Abstract
Background
Enhancer of Zeste homologue 2 (EZH2) is a polycomb group gene and an epigenetic regulator that inhibits transcription, a modification associated with gene silencing. EZH2 plays an essential role in humoral and cell-mediated adaptive immunity. The purpose of the current study is to investigate the prognostic potential of EZH2 and to comprehensively analyse the correlation between EZH2 and immune infiltration in multiple cancer cases, especially liver hepatocellular carcinoma.
Methods
EZH2 expression across cancers was explored through Oncomine, HPA, and GEPIA2. Additionally, the prognostic value of EZH2 analysis across cancers was based on the GEPIA2, TCGA portal, Kaplan–Meier Plotter, and LOGpc databases. Based on GO and KEGG analyses, GSEA helped demonstrate the biological processes through which EZH2 might lead to HCC development. GEPIA and TIMER were adopted to detect the possible relationship of EZH2 expression with tumour-infiltrating immune cells (TIICs).
Results
EZH2 overexpression levels were associated with poor prognosis of cancer, especially hepatocellular carcinoma. A high EZH2 expression level is related to a poor prognosis of HCC, especially in disease histology and stage III. The EZH2 expression level was positively correlated with critical gene markers of TAMs, M2 macrophages, M1 macrophages, and monocytes. Further analysis revealed that EZH2 genes were mainly related to DNA recombination, mitotic cell cycle phase transition, and chromosome segregation.
Conclusion
EZH2 plays an essential role in the immune microenvironment and is a potential prognostic marker and immunotherapy target for hepatocellular carcinoma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12865-022-00502-7.
Keywords: Immunotherapy, Immune infiltration, Liver hepatocellular carcinoma, T cells exhaustion, Prognosis, EZH2
Introduction
Immunotherapy for cancer is becoming a critical approach that manages cancer cells via the immune system [1–3]. Several studies in some preclinical models and advanced tumour patients have proven that epigenetic modulators have immunomodulatory properties, providing a theoretical basis for combining epigenetics with immunotherapy [4–6]. Liver hepatocellular carcinoma (LIHC) is the most common cancer globally, and it is the third primary cause of tumour-related deaths [7, 8]. Over the past decade, efforts have been made to develop novel drugs and therapeutic strategies for HCC [9, 10]. However, the efficacy of anti-LIHC therapy is compromised due to unclear carcinogenesis and progression mechanisms at the molecular level [11]. Serum alpha-fetoprotein (AFP) detection, B ultrasound, and CT scans can be used to diagnose liver cancer. However, the misdiagnosis rate is high [12, 13]. Currently, biomarkers for liver cancer are rapidly advancing, but the 5-year survival remains low [14, 15]. Therefore, more sensitive biomarkers and novel therapeutic strategies must be explored for HCC treatment.
EZH2 is a polycomb group gene (PcG) and an epigenetic regulator that inhibits transcription. Polycomb repression complex 2 (PRC2) in the PcG protein core complex can regulate chromatin structure-mediated gene silencing [16]. EZH2 acts as an enzyme catalytic subunit of PRC2, mediating trimethylation of Lys27 in histone 3 (H3K27me3) and gene silencing [17]. H3K27me3 inhibits gene expression [18]. In addition to H3K27me3, PRC2 also methylates nonhistone substrates [19]. EZH2 activates downstream genes by methylating nonhistone targets in a PRC2-independent manner [20–22]. As a result, EZH2 crucially affects cell lineage determination and signalling pathways and is a master regulator of cell cycle progression, autophagy, apoptosis, DNA damage repair, and cellular senescence inhibition [23–26]. Studies have revealed that EZH2, a modifier associated with epigenetic regulation and immune function, can promote local and systemic anticancer immune responses by modulating TH-1 chemokine expression, affecting marrow-derived suppressor cells (MDSCs) or CD8 + T-cell infiltration [27–31]. However, the mechanisms of human tumours must be explained. These pieces of evidence indicate that EZH2 may be a modifier involved in epigenetic regulation and immune function.
The work presented in the current study provides an analysis of the role of EZH2 levels in LIHC and explores the function of EZH2 in tumour immunity, which was derived from publicly accessible databases. Our study confirmed that EZH2 upregulation could predict poor overall survival (OS) in HCC patients. This illustrated the possible association and regulatory mechanism of EZH2-associated genomic alterations and functional networks within LIHC and helped identify novel diagnostic and therapeutic LIHC targets.
Methods
Oncomine analysis
EZH2 gene expression levels in tumour and normal tissues was obtained from the Oncomine database (http://www.oncomine.org), a web-based data mining platform for collecting, analysing, and offering tumour microarray information [32].
Comprehensive correlation analysis in tumour-infiltrating immune cells
Tumour Immune Estimation Resource (TIMER), a free database containing 32 TCGA-derived cancers involving 10,897 samples, can assess inner immune infiltrate levels (http://cistrome.org/TIMER/). We evaluated the association of EZH2 levels with six immune cell types within LIHC using the TIMER database [33–36]. Similarly, we investigated the association of EZH2 expression with tumour purity.
Analysis based on the OnCoLnc Database
The OncoLnc database contains 21 pieces of TCGA cancer survival information (http://www.oncolnc.org/). In addition, the current study analysed EZH2’s prognostic value in 21 cancers. In accordance with the obtained results, the EZH2 expression level was remarkably related to survival in eight cancers.
Gene expression profiling interactive analysis
GEPIA2 (The Gene Expression Profiling Interactive Analysis 2) database is a comprehensive analytical tool that analyses customizable functions, such as the interaction function and genes’ prognostic significance in cancer and noncarcinoma samples [37] (http://gepia2.cancer-pku.cn/). The current work used GEPIA to detect EZH2 mRNA levels in LIHC and its prognostic value and analysed gene expression correlations.
Kaplan–Meier (K–M) survival curve analysis
K-M Plotter can be adopted for evaluating the connection between gene expression and 21 cancer prognoses (http://kmplot.com/). In the current study, we adopted K-M Plotter to detect the connection between EZH2 levels and HRs, P values (upon log-rank test), OS, and RFS in LIHC patients. More than 50,000 samples from the database were evaluated using gene array and RNA sequencing [38].
TISIDB database analysis
TISIDB is a public database used for analysing the interactions between the immune system and cancers (http://cis.hku.hk/TISIDB). It combines different cancer immunology data sources [39]. Based on the TISIDB database, we explored the Spearman correlation between EZH2 expression level and tumour-infiltrating cell level and subtype.
Human protein atlas database analysis
The HPA database includes the gene expression profiles and pathology, which can be adopted to validate prognostic gene immunohistochemistry (IHC) [40] (https://www.proteinatlas.org/). IHC images from HPA can be directly accessed at EZH2 in tumour tissue (https://www.proteinatlas.org/ENSG00000148773-EZH2/pathology/liver+cancer#img) and EZH2 in normal tissue (https://www.proteinatlas.org/ENSG00000148773-EZH2/tissue/liver).
Coexpression gene prediction and GSEA
LinkedOmics is an open platform that includes multiomics data for 32 cancers derived from TCGA (http://www.linkedomics.org/) [41]. We screened for EZH2-related differentially expressed genes in the TCGA LIHC cohort (n = 371) using LinkFinder's LinkedOmics, and their correlations were analysed using Pearson's correlation. The LinkInterpreter module was employed for network and pathway analyses of DEGs. Kyoto Encyclopedia of Genes and Genomes is a database (https://www.kegg.jp/) that relates genomic information to higher-order functional information for analysing gene function [42–44]. KEGG pathway and GO analyses were performed using GSEA tools.
