Abstract
Insertion of new material into the Escherichia coli peptidoglycan (PG) sacculus between the cytoplasmic membrane and the outer membrane requires a well-organized balance between synthetic and hydrolytic activities to maintain cell shape and avoid lysis. Since most bacteria carry multiple enzymes carrying the same type of PG hydrolytic activity, we know little about the specific function of given enzymes. Here we show that the DD-carboxy/endopeptidase PBP4 localizes in a PBP1A/LpoA and FtsEX dependent fashion at midcell during septal PG synthesis. Midcell localization of PBP4 requires its non-catalytic domain 3 of unknown function, but not the activity of PBP4 or FtsE. Microscale thermophoresis with isolated proteins shows that PBP4 interacts with NlpI and the FtsEX-interacting protein EnvC, an activator of amidases AmiA and AmiB, which are needed to generate denuded glycan strands to recruit the initiator of septal PG synthesis, FtsN. The domain 3 of PBP4 is needed for the interaction with NlpI and EnvC, but not PBP1A or LpoA. In vivo crosslinking experiments confirm the interaction of PBP4 with PBP1A and LpoA. We propose that the interaction of PBP4 with EnvC, whilst not absolutely necessary for mid-cell recruitment of either protein, coordinates the activities of PBP4 and the amidases, which affects the formation of denuded glycan strands that attract FtsN. Consistent with this model, we found that the divisome assembly at midcell was premature in cells lacking PBP4, illustrating how the complexity of interactions affect the timing of cell division initiation.
Author summary
Peptidoglycan biosynthesis is a major target for antibacterials. The covalently closed peptidoglycan mesh, called sacculus, protects the bacterium from lysis due to its turgor. Sacculus growth is facilitated by the balanced activities of synthases and hydrolases, and disturbing this balance leads to cell lysis and bacterial death. Because of the large number and possible redundant functions of peptidoglycan hydrolases, it has been difficult to decipher their individual functions. In this paper we show that the DD-endopeptidase PBP4 localizes at midcell during septal peptidoglycan synthesis in Escherichia coli and is involved in the timing of the assembly and activation of the division machinery. This shows that inhibition of certain hydrolases could weaken the cells and might enhance antibiotic action.
Introduction
The peptidoglycan (PG) layer is sandwiched between the cytoplasmic membrane (CM) and the outer membrane (OM) of the Gram-negative bacterium Escherichia coli forming a covalently closed network of glycan strands that are interconnected by short peptides [1]. The PG layer maintains the shape of the bacterium by stabilizing the cell against its high internal osmotic pressure. To proliferate, the rod-shaped bacterium grows in length and then divides by binary fission into two equally sized daughter cells, with nascent PG inserted into the lateral cell wall and septum.
The basic precursor (lipid II) for PG synthesis is the β-(1,4) linked disaccharide N-acetylglucosamine-N-acetylmuramic acid (GlcNAc-MurNAc) with an undecaprenol-pyrophosphate linked to C1 of MurNAc and a stem peptide with the sequence L-alanine-D-iso-glutamate-diaminopimelic acid (Dap)-D-alanine-D-alanine coupled to the C3 of MurNAc. Lipid II is used by glycosyltransferases (GTases), which polymerize the glycan chains, and TPases, which crosslink the stem peptides. The main PG synthases in E. coli are the class A PBPs (PBP1A and PBP1B) with GTase and TPase activities [2–6], the integral membrane proteins RodA and FtsW with GTase activity [7–9] and their partner TPases PBP2 and PBP3, respectively [10–13].
Binary fission in E. coli is initiated by the assembly of the Z-ring at midcell, which consists of a network of dynamic FtsZ filaments that are anchored to the cytoplasmic membrane by FtsA and ZipA [14,15]. The Z-ring also recruits the PG synthases PBP1A and PBP1B to midcell [2,16]. A number of Z-associated proteins play a role in the organization and dynamics of the ring [17–22]. Of these, the integral membrane protein FtsX interacts with FtsA [23] and its cytoplasmic ATPase partner protein FtsE interacts with FtsZ [24]. FtsE and FtsX localize early to midcell, together with the Z-ring [25]. The recruitment of the other cell division proteins occurs with a time delay of about 20% of the cell division cycle, depending on the growth condition [26–28]. FtsK, FtsBLQ, FtsW, PBP3 and FtsN localize in an interdependent fashion. The FtsBLQ complex inhibits the PG synthase activities of PBP3, FtsW and PBP1B until it is outcompeted by the accumulation of FtsN that relieves this inhibition and initiates septum synthesis [29–32].
The maintenance of the PG sacculus is a dynamic process, which is achieved by the balanced activities of PG synthases and hydrolases that ensures a safe extension of the PG layer without defects that would cause cell lysis [33]. Beta-lactam antibiotics disturb this balance by inactivating the transpeptidase (TPase) activity of the penicillin binding proteins (PBP)s causing lysis of growing cells. We currently know more than twenty PG hydrolases that can cleave either in the glycan chains or peptides within the PG layer, but how their activities are regulated is poorly understood. This is particularly true for the PG endopeptidases [33].
The hydrolases are present in greater redundancy than the synthases [33,34]. The carboxypeptidases (CPase) PBP5, PBP6A and PBP6B remove the terminal D-Ala from nascent pentapeptides [35–38]. The endopeptidases (EPases) PBP4, PBP7, MepA, MepM, MepH, and MepS cleave the crosslinks in the PG [39–41]. The amidases AmiA-D hydrolyze the bond between MurNAc and L-alanine at position 1 of the stem peptide [25,42–44]. During cell division, as a complex, FtsEX recruits EnvC, which activates AmiA and AmiB that are needed for septum cleavage [43,45]. FtsE versions without ATPase activity still localize at midcell and recruit as FtsEX-EnvC complex but the latter is not able to activate the amidases [46]. Presumably an ATPase dependent conformational change in FtsEX induces a conformational change in EnvC, which is then able to relieve an autoinhibitory alpha-helix from the active site of AmiA and AmiB, activating them [47]. The third amidase AmiC is activated by the OM-anchored lipoprotein NlpD [25,48] and all three amidases AmiA-C are activated under certain stress conditions by the recently identified ActS [49,50]. Finally, the lytic transglycosylases Slt70 and MltA-G cleave the glycan strands to release 1,6-anhydroMurNAc containing turnover products [51–53]. Although the large number of hydrolases may reflect the need to adapt to various environmental conditions [16,50], this still begs the question of how the cell controls their potentially dangerous activities to avoid cell lysis.
Recently, the OM bound lipoprotein NlpI was shown to interact with similar affinity with several endopeptidases, suggesting that NlpI acts as an interaction hub for endopeptidases to regulate their activities by competing protein-protein interactions. [54,55].
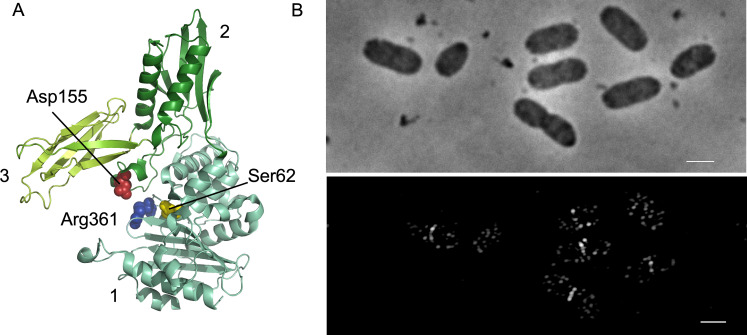
While investigating the specific function of various PG hydrolases, we found that PBP4 localizes specifically at midcell as part of the division machinery. PBP4 is a periplasmic endopeptidase [56] with a C-terminal amphipathic alpha-helix that associates with membranes [57] and has three domains that are assembled in an unusual way [58] (Fig 1A). A non-catalytical domain of unknown function (domain 2), which is inserted into the transpeptidase domain 1, and a third domain (domain 3), which is inserted into domain 2. Domain 3 is positioned above the active site (Serine 62, in domain 1), and might be involved in substrate binding or regulation [58,59] (Fig 1A). Site directed mutagenesis suggested that residues from domain 1 (R361) and 2 (D155) are important for the endopeptidase activity [60]. Deletion of dacB, which encodes PBP4, does not have morphological consequences but causes a decrease in the percentage of PG crosslinks and of PG-attached lipoprotein [61] and an increased sensitivity to bile salts [62]. A dacB deletion aggravates the morphological defects of a dacA (PBP5) deletion mutants [40], and the chaining phenotype of the ΔamiC strain [45]. PBP4 has also been reported to be involved in exopolysaccharide synthesis regulation [63]. Overproduction of PBP4 is toxic [64] presumably because the enhanced cleavage of peptide cross-links weakens the PG mesh and eventually causes lysis. There is litttle knowledge about the spatio-temporal regulation of PBP4 activity and its coordination with cell cycle events. Here we determined the cellular localization of PBP4 during the cell cycle of E. coli. The timing of localization at midcell and interactions of PBP4 support a role in remodeling of PG synthesized by the divisome.
Fig 1. Structure of PBP4 and its localization at midcell and in the lateral wall during fast growth.
A. Crystal structure of PBP4 in which the three domains (numbers 1–3) and some of the residues that are essential for its endopeptidase activity are indicated. B. MC4100 cells were grown exponentially in TY medium at 37°C to an OD600 of 0.3 and fixed and immunolabeled with BW25113ΔdacB pre-adsorbed anti-PBP4. Upper panel phase contrast image corresponding to the SIM fluorescence image in the lower panel. Scale bar equals 2 μm.
Results
PBP4 localizes to the lateral wall and at midcell
To determine the localization of PBP4 in E. coli cells as a function of the bacterial cell cycle, we generated an antiserum against PBP4 and removed unspecific antibodies by adsorption to cells of a dacB deletion mutant that lacks PBP4. The purified antibodies were specific for PBP4 (S1 Fig) and used to immunolabel the wild-type strain MC4100 grown in rich (TY) medium at 37°C. PBP4 localized strongly at midcell and to a lesser extent in the lateral wall in wild-type cells (Figs 1 and 2).
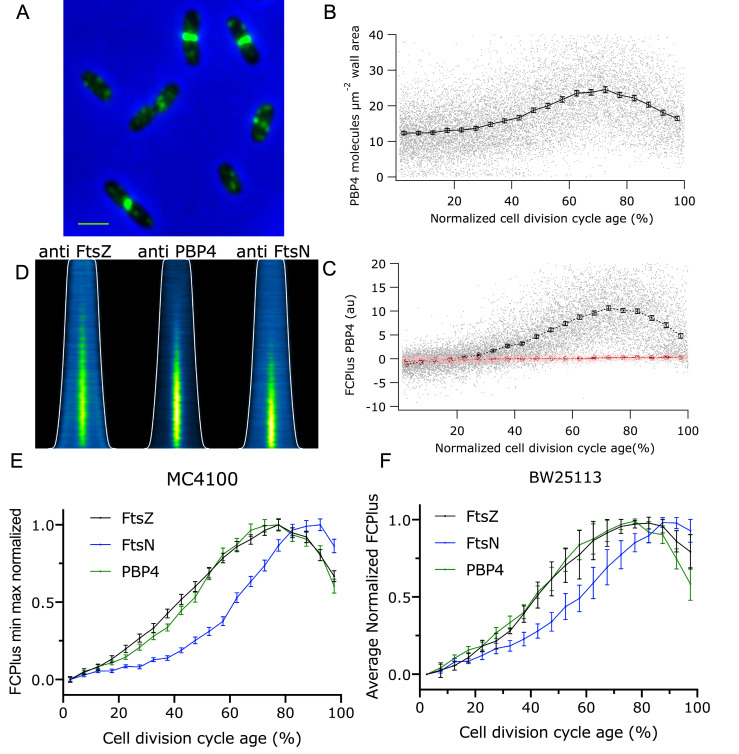
Fig 2. Cell division cycle timing of the localization of PBP4.
(A) MC4100 cells were grown to steady state in minimal glucose medium at 28°C and immunolabeled with antibodies specific for PBP4. An overlay of a phase contrast and its corresponding fluorescence image is shown. Scale bar equals 2 μm. (B) The potential number of PBP4 molecules in the cell wall area (see results section), calculated from the volume of the cells is plotted as function of the cell division cycle age. The dots are the values for the individual cells and the markers are 5% age bins with 95% confidence. (C) The extra fluorescence at midcell compared to the rest of the cell (FCPlus) is plotted as function of the normalized cell division cycle age. The dots (grey for MC4100 and light pink for the BW25113 ΔdacB strain) are the values measured for the individual cells and the markers with bars (black for the wild-type MC4100, red for the BW25113 ΔdacB) are 5% age bins with 95% confidence. (D) Demographs of the localization of FtsZ, PBP4 and FtsN in the cells sorted according to cell length. The white line indicates the length of the cells. (E) Comparison of the timing of the localization at midcell of FtsZ (black), PBP4 (green) and FtsN (blue) during the cell division cycle age. The FCPlus values are min-max normalized to enable timescale comparison, despite differences in molecule number and antibody affinities. (F) BW25113 wild-type cells were grown to “steady state” in minimal glucose medium at 28°C and immunolabeled with antibodies specific for FtsZ, PBP4 and FtsN. The FCPlus of three independent biological experiments in 5% age bins was determined. The binned FCPlus values were min-max normalized. The average min-max normalized FCPlus values of the three experiments were subsequently plotted as function of the cell division age cycle.
PBP4 localizes close to the peptidoglycan layer and the outer membrane
Although it was reported that PBP4 has a stretch of weakly amphipathic amino acids at its C-terminus [57], it was not known whether PBP4 freely diffuses in the periplasm or associates with the membrane or other proteins. It has been previously observed that chemically fixing freely diffusing periplasmic proteins can shock them into the cell poles [10,65]. The fact that we did not observe any polar localization of PBP4 after fixation (Figs 1B, 1C and S1), suggests that it is either associated with the membrane or other proteins. Next, we investigated whether PBP4 was accessible to anti-PBP4 in cells with an intact PG layer. For this, wild-type cells were grown in TY at 37°C and immunolabeled without the typically used Triton X-100-mediated permeabilization of the OM or without digesting the PG mesh with lysozyme. PBP4 was not accessible without permeabilization of the OM but was fully accessible in cells with an intact PG (S1D Fig). In contrast, cytoplasmic membrane bound proteins like FtsN and PBP3 were not accessible without degradation of the PG layer by lysozyme, whereas the outer membrane lipoprotein LpoB is accessible with Triton only (S1D Fig). This suggests that PBP4 resides close to the OM and the PG layer.
PBP4 localizes as early divisome protein and its concentration in the envelope is not constant
Since PBP4 localizes at midcell it could be part of or associated with the divisome. The division machinery is assembled in two successive steps [26,66]. First, FtsZ and its membrane bound partners ZipA and FtsA in conjunction with other Z-ring associated proteins form the proto-ring that localizes at midcell [67]. Second, after approximately 20% of the cell division cycle the remaining proteins assemble subsequently at the division site. To obtain more details on PBP4 localization in relation to other cell division proteins, MC4100 cells were grown to steady state in minimal glucose medium at 28°C. The cells were immunolabeled with antibodies against PBP4, FtsZ (early localizing), or FtsN (late localizing). Interestingly, the concentration of PBP4 was not constant during the cell division cycle but increased simultaneously with its enhanced localization at midcell (Fig 2B and 2C). Using the ribosome profiling data from [68] on the average number of 133 PBP4 molecules per cell in a culture grown in MOPS minimal medium, we calculated from the total fluorescence per average cell in the culture and the amount of fluorescence at midcell (FCPlus) that about 100 PBP4 molecules are present at midcell during the process of septal PG synthesis. The strong localization bias of PBP4 at the midcell (Fig 2), suggests a dedicated function in the cell division process. To determine whether PBP4 belongs to the early or late localizing proteins of the divisome, we analyzed the timing of its midcell localization. To this end, we plotted the extra fluorescence (FCPlus) present at midcell in comparison to the amount of fluorescence in the rest of the cell (Fig 2D, 2E, and 2F). PBP4 localization coincided with that of FtsZ, indicating that PBP4 localizes relatively early in the cell cycle to midcell.
