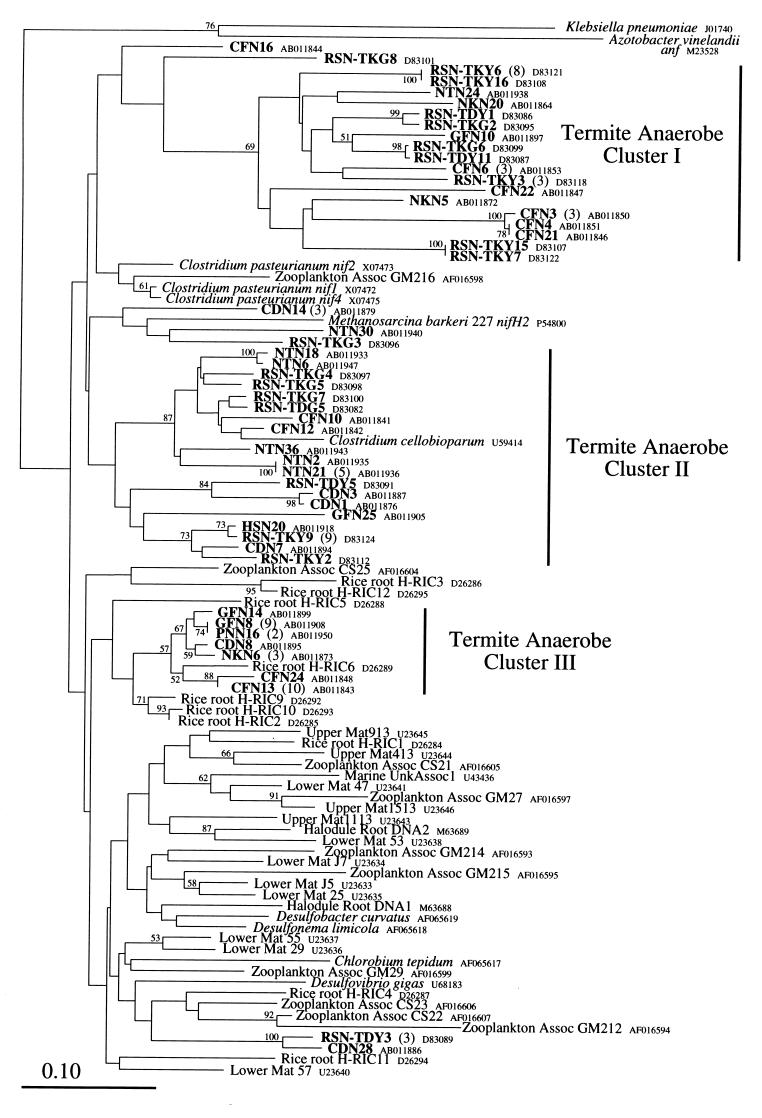
Anaerobe nif group.
Figure 3 shows the phylogenetic relationships of the nifH sequences in the anaerobic group, which includes sequences from clostridia, sulfate reducers, and Methanosarcina barkeri 227. The termite-derived nifH sequences formed three clusters (clusters I to III). Two large clusters (I and II), which corresponded to termite clusters I and II in the previous analysis of nifH sequences derived from R. speratus, respectively (28), are related to sequences from clostridia, Clostridium pasteurianum and Clostridium cellobioparum. Cluster II, especially, includes the sequence from C. cellobioparum, suggesting that the sequences belonging to this cluster may be derived from clostridia. Cluster I, however, consists only of the termite sequences and forms a distinct lineage within the anaerobe nif group, indicating the presence of unique nitrogen-fixing microorganisms in termites. Also, the third cluster (III) includes no sequences related to those of cultivated organisms. The sequences in cluster III are related to a sequence from rice roots that was amplified by PCR without cultivation of the resident microorganism (39), suggesting the presence of similar diazotrophic habitats in both ecosystems. Other than the sequences within these three clusters, there were also two minor clusters consisting only of a few sequences. The sequences CDN28 and RSN-TDY3 clustered together and are related to that of Desulfovibrio gigas (83.9 and 83.5% amino acid identity, respectively). These two sequences are somewhat related to those from marine environments, especially zooplankton (copepod)-associated sequences (3, 42). As discussed previously (3, 42), similar diazotrophic anaerobes might inhabit the guts of both invertebrates. Three sequences derived from termites, CDN14, NTN30, and RSN-TKG3, were grouped together with nifH2 of M. barkeri 227, although the grouping was not supported by bootstrap analysis. The presence of methanogenic archaea in the termite gut is well known (19, 20, 26, 27, 33); thus, these sequences are presumed to originate from gut methanogens.