Results
Expression level of EZH2 across cancers
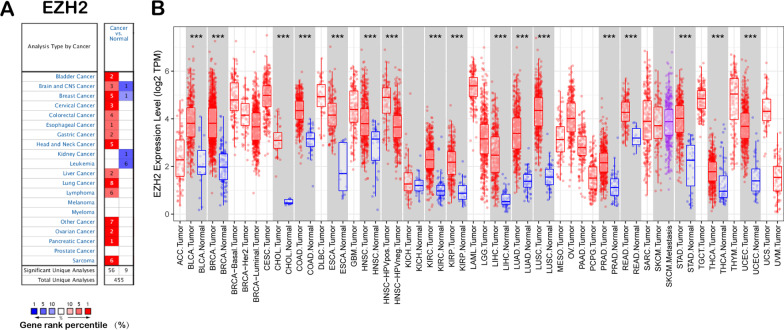
The current study used Oncomine database to analyse the difference in EZH2 expression between carcinomas and normal samples. Our analysis revealed that EZH2 is highly expressed in breast, bladder, head and neck, sarcoma, pancreatic, cervical, liver, and other cancers compared with in normal tissues (Fig. 1A). Additionally, EZH2 downregulation was detected in prostate cancer, myeloma, melanoma, kidney cancer, and leukaemia cancers. Additional file 2: Table S1 offers more detailed results of EZH2 expression across cancers. TIMER was used to detect RNA sequencing data in TCGA to evaluate EZH2 expression across cancers. Differential EZH2 expression between cancer and healthy samples is displayed in Fig. 1B. EZH2 expression in SKCM was significantly lower than that in healthy samples. However, EZH2 expression was upregulated within the BRCA, BLCA, COAD, CHOL, HNSC, ESCA, KIRP, KIRC, KICH, LIHC, PRAD, LUSC, LUAD, STAD, READ, UCEC, and THCA.
Fig. 1.
EZH2 levels within pan-cancers. A High or low EZH2 expression within pan-cancers relative to healthy samples from the Oncomine database. B EZH2 levels within pan-cancers in TCGA database measured through TIMER. *P < 0.01, **P < 0.001, ***P < 0.0001
Prognostic potential value of EZH2 across cancers
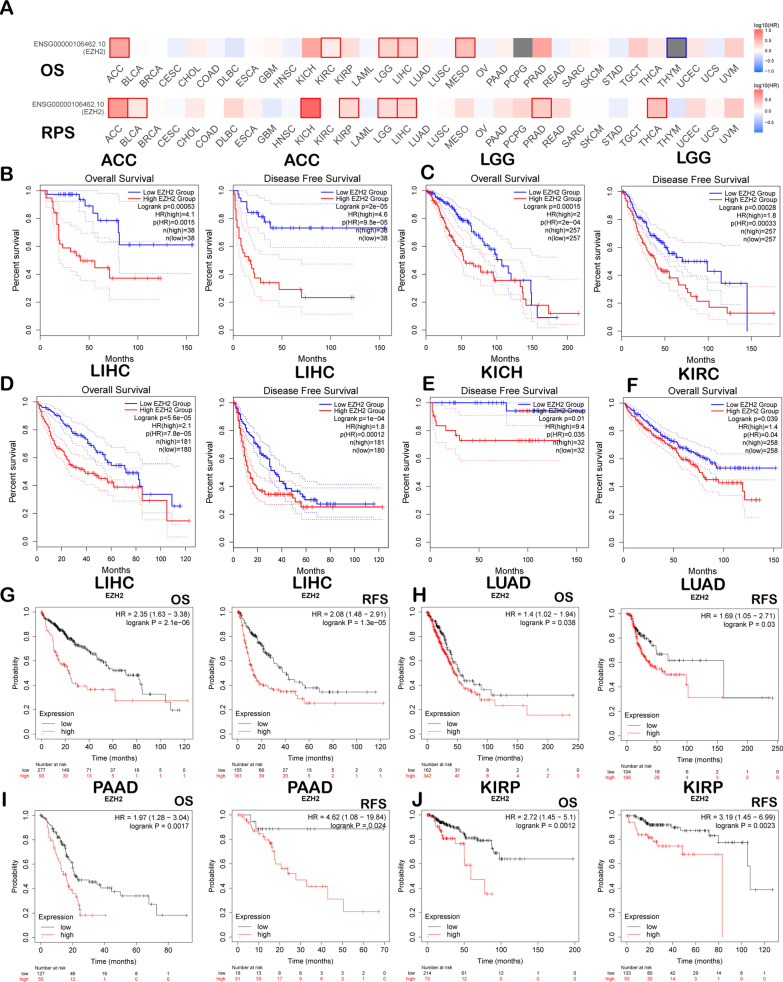
The current study analysed EZH2’s effect on cancers prognosis from three databases. Table 1 presents the correlation of EZH2 levels with cancers survival from the OncoLnc database. We assessed the correlation between EZH2 expression level and prognosis by using GEPIA2. Figure 2A presents the influence of EZH2 levels on survival in cancer. Poor prognosis was associated with high EZH2 expression levels in ACC (OS: (P = 6.3e-04, HR = 2); PFS: P = 2.8e-04, HR = 1.8); LGG (OS: P = 1.5e-04, HR = 2); PFS: P = 2.8e-04, HR = 1.8); LIHC (OS: P = 5.6e-5, HR = 2.1); PFS: P = 1.0e-04, HR = 1.8); KICH (OS: P = 0.01, HR = 9.8); and KIRC (OS: P = 0.039, HR = 1.4) (Fig. 2B–F). Then, we assessed EZH2-related survival (OS and RFS) by using Kaplan–Meier Plotter. Similarly, we discovered that EZH2 is a detrimental prognostic factor for LIHC (OS: P = 2.1e-06, HR = 2.35 [1.63–3.38]; PFS: P = 1.3e-05, HR = 2.08 [1.48–2.91]), LUAD (OS: P = 0.038, HR = 1.4 [1.02–1.94]; PFS: P = 0.03, HR = 1.69 [1.05–2.71]), PAAD (OS: P = 1.7e-03, HR = 1.97 [1.28–3.04]; PFS: P = 0.024, HR = 4.62 [1.08–19.84]), KIRP (OS: P = 1.2e-03, HR = 2.72 [1.45–5.1]; and PFS: P = 2.3e-03, HR = 3.19 [1.45–6.99]) (Fig. 2G–J).
Table 1.
Prognostic role of EZH2 in diverse cancers through OncoLnc
| Cancer | Cox | P-value | FDR | Rank | Median | Mean |
|---|---|---|---|---|---|---|
| BLCA | 0.032 | 7.00e-01 | 8.58e-01 | 13,285 | 475.71 | 599.61 |
| BRCA | 0.082 | 3.50e-01 | 7.03e-01 | 8182 | 395.74 | 504.19 |
| CESC | − 0.327 | 1.60e-02 | 2.05e-01 | 1239 | 1033.8 | 1118.51 |
| COAD | − 0.147 | 1.50e-01 | 5.58e-01 | 4290 | 657.07 | 697.51 |
| ESCA | 0.001 | 9.90e-01 | 9.95e-01 | 16,624 | 482.73 | 573.25 |
| GBM | − 0.037 | 6.90e-01 | 9.58e-01 | 12,003 | 598.43 | 689.2 |
| HNSC | − 0.158 | 2.40e-02 | 2.46e-01 | 1590 | 479.81 | 577.87 |
| KIRC | 0.292 | 2.30e-04 | 1.62e-03 | 2346 | 126.98 | 149.04 |
| KIRP | 0.527 | 7.90e-04 | 8.76e-03 | 1477 | 123.29 | 140.76 |
| LAML | − 0.028 | 8.20e-01 | 9.43e-01 | 13,200 | 1041.46 | 1043.88 |
| LGG | 0.277 | 6.20e-03 | 1.62e-02 | 6414 | 216.22 | 286.97 |
| LIHC | 0.478 | 2.80e-06 | 1.23e-03 | 36 | 248.55 | 306.71 |
| LUAD | 0.097 | 1.80e-01 | 4.10e-01 | 7248 | 320.93 | 397.7 |
| LUSC | − 0.088 | 1.90e-01 | 7.01e-01 | 4467 | 629.89 | 713.28 |
| OV | − 0.003 | 9.70e-01 | 9.93e-01 | 16,448 | 503.56 | 601.14 |
| PAAD | 0.228 | 4.10e-02 | 1.63e-01 | 4299 | 207.07 | 238.52 |
| READ | − 0.188 | 3.80e-01 | 9.57e-01 | 6463 | 643.34 | 675.6 |
| SARC | 0.133 | 2.00e-01 | 4.83e-01 | 6586 | 440.14 | 578.62 |
| SKCM | 0.052 | 4.40e-01 | 6.36e-01 | 11,030 | 447.42 | 528.58 |
| STAD | − 0.169 | 5.30e-02 | 3.49e-01 | 2548 | 483.32 | 521.53 |
| UCEC | 0.121 | 3.00e-01 | 9.88e-01 | 4955 | 460.41 | 528.01 |
Significantly different results are displayed in this table (P < 0.05)
Fig. 2.