Midcell localization of PBP4 depends on the FtsEX complex
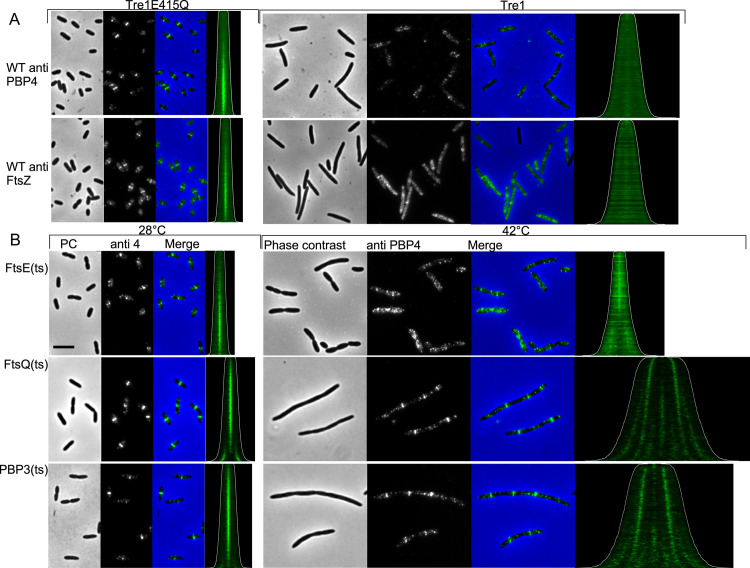
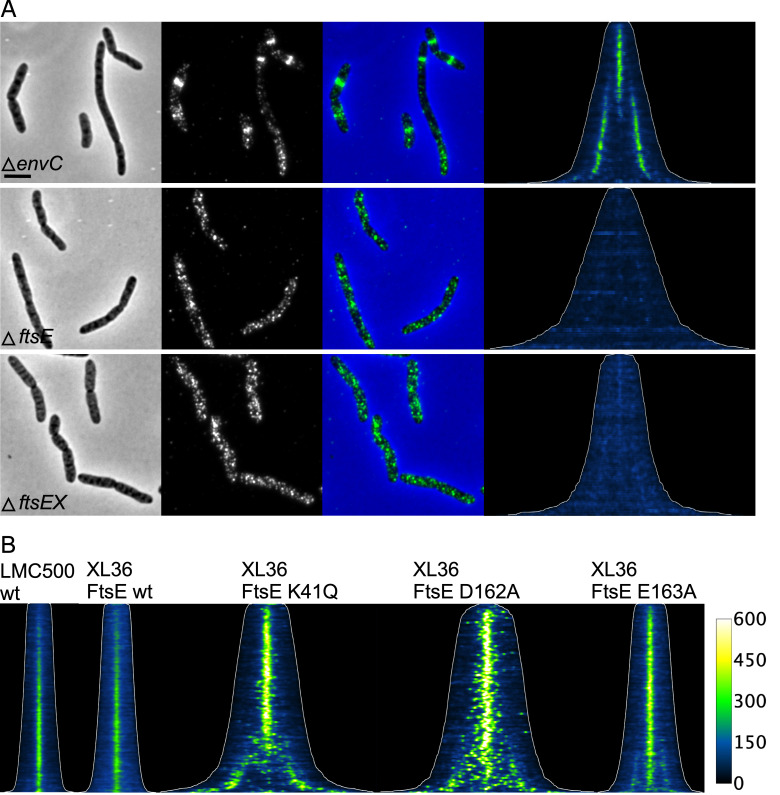
To investigate if PBP4 localization is dependent on the presence of the Z-ring or the later localizing division associated proteins we localized PBP4 in various division defective strains. First, the localization of PBP4 was assessed in a strain that expressed Tre1 that ADP-ribosylates 6 proteins including FtsZ at R174, which renders it unable to participate in protofilament formation [69,70]. The inactive E415Q variant of Tre1 was used as negative control. PBP4 was unable to localize in cells with insufficient FtsZ protofilaments to form division Z-rings (Fig 3A), whereas it localized as in WT cells in the presence of the TreE415Q variant, proving that a Z-ring is needed for PBP4 localization. Next, strains harboring temperature sensitive alleles of FtsE, FtsQ and PBP3 (FtsI) were grown in minimal glucose medium at 28°C to an OD450 of 0.2, diluted 1:4 in prewarmed medium of 28°C (permissive temperature) or 42°C (non-permissive temperature) and grown for two mass doublings before PBP4 immunolabelling. PBP4 localized at stalled division sites in filamentous cells with thermo-labile FtsQ or PBP3 indicating that these proteins are not needed to recruit PBP4 (Fig 3B). However, PBP4 localized very diffuse around potential division sites in the filamentous ftsE(ts) cells (Fig 3B). This suggests that a functional FtsE is important for PBP4 localization. The FtsEX/EnvC complex assists in the assembly of the divisome [71,72] and is essential to activate the amidase function of AmiA and AmiB to i) generate denuded glycan strands for the recruitment of the septal PG synthesis activator FtsN and ii) cleave PG for cell separation during constriction [46]. To investigate a possible link between PBP4 and the amidase activation complex we further looked for the localization requirement of PBP4 in a ΔenvC strain, as EnvC bridges FtsEX with AmiA and AmiB, and is required for their activation [43,45]. EnvC was not necessary for PBP4 localization (Fig 4A), although PBP4 failed to localize in a ΔftsEX strain (Fig 4A) and a newly constructed ΔftsE single mutant strain. The ftsE and ftsX genes reside in an operon and the expression of ftsX requires the ftsE gene [73]. We therefore constructed a ΔftsE strain with a weakened pTrc99A promoter placed upstream of ftsX to ensure sufficient expression of FtsX. The resulting strain, XL36 grew with a mild filamentous phenotype in rich medium that could be complemented by the expression of FtsE(wt) (S2 Fig). PBP4 did not localize at septal positions in the ΔftsE strain XL36 (Fig 4A) but localized normally when FtsE was expressed to complement this strain (Fig 4B). This suggests that the periplasmic part of FtsX enhances the septal localization of PBP4 and that this requires FtsE.
Fig 3. PBP4 localization is dependent on the presence of the proto-ring.
A. MC4100 was transform with a plasmid that expressed either Tre1 that abolished the ability of FtsZ to polymerize by ADP-ribosylation of residue R174 or with the inactive variant Tre1(E415Q). Cells were grown in Gb1 medium at 28°C and expression of the inhibitor was induced for 2 mass doublings (MD) with 0.15% arabinose. Subsequently, the cells were fixed and immunolabeled with antibodies against FtsZ or PBP4. From left to right: the phase contrast (PC), corresponding fluorescence image of the PBP4 labeling and the merged images are shown. In the demographs the cells are sorted according to cell length (contours in white). The number of cells analyzed were 3694 and 3756 for Tre1E415Q and 1275 and 1621 for Tre1 for anti FtsZ and anti PBP4, respectively. B. Isogenic strains of MC4100 producing different temperature sensitive (ts) versions of cell division proteins were grown to steady state in Gb1 medium at 28°C and split in two parts. These were 1:4 diluted in prewarned medium and grown for two MD at either 28°C or 42°C. The cells were fixed, labeled with antibodies specific for PBP4 (anti4). From left to right: the phase contrast (PC), corresponding fluorescence image of the PBP4 labeling and the merged images at the permissive temperature, demograph of PBP4 fluorescence localization where cells are sorted according to their cell length (contours in white). This is followed by the same series from the non-permissive temperature samples. The number of cells analyzed were 2809 and 1576 for LMC515 FtsE(ts), 3833 and 1203 for LMC531 FtsQ (ts) and 3876 and 926 for LMC510 PBP3(ts), for the cells grown at 28°C and 42°C, respectively. The scale bar equals 5 μm. Within one antibody staining the brightness and contrast of the samples is identical and therefore directly comparable.
Fig 4. PBP4 is recruited by the septal cleavage complex FtsEX.
(A) Cells of BW25113 expressing the proteins indicated were grown in LB at 37°C to OD600 = 0.3, fixed and labeled with specific antibodies against PBP4. From left to right, the phase contrast, corresponding fluorescence image of the PBP4 labeling, and the merged former two images, demograph of fluorescence with the cell lengths outlined (with the same contrast and brightness) for all demographs of PBP4 localization where cells are sorted according to their cell length are shown. The number of cells analyzed were 687 for ΔenvC, 434 for ΔftsE, and 346 for ΔftsEX. The scale bar equals 5 μm. (B) Cells of LMC500 or XL36 (LMC500ΔftsE::pTrc99Adown ftsX) expressing wild-type or active site versions of FtsE were immunolabeled with anti-PBP4 antibodies. Cells were grown in TY at 30°C and FtsE expression from plasmid was induced for 2 mass doubling with 30 μM IPTG, and cell were fixed and harvested at an OD600 of 0.3. The demographs of PBP4 fluorescence with the cell lengths outlined is shown for the indicated strains. Number of cells analyzed: LMC500 (2462), wild-type (2588), K41Q (728), D162A (554), and E163E (1052).
To determine whether the activity of FtsE was needed for the localization of PBP4, we expressed three inactive FtsE variants that had been shown to localize to midcell [73]. The K41Q and D162A versions of FtsE cannot bind ATP and FtsE(E163A) binds ATP but cannot hydrolyze it. PBP4 was able to localize at the division site in cells that expressed any of these inactive variants of FtsE as the only copy (Fig 4B), demonstrating that the activity of FtsE is not required for the localization of PBP4.
Amidases affect the localization of PBP4
Interestingly, PBP4 localized strongly at division sites in a strain lacking the three amidases or lacking amiA and B but more diffuse in strains that lacked amiC or nlpD (S3 and S4 Figs). The absence of amiAB or amiABC also resulted in a 2- or 3-times enhanced expression of PBP4 compared to the parental wild-type strain. Furthermore, PBP4 localized as in wild-type in a strain lacking essential components of the Tol-Pal system, which is involved in the coordination of OM constriction and PG invagination during cell division (S4 Fig).
PBP1A/LpoA contribute to midcell localization of PBP4
PBP4 was recently reported to interact with PBP1A, LpoA and the EPase adaptor protein NlpI [55]. To investigate whether PBP1A and/or LpoA proteins were needed for midcell localization of PBP4, strains deleted for mrcA (PBP1A), lpoA, mrcB (PBP1B) or lpoB were grown in rich medium at 37°C and immunolabeled with anti-PBP4. The amount of PBP4 that localized at midcell was identical for the parental wild-type strain BW25113, ΔmrcB and ΔlpoB, but reduced in the ΔmrcA and ΔlpoA strains (even after correction for the smaller diameter of these cells, [74], whereas the PBP4 concentration was similar for all strains (S5 Fig). This indicates that PBP1A/LpoA could assist in the localization of PBP4 at midcell.
NlpI seems not to be involved in midcell localization of PBP4
As NlpI was reported to interact with PBP4 [55], we next tested if the interaction with the OM anchored NlpI affected the localization of PBP4. In the ΔnlpI cells the amount of PBP4 at midcell per μm average cell circumference was reduced by 42 ± 6.7% (n = 3) of the wild-type and the protein localized later at midcell than in the wild-type cells (S6 Fig). However, the PBP4 concentration in the ΔnlpI strain was also lower than in the wild-type cells (64.8 ± 17.6%, n = 3). This could indicate that the cells down-regulate PBP4 expression or that PBP4 becomes unstable in the ΔnlpI strain. Labeling of PBP4 in a ΔnlpI strain did not require lysozyme (S6 Fig), suggesting that NlpI is not needed to maintain PBP4 localized outside the PG layer, which could be facilitated by additional interactions of PBP4.
In conclusion, the midcell localization of PBP4 depends on the presence of the Z-ring and FtsEX, its distribution in the lateral wall and at midcell is affected by PBP1A, LpoA, and the amidases. Its concentration during the cells cycle is not constant and mutants have either less (ΔnlpI) or more ΔamiABC) PBP4 per cell than wild-type cells.
PBP4 localizes at inactive divisomes
To further dissect whether the localization of PBP4 at the division site is dependent on the availability of its substrate or the presence of other proteins at the division site, cell division was inhibited through the specific inactivation of PBP3 by aztreonam [75]. Aztreonam treatment for 1–3 mass doublings stalls the division machinery at midcell resulting in filamentous growth. In the longer filaments new division machineries are formed and localize at future division sites [3]. In these filaments PBP4 still localized albeit somewhat weaker than in untreated cells at all possible division sites (S7 Fig), confirming that the activity of PBP3 was not required for the recruitment of PBP4. While PBP3 is inhibited, preseptal PG synthesis [76] still occurs at the potential division sites in these filamenting cells [16,36,77]. The localization of PBP4 mimics the localization of PBP1A in cells treated with aztreonam, which was previously shown to be recruited to preseptal PG synthesis sites by early cell division proteins [2,16].
Activity of PBP4 is not needed for midcell localization
Substrate binding can be a key determinant in the localization of proteins [36,78,79]. To test whether PBP4 substrate binding is needed to localize at division sites we expressed the active site mutants PBP4(S62G) and (S62A) [59] that fail to bind β-lactams and are not able to hydrolyze the PG stem peptide (S8 and S9 Figs). A plasmid encoding the mutant protein was constitutively expressed in a ΔdacB background by basal activity of a weakened pTrc99A promoter. Interestingly, the wild-type and active site mutant when expressed from plasmid were both able to localize at midcell, although the S62G variant localized more diffusely (Fig 5). This indicates that although PBP4(S62G) cannot hydrolyze its substrate or bind β-lactams covalently (S8C and S9 Figs), it is either still able to interact with peptidoglycan or it localizes through protein interactions.
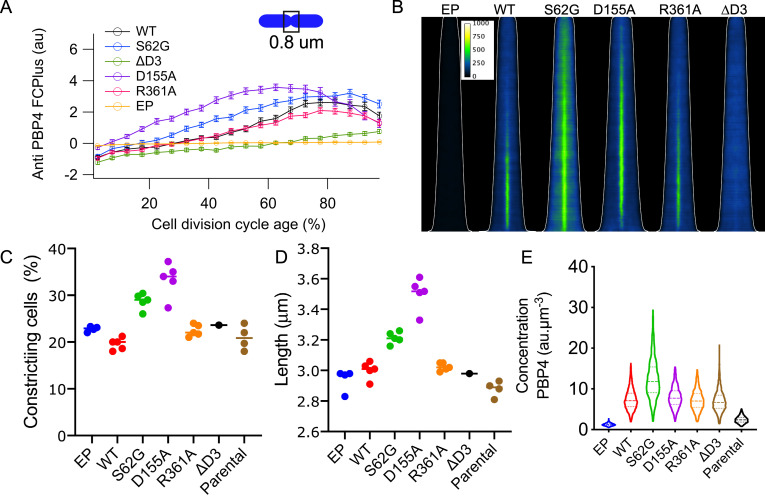
Fig 5. Localization of PBP4 depends on the presence of domain 3 but not on activity.
Cells were grown in minimal glucose medium to steady state at 28°C, fixed and immunolabeled with antibodies against PBP4. (A) The extra fluorescence at midcell (FCPlus) in the ΔdacB strain transformed with the empty plasmid (EP, yellow), or plasmids expressing wild-type PBP4 (black), PBP4S63G (blue), PBP4D155A, (purple) PBP4R361A (red), PBP4ΔD3 (green) was determined and plotted as function of the cell division cycle age as in bins of 5% age classes with the error bar indicating the 95% confidence interval. (B) Demographs of the localization fluorescence pattern of the PBP4 variants shown in (A) with the cells sorted according to their cell length. The white line indicates the length of the cells. Intensity scaling is identical for all demographs. Number of cells analyzed for each immunolabeling was at least 2000 cells. Graphs with the percentage of constricting cells (C) and the average cell length in μm (D) for the various mutants expressed from plasmid without induction in the ΔdacB strain and of the parental strain BW24113 (n = 4). (E) The concentration of PBP4 in fluorescence units per μm3 in these cells for a representative experiment (out of the four repeats).
Since PBP4(S62G) could still interact with its PG substrate, we mutagenized the gene to express PBP4 versions in which residues D155 or R361 involved in the binding of the DAP residue in the stem peptide [60] were replaced by alanine. The genes encoding PBP4(D155A) or PBP4(R361A) were expressed again from same plasmid (pTrc99A) in a ΔdacB background, and both PBP4 versions localized at midcell (Fig 5). An immunoblot analysis showed that all mutants were expressed to similar levels and that PBP4(R361A) and PBP4(D155A) were able to bind the fluorescent β-lactam Bocilin-FL (S8 Fig). Therefore, all three active site protein variants are potentially able to interact with substrate as an inactive protein.
A PBP4 version lacking domain 3 (PBP4ΔD3) was still able to bind β-lactams (S8B and S8 Fig) but did not have DD-carboxypeptidase or DD-endopeptidase activity (S9 Fig). Analytical ultracentrifugation (S10A Fig) and circular dichroism analysis (S10B Fig) of isolated PBP4ΔD3 showed that the removal of domain 3 did not affect the dimerization of the rest of the protein. The truncated protein interacted with PG sacculi, albeit somewhat weaker than the native protein and perhaps non-specific, (S10C Fig). However, although the inactive PBP4ΔD3 was capable of binding substrate, it did not localize at midcell (Fig 5). Hence, we conclude that the midcell localization of PBP4 is driven by protein-protein interactions and/or by substrate interaction but is independent from its own activity.
Although the PBP4 variants were not induced, the leakiness of the plasmids was sufficient to produce PBP4 in 3 x excess to the wild-type strain, which clearly affected their morphological parameters (Fig 5C and 5D). The timing of the PBP4 localization of these mutants was also quite variable (Fig 5A). To verify whether the timing of the rest of the divisome had adapted itself to the timing of PBP4, cells grown to steady state in minimal medium at 28°C, were fixed, divided into three aliquots, and labeled with antibodies against FtsZ, PBP4 or FtsN. The timing of the arrival of these proteins at midcell was determined by analyzing fluorescence signals of several thousands of cells [80]. The initiation of localization at midcell was defined as the timepoint during the cell division cycle age where more fluorescence is detected at mid cell than in the rest of the cell (t0) and the time it would take to reach half of the maximum value of the fluorescence at midcell (t1/2) was determined by dividing the maximum FCPlus by two and noting the corresponding timepoint (see for a graphical explanation S11A or 6A Figs). We decided against using the maximum because this time point was more difficult to determine as the maximum FCPlus often reached a plateau before decreasing at the end of the cell cycle. Indeed, the variation in the timing of the PBP4 variants localization reflected differences in the timing of the assembly of the divisome (S11 Fig).
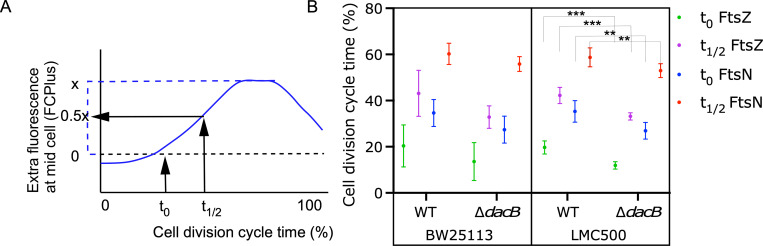
Fig 6. The absence of PBP4 advances divisome assembly.
Cells were grown in minimal glucose medium to steady state at 28°C, fixed and immunolabeled against FtsZ and FtsN. (A) Graphical illustration of the meaning of t0 and t1/2. (E). Cell division cycle age timing of FtsZ and FtsN for the ΔdacB strain (10 and 6 replicates, respectively) and its parental BW25113 (7 and 6 replicates, respectively) and the ΔdacB strain (6 and 6 replicates, respectively) and its parental LMC500 (13 and 12 replicates, respectively). Using an unpaired T-test: P 0.0001 = ***, P 0.001 = **.
PBP4 affects the timing of the divisome assembly
Because the PBP4 variants affected the morphology and the timing of the divisome assembly (Figs 5 and S11), we hypothesized a role for PBP4 in the divisome assembly. To verify this, we measured the timing of the divisome assembly in the absence of PBP4 in the ΔdacB strain and in its parental BW25113. A difference was observed between the two strains, which was not significant due to the large variation in the localization timing of FtsZ and FtsN in both strains (Fig 6B). We inferred that BW25113 is not able to reach a reproducible steady state, possibly due to a known frameshift in the rph gene, which leads to pyrimidine starvation and hence irregular DNA replication [81]. The uracil and thymine that we add as standard to the minimal medium for this strain is evidently unable to fully compensate the deficiency. To solve this possible issue, the dacB gene was deleted from the wild-type strain MC4100 and the cells were grown to steady state in minimal medium in the absence of an antibiotic. The results reproduced the difference between the ΔdacB and wild-type strains but now with a significance of P = 0.0001 (Fig 6B). No significant difference between the ΔdacB and its parental in other parameters that could account for a change in cell cycle such as mass doubling time, cell length, diameter or the percentage of constricting cells was detected (S1 Table). However, FtsZ started to localize (t0) 8% of the cell division cycle time earlier in the ΔdacB strain and reached 50% of its maximum intensity (t1/2) 9% earlier compared to the parental strain. FtsN localized 8.9% (t0) and 6.5% (t1/2) earlier (see for an explanation of t0 and t1/2 Fig 6A). These results show that the cells initiated divisome assembly significantly earlier in the absence of PBP4.
The absence of PBP4 exacerbates filamentation of ΔenvC
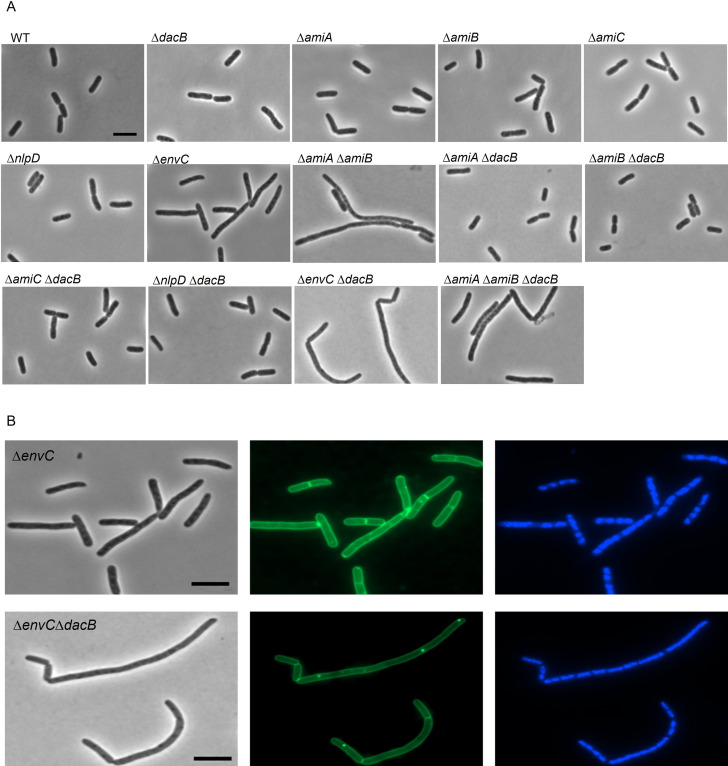
The early localization, the delay in divisome assembly, and the dependence on FtsEX for midcell localization in combination with the enhanced expression of PBP4 in a ΔamiAB strain, suggested a connection between the activity of PBP4 and the start of cell division. To test the hypothesis that PBP4 could help AmiAB to provide denuded glycan strands that would attract FtsN, the morphology of a series of deletion combinations was analyzed (S2 Table). Interestingly, ΔenvC cell chains became twice as long when dacB was also deleted (Fig 7). In contrast, the morphology of mutants lacking single amidase genes or a double amiAB deletion mutant was not affected by the additional deletion of dacB (S2 Table). From these results we conclude that EnvC and PBP4 are both needed for the correct function of AmiA and/or AmiB.
Fig 7. Morphology of deletetion strains lacking amidase or their regulators.
(A) Phase contrast microscopy of WT, ΔdacB, ΔnlpD, ΔenvC, ΔamiA, ΔamiB, ΔamiC, ΔamiAΔamiB, ΔnlpDΔdacB, ΔenvCΔdacB, ΔamiAΔdacB, ΔamiBΔdacB, ΔamiCΔdacB, and ΔamiAΔamiBΔdacB cells in early-exponential phase growth (OD600 = 0.2) in LB medium at 37°C. (B) Microscopy of ΔenvC and ΔenvCΔdacB cells. Cells were grown to early-exponential phase (OD600 = 0.2) in TY at 37°C, stained with the membrane dye FM1-43FX, fixed, and stained with DAPI. Cells were visualized by phase contrast microscopy and fluorescence microscopy. Scale bar equals 5 μm.
PBP4 domain 3 is needed for interaction with NlpI but not PBP1A/LpoA
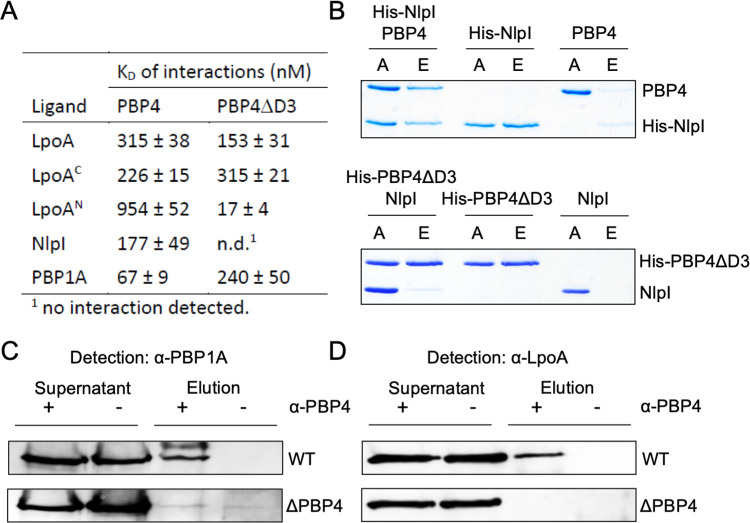
We next used microscale thermophoresis (MST) in which one protein is labelled with a fluorophore and titrated with an unlabeled partner protein to analyze the interactions between PBP4 and its known and putative interaction partners. Purified proteins were assayed for direct protein-protein interactions. We confirmed our previous result showing that wild-type PBP4 interacted with PBP1A, LpoA and NlpI [55], and we could now cross-link PBP1A to PBP4 in cells, followed by co-immunoprecipitation (Fig 8C). Interestingly, PBP4 lacking domain 3 (PBP4ΔD3) did not interact with NlpI (Fig 8).
Fig 8. PBP4 interacts with PBP1A, LpoA and NlpI.
(A) Summary of apparent KD values of the interactions of PBP4 or a version lacking domain 3 (PBP4ΔD3) with LpoA, the C-terminal domain of LpoA (LpoAC), the N-terminal domain of LpoA (LpoAN), NlpI and PBP1A determined by microscale thermophoresis (MST). Corresponding binding curves are shown in S11 Fig (B) Pulldown experiment showing that domain 3 of PBP4 is required for interaction with NlpI. Oligo-histidine tagged NlpI (His-NlpI) pulled down PBP4 to Ni-NTA beads. His-tagged PBP3ΔD3 did not pull down untagged NlpI. A, applied sample; E, eluted sample. (C) In vivo cross-linking/co-immunoprecipitation showing interaction between PBP1A and PBP4. Growing cells of BW25113 (wt) or BW25113ΔdacB (ΔPBP4) were chemically cross-linked with DTSSP and cell extract was immunoprecipitated with purified anti-PBP4 antibody. Control samples did not receive anti-PBP4 antibody. The cross-linker was cleaved by reducing agent and proteins were separated by SDS-PAGE and transferred to a membrane, followed by detection of PBP1A with specific antibodies. PBP1A was detected in the elution of the sample from wild-type and not from ΔPBP4. (D) In vivo cross-linking/co-immunoprecipitation (as in panel C) showing that PBP4 and be cross-linked with LpoA in cells.
We found that PBP4 and PBP4ΔD3 interacted with full length LpoA with apparent KD values of 315 ± 38 nM and 153 ± 31 nM, respectively (Figs 8A and S12) suggesting that domains 1 or 2 of PBP4 are sufficient for the interaction with LpoA. MST assays of PBP4 or PBP4ΔD3 with LpoA’s C-terminal (residues N257-S678; LpoAC) or N-terminal (LpoA residues G28-T256; LpoAN) domains also yielded positive binding curves (Figs 8A and S12) [82–84]. LpoAC and the full length LpoA version had comparable affinities for both PBP4 versions, suggesting that the C-terminal domain of LpoA is sufficient for interaction with PBP4 (Fig 8A). Interestingly, the removal of domain 3 from PBP4 resulted in an increased affinity of LpoAN for PBP4 (app. KD decreased by ~50 fold) (Figs 8A and S12). Hence, the interaction between the N-terminal domain of LpoA and PBP4 possibly occurs via a different mechanism and may be enhanced by a conformational rearrangement of domain 3 of PBP4. In conclusion PBP4 robustly interacts with PBP1A/LpoA independent of its domain 3.
LpoA has a small effect on PBP4 activity
We found previously that NlpI does not affect the activity of PBP4 [55] but it was possible that PBP1A/LpoA have an effect. We first verified that NlpI or PBP4 did not alter the activity of PBP1A in the presence or absence of LpoA using an in vitro glycosyltransferase assay (S13 Fig). We then tested if PBP1A or LpoA affected the activity of PBP4 using different in vitro assays. LpoA decreased the DD-CPase activity of PBP4 by about 30% in a continuous assay with the soluble UDP-MurNAc pentapeptide substrate (S14B Fig) and its DD-EPase activity in an endpoint PG digestion assay (S14C Fig).
Amidase activator EnvC interacts with PBP4 and NlpI
PBP4 localizes at midcell in cells lacking EnvC but not in cells lacking membrane anchored FtsEX. To begin investigating interactions of PBP4 with members of the amidase activation pathway, we first tested for the interactions with EnvC, a periplasmic protein with a coiled-coil and a LytM domain that activates AmiA and B upon interaction with FtsEX [85]. MST experiments showed that full-length EnvC interacted with full-length PBP4 (KD = 307±45 nM), but substantially weaker with PBP4ΔD3 (KD > 5900±2823 nM) (S14 Fig). The LytM domain of EnvC did not interact with PBP4 suggesting that the interaction between both proteins is mediated by domain 3 of PBP4 and the coiled-coil domain of EnvC. Hence domain 3 of PBP4 is needed for midcell localization (Fig 6) and is also used to bind to EnvC (Fig 8) and NlpI (S15 Fig). Yet the absence of none of these proteins alone is enough to abrogate the PBP4 midcell localization (Figs 3, 4 and S6).
During these studies, we also discovered an interaction between EnvC with NlpI, via the LytM domain of EnvC (S15 Fig). The different binding domains in EnvC for PBP4 and NlpI suggested that a trimeric complex could be formed. Indeed, a pulldown assay with oligo-histidine-tagged NlpI and untagged PBP4 and EnvC supported the formation of a trimeric complex. In summary, we show that PBP4 interacts with a component of the amidase-activation pathway, EnvC and with NlpI in a manner that can imply the formation of dynamic/different tripartite complexes that requires further investigation.
Discussion
E. coli contains a large number of seemingly redundant PG hydrolases, which possibly fine tune peptidoglycan biogenesis and remodelling in response to environmental parameters [34]. Apart from their enzymatic activities, little is known about the function of most PG hydrolases. Here we report that the DD-carboxy/endopeptidase PBP4 localizes at midcell during septal PG synthesis in E. coli and is important for the timing of the assembly of the division machinery.
PBP4 localizes at midcell during cell division in an FtsEX dependent manner
PBP4 localizes predominantly at midcell during cell division in an FtsEX dependent manner. Inactive PBP4 variants were able to localize at midcell indicating that substrate hydrolysis is not a requirement for localization. However, a version of PBP4 lacking the non-catalytic domain 3 (PBP4ΔD3), which is required for activity but not β-lactam binding, did not localize to midcell. Domain 3 is also required for the interaction with EnvC and NlpI, but not with PBP1A/LpoA. From these results we infer that the midcell localization of PBP4 requires multiple protein interactions, but not substrate hdyrolysis.
PBP4 might be initially recruited by preseptal PG synthesis
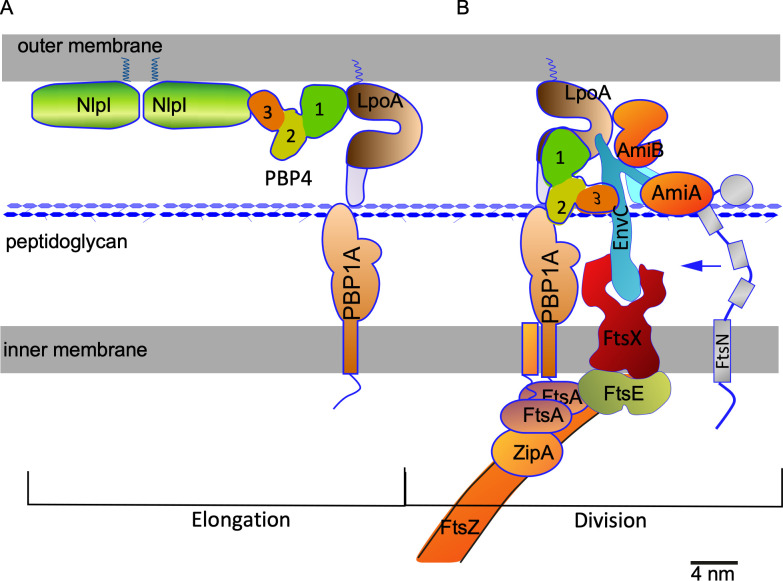
PBP4 interacts with NlpI, PBP1A and LpoA and these proteins can form a ternary complex ([55] and (Fig 8). PBP4ΔD3 interacted with PBP1A and LpoA but not with NlpI or EnvC, suggesting that NlpI and EnvC bind to domain 3. The interaction with PBP1A and/or LpoA may contribute to the recruitment of PBP4 at midcell, as the amount of PBP4 at midcell was slightly reduced in ΔmrcA and ΔlpoA strains. PBP1A and its partner LpoA are involved in preseptal PG synthesis [16,80]. Although somewhat weaker, PBP4 still localizes at putative division sites in cell filaments generated with aztreonam that have lost septal synthesis but not preseptal synthesis activity. Possibly, some PBP4 enriches at preseptal PG synthesis sites together with PBP1A/LpoA, which localize through the interaction of PBP1A with ZipA [16], and preseptal PG synthesis may provide initially the substrate for PBP4. The interaction between NlpI and PBP4 could function to sequester PBP4 in the lateral wall to prevent its interaction with potential substrate (Fig 9).
Fig 9. Model of the organization of PBP4 in the periplasm of E. coli.
(A) During elongation PBP4 (or a fraction of the PBP4 molecules) is kept away from its substrate through its interaction with NlpI and possibly LpoA. (B) PBP1A and LpoA associate with ZipA and the Z-ring to assist in preseptal PG synthesis. Because of their presence, the absence of NlpI and the newly synthesized PG, PBP4 is attracted by PBP1A and LpoA at the septal synthesis site, where it interacts with the FtsEXEnvC complex. Preseptal PG synthesis might provide substrate for PBP4 and the activated AmiA and B to produce denuded strand that attract FtsN. All proteins apart from the FtsZ polymer have been drawn approximately according to their crystal structure shape and hydrated crystal structure sizes (scale bar equals 4 nm). The membranes are assumed to be 2 nm and the distance between the outer and the inner membrane 21 nm.
Why does the loss of PBP4 cause premature divisome assembly?
The absence of PBP4 interferes with the timing of divisome assembly. In most bacteria studied thus far, the Z-ring and associated early division proteins localize first to prepare the future cell division site, perhaps by generating the border between the side wall and the future septum [16,76,86]. After a time-delay, the proteins that synthesize the bulk of the septal PG arrive and septation starts [26–28]. The absence of PBP4 causes a premature initiation of the assembly of the divisome for yet unknown reasons. The absence of PBP4 might enable NlpI to bind other EPases [54,55], coordinating their activity for remodelling the PG layer. However, the robust localization of PBP4 at midcell suggests a more direct role in modulating the timing of division initiation. We hypothesize that during cell elongation sufficient PBP4 molecules are retained by NlpI in the lateral wall above the PG layer to avoid promiscuous PBP4 activity (Fig 9), consistent with the reduced amount of PBP4 in cells lacking NlpI (S6 Fig and [55]). PBP4 might be initially recruited to midcell through interactions with PBP1A and LpoA, and the FtsEX-EnvC complex (Fig 9). The interaction of PBP4 with EnvC (S15 Fig) might ensure a concerted endopeptidase/amidase activity as EnvC activates AmiA and AmiB, which generate denuded glycan chains that attract FtsN, the trigger of septal PG synthesis. This is also suggested by the observation that a double ΔdacB ΔamiC mutant forms longer cell chains than a single ΔamiC strain [45] and by the observation that a ΔenvC ΔdacB forms longer filaments than a ΔenvC strain (Fig 7), whereas the deletion of dacB in an amiA and amiB double deletion strain has no exacerbating effect. As deleting dacB does not prevent the assembly of the divisome, it is plausible that other endopeptidases either contribute and/or partially take over the function of PBP4. Once septal peptidoglycan synthesis starts by the mature divisome, PBP4 remains at midcell as shown by the immunolocalization experiments (Fig 2). This could suggest that continued PBP4 activity is needed in cooperation with the amidases and other endopeptidase to create sufficient binding substrate for FtsN.
Materials and methods
Culturing conditions
E. coli K12 cells were grown to steady state [80] in glucose minimal medium containing 6.33 g of K2HPO4×3H2O, 2.95 g of KH2PO4, pH 7.0, 1.05 g of (NH4)2, 0.10 g of MgSO4×7 H2O, 0.28 mg of FeSO4×7 H2O, 7.1 mg of Ca(NO3)2×4 H2O, 4 mg of thiamine and 4 g of glucose per L. For strain MC4100 and its derivatives 50 μg lysine (Gb1) and for BW25114 and MG1655 derivatives 2 μg uracil, 20 μg thymidine, 50 μg arginine and glutamine (Gb4) per liter at 28°C or 37°C were added. Absorbance was measured at 450 nm with a 300-T-1 spectrophotometer (Gilford Instrument Laboratories Inc.). Alternatively, cells were grown in TY (5g yeast extract, 10 g bacto trypton and 85 mM NaCl, pH 7.0), or LB, which is TY with 170 mM NaCl and absorbance was measured at 600 nm.
Strain construction
Strain XL35, the FtsE partial deletion strain, was constructed as follows: Primers priXL221 and priXL222 were used to amplify the upstream homologous sequences (UHS) from the ftsE start codon, and primers priXL223 and priXL224 were used to amplify the downstream homologous sequence after the FtsE 151th amino acid (the potential promoter of ftsX), from the MC4100 genome. Primers priXL51 and priXL54 were used to amplify the kanamycin resistance cassette from plasmid pKD4 [87]. The amplified overlap PCR product (UHS-FRT-KAN-FRT-DHS) with primers priXL221 and priXL224 was used to construct the recombinant strain XL34 in which the first 150 amino acids encoding sequence of ftsE was deleted [87]. Subsequently, the kanamycin selection cassette was removed with plasmid pCP20 [87] yielding XL35.
Strain XL36, the ftsE clean deletion strain, was constructed similarly as XL35. Primers priXL221 and priXL222 were used to amplify the upstream homologous sequences from the ftsE start codon, and primers priXL225 and priXL226 were used to amplify the downstream homologous sequences after the ftsE stop codon, from the MC4100 genome. In order to restore the FtsX expression, a pTrc99Adown promoter [88] was amplified from plasmid pSAV057 [89] with primers priXL77 and priXL227 and a chloramphenicol selection cassette was amplified from plasmid pKD3 with primers priXL52 and priXL54. The amplified overlap PCR product (UHS-FRT-CAM-FRT-DHS) with primers priXL221 and priXL226 was used to construct the recombinant strain XL33 in which the entire ftsE coding sequence was deleted and FtsX was expressed from the chromosomally encoded pTrc99Adown promoter. Subsequently, the chloramphenicol selection cassette was removed with plasmid pCP20 yielding XL36 (Table 1). Bacterial strains construction for ΔdacB and amidases/regulators: E. coli BW25113 was the parental strain used for these experiments. E. coli amiA::kan, amiB::kan, amiC::kan, envC::kan, nlpD::kan, dacB::kan mutants were obtained from the KEIO library [90] and the mutations transduced into a clean parental strain. The strain dacB::cam was obtained from a previous study [55]. Double mutants were created by P1 transduction as described previously [91]. The chromosomal modification was confirmed by PCR when a CamR or KanR cassette was transduced.
Table 1. Strain and plasmids used in this study.
| Strain | Protein affected | Genotype | Source |
| LMC500 (MC4100) | Wild-type | F-, araD139, Δ(argF-lac)U169, deoC1, flbB5301, lysA1, ptsF25, rbsR, relA1, rpsL150 | [92] |
| BL21(DE3) | Overexpression strain | F- ompT, dcm lon hsdS hsdSB(rB-mB-) λ(DE3) | Novagen |
| BW25113 | Keio background strain | F-, DE(araD-araB)567, lacZ4787(del)::rrnB-3, LAM-, rph-1, DE(rhaD-rhaB)568, hsdR514 | [87] |
| DH5α | Storage strain | F- endA1 glnV44 thi-1 recA1 ϕ80ΔlacZΔM15, λ- | Invitrogen |
| MC1061 | Laboratory strain | Δ(araA-leu)7697 Δ(lac)X74 galK16 galE15(GalS) λ-e14- mcrA0 relA1 rpsL150(strR) spoT1 mcrB1 hsdR2 | [105] |
| LMC509 | FtsZ (ts) | MC4100 ftsZ184 | [92] |
| LMC511 | FtsA (ts) | MC4100 ftsA10 | [92] |
| LMC512 | FtsA (ts) | MC4100 ftsA1882 | [92] |
| LMC515 | FtsE (ts) | MC4100 ftsE1181 | [92] |
| LMC531 | FtsQ (ts) | MC4100 ftsQ(ts) | [92] |
| LMC2487 | FtsW (ts) | JLB17; F- thr, trp, his, thy, ara, lac, gal, xyl, mtl, rspL, tonA, ftsW amino acid replacement G311D | [106] |
| LMC510 | PBP3 (ts) | MC4100 ftsI 2158 | [92] |
| BCB677 | ΔPBP1A | BW25113ΔmrcA | [90] |
| BCB678 | ΔPBP1B | BW25113ΔmrcB | [90] |
| BCB192 | ΔLpoA | BW25113ΔlpoA | [90] |
| LMC2217 | ΔLpoB | BW25113ΔlpoA | [90] |
| BCB655 | ΔPBP4 | BW25113ΔdacB::kan | [90] |
| BCB1433 | ΔPBP4 | XWL010 MC4100ΔdacB::kan | This work |
| BCB904 | ΔNlpI | BW25113ΔnlpI::frt pCP20 (NT10087) | This work |
| BCB197 | ΔPal | BW25113Δpal::kan (CAG70150) | [90] |
| BCB195 | ΔTolA | BW25113ΔtolA::kan (CAG70204) | [90] |
| BCB742 | ΔEnvC | BW25113ΔenvC:: frt pPC20 (NT10086) | This work |
| BCB968 | ΔAmiC | BW25113ΔamiC::cam (NT10090) | This work |
| BCB970 | ΔAmiCΔNlpI | BW25113ΔamiCΔnlpI::tet (NT10266) | This work |
| BCB1153 | ΔAmiAB | MC1061ΔamiAB:cam | [42] |
| BCB021 | ΔAmiABC | MC1061ΔamiABC:cam, kan | [42] |
| BCB744 | ΔFtsE | BW25113ΔftsE kanR CAG60397 | This work |
| BCB746 | ΔFtsEX | BW25113ΔftsEX catR CAG60399 | This work |
| BCB1081 | ΔFtsE 1–150 | XL34 LMC500ΔftsE 1–150::kan-ftsX | This work |
| BCB849 | ΔFtsE 1–150 | XL35 LMC500ΔftsE 1–150::ftsX | This work |
| BCB1082 | ΔFtsE | XL33 LMC500ΔftsE::-cam-pTrc99Adown ftsX | This work |
| BCB850 | ΔFtsE | XL36 LMC500ΔftsE::pTrc99Adown ftsX | This work |
| NT10022 | ΔPBP4 | BW25113ΔdacB::cam | [55] |
| MB01119 | ΔAmiA | BW25113ΔamiA::kan | [90] |
| MB01120 | ΔAmiB | BW25113ΔamiB::kan | [90] |
| MB01121 | ΔAmiC | BW25113ΔamiC::kan | [90] |
| MB01051 | ΔNlpD | BW25113ΔnlpD::kan | [90] |
| MB01122 | ΔEnvC | BW25113ΔenvC::kan | [90] |
| NT10274 | ΔAmiAΔAmiB | BW25113ΔamiA::kanΔamiB::kan | This work |
| MB01154 | ΔAmiAΔPBP4 | BW25113ΔamiA::kanΔdacB::cam | This work |
| MB01155 | ΔAmiBΔPBP4 | BW25113ΔamiB::kanΔdacB::cam | This work |
| MB01156 | ΔAmiCΔPBP4 | BW25113ΔamiC::kanΔdacB::cam | This work |
| MB01157 | ΔEnvCΔPBP4 | BW25113ΔenvC::kanΔdacB::cam | This work |
| MB01158 | ΔNlpDΔPBP4 | BW25113ΔnlpD::kanΔdacB::cam | This work |
| MB01159 | ΔAmiAΔAmiBΔPBP4 | BW25113ΔamiAΔamiB::kanΔdacB::cam | This work |
| Plasmid | Name | Characteristics | Source |
| pET21b | pET21b-PBP4(S62A)Δ1–60 | Inactive PBP4 lacking residues 1–60, AmpR | [55] |
| pET21b | pET21b-His-PBP4(S62A)Δ1–60 | For purification of His-PBP4 (active site mutant), AmpR | [55] |
| pET21b | pET21b-PBP4Δ1–60 | Native PBP4 lacking residues 1–60, AmpR | [55] |
| pET28a | pET28a-His-LpoA(sol) | Soluble LpoA (LpoAΔ1–27) construct, N-terminal His-tag, KanR | [82] |
| pET28a | pET28-His-LpoAN | Purification of His-LpoAN, KanR | [82] |
| pET28a | pET28-His-LpoAC | Purification of His-LpoAC, KanR | [82] |
| pTK1A-His | pTK1A-His | Full length His-PBP1A, KanR | [82] |
| pET28a | pET28a-His-NlpI | Soluble NlpI construct, N-terminal His-tag, KanR | [4] |
| pBAD18 | pBAD18-His-PBP4ΔD3 | for purification of His-PBP4 lacking domain 3, AmpR | This work |
| pET28a | pET28a-His-EnvC | for purification of His-EnvC, KanR | This work |
| pBAD18 | Tre1 | Arabinose inducible Tre1, AmpR | [69] |
| pBad18 | Tre1 E415Q | TreE415Q inactive Tre1 variant, AmpR | [69] |
| pXL86 | pTHV mCh-FtsE wt | pTrc 99A down expressing mCh-FtsE(wt) colE1 ori, AmpR | This work |
| pXL89 | pTHV mCh-FtsE ts | pTrc 99A down expressing mCh-FtsE(ts) (P135S) colE1 ori, AmpR | This work |
| pXL110 | pSAV mNG-NNN-FtsE | pTrc 99A down expressing mNG-FtsE, p15A origin, CamR | This work |
| pXL133 | pSAV-PBP4 | pTrc 99A down expressing PBP4, p15A origin, CamR | This work |
| pXL134 | pSAV-PBP4 S62G | pTrc 99A down expressing PBP4 S62G, p15A origin, CamR | This work |
| pXL135 | pSAV-FtsE | pTrc 99A down expressing FtsE, p15A origin, CamR | This work |
| pXL137 | pSAV-FtsE K41Q | pTrc 99A down expressing FtsE K41Q, p15A origin, CamR | This work |
| pXL138 | pSAV-FtsE D162A | pTrc 99A down expressing FtsE D162A, p15A origin, CamR | This work |
| pXL139 | pSAV-FtsE E163A | pTrc 99A down expressing FtsE E163A, p15A origin, CamR | This work |
| pXL140 | pSAV-PBP4-ΔD3 | pTrc 99A down expressing PBP4Δ173–247 (domain 3), p15A origin, CamR | This work |
| pXL150 | pSAV-PBP4-D155A | pTrc 99A down expressing PBP4 D155A, p15A origin, CamR | This work |
| pXL151 | pSAV-PBP4-R361A | pTrc 99A down expressing PBP4 R361A, p15A origin, CamR | This work |
AmpR, KanR and CamR are ampicillin, kananamycin and chloramphenicol resistance, respectively
Plasmid construction
The ftsE gene was amplified from the chromosome of LMC500 [92] using the forward primer priXL187 and the reverse primer priXL188, and sequenced to characterize the mutation. The wild-type ftsE gene was cloned into pSAV057, p15A origin and Cam resistant [89] to produce the plasmid pXL135. FtsE was expressed under the control of the pTcr99A down promoter. QuickChange site directed mutagenesis (Agilent technologies, Santa Clara, CA) and Gibson assembly [93] approaches were applied afterwards to construct the inactive ftsE mutant plasmids pXL137, pXL138 and pXL139, using the primers listed in Table 2.
