Abstract
Owing to the high heterogeneity of pancreatic cancer, patient-derived xenografts (PDX) can compensate for the defects of cell line-derived xenografts (CDX) and also better preserve the heterogeneity and tumor microenvironment of primary tumors. Further, gemcitabine, which is used for the treatment of various cancers, is prone to tumor drug resistance, and this limits its sustained efficacy. Therefore, in this study, our objective was to screen appropriate individual therapeutic drugs for pancreatic cancer. To this end, we established pancreatic cancer PDX models from different patients and screened gemcitabine sensitivity regulatory molecules via high-throughput transcriptome sequencing and bioinformatics analysis. Based on the results obtained, gemcitabine was identified as the most suitable chemotherapeutic drug in a variety of PDX models. Additionally, our results indicated that Lipocalin 2 (LCN 2) may play an important role in the sensitivity of pancreatic cancer to gemcitabine treatment. Thus, the study provides a new potential intervention target for the treatment of pancreatic cancer in clinical practice.
Keywords: Lipocalin 2, Gemcitabine, Chemosensitivity, Pancreatic cancer, patient-derived xenografts
Highlights
-
•
PDX model plays an important role in the screening of chemotherapeutic agents for pancreatic cancer.
-
•
Gemcitabine is the most suitable chemotherapeutic drug in a variety of PDX models of pancreatic cancer.
-
•
Lcn2 may be involved in the sensitivity of gemcitabine in the treatment of pancreatic cancer and the change of Lipocalin 2 levels determines the gemcitabine therapeutic output.
1. Introduction
Pancreatic cancer is a malignant tumor of the digestive system with a poor prognosis. Epidemiological data shows that the overall 5-year survival rate of patients with pancreatic cancer is < 7%, and the disease ranks as the fourth and sixth cause of malignant tumor-related deaths in the United States and China, respectively [1]. Presently, surgical resection followed by adjuvant chemotherapy is still the preferred treatment strategy for pancreatic cancer; however, the success rate of radical resection is only 15–20%, and this can be attributed to a locally advanced stage of the disease or distant metastasis at first diagnosis [2]. Chemotherapy is an important treatment strategy for improving the prognosis of patients with locally advanced and metastatic pancreatic cancer. Current chemotherapeutic agents in this regard include gemcitabine, docetaxel, irinotecan, oxaliplatin, and pemetrexed. Among them, docetaxel, which is an antineoplastic agent that has a unique mechanism of action, as an inhibitor of cellular mitosis, plays a central role in the therapy of several solid tumors, such as breast and lung cancer [3]. Further, irinotecan is a semisynthetic derivative of the plant alkaloid, camptothecin, and is used as an antineoplastic agent in the treatment of colorectal, ovarian, and non-small cell lung cancers [4]. Furthermore, oxaliplatin is an intravenously administered platinum-containing alkylating agent used for treating advanced colorectal cancer [5]. Pemetrexed is a cytostatic antifolate drug and a cornerstone treatment for lung cancer [6], and gemcitabine, which displays extensive and exact therapeutic effects, is considered the first-choice treatment in pancreatic cancer management [7]. However, patients with pancreatic cancer tend to be resistant to gemcitabine, failing to maintain the efficacy of the drug [8]. Thus, strategies to overcome such drug resistance and improve gemcitabine sensitivity have become a challenging and topical issue in preclinical research.
The use of cell line-derived xenograft (CDX) models for drug screening is questionable owing to the lack of tumor stromal components and the single cell type-characteristics of the cell lines. Thus, patient-derived xenograft (PDX) models, which offer the possibility to overcome the above limitations of CDX models and better retain the heterogeneity of primary tumors and the tumor microenvironments, are now used to screen appropriate individualized therapeutic drugs [9].
Thus, in this study, we selected a variety of pancreatic cancer PDX models to screen for suitable chemotherapeutic agents. RNA-seq was performed on gemcitabine sensitive and control samples [10]. Thereafter, gene data sets were utilized to identify regulatory molecules affecting gemcitabine sensitivity. Based on the results obtained, gemcitabine was identified as the most suitable chemotherapeutic drug in a variety of pancreatic cancer PDX models, and we also observed that Lipocalin 2 (LCN2) possibly plays a key role in pancreatic cancer-related biological processes, such as cell cycle regulation, cell movement, and cell migration regulation. Recently, it was observed that LCN2 expression can distinguish tumor tissues from normal tissues. Notably, LCN2 is primarily expressed in PanIN-1 and PanIN-2 lesions and shows a higher expression level in the PanIN fractions than in other fractions. Further, patients with pancreatic cancer accompanied by diabetes show elevated serum LCN2 levels, which decrease after the surgical resection of pancreatic cancer. Furthermore, patients with pancreatic cancer show higher plasma LCN2 levels than healthy individuals [11]. This study preliminarily demonstrated that LCN2 may be involved in pancreatic cancer sensitivity to gemcitabine treatment and that changes in its level determine gemcitabine therapeutic output. Thus, it provides a novel potential intervention target for clinical treatment.
2. Material and methods
2.1. Tumor specimens and ethical review
Pancreatic cancer specimens (D42113, D37774, and E32051), specimens from adjacent tissues (3 cases), and normal tissues were obtained from the Hepatobiliary, Pancreatic, and Splenic Surgery of Xijing Hospital, China, with written informed consent from the patients and their family members. The characteristics of the patients were showed in Supplemental Table 1. The experiments involving these human specimens were approved by the medical ethics committee of Xijing Hospital (approval number 2015432).
2.2. Establishment and evaluation of pancreatic cancer PDX model
Six-to seven-week-old male BALB/c nude mice were purchased from Changzhou Cavens Lab Animal Co., Ltd. and housed in a pathogen-free system at the Laboratory Animal Center of the Fourth Military Medical University (FMMU, Xi'an, China). At this facility, the mice were anesthetized with ketamine (100 mg/mL, i.p.) and xylazine (20 mg/mL, i.p.) and maintained under isoflurane during surgery. All the animal experiments were approved by the Institutional Animal Care and Use Committee of FMMU (protocol number 16013).
Fresh pancreatic cancer specimens obtained from the patients at Xijing Hospital, named the P0 generation, were placed in a serum free RPMI-1640 medium (GIBCO, USA). Thereafter, these tumor tissues were cut into 1–3 mm3 pieces, mixed with matrigel, and transplanted into the nude mice. Next, the length (L) and width (W) of the subcutaneous tumor tissues were measured and tumor volume was calculated using the formula, V = 1/2 × (w2 × l). When the volume of the tumors reached 800–1000 mm3, the mice were euthanized. Thereafter, the tumor tissues were harvested and inoculated subcutaneously into different mice, which were named the P1 generation. This procedure was repeated until the P3 generation was obtained.
2.3. Experimental grouping and chemotherapy drug treatment
P3 generation PDX tumors (D37774, D42113, and E32051) were cut into 1 mm3 blocks and inoculated subcutaneously into 30 nude mice. When tumor tissues grew to 100–150 mm3, the mice were randomly divided into six groups (n = 5), namely, the control group (control), oxaliplatin treatment group (5 mg/kg), docetaxel treatment group (5 mg/kg), irinotecan treatment group (50 mg/kg), pemetrexed (100 mg/kg), and gemcitabine treatment group (100 mg/kg). All the mice in the different groups were administered the respective drugs via intraperitoneal injection twice a week.
2.4. Immunohistochemical and immunofluorescence staining
Tumor tissues were fixed with 4% paraformaldehyde, embedded in paraffin, sliced, and hydrated with xylene and gradient alcohol. After antigen retrieval via microwave heating, endogenous peroxidase was blocked. Goat serum was also blocked at 37 °C for 10 min, and the tissues were incubated overnight with primary antibodies (CA19-9 antibody, Abcam, dilution ratio 1:100; Ki-67 antibody, Abcam, dilution ratio 1:100) at 4 °C. The tissues were incubated with biotin-labeled goat anti-rabbit secondary antibody at room temperature in the dark for 20 min. Next, 3,3′-diaminobenzidine staining, hematoxylin and eosin (H&E) staining, hydrochloric acid alcohol differentiation, dehydration, sealing, and neutral gum sealing were performed, and the proportion of positive cells in the visual field were quantitatively analyzed using Image Pro Plus 6.0 software. Some of the tumor tissues were fixed with 4% formalin for apoptosis assay using the TUNEL assay kit (Abcam, dilution ratio 1:100). Thereafter, the proportion of apoptotic cells in the visual field was quantitatively analyzed via confocal microscopy.
2.5. CA19-9 detection in plasma
Cardiac blood was collected and centrifuged at 5000 rpm/min for 10 min. Thereafter, the supernatant was sent to the laboratory department of Xijing Hospital for CA19-9 detection.
2.6. High-throughput sequencing
The control group samples (D37774, D42113, and E32051 tissues not treated with gemcitabine) and the gemcitabine-treated D37774, D42113, and E32051 samples were sequenced via high-throughput transcriptome sequencing at Shanghai Kangcheng Bioengineering Company. The first mock experiment was performed at a resolution that was 1.5 times higher than that employed for the experiments involving the control and gemcitabine specimens of the same model. Thereafter, the average of the three groups of differentially expressed genes was considered to establish the differential expressed gene dataset. Finally, the heat map for these differentially expressed genes was then drawn using a visualization software.
2.7. Bioinformatics analysis and screening of target genes
2.7.1. Principal component analysis (PCA)
PCA was based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases, which can be accessed at http://www.oncomine.org and http://gepia.cancer-pku.cn, respectively. Using the visual analysis tool of the website, the differential gene dataset for PCA analysis was entered, and the differences between tumor and normal tissues with respect to the differentially expressed genes were determined and verified.
2.7.2. Protein-protein interaction (PPI) analysis
PPI analysis was performed based on the STRING database, using the http://string-db.org website visualization tools, topology optimization, the differential gene datasets, and further screening of core datasets.
2.7.3. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
Through GO and KEGG pathway enrichment analyses (https://www.cbioportal.org), the biological functions, regulatory pathways, and signaling pathways associated with the genes in the core dataset were predicted.
2.8. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using an RNA Extraction Kit (Tiangen Biochemical Company) and reverse transcribed using an RNA Reverse Transcription Kit (Takara, Japan). Finally, qRT-PCR was performed according to the manufacturer's instructions.
2.9. Statistical analyses
SPSS software version 21.0 was used for statistical analyses, and countable data were presented as the mean ± SD. One way ANOVA was used for comparison among multiple groups of countable data followed by Tukey test. Further, t-test was performed to compare two groups, and Pearson's correlation tests were performed to determine the degree of linear correlation between two continuous variables. Statistical significance was set at P < 0.05.
3. Results
3.1. Screening individual chemotherapy drugs
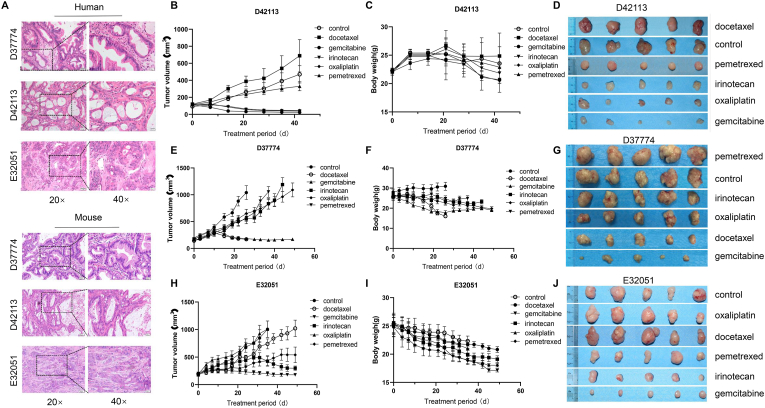
The three PDX models (D37774, D42113, and E32051) were successfully established by subcutaneously transplanting fresh human pancreatic cancer specimens into nude mice. Based on H&E staining, the histology of the PDX-derived tumor tissues was identical to that of the original samples obtained from the patients with pancreatic cancer (Fig. 1A).
Fig. 1.
Screening of individual chemotherapy drugs.
A. H&E analyses of tumor tissues derived from different PDX models and patient samples; B. Tumor volume change trend in each D42113 model treatment group; C. Weight change trend of nude mice in each D42113 model treatment group; D. Comparison of the tumor sizes and morphologies corresponding to the different D42113 model groups at the treatment end point; E. Tumor volume change trend in each D37774 model treatment group; F. Weight change trend of nude mice in each D37774 model treatment group; G. Comparison of tumor size and morphology in each D37774 model group at the end point of treatment; H. Tumor volume change trend in each treatment E32051 model group; I. Weight change trend of nude mice in each E32051 model treatment group; J. Comparison of the tumor sizes and morphologies corresponding to the different E32051 model groups at the treatment end point (n = 5).
These PDX models were further selected to evaluate the therapeutic effects of different chemotherapy drugs. As shown in Fig. 1B and C, the growth rate of the D42113 model was relatively low, and after 45 d of treatment, none of the tumor samples had volume > 1000 mm3, and the average body weight of the mice in each of these D42113 groups was not < 20 g. Further, the D42113 model that received docetaxel treatment showed a higher tumor growth rate than the control group; no tumor inhibition was observed. Conversely, the gemcitabine, oxaliplatin, and irinotecan groups showed significant tumor growth inhibition effects (P < 0.01), indicating that the D42113 model was sensitive to gemcitabine, oxaliplatin, and irinotecan (Fig. 1D). Regarding the D37774 model group, the control group showed the fastest tumor growth rate, with an average tumor volume > 1000 mm3 by day 26 (Fig. 1E and F). At the end of the treatment period, the volumes of the tumors corresponding to the gemcitabine and docetaxel groups were < 100 mm3 (P < 0.01 and P < 0.05, respectively), and all the tumor tissues showed high sensitivity to these drugs. Notably, the tumor tissues also showed different chemosensitivities to the other treatments, which failed to inhibit tumor growth (P > 0.05) (Fig. 1G). Further, based on our observations, gemcitabine was identified as the most suitable chemotherapeutic drug for the E32051 PDX model. The relative results in this regard are shown in Fig. 1H, I, and J.
Ki-67 expression in all PDX model tissues that showed a definite response to the therapeutic effect of the different drugs was significantly lower than that in the control tissues (P < 0.01, Supplemental Figs. 1–4). However, there was no significant difference in Ki-67 expression between tissues with an unclear response to the therapeutic effects of the different treatments and the control groups (P > 0.05; Supplemental). Additionally, TUNEL assays (P < 0.01, Supplemental Fig. 5) clearly revealed that gemcitabine treatment resulted in more obvious tumor cell apoptosis. The plasma levels of CA19-9 corresponding to the gemcitabine, irinotecan, and oxaliplatin groups were lower than the clinical reference values, and those corresponding to other groups were higher than this reference value (P < 0.01, Supplemental Figs. 1–4). These findings demonstrated that gemcitabine was the most suitable chemotherapeutic drug in all the PDX models.
3.2. High-throughput sequencing analysis of differentially expressed genes
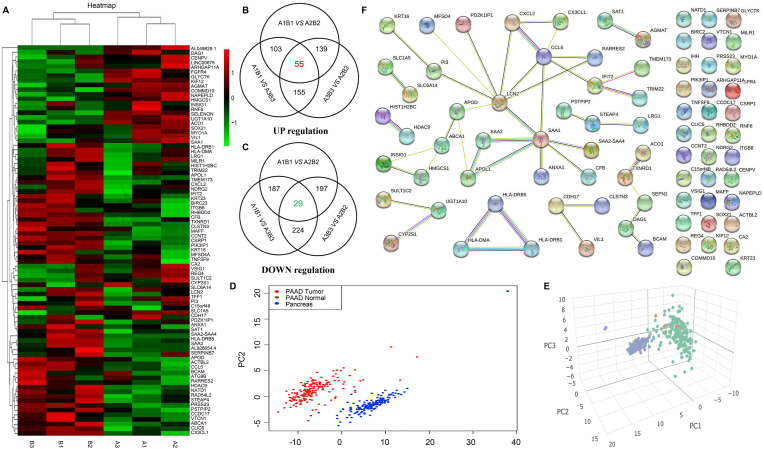
High-throughput transcriptome sequencing demonstrated that 1192 differentially expressed genes were upregulated and 954 were downregulated, while 10,625 were not significantly different between the D37774 groups (Fig. 2A). The averages corresponding to the three groups of data were used and the differentially expressed gene dataset was screened out. The screening strategy is as shown in Fig. 2B and C. After screening, 55 and 29 upregulated and downregulated genes, respectively, were identified. These genes were selected from TCGA and GTEx databases, and based on the pancreatic cancer database, PCA revealed that the genes in the dataset could better distinguish tumor tissues from normal tissues (Fig. 2D and E). To further narrow the scope and screen key genes, the core dataset of the differentially expressed genes was entered through the STRING database, the PPI was constructed, and the filtering of isolated genes was optimized via complementary analysis (Fig. 2F).
Fig. 2.
High-throughput sequencing analysis of differentially expressed genes.
A. Heat map of three groups of differentially expressed genes; B. Upregulated gene screening strategy and results; C. Downregulated gene screening strategy and results; D. Principal component analysis results (PCA; 2D view) distinguishing pancreatic cancer tissue from normal tissue. E. PCA (3D view) distinguishing pancreatic cancer tissue and normal tissue. F. Extension and optimization of the PPI network gene dataset and screening of the core gene dataset.
3.3. Analysis of differentially expressed genes
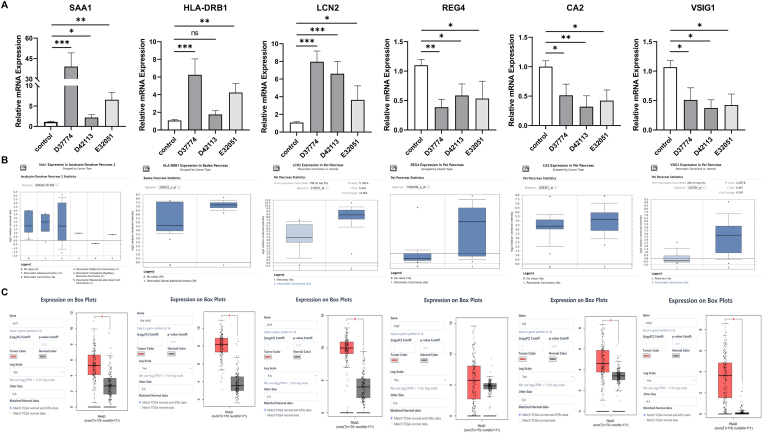
The top three upregulated genes and top three downregulated genes are shown in Supplementary Tables 2 and 3, respectively. qRT-PCR was used to detect the expression levels and trend of these upregulated (SAA1, HLA-DRB1, and LCN2) and downregulated (REG4, CA2, and VSIG1) genes. Further, the sequencing results are shown in Fig. 3A, from which it is evident that the expression levels and trends of the upregulated and downregulated genes were consistent with the results of the sequencing analysis. Furthermore, analysis based on the PEI Pancreas Statistics database showed that the expression levels of REG4, CA2, VSIG1, HLA-DRB1, and LCN2 in pancreatic cancer tissues were higher than those in adjacent tissues. However, no significant difference was observed between the pancreatic cancer tissues and normal tissues with respect to SAA1 expression of (Fig. 3B). Analysis based on the TCGA and GTEx databases indicated that pancreatic cancer tissues showed higher REG4, CA2, VSIG1, HLA-DRB1, and LCN2 expression levels than normal tissues (Fig. 3C).
Fig. 3.
Analysis of differentially expressed genes.
A. Expression trend of six genes detected via qRT-PCR; B. Comparison of the expression of genes in pancreatic cancer (n = 36) and adjacent tissues (n = 16) (data source: PEI Pancreas Statistics). C. Comparison of the expression of genes in pancreatic cancer (n = 179) and normal tissues (n = 171) (data sources: TCGA and GTEx).
3.4. Potential role of LCN2 in pancreatic cancer
PPI results suggested that LCN2 is located in the central regulatory region, and possibly plays a key regulatory role in the gene dataset. Data analysis also showed that LCN2 was significantly (P < 0.05) upregulated in pancreatic cancer tissues; thus, it was our primary focus in this study.
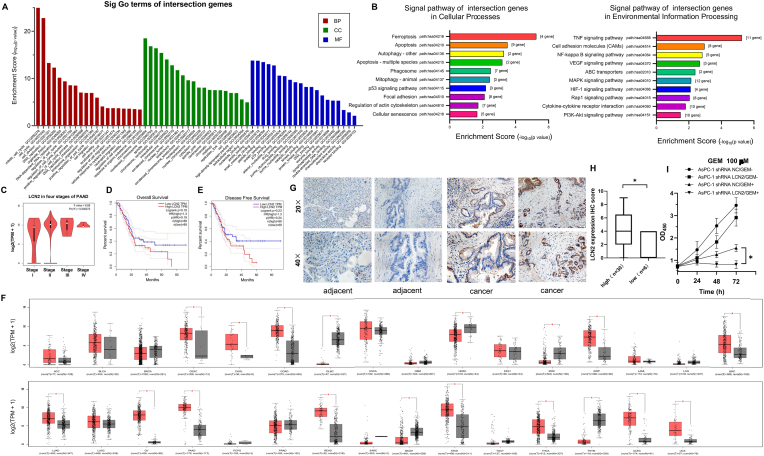
GO enrichment analysis (Fig. 4A) showed that the core gene sets that were enriched were those associated with biological processes, cell components, and molecular function cycle (GO: 0007049), cell division (GO:0051,301), DNA replication (GO: 0006260), cell cycle regulation (GO:0051,726), cell migration (GO: 0016,477), cell motility (GO: 0031,347), cell migration regulation (GO: 0030,334), cell motility regulation (GO: 2000145), drug binding (GO: 0008144), and chemoattractant activity (GO: 0042,056). Additionally, the results obtained suggested that the LCN2 core gene set possibly plays a key role in biological processes, such as cell cycle, DNA replication, cell cycle regulation, cell migration, cell movement, cell migration regulation, drug binding, and chemical inducer activity. KEGG pathway enrichment analysis showed that the LCN2 core gene set was primarily enriched in ferroptosis (path: hsa04216), apoptosis (path: hsa04210), cell adhesion molecules (path: hsa04064), the NF-κB signaling pathway (path: hsa04064), VEGF signaling pathway (path: hsa04370), and PI3K/Akt signaling pathway (path: hsa04151). The results also suggested that the LCN2 core gene set may regulate apoptosis, cell adhesion molecule, and NF-κB signaling pathways. Further, the VEGF and PI3K/Akt signaling pathways regulated the occurrence and development of pancreatic cancer (Fig. 4B), and analyses based on the TCGA and GTEx databases showed that the four stages of pancreatic cancer showed no differences with respect to LCN2 expression (P > 0.05) (Fig. 4C). Overall survival (OS) and disease free survival (DFS) analyses showed that the 5-year survival of patients with high LCN2 expression levels was significantly lower than that of patients with low LCN2 expression levels (Fig. 4D and E). Based on a comparison between TCGA and GTEx databases, LCN2 expression in tumor tissues (18/31, 58%) significantly increased. Further, LCN2 expression level in tumor tissues, such as stomach, pancreas, and intestinal tumor tissues, was higher than that in normal tissues, suggesting that LCN2 might be a potential biomarker for tumor prognosis. From Fig. 4F, which shows the immunohistochemical staining of pancreatic cancer tissues in the PDX model library, it is evident that LCN2 expression was negative in adjacent tissues (n = 6). Most of the tumor tissues (n = 30) showed positive LCN2 expression (26/30), and the statistical results were negative, positive, moderately positive, and strongly positive for n = 4, n = 12, n = 10, and n = 4, respectively (Fig. 4G and H).
Fig. 4.
Potential role of LCN2 in pancreatic cancer.
A. Biological function of LCN2 predicted via GO enrichment analysis based on the DAVID database. A1 and B1 are D37774 samples, A2 and B2 are D42113 samples, and A3 and B3 are E32051 samples. B. LCN2 core dataset based on the use of DAVID database KEGG enrichment analysis to predict the regulation pathway; C. Comparison of LCN2 expression levels corresponding to different stages of pancreatic cancer (data source: TCGA and GTEx); D. Correlation between LCN2 expression and overall survival; E. Correlation between LCN2 expression and disease free survival; F. Differential expression of LCN2 RNA level in normal tissue (n = 5) and tumor tissue (n = 5); G. Immunohistochemical staining results showing LCN2 expression in pancreatic tumor tissues; H. Immunohistochemical staining analysis of LCN2 expression (*, P < 0.05; n = 5). I. Proliferation characteristics of AsPC-1 cell lines with LCN2 expression knockdown under GEM ±conditions (*, P < 0.05).
To verify the potential role of LCN2 in pancreatic cancer, we selected the gemcitabine-resistant cell line, AsPC-1/GEM-, and the gemcitabine-sensitive cell line, AsPC-1 GEM+, to knockdown LCN2 and test the sensitivity of its expression to this treatment. The results showed that gemcitabine sensitivity was enhanced after LCN2 knockdown (Fig. 4I), confirming that Lipocalin 2 levels can determine chemotherapeutic output.
4. Discussion
PDX models maintain the majority of the key genes and signaling pathways in primary tumors. Thus, they can better simulate the parental genetic characteristics of patients with tumors [12]. Except for hormone-sensitive tumors, the sex of immunodeficient animals has no significant effect on PDX models of other tumors. Further, PDX and biological tumor models from different patients will have different biological and genetic characteristics. Thus, the effective target drugs for one model are not suitable for other models from different sources [13]. These differences in characteristics make targeted drug screening and individualized treatment necessary, and provide a good experimental basis for screening suitable drugs for different patients with malignant tumors. Thus, optimized treatment schemes can be made available, thereby overcoming the problem of tumor drug resistance. Additionally, the PDX model has unique advantages in tumor biology experiments [14].
By measuring changes in tumor volume, we observed that gemcitabine significantly inhibited tumor growth in different PDX models and showed the most suitable chemotherapeutic effect. Furthermore, there are various tumor markers that are regarded as important indicators for evaluating the therapeutic effect of chemotherapeutic drugs. Specifically, plasma CA19-9 level is an auxiliary index for the clinical diagnosis and prognosis of pancreatic cancer [15]. In this study, we found that after gemcitabine treatment, the plasma CA19-9 levels corresponding to the three model groups were the lowest, indicating that these tumor cells showed the lowest CA19-9 secretion abilities. Our histological staining results also showed fewer ductal gland structures as well as a normal cell morphology and prognostic trend for the gemcitabine treatment group. In pathological diagnosis, Ki-67 is an important tumor proliferation marker [16]. In this study, gemcitabine treatment significantly downregulated Ki-67 expression in each group. Considering the toxic and side effects of drugs, tumor growth inhibition, and CA19-9 and Ki-67 auxiliary diagnostic indices, the most suitable chemotherapeutic drugs for each model were selected. For the D37774 model, gemcitabine was identified as the most suitable chemotherapy agent, followed by docetaxel. Thus, the use of these three chemotherapeutic drugs for the D42113 model is recommended. For the E32051 model, gemcitabine was found to be most suitable, followed by irinotecan. Further, we observed that although there were significant differences in the performance of the chemotherapy drugs in the three models, the efficacy of gemcitabine was significant in all three cases; this is consistent with the results of previous studies [17]. Accordingly, we selected the gemcitabine treatment group and the control group samples as screening items to perform gemcitabine sensitivity-related research.
We screened 84 differentially expressed genes via high-throughput RNA-seq sequencing of gemcitabine-sensitive PDX model samples and control samples. Based on these differentially expressed gene datasets, PCA based on the TCGA pancreatic cancer database revealed that the genes in the dataset could better distinguish tumor tissues from normal tissues, indicating that the differentially expressed genes possibly play an important role in the occurrence and development of pancreatic cancer. Additionally, the PPI network was constructed using the STRING database, and the isolated genes were supplemented and optimized to further narrow the scope and screen out gene sets with LCN2 as the regulatory center, which may play a key role in regulating the sensitivity of gemcitabine. Thereafter, GO and KEGG pathway enrichment analyses were performed to predict the biological function of the LCN2 core dataset. The results thus obtained showed that the LCN2 core gene set possibly plays a key role in DNA replication, cell cycle regulation, cell movement, cell migration regulation, drug binding, chemical inducer activity, and other biological processes.
Moreover, pathway enrichment analysis showed that apoptosis, cell adhesion-related molecules as well as the NF- κB, VEGF, and PI3K/Akt pathways may regulate the biological characteristics of pancreatic cancer PDX models. Stoesz [18] reported for the first time that LCN2 is highly expressed in primary breast cancer. Consistent with this finding, our immunohistochemical staining results showed that 39% of the samples showed positive cytoplasm LCN2 staining. In a previous study, out of 250 breast cancer tissues analyzed via western blotting, 109 (44%) showed LCN2 expression. Other studies have also shown that LCN2 is expressed in urine and breast cancer cell line samples from patients with invasive breast cancer tissues, and patients with metastatic breast cancer [19]. Our analysis also showed an association between an increase in LCN2 protein level and a decline of DFS rate, disease-specific survival rate, and OS rate.
