Abstract
Expression of the nitrogen fixation gene, nifH, in the gut of the termite Neotermes koshunensis was characterized without cultivation. nifH cDNA was directly amplified from mRNA of the mixed microbial population in the gut by reverse transcription (RT)-PCR. Analyses of the RT-PCR products revealed that, among the diverse nifH sequences, only a few corresponding to an alternative nitrogenase (encoded by the anf gene) were preferentially transcribed in the termite gut. Expression of the anf gene was further investigated quantitatively under several termite feeding conditions by competitive PCR. The levels of expression of the anf gene were largely congruent with the nitrogen fixation activity displayed by the termite. The amounts of the genomic anf gene in the population showed no significant change, indicating that the level of expression was critical for nitrogen fixation activity. Interestingly, no significant decrease in the expression level was observed when the diet contained molybdenum (Mo), which represses ordinary anf genes. A 3.6-kb DNA region downstream of the anf gene was isolated and found to contain reading frames homologous to anfH, anfD, and anfG of the Bacteria domain which encode subunits of an alternative nitrogenase having no Mo as a cofactor. This DNA region also contained reading frames encoding glnB-like proteins, which is a common feature of the nitrogenase genes of the Archaea domain. These results indicate that the anf group of nitrogenase genes is the most important group of genes responsible for nitrogen fixation in N. koshunensis and that the anf gene possesses novel features with respect to the regulation of its expression and its gene organization.
Nitrogen fixation by the symbiotic microorganisms inhabiting the gut of termites is important, since termites thrive on a nitrogen-poor diet (3, 7, 8). The identity of the microorganisms responsible has been investigated in evolutionarily diverse termites based on comparisons of the sequences of the nitrogen fixation gene, nifH, directly amplified by PCR and cloned from the microbial community in the termite gut (26, 27). These culture-independent studies have revealed the presence of a phylogenetically diverse and hitherto unrecognized population of potential nitrogen-fixing microorganisms in the gut of termites. This kind of molecular approach, as opposed to conventional microbiological approaches, is beneficial and is increasingly being used to study natural microbial communities, to avoid the largely unrepresentative nature of microbial cultivation (1).
However, since there are some differences in the efficiencies of DNA extraction, PCR amplification, and cloning, the distribution of nifH sequences as final clones may not reflect the real distribution of nifH genes in the original microbial community. Furthermore, it must be emphasized that the existence of nifH sequences does not always mean that nitrogen-fixing activity is being expressed by the corresponding microorganisms, since the nitrogenase genes are regulated at the transcriptional and posttranslational levels (4, 12, 19). In fact, in spite of the existence of remarkably diverse nifH sequences in the gut microbial community, some termite species exhibit only a slight level of nitrogen fixation activity (26). In order to understand the nature of nitrogen fixation in termites, it is necessary to clarify the real distribution and contribution of the nitrogen fixation genes and the corresponding microorganisms.
Sequence analysis of PCR products amplified from mixed communities has proved time-consuming and is potentially biased due to the requirement for cloning prior to analysis. However, methods of resolving the diversity of the amplified products in a single electrophoretic profile, such as fluorescently labeled terminal-restriction fragment length polymorphism (FLT-RFLP) analysis (9, 18) and denaturing gradient gel electrophoresis (23), have been successfully developed to study natural microbial communities. FLT-RFLP analysis, especially, is expected to be beneficial once the sequences of the amplified products have been determined, since the identity and affiliation of the profiles obtained are predictable. FLT-RFLP analysis of PCR-amplified nifH genes has already been applied to comparing the diazotrophic inhabitants in the gut among several termite species (26). As stated above, the existence of the nif genes does not always mean that biological activity is being expressed. An elegant method of monitoring microbial activity in situ would be detection of the mRNA. For this purpose, the reverse transcription (RT)-PCR method is advantageous because of its high sensitivity and specificity, and it has been used to detect mRNA in environmental samples (5, 20, 36). These molecular approaches depend on the PCR technique, and conventional PCR serves to amplify target DNA exponentially, making it difficult to use the technique in a quantitative manner. However, competitive PCR (15, 33, 37), in which an internal DNA standard has been added as a control to correct for the variation among reactions, allows reliable PCR quantification and has been applied to environmental samples (2, 14, 17, 20, 29).
Biological nitrogen fixation is catalyzed by a nitrogenase complex (12). A typical nitrogenase is a molybdenum (Mo)-containing enzyme encoded by the nifHDK operon. Mo-independent nitrogenases have cofactors that coordinate vanadium in place of Mo (V nitrogenase), or they have neither Mo nor vanadium (alternative nitrogenase); these nitrogenases are encoded by the vnfH-vnfDGK operon and the anfHDGK operon, respectively. There is an especially high degree of sequence conservation in the nifH, vnfH, and anfH genes, and for this reason, the nifH gene is usually used to detect nitrogen fixation genes in natural environments (6, 26, 27, 34, 35, 38, 39). The genes within the single operon are regulated simultaneously, but the three operons, nif, vnf, and anf, are regulated differentially. Although the transcription of all three types of nitrogenase operons is strictly regulated by the availability of fixed nitrogen, the availability of Mo differentially affects the expression of nitrogenase genes at the transcriptional level (4, 19). In the absence of Mo, the nif operon is repressed, whereas in the presence of Mo, the vnf and anf operons are repressed.
The dry-wood termite Neotermes koshunensis (order, Isoptera; family, Kalotermitidae) shows high nitrogen fixation activity (26), and stable isotope measurements have shown that more than half of the fixed nitrogen in the case of this termite is derived from atmospheric N2 (31). The presence of diverse nifH genes has been demonstrated in the gut of N. koshunensis (26). In this study, we examined the expression of the nitrogen fixation genes in the gut microbial community of N. koshunensis by culture-independent molecular methods. The mRNA of nifH genes was detected by RT-PCR, and the RT-PCR products were analyzed. A preferentially transcribed nitrogen fixation gene was characterized with respect to the regulation of its expression and the gene organization.