Comparison of EZH2 expression within diverse cancers (A–F) by K–M survival curves based on GEPIA2 database as well as K–M Plot (G–J). A Survival heatmap showing EZH2 within 33 cancers obtained from the TCGA database. The rectangular box indicates the significance of prognostic analyses, and the blue and red squares indicate low and high expression, respectively. DFS and OS curves for B ACC (n = 38), C LGG (n = 257), and D LIHC (n = 180); DFS curves for E KICH (n = 32); and OS curves for F KIRC (n = 258). OS and PFS survival curves in G LIHC (n = 370, n = 316), H LUAD (n = 504, n = 300), I PAAD (n = 177, n = 69), and J KIRP (n = 287, n = 183). OS-overall survival, RFS-relapse-free survival, DFS-disease-free survival, and PFS-progression-free survival
Relationship between EZH2 expression and Immune Infiltration
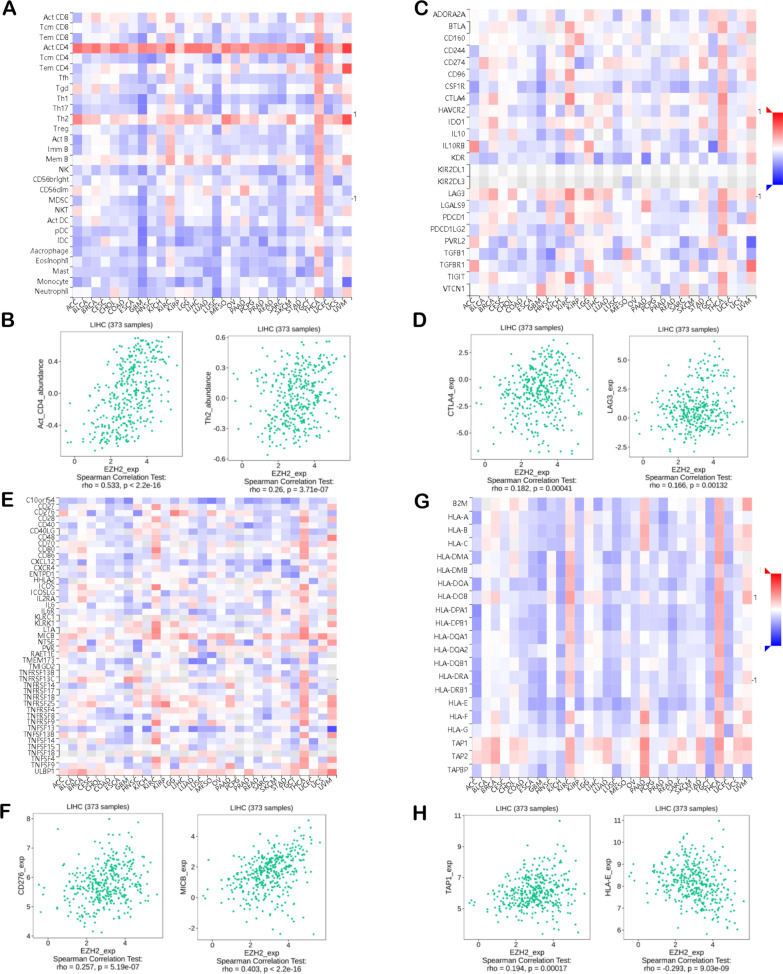
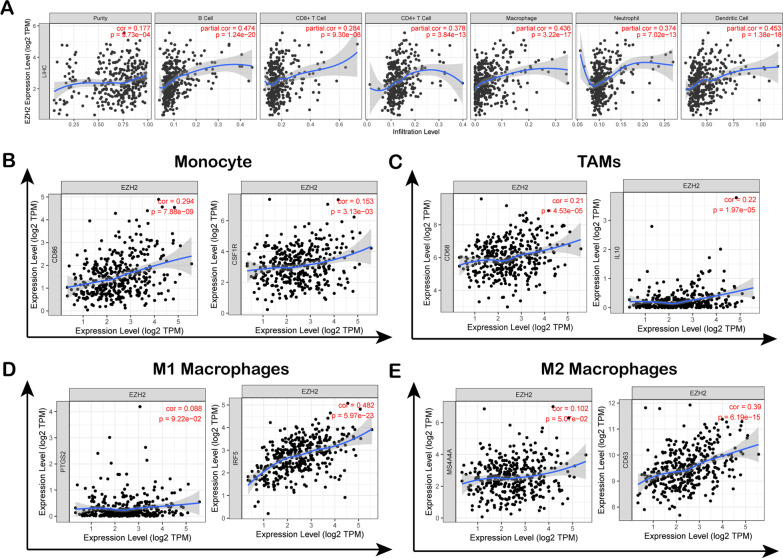
Cancer survival and lymph node metastasis (LNM) might be predicted according to tumour-infiltrating immune cell (TIIC) levels [45–47]. The current study analysed the connection between EZH2 levels and immunomodulators and lymphocytes within LIHC based on the TISIDB database (Fig. 3). Figure 3A illustrates the connection between EZH2 expression and 28 TIICs across cancers. The EZH2 level revealed a positive relationship with type 2 T helper cells (Th2; Spearman: ρ = 0.26, P = 3.17e − 07) and activated CD4 + T cells (Act-cd4; Spearman: ρ = 0.533, P < 2.2e − 16) (Fig. 3B). Immunomodulators are immune inhibitors, immunostimulators, or major histocompatibility complex molecules (MHC). Figure 3C presents the analyses of the correlation between EZH2 expression and 24 types of human tumour immunoinhibitors. EZH2 expression was positively correlated with LAG3 (Spearman: ρ = 0.166, P = 1.32e − 03) and CTLA4 (Spearman: ρ = 0.182, P = 4.1e − 04) (Fig. 3D). Figure 3E reveals the correlation between EZH2 levels and 45 types of immunostimulators. In LIHC, EZH2 expression was positively correlated with MICB (Spearman: ρ = 0.403, P < 2.2e − 16) and CD276 (Spearman: ρ = 0.257, P = 5.19e − 07) (Fig. 3F). As shown in Fig. 3G, EZH2 levels were correlated with 21 MHC molecules across cancers. EZH2 was positively related to TAP1 and HLA-E in LIHC (Fig. 3H). Subsequently, we explored the relationship between EZH2 expression and immune infiltration in 39 tumour types by using the TIMER database (Figure. S1). Cancer survival and LNM were predicted according to TIIC levels [45–47]. EZH2 expression revealed a positive relationship with infiltrating levels of CD8 + T cells in 20 cancer cases, those of macrophages for 12 cancers, those of CD4 + T cells for 18 cancers, those of DCs for 19 cancers, and those of neutrophils for 24 cancers. The results showed that in LIHC, EZH2 expression levels correlated positively with the infiltration degrees of CD4 + T cells (R = 0.378, P = 3.84e-13), CD8 + T cells (R = 0.284, P = 9.30e-08), DCs (R = 0.453, P = 1.38e-18), neutrophils (R = 0.374, P = 7.02e-13), macrophages (R = 0.436, P = 3.22e-17), and B cells (R = 0.474, P = 1.24e-20) (Fig. 4A). The current study suggested that EZH2 stimulated TIIC infiltration within LIHC, especially B cells, neutrophils, CD4 + T cells, DCs, and CD8 + T cells.