Table 2. Primers used in this study.
| Primer | Sequence 5’-3’ | Purpose |
|---|---|---|
| priXL51 | ACCATGGCTAATTCCCATGTCAG | GA_pkd3_F |
| priXL54 | GTGTAGGCTGGAGCTGCTTC | PCR_pkd3_R |
| priXL187 | CCGGCCGAATTCAACAACAACTCGATGATTCGCTTTGAACATGTCAGC | FtsE-EcoRI fw |
| priXL188 | GGCCAAGCTTTTATTCATGGCCCACGCCTCC | FtsE-EcoRI rv |
| priXL221 | CATGAAGCCGTTGGCTTAACC | ΔFtsE-FA-F |
| priXL222 | TAGGAACTTCGAAGCAGCTCCAGCCTACACTGTTAATCCTCTCGGGCAAAAAG | ΔFtsE -FA-R |
| priXL223 | GGATATTCATATGGACCATGGCTAATTCCCATGTCAGGCGGTGGTGAACAAGCCCGC | ΔFtsE- 1–150 |
| priXL224 | GGTGAGCATGCGATAGGAACG | ΔFtsE-1:150 |
| priXL225 | TGAGCGGATAACAATTTCACACAGGAAACAGACCATGAATAAGCGCGATGCAATC | ΔFtsE-clean |
| priXL226 | CTTTTCAGATCCTGCAATGCG | ΔFtsE-clean |
| priXL230 | ATTCCGGCGCAGGGCAAAGTACTCTCCTGAAGCTG | FtsE-K41Q-F |
| priXL231 | GATCAGCTTCAGGAGAGTACTTTGCCCTGCGCCGGA | FtsE-K41Q-R |
| priXL232 | CGGTACTGCTAGCGGCTGAACCGACTGGTAACCTG | FtsE-D162A-F |
| priXL233 | AGGTTACCAGTCGGTTCAGCCGCTAGCAGTACCGC | FtsE-D162A-R |
| priXL234 | GTACTGCTAGCGGACGCTCCGACTGGTAACCT | FtsE-E163A-F |
| priXL235 | AGGTTACCAGTCGGAGCGTCCGCTAGCAGTACCGC | FtsE-E163A-R |
| priXL236 | AGATGGCGCTGCCTGCCGGCACCCAGAAAG | PBP4-S62G-F |
| priXL244 | GCGCGAATTCatgCGATTTTCCAGATTTATCA | EcoRI-PBP4-F |
| priXL245 | GCGCAAGCTTctaATTGTTCTGATAAATATC | HindIII-PBP4-R |
| priXL250 | GTTGACCGCAACGGTGGCGGAAGCGGGTCTGTGCAGGATGGAGCCAGCTATG | PBP4ΔD3-F |
| priXL251 | TCCATCCTGCACAGACCCGCTTCCGCCACCGTTGCGGTCAACTATGGCGGC | PBPΔD3-R |
| priXL266 | CCGGCTGGCCATGGAATGCCATGACACAATGCTTTAGCGCTC | PBP4-D155A-F |
| priXL267 | GGAGCGCTAAAGCATTGTGTCATGGCATTCCATGGCCAGCCGG | PBP4-D155A-R |
| priXL268 | GCCGATGGTTCAGGGCTTTCGGCGCATAACCTGATTGCCCCCGCC | PBP4-R361A-F |
| priXL269 | GGCGGGGGCAATCAGGTTATGCGCCGAAAGCCCTGAACCATCGGC | PBP4-R361A-R |
| prXW5-F | TAGTATGACGGCTCGATTCCAGGTTGTTAGCGCGAGATTgtgtaggctggagctgcttc | LMC500ΔdacB |
| priXW6-R | AATCTGAAGCCCCGGCCATGTGCCGGGGTTTCTTTTTGAatgggaattagccatggtcc | LMC500ΔdacB |
| Env-F | GGAATTC GATGAGCGTGACCAACTC | pET28a-His-EnvC |
| Env-R | CCCttaTCTTCCCAACCACGGC | pET28a-His-EnvC |
To construct the PBP4 expression plasmids, the wild-type dacB gene was firstly amplified from the E. coli MG1655 genome using primers priXL244 and priXL245, and subsequently cloned into plasmid pSAV057 and pSAV057-dsbass-mCherry-PBP5 (pNM010 [38]) with EcoRI and HindIII, to generate the PBP4 plasmids without and with mCherry fusion. Similarly, QuickChange site directed mutagenesis and Gibson assembly approaches were applied afterwards to construct the inactive dacB mutant plasmids from these two wild-type plasmids, using the primers listed in Table 2. Tre1 and Tre1E415Q were expressed from the arabinose inducible promoter of pBAD18 as described [69].
Immunolabeling
After reaching steady state for minimal glucose grown cells or the desired OD for rich medium grown cells, the cells were fixed for 15 min by addition of a mixture of formaldehyde (f. c. 2.8%) and glutaraldehyde (f. c. 0.04%) to the cultures in the shaking water bath and immunolabeled as described [94] with Rabbit polyclonal antibodies against PBP4, NlpI [55] preabsorbed against ΔdacB or ΔnlpI, strains, respectively, or against FtsZ or FtsN [26]. As secondary antibody, donkey anti-rabbit conjugated to Cy3 or to Alexa488 (Jackson Immunochemistry, USA) diluted 1:300 in blocking buffer (0.5% (wt/vol) blocking reagents (Boehringer, Mannheim, Germany) in PBS) was used, and the samples were incubated for 30 minutes at 37°C. For immunolocalization, cells were immobilized on 1% agarose in PBS slabs coated object glasses as described [70] and photographed with an Orca Flash 4.0 (Hamamatsu, Japan) CCD camera mounted on an Olympus BX-60 (Japan) fluorescence microscope through a 100x/N.A. 1.35 oil objective. Images were taken using the program ImageJ with MicroManager (https://www.micro-manager.org).
SIM images were obtained with a Nikon Ti Eclipse microscope (Japan) and captured using a Hamamatsu Orca-Flash 4.0 LT camera. Phase contrast images were acquired with a Plan APO 100x/1.45 Ph3 oil objective. SIM images were obtained with a SR APO TIRF 100x/1.49 oil objective, using 3D-SIM illumination with a 561nm laser, and were reconstructed with Nikon-SIM software using the values 0.20–0.25–0.20 for the parameters Illumination Modulation Contrast (IMC), High Resolution Noise suppression (HNS) and Out of focus Blur Suppression (OBS), respectively.
Image analysis
Phase contrast and fluorescence images were combined into hyperstacks using ImageJ (http://imagej.nih.gov/ij/) and these were linked to the project file of Coli-Inspector running in combination with the plugin ObjectJ (https://sils.fnwi.uva.nl/bcb/objectj/). The images were scaled to 15.28 pixel per μm. The fluorescence background has been subtracted using the modal values from the fluorescence images before analysis. Slight misalignment of fluorescence with respect to the cell contours as found in phase contrast was corrected using Fast-Fourier techniques as described [80]. Data analysis was performed as described [80]. In brief, midcell was defined as the central part of the cell comprising 0.8 μm of the axis. From either cell part, midcell and remaining cell, the volume, the integrated fluorescence, and thus the concentration of fluorophores was calculated. The difference of the two concentrations is multiplied with the volume of midcell. It yields FCPlus (surplus of fluorescence) at midcell. For age calculation, all cell lengths are sorted in ascending order. Then the equation
is used, where rank is a cell’s index in the sorted array, nCells is the total number of cells, and age is the cell’s age expressed in the range 0.. 1. To analyze the FCPlus or the total concentration of fluorescence as a function of cell division cycle age, the data points of the individual cells were binned in 5% age classes and plotted with the 95% confidence interval indicated as error bars. To determine the timing of the divisome assembly, the moment that more fluorescent material was present at midcell (FCPlus) than in the rest of the cell was taken as timepoint zero of arrival (t0) and then the maximum FCPLus value (usually a small plateau) was divided by half and the corresponding timepoint was noted as the half maximum fluorescent intensity time value (t1/2).
ΔdacB and amidases/regulators fluorescence imaging
To image membrane and DNA compartmentalisation of the cells, FM1-43FX dye (Invitrogen) and DAPI (Stratech Scientific) were used, respectively. Overnight cultures were adjusted to initial OD600 = 0.01 in LB. The cells were incubated at 37°C until OD600 = ~0.2. A volume of 500 μl of the culture was stained with 5 μg ml-1 of FM1-43FX at room temperature for 10 min. The cells were adjusted to 33 mM sodium phosphate pH 7.4 and fixed by addition of 2.4% formaldehyde and 0.04% glutaraldehyde. Fixed cells were inoculated on agarose pads, which were prepared with 1.5% agarose in PBS and set in Gene Frames (Thermo Scientific). Cells were imaged using a Zeiss AxioObserver equipped with a Plan-Apochromat 100x/Oil Ph3 objective and illumination from HXP 120V for phase contrast images. For FM1-43FX images Zeiss filter set 38 (Ex: 470/40 nm, beamsplitter 495 nm, Em: 525/50 nm) was used. DAPI images were captured using the Zeiss filter set 96 (Ex: 390/40 nm, beamsplitter 420, Em: 450/40 nm). For phenotype analysis in S2 Table we used MicrobeJ plugin for Fiji 600 [95].
Proteins
Proteins were purified from overexpression strains according to published procedures: NlpI, PBP4, PBP4(A62) and PBP4ΔD3 [55]. LpoA, LpoAN and LpoAC [82]; PBP1A [4]. EnvC was purified from BL21(DE3) pET28a-His-EnvC. The overexpression plasmid pET28a-His-EnvC was constructed by amplifying a soluble EnvC version without the N-terminal, 34 amino acid long, signal peptide into the pET28a (+) vector using the restriction enzymes NdeI and HindIII. To purify recombinant EnvC, cells which were grown in 3 L LB with 50 μg/ml kanamycin to OD578 0.5–0.6, at 37°C. Protein overproduction was induced with 1 mM IPTG for 3 h at 30°C before cells were harvested by centrifugation and lysed by sonication before ultracentrifugation at 14,000 × ɡ, 1 h, 4°C. The cell lysate was centrifuged and the supernatant was applied to a 5 ml HisTrap HP column (GE healthcare). The column was washed with 4 column volumes of 25 mM Tris/HCl, 300 mM NaCl, 20 mM imidazole, pH 7.5 at a flow rate of 2 ml/min. The oligohistidine-tagged EnvC was eluted using 25 mM Tris/HCl, 300 mM NaCl, 400 mM imidazole, 10% glycerol, pH 7.5. Protein purity and yield was analysed by SDS-PAGE and fractions of interest were pooled. The oligohistidine tag was removed by incubating the samples with 1 unit/ml of restriction grade thrombin (Novagen), overnight at 4°C, during dialyses against 25 mM Tris/HCl, 200 mM NaCl, pH 8.0. The sample was concentrated to ~ 5 ml using Vivaspin concentrator spin columns (Sartorius) at 4,500 × ɡ at 4°C. The sample was then applied to a HiLoad 16/600 Superdex 200 column (GE healthcare) at 1 ml/min pre-equilibrated with 25 mM HEPES/NaOH, 300 mM NaCl, 10%, glycerol, pH 7.5. Fractions were analysed by SDS-PAGE and those with the highest protein purity and yield were pooled and stored at -80°C.
Ni2+-NTA pulldown assays
Nickel-Nitrilotriacetic acid (Ni-NTA) beads (100μl) were pre-equilibrated with dH2O and binding buffer (10 mM HEPES/NaOH, 10 mM MgCl2, 150 mM NaCl, 0.05% Triton X-100, pH 7.5) by centrifugation at 4000 × g, 4 min at 4°C, and then incubated with proteins of interest. Equimolar amounts (1–2 μM) of His6-tagged and untagged proteins were incubated alone (control) and in combination, for 10 min at 4°C in a 200 μl reaction volume, containing 10 mM binding buffer. An aliquot of this mixture is taken as an ‘applied’ sample. The protein samples were then added to pre-equilibrated Ni-NTA beads and incubated overnight on a spinning plate at 4°C with 1.3 ml of binding buffer.
Beads were centrifuged at 4000 × g, 4 min, 4°C and washed a further 4–6 times with 1 ml of washing buffer (binding buffer with 30 mM Imidazole) before re-suspending in 250 μl washing buffer and transferring beads to Proteus spin columns (Generon), centrifuging as described above. Bound proteins were eluted by the addition of 50 μl of SDS-PAGE loading buffer and boiling at 100°C for 5 min. Spin columns were centrifuged a final time at 1500 × g, 5 min, RT, to collect ‘eluted’ protein before separation by SDS-PAGE. Eluted samples were run alongside applied samples for comparison.
Microscale thermophoresis
Microscale thermophoresis (MST) is an immobilisation-free method which allows the detection of biomolecular interactions in solution. This technique is based on the specific directed movement of a protein along a heat gradient (thermophoresis), an observation first reported by Carl Ludwig in 1856 [96]. The thermophoresis of a protein changes upon ligand binding due to one or more changes to size, charge and/or hydration shell. In MST, a localized heat gradient is initiated by an IR-laser (wavelength 1470 nm). A protein of interest is fluorescently labelled and the change in its thermophoretic mobility, in the presence of an unlabelled ligand, is measured and expressed as a change in the normalised fluorescence (FNorm).
Proteins of interest (10–20 μM) were fluorescently labelled with an amine reactive dye (NT-647- N-hydroxysuccinimide (NHS)), cysteine reactive dye (NT-647 maleimide) or a histidine reactive dye (NT-647-Tris-NTA), according to manufacturer’s instructions (Nanotemper). Fluorescently labelled proteins were diluted to an appropriate concentration and a fixed concentration was titrated against a two-fold serial dilution of unlabelled ligand, across 16 samples, in MST running buffer (25 mM HEPES/NaOH, 150 mM NaCl, 0.05% Triton X-100, pH 7.5).
MST measurements were carried out as described in [97], using standard or premium capillaries on a Monolith NT.115 MST machine (Nanotemper). Binding curves and Kinetic parameters were plotted and estimated using manufacturer provided software (NT Analysis 1.5.41 and MO. Affinity Analysis (x64)). Capillary scans were carried out prior to all measurements to check for consistent fluorescence counts, confirming that any subsequent change in fluorescence was due to ligand binding and not due to inaccurate pipetting or adsorption and dilution effects.
SDS-denaturation (SD) test
In instances where capillary scans showed a ligand concentration dependant change in raw fluorescence, we investigated whether this was a property of the binding interaction by carrying out an SD-test.
Samples (10 μL) with the highest and lowest concentration of unlabelled ligand were centrifuged 10 000 × g, 5 min, RT and mixed 1:1 volume ratio with SD-test buffer (40 mM DTT, 4% SDS). Mixtures were boiled at 100°C for 10 min, to abolish ligand binding, before being spun down and subjected to another capillary scan. If the fluorescence between samples that contained the highest and lowest concentration of ligand, after SDS treatment, were now back to ± 10% of each other, the initial observations were a property of ligand binding, and a binding curve was plotted from the raw fluorescence data. If the ligand concentration dependent change in fluorescence was still observed, then this indicated that the fluorescently labelled protein was aggregated, and assay and buffer conditions were optimised accordingly.
In vivo cross-linking / co-immunoprecipitation assays
Method is described in, and adapted from [98]. An overnight culture of E. coli BW25113 cells and an appropriate mutant strain was used to inoculate 150 ml of Lennox LB (Fisher Scientific) and was cultivated to an OD578 of 0.5–0.6 at 37°C before harvesting by centrifugation (4500 × g, 4°C, 25 min). Cells were resuspended in 6 ml of CL buffer 1 (50 mM NaH2PO4, 20% sucrose, pH 7.4). The amine reactive cross-linker, DTSSP (3,3’-dithiobis (sulfosuccinimidylpropionate) (ThermoFisher), was freshly dissolved (20 mg/ml in dH2O) and added to the isolated cell suspension and incubated at 4°C with agitation for 1 h. Cross-linked cells were then harvested by centrifugation (4500 × g, 4°C, 25 min) and resuspended in 6 ml CL buffer 2 (100 mM Tris/HCl, 10 mM MgCl2, 1 M NaCl, pH 7.5). DNase, protease inhibitor cocktail and phenylmethylsulfonly fluoride were added prior to sonication at low levels before ultracentrifugation of the lysate (140,000 × g, 4°C, 1 h). The membrane pellet was resuspended in 2.5 ml of CL buffer 3 (25 mM Tris/HCl, 10 mM MgCl2, 1 M NaCl, 1% Triton X-100, 20% glycerol, pH 7.5) and the solubilised membrane extracted o/n with stirring at 4°C.
Samples were ultracentrifuged (140,000 × g, 4°C, 1 h) to remove debris, before removing 2 × 1.2 ml of each supernatant to be subsequently diluted with 0.6 ml of CL buffer 4 (75 mM Tris/HCl, 10 mM MgCl2, 1 M NaCl, pH 7.5). One sample was incubated with an optimised concentration of specific antibody with the other used as a negative control. Both samples were incubated at 4°C with agitation for 5 h. For the isolation of antibodies, and thus cross-linked interaction partners, 100 μl of protein G-coupled agarose bead resin (Roche) were washed (2 × CL buffer 4, 2 × CL wash buffer [2:1 CL buffer 3 and CL buffer 4]) and added to each sample, and incubated o/n at 4°C with agitation.
Samples were centrifuged and the supernatant retained before washing the beads 10 × 1 ml with CL wash buffer. After the final wash, beads were resuspended in 250 μl CL wash buffer and transferred to 2 ml spin dry columns and centrifuged to isolate the beads. These were then resuspended in 50 μl of fresh SDS-loading buffer and boiled to elute bound proteins, and reverse cross-linkage, and were collected by centrifugation (10,000 × g, RT, 5 min). Supernatant and elution samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane by Western blotting to detect for specific interaction partners using purified antibodies. The secondary antibody used here is Trueblot Anti-Rabbit IgG-HRP specific for native antibodies.
Peptidoglycan digestion assays
To test for hydrolase activity on PG, 10 μl of sacculi isolated from strains of interest (usually E. coli strains MC1061, BW25113 or D456) were incubated with 1–10 μM of respective enzymes, 37°C for between 1 h and overnight, as indicated for respective proteins. The standard reaction conditions were 10 mM HEPES/NaOH, 10 mM MgCl2, 150 mM NaCl, 0.05% Triton X-100, pH 7.5, in 100 μl reaction volume. Following incubation, samples were boiled at 100°C, 10 min, to terminate reactions before digesting remaining PG overnight at 37°C, with 1 μM cellosyl. The samples were centrifuged at 10,000 × g for 5 min, RT, to obtain digested muropeptide products in the supernatant.
To test for activity against soluble muropeptides; first, 100 μl of intact sacculi were incubated overnight at 37°C with 1 μM cellosyl and cellosyl digestion buffer (20 mM NaPO4, pH 4.8). Next, samples were boiled at 100°C, 10 min, before centrifugation at 10,000 × g for 5 min, RT, to obtain the soluble muropeptides in the supernatant [99]. Ten μl of the supernatant was then used as muropeptide substrate for incubating with enzymes of interest, for the appropriate incubation time, at 37°C. Reactions were then terminated by boiling at 100°C for 10 min.