MATERIALS AND METHODS
Collection and culture of termites.
The termite N. koshunensis was collected in the Okinawa prefecture, Japan, in October 1997, January 1998, and August 1998. Fifty to 100 termites were fed a sterile diet as follows. One gram of filter paper (Toyo-roshi no. 5) was moistened with 0.6 ml of sterile water with or without 2% (NH4)2SO4. Sodium molybdate was added to the water solution at 1 mM, when appropriate. After 5 days, the worker-like larvae were removed for DNA and RNA extraction and for measurement of nitrogen fixation activity. Nitrogen fixation activity was measured by the acetylene reduction assay as described previously (26).
DNA and RNA extraction.
Approximately 30 termites were collected, and after their exterior surfaces had been washed with distilled water, their entire guts were removed with forceps and gently squeezed. DNA from the intestinal mixed population was extracted as described previously (25). Alternatively, the DNA was extracted by using a DNA purification system (Qiagen) according to the supplier’s instructions. Total RNA was extracted from the gut mixed population as follows. The gut microflora was treated with a solution (0.5% sodium dodecyl sulfate, 20 mM sodium acetate, 10 mM EDTA; adjusted to pH 5.5) to induce cell lysis, an equal volume of phenol (equilibrated with the above solution) was added, and then the mixture was incubated at 60°C for 10 min. After centrifugation, the aqueous phase was transferred to a new tube, and then the total RNA was precipitated with ethanol. The RNA was further purified by precipitation with 2.5 M LiCl. Purified RNA was incubated with RNase-free DNase I (Boehringer Mannheim) and with anti-RNase (Ambion, Inc.). Residual DNase activity was heat inactivated at 95°C for 5 min.
RT.
The total RNA (10 μg) and 10 pmol of YAA primer (27) in a total volume of 9 μl was heat denatured at 70°C for 10 min. The reverse transcriptase (RTase) used was SuperScriptII RNase H− RTase (Life Technologies). After heat denaturation, an RT reaction mixture was prepared as specified by the manufacturer’s instructions concerning the RTase. The RT mixture was preheated to 42°C for 2 min, and then 1 μl (200 U) of RTase was added. After incubation at 42°C for 60 min, the RTase was heat inactivated at 70°C for 10 min and the reaction products were precipitated by ethanol. The resulting cDNA was used as a template for the subsequent PCR amplification.
PCR amplification for FLT-RFLP analysis.
The nifH genes were amplified from the cDNA and from DNA extracted from the termite gut by PCR with Ex Taq DNA polymerase (Takara) according to the manufacturer’s instructions. The PCR primer corresponded to amino acid positions 11 to 16 (Klebsiella pneumoniae nifH numbering) in the case of the forward primer IGK (27) and positions 132 to 137 in the case of the reverse primer VCG (5′-GCRAANCCNCCRCANAC-3′). The forward primer was labeled at the 5′ end with a fluorescent dye, Cy5. All of the primers used in this study were synthesized by and purchased from Pharmacia. When the IGK-YAA primer combination was used for PCR with the cDNA as template, nonspecific amplification of sequences other than nifH was observed. Thus, we used the internal VCG primer instead of the YAA primer. The reaction conditions were 35 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 2 min. The PCR products were purified on a low-melting-point agarose gel by means of the Wizard PCR prep DNA purification system (Promega). The purified PCR products were digested with HhaI (Takara), and the lengths of the fluorescently labeled terminal restriction fragments of the PCR products were determined by electrophoresis by using an automatic sequencer, ALFred Express (Pharmacia), and analyzed by means of Fragment Manager software (Pharmacia).
Cloning of RT-PCR products and nucleotide sequencing.
The primers used for amplification of the nifH gene from the cDNA were IGK and YAA. The reaction conditions were as follows: 35 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 2 min. The PCR products of the expected size were purified on a low-melting-point agarose gel by means of the Wizard PCR prep DNA purification system (Promega). Purified PCR products were cloned into the vector pGEM-T (Promega) according to the manufacturer’s instructions. The insertion of DNA fragments of the appropriate sizes was confirmed by PCR amplification with universal and reverse primers (Takara) which corresponded to both sides of the cloning site on the vector. The amplified product was digested with either HhaI or RsaI, examined for RFLP, and then sorted. Plasmid DNA was prepared from a clone representative of each RFLP group by means of the Wizard mini prep DNA purification system (Promega) and used as a template for sequencing performed by using the Dye Terminator Cycle Sequencing Kit and an automatic sequence analyzer (Applied Biosystems model 377). Sequences showing no relatedness to nifH and the corresponding RFLP groups were excluded and not counted.
Quantitative PCR.
For the quantification of mRNA, a competitive quantitative PCR approach (15, 33, 37) was applied. Two primers, NKN-QQV (5′-CAACAGGTGTTCATACAC-3′) and NKN-THG (5′-CGTATAGGCTCCGTGCGT-3′), were used for specific amplification of the nifH gene of the termite alternative nif methanogen (anf-methano) cluster I. This primer pair was tested for its specificity by using plasmid DNA from various nifH clones of termite gut origin as templates. For the quantitative PCR, a competitor plasmid (pNK-N) was constructed as follows. A 310-bp DNA region of λ phage DNA (nucleotide positions 5205 to 5514; database accession no. X00906) was PCR amplified with primers QQVFIH-LAM (5′-CAACAGGTGTTCATACACATACCGAGGCTGACGT-3′) and THGAYT-LAM (5′-CGTATAGGCTCCGTGCGTGTTGAGGATCCCCATAA-3′), which contain the sequences of the primers NK-QQV and NK-THG (underlined). The PCR product was cloned into the vector pGEM-T (Promega) according to the manufacturer’s instructions.
A set of standard samples containing a certain amount of competitor DNA (from 1 to 100 pg) was prepared. To each reaction mixture, a constant amount of the cDNA was added. The reaction conditions were 35 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 2 min. The PCR products were separated on a 2% agarose gel, the gel was stained with ethidium bromide, and the stained products were visualized by means of a fluorescence imaging analyzer, FMBIO II Multi-view (Hitachi). Quantification of the PCR products was performed by using FMBIO Analysis, version 6.0 (Hitachi). To adjust for the difference in fluorescence based on the size of the fragment, the intensity of the internal control was multiplied by the ratio of the target size to the competitor size (256/346). Standard curves for the competitive PCR were made by using data from a series of 10-fold dilutions of a known amount of the competitor pNK-N with 10 pg of plasmid DNA of the clone NKN4 used as the target. In ordinary experiments, cDNA corresponding to 2.6 μg of total RNA gave a linear curve and this curve was used for the quantification. When linearity was not obtained, the cDNA was diluted and the cDNA corresponding to 0.5 μg of total RNA gave the linearity and was used. The mRNA concentrations were expressed as femtomol per milligram of total RNA, assuming that the RTase reaction was complete. For the quantification of the genomic nifH gene, 50 pg of total DNA of the microbial community was used.
Cloning, sequencing, and phylogenetic analysis of the nitrogenase structural gene cluster.
The DNA region downstream of the nifH gene in termite anf-methano cluster I was isolated and characterized. Entire termite guts were squeezed in 0.4% NaCl solution and used as a template for PCR. The primers used for the amplification were NKN-QQV and ANFK (5′-GGYTGRCAXGTRAADATXGG-3′). The latter primer corresponded to a conserved amino acid sequence in anfK (the Azotobacter vinelandii anfK amino acid positions 16 to 22 [database accession no. M23528]). The reaction conditions were 35 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 2 min. The PCR products were purified on an agarose gel and cloned into the vector pGEM-T. Deletion derivatives of the clones were constructed by means of a kilosequence deletion kit (Takara). The plasmid DNA was prepared and used as templates in sequencing as described above. Open reading frames (ORFs) in the determined nucleotide sequence were analyzed by means of GENETYX software (Software Development), and the amino acid sequences were aligned by using the CLUSTAL W package (32). The PHYLIP version 3.5c phylogeny inference software package (13) was used to infer the protein phylogeny. PROTDIST with the Dayhoff PAM matrix option was used to calculate evolutionary distances. Phylogenetic trees were constructed from the evolutionary distance data by the neighbor-joining method, implemented through the program NEIGHBOR. A total of 100 bootstrapped replicate resampling data sets for PROTDIST were generated with the program SEQBOOT to provide confidence estimates for tree topologies.
Nucleotide sequence accession numbers.
The sequences determined in this study will appear in the nucleotide sequence databases under accession no. AB027742 to AB027751.
RESULTS
Comparison of the transcripts and genomic DNA of nifH genes by FLT-RFLP.
In order to investigate the contribution of each nifH gene in the gut of N. koshunensis to nitrogen fixation in the termite, expression of the nifH gene was investigated. The nifH mRNA was amplified from total RNA extracted from the gut microflora by RT-PCR to obtain products of the anticipated size. In the absence of the RTase reaction step, no amplification was detected. This result indicated that the RT-PCR could specifically detect nifH mRNA, and the products were not derived from the genomic DNA.
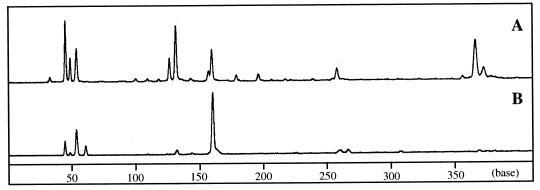
The composition of the amplified nifH sequences was examined by FLT-RFLP analysis comparing those derived from the nifH mRNA and those derived from nifH genes in genomic DNA. As shown in Fig. 1, the diversity of the nifH genes in the DNA extracted from the gut mixed population was confirmed. In contrast, only limited heterogeneity was detected in the mRNA from the gut. The terminal restriction fragment (T-RF) of 161 bases, especially, gave a strong fluorescent signal (more than 60% of the total fluorescence intensity detected). However, the fluorescence intensity of the 161-base-sized T-RF was less than 10% of the total in the FLT-RFLP profile of the PCR products amplified from the genomic DNA. Regardless of the length of time the termites were kept in the laboratory, the feeding conditions, or the termite colonies examined, the heterogeneity of the T-RFs detected in the RT-PCR products was less than that in the case of the PCR products amplified from genomic DNA. The T-RF of 161 bases was always a major component in the FLT-RFLP profiles of the RT-PCR products. In the case of some colonies, a T-RF of 54 bases also showed strong fluorescence intensity, up to 40% of the total (Fig. 2D). These results suggested that among the diverse nifH genes in the termite gut only a few represented by the T-RFs of 54 and 161 bases were preferentially expressed.
FIG. 1.
A comparison of the nifH constituents of the RT-PCR and PCR products derived from the gut microflora of the termite N. koshunensis. Electropherograms of the HhaI-digested nifH genes amplified from the genomic DNA by PCR (A) and from the mRNA by RT-PCR (B) are shown. The termites were fed filter paper for 5 days before nucleic acid extraction. The nifH genes were amplified with the 5′-fluorescently labeled primer IGK and the primer VCG. Numbers below the electropherograms show base lengths.
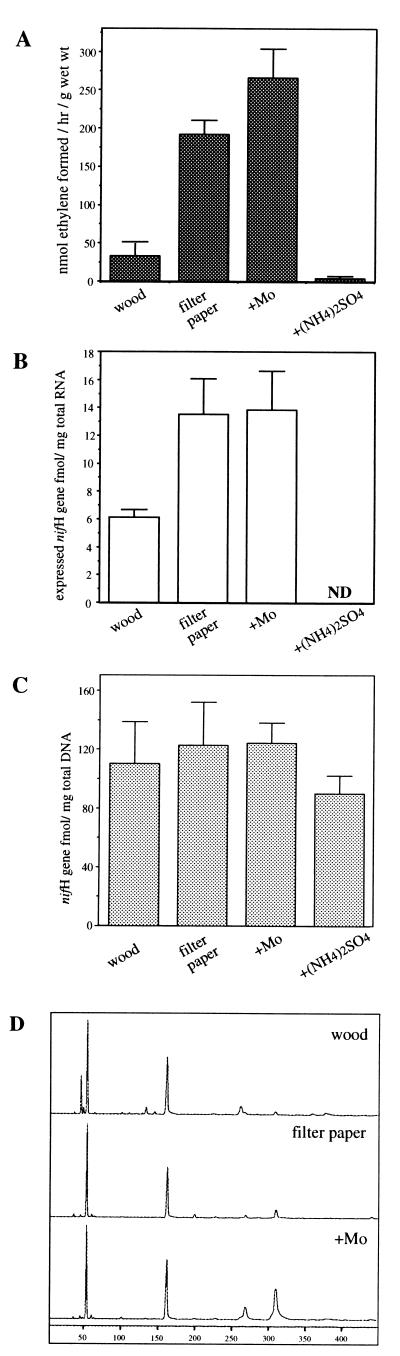
FIG. 2.
Effects of termite feeding conditions on nitrogen fixation activity, the levels of nifH mRNA, and the amount of DNA of the termite anf gene. The termites were shifted from a diet of infested wood to filter paper moistened with distilled water or filter paper moistened with an aqueous solution of either 1 mM sodium molybdate (+Mo column) or 2% ammonium sulfate [+(NH4)2SO4 column]. (A) C2H2 reduction activity of live termites. (B) Quantification of the amount of mRNA of the termite anf gene. The RT-PCR product was not detected (ND), and thus the amount of mRNA was below the detection limit. (C) Quantification of the termite anf gene in the genomic DNA of the gut microflora. Results in panels A to C are means of two to six determinations, and the error bars indicate standard deviations. (D) FLT-RFLP analysis of the nifH RT-PCR products amplified from the mRNA of the gut microflora. Since no amplification by RT-PCR was observed, the profile for the termites fed filter paper with ammonium sulfate is not shown.
Identification of the preferentially expressed genes.
Based on the sequence information on nifH genes previously isolated from the gut microbial community of N. koshunensis (26), it is possible to predict the sequence identity for most profiles obtained in the FLT-RFLP analysis. Among the isolated nifH clones, only the sequences of termite anf-methano cluster I were predicted to correspond to the T-RF of 161 bases. The T-RF of 54 bases corresponds to the sequence of either termite anf-methano cluster I or termite pseudo nif cluster I.
In order to determine their exact identity, the RT-PCR products were clonally isolated and analyzed. A termite colony showing a significant amount of both the 54- and 161-base-sized T-RFs in the FLT-RFLP profile of the RT-PCR products was used. The 21 nifH clones isolated were sorted into nine RFLP groups, and representatives of these groups were analyzed by nucleotide sequencing, to obtain five different amino acid sequences. Table 1 shows the results of analysis of the RT-PCR clones. A large proportion of the clones (18 of the 21 clones) consisted of clones sharing close similarity to each other, and these were affiliated with termite anf-methano cluster I. The predicted T-RF size of these sequences was either 54 or 161 bases. This finding was congruent with the results of FLT-RFLP analysis of the RT-PCR products, although more of the 54-base-sized T-RFs were isolated than in the case of the 161-base-sized T-RFs, probably reflecting a difference in cloning efficiency. The results indicated that the gene of termite anf-methano cluster I was preferentially expressed and thus might be critical for nitrogen fixation in N. koshunensis.
TABLE 1.
Number of nifH clones obtained from RT-PCR products
| Clone | Affiliationa | Most similar sequenceb (% amino acid identity) | T-RF size | No. of clonesc | Accession no. |
|---|---|---|---|---|---|
| NKN-RT2-7 | anf-methano I | NKN9 (99.3) | 54 | 6 | AB027744 |
| NKN-RT23 | anf-methano I | NKN9 (100) | 54 | 5 | AB027747 |
| NKN-RT2-2 | anf-methano I | NKN9 (100) | 54 | 1 | AB027750 |
| NKN-RT2-8 | anf-methano I | NKN9 (100) | 54 | 1 | AB027745 |
| NKN-RT11 | anf-methano I | NKN9 (100) | 161 | 4 | AB027742 |
| NKN-RT26 | anf-methano I | NKN9 (100) | 161 | 1 | AB027746 |
| NKN-RT6 | Anaerobe III | NKN9 (97.9) | 158 | 1 | AB027749 |
| NKN-RT16 | Anaerobe III | CDN28 (99.3) | 132 | 1 | AB027743 |
| NKN-RT27 | Pseudo nif I | NKN24 (100) | 43 | 1 | AB027748 |
Quantification of mRNA and effect of termite feeding conditions.
Since a gene belonging to the termite anf-methano cluster I was suggested to be responsible for the most abundant type of nifH transcripts in the gut microbial community, its expression was investigated in a quantitative manner. For quantification, competitive PCR targeting the product of RT was applied. In order to detect the sequences specifically, specific primers for the termite anf gene were designed. An internal DNA standard (competitor) for the competitive PCR which contained the same primer binding sites as the target and from which PCR products of a different size were produced was constructed. The relative amplification efficiency of the target compared to that of the competitor was determined from a plot of the log ratio of target intensity to competitor intensity against the log ratio of the concentrations of target to input competitor. Coamplification of the sequence from a clone of termite anf-methano cluster I with a series of dilutions of the competitor resulted in a line with a slope of 0.595 and a regression value of 0.983, indicating that the standard curve prepared could be used for quantification of samples within this range.
To investigate the relationship between nitrogen-fixing activity and the amount of mRNA, the termite N. koshunensis was fed under different conditions (Fig. 2). The termites were shifted from a diet of infested wood to filter paper moistened with water with or without the inclusion of a nitrogen source. The termites fed filter paper without any added nitrogen source showed more than fivefold greater levels of C2H2 reduction activity than the termites fed wood (Fig. 2A). The filter paper probably contained a lower amount of combined nitrogen than the wood. The termites fed the diet with a nitrogen source added showed diminished activity, to the extent of 0.1-fold, as compared to those fed wood. A similar fluctuation of nitrogen fixation activity has been demonstrated in the case of the termite Coptotermes formosanus (8).
Quantitative analysis of the levels of mRNA expressed from the termite anf gene showed that the total RNA extracted from termites fed filter paper contained a 2.2-fold higher concentration of this mRNA than that from termites fed wood (Fig. 2B). In the case of feeding the diet with a nitrogen source added, the level of mRNA expressed was below the detection limit. The nitrogen-fixing activity and the amount of mRNA from the termite anf gene were well correlated. Such a correlation was also observed with another termite colony with a different feeding regiment. Under these feeding conditions, mRNAs derived from the termite anf gene were the major mRNA species detected by RT-PCR and the following FLT-RFLP analysis (Fig. 2D). Thus, the amount of nitrogen fixation activity in N. koshunensis appears to be determined by the levels of mRNA expressed from the termite anf gene. As shown in Fig. 2D, a minor signal corresponding to a T-RF of 43 bases, which was predicted to correspond to a nifH gene of termite pseudo nif cluster I, was observed when the diet was wood. However, in spite of the increased activity, the signal disappeared when the termites were fed filter paper.
It is known that expression of nitrogenases of the anf group is repressed at the transcriptional level in the presence of a trace amount of Mo (4, 19). The effect of Mo on the levels of nitrogen fixation activity and expression of the nifH gene was investigated. The termites were fed filter paper moistened with a Mo-containing solution without any added nitrogen source. The C2H2 reduction activity was slightly increased (around 1.4-fold) compared with that of the termites fed filter paper without Mo (Fig. 2A). Interestingly, the Mo in the diet had no effect on the amount of mRNA detected (Fig. 2B). This finding indicates that expression of the termite anf gene is independent of the presence of Mo, as opposed to the ordinary anf genes. FLT-RFLP analysis of the mRNA produced in the presence of Mo revealed a significant amount (up to 50% of the total fluorescence intensity) of expression of mRNA derived from a non-anf group of the nifH sequences represented by T-RFs which were 266 and 307 bases in size (Fig. 2D). The corresponding nifH sequences for both were predicted to be those affiliated with termite anaerobe cluster III (26). This increased expression was probably responsible for the slight increase in nitrogen-fixing activity observed.
Whether the population of microorganisms possessing the termite anf gene had changed or not was examined by means of the quantitative competitive PCR targeting these genes in genomic DNA extracted from the gut microbial community. In contrast to the C2H2 reduction activity and the amount of mRNA detected, only a slight fluctuation of the nifH copy number per unit amount of genomic DNA was observed (Fig. 2C). This result indicates that the nitrogen fixation activity observed was not influenced by a change in the population of microorganisms. Rather, the activity observed reflected the expression level per cell in the population of microorganisms.
Cloning and phylogenetic analysis of the anf gene cluster.
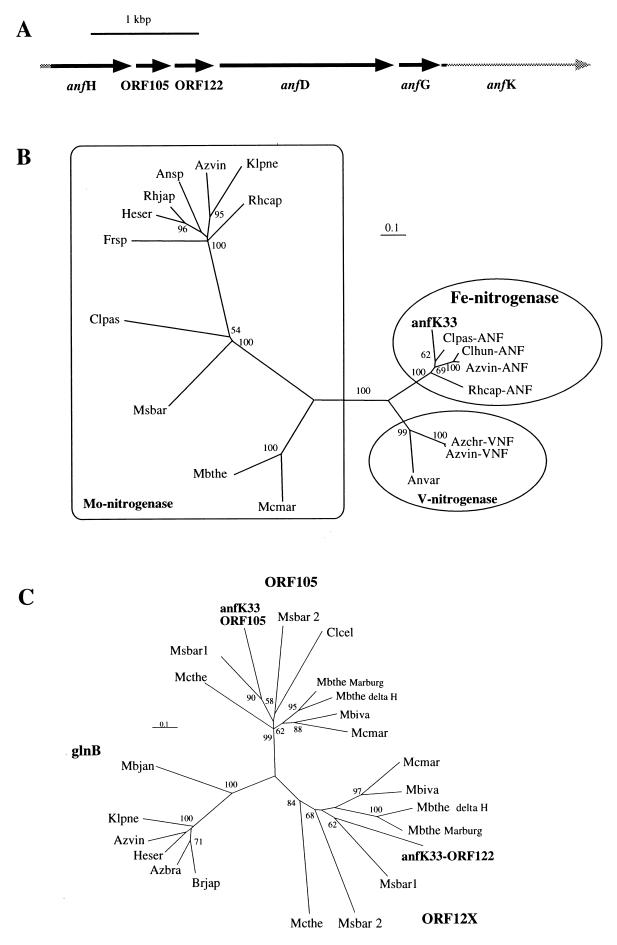
The structural features of the nitrogenase gene cluster which is responsible for nitrogen fixation in N. koshunensis were further characterized. Since nifH, nifD, and nifK usually occur in this order in nitrogenase operons, the entire region encoded by nifD was amplified by PCR. One of the primers used was specific for the nifH genes of termite anf-methano cluster I, and the other was a degenerate primer corresponding to a conserved region of nifK. A single band, 3.6 kb in size, was detected upon electrophoresis of the PCR products and was then cloned. Nucleotide sequences corresponding to the nifH region were determined for several clones. They showed high sequence similarity, sharing more than 97% amino acid identity and showing around 98% amino acid identity with the RT-PCR clone NKN-RT11. The 3.6-kb DNA region of a clone designated anfK33 was completely sequenced, and putative genes for the alternative nitrogenase were identified: anfH, ORF105, ORF122, anfD, anfG, and anfK, in this order (Fig. 3A).