Fig. 3.
Spearman’s correlation for the EZH2 level and TIIC infiltrating degrees (TISIDB). A Connection of TILs levels with EZH2 expression. B In Spearman analysis, acT-CD4 and Th2 of TIL has the strongest correlation with the EZH2 level. C Relationship between immunosuppressant and the EZH2 level. D In Spearman analysis, CTLA4 and LAG3 of immune inhibitors has the strongest correlation with the EZH2 level. E Relationship between immunostimulators and the EZH2 level. F In Spearman analysis, CD276 and MICB of immunostimulators has the strongest correlation with EZH2 level; G Association of MHC molecules with the EZH2 level. H In Spearman analysis, TAP1 and HLA-E of MHC molecules has the strongest correlation with the EZH2 level. Negative and positive correlations are represented by blue and red, respectively. Colour intensity is directly proportional to correlation strength. MHC-major histocompatibility complex; TILs-tumour-infiltrating lymphocytes
Fig. 4.
Connection between EZH2 and TIIC infiltrating levels in LIHC. A EZH2 expression showed a positive relation with infiltrating degrees of CD8 + T cells, B cells, neutrophils macrophages, and DCs within LIHC based on TIMER database; B–E Scatter plots showing the connection of EZH2 expression with TIIC markers
Association of EZH2 expression with immune markers
Subsequently, an association between EZH2 levels with immune infiltration was analysed in 39 cancers derived from the TIMER database. EZH2 expression revealed a positive relationship with the degree of CD8 + T cell infiltration in 20 cancers, macrophages in 12 cancers, CD4 + T cells in 18 cancers, DCs in 19 cancers, and neutrophils in 24 cancers (Additional file 1: Figure S1). Figure 4A showed that EZH2 expression levels correlated positively with the infiltration degrees of CD4 + T cells (R = 0.378, P = 3.84e-13), CD8 + T cells (R = 0.284, P = 9.30e-08), DCs (R = 0.453, P = 1.38e-18), neutrophils (R = 0.374, P = 7.02e-13), macrophages (R = 0.436, P = 3.22e-17), and B cells (R = 0.474, P = 1.24e-20) in LIHC. Moreover, in some cancer types, including THYM, BRCA, HNSC, and KIRC, the immune infiltration degrees were markedly associated with EZH2 (Additional file 1: Figure S1). The results confirmed that EZH2 expression was related to immune markers and the levels of different T cells, TAMs, M1/M2 macrophages, monocytes, and DCs in LIHC. The scatter plot in Fig. 4B–E shows the correlation of EZH2 levels with macrophage phenotype markers (MS4A4A, CD63 for M2 macrophages; IFR5, COX2 for M1 macrophages; IL10, CD68 for TAMs) and monocytes (such as CSF1R and CD86) within LIHC. The TIMER database confirmed that EZH2 expression was markedly related to marker expression in TAMs, M2 macrophages, M1 macrophages, and monocytes (Fig. 4B–E). To investigate the relationship between EZH2 and TIIC levels in LIHC, the TIMER and GEPIA databases were adopted to analyse the connection between EZH2 and immune markers (Tables 2, 3). As shown in Table 2, after purity adjustment, EZH2 expression exhibited a positive relationship with marker expression in T cells and immune cells in HCC. EZH2 upregulation in M2 macrophages was associated with CD8 + T-cell and DC infiltration in LIHC. DCs can cause cancer migration by reducing CD8 + T-cell toxicity and elevating the number of Tregs [48]. In addition, the EZH2 level revealed a positive relationship with markers for Tregs and exhausted T cells (CTLA4, TIM-3, LAG3, Act-CD4, CD276, and PD-1). As shown in Table 3, we verified a similar correlation between EZH2 levels and monocytes, M2 macrophages, and TAM markers in the GEPIA database in LIHC. As a result, immune markers of diverse TAMs, T cells, DCs, monocytes, and M1/M2 macrophages were related to the EZH2 level in LIHC.
Table 2.
The correlation analysis between EZH2 and related immune cell genes and markers of immune cells in TIMER
| Immune cell | Gene markers | None | Purity | ||
|---|---|---|---|---|---|
| Cor | P-value | Cor | P-value | ||
| CD8 + T cell | CD8A | 0.180 | ** | 0.286 | *** |
| CD8B | 0.160 | * | 0.272 | *** | |
| T cell | CD6 | 0.161 | * | 0.305 | *** |
| CD3D | 0.235 | *** | 0.350 | *** | |
| CD3E | 0.161 | * | 0.214 | *** | |
| SH2D1A | 0.147 | * | 0.265 | *** | |
| TRAT1 | 0.141 | * | 0.267 | *** | |
| CD3G | 0.226 | *** | 0.324 | *** | |
| CD2 | 0.182 | ** | 0.323 | *** | |
| B cell | CD19 | 0.244 | *** | 0.318 | *** |
| FCRL2 | 0.173 | ** | 0.303 | *** | |
| KIAA0125 | 0.118 | 0.023 | 0.226 | *** | |
| TNFRSF17 | 0.130 | 0.012 | 0.264 | *** | |
| SPIB | 0.373 | *** | 0.464 | *** | |
| PNOC | 0.166 | * | 0.279 | *** | |
| CD79A | 0.131 | 0.012 | 0.245 | *** | |
| Monocyte | CD86 | 0.294 | *** | 0.449 | *** |
| CD115(CSF1R) | 0.153 | * | 0.296 | *** | |
| TAM | CD68 | 0.210 | *** | 0.301 | *** |
| IL10 | 0.220 | *** | 0.335 | *** | |
| M1 macrophage | IRF5 | 0.482 | *** | 0.487 | *** |
| COX2(PTGS2) | 0.088 | 0.092 | 0.212 | *** | |
| M2 macrophage | CD163 | 0.101 | 0.052 | 0.218 | *** |
| MS4A4A | 0.102 | 0.051 | 0.231 | *** | |