After digestion of respective substrates, products were reduced with NaBH4, adjusted to pH 4–5 and separated for analysis by reversed-phase HPLC as described below in HPLC analysis.
Reduction of muropeptides with sodium borohydride
PG digestion samples were transferred to 2 ml vials following centrifugation. Muropeptides were reduced in a 1:1 volume ratio of sodium borohydride buffer (0.5 M sodium borate, pH 9.0) and a small spatula of sodium borohydride pellets, centrifuging at 3000 × g, 30 min, RT as described in [99]. The samples were adjusted to pH 4–5 using HPLC grade phosphoric acid before separation and analysis by reversed-phase HPLC.
Reversed-phase HPLC analysis of muropeptides
Following the protocol of [99], reduced muropeptides were separated for analysis on reversed-phase HPLC systems with a Prontosil 120-3-C18-AQ 3 μm reversed-phase HPLC column (Bischoff). A linear gradient of solvent A (50 mM sodium phosphate, pH 4.31 supplemented with 0.2% NaN3) to 100% solvent B (75 mM sodium phosphate, pH 4.95, 15% methanol) over 90 or 180 min was used to separate muropeptides at 52°C. Unlabelled muropeptides were detected at UV absorbance 205 nm. In assays where 14C-radiolabelled muropeptides were used, detection was achieved by flowing scintillation cocktail along with standard buffers to give a radioisotope scintillation count (radioactivity CPM). Muropeptide profiles were recorded and analysed using Laura V4.2.11.129 (LabLogic Systems Ltd.).
Spectrophotometric D-alanine release assay (DD-carboxypeptidase assay)
This protocol was adapted from [60]. The carboxypeptidase (CPase) activity of PG hydrolases results in the release of the terminal D-Ala residue from the pentapeptide stem of PG precursors. Using UDP-MurNAc pentapeptide as a substrate, and in this case PBP4, it was possible to spectrophotometrically measure the release of D-Ala.
Each reaction sample consisted of 200 μl of CPase buffer (50 mM HEPES/NaOH, 10 mM MgCl2, pH 7.6), 3 units of D-amino acid oxidase (Sigma), 6 units of horseradish peroxidase (HRP) (Sigma), Amplex Red (Sigma), and an optimised concentration of protein.
All constituents of the reaction were added and mixed directly in a quartz cuvette (Hellma, 10 mm light path, 15 mm centre), before the addition, and brief mixing by pipette, of purified UDP-MurNAc pentapeptide (BACWAN, Warwick University) to begin the reaction. The released D-Ala residues from the CPase activity of PBP4 are oxidatively deaminated by the action of D-amino acid oxidase to produce pyruvate and hydrogen peroxide (H2O2). The released H2O2 is reduced to H2O by HRP using Amplex Red as an electron donor. Oxidised Amplex Red produces resorufin, which has an intense pink colour and the production of which was measured spectrophotometrically using a Cary 100 Bio UV-visible spectrophotometer (wavelength 555 nm). The change in absorption over 10 min was measured and analysed using the complementing software.
Circular dichroism
Proteins were dialysed overnight against 10 mM NaPO4, pH 7.5 and concentrated/diluted to 0.4 mg/ml. CD measurements were taken using a Jasco J-810 spectropolarimeter (Jasco, Tokyo, Japan) using a wavelength range of 180–250 nm. The average of 10 runs was taken for each protein with a buffer control subtraction. For a direct comparison, correcting for the differing amino acid sequences, the collected data were converted to molecular CD and plotted against wavelength (nm). The resulting CD spectra are compared in S4 Fig and show that PBP4 lacking domain 3 is folded, consisting of both α-helices (~190, 208 and 222 nm) and β-sheets (~210 nm).
Analytical ultracentrifugation
Purified PBP4 and PBP4ΔD3 were dialyzed over night against 25 mM HEPES/NaOH, 150 mM NaCl, pH 7.5, in preparation for AUC experiments. AUC sedimentation velocity (SV) experiments were carried out in a Beckman Coulter (Palo Alto, CA, USA) ProteomeLab XL-I analytical ultracentrifuge using absorbance at 280 nm and interference detection. The AUC runs were executed at the rotation speed of 45,000 rpm and the temperature of 20°C using an 8-hole AnTi50 rotor and double-sector aluminium-Epon centerpieces. The sample volume was 400 μl and the sample concentrations ranged between approximately 0.25 and 1.4 mg/ml. The partial specific volumes () of the proteins were calculated from their amino acid sequence, using the program SEDNTERP [100]. Sedimentation velocity boundaries were analyzed using the size-distribution c(s) model implemented in the program SEDFIT http://www.analyticalultracentrifugation.com [101]. The experimental values of the sedimentation coefficient were converted to the standard conditions (s20,w), which is the value of sedimentation coefficient in water at 20°C. The size-distribution peaks were integrated to obtain the weight-averaged values for sedimentation coefficient and molecular mass.
The atomic coordinates from the published high-resolution structure of [58] (pdb accession code 2EX2) were used to calculate the sedimentation coefficient values for the monomeric and dimeric forms of PBP4 /PBP4ΔD3 using the program SoMo [102]. PBP4 /PBP4ΔD3 crystallographic dimer was built using program PyMol (https://pymol.org/2/).
PG binding assay
To assay for interactions of respective proteins with purified PG, we used a PG binding protocol as described in [103]. Briefly, ~100 μg purified PG was pelleted by centrifugation (10,000 × g, 10 min, 4°C) and resuspended in binding buffer (10 mM Tris/Maleate, 10 mM MgCl2, 50 mM NaCl, pH 6.8). PG was incubated with 10 μg of desired protein before incubating on ice for 30 min and centrifuging at 10,000 × g, 10 min, 4°C. An aliquot of the supernatant (S) was taken for SDS-PAGE. The pelleted PG was washed with 200 μl binding buffer by centrifugation as before and the wash supernatant was taken as sample W for SDS-PAGE. The pelleted PG was then resuspended in 100 μl of 2% SDS and incubated for 1 h on a stirring plate, 4°C. Mixtures were centrifuged a final time at 10,000 × g, 10 min, 4°C, with the supernatant, now containing any initially bound protein, taken as the pellet sample (P). A negative control sample with no PG was assayed in parallel to determine that binding of protein was specific. Samples, along with corresponding controls, were analysed by SDS-PAGE and visualised by Coomassie staining. Presence of protein band in pellet sample (P) indicated binding to PG, whilst presence in supernatant sample (S) indicated no binding.
Bocillin binding assay
A fluorescent-bocillin binding assay was used to determine whether a PBP was purified with a correctly folded active site. Bocillin FL is a commercially available dye-β-lactam conjugate, synthesised from penicillin V and BODIPY (Molecular Probes, In., Eugene, Oreg.). Ten μg of purified PBP was incubated for 10 min at 37°C, with 20 ng of Bocillin FL in a 50 μl volume with 10 mM HEPES/NaOH, 10 mM MgCl2, pH 7.5. A negative control sample was pre-incubated with 1 mM ampicillin for 30 min, 37°C, to block the PBP active site, prior to incubation with Bocillin FL. After incubation, samples were boiled at 100°C for 10 min with 40 μl of SDS-PAGE loading buffer and resolved by SDS-PAGE. Fluorescent signal was observed using Typhoon Fluorescence-imager (Excitation; 488 nm, emission; 520 nm, BP20 PMT 400–600 V), followed by visualisation of gels by Coomassie staining. Proteins incubated with ampicillin should have no detectable binding of Bocillin FL. For whole cells, a culture was grown in 5 mL TY and induced with 10 μM IPTG for 3 mass doublings to an OD600 of 0.5 and the cells were pelleted and washed once with PBS (8 g NaCl, 1.8 g Na2HPO4, 0.24 g KH2PO4 per liter, adjusted to pH 7.4) at 4°C and resuspended in 100 μl PBS with 5μg/ml Bocilin FL and incubated for 20 min at RT. To remove the excess Bocillin FL, the samples were pelleted and resuspended in 100 μl PBS with cOmplete Mini Protease Inhibitor Cocktail (Sigma) and 2 U DNAse I (NEB). Then sonicated on ice for 10 sec and boiled 15’ at 99°C after adding 25 μL of SDS-PAGE 5x loading buffer and resolved by SDS-PAGE. Fluorescent signal was observed using a home-made imager for Midori green, followed by visualisation of gels by Coomassie staining.
In vitro PG synthesis assay with radiolabelled lipid II
The assay was published by Bertsche et al., [104]. Lipid II (1.2 nmol, 11,000 dpm) was vacuum dried and dissolved in 5 μl of methanol. The reaction was performed in a total volume of 50 μl in 25 mM Tris/HCl, 10 mM MgCl2, 100 mM NaCl, 0.05% Triton X-100, pH 7.5 (buffer B), with 0.5 μM PBP1A, 1 μM LpoA, 1 μM PBP4(S62A), 2 μM NlpI. The reaction was performed for 1 h at 30°C. The pH of the samples was adjusted to 4.8 prior to boiling for 5 min followed by a digestion with 50 g/ml cellosyl for 1 h at 37°C and boiling for 5 min.
Supporting information
(A) Demograph with cells sorted according to length of PBP4 fluorescence of the BW25113ΔdacB (PBP4) strain, BW245113 (wt) strain before purification of the antibody and the BW25113ΔdacB strain and BW245113 strain after purification of the antibody. (B) Absolute fluorescence average profiles of cells from the BW25113 ΔdacB strain and the BW245113 strain before purification of the antibody and from the BW245113 strain after purification of the antibody (C) Phase contrast and corresponding fluorescence image of anti PBP4 immunolabeled BW25113ΔdacB (left) and wild-type cells using the supernatant of BW25113ΔdacB pre-adsorbed antibodies. The scale bar equals 5 μm. (D) BW25113 wild-type cells were grown in TY at 37°C and harvested in the exponential phase at an OD600 of 0.3 and fixed. Samples were divided into two aliquots and cells were immunolabeled with antibodies against PBP3, FtsN, LpoB and PBP4. The first aliquot was immunolabeled after permeabilizing the cell membrane with Triton X-100 (right: phase contrast and fluorescence images) and the second after permeabilizing cells with Triton X-100 and lysozyme that cleaves the glycan strands of the PG layer (left: phase contrast and fluorescence images). The scale bar equals 5 μm. PBP4 and LpoB, but not PBP3 or FtsN, are accessible without degrading the peptidoglycan layer.
(TIF)
Strains were grown in LB at 37°C to an OD of 0.3 in the presence of 30 μM IPTG to induce expression of mNG-FtsE then fixed and immunolabeled with antibodies specific for PBP4. (A) LMC500 (1108 cells analyzed). (B) XL36 (LMC500::pTrc99downftsX, ΔftsE) with pXL110 mNG-FtsE(wt) (1189 cells analyzed). From left to right, phase contrast image, anti-PBP4 immunolabeling, mNG-FtsE, and the corresponding demograph of the diameter and the demograph of fluorescence of cells sorted according to length. Scale bar equals 5 μm.
(TIF)
Isogenic strains of the wild-type strain MC1061 were grown in LB at 37°C to an OD600 of 0.3, fixed and labeled with specific antibodies against PBP4. (A) Demographs of the fluorescence distribution in cell sorted according to their length. The numbers above the graph indicate the adjustment of the brightness and the contrast, which had to be different for the double and triple-ami deletion strain as they produced more PBP4 than the other strains. (B) peak normalized fluorescence along the cell length of all cells in the demograph. (C) Images showing examples of the localization of PBP4 (green) overlayed with the phase contrast of the corresponding cells. The number of cells analyzed were 3311 for MC1061 (wt), 1691 for ΔamiC, 1865 for ΔnlpI AmiC, 926 for ΔamiAB for 484 for ΔamiABC. The scale bar equals 2 μm.
(TIF)
Isogenic strains of the wild-type strain BW25113 were grown in LB at 37°C to an OD600 of 0.3, fixed and labeled with specific antibodies against PBP4. From left to right, the phase contrast, corresponding fluorescence image of the PBP4 labeling, and the merged former two images, demograph of diameters (Dia) and demograph of fluorescence (Fluor) PBP4 localization where cells are sorted according to their cell length are shown. The number of cells analyzed were 1053 for BW25113 (wt), 918 for ΔtolA, 1314 for Δpal, and 1600 for ΔnlpD. The scale bar equals 5 μm.
(TIF)
Cells were grown exponentially to an OD of 0.3 in TY at 37°C, then fixed and immunolabeled with antibodies against PBP4. Because fluorescence imaging by microscopes is usually not directly comparable between different experiments, all results in each experiment were normalized to the parental strain BW25113. (A) PBP4 fluorescence at midcell per μm circumference of the cell. (B) Concentration of PBP4 in the cells. (C) Example of demographs showing fluorescence of PBP4 and its localization in cells sorted according to length of one experiment. The white line gives the limit of the cell lengths. (D) length of the cells. (E) Diameter of the cells. ΔlpoA (n = 5), ΔlpoB (n = 5), ΔmrcA (PBP1A, n = 2), ΔmrcB (PBP1B, n = 2). Each point is the average of 1000–2000 cells. Based on the one-way Anova the difference in midcell localization is significant (P = 0.009) while the difference in concentration of PBP4 is not significant (P = 0.0484).
(TIF)
(A) BW 25113 ΔnlpI cells were grown exponentially in TY medium at 37°C and fixed while shaking when at an OD600 of 0.3. The cells were harvested and split in three portions of which one was directly immunolabeled with anti-PBP4 antibodies that were pre-adsorbed to ΔdacB cells, the second was first treated with Triton X-100 and the last was treated with Triton X-100 and lysozyme and then immunolabeled. The scale bar equals 2 μm. (B) Map of PBP4 fluorescence sorted according to cell length of wild-type cells and ΔnlpI cells displayed at the same brightness and contrast values.
(TIF)
MC4100 cells were grown to steady state in minimal glucose medium at 28°C and split in two parts. One part was 1:4 diluted in prewarmed medium without aztreonam, and the other part was 1:4 diluted in medium with 10 μg/ml aztreonam. The cells continued to grow for 0, 1, 2 or 3 mass doublings (MD) and were fixed and immunolabeled with antibodies specific for FtsZ or PBP4. Demographs with identical brightness and contrast of the FtsZ or the PBP4 fluorescence of cells grown in the absence or presence of aztreonam sorted according to length. The white line shows the borders of the cells. Number of analyzed cells per demograph for FtsZ and PBP4 were, 5084 and 3448 (0), 1430 and 1858 (1), 857 and 1630 (2) and 659 and 595 (3), respectively.
(TIF)
Mutants expressed from plasmid without induction in the ΔdacB strain grown in TY at 37°C. (A) An immunoblot of membranes of PBP4 wild-type and variants. *This extra band is due to non-specificity of the primary antibody, which was for the purpose of immunoblotting not affinity purified. (B) Immunoblot of supernatant after pelleting the membrane of PBP4 wild-type and variants. (C) The corresponding gel where binding of the fluorescent β-lactam Bocillin FL is visible for all mutants apart from S62G. EP is empty plasmid, WT is wild-type. The other samples are PBP4 variants.
(TIF)
(A) PBP4 versions were incubated with the fluorescent β-lactam Bocillin FL with or without pre-incubation with Penicillin G (PenG), followed by SDS-PAGE analysis and detection of covalent Bocillin-PBP4 adducts by fluorescence scanner. PBP4 and PBPP4ΔD3, but not catalytically inactive PBP4(S62A), bound Bocillin FL. (B) Only wild-type PBP4, but not PBPP4ΔD3 or PBP4(S62A), was active in a DD-carboxypeptidase assay with UDP-MurNAc pentapeptide substrate. (C) PBP4 versions were incubated with muropeptides from BW25113 prior to their separation by high-performance liquid chromatography (HPLC). A control sample contained no enzyme. PBP4 DD-endopeptidase activity is demonstrated by the reduction in the dimer (TetraTetra) substrate peak and increase in the monomer (Tetra) product peak. PBPP4ΔD3 and PBP4(S62G) were inactive. (D) Quantification of the Tetra and TetraTetra peaks shown in panel C (top) and quantification of a similar analysis with PG sacculi (incubated with PBP4 versions or no enzyme), followed by generation of muropeptides and HPLC analysis (bottom).
(TIF)
(A) Analytical ultracentrifugation sedimentation velocity experiment of PBP4 and PBP4ΔD3 shows that both proteins are mainly dimers. The determined sedimentation coefficients (sexp) for the dimers fit well with the theoretical values (scalc) that were calculated from atomic coordinates of both protein dimers (pdb accession code for monomer is 2EX2). (B) PBP4 and PBP4ΔD3 show similar far UV circular dichroism spectra. (C) PBP4 and PBP4ΔD3 co-sediment with PG sacculi from BW25113 cells, but did not sediment in control samples without sacculi, demonstrating binding to PG. S, supernatant; W, wash fraction; P, pellet fraction.