FIG. 3.
Phylogenetic relationships of nifH sequences within the anaerobe group. The tree was constructed by the neighbor-joining method based on 90-amino-acid alignment positions corresponding to positions 45 to 129 in the K. pneumoniae nifH protein. Bootstrap values above 50 from 100 resamplings are shown for each node. The sequences of K. pneumoniae and Azotobacter vinelandii anfH were used as outgroups. The scale bar shows 0.10 substitutions per site. Three clusters (I, II, and III) consisting of sequences from termites are indicated. Numbers in parentheses denote numbers of clones having identical amino acid sequences from a single termite species (clones with unique sequences are not shown).
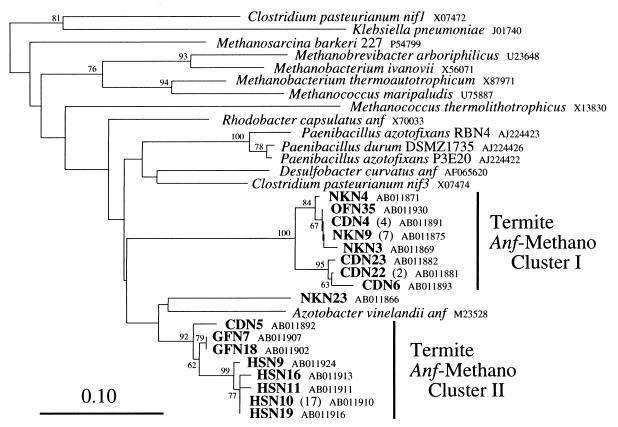
anf-methano group.
Figure 4 shows the phylogenetic relationships of the nifH sequences in the anf-methano group. Most of the sequences in this group are derived from termites of the families Kalotermitidae and Termopsidae. Two clusters (designated as anf-methano clusters I and II) comprised only of termite-derived sequences are present in this group, and the clustering was supported by high bootstrap values (100 and 92%, respectively). Except for NKN23, all of the termite-derived sequences in the anf-methano group belong to one of these two clusters. The members of anf-methano clusters I and II share more than 94 and 91% amino acid identity, respectively, in clear contrast with the lower rates of relatedness among members of the clusters in the anaerobe groups. This observation indicates that these sequences are derived from closely related organisms and that they are shared among several termite species. The sequences from organisms in the domain Bacteria seem to form a monophyletic lineage in the anf-methano group, although the monophyly was not supported by bootstrap analysis. This lineage contains all of the termite-derived sequences in the anf-methano group, suggesting their eubacterial origin. However, the identity of the corresponding nitrogen-fixing microorganisms could not be predicted because the branching order was unstable and not supported by bootstrap analysis. anf-methano cluster II includes most of the sequences derived from H. sjoestedti (22 of 24), indicating that the microorganisms represented by them are a major population in diazotrophic habitats in the gut and thus are responsible for nitrogen fixation in H. sjoestedti.
FIG. 4.
Phylogenetic relationships of nifH sequences within the anf-methano group. The tree was constructed by the neighbor-joining method based on 112-amino-acid alignment positions corresponding to positions 45 to 153 in the K. pneumoniae nifH protein. Bootstrap values above 50 from 100 resamplings are shown for each node. The sequences of K. pneumoniae and C. pasteurianum nifH1 were used as outgroups. The scale bar shows 0.10 substitutions per site. Two clusters (I and II) consisting of sequences from termites are indicated. Numbers in parentheses denote numbers of clones having identical amino acid sequences from a single termite species (clones with unique sequences are not shown).