| Neutrophils | FPR1 | 0.218 | *** | 0.35 | *** |
| SIGLEC5 | 0.271 | *** | 0.402 | *** | |
| CSF3R | 0.285 | *** | 0.428 | *** | |
| FCGR3B | 0.175 | ** | 0.206 | ** | |
| CEACAM3 | 0.16 | 0.041 | 0.186 | ** | |
| CD116(ITGAM) | 0.289 | *** | 0.387 | *** | |
| Dendritic cell | CCL13 | 0.137 | * | 0.196 | ** |
| CD209 | 0.127 | 0.014 | 0.212 | *** | |
| HSD11B1 | 0.332 | *** | − 0.336 | *** | |
| HLA-DPB1 | 0.154 | * | 0.271 | *** | |
| HLA-DQB1 | 0.146 | * | 0.256 | *** | |
| HLA-DRA | 0.167 | * | 0.287 | *** | |
| HLA-DPA1 | 0.158 | * | 0.283 | *** | |
| BCDA-1(CD1C) | 0.114 | 0.029 | 0.207 | ** | |
| BDCA-4(NRP1) | 0.263 | *** | 0.291 | *** | |
| CD11c(ITGAX) | 0.348 | *** | 0.484 | *** | |
| Natural killer cell | XCL2 | 0.17 | ** | 0.258 | *** |
| KIR2DL1 | 0.039 | 0.455 | 0.018 | 0.742 | |
| KIR2DL3 | 0.168 | * | 0.205 | ** | |
| KIR2DL4 | 0.211 | *** | 0.241 | *** | |
| Mast cell | TPSB2 | − 0.037 | 0.479 | 0.001 | 0.986 |
| HDC | − 0.149 | * | − 0.121 | 0.025 | |
| Th1 | IFN-γ(IFNG) | 0.249 | *** | 0.333 | *** |
| TNF-α(TNF) | 0.282 | *** | 0.414 | *** | |
| STAT4 | 0.238 | *** | 0.303 | *** | |
| STAT1 | 0.387 | *** | 0.246 | *** | |
| Th2 | GATA3 | 0.172 | ** | 0.309 | *** |
| STAT6 | 0.193 | ** | 0.179 | ** | |
| STAT5A | 0.329 | *** | 0.389 | *** | |
| Tfh | BCL6 | 0.201 | *** | 0.205 | ** |
| IL21 | 0.147 | * | 0.188 | ** | |
| Th17 | STAT3 | 0.168 | * | 0.209 | *** |
| IL17A | 0.072 | 0.168 | 0.075 | 0.164 | |
| Effector T cell | CX3CR1 | 0.187 | ** | 0.242 | *** |
| FGFBP2 | − 0.085 | 0.103 | − 0.07 | 0.195 | |
| FCGR3A | 0.295 | *** | 0.383 | *** | |
| Effector memory T cell | PD-1 (PDCD1) | 0.295 | *** | 0.391 | *** |
| DUSP4 | 0.256 | *** | 0.363 | *** | |
| Central memory T cell | CCR7 | 0.083 | 0.111 | 0.219 | *** |
| SELL | 0.196 | ** | 0.308 | *** | |
| IL7R | 0.093 | 0.073 | 0.201 | ** | |
| Resident memory T cell | CD69 | 0.109 | 0.035 | 0.222 | *** |
| ITGAE | 0.279 | *** | 0.280 | *** | |
| CXCR6 | 0.156 | * | 0.288 | *** | |
| MYADM | 0.354 | *** | 0.388 | *** | |
| Exhausted T cell | TIM-3 (HAVCR2) | 0.310 | *** | 0.472 | *** |
| TIGIT | 0.262 | *** | 0.385 | *** | |
| LAG3 | 0.287 | *** | 0.332 | *** | |
| CXCL13 | 0.264 | *** | 0.342 | *** | |
| LAYN | 0.185 | ** | 0.257 | *** | |
| Resting treg T cell | FOXP3 | 0.191 | ** | 0.262 | *** |
| IL2RA | 0.252 | *** | 0.377 | *** | |
| Effector treg T cell | CTLA4 | 0.296 | *** | 0.404 | *** |
| CCR8 | 0.383 | *** | 0.479 | *** | |
Cor, R-value of Spearman correlation; Purity, tumor purity-adjusted correlation. None, correlation without adjustment. *P < 0.01; **P < 0.001; ***P < 0.0001
Table 3.
Correlation analysis between EZH2 markers of immune cells in GEPIA2
| Immune cell | Gene markers | Tumor | Tumor-sum | Normal | Normal-sum | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cor | P-value | Cor | P-value | Cor | P-value | Cor | P-value | ||
| CD8 + T cell | CD8A | 0.2 | *** | 0.18 | ** | 0.71 | *** | 0.69 | *** |
| CD8B | 0.15 | *** | 0.62 | *** | |||||
| T cell | CD6 | 0.029 | 0.11 | 0.17 | * | 0.49 | ** | 0.71 | *** |
| CD3D | 0.23 | *** | 0.65 | *** | |||||
| CD3E | 0.13 | 0.01 | 0.59 | *** | |||||
| SH2D1A | 0.13 | 0.011 | 0.68 | *** | |||||
| TRAT1 | 0.034 | 0.51 | 0.65 | *** | |||||
| CD3G | 0.24 | *** | 0.63 | *** | |||||
| CD2 | 0.15 | * | 0.52 | ** | |||||
| B cell | CD19 | 0.11 | 0.03 | 0.21 | *** | 0.39 | * | 0.36 | *** |
| FCRL2 | 0.076 | 0.14 | 0.57 | *** | |||||
| KIAA0125 | 0.15 | * | 0.53 | *** | |||||
| TNFRSF17 | 0.094 | 0.071 | 0.6 | *** | |||||
| SPIB | 0.14 | * | 0.43 | * | |||||
| PNOC | 0.1 | 0.047 | 0.59 | *** | |||||
| CD79A | 0.083 | 0.11 | 0.51 | ** | |||||
| Monocyte | CD86 | 0.32 | *** | 0.27 | *** | 0.55 | *** | 0.55 | *** |
| CD115(CSF1R) | 0.22 | *** | 0.49 | ** | |||||
| TAM | CD68 | 0.23 | *** | 0.26 | *** | 0.53 | *** | 0.57 | *** |
| IL10 | 0.21 | *** | 0.43 | * | |||||
| M1 Macrophage | IRF5 | 0.44 | *** | 0.37 | *** | 0.44 | * | 0.44 | * |
| COX2(PTGS2) | 0.035 | 0.5 | 0.27 | 0.055 | |||||
| M2 Macrophage | CD163 | 0.16 | * | 0.12 | 0.017 | 0.38 | * | 0.47 | ** |
| MS4A4A | 0.16 | * | 0.48 | ** | |||||
| Neutrophils | FPR1 | 0.24 | *** | 0.35 | *** | 0.31 | 0.028 | 0.49 | ** |
| SIGLEC5 | 0.27 | *** | 0.17 | 0.24 | |||||
| CSF3R | 0.21 | *** | 0.5 | ** | |||||
| FCGR3B | 0.03 | 0.56 | 0.32 | 0.021 | |||||
| CEACAM3 | 0.15 | * | 0.25 | 0.078 | |||||
| CD116(ITGAM) | 0.34 | *** | 0.69 | *** | |||||
| Natural killer cell | XCL2 | 0.049 | 0.35 | 0.15 | * | 0.67 | *** | 0.6 | *** |
| KIR2DL1 | 0.11 | 0.038 | 0.62 | *** | |||||
| KIR2DL3 | 0.21 | *** | 0.22 | 0.12 | |||||
| KIR2DL4 | 0.2 | *** | 0.32 | 0.025 | |||||
| Dendritic cell | CCL13 | 0.065 | 0.22 | 0.068 | 0.19 | 0.41 | * | 0.64 | *** |
| CD209 | 0.16 | * | 0.44 | * | |||||
| HSD11B1 | − 0.22 | *** | 0.094 | 0.51 | |||||
| HLA-DPB1 | 0.22 | *** | 0.58 | *** | |||||
| HLA-DQB1 | 0.088 | 0.09 | 0.52 | *** | |||||
| HLA-DRA | 0.22 | *** | 0.59 | *** | |||||
| HLA-DPA1 | 0.2 | ** | 0.54 | *** | |||||