(TIF)
Cells were grown in minimal glucose medium at 28°C, fixed and immunolabeled against FtsZ and FtsN. (A) Graphical illustration of the meaning of t0 and t1/2. (B). Cell division cycle age timing of FtsZ and FtsN for the ΔdacB strain expressing PBP4 variants from plasmid without induction and its parental BW25113. Because BW25113 cannot be grown to steady state, the difference in divisome assembly of the PBP4 variant should be interpretated as not identical to that of the wild type in the case of S63G and D155A, ignoring the absolute numbers.
(TIF)
MST binding curves for interactions using different fluorescently labelled proteins (LpoA, LpoAC, LpoN, NlpI or PBP1A) titrated against fixed concentrations of PBP4 or PBPP4ΔD3. Apparent KD values are mean ± SD of three independent experiments and summarized in Fig 8A.
(TIF)
Representative HPLC chromatograms of samples from an in vitro PG synthesis assay of PBP1A and radiolabelled Lipid II, in the presence of proteins indicated. The main PG products upon digestion with the muramidase cellosyl are the disaccharide pentapeptide (Penta) and the bis-disaccharide tetrapentapeptide (TetraPenta). The % peptides present in cross-links is quantified on the right side and presented as mean ± variation of two independent experiments.
(TIF)
(A) Summary of the results from an in vitro PG synthesis assay with PBP1A (in the presence or absence of LpoA) and radio-labelled lipid II substrate. The presence of NlpI, catalytically inactive PBP4(S62A) or both together, does not affect the cross-linkage of the PG product of a PBP1A or PBP1A/LpoA reaction (left side), and NlpI, PBP4(S62A) or both together, do not induce enhanced carboxypeptidase activity of PBP1A, which would be seen as a higher content of tetrapeptides (right side). Values are mean ± variation of two independent repeats. Example chromatograms are shown in S12B Fig. (B) The presence of LpoA modestly reduces the activity of PBP4 in a DD-carboxypeptidase assay using the substrate UDP-MurNAc pentapeptide (C) LpoA also reduces slightly the activity of PBP4 against PG sacculi from BW25113, as seen by the reduced digestion of TetraTetra upon a 30 min incubation period.
(TIF)
(A) Interaction assays by MST with fluorescently labelled EnvC (Fl-EnvC), fluorescently labelled NlpI (Fl-NlpI), NlpI, PBP4, PBP4ΔD3 and LytM domain of EnvC (EnvC-LytM). The apparent KD values are indicated. (B) Summary of interaction data. (C) Pulldown to Ni-NTA beads using oligohistidine-tagged-NlpI (His-NlpI) and untagged PBP4 and EnvC. Proteins were detected on a Coomassie Blue-stained SDS-PAGE. A, applied sample; E, elution sample.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank David Roper and Smita Chauhan (University of Warwick) for help with the carboxypeptidase assay, Helen Waller (Newcastle University) for help with circular dichroism methodology, Lisa Atkinson and Daniela Vollmer (Newcastle University) for preparation of PG sacculi, and Thomas van Wingen (University of Amsterdam) for with help the Tre1 assay. We thank See-Yeun Ting for the plasmids expressing Tre1 and Tre1E415Q (University of Washington School of Medicine, Seattle, USA).
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The work was supported by funding from the Wellcome Trust (101824/Z/13/Z), to WV). Xiaolong Liu (File No.201406220123) and Xinwei Liu (File No. 201804910650) were supported by the China Scholarship Council fellowship. This work was supported by a UKRI Future Leaders Fellowship to MB [MR/V027204/1]. GB was supported by a Darwin PhD Fellowship. The funders had no role in studydesign, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Vollmer W, Blanot D, Pedro MAD. Peptidoglycan structure and architecture. FEMS microbiol. rev. 2008;32: 149–167. doi: 10.1111/j.1574-6976.2007.00094.x [DOI] [PubMed] [Google Scholar]
- 2.Gray AN, Egan AJF, Veer ILV, Verheul J, Colavin A, Koumoutsi A, et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Storz G, editor. eLife. 2015;4: e07118. doi: 10.7554/eLife.07118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman MEG, Kannenberg K, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 2006;61: 675–690. doi: 10.1111/j.1365-2958.2006.05280.x [DOI] [PubMed] [Google Scholar]
- 4.Born P, Breukink E, Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. Biol. Chem. 2006;281: 26985–26993. doi: 10.1074/jbc.M604083200 [DOI] [PubMed] [Google Scholar]
- 5.Egan AJF, Biboy J, van’t Veer I, Breukink E, Vollmer W. Activities and regulation of peptidoglycan synthases. Philosophical transactions of the Royal Society of London Series B, Biol. Sci. 2015;370. doi: 10.1098/rstb.2015.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pazos M, Vollmer W. Regulation and function of class A Penicillin-binding proteins. Curr Opin Microbiol. 2021;60: 80–87. doi: 10.1016/j.mib.2021.01.008 [DOI] [PubMed] [Google Scholar]
- 7.Rohs PDA, Buss J, Sim SI, Squyres GR, Srisuknimit V, Smith M, et al. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genetics. 2018;14: e1007726. doi: 10.1371/journal.pgen.1007726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537: 634–638. doi: 10.1038/nature19331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol. 2019;4: 587–594. doi: 10.1038/s41564-018-0345-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ploeg R van der, Verheul J, Vischer NOE, Alexeeva S, Hoogendoorn E, Postma M, et al. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol Microbiol. 2013;87: 1074–1087. doi: 10.1111/mmi.12150 [DOI] [PubMed] [Google Scholar]
- 11.Datta P, Dasgupta A, Singh AK, Mukherjee P, Kundu M, Basu J. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol Microbiol. 2006;62: 1655–1673. doi: 10.1111/j.1365-2958.2006.05491.x [DOI] [PubMed] [Google Scholar]
- 12.Fraipont C, Alexeeva S, Wolf B, van der Ploeg R, Schloesser M, Blaauwen T den, et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology. 2011;157: 251–259. doi: 10.1099/mic.0.040071-0 [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Biboy J, Consoli E, Vollmer W, Blaauwen T den. MreC and MreD balance the interaction between the elongasome proteins PBP2 and RodA. PLoS Genetics. 2020;16: e1009276. doi: 10.1371/journal.pgen.1009276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Addinall SG, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 1996;22: 231–237. doi: 10.1046/j.1365-2958.1996.00100.x [DOI] [PubMed] [Google Scholar]
- 15.Raychaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18: 2372–2383. doi: 10.1093/emboj/18.9.2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazos M, Peters K, Casanova M, Palacios P, VanNieuwenhze M, Breukink E, et al. Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat Commun. 2018;9: 5090–12. doi: 10.1038/s41467-018-07559-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galli E, Gerdes K. FtsZ-ZapA-ZapB Interactome of Escherichia coli. J. Bacteriol. 2012;194: 292–302. doi: 10.1128/JB.05821-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand-Heredia JM, Yu HH, Carlo SD, Lesser CF, Janakiraman A. Identification and Characterization of ZapC, a Stabilizer of the FtsZ Ring in Escherichia coli. J. Bacteriol. 2011;193: 1405–1413. doi: 10.1128/JB.01258-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 2011;193: 1393–1404. doi: 10.1128/JB.01245-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roach EJ, Wroblewski C, Seidel L, Berezuk AM, Brewer D, Kimber MS, et al. Structure and mutational analyses of Escherichia coli ZapD reveal charged residues involved in FtsZ filament bundling. DiRita VJ, editor. J. Bacteriol. 2016;198: 1683–1693. doi: 10.1128/JB.00969-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, Touqui L, et al. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio. 2014;5: e00022–14. doi: 10.1128/mBio.00022-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehla J, Liechti G, Morgenstein RM, Caufield JH, Hosseinnia A, Gagarinova A, et al. ZapG (YhcB/DUF1043), a novel cell division protein in gamma-proteobacteria linking the Z-ring to septal peptidoglycan synthesis. J Biol Chem. 2021;296: 100700. doi: 10.1016/j.jbc.2021.100700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du S, Pichoff S, Lutkenhaus J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Nat. Acad. Sci. USA. 2016;113: E5052–61. doi: 10.1073/pnas.1606656113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbin BD, Wang Y, Beuria TK, Margolin W. Interaction between Cell Division Proteins FtsE and FtsZ. J. Bacteriol. 2007;189: 3026–3035. doi: 10.1128/JB.01581-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J. Bacteriol. 2011;193: 4973–4983. doi: 10.1128/JB.00316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, Blaauwen T den. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 2005;55: 1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x [DOI] [PubMed] [Google Scholar]
- 27.Perez AJ, Cesbron Y, Shaw SL, Villicana JB, Tsui H-CT, Boersma MJ, et al. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Nat. Acad. Sci. USA. 2019;116: 3211–3220. doi: 10.1073/pnas.1816018116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature. 2018;554: 528–532. doi: 10.1038/nature25506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsang M-J, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 2014. doi: 10.1111/mmi.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 2007;64: 1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boes A, Olatunji S, Breukink E, Terrak M. Regulation of the Peptidoglycan Polymerase Activity of PBP1b by Antagonist Actions of the Core Divisome Proteins FtsBLQ and FtsN. Blaauwen T den, Salama NR, editors. mBio. 2019;10: e01912–18. doi: 10.1128/mBio.01912-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaauwen T den, Luirink J. Checks and Balances in Bacterial Cell Division. mBio. 2019;10. doi: 10.1128/mBio.00615-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egan AJF, Errington J, Vollmer W. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol. 2020;18: 446–460. doi: 10.1038/s41579-020-0366-3 [DOI] [PubMed] [Google Scholar]
- 34.Pazos M, Peters K, Vollmer W. Robust peptidoglycan growth by dynamic and variable multi-protein complexes. Curr. Opin. Microbiol. 2017;36: 55–61. doi: 10.1016/j.mib.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 35.Sarkar SK, Dutta M, Chowdhury C, Kumar A, Ghosh AS. PBP5, PBP6 and DacD play different roles in intrinsic -lactam resistance of Escherichia coli. Microbiology (Reading, England). 2011;157: 2702–2707. doi: 10.1099/mic.0.046227-0 [DOI] [PubMed] [Google Scholar]
- 36.Potluri L, Karczmarek A, Verheul J, Piette A, Wilkin J-M, Werth N, et al. Septal and lateral wall localization of PBP5, the major D,D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol. Microbiol. 2010;77: 300–323. doi: 10.1111/j.1365-2958.2010.07205.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hugonnet J-E, Mengin-Lecreulx D, Monton A, Blaauwen T den, Carbonnelle E, Veckerlé C, et al. Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. eLife. 2016;5: 2006. doi: 10.7554/eLife.19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meiresonne NY, Ploeg R van der, Hink MA, Blaauwen T den. Activity-Related Conformational Changes in d,d-Carboxypeptidases Revealed by In Vivo Periplasmic Förster Resonance Energy Transfer Assay in Escherichia coli. Søgaard-Andersen L, editor. mBio. 2017;8: e01089–17. doi: 10.1128/mBio.01089-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh SK, Saisree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol. Microbiol. 2012;86: 1036–1051. doi: 10.1111/mmi.12058 [DOI] [PubMed] [Google Scholar]
- 40.Meberg BM, Paulson AL, Priyadarshini R, Young KD. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 2004;186: 8326–8336. doi: 10.1128/JB.186.24.8326-8336.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters K, Kannan S, Rao VA, Biboy J, Vollmer D, Erickson SW, et al. The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia coli. mBio. 2016;7. doi: 10.1128/mBio.00819-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 2001;41: 167–178. doi: 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- 43.Bernhardt TG, Boer PAJD. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 2003;48: 1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29: 1412–1422. doi: 10.1038/emboj.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Priyadarshini R, Popham DL, Young KD. Daughter Cell Separation by Penicillin-Binding Proteins and Peptidoglycan Amidases in Escherichia coli. J. Bacteriol. 2006;188: 5345–5355. doi: 10.1128/JB.00476-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Nat. Acad. Sci. USA. 2011;108: E1052–60. doi: 10.1073/pnas.1107780108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol. Microbiol. 2012;85: 768–781. doi: 10.1111/j.1365-2958.2012.08138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsang M-J, Yakhnina AA, Bernhardt TG. NlpD links cell wall remodeling and outer membrane invagination during cytokinesis in Escherichia coli. PLoS Genetics. 2017;13: e1006888. doi: 10.1371/journal.pgen.1006888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano CKG, Winkle M, Martorana AM, Biboy J, Morè N, Moynihan P, et al. ActS activates peptidoglycan amidases during outer membrane stress in Escherichia coli. Mol Microbiol. 2021. doi: 10.1111/mmi.14712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mueller EA, Iken AG, Öztürk MA, Winkle M, Schmitz M, Vollmer W, et al. The active repertoire of Escherichia coli peptidoglycan amidases varies with physiochemical environment. Mol Microbiol. 2021;116: 311–328. doi: 10.1111/mmi.14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Straaten KE van, Dijkstra BW, Vollmer W, Thunnissen A-MWH. Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold. J. Mol. Biol. 2005;352: 1068–1080. doi: 10.1016/j.jmb.2005.07.067 [DOI] [PubMed] [Google Scholar]
- 52.Heijenoort J van. Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev.: MMBR. 2011;75: 636–663. doi: 10.1128/MMBR.00022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yunck R, Cho H, Bernhardt TG. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol. Microbiol. 2015. doi: 10.1111/mmi.13258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh SK, Parveen S, Saisree L, Reddy M. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc. Nat. Acad. Sci. USA. 2015;112: 10956–10961. doi: 10.1073/pnas.1507760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banzhaf M, Yau HCL, Verheul J, Lodge A, Kritikos G, Mateus A, et al. Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO J. 2020; e102246. doi: 10.15252/embj.2019102246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korat B, Mottl H, Keck W. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol. Microbiol. 1991;5: 675–684. doi: 10.1111/j.1365-2958.1991.tb00739.x [DOI] [PubMed] [Google Scholar]
- 57.Harris F, Brandenburg K, Seydel U, Phoenix D. Investigations into the mechanisms used by the C-terminal anchors of Escherichia coli penicillin-binding proteins 4, 5, 6 and 6b for membrane interaction. Eur. J. Biochem. 2002;269: 5821–5829. doi: 10.1046/j.1432-1033.2002.03295.x [DOI] [PubMed] [Google Scholar]
- 58.Kishida H, Unzai S, Roper DI, Lloyd A, Park S-Y, Tame JRH. Crystal Structure of Penicillin Binding Protein 4 (dacB) from Escherichia coli, both in the Native Form and Covalently Linked to Various Antibiotics. Biochemistry. 2006;45: 783–792. doi: 10.1021/bi051533t [DOI] [PubMed] [Google Scholar]
- 59.Thunnissen MMGM, Fusetti F, Boer BD, Dijkstra BW. Purification, crystallisation and preliminary X-ray analysis of penicillin binding protein 4 from Escherichia coli, a protein related to class a β-lactamases. J. Mol. Biol. 1995;247: 149–153. doi: 10.1006/jmbi.1994.0128 [DOI] [PubMed] [Google Scholar]
- 60.Clarke TB, Kawai F, Park S-Y, Tame JRH, Dowson CG, Roper DI. Mutational analysis of the substrate specificity of Escherichia coli penicillin binding protein 4. Biochemistry. 2009;48: 2675–2683. doi: 10.1021/bi801993x [DOI] [PubMed] [Google Scholar]
- 61.Pedro MAD, Schwarz U. Heterogeneity of newly inserted and preexisting murein in the sacculus of Escherichia coli. Proc. Nat. Acad. Sci. USA. 1981;78: 5856–5860. doi: 10.1073/pnas.78.9.5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwaya M, Strominger JL. Simultaneous deletion of D-alanine carboxypeptidase IB-C and penicillin-binding component IV in a mutant of Escherichia coli K12. Proc Nat. Acad Sci.USA. 1977;74: 2980–2984. doi: 10.1073/pnas.74.7.2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mallick S, Kiran S, Maiti TK, Ghosh AS. PBP4 and PBP5 are involved in regulating exopolysaccharide synthesis during Escherichia coli biofilm formation. Microbiology+. 2021;167. doi: 10.1099/mic.0.001031 [DOI] [PubMed] [Google Scholar]
- 64.Nelson DE, Young KD. Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 2001;183: 3055–3064. doi: 10.1128/JB.183.10.3055-3064.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hocking J, Priyadarshini R, Takacs CN, Costa T, Dye NA, Shapiro L, et al. Osmolality-Dependent Relocation of Penicillin-Binding Protein PBP2 to the Division Site in Caulobacter crescentus. J. Bacteriol. 2012;194: 3116–3127. doi: 10.1128/JB.00260-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol.: CB. 2005;15: R514–26. doi: 10.1016/j.cub.2005.06.038 [DOI] [PubMed] [Google Scholar]
- 67.Blaauwen T den, Hamoen LW, Levin PA. The divisome at 25: the road ahead. Curr. Op. Microbiol. 2017;36: 85–94. doi: 10.1016/j.mib.2017.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157: 624–635. doi: 10.1016/j.cell.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ting S-Y, Bosch DE, Mangiameli SM, Radey MC, Huang S, Park Y-J, et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell. 2018;175: 1380–1392.e14. doi: 10.1016/j.cell.2018.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koppelman C-M, Aarsman MEG, Postmus J, Pas E, Muijsers AO, Scheffers D-J, et al. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 2004;51: 645–657. doi: 10.1046/j.1365-2958.2003.03876.x [DOI] [PubMed] [Google Scholar]