Pseudo nif group.
Figure 5 shows the phylogenetic relationships of the nifH sequences in the pseudo nif group, which is deeply branched in the large nifH phylogenetic tree (Fig. 1). The known members of this group were derived from methanogenic archaea and were considered to function in some process other than nitrogen fixation. Most of the sequences derived from the higher termites, especially those from O. formosanus and P. nitobei, were assigned to the archaea group. The sequences from the termites form four clusters within this group (designated as pseudo nif clusters I to IV), which were significantly supported by bootstrap analysis (62, 100, 77, and 98% support, respectively). Most of the members of cluster I are sequences derived from lower termites. All four sequences in cluster II are derived from the higher termites O. formosanus and P. nitobei. A majority of the sequences from O. formosanus (17 of 24) were found to be identical to OFN1 in cluster II. Clusters III and IV seem to be somewhat related; however, the grouping was not supported by bootstrap analysis. Cluster IV consists of the most diverse sequences but includes the nifH sequence of M. barkeri DSM800, suggesting that the sequences in this cluster may have originated from Methanosarcina-related organisms.
FIG. 5.
Phylogenetic relationships of nifH sequences within the pseudo nif group, probably functioning in some process other than nitrogen fixation. The tree was constructed by the neighbor-joining method based on 118-amino-acid alignment positions corresponding to positions 45 to 153 of the K. pneumoniae nifH protein. Bootstrap values above 50 from 100 resamplings are shown for each node. Two chlorophyll iron protein sequences, that of Rhodobacter capsulatus bchL and that of Plectonema boryanum frxC, were used as outgroups. The scale bar shows 0.20 substitutions per site. Four clusters (I, II, III, and IV) consisting of sequences from termites are indicated. Numbers in parentheses denote numbers of clones having identical amino acid sequences from a single termite species (clones with unique sequences are not shown).
DISCUSSION
The nitrogen fixation gene nifH was isolated from members of the symbiotic microbial community in the gut of evolutionarily diverse termites by a culture-independent approach and analyzed phylogenetically. Remarkably diverse nifH sequences were isolated from each termite species, and most of the nifH sequences were found to be novel and distantly related to those of cultivated organisms or as yet unidentified organisms detected in other environments (3, 34, 39, 40, 42, 43). The results indicate the presence of potential diazotrophic habitats of unexpected diversity in the gut of termites, which are as yet unidentified and uncharacterized. Notably, identical nifH amino acid sequences were scarcely isolated from different termite species (only four times). The more termite species we investigated, the more distinct were the nifH sequences isolated. Given the existence of more than 2,000 described species on the earth, termites may be a rich reservoir of novel and diverse microorganisms that potentially fix nitrogen.
Several species of nitrogen-fixing bacteria, including Citrobacter freundii, Enterobacter agglomerans, and Desulfovibrio spp., have been isolated from the gut of termites (12, 18, 31). The first two belong to the γ subclass of proteobacteria, and the termite-derived sequences RSN-TKY17 and NKN19 were assigned to proteobacteria nifH clusters. The sequences RSN-TDY3 and CDN28 are related to the nifH sequence of D. gigas. Although the nifH genes of bacterial isolates from termites have not been characterized, these sequences may originate from organisms related to them. However, the number of clones found to have these sequences was relatively few, suggesting that these represent minor populations in the termite gut. On the other hand, the organisms presumably corresponding to the remaining clusters and/or sequences, which comprise the majority of the isolated sequences, have not yet been identified or cultivated from termites as nitrogen fixers. The isolation of organisms related to clostridia and methanogens from the gut of termites has been reported (13, 15, 19, 20), but their nitrogen-fixing ability has not been reported. Thus, we have little knowledge of the organisms responsible for nitrogen fixation in termites.