| BCDA-1(CD1C) | 0.17 | ** | 0.63 | *** | |||||
| BDCA-4(NRP1) | 0.28 | *** | 0.41 | * | |||||
| CD11c(ITGAX) | 0.25 | *** | 0.5 | *** | |||||
| Mast cell | TPSB2 | − 0.042 | 0.42 | 0.031 | 0.56 | 0.096 | 0.51 | − 0.097 | 0.5 |
| HDC | − 0.059 | 0.26 | − 0.083 | 0.57 | |||||
| Th1 | IFN-γ(IFNG) | 0.21 | *** | 0.29 | *** | 0.65 | *** | 0.84 | *** |
| TNF-α(TNF) | 0.11 | .031 | 0.33 | 0.02 | |||||
| STAT4 | 0.081 | 0.12 | 0.61 | *** | |||||
| STAT1 | 0.21 | *** | 0.8 | *** | |||||
| Th2 | GATA3 | 0.18 | ** | 0.29 | *** | 0.19 | 0.19 | 0.62 | *** |
| STAT6 | 0.18 | ** | 0.57 | *** | |||||
| STAT5A | 0.22 | *** | 0.69 | *** | |||||
| Tfh | BCL6 | 0.1 | 0.054 | 0.17 | ** | 0.011 | 0.94 | 0.11 | 0.46 |
| IL21 | 0.12 | 0.023 | 0.31 | 0.031 | |||||
| Th17 | STAT3 | 0.29 | *** | 0.26 | *** | 0.11 | 0.44 | 0.2 | 0.16 |
| IL17A | 0.0028 | 0.96 | 0.25 | 0.085 | |||||
| Effector T cell | CX3CR1 | 0.14 | * | 0.27 | *** | 0.22 | 0.13 | 0.45 | * |
| FGFBP2 | − 0.098 | 0.06 | 0.074 | 0.61 | |||||
| FCGR3A | 0.29 | *** | 0.5 | *** | |||||
| Effector memory T cell | PD-1 (PDCD1) | 0.22 | *** | 0.39 | *** | 0.73 | *** | 0.72 | *** |
| DUSP4 | 0.29 | *** | 0.54 | *** | |||||
| Central memory T cell | CCR7 | 0.064 | 0.22 | 0.12 | 0.023 | 0.58 | *** | 0.68 | *** |
| SELL | 0.12 | 0.024 | 0.54 | *** | |||||
| IL7R | 0.029 | 0.58 | 0.56 | *** | |||||
| Resident memory T cell | CD69 | 0.039 | 0.46 | 0.28 | *** | 0.63 | *** | 0.64 | *** |
| ITGAE | 0.23 | *** | 0.092 | 0.52 | |||||
| CXCR6 | 0.14 | * | 0.44 | * | |||||
| MYADM | 0.29 | *** | 0.24 | 0.1 | |||||
| Exhausted T cell | TIM-3(HAVCR2) | 0.13 | 0.014 | 0.3 | *** | 0.53 | *** | 0.65 | *** |
| PD-1 (PDCD1) | 0.22 | *** | 0.73 | *** | |||||
| TIGIT | 0.22 | *** | 0.61 | *** | |||||
| LAG3 | 0.2 | ** | 0.48 | ** | |||||
| CXCL13 | 0.097 | 0.064 | 0.31 | 0.028 | |||||
| LAYN | 0.087 | 0.095 | 0.52 | *** | |||||
| Resting treg T cell | FOXP3 | − 0.002 | 0.96 | 0.15 | * | 0.42 | * | 0.6 | *** |
| IL2RA | 0.19 | ** | 0.37 | * | |||||
| Effector treg T cell | CTLA4 | 0.23 | *** | 0.25 | *** | 0.65 | *** | 0.68 | *** |
| CCR8 | 0.21 | *** | 0.51 | ** | |||||
| TNFRSF9 | 0.015 | 0.77 | 0.65 | *** | |||||
Tumour, correlation analysis in tumour tissue of TCGA; Normal, correlation analysis in normal tissue of TCGA. *P < 0.01; **P < 0.001; ***P < 0.0001
Expression levels of EZH2 in LIHC
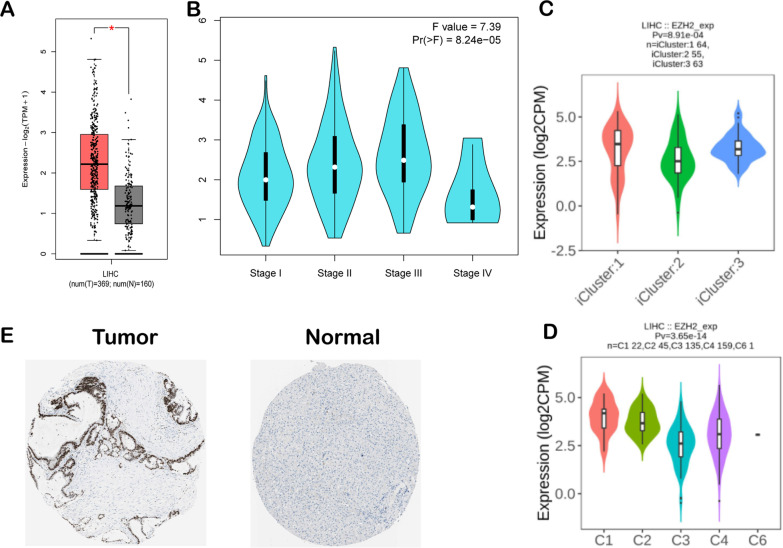
The GEPIA database demonstrated that the expression level of EZH2 in hepatocellular carcinoma samples increased in relation to healthy controls (Fig. 5A). Furthermore, EZH2 was related to the tumour immune microenvironment (TIME). According to the GEPIA database, compared with different stages of LIHC, the EZH2 expression level was higher in stage III and lower in stage IV (Fig. 5B). Vesteinn Thorsson's study clustered six immune subtypes for cancer and revealed that the immune subtypes of cancer might play a key role in predicting disease outcome [49]. According to our results, we detected that EZH2 expression was the highest in C1 (wound healing) and C2 (IFN-γ), whereas it was the lowest in C3 (inflammation) in TISIDB (Fig. 5D). Moreover, we analysed the correlation between EZH2 expression levels and different molecular subtypes in hepatocellular carcinoma. We discovered EZH2 expression, which was highest in iCluster: 1 type and lowest in iCluster: 2 type, in four molecular types (Fig. 5C). According to the HPA database, the increased staining intensity of the EZH2 protein level was detected in tumour tissues compared with noncarcinoma tissues (Fig. 5E).
Fig. 5.
EZH2 expression levels in LIHC. A LIHC and healthy samples from the GEPIA database; B EZH2 expression in diverse liver hepatocellular carcinoma stages in the GEPIA database; C EZH2 levels within diverse LIHC molecular subtypes derived from the TISIDB database; D EZH2 expression within diverse LIHC immune subtypes in the TISIDB database; E EZH2 protein expression within LIHC samples in relation to healthy samples in the Human Protein Atlas data. T: EZH2 protein expression within cancer samples (quantity: 75–25%; intensity: strong; staining: high) https://www.proteinatlas.org/ENSG00000106462-EZH2/pathology/liver+cancer#img; N: Protein levels of EZH2 in normal tissue (staining: Not detected; intensity: weak, quantity: < 25%) https://www.proteinatlas.org/ENSG00000106462-EZH2/tissue/liver#img;
Coexpression genes were correlated with EZH2 in LIHC
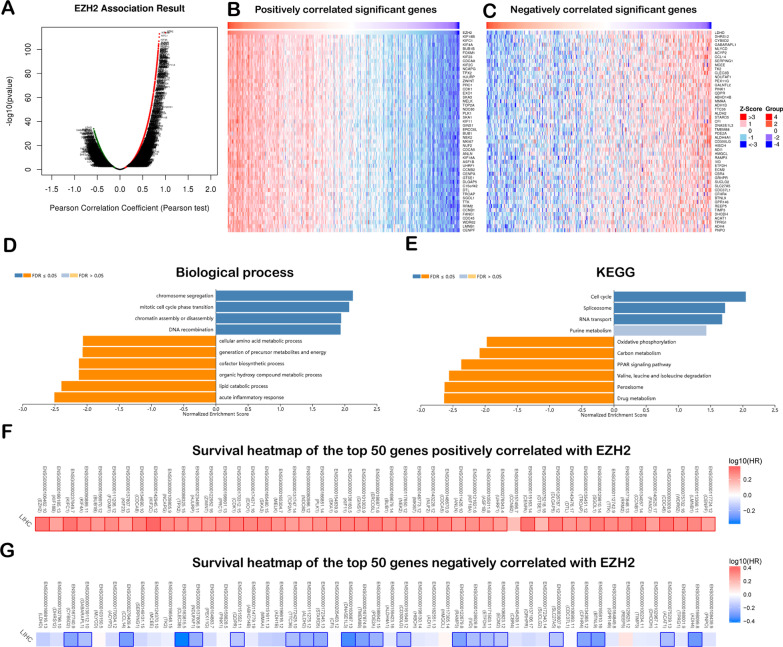
The biological effect of EZH2 on LIHC was probably connected with the neighbouring gene expression within LIHC. EZH2 coexpression profiles were checked through the ‘LinkFinder’ module LinkedOmics. The EZH2 coexpression gene levels within 371 LIHC cases were analysed through the LinkedOmics database (Additional file 3: Table S2). We discovered that 12,451 genes were positively related to EZH2, whereas 7,471 were negatively correlated with EZH2 (Fig. 6A). The heatmap presented 50 gene sets positively and negatively correlated with EZH2 (Fig. 6B, C).