- 71.Pichoff S, Du S, Lutkenhaus J. Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc. Nat. Acad. Sci. USA. 2018;115: E6855–E6862. doi: 10.1073/pnas.1806450115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du S, Henke W, Pichoff S, Lutkenhaus J. How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol. Microbiol. 2019. doi: 10.1111/mmi.14324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arends SJR, Kustusch RJ, Weiss DS. ATP-binding site lesions in FtsE impair cell division. J. Bacteriol. 2009;191: 3772–3784. doi: 10.1128/JB.00179-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Banzhaf M, Saparoea B van den B van, Terrak M, Fraipont C, Egan A, Philippe J, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 2012;85: 179–194. doi: 10.1111/j.1365-2958.2012.08103.x [DOI] [PubMed] [Google Scholar]
- 75.Sykes RB, Bonner DP, Bush K, Georgopapadakou NH. Azthreonam (SQ 26,776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob Agents Ch. 1982;21: 85–92. doi: 10.1128/aac.21.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nanninga N. Cell division and peptidoglycan assembly in Eschenchia coli. Mol. Microbiol. 1991;5: 791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x [DOI] [PubMed] [Google Scholar]
- 77.Pedro MAD, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli. J. Bacteriol. 1997;179: 2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu X, Meiresonne NY, Bouhss A, Blaauwen T den. FtsW activity and lipid II synthesis are required for recruitment of MurJ to midcell during cell division in Escherichia coli. Mol. Microbiol. 2018;109: 855–884. doi: 10.1111/mmi.14104 [DOI] [PubMed] [Google Scholar]
- 79.Alcorlo M, Dik DA, Benedetti SD, Mahasenan KV, Lee M, Domínguez-Gil T, et al. Structural basis of denuded glycan recognition by SPOR domains in bacterial cell division. Nat Commun. 2019;10: 5567–13. doi: 10.1038/s41467-019-13354-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vischer NOE, Verheul J, Postma M, Saparoea B van den B van, Galli E, Natale P, et al. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front. Microbiol. 2015;6: 586. doi: 10.3389/fmicb.2015.00586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175: 3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jean NL, Bougault CM, Lodge A, Derouaux A, Callens G, Egan AJF, et al. Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: implications for PBP1A stimulation. Structure (London, England: 1993). 2014;22: 1047–1054. doi: 10.1016/j.str.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jean NL, Bougault C, Derouaux A, Callens G, Vollmer W, Simorre J-P. Backbone and side-chain (1)H, (13)C, and (15)N NMR assignments of the N-terminal domain of Escherichia coli LpoA. Biomol. NMR Assign. 2015;9: 65–69. doi: 10.1007/s12104-014-9546-2 [DOI] [PubMed] [Google Scholar]
- 84.Typas A, Banzhaf M, Saparoea B van den B van, Verheul J, Biboy J, Nichols RJ, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143: 1097–1109. doi: 10.1016/j.cell.2010.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cook J, Baverstock TC, McAndrew MBL, Stansfeld PJ, Roper DI, Crow A. Insights into bacterial cell division from a structure of EnvC bound to the FtsX periplasmic domain. Proc. Nat. Acad. Sci. USA. 2020;117: 28355–28365. doi: 10.1073/pnas.2017134117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Potluri L-P, Kannan S, Young KD. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J. Bacteriol. 2012;194: 5334–5342. doi: 10.1128/JB.00859-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Nat. Acad. Sci. USA. 2000;97: 6640–6645. doi: 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen JC, Weiss DS, Ghigo J-M, Beckwith J. Septal Localization of FtsQ, an Essential Cell Division Protein in Escherichia coli. J. Bacteriol. 1999;181: 521–530. doi: 10.1128/JB.181.2.521-530.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alexeeva S, Gadella TWJ, Verheul J, Verhoeven GS, Blaauwen T den. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol. Microbiol. 2010;77: 384–398. doi: 10.1111/j.1365-2958.2010.07211.x [DOI] [PubMed] [Google Scholar]
- 90.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Sys. Biol. 2006;2: 2006.0008. doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomason LC, Costantino N, Court DL. E. coli Genome Manipulation by P1 Transduction. Curr Protoc Mol Biol. 2007;79: 1.17.1–1.17.8. doi: 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- 92.Taschner PE, Huls PG, Pas E, Woldringh CL. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J. Bacteriol. 1988;170: 1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Chem.Biol. 2009;6: 343–345. doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 94.Buddelmeijer N, Aarsman M, Blaauwen T den. Immunolabeling of proteins in situ in Escherichia coli K12 strains. bio-protocol. 2013;3: 1–5. [Google Scholar]
- 95.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1: 16077–16077. doi: 10.1038/nmicrobiol.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ludwig C. Diffusion between unequally heated regions of initially uniform solutions. Sitzungsber Akad Wiss Wien Math-Naturwiss K1. 1856;20: 593. [Google Scholar]
- 97.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay and drug development technologies. 2011;9: 342–353. doi: 10.1089/adt.2011.0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Müller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, et al. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. Biol. Chem. 2007;282: 36394–36402. doi: 10.1074/jbc.M706390200 [DOI] [PubMed] [Google Scholar]
- 99.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Analyt. Biochem. 1988;172: 451–464. doi: 10.1016/0003-2697(88)90468-x [DOI] [PubMed] [Google Scholar]
- 100.laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided interpretation of analytical sedimentation data for proteins. Roy. Soc. Chem. 1992;90: 125. [Google Scholar]
- 101.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78: 1606–1619. doi: 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brookes E, Demeler B, Rosano C, Rocco M. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur. Biophys. J.: EBJ. 2010;39: 423–435. doi: 10.1007/s00249-009-0418-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ursinus A, Ent F van den, Brechtel S, Pedro M de, Höltje J-V, Löwe J, et al. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J. Bacteriol. 2004;186: 6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bertsche U, Breukink E, Kast T, Vollmer W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. Biol. Chem. 2005;280: 38096–38101. doi: 10.1074/jbc.M508646200 [DOI] [PubMed] [Google Scholar]
- 105.Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. Journal of Mol. Biol. 1980;138: 179–207. doi: 10.1016/0022-2836(80)90283-1 [DOI] [PubMed] [Google Scholar]
- 106.Ishino F, Jung HK, Ikeda M, Doi M, Wachi M, Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J. Bacteriol. 1989;171: 5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Demograph with cells sorted according to length of PBP4 fluorescence of the BW25113ΔdacB (PBP4) strain, BW245113 (wt) strain before purification of the antibody and the BW25113ΔdacB strain and BW245113 strain after purification of the antibody. (B) Absolute fluorescence average profiles of cells from the BW25113 ΔdacB strain and the BW245113 strain before purification of the antibody and from the BW245113 strain after purification of the antibody (C) Phase contrast and corresponding fluorescence image of anti PBP4 immunolabeled BW25113ΔdacB (left) and wild-type cells using the supernatant of BW25113ΔdacB pre-adsorbed antibodies. The scale bar equals 5 μm. (D) BW25113 wild-type cells were grown in TY at 37°C and harvested in the exponential phase at an OD600 of 0.3 and fixed. Samples were divided into two aliquots and cells were immunolabeled with antibodies against PBP3, FtsN, LpoB and PBP4. The first aliquot was immunolabeled after permeabilizing the cell membrane with Triton X-100 (right: phase contrast and fluorescence images) and the second after permeabilizing cells with Triton X-100 and lysozyme that cleaves the glycan strands of the PG layer (left: phase contrast and fluorescence images). The scale bar equals 5 μm. PBP4 and LpoB, but not PBP3 or FtsN, are accessible without degrading the peptidoglycan layer.
(TIF)
Strains were grown in LB at 37°C to an OD of 0.3 in the presence of 30 μM IPTG to induce expression of mNG-FtsE then fixed and immunolabeled with antibodies specific for PBP4. (A) LMC500 (1108 cells analyzed). (B) XL36 (LMC500::pTrc99downftsX, ΔftsE) with pXL110 mNG-FtsE(wt) (1189 cells analyzed). From left to right, phase contrast image, anti-PBP4 immunolabeling, mNG-FtsE, and the corresponding demograph of the diameter and the demograph of fluorescence of cells sorted according to length. Scale bar equals 5 μm.
(TIF)
Isogenic strains of the wild-type strain MC1061 were grown in LB at 37°C to an OD600 of 0.3, fixed and labeled with specific antibodies against PBP4. (A) Demographs of the fluorescence distribution in cell sorted according to their length. The numbers above the graph indicate the adjustment of the brightness and the contrast, which had to be different for the double and triple-ami deletion strain as they produced more PBP4 than the other strains. (B) peak normalized fluorescence along the cell length of all cells in the demograph. (C) Images showing examples of the localization of PBP4 (green) overlayed with the phase contrast of the corresponding cells. The number of cells analyzed were 3311 for MC1061 (wt), 1691 for ΔamiC, 1865 for ΔnlpI AmiC, 926 for ΔamiAB for 484 for ΔamiABC. The scale bar equals 2 μm.
(TIF)
Isogenic strains of the wild-type strain BW25113 were grown in LB at 37°C to an OD600 of 0.3, fixed and labeled with specific antibodies against PBP4. From left to right, the phase contrast, corresponding fluorescence image of the PBP4 labeling, and the merged former two images, demograph of diameters (Dia) and demograph of fluorescence (Fluor) PBP4 localization where cells are sorted according to their cell length are shown. The number of cells analyzed were 1053 for BW25113 (wt), 918 for ΔtolA, 1314 for Δpal, and 1600 for ΔnlpD. The scale bar equals 5 μm.
(TIF)
Cells were grown exponentially to an OD of 0.3 in TY at 37°C, then fixed and immunolabeled with antibodies against PBP4. Because fluorescence imaging by microscopes is usually not directly comparable between different experiments, all results in each experiment were normalized to the parental strain BW25113. (A) PBP4 fluorescence at midcell per μm circumference of the cell. (B) Concentration of PBP4 in the cells. (C) Example of demographs showing fluorescence of PBP4 and its localization in cells sorted according to length of one experiment. The white line gives the limit of the cell lengths. (D) length of the cells. (E) Diameter of the cells. ΔlpoA (n = 5), ΔlpoB (n = 5), ΔmrcA (PBP1A, n = 2), ΔmrcB (PBP1B, n = 2). Each point is the average of 1000–2000 cells. Based on the one-way Anova the difference in midcell localization is significant (P = 0.009) while the difference in concentration of PBP4 is not significant (P = 0.0484).
(TIF)
(A) BW 25113 ΔnlpI cells were grown exponentially in TY medium at 37°C and fixed while shaking when at an OD600 of 0.3. The cells were harvested and split in three portions of which one was directly immunolabeled with anti-PBP4 antibodies that were pre-adsorbed to ΔdacB cells, the second was first treated with Triton X-100 and the last was treated with Triton X-100 and lysozyme and then immunolabeled. The scale bar equals 2 μm. (B) Map of PBP4 fluorescence sorted according to cell length of wild-type cells and ΔnlpI cells displayed at the same brightness and contrast values.
(TIF)
MC4100 cells were grown to steady state in minimal glucose medium at 28°C and split in two parts. One part was 1:4 diluted in prewarmed medium without aztreonam, and the other part was 1:4 diluted in medium with 10 μg/ml aztreonam. The cells continued to grow for 0, 1, 2 or 3 mass doublings (MD) and were fixed and immunolabeled with antibodies specific for FtsZ or PBP4. Demographs with identical brightness and contrast of the FtsZ or the PBP4 fluorescence of cells grown in the absence or presence of aztreonam sorted according to length. The white line shows the borders of the cells. Number of analyzed cells per demograph for FtsZ and PBP4 were, 5084 and 3448 (0), 1430 and 1858 (1), 857 and 1630 (2) and 659 and 595 (3), respectively.
(TIF)
Mutants expressed from plasmid without induction in the ΔdacB strain grown in TY at 37°C. (A) An immunoblot of membranes of PBP4 wild-type and variants. *This extra band is due to non-specificity of the primary antibody, which was for the purpose of immunoblotting not affinity purified. (B) Immunoblot of supernatant after pelleting the membrane of PBP4 wild-type and variants. (C) The corresponding gel where binding of the fluorescent β-lactam Bocillin FL is visible for all mutants apart from S62G. EP is empty plasmid, WT is wild-type. The other samples are PBP4 variants.
(TIF)
(A) PBP4 versions were incubated with the fluorescent β-lactam Bocillin FL with or without pre-incubation with Penicillin G (PenG), followed by SDS-PAGE analysis and detection of covalent Bocillin-PBP4 adducts by fluorescence scanner. PBP4 and PBPP4ΔD3, but not catalytically inactive PBP4(S62A), bound Bocillin FL. (B) Only wild-type PBP4, but not PBPP4ΔD3 or PBP4(S62A), was active in a DD-carboxypeptidase assay with UDP-MurNAc pentapeptide substrate. (C) PBP4 versions were incubated with muropeptides from BW25113 prior to their separation by high-performance liquid chromatography (HPLC). A control sample contained no enzyme. PBP4 DD-endopeptidase activity is demonstrated by the reduction in the dimer (TetraTetra) substrate peak and increase in the monomer (Tetra) product peak. PBPP4ΔD3 and PBP4(S62G) were inactive. (D) Quantification of the Tetra and TetraTetra peaks shown in panel C (top) and quantification of a similar analysis with PG sacculi (incubated with PBP4 versions or no enzyme), followed by generation of muropeptides and HPLC analysis (bottom).
(TIF)
(A) Analytical ultracentrifugation sedimentation velocity experiment of PBP4 and PBP4ΔD3 shows that both proteins are mainly dimers. The determined sedimentation coefficients (sexp) for the dimers fit well with the theoretical values (scalc) that were calculated from atomic coordinates of both protein dimers (pdb accession code for monomer is 2EX2). (B) PBP4 and PBP4ΔD3 show similar far UV circular dichroism spectra. (C) PBP4 and PBP4ΔD3 co-sediment with PG sacculi from BW25113 cells, but did not sediment in control samples without sacculi, demonstrating binding to PG. S, supernatant; W, wash fraction; P, pellet fraction.
(TIF)
Cells were grown in minimal glucose medium at 28°C, fixed and immunolabeled against FtsZ and FtsN. (A) Graphical illustration of the meaning of t0 and t1/2. (B). Cell division cycle age timing of FtsZ and FtsN for the ΔdacB strain expressing PBP4 variants from plasmid without induction and its parental BW25113. Because BW25113 cannot be grown to steady state, the difference in divisome assembly of the PBP4 variant should be interpretated as not identical to that of the wild type in the case of S63G and D155A, ignoring the absolute numbers.
(TIF)
MST binding curves for interactions using different fluorescently labelled proteins (LpoA, LpoAC, LpoN, NlpI or PBP1A) titrated against fixed concentrations of PBP4 or PBPP4ΔD3. Apparent KD values are mean ± SD of three independent experiments and summarized in Fig 8A.
(TIF)
Representative HPLC chromatograms of samples from an in vitro PG synthesis assay of PBP1A and radiolabelled Lipid II, in the presence of proteins indicated. The main PG products upon digestion with the muramidase cellosyl are the disaccharide pentapeptide (Penta) and the bis-disaccharide tetrapentapeptide (TetraPenta). The % peptides present in cross-links is quantified on the right side and presented as mean ± variation of two independent experiments.
(TIF)
(A) Summary of the results from an in vitro PG synthesis assay with PBP1A (in the presence or absence of LpoA) and radio-labelled lipid II substrate. The presence of NlpI, catalytically inactive PBP4(S62A) or both together, does not affect the cross-linkage of the PG product of a PBP1A or PBP1A/LpoA reaction (left side), and NlpI, PBP4(S62A) or both together, do not induce enhanced carboxypeptidase activity of PBP1A, which would be seen as a higher content of tetrapeptides (right side). Values are mean ± variation of two independent repeats. Example chromatograms are shown in S12B Fig. (B) The presence of LpoA modestly reduces the activity of PBP4 in a DD-carboxypeptidase assay using the substrate UDP-MurNAc pentapeptide (C) LpoA also reduces slightly the activity of PBP4 against PG sacculi from BW25113, as seen by the reduced digestion of TetraTetra upon a 30 min incubation period.
(TIF)
(A) Interaction assays by MST with fluorescently labelled EnvC (Fl-EnvC), fluorescently labelled NlpI (Fl-NlpI), NlpI, PBP4, PBP4ΔD3 and LytM domain of EnvC (EnvC-LytM). The apparent KD values are indicated. (B) Summary of interaction data. (C) Pulldown to Ni-NTA beads using oligohistidine-tagged-NlpI (His-NlpI) and untagged PBP4 and EnvC. Proteins were detected on a Coomassie Blue-stained SDS-PAGE. A, applied sample; E, elution sample.
(TIF)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.