FIG. 3.
(A) Schematic representation of the genes identified in the DNA region downstream of the termite anf gene. A 3.6-kb DNA region of the clone anfK33 was completely sequenced, and presumptive coding regions were identified by comparison with nif operons in the DNA sequence databases. (B) Unrooted phylogenetic tree for nifD amino acid sequences. The tree was inferred from 201 unambiguously aligned amino acid positions by the neighbor-joining method. Bootstrap values above 50 from 100 resamplings are shown for each node. The scale bar denotes 0.1 substitutions per site. Abbreviations, with accession numbers in parentheses: Ansp, Anabaena sp. strain PCC7120 (V01482-2); Anvar, Anabaena variabilis (L20472); Azchr-VNF, A. chroococcum (X51756); Azvin, A. vinelandii (X06886); Azvin-VNF, A. vinelandii vnf (M32371); Azvin-ANF, A. vinelandii anf (M23528); Clhun-ANF, C. hungatei (U59415); Clpas, C. pasteurianum (M21537); Clpas-ANF, C. pasteurianum (L09762); Frsp, Frankia alni ArI3 (L41344); Heser, Herbaspirillum seropedicae (Z54207); Klpne, K. pneumoniae (X07748); Mcmar, Methanococcus maripaludis (U75887); Mbthe, Methanobacterium thermoautotrophicum (X87971); Msbar, M. barkeri nifH2 (U11291); Rhcap, R. capsulatus (M15270); Rhcap-ANF, R. capsulatus anfD (X70033); Rhjap, Rhizobium japonicum (X01045). (C) Unrooted tree for glnB, ORF105, and ORF12X (X represents 2 to 8). The tree was inferred from 133 unambiguously aligned amino acid positions by the neighbor-joining method. Bootstrap values above 50 from 100 resamplings are shown for each node. The scale bar denotes 0.1 substitutions per site. Abbreviations: Klpne, K. pneumoniae (X14012); Azvin, A. vinelandii (U91902); Heser, H. seropedicae glnB (Z54207); Azbra, Azospirillum brasilense (X51499); Brjap, Bradyrhizobium japonicum BJ110d (M26753); Mbjan, Methanobacterium jannassu (U67464); Clcel, C. cellobioparum (U59414); Mcthe, Methanococcus thermolithotrophicus (X13830); Mbthe delta H, M. thermoautotrophicum delta H (X87971); Mbthe Marburg, M. thermoautotrophicum Marburg (AE00916); Msbar1, M. barkeri (X56072); Msbar2, M. barkeri (X56073); Mcmar, Methanococcus maripaludis (U75887); Mbiva, Methanobacterium ivanobii (X56071).
Phylogenetic analysis based on the sequence of nifD has shown that Mo-independent nitrogenases form monophyletic groupings separate from the primary Mo nitrogenase (10, 11, 16). When the anfD sequence isolated here was included in the analysis (Fig. 3B), it was clearly in a cluster together with the members of the alternative nitrogenase group (Fe nitrogenases). A bootstrap value of 100% at the node supported this clustering. anfD in the isolated clone shared 84% amino acid identity with the anfD genes of Clostridium pasteurianum and A. vinelandii, respectively. More than 78% amino acid identity of the anfD gene in the isolated clone to other anfD genes was evident, whereas it showed less than 62% and less than 44% amino acid identity to the vnfD and nifD genes, respectively. The genes for the third subunit of dinitrogenases, anfG and vnfG, are only known to be present in the gene clusters encoding Mo-independent nitrogenases. The anfG sequence in the isolated clone was the most similar to anfG of C. pasteurianum (48% amino acid identity), and it showed more than 38% amino acid sequence identity to anfG from other organisms but less than 27% identity to the product encoded by vnfG. Although the anfK sequence was too short to characterize, the phylogenetic positions and sequence comparisons of anfD and anfG suggested that the gene cluster representing termite anf-methano cluster I may encode an alternative nitrogenase having neither Mo nor V as a cofactor.
The isolated gene cluster was also found to contain two ORFs, ORF105 and ORF122, inserted between anfH and anfD that showed significant amino acid similarity to the glnB-like PII proteins. In enteric bacteria, glnB encodes the PII protein which participates in the nitrogen regulatory cascade (21). The amino acid sequences encoded by these ORFs were grouped together with those of ORF105 and ORF12X (X represents 2 to 8) reported in the nif gene cluster of the Archaea domain. The products of ORF105 and ORF122, especially, showed the highest amino acid identities, 61 and 55%, respectively, with those located downstream of nifH1 of Methanosarcina barkeri 227. The tyrosyl residue (amino acid position 49), the site of uridylylation of the PII protein in enterobacteria (21), was present in ORF105 but not in ORF122.
DISCUSSION
In the symbiotic microbial community in the gut of N. koshunensis, the anf gene affiliated with termite anf-methano cluster I has been shown to be the most critical gene responsible for nitrogen fixation in the termite. Transcripts of the anf gene were the major mRNA species derived from nifH detected in the gut community by RT-PCR, and the amount of anf mRNA was well correlated with the level of nitrogen fixation activity in the termite. The results of feeding experiments performed under several conditions suggested that the level of expression of the anf gene determines the level of nitrogen fixation activity in the termite. Generally, in order to investigate a certain biological activity in natural environments, a survey using genes for key functional enzymes may be beneficial. However, as described in this study, monitoring the expression of the genes detected is necessary to evaluate their real contributions.