The nifH sequences isolated from the termites form several unique clusters in the phylogenetic trees. They are not randomly distributed over the nifH taxa. Some particular types of nitrogen-fixing microorganisms probably inhabit the gut of termites. Notably, sequences affiliated with the proteo-cyano group were found to occur very rarely in the termite gut. Since the proteobacteria are believed to comprise a substantial proportion of the gut microbial community (24) and since most of the nitrogen-fixing organisms isolated from the gut of termites are proteobacteria (12, 31), the extremely low abundance of their nifH sequences was unexpected. The finding that the minority of termite-derived nifH sequences were clustered in the proteo-cyano group is in striking contrast to the results of studies on nifH sequences derived from other natural environments, such as those from the picoplankton-size fraction of oligotrophic oceans (42) and those from soil and litter in a Douglas fir forest (40), where nifH sequences of the proteo-cyano group are rather predominant. The presence of large numbers of heterogeneous sequences clustering in the anaerobe group is common in several environments, such as in the termite gut (this study and reference 28), rice roots (39), marine cyanobacterial mats (43), and enrichment cultures initiated with marine zooplankton (3), though clustering in the proteo-cyano group was observed also in the study of rice roots. However, a majority of the termite-derived sequences in the anaerobe group form lineages distinct from those derived from other environments (e.g., termite anaerobe clusters I and II). These features may reflect differences in diazotrophic habitats dependent on the microbial ecosystems. Above all, the most striking and distinctive feature of the sequences from the other environments was the presence of those affiliated with the anf-methano group in the gut of some termites. The anf sequences have never been found in any other environment. The alternative nitrogenase encoded by the anf gene differs from conventional nitrogenases in terms of its metal components serving as cofactors (9). The alternative nitrogenase contains neither molybdenum nor vanadium and is expressed under conditions of molybdenum depletion. Metal availability probably is a key factor determining the presence of nitrogen fixation genes of the anf-methano group (discussed also in reference 23).
Only low levels of nitrogen fixation activity were detected in higher termites (Table 1). Of course, our experimental conditions might not be adequate to obtain optimal activity. For example, there was an interval of several days between the time of sample collection (the removal of termites from their nests) and the assay, and they cannot be kept alive in vitro for a long time after removal from their nests. In fact, a significant level of C2H2 reduction activity (up to 50 nmol of C2H4 formed per h per g [wet weight]) has been demonstrated in the case of the wood-feeding higher termite N. takasagoensis (22). However, little activity was found in the soil-feeding termite P. nitobei or the fungus-growing termite O. formosanus (22). Based on the results of stable isotope analyses in studies of both soil-feeding and fungus-growing higher termites, nitrogen fixation appears to contribute less to their nitrogen economy than in the case of wood-feeding termites (35–37). As discussed previously, the feeding habits and foraging preferences may obviate the need for nitrogen fixation simply because their diet contains an adequate amount of combined nitrogen (4).
In spite of the low levels of nitrogen fixation activity displayed by the higher termites examined here, various nifH sequences were isolated from them. A large proportion of the sequences isolated from the higher termites, especially from P. nitobei and O. formosanus, were assigned to the pseudo nif group. The results suggest that the product of the nifH gene in the pseudo nif group may not be a functional nitrogenase. It has been suggested previously that it may encode a product that is not a nitrogenase based on the following criteria: their high degree of divergence relative to other nifH groups; their lack of nifD- or glnB-like open reading frames, found downstream from them; the inability by some members in this group (Methanococcus voltae and Methanococcus jannaschii) to detect nitrogen fixation; their expression in Methanococcus thermolithotrophicus; and their significant sequence similarity with the iron proteins involved in bacteriochlorophyll synthesis (reference 8 and the references therein). The variation in the nifH sequences can be simply explained by the variation in the methanogen species present, since it has been reported that phylogenetically there is a greater variety of methanogen species in the gut of higher termites than in the gut of lower termites (27). The dominance of the clones in this group probably reflects the absence of functional nitrogenase genes within the gut community.
