Graphical abstract
Keywords: CTCF, Tumor suppressor, 3D genome organization, TADs, Topologically associating domains, Enhancer hijacking, Cancer genomes, Computational biology
Abbreviations: 3C, chromosome conformation capture; AS, alternative splicing; CBS, CTCF binding site; CNV, copy number variation; CRE, cis-regulatory element; CT, chromosome territory; DSB, DNA double strand break; FISH, fluorescence in situ hybridization; GIST, gastrointestinal stromal tumors; Hi-C, high-throughput 3C; RBR, RNA binding region; SE, super enhancer; T-ALL, T cell acute lymphoblastic leukemia; TAD, topologically associating domain; ZF, zinc finger
Abstract
The emergence and progression of cancers is accompanied by a dysregulation of transcriptional programs. The three-dimensional (3D) organization of the human genome has emerged as an important multi-level mediator of gene transcription and regulation. In cancer cells, this organization can be restructured, providing a framework for the deregulation of gene activity. The CTCF protein, initially identified as the product from a tumor suppressor gene, is a jack-of-all-trades for the formation of 3D genome organization in normal cells. Here, we summarize how CTCF is involved in the multi-level organization of the human genome and we discuss emerging insights into how perturbed CTCF function and DNA binding causes the activation of oncogenes in cancer cells, mostly through a process of enhancer hijacking. Moreover, we highlight non-canonical functions of CTCF that can be relevant for the emergence of cancers as well. Finally, we provide guidelines for the computational identification of perturbed CTCF binding and reorganized 3D genome structure in cancer cells.
1. Introduction
Cancer cells display profoundly reorganized gene expression programs, which are modulated by changes in transcription factor binding at cis-regulatory elements (CREs: promoters and enhancers) and differences in promoter enhancer contacts [1]. As a consequence, oncogenes may become activated and tumor-suppressor genes may become repressed. Whereas the genes whose activity is changed can vary between different types of cancer cells, the underlying cause of gene deregulation usually incorporates similar genomic changes. These genomic changes can be classified into two non-exclusive mechanisms: genetic changes and epigenetic changes. Genetic changes directly affect the DNA sequence and can encompass anything from single nucleotide substitutions to large-scale structural variation like chromosome copy number variation (CNV) or chromosomal rearrangements. The consequences of these changes can be diverse, ranging from changes in gene dose, fusions between genes, changes to protein coding sequence or changes to the gene regulatory information in the genome. Epigenetic changes include modifications to DNA methylation patterns and the redistribution of activating and repressive histone modifications. The outcome of these epigenetic changes is generally more restricted to changes in transcriptional output. Nonetheless, both types of changes are well-established as causes for the emergence and progression of various cancers [2]. Importantly, these mechanisms are not mutually exclusive, with late-stage cancer cells often displaying profound reorganization of both their genome and epigenome [3].
Interestingly, the ever-evolving advances in genomics technologies have identified an intimate relationship between the three-dimensional (3D) organization of the genome and transcriptional activity, with reorganization of genome structure observed in many cancer types. An important DNA-binding factor, the CTCF insulator protein (CCCTC-binding factor), has emerged as a jack-of-all-trades in 3D genome organization [4], [5], [6], [7]. Increasing evidence is accumulating that various perturbations of CTCF function, including reorganized DNA binding, can act as key factors in cancer-associated transcriptional reorganization.
In this review, we will first discuss global insights from recent studies on 3D genome organization, with a particular focus on the essential functions of the CTCF protein. Next, we will detail the different categories of CTCF perturbations that are observed in cancer cells, and how they may lead to cancerous transformation. In the last part, we provide guidelines on how genomics studies can be performed to determine the impact of CTCF perturbations on the 3D organization of cancer genomes.
2. 3D organization of the genome in the mammalian cell nucleus
Besides the coding sequence of genes, the genome also contains the regulatory instructions for the timing (when), the location (where) and the level (how much) of gene activity. These regulatory instructions are encoded within CREs, which include promoters and enhancers. In normal human cells, this regulatory information is contained within a diploid genome that is made up of 46 chromosomes, which in turn is nearly two meters in length. To fit these chromosomes within the interphase cell nucleus of around 10 µm in diameter, a large degree of compaction is needed. A further prerequisite is that these compacted chromosomes can faithfully maintain essential functions such as gene expression, DNA replication and repair. During interphase, a trade-off must therefore be achieved between DNA compaction and accessibility (as compared to the extremely compacted state of chromosomes during mitosis).
Evolving technological innovations have made it possible to uncover the 3D organization of chromosomes at ever-improving resolution. Super-resolution and live-cell imaging can nowadays reveal 3D genome organization at the nanometer-scale within individual cells, yet these approaches require the pre-selection of probes to target loci or structures of interest. As a result, these assays are less suited for explorative and genome-wide analyses. Genome-wide explorations, i.e. studies without the need for pre-selection of the genomic regions included in the assay, can be performed using genomics-based assays from the chromosome conformation capture (3C) family [8]. 3C technology is based on proximity ligation, whereby DNA fragments that are in close proximity are cross-linked, followed by DNA fragmentation and enzymatic ligation of fragments that remained close due to their crosslinks. The frequency of detection of the resulting chimeric DNA molecules subsequently serves as a proxy for initial proximity in the nucleus. Initially these chimeras were characterized using PCR, Sanger sequencing or microarrays. Nowadays the state-of-the-art is to use high-throughput sequencing, with developments of a wide range of 3C-derivative techniques in recent years. These advances are generally aimed at expanding the interaction throughput (up to the genome-wide level), at increasing resolution, at focusing on specific aspects of genome organization or at reducing costs of the experiments (see for review e.g. [9], [10], [11]). Particularly the development of Hi-C (high-throughput 3C) has been instrumental in our understanding of structure-function relationships in 3D genome organization, as it allows the detection of genomic interactions at high resolution between regions anywhere in the genome (all-vs-all interactions) [12].
Whereas 3C assays have tremendously advanced our understanding of the principles that govern 3D genome organization, these observations were mostly made from studies on large populations of cells. Most insights are therefore based on the description of averages in the population, which are not always reconciled by imaