Abstract
Pancreatic ductal adenocarcinoma (PDAC) and cholangiocarcinoma (CCA) are malignant tumors with poor prognosis because of the limited effectiveness of traditional chemotherapy and few effective molecular therapeutic agents. Here, we determined the essential roles of Zic family member 5 (ZIC5) in the survival of PDAC and CCA cells. The results showed that ZIC5 is strongly expressed in PDAC and CCA tissues, while ZIC5 expression is barely observed in most normal human adult tissues. Furthermore, ZIC5 expression is related to poor prognosis of patients with PDAC. ZIC5 knockdown via small interfering RNA decreased the phosphorylation of signal transducer and activator of transcription 3 (STAT3), a protein that is associated with PDAC and CCA aggressiveness. However, ZIC5 knockdown induced cell death regardless of STAT3 activation, which is promoted by interleukin (IL) −6, a factor associated with inflammation. Furthermore, knockdown of ZIC5 in PDAC and CCA cells additively or synergistically induced apoptosis with the anti-cancer drug gemcitabine. Thus, ZIC5 constitutes a potential therapeutic target for the treatment of PDAC and CCA.
Keywords: ZIC5, Pancreatic ductal adenocarcinoma, Cholangiocarcinoma, STAT3, Apoptosis, Gemcitabine
Highlights
-
•
ZIC5 is expressed in PDAC and CCA, while barely observed in normal adult tissues.
-
•
ZIC5 expression is related to poor prognosis of patients with PDAC.
-
•
ZIC5 knockdown induces apoptosis in several PDAC and CCA cell lines.
-
•
Knockdown of ZIC5 additively or synergistically induces apoptosis with gemcitabine.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- CCA
cholangiocarcinoma
- IL
interleukin
- STAT3
signal transducer and activator of transcription 3
- ZIC5
Zic family member 5
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) and cholangiocarcinoma (CCA) are some of the most lethal malignancies; there are few effective therapies for these diseases. Despite advances in understanding the cancer biology of PDAC and CCA, the clinical outcomes of both of these diseases are still poor [1]. Because the majority of patients have advanced disease at the time of diagnosis and given the limited effectiveness of traditional chemotherapy, the development of effective molecular therapeutic agents with activity against these disease is urgently needed.
Previously, our screening identified Zic family member 5 (ZIC5) as a critical transcription factor for melanoma drug resistance [2]. ZIC5 activates signal transducer and activator of transcription 3 (STAT3) by promoting expression of platelet-derived growth factor D (PDGFD), rendering melanoma cells resistant to BRAF inhibitors and cytotoxic reagents [2]. ZIC5 expression also is enhanced and associated with tumor pathological stage in colorectal and prostate cancer, while ZIC5 expression is rarely detected in relevant non-tumor tissues [3]. In colorectal and prostate cancer cell lines, ZIC5 positively regulates PDGFD expression, STAT3 activation, proliferation, survival, and primary drug resistance. Furthermore, ZIC5 regulates the malignant phenotype of cancer cells not only by regulating the expression of PDGFD and STAT3, but also by regulating the expression of other factors such as cyclin dependent kinase inhibitor 1B (CDKN1B) and heat shock protein family D member 1 (HSPD1) [3].
ZIC5 is strongly expressed in many other cancer types (as evident in the TCGA database). ZIC5 has been shown to promote the malignant phenotype in non-small cell lung cancer (NSCLC), hepatocellular carcinoma (HCC), and glioma [[4], [5], [6]]. In NSCLC, ZIC5 knockdown significantly inhibits both proliferation (causing cell cycle arrest) and tumor growth in vivo [4]. In HCC, ZIC5 promotes the proliferation, migration, and invasion of HCC cell lines by regulating β-catenin signaling [5]. In glioma, ZIC5 has been shown to be a target of miR-761, which is inhibited by hsa_circ_0007534, a circular RNA highly expressed in glioma tissues [6]. However, to our knowledge, the role of ZIC5 in PDAC and CCA has not been elucidated. Therefore, in the present study, we investigated the role of ZIC5 in PDAC and CCA.
2. Material and methods
2.1. Cells and cultures
AsPC-1, PANC-1, and MiaPaca-2 (PDAC) cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). The RBE (CCA) cell line was obtained from the RIKEN Cell Bank (Tsukuba, Japan). Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum.
2.2. RNA isolation, cDNA synthesis, and quantitative real-time PCR
These were performed as described previously [3]. Quantitative real-time PCR (qPCR) was performed using the following primers: ZIC5, F: AACCTCAAGATCCACAAGCGT and R: CACTGGTGTGGACATGGGAA; ACTB, F: GCCCTGGCACCCAGCACAAT and R: GGAGGGGCCGGACTCGTCAT; and GAPDH, F: AGC CTC CCG CTT CGC TCTCT and R: CCA GGC GCC CAA TAC GACCA. ACTB (encoding β-actin) and GAPDH (encoding the housekeeping protein glyceraldehyde phosphate dehydrogenase) were used as an internal control for normalization of transcript levels.
2.3. Small interfering RNA (siRNA) and plasmid transfection
The negative control siRNA and siRNA for ZIC5 was previously described [2]. Transient transfections with siRNA were performed using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's protocol. In each experiment, the total amount of transfected siRNA was adjusted with a relevant negative control siRNA. The ZIC5 expression vector was as described previously [2]. To obtain stable cells, transfected cells were selected with 1 mg/ml G418 (10131035, Thermo) for 12 days, and the resulting bulk cells were analyzed.
2.4. Cell proliferation assay
Cell proliferation assay was performed as previously described [2].
2.5. Apoptosis assays
Cells were incubated with CellEvent Caspase-3/7 Green Detection Reagent (Invitrogen) and Hoechst33342 (Dojindo) for 30 min, after which the cells were analyzed using the In Cell Analyzer 2000 with 4′,6-diamidino-2-phenylindole (DAPI) and fluorescein isothiocyanate (FITC) filters. The ratio of Caspase-3/7-positive cells to Hoechst33342-positive cells was determined with the In Cell Analyzer Workstation 3.7 software (GE Healthcare). Experiments were performed a minimum of 3 times.
2.6. Reagents
Gemcitabine (ChemScene, Monmouth Junction, NJ) was used at the indicated concentrations to treat PDAC or CCA cells.
2.7. Western blot analysis
Western blotting was performed as previously described [3]. Primary antibodies for STAT3 (BD Biosciences), GAPDH (Santa Cruz Biotechnology), and phospho-STAT3 (Tyr705) (Cell Signaling, Danvers, MA, USA) were used. GAPDH and β-actin were employed as loading controls. Images were obtained using a LuminoGraph I (ATTO, Tokyo, Japan). Signal intensity was quantified using the CS Analyzer (ATTO).
2.8. Statistical analysis
The expression value of ZIC5 in PDAC and CCA tissues was obtained from mRNA sequence data in The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/), and statistical analysis was performed using the two-tailed Mann–Whitney U test or Fisher's exact test. Kaplan-Meier analysis and the other statistical analyses were performed using the JMP software (Pro 15.1.0) or R statistical software package (v. 4.0.3). Data are presented as mean ± standard deviation (SD) in the bar graphs unless otherwise indicated. Box-plots summarize the median and the values between the 25th and 75th percentile. The significance of differences was determined by the statistical tests indicated in the individual figure legends. P < 0.05 was considered statistically significant.
3. Results and discussion
3.1. ZIC5 expression in PDAC, CCA, and normal adult human tissues
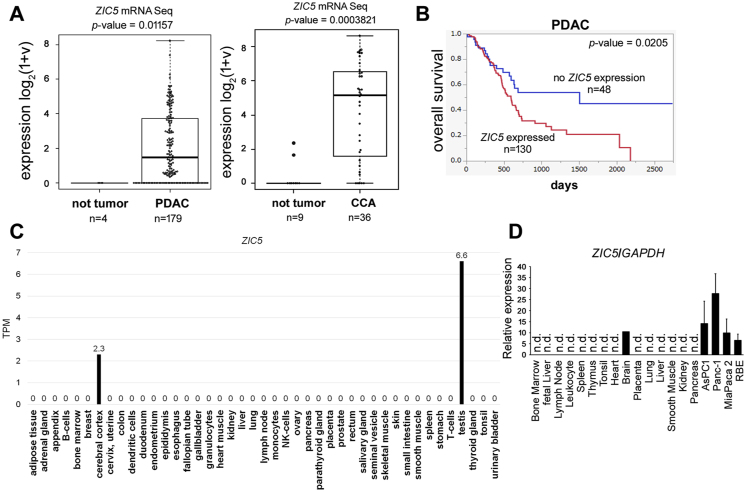
First, we confirmed that ZIC5 was highly expressed in PDAC and CCA tissues compared to the relevant non-tumor tissues, as determined by the statistical analysis of RNA-Seq data from the TCGA database (Fig. 1A). Summaries of clinicopathological parameters of patients with PDAC or CCA and ZIC5 expression are shown in Supplementary Tables 1 and 2 ZIC5 expression correlated with pathologic stage of PDAC (Supplementary Table 1).
Fig. 1.
ZIC5 expression in PDAC, CCA, and normal human tissues. (A) RNA sequence values of ZIC5 expression in pancreatic adenocarcinoma (left; PDAC) and cholangiocarcinoma (right; CCA) tissues, as well as in relevant non-tumor tissues, were obtained from The Cancer Genome Atlas (TCGA) database. Statistical comparisons between disease and normal tissues (for each disease) were performed using the two-tailed Mann–Whitney U test. (B) The relationship between ZIC5 expression and survival was determined and plotted using the Kaplan–Meier method. The overall survival rates for patients with or without detectable ZIC5 expression are plotted as red and blue lines, respectively. (C) RNA-Seq values of ZIC5 expression in normal human adult tissues were obtained from The Protein Atlas database. (D) ZIC5 expression was assessed by quantitative reverse transcription-PCR with cDNA samples from normal human tissues and from AsPC-1, PANC-1, MiaPaca-2 (PDAC), and RBE (CCA) cell lines. The values were normalized to the expression (in the respective samples) of the housekeeping gene GAPDH. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
It was reported that ZIC5 is a target of miR-761, which is inhibited by circ_0007534 [6], and circ_0007534 expression is upregulated in PDAC tissues [7]. Moreover, both miR-625 and miR-892b, which are sponged by circ_0007534, are downregulated in the PDAC tumor tissues compared with those in the paired normal tissues [7]. To assess whether miR-761 is involved in ZIC5 suppression in PDAC and/or CCA, the expression of miR-761 was assessed based on the OncoMir Cancer Database [8]. The expression of miR-761 was not significantly different between the normal and tumor samples in both PDAC and CCA. Moreover, the expression of miR-761 was not correlated with the expression of ZIC5 (OncoMir Cancer Database). From these data, we speculate that neither miR-761 nor circ_0007534 is involved in ZIC5 regulation in these tumors.
To determine whether ZIC5 expression affects prognosis, we evaluated the gene expression in patients with PDAC or CCA, and constructed survival curves using the Kaplan–Meier method. The incidence of overall survival rate was significantly lower in PDAC patients with tumors with ZIC5 expression compared to that in patients with no detectable ZIC5 expression (Fig. 1B). In CCA patients, the overall survival rates did not differ significantly between these two groups (data not shown).
RNA-Seq data suggests that ZIC5 expression in normal adult human tissues is limited in testis and cerebral cortex (The Human Protein Atlas) [9] (Fig. 1C). To confirm this observation, cDNA from human normal tissues (Human Multiple Tissue cDNA Panels I and Human Immune System MTC Panel; TaKaRa, Japan) was assessed for ZIC5 expression using qPCR analysis. ZIC5 transcript was detected in brain as well as in PDAC and CCA cell lines, but not in bone marrow, fetal liver, lymph node, leukocyte, spleen, thymus, tonsil, heart, placenta, lung, liver, smooth muscle, kidney, or pancreas (Fig. 1D). These results suggested that the ZIC5 transcript exhibits strong expression specificity in PDAC and CCA.
3.2. ZIC5 positively regulates proliferation of PDAC and CCA cells
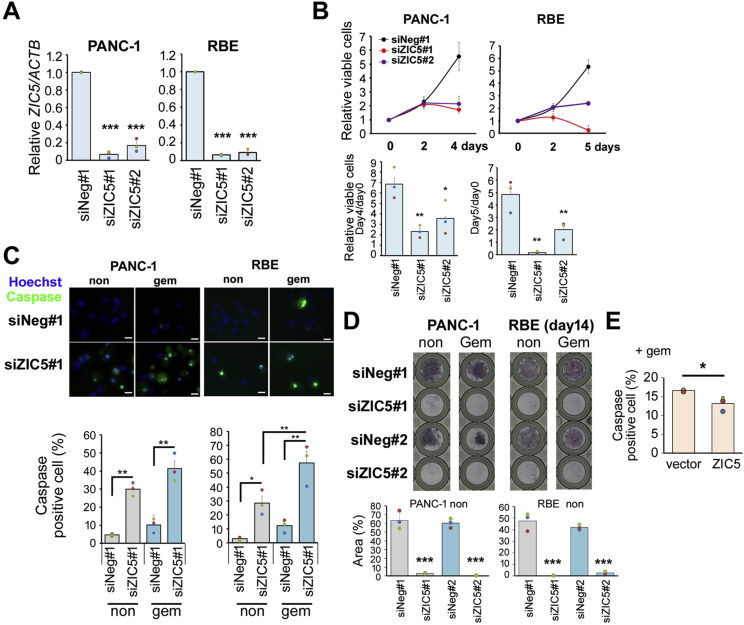
To elucidate the roles of ZIC5 in PDAC and CCA cells, we performed siRNA-mediated knockdown of ZIC5 in several cell lines (Fig. 2A and Supplementary Fig. 1A). Cell proliferation assay revealed that ZIC5 knockdown significantly reduced the cell numbers of PDAC (AsPC-1, PANC-1, and MiaPaca-2) and CCA (RBE) cell lines (Fig. 2B and Supplementary Fig. 1B) compared to the effect of a control siRNA. These results indicated that ZIC5 positively regulates the growth of these cells.
Fig. 2.
ZIC5 positively regulates survival of PDAC and CCA cells. PANC-1 (PDAC cell line) and RBE (CCA cell line) were transfected with a negative control (siNeg) or ZIC5 (siZIC5) siRNA. The target sequence of the siRNA differed between #1 and #2. (A) At 2 days after transfection, ZIC5 mRNA expression was assessed by qPCR. The relative expression level was normalized to that of the housekeeping gene ACTB as an internal control. Statistical analysis was performed using Dunnett's multiple comparison of means test. (B) Cell proliferation was assessed at the indicated times. Relative number of viable cells at Day 4 or 5 (compared to that at Day 0) from three independent experiments is indicated in the bar graphs. Statistical analysis was performed using Dunnett's multiple comparison of means test (***P < 0.001, **P < 0.01, *P < 0.05). (C) PANC-1 and RBE cells were transfected with a negative control (siNeg) or ZIC5 (siZIC5) siRNA. After 24 h, cells were treated with gemcitabine (Gem) (0.2 μM) as indicated. After 2 days, the cells were incubated with Caspase-3/7 Green Detection Reagent (green) and Hoechst33342 (blue). The percentage of caspase-positive cells from three independent experiments was determined. Statistical analysis was performed using Tukey's multiple comparison test (***P < 0.001, **P < 0.01, *P < 0.05). (D) After 14 days of gemcitabine treatment (0.1 μM), cells were stained with Giemsa solution. Stained area was quantified using Image J. Statistical analysis was performed using Tukey's multiple comparison test (***P < 0.001). (E) MiaPaca-2 cells stably overexpressed ZIC5 and control cells (vector) were treated with gemcitabine (Gem) (0.2 μM) for 2 days, and then apoptosis assays were performed. Statistical analysis was performed using the two-tailed non-paired Student's t-test (*P < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. ZIC5 positively regulates survival of PDAC and CCA cells
To determine whether ZIC5 is involved in the survival of PDAC and CCA cells, we next assessed apoptosis induction upon ZIC5 knockdown. ZIC5 knockdown induced apoptosis rates of approximately 15–30% in these cell lines on day 3 after transfection (Fig. 2C and Supplementary Fig. 1C). When cells also were treated with gemcitabine, a therapeutic drug used clinically to treat PDAC and CCA, ZIC5 knockdown additively or synergistically induced apoptosis to approximately 30–60% at 48 h after gemcitabine treatment (Fig. 2C and Supplementary Fig. 1C). To further assess the surviving cells and their growth, we cultured cells for 14 days after treatment with low-dose gemcitabine. Giemsa staining showed that PANC-1 and RBE cells were eradicated by ZIC5 knockdown alone (Fig. 2D), while AsPC-1 and MiaPaca-2 were eradicated only by the combination of gemcitabine and ZIC5 knockdown (Supplementary Fig. 1D). When ZIC5 was overexpressed, gemcitabine-induced apoptosis was reduced in MiaPaca-2 (Fig. 2E), however, ZIC5 overexpression in PANC-1 or RBE cells did not affect their survival (data not shown). We speculated that the effect of ZIC5 overexpression was not evident because ZIC5 expression at an endogenous level in these cells was sufficient to exert its biological effects. These results suggested that endogenous ZIC5 promotes survival in PDAC and CCA cells and ZIC5 suppression renders cancer cells more susceptible to anti-cancer drugs. Improving drug sensitivity is important for preventing the drug resistance of cancer cells.
3.4. ZIC5 knockdown decreases STAT3 phosphorylation in PDAC and CCA cells
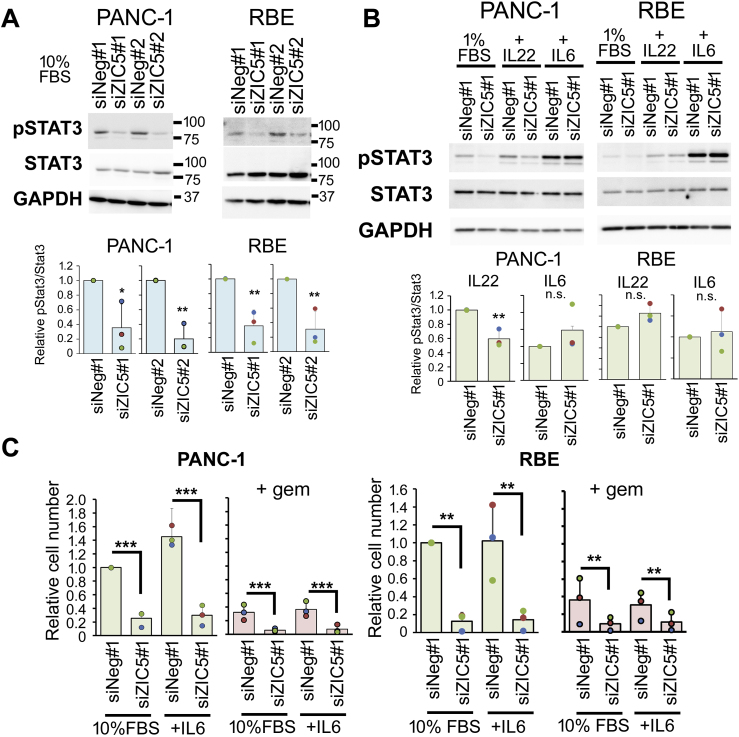
STAT3 activation promotes the expression of anti-apoptotic genes and is associated with the aggressive phenotypes of PDAC and CCA [[10], [11], [12], [13]]. We previously reported that ZIC5 is involved in the activation of STAT3 in melanoma, colorectal, and prostate cancer cells [2,3]. Therefore, to determine whether ZIC5 contributes to STAT3 activation in PDAC and CCA cells, the phosphorylation level of STAT3 was examined. Because of low STAT3 expression in AsPC-1 cells [10], we performed further experiments using PANC-1, MiaPaca-2, and RBE cells. ZIC5 knockdown significantly decreased the amount of phosphorylated STAT3 (Tyr705) in PANC-1, MiaPaca-2, and RBE cells (Fig. 3A and Supplementary Fig. 2A). These results and our previous research indicated that ZIC5 regulates STAT3 activity in most of the tested cancer cell lines (Fig. 3A and Supplementary Fig. 2A) [2,3]. Given that ZIC5 knockdown induced apoptosis in AsPC-1 cells, which has very low STAT3 phosphorylation [10], ZIC5-regulated factors other than STAT3 must be important for survival of AsPC-1 cells.
Fig. 3.
ZIC5 positively regulates the phosphorylation of STAT3, although ZIC5 knockdown attenuates survival of PDAC and CCA cells despite interleukin (IL) -6-mediated activation of STAT3. (A) The levels of phosphorylated STAT3 (Tyr705) (pSTAT3) as well as of total STAT3 and GAPDH were determined in PANC-1 and RBE cells transfected with a negative control (siNeg) or ZIC5 (siZIC5) siRNA. Quantification of phosphorylated STAT3 levels normalized to total STAT3 are shown as mean ± SD (n = 3) in the bar graphs. Statistical analysis was performed using the two-tailed non-paired Student's t-test (**P < 0.01, *P < 0.05). (B) PANC-1 and RBE cells were transfected with siNeg or siZIC5. At 2 days after transfection, the cells were treated with 1% FBS with or without IL-6 (20 ng/mL) or IL-22 (50 ng/mL). After 24 h, the cells were harvested to assess the phosphorylation of STAT3. (C) The relative cell number was determined after 6 days of IL-6 treatment (20 ng/mL) with or without gemcitabine (gem) (0.1 μM) from three independent experiments. Statistical analysis was performed using the two-tailed Tukey's multiple comparison test (***P < 0.001, **P < 0.01, *P < 0.05).
ZIC5 is known to regulate PDGFD expression in melanoma, colorectal cancer, and prostate cancer cell lines [2]. Therefore, we also assessed PDGFD expression in cells subjected to ZIC5 knockdown. However, the level of pro-PDGFD was not significantly reduced by ZIC5 knockdown in PDAC and CCA cell lines (data not shown). Moreover, expression of E-cadherin, CDKN1B and HSPD1, each of which has been shown to be regulated by ZIC5 in melanoma or prostate cancer [2,3], also was unchanged in PDAC and CCA cell lines (data not shown), suggesting divergent target genes for ZIC5 in various cancer cells.
3.5. ZIC5 knockdown suppresses survival of PDAC and CCA cells despite interleukin (IL) -6-mediated activation of STAT3
PDAC and CCA often are associated with chronic inflammatory conditions such as pancreatitis or cholangitis, and the inflammatory mediators IL-6 and IL-22 have been implicated in the progression of PDAC and CCA [[11], [12], [13], [14]]. Because both IL-6 and IL-22 cause STAT3 activation, we next examined whether ZIC5 knockdown attenuated STAT3 phosphorylation and survival of cancer cells even in the presence of IL-6 or IL-22. In PANC-1 and RBE cells, ZIC5 knockdown had no significant effect on the level of STAT3 phosphorylation induced by IL-6 (Fig. 3B), while decreasing the level of IL-6- or IL-22-induced STAT3 phosphorylation in MiaPaca-2 (Supplementary Fig. 2B). Furthermore, ZIC5 knockdown significantly decreased the survival of PANC-1 and RBE cells exposed to IL-6 as well as 10% FBS (Fig. 3C). Because ZIC5 knockdown could not suppress phosphorylation of STAT3 induced by IL-6 (Fig. 3B), while suppressed that induced by 10% FBS in PANC-1 and RBE cells (Fig. 3A), ZIC5 knockdown induced cell death regardless of STAT3 activation. ZIC5-regulated factors other than STAT3 also must be important for survival of these cells.
In the present study, we demonstrated that ZIC5 promotes the survival of PDAC and CCA cells via unknown mechanisms. Recently, ZIC activity was shown to be influenced by WNT dependent SUMOylation during neural crest development [15]. In a high-WNT environment, ZIC5 is SUMOylated, which decreases formation of the transcription factor 7 like 2/ZIC co-repressor complex and shifts the balance towards ZIC transcription factor function [15]. Thus, cellular context (e.g., post-transcriptional regulation and co-factors) may render a variety of transcriptional regulatory targets available to ZIC5. Nevertheless, ZIC5 knockdown showed additive or synergistic effects in inducing apoptosis with gemcitabine treatment in all of the tested cell lines, suggesting that ZIC5 may serve as an attractive therapeutic target for the treatment of PDAC and CCA.
4. Conclusions
In this study, we confirmed that ZIC5 is strongly expressed in PDAC and CCA tissues, whereas ZIC5 expression is barely observed in most normal human adult tissues. ZIC5 knockdown induced PDAC and CCA cell death. Furthermore, the knockdown of ZIC5 and the anti-cancer drug gemcitabine additively or synergistically induced apoptosis in PDAC and CCA cells. Thus, ZIC5 constitutes a potential therapeutic target for the treatment of PDAC and CCA.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by a Grants-in-Aid for Young Scientists (Japan Society for the Promotion of Science KAKENHI Grant Nos. 17H05056) to R.S. This research was also supported by Japan Agency for Medical Research and Development under Grant Number JP21cm0106176h0002. This work was also supported by a Research Grants of the Princess Takamatsu Cancer Research Fund to R.S.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101289.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
I have shared the data in https://data.mendeley.com/datasets/wyhj26s4ny/draft?a = cf2878d2-3d20-4310-9dfa-5f1e4e8f58c4.