The results indicating that only the anf gene was preferentially expressed are surprising because there are diverse nifH sequences in the DNA extracted from the gut microbial community of N. koshunensis. Other than the anf gene, nifH genes affiliated with the anaerobe group and the proteobacteria-cyanobacteria group were isolated (26). However, only small amounts of the transcripts of these non-anf genes were detected by RT-PCR, and thus they seem to be nonfunctional under the standard experimental conditions employed here. However, the transcription of some of these genes was induced when an excess amount of Mo was present (Fig. 2D). Simultaneously, nitrogen fixation activity also increased slightly but significantly, suggesting that these genes are functionable when they are expressed. Thus, the non-anf genes appear to encode potentially active nitrogenases. These results imply that the metal supply may not be enough to induce the expression of these non-anf genes under natural states of the termite. On the other hand, not all the non-anf genes were found to be induced in the presence of Mo. There may be some reason, other than Mo availability, for the repression of their expression.
The presence of diverse nifH sequences in the gut community has been reported for various termites, and the nifH genes of the anf group were not always present in all of the termites investigated (26, 27). The existence of anf genes can be explained simply as being due to inadequate amounts of Mo in their diets, as the ordinary Mo-dependent nitrogenases need Mo as a cofactor for expression of nitrogen-fixing activity. The termite species possessing no anf genes in the gut community can probably obtain sufficient Mo from their food.
Transcription of the anf gene in the gut community of the termite was found to be repressed upon the addition of fixed nitrogen to the termite diet. The repression of transcription by fixed nitrogen is a common feature of the regulation of nitrogenase expression. In general, expression of anf genes is known to be repressed in the presence of Mo. Depending on the availability of this metal as a cofactor, expression of the Mo-dependent primary nitrogenase and expression of the alternative nitrogenase are switched on and off at the transcriptional level and are exchanged with each other, to express the activity (4, 19). However, the regulation of expression of the termite anf gene was found to be independent of the presence of Mo (Fig. 2B). Since the amount of Mo added was excessive (around 100-fold compared to the concentrations repressing ordinary anf genes [4, 19]) and since the induction of some non-anf genes was observed, the Mo concentration in the gut is believed to be sufficient to regulate gene expression. The Mo-independent regulatory feature is unique to the anf gene in the termite symbiotic system. One possible explanation for the Mo-independent expression is that the Mo availability may be limited in the termite gut, and due to the long-term symbiotic relationship within the termite gut, the gene may have lost the regulatory mechanism sensing Mo. Alternatively, the symbiont possessing the anf gene may have lost the gene encoding the Mo-dependent primary nitrogenase, and thus, under diazotrophic conditions, the organisms evolved to constitutively express the Mo-independent anf nitrogenase regardless of the presence of Mo. Further analyses are necessary to clarify these possibilities.
Some methanogenic archaea possess nifH genes affiliated with the anf-methano group of nifH. However, these nitrogenases contain Mo as a cofactor, and their nifD genes are phylogenetically grouped with those of the Mo-dependent nitrogenases (16) (database accession no. X87971). The Mo-independent regulation of gene expression in the case of the termite anf gene might suggest a Mo-dependent ordinary enzyme encoded by it as in methanogens. However, this is probably not the case for the anf gene cluster of the termite. The results of sequencing and phylogenetic analysis indicated that anfH, anfD, and anfG in the anf gene cluster identified here encode an alternative nitrogenase that is both Mo and V independent.
The gene organization and sequence features of the termite anf gene were found to be distantly related to those of well-characterized organisms. Thus, information about the taxonomy of organisms possessing the anf gene is limited. Except for the presence of two ORFs homologous to glnB, the phylogenetic character of anfD and anfG described here and also that of anfH reported previously (26) suggests that particular species in the domain Bacteria possess the anf gene cluster. In the Archaea domain, the presence of a Mo-independent alternative nitrogenase and the existence of anfD and anfG orthologous genes have never been reported. However, the anfH gene was distantly related to any known bacterial anfH gene. In the case of anfD analysis (Fig. 3B), taxonomy of the organism possessing the termite anf gene could not be predicted because anfD sequences of phylogenetically related organisms did not form clusters within the Fe nitrogenase group (C. pasteurianum-Clostridium hungatei and A. vinelandii-Rhodobacter capsulatus). The presence of the two ORFs between the nifH and nifD genes seems to be a common feature within the Archaea domain (11, 30). All diazotrophic methanogenic archaea share this feature of gene organization. However, the sequence homologous to ORF105 has recently been reported for Clostridium cellobioparum (sequence database accession no. U59414). Because the nucleotide sequence of the DNA region corresponding to ORF122 has not yet been reported, the existence of ORF122-like genes is now not certain. Nevertheless, the presence of ORF105 in the genome of an organism in the Bacteria domain indicates that its presence is no longer characteristic of the Archaea domain. Still, the two ORFs in the termite anf gene cluster are most closely related to those of a methanogen, M. barkeri. An organism of yet-uncharacterized diazotroph probably possesses the termite anf gene. Further investigations are necessary to taxonomically identify the organism that possesses the anf gene and thus the organism most responsible for nitrogen fixation in the termite.
The molecular approaches described in this study have been shown to be powerful tools to investigate and characterize the expression of a certain gene within a mixed microbial community. Of course, for more detailed taxonomic, physiological, and genetic analyses, isolation and cultivation of the responsible microorganisms are important. Our preliminary attempts to isolate microorganisms harboring the anf gene by enrichment, however, have failed, though a variety of the enrichment conditions have been used (24). Although diazotrophic bacterial growth has been observed under some conditions, it has never been shown whether the bacteria possess the anf gene. Isolation of such bacteria is thought to be difficult under ordinary conditions. An in situ hybridization technique with a specific probe may be useful for identification of the cell types expressing this gene (1, 22, 28). Even if isolation of these cells is successful, the cultivation conditions may not represent the natural state of the organisms, since complicated symbiotic relationships with the termite and with other members of the community may be involved. Mimicry of the environment within the termite gut in vitro cannot be expected to be achieved. For these reasons, culture-independent approaches are believed to be beneficial and advantageous in order to understand the real nature of the microbial community consisting of a mixed population.
ACKNOWLEDGMENTS
This work was partially supported by grants from the Biodesign Research Program, the Genome Research Program, and the Eco Molecular Science Research Program from RIKEN and by a grant from the International Cooperative Research Project (Bio-Recycle Project) from Japan Science and Technology Corporation. S.N. was supported by a grant from the Junior Research Associate Program from RIKEN.