Fig. 6.
Co-expression genes of EZH2 within LIHC (LinkedOmics). A Connection of EZH2 with DEGs in LIHC analysed by the Spearman test. Red and green indicate the positive and negative correlation, respectively; B and C Heatmap shows the 50 most significantly positive or negative genes within LIHC; D and E GSEA for analysing the GO annotation and KEGG pathway enriched with EZH2 co-expressed genes in LIHC; F Survival heatmaps show the 50 most significantly positive or negative genes within LIHC
GO and KEGG analysis of EZH2-related coexpressed genes in LIHC
GO analysis conducted using GSEA in LinkedOmics revealed that EZH2 coexpressed genes were mainly related to DNA recombination, mitotic cell cycle phase transition, and chromosome segregation (Fig. 6D). Through KEGG pathway analysis, coexpressed genes revealed a major enrichment of microRNAs in cancer, the cell cycle, the spliceosome, and pyrimidine metabolism (Fig. 6E). The top 50 most remarkably positive genes were the high-risk factors for LIHC, and 49 of them had large HRs (Fig. 6F). By contrast, 22 markedly negative genes had low HRs (Fig. 6G).
Discussion
EZH2 is a member of PcGs and an epigenetic regulator that can inhibit transcription [16]. EZH2 plays a vital role in cell lineage determination and related signalling pathways and is a master regulator of cell cycle progression, autophagy, apoptosis, DNA damage repair, and cellular senescence inhibition [23–26]. Recent studies have revealed that EHZ2, as a modifier associated with epigenetic regulation and immune function, can promote local and systemic anticancer immunity by modulating TH-1 chemokine levels and affecting MDSC CD8 + T-cell infiltration [27–31]. Hence, our study illustrates that EZH2 expression predicts HCC survival. EZH2 upregulation indicates dismal survival. Consequently, the current study sheds more light on EZH2’s regulatory function in LIHC through comprehensive and systematic analysis and studies.
In the current study, the EZH2 level exhibited a remarkable relationship with lymphocyte infiltration and immune responses in LIHC. Figure 4A shows that EZH2 expression was positively related to the TIIC infiltrating levels of CD4 + T cells, macrophages, CD8 + T cells, B cells, neutrophils, and DCs in LIHC. EZH2 expression was related to gene markers for diverse T cells, M1/M2 macrophages, TAMs, DCs, and monocytes in LIHC (Tables 2, 3). T-cell exhaustion is an important cause of poor antitumor immunity; hence, suppressing such exhaustion is a crucial immunotherapeutic strategy to manage cancer [50–52]. According to the obtained results, EZH2 upregulation revealed a positive correlation with several critical genes associated with exhausted T cells, such as TIM-3, PD-1, and LAG3. These T cells play the role of therapeutic targets for immunotherapy [53, 54]. EZH2 upregulation in M2 macrophages was associated with CD8 + T-cell and DC infiltration in LIHC. DCs can cause cancer migration by decreasing CD8 + T-cell toxicity and elevating the number of Tregs [48]. Our analysis revealed that EZH2 regulates the tumour immune microenvironment in LIHC, which is related to the activation and regulation of B-cell, DC, and T-cell immune responses.
EZH2 mRNA expression levels in normal tissues and liver hepatocellular carcinoma were analysed in detail based on GEPIA, Oncomine, and TISIDB online datasets. The expression level of EZH2 in LIHC was higher than that in normal tissues. Figure 3E displays the correlation between EZH2 levels and 45 types of immunostimulators. The current article assessed the connection between EZH2 and immunity based on the TISIDB database. As a result, EZH2 was closely associated with immunostimulators (MICB, CD276), lymphocytes (activated CD4 T cells, Th2), MHC molecules (including TAP1, HLA-E), and immunoinhibitors (CTLA4, LAG3). However, LIHC can be subdivided into several molecular subtypes. Based on the aforementioned analysis, the TISIDB database revealed that EZH2 exhibited the greatest expression in iCluster: 1, followed by iCluster: 3, and less in iCluster: 2 (Fig. 5D). The EZH2 expression level is a different indistinct immune subtype of hepatocellular carcinoma, and C1 (wound healing) and C2 (IFN-γ) exhibited the highest expression in relation to the remaining four subtypes (Fig. 5C). According to our comprehensive analysis of EZH2 gene expression in LIHC and different databases of different subtypes, EZH2 might be closely related to immunological properties in the microenvironment.
Based on the GEPIA and K-Meier plotter databases, we further analysed EZH2-related survival (OS and RFS) and discovered that EZH2 is a detrimental prognostic factor for LIHC. High EZH2 expression was correlated with a poor prognosis in LIHC. The EZH2 expression level of stage III was higher and that of stage IV, indicating the possible role of EZH2 in liver hepatocellular carcinoma prognosis.
In conclusion, the upregulation of EZH2 was markedly related to TIIC infiltration levels (B cells, CD8 + T cells, CD4 + T cells, DCs, neutrophils, and many functional T cells). EZH2 has an immune-stimulating effect, which may be a critical factor that promotes T-cell exhaustion within LIHC. According to these results, EZH2 has a critical function within the immune microenvironment and deserves to be regarded as a prognostic marker and immunotherapeutic target for hepatocellular carcinoma.
Supplementary Information
Additional file1: Figure S1. Correlation of EZH2 expression with immune infiltration level in different tumor types via TIMER database.
Additional file2: Table S1. EZH2 expression in cancers verus normal tissue in oncomine database.
Additional file3: Table S2. The co-expressed genes of EZH2 in LIHC.
Acknowledgements
We acknowledge TCGA, TIMER, GEPIA2, KEGG and so on databases for providing their platforms and contributors for uploading their meaningful datasets. The authors sincerely thank all participants involved in this study.
Abbreviations
- EZH2
Enhancer of zeste homolog 2
- LIHC
Liver hepatocellular carcinoma
- KM
Kaplan–Meier
- DC
Dendritic cell
- SCNA
Somatic copy number alteration
- FDR
False discovery rate
- FC
Fold-change
- LNM
Lymph node metastasis
- TME
Tumor microenvironment
Author contributions
SYW designed the project. DAW, JD and HTS was responsible for supervision the work. DAW, ZYX, LYY, XFL and YZ collected and analyzed the data. SYW and DAW interpreted and composed the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The data that support the findings of this study are available in TIMER (https://cistrome.shinyapps.io/timer/), OnCoLnc Database (http://www.oncolnc.org/), Kaplan–Meier Plotter (http://kmplot.com/), GEPIA2 (http://gepia2.cancer-pku.cn/), KEGG database (https://www.kegg.jp/), The HPA database (https://www.proteinatlas.org/) and LinkedOmics Database (http://www.linkedomics.org).
Declarations
Ethics approval and consent to participate
All methods were carried out in accordance with relevant guidelines and regulations. TCGA and TIME belong to public databases. Users can download relevant data for free for research and publish relevant articles. Our study is based on open-source data, and does not involve animal experiments, human specimens, no ethics-related issues, and other conflicts of interest.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Da-an Wang, Email: wangdaan2021@163.com.
Jie Dong, Email: dongjie202107@163.com.