References
- 1.Kato S. Tumour-agnostic therapy for pancreatic cancer and biliary tract cancer. Diagnostics. 2021;11 doi: 10.3390/diagnostics11020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satow R., Nakamura T., Kato C., Endo M., Tamura M., Batori R., Tomura S., Murayama Y., Fukami K. ZIC5 drives melanoma aggressiveness by PDGFD-mediated activation of FAK and STAT3. Cancer Res. 2017;77:366–377. doi: 10.1158/0008-5472.CAN-16-0991. [DOI] [PubMed] [Google Scholar]
- 3.Satow R., Inagaki S., Kato C., Shimozawa M., Fukami K. Identification of zinc finger protein of the cerebellum 5 as a survival factor of prostate and colorectal cancer cells. Cancer Sci. 2017;108:2405–2412. doi: 10.1111/cas.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q., Shi R., Wang X., Li D., Wu H., Ren B. Overexpression of ZIC5 promotes proliferation in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2016:479. doi: 10.1016/j.bbrc.2016.09.098. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., Hu X., Sun D., Wu Y., Zhao Z. ZIC5 facilitates the growth of hepatocellular carcinoma through activating Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018;503 doi: 10.1016/j.bbrc.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Li G.F., Li L., Yao Z.Q., Zhuang S.J. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem. Biophys. Res. Commun. 2018;499 doi: 10.1016/j.bbrc.2018.03.219. [DOI] [PubMed] [Google Scholar]
- 7.Hao L., Rong W., Bai L., Cui H., Zhang S., Li Y., Chen D., Meng X. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J. Cell. Biochem. 2019;120:3780–3789. doi: 10.1002/jcb.27658. [DOI] [PubMed] [Google Scholar]
- 8.Wong N.W., Chen Y., Chen S., Wang X. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics. 2018;34:713–715. doi: 10.1093/bioinformatics/btx627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlen M., Uhlén M., Fagerberg L., Hallström B.M., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Sivertsson A., Kampf C., Sjöstedt E., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A.-K., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., von Heijne G., Nielsen J., Ponten F., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015:347. doi: 10.1126/science.1260419. 1979. [DOI] [PubMed] [Google Scholar]
- 10.Nagathihalli N.S., Castellanos J.A., Lamichhane P., Messaggio F., Shi C., Dai X., Rai P., Chen X., VanSaun M.N., Merchant N.B. Inverse correlation of STAT3 and MEK signaling mediates resistance to Ras pathway inhibition in pancreatic cancer. Cancer Res. 2018;78:6235–6246. doi: 10.1158/0008-5472.CAN-18-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He W., Wu J., Shi J., Huo Y.M., Dai W., Geng J., Lu P., Yang M.W., Fang Y., Wang W., Zhang Z.G., Habtezion A., Sun Y.W., Xue J. IL22RA1/STAT3 signaling promotes stemness and tumorigenicity in pancreatic cancer. Cancer Res. 2018;78:3293–3305. doi: 10.1158/0008-5472.CAN-17-3131. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda A., Wang S.C., Morris J.P., Folias A.E., Liou A., Kim G.E., Akira S., Boucher K.M., Firpo M.A., Mulvihill S.J., Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isomoto H., Mott J.L., Kobayashi S., Werneburg N.W., Bronk S.F., Haan S., Gores G.J. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132:384–396. doi: 10.1053/j.gastro.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouassier L., Marzioni M., Afonso M.B., Dooley S., Gaston K., Giannelli G., Rodrigues C.M.P., Lozano E., Mancarella S., Segatto O., Vaquero J., Marin J.J.G., Coulouarn C. Signalling networks in cholangiocarcinoma: molecular pathogenesis, targeted therapies and drug resistance. Liver Int. 2019;39:43–62. doi: 10.1111/liv.14102. [DOI] [PubMed] [Google Scholar]
- 15.Ali R.G., Bellchambers H.M., Warr N., Ahmed J.N., Barratt K.S., Neill K., Diamand K.E.M., Arkell R.M. WNT-responsive SUMOylation of ZIC5 promotes murine neural crest cell development, having multiple effects on transcription. J. Cell Sci. 2021;134 doi: 10.1242/jcs.256792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
I have shared the data in https://data.mendeley.com/datasets/wyhj26s4ny/draft?a = cf2878d2-3d20-4310-9dfa-5f1e4e8f58c4.