In N. takasagoensis, nifH sequences in the anaerobe group as well as those in the pseudo nif group were present, although the level of nitrogen fixation activity was very low. The results imply that the existence of nifH sequences does not simply lead to active nitrogen fixation. Since nitrogenases are strictly regulated at the transcriptional and posttranslational levels (9), further analysis of the expression of nifH will be necessary in order to determine whether the gene is functional in these organisms and their contribution to nitrogen fixation in termites. Even in those termites showing high levels of activity, whether the nifH sequences detected are really responsible for nitrogen fixation in termites remains to be clarified. In fact, we have shown that only restricted groups of the nifH sequences are preferentially expressed in N. koshunensis, as determined by analyzing the levels of nifH mRNA in the microbial population in the gut (23). Still, the potential nifH phylotypes described here will serve as an important basis for further studies.
Interestingly, some phylogenetic relationships between the termite families and the nifH groups of symbiotic microorganisms are evident (Table 2). In the higher termites, which are phylogenetically related and assembled into a single termite family, Termitidae, the majority of the nifH sequences were assigned to the pseudo nif group. In the lower termites, the number of clones of nifH sequences belonging to the pseudo nif group was few. Many of the nifH sequences assigned to the anaerobe group were isolated from termites of the family Rhinotermitidae, and those belonging to the anf-methano group were never found in this termite family. From the three members of the family Kalotermitidae, sequences assigned to either the anaerobe group or the anf-methano group were isolated in large numbers. The sequences in the anf-methano group are exclusively derived from termites of either the family Kalotermitidae or the family Termopsidae. Surprisingly, all of the nifH sequences from H. sjoestedti (family Termopsidae) were exclusively assigned to cluster II of the anf-methano group, with only one exception. Of course, more analyses with more diverse termites are necessary to reach any definitive conclusion. Nevertheless, these relationships are suggestive of the evolution of the symbiosis between termites and their nitrogen-fixing inhabitants. Alternatively, these relationships can be simply explained in terms of the nutritional ecology of the termites, since their feeding behavior differs somewhat. The three termites of the family Kalotermitidae feed on dry and sound wood. The termites of the family Rhinotermitidae are known to be subterranean termites, whereas the termites of the family Termopsidae are known as damp wood termites. The factors affecting the choice of termites as diazotrophic habitats by symbionts are as yet uncertain and remain to be clarified.
The culture-independent approach applied here has revealed that the major population responsible for nitrogen fixation in the gut of termites is a population of as yet uncharacterized microorganisms. The nifH sequence was found to be a useful means of detecting them and predicting their taxonomy. Since cloning and sequencing are laborious tasks, FLT-RFLP analysis may serve as a simpler but significantly informative technique for surveying community structures as demonstrated in this study. Now that we have sequence data on nifH genes from nine evolutionarily diverse termites, we can predict the presence of a particular class of nitrogenase genes within the microbial community in the termite gut, depending on the presence of certain T-RFs in the FLT-RFLP analysis. For example, the T-RFs of 161 and 172 bases are exclusively derived from the nifH sequences in the anf-methano group, and the T-RFs of 259 and 307 bases are derived from those in the anaerobe group. However, since PCR amplification may introduce some biases with respect to the gene composition of the products, a quantitative approach is necessary to measure real populations in the original sample. The nifH sequence data described in this study will allow us to design sequence-specific probes and/or primers for specific detection, hybridization, and quantitative experiments. Although isolation and cultivation of the corresponding microorganisms are advantageous for taxonomic and physiological characterization in detail, culture-independent approaches will provide valuable information about the nitrogen economy and the ecology within the symbiotic community in the gut of termites.
ACKNOWLEDGMENTS
This work was partially supported by grants from the Biodesign Research Program, the Genome Research Program, and the Eco Molecular Science Research Program from RIKEN and by a grant from the International Cooperative Research Project (Bio-Recycle Project) from Japan Science and Technology Corporation. S.N. was supported by a grant from the Junior Research Associate Program from RIKEN.
We thank F. Aoki for assistance and I. Yasuda for advice on termite collection.
REFERENCES
- 1.Benemann J R. Nitrogen fixation in termites. Science. 1973;181:164–165. doi: 10.1126/science.181.4095.164. [DOI] [PubMed] [Google Scholar]
- 2.Berchtold M, König H. Phylogenetic analysis and in situ identification of uncultivated spirochetes from the hindgut of the termite Mastotermes darwiniensis. Syst Appl Microbiol. 1996;19:66–73. [Google Scholar]
- 3.Braun S T, Proctor L M, Zani S, Mellon M T, Zehr J P. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol Ecol. 1999;28:273–279. [Google Scholar]
- 4.Breznak J A. Intestinal microbiota of termites and other xylophagous insects. Ann Rev Microbiol. 1982;36:323–343. doi: 10.1146/annurev.mi.36.100182.001543. [DOI] [PubMed] [Google Scholar]
- 5.Breznak J A, Brill W J, Mertins J W, Coppel H C. Nitrogen fixation in termites. Nature. 1973;244:577–580. doi: 10.1038/244577a0. [DOI] [PubMed] [Google Scholar]
- 6.Breznak J A, Brune A. Role of microorganisms in the digestion of lignocellulose by termites. Ann Rev Entomol. 1994;39:453–487. [Google Scholar]
- 7.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien Y-T, Zinder S H. Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD1 from the eubacterium Clostridium pasteurianum. J Bacteriol. 1994;176:6590–6598. doi: 10.1128/jb.176.21.6590-6598.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacy G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834. [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP—phylogeny inference package version 3.5. Cladistics. 1989;5:164–166. [Google Scholar]
- 12.French J R J, Turner G L, Bradbury J F. Nitrogen fixation by bacteria from the hindgut of termite. J Gen Microbiol. 1976;95:202–206. doi: 10.1099/00221287-95-2-202. [DOI] [PubMed] [Google Scholar]
- 13.Hethener P, Brauman A, Garcia J-L. Clostridium termitidis sp. nov., a cellulolytic bacterium from the gut of the wood-feeding termite, Nasutitermes lujae. Syst Appl Microbiol. 1992;15:52–58. [Google Scholar]
- 14.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenase of Azoarcus spp., proteobacteria of the β subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane M D, Brauman A, Breznak J A. Clostridium mayombei sp. nov., an H2/CO2 acetogenic bacterium from the gut of the African soil-feeding termite, Cubitermes speciosus. Arch Microbiol. 1991;156:99–104. [Google Scholar]
- 16.Kirshtein J D, Paerl H W, Zehr J. Amplification, cloning, and sequencing of a nifH segment from aquatic microorganisms and natural communities. Appl Environ Microbiol. 1991;57:2645–2650. doi: 10.1128/aem.57.9.2645-2650.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishna K. Taxonomy, phylogeny and distribution of termites. In: Krichna K, Weesner F M, editors. Biology of termites. Vol. 2. New York, N.Y: Academic Press, Inc.; 1970. pp. 127–152. [Google Scholar]
- 18.Kuhniugk T, Krekeler J B D, Cypionka H, König H. A feasible role of sulfate-reducing bacteria in the termite gut. Syst Appl Microbiol. 1996;19:139–149. [Google Scholar]
- 19.Leadbetter J R, Breznak J A. Physiological ecology of Methanobrevibacter cuticularis sp. nov., and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl Environ Microbiol. 1996;62:3620–3631. doi: 10.1128/aem.62.10.3620-3631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leadbetter J R, Crosby L D, Breznak J A. Methanobrevibacter filiformis sp. nov., a filamentous methanogen from termite hindguts. Arch Microbiol. 1998;169:287–292. doi: 10.1007/s002030050574. [DOI] [PubMed] [Google Scholar]
- 21.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura, T., and K. Yara. Unpublished data.