Hong-tao Sun, Email: sunhongtao2019@163.com.
References
- 1.Naoum GE, Morkos M, Kim B, Arafat W. Novel targeted therapies and immunotherapy for advanced thyroid cancers. Mol Cancer. 2018;17:51. doi: 10.1186/s12943-018-0786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24:1518–1524. doi: 10.1158/1078-0432.CCR-17-0289. [DOI] [PubMed] [Google Scholar]
- 3.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 4.Dueñas-González A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer. 2005;4:38. doi: 10.1186/1476-4598-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weintraub K. Take two: combining immunotherapy with epigenetic drugs to tackle cancer. Nat Med. 2016;22:8–10. doi: 10.1038/nm0116-8. [DOI] [PubMed] [Google Scholar]
- 6.Emran AA, Chatterjee A, Rodger EJ, Tiffen JC, Gallagher SJ, Eccles MR, Hersey P. Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. 2019;40:328–344. doi: 10.1016/j.it.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Cronin KA, Lake AJ, Scott S, Sherman RL, Noone A-M, Howlader N, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2018;124:2785–2800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 9.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Makarova-Rusher OV, Medina-Echeverz J, Duffy AG, Greten TF. The yin and yang of evasion and immune activation in HCC. J Hepatol. 2015;62:1420–1429. doi: 10.1016/j.jhep.2015.02.038. [DOI] [PubMed] [Google Scholar]
- 11.DiStefano JK, Davis B. Diagnostic and prognostic potential of AKR1B10 in human hepatocellular carcinoma. Cancers (Basel) 2019 doi: 10.3390/cancers11040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thein H-H, Qiao Y, Zaheen A, Jembere N, Sapisochin G, Chan KKW, et al. Cost-effectiveness analysis of treatment with non-curative or palliative intent for hepatocellular carcinoma in the real-world setting. PLoS ONE. 2017;12:e0185198. doi: 10.1371/journal.pone.0185198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu AX, Kang Y-K, Yen C-J, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20:282–296. doi: 10.1016/S1470-2045(18)30937-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Ha J, Lopez A, Bhuket T, Liu B, Wong RJ. Medicaid and uninsured hepatocellular carcinoma patients have more advanced tumor stage and are less likely to receive treatment. J Clin Gastroenterol. 2018;52:437–443. doi: 10.1097/MCG.0000000000000859. [DOI] [PubMed] [Google Scholar]
- 15.Kulik L, Heimbach JK, Zaiem F, Almasri J, Prokop LJ, Wang Z, et al. Therapies for patients with hepatocellular carcinoma awaiting liver transplantation: a systematic review and meta-analysis. Hepatology. 2018;67:381–400. doi: 10.1002/hep.29485. [DOI] [PubMed] [Google Scholar]
- 16.Pasini D, Di Croce L. Emerging roles for polycomb proteins in cancer. Curr Opin Genet Dev. 2016;36:50–58. doi: 10.1016/j.gde.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Yin X, Yang S, Zhang M, Yue Y. The role and prospect of JMJD3 in stem cells and cancer. Biomed Pharmacother. 2019;118:109384. doi: 10.1016/j.biopha.2019.109384. [DOI] [PubMed] [Google Scholar]
- 19.He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E, Kim M, Woo D-H, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Lee Y, Lu X, Song B, Fong K-W, Cao Q, et al. Polycomb- and methylation-independent roles of EZH2 as a transcription activator. Cell Rep. 2018;25:2808–2820.e4. doi: 10.1016/j.celrep.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nutt SL, Keenan C, Chopin M, Allan RS. EZH2 function in immune cell development. Biol Chem. 2020;401:933–943. doi: 10.1515/hsz-2019-0436. [DOI] [PubMed] [Google Scholar]
- 24.Yao Y, Hu H, Yang Y, Zhou G, Shang Z, Yang X, et al. Downregulation of enhancer of Zeste homolog 2 (EZH2) is essential for the Induction of autophagy and apoptosis in colorectal cancer cells. Genes (Basel) 2016 doi: 10.3390/genes7100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T, Teo YV, Evans SA, Neretti N, Sedivy JM. Regulation of cellular senescence by polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep. 2018;22:3480–3492. doi: 10.1016/j.celrep.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batool A, Jin C, Liu Y-X. Role of EZH2 in cell lineage determination and relative signaling pathways. Front Biosci (Landmark Ed) 2019;24:947–960. doi: 10.2741/4760. [DOI] [PubMed] [Google Scholar]
- 27.Ennishi D, Takata K, Béguelin W, Duns G, Mottok A, Farinha P, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 2019;9:546–563. doi: 10.1158/2159-8290.CD-18-1090. [DOI] [PubMed] [Google Scholar]
- 28.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagarsheth N, Peng D, Kryczek I, Wu K, Li W, Zhao E, et al. PRC2 Epigenetically silences Th1-type chemokines to suppress effector T-Cell trafficking in colon cancer. Cancer Res. 2016;76:275–282. doi: 10.1158/0008-5472.CAN-15-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Liu M, Sun H, Feng Y, Xu L, Chan AWH, et al. Hepatoma-intrinsic CCRK inhibition diminishes myeloid-derived suppressor cell immunosuppression and enhances immune-checkpoint blockade efficacy. Gut. 2018;67:931–944. doi: 10.1136/gutjnl-2017-314032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S, Wang Z, Zhou J, Huang J, Zhou L, Luo J, et al. EZH2 Inhibitor GSK126 suppresses antitumor immunity by driving production of myeloid-derived suppressor cells. Cancer Res. 2019;79:2009–2020. doi: 10.1158/0008-5472.CAN-18-2395. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharya S, Dunn P, Thomas CG, Smith B, Schaefer H, Chen J, et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci Data. 2018;5:180015. doi: 10.1038/sdata.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 36.Nirmal AJ, Regan T, Shih BB, Hume DA, Sims AH, Freeman TC. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res. 2018;6:1388–1400. doi: 10.1158/2326-6066.CIR-18-0342. [DOI] [PubMed] [Google Scholar]
- 37.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lánczky A, Nagy Á, Bottai G, Munkácsy G, Szabó A, Santarpia L, Győrffy B. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 39.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 40.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 41.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravelli A, Roviello G, Cretella D, Cavazzoni A, Biondi A, Cappelletti MR, et al. Tumor-infiltrating lymphocytes and breast cancer: beyond the prognostic and predictive utility. Tumour Biol. 2017;39:1010428317695023. doi: 10.1177/1010428317695023. [DOI] [PubMed] [Google Scholar]
- 46.Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30:2678–2683. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 47.Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PMC free article] [PubMed] [Google Scholar]
- 48.Sawant A, Hensel JA, Chanda D, Harris BA, Siegal GP, Maheshwari A, Ponnazhagan S. Depletion of plasmacytoid dendritic cells inhibits tumor growth and prevents bone metastasis of breast cancer cells. J Immunol. 2012;189:4258–4265. doi: 10.4049/jimmunol.1101855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 51.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zarour HM. Reversing T-cell dysfunction and exhaustion in cancer. Clin Cancer Res. 2016;22:1856–1864. doi: 10.1158/1078-0432.CCR-15-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file1: Figure S1. Correlation of EZH2 expression with immune infiltration level in different tumor types via TIMER database.
Additional file2: Table S1. EZH2 expression in cancers verus normal tissue in oncomine database.
Additional file3: Table S2. The co-expressed genes of EZH2 in LIHC.
Data Availability Statement
The data that support the findings of this study are available in TIMER (https://cistrome.shinyapps.io/timer/), OnCoLnc Database (http://www.oncolnc.org/), Kaplan–Meier Plotter (http://kmplot.com/), GEPIA2 (http://gepia2.cancer-pku.cn/), KEGG database (https://www.kegg.jp/), The HPA database (https://www.proteinatlas.org/) and LinkedOmics Database (http://www.linkedomics.org).