We thank I. Yasuda for advice on termite collection.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baek J-M, Kenerly C M. Detection and enumeration of a genetically modified fungus in soil environments by quantitative competitive polymerase chain reaction. FEMS Microbiol Ecol. 1998;25:419–428. [Google Scholar]
- 3.Benemann J R. Nitrogen fixation in termites. Science. 1973;181:164–165. doi: 10.1126/science.181.4095.164. [DOI] [PubMed] [Google Scholar]
- 4.Bishop R E, Premakumar R. Alternative nitrogen fixation systems. In: Stacy G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 736–762. [Google Scholar]
- 5.Bogan B W, Schoenike B, Lamar R T, Cullen D. Manganase peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl Environ Microbiol. 1996;62:2381–2386. doi: 10.1128/aem.62.7.2381-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun S T, Proctor L M, Zani S, Mellon M T, Zehr J P. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol Ecol. 1999;28:273–279. [Google Scholar]
- 7.Breznak J A. Intestinal microbiota of termites and other xylophagous insects. Ann Rev Microbiol. 1982;36:323–343. doi: 10.1146/annurev.mi.36.100182.001543. [DOI] [PubMed] [Google Scholar]
- 8.Breznak J A, Brill W J, Mertins J W, Coppel H C. Nitrogen fixation in termites. Nature. 1973;244:577–580. doi: 10.1038/244577a0. [DOI] [PubMed] [Google Scholar]
- 9.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien Y-T, Zinder S H. Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD1 from the eubacterium Clostridium pasteurianum. J Bacteriol. 1994;176:6590–6598. doi: 10.1128/jb.176.21.6590-6598.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien Y-T, Zinder S H. Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archaeon Methanosarcina barkeri 227. J Bacteriol. 1996;178:143–148. doi: 10.1128/jb.178.1.143-148.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacy G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834. [Google Scholar]
- 13.Felsenstein J. PHYLIP—phylogeny inference package version 3.5. Cladistics. 1989;5:164–166. [Google Scholar]
- 14.Gettemy J M, Ma B, Alic M, Gold M H. Reverse transcription-PCR analysis of the regulation of the manganese peroxidase gene family. Appl Environ Microbiol. 1998;64:569–574. doi: 10.1128/aem.64.2.569-574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilliland G, Perrin S, Bunn H F. Competitive PCR for quantitation of mRNA. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 60–69. [Google Scholar]
- 16.Kessler P S, McLarnan J, Leigh J A. Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J Bacteriol. 1997;179:541–543. doi: 10.1128/jb.179.2.541-543.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masepohl B, Klipp W. Organization and regulation of genes encoding the molybdenum nitrogenase and the alternative nitrogenase in Rhodobacter capsulatus. Arch Microbiol. 1996;165:80–90. [Google Scholar]
- 20.Meckenstock R, Steinle P, van der Meer J R, Snozzi M. Quantification of bacterial mRNA involved in degradation of 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 from liquid culture and from river sediment by reverse transcriptase PCR (RT/PCR) FEMS Microbiol Lett. 1998;167:123–129. doi: 10.1111/j.1574-6968.1998.tb13217.x. [DOI] [PubMed] [Google Scholar]
- 21.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriya S, Ohkuma M, Kudo T. Phylogenetic position of symbiotic protist Dinenympha exilis in the hindgut of the termite Reticulitermes speratus inferred from the protein phylogeny of elongation factor 1α. Gene. 1998;210:221–227. doi: 10.1016/s0378-1119(98)00078-x. [DOI] [PubMed] [Google Scholar]
- 23.Muyzer G E, de Waal C, Uitterlineden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noda, S., M. Ohkuma, and T. Kudo. Unpublished data.
- 25.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkuma M, Noda S, Kudo T. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl Environ Microbiol. 1999;65:4926–4934. doi: 10.1128/aem.65.11.4926-4934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohkuma M, Ohtoko K, Grunau C, Moriya S, Kudo T. Phylogenetic identification of the symbiotic hypermastigote Trichonympha agilis in the hindgut of the termite Reticulitermes speratus based on small-subunit rRNA sequence. J Eukaryot Microbiol. 1998;45:439–444. doi: 10.1111/j.1550-7408.1998.tb05096.x. [DOI] [PubMed] [Google Scholar]
- 29.Reilly K, Attwood G T. Detection of Clostridium proteoclasticum and closely related strains in the rumen by competitive PCR. Appl Environ Microbiol. 1998;64:907–913. doi: 10.1128/aem.64.3.907-913.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sibold L, Henriquet M, Possot O, Aubert J-P. Nucleotide sequence of nifH regions from Methanobacterium ivanovii and Methanosarcina barkeri 227 and characterization of glnB-like genes. Res Microbiol. 1991;142:5–12. doi: 10.1016/0923-2508(91)90091-n. [DOI] [PubMed] [Google Scholar]
- 31.Tayasu I, Sugimoto A, Wada E, Abe T. Xylophagous termites depending on atmospheric nitrogen. Naturwissenschaften. 1994;81:229–231. [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai S-J, Wiltbank M C. Quantification of mRNA using competitive RT-PCR with standard curve methodology. BioTechniques. 1996;21:862–866. doi: 10.2144/96215st04. [DOI] [PubMed] [Google Scholar]
- 34.Ueda T, Suga Y, Yahiro N, Matuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widmer F, Shaffer B T, Porteous L A, Seidler R J. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl Environ Microbiol. 1999;65:374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson M S, Bakermans C, Madsen E L. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl Environ Microbiol. 1999;65:80–87. doi: 10.1128/aem.65.1.80-87.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarchar V, Thomas R A, Goustin A S. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;21:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehr J P, Mellon M T, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zehr J P, Mellon M, Braun S, Litaker W, Steppe T, Paerl H W. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]