- 23.Noda S, Ohkuma M, Usami R, Horikoshi K, Kudo T. Culture-independent characterization of a gene responsible for nitrogen fixation in the symbiotic microbial community in the gut of the termite Neotermes koshunensis. Appl Environ Microbiol. 1999;65:4935–4942. doi: 10.1128/aem.65.11.4935-4942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkuma M, Kudo T. Phylogenetic analysis of the symbiotic intestinal microflora of the termite Cryptotermes domesticus. FEMS Microbiol Lett. 1998;164:389–395. [Google Scholar]
- 26.Ohkuma M, Noda S, Horikoshi K, Kudo T. Phylogeny of symbiotic methanogens in the gut of the termite Reticulitermes speratus. FEMS Microbiol Lett. 1995;134:45–50. doi: 10.1111/j.1574-6968.1995.tb07912.x. [DOI] [PubMed] [Google Scholar]
- 27.Ohkuma M, Noda S, Kudo T. Phylogenetic relationships of symbiotic methanogens in diverse termites. FEMS Microbiol Lett. 1999;171:147–153. doi: 10.1111/j.1574-6968.1999.tb13425.x. [DOI] [PubMed] [Google Scholar]
- 28.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paster B J, Dewhirst F E, Cooke S M, Fussing V, Poulsen L K, Breznak J A. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl Environ Microbiol. 1996;62:347–352. doi: 10.1128/aem.62.2.347-352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postgate J R. The acetylene reduction test for nitrogen fixation. In: Norris J R, Ribbons D W, editors. Methods in microbiology. 6B. New York, N.Y: Academic Press, Inc.; 1972. pp. 343–356. [Google Scholar]
- 31.Potrikus C J, Breznak J A. Nitrogen-fixing Enterobacter agglomerans isolated from guts of wood-eating termites. Appl Environ Microbiol. 1977;33:392–399. doi: 10.1128/aem.33.2.392-399.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saiton N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Shinzato N, Matsumoto T, Yamaoka I, Oshima T, Yamagishi A. Phylogenetic diversity of symbiotic methanogens living in the hindgut of the lower termite Reticulitermes speratus analyzed by PCR and in situ hybridization. Appl Environ Microbiol. 1999;65:837–840. doi: 10.1128/aem.65.2.837-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steppe T F, Olson J B, Paerl H W, Litaker R W, Belnap J. Consortial N2 fixation: a strategy for meeting nitrogen requirements of marine and terrestrial cyanobacterial mats. FEMS Microbiol Ecol. 1996;21:149–156. [Google Scholar]
- 35.Tayasu I, Abe T, Eggleton P, Bignell D E. Nitrogen and carbon isotope ratios in termites: an indicator of trophic habit along the gradient from wood-feeding to soil-feeding. Ecol Entomol. 1997;22:343–351. [Google Scholar]
- 36.Tayasu I, Inoue T, Miller L R, Sugimoto A, Takeichi S, Abe T. Confirmation of soil-feeding termites (Isoptera; Termitidae; Termitinae) in Australia using stable isotope ratios. Funct Ecol. 1998;12:536–542. [Google Scholar]
- 37.Tayasu I, Sugimoto A, Wada E, Abe T. Xylophagous termites depending on atmospheric nitrogen. Naturwissenschaften. 1994;81:229–231. [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda T, Suga Y, Yahiro N, Matuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Widmer F, Shaffer B T, Porteous L A, Seidler R J. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl Environ Microbiol. 1999;65:374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood T G, Sand W A. The role of termites in ecosystems. In: Brian M V, editor. Production ecology of ants and termites. Cambridge, England: Cambridge University Press; 1978. pp. 245–292. [Google Scholar]
- 42.Zehr J P, Mellon M T, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zehr J P, Mellon M, Braun S, Litaker W, Steppe T, Paerl H W. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]