The analysis of the gene expression profiles of 19 pancreatic cancer cell lines and pancreatic ductal epithelial cell lines revealed that LCN2 is one of the genes with significantly increased expression in pancreatic cancer [20]. In a study on colon cancer, Nielsen [21] selected 14 colorectal adenocarcinoma and normal epithelial tissues surrounding the cancer tissue for immunohistochemical analysis. The results of this previous study indicated that compared with LCN2 expression in normal epithelial cells, LCN2 staining in all the rectal adenocarcinoma samples was strongly positive. Immunohistochemical staining results corresponding to different thyroid tissues also displayed that the expression level of LCN2 in papillary, follicular, and anaplastic thyroid cancer is significantly higher than that in normal thyroid tissues. Further studies have also demonstrated that the increase in LCN2 expression level is directly proportional to the malignant phenotype of these tumors, with the intermediate degenerative thyroid cancer showing the highest LCN2 expression level [22]. Based on the TCGA database, LCN2 expression in tumor tissues (18/31, 58%) is significantly increased compared with the GTEx database, suggesting that LCN2 may be used as a potential biomarker for tumor prognosis [23]. In this study, immunohistochemical staining also showed upregulated LCN2 expression in tumor tissues. Therefore, we speculated that LCN2 may affect gemcitabine sensitivity via a variety of molecular and signaling pathways.
Most previous studies have shown that LCN2 has a variety of biological characteristics and plays an important role in a variety of disease-related processes. Additionally, LCN2 has been described as a potential biomarker for cancer progression given its presence in urine and serum samples from patients with breast cancer and pancreatic ductal adenocarcinoma (PDAC), respectively. Thus, it may represent a new biomarker for early diagnosis, prognostication, and therapeutic targeting in PDAC [24]. Reportedly, LCN2 can be used as a predictor for early cancer screening [22]. Moreover, it can regulate the occurrence and development of several tumor types, especially pancreatic cancer, given that it regulates apoptosis, invasion, angiogenesis, as well as the EMT process [25]. Further, reports on the effect of LCN2 on gemcitabine sensitivity are rare. We will further explore the role of LCN2 in gemcitabine resistance as this may provide a new therapeutic target for drug-resistant pancreatic cancer.
Author contributions
All authors contributed to the writing of the manuscript and have approved the final version.
Funding
This work was supported by the National Natural Science Foundation Program of China (grant numbers 31572340 and 81372606).
Declaration of competing interest
All the authors declare that they have no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101291.
Contributor Information
Jiaze An, Email: anchen@fmmu.edu.cn.
Changhong Shi, Email: changhong@fmmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Tang S., Hang Y., Ding L., Tang W., Yu A., Zhang C., Sil D., Xie Y., Oupický D. Intraperitoneal siRNA nanoparticles for augmentation of gemcitabine efficacy in the treatment of pancreatic cancer. Mol. Pharm. 2021 doi: 10.1021/acs.molpharmaceut.1c00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin Q., Qian Z., Jusko W.J., Mager D.E., Ma W.W., Straubinger R.M. Synergistic pharmacodynamic effects of gemcitabine and fibroblast growth factor receptor inhibitors on pancreatic cancer cell cycle kinetics and proliferation. J. Pharmacol. Exp. Therapeut. 2021;377:370–384. doi: 10.1124/jpet.120.000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Docetaxel . LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda (MD): 2020. October 13. [PubMed] [Google Scholar]
- 4.Irinotecan . LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda (MD): 2018. April 27. [PubMed] [Google Scholar]
- 5.Oxaliplatin . LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda (MD): 2020. September 12. [PubMed] [Google Scholar]
- 6.de Rouw N., Piet B., Derijks H.J., van den Heuvel M.M., Ter Heine R. Mechanisms, management and prevention of pemetrexed-related toxicity. Drug Saf. 2021;44(12):1271–1281. doi: 10.1007/s40264-021-01135-2. [DOI] [PubMed] [Google Scholar]
- 7.Kieler M., Unseld M., Bianconi D., Schindl M., Kornek G.V., Scheithauer W., Prager G.W. Impact of new chemotherapy regimens on the treatment landscape and survival of locally advanced and metastatic pancreatic cancer patients. J. Clin. Med. 2020;9 doi: 10.3390/jcm9030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai H., Wang R., Guo X., Song M., Yan F., Ji B., Liu Y. Combining gemcitabine-loaded macrophage-like nanoparticles and erlotinib for pancreatic cancer therapy. Mol. Pharm. 2021;18:2495–2506. doi: 10.1021/acs.molpharmaceut.0c01225. [DOI] [PubMed] [Google Scholar]
- 9.Yang G., Guan W., Cao Z., Guo W., Xiong G., Zhao F., Feng M., Qiu J., Liu Y., Zhang M.Q., You L., Zhang T., Zhao Y., Gu J. Integrative genomic analysis of gemcitabine resistance in pancreatic cancer by patient-derived xenograft models. Clin. Cancer Res. 2021;27:3383–3396. doi: 10.1158/1078-0432.ccr-19-3975. [DOI] [PubMed] [Google Scholar]
- 10.Leung L., Radulovich N., Zhu C.Q., et al. Lipocalin2 promotes invasion, tumorigenicity and gemcitabine resistance in pancreatic ductal adenocarcinoma. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gumpper K., Dangel A.W., Pita-Grisanti V., et al. Lipocalin-2 expression and function in pancreatic diseases. Pancreatology. 2020;20(3):419–424. doi: 10.1016/j.pan.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamska A., Elaskalani O., Emmanouilidi A., Kim M., Abdol Razak N.B., Metharom P., Falasca M. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv. Biol. Regul. 2018;68:77–87. doi: 10.1016/j.jbior.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Waheed A., Purvey S., Saif M.W. Masitinib in treatment of pancreatic cancer. Expet Opin. Pharmacother. 2018;19:759–764. doi: 10.1080/14656566.2018.1459566. [DOI] [PubMed] [Google Scholar]
- 14.Golan T., Atias D., Stossel C., Raitses-Gurevich M. Patient-derived xenograft models of BRCA-associated pancreatic cancers. Adv. Drug Deliv. Rev. 2021;171:257–265. doi: 10.1016/j.addr.2021.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Kim J., Bamlet W.R., Oberg A.L., Chaffee K.G., Donahue G., Cao X.J., Chari S., Garcia B.A., Petersen G.M., Zaret K.S. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aah5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Striefler J.K., Sinn M., Pelzer U., Jühling A., Wislocka L., Bahra M., Sinn B.V., Denkert C., Dörken B., Oettle H., Riess H., Bläker H., Lohneis P. P53 overexpression and Ki67-index are associated with outcome in ductal pancreatic adenocarcinoma with adjuvant gemcitabine treatment. Pathol. Res. Pract. 2016;212:726–734. doi: 10.1016/j.prp.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Garrido-Laguna I., Uson M., Rajeshkumar N.V., Tan A.C., de Oliveira E., Karikari C., Villaroel M.C., Salomon A., Taylor G., Sharma R., Hruban R.H., Maitra A., Laheru D., Rubio-Viqueira B., Jimeno A., Hidalgo M. Tumor engraftment in nude mice and enrichment in stroma- related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clin. Cancer Res. 2011;17:5793–5800. doi: 10.1158/1078-0432.ccr-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoesz S.P., Friedl A., Haag J.D., Lindstrom M.J., Clark G.M., Gould M.N. Heterogeneous expression of the lipocalin NGAL in primary breast cancers. Int. J. Cancer. 1998;79:565–572. doi: 10.1002/(sici)1097-0215(19981218)79:6<565::aid-ijc3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 19.Bauer M., Eickhoff J.C., Gould M.N., Mundhenke C., Maass N., Friedl A. Neutrophil gelatinase-associated lipocalin (NGAL) is a predictor of poor prognosis in human primary breast cancer. Breast Cancer Res. Treat. 2008;108:389–397. doi: 10.1007/s10549-007-9619-3. [DOI] [PubMed] [Google Scholar]
- 20.Missiaglia E., Blaveri E., Terris B., Wang Y.H., Costello E., Neoptolemos J.P., Crnogorac-Jurcevic T., Lemoine N.R. Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int. J. Cancer. 2004;112:100–112. doi: 10.1002/ijc.20376. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen B.S., Borregaard N., Bundgaard J.R., Timshel S., Sehested M., Kjeldsen L. Induction of NGAL synthesis in epithelial cells of human colorectal neoplasia and inflammatory bowel diseases. Gut. 1996;38:414–420. doi: 10.1136/gut.38.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Celestino R., Nome T., Pestana A., Hoff A.M., Gonçalves A.P., Pereira L., Cavadas B., Eloy C., Bjøro T., Sobrinho-Simões M., Skotheim R.I., Soares P. CRABP1, C1QL1 and LCN2 are biomarkers of differentiated thyroid carcinoma, and predict extrathyroidal extension. BMC Cancer. 2018;18:68. doi: 10.1186/s12885-017-3948-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung L., Radulovich N., Zhu C.Q., et al. Lipocalin2 promotes invasion, tumorigenicity and gemcitabine resistance in pancreatic ductal adenocarcinoma. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago-Sánchez G.S., Pita-Grisanti V., Quiñones-Díaz B., Gumpper K., Cruz-Monserrate Z., Vivas-Mejía P.E. Biological functions and therapeutic potential of lipocalin 2 in cancer. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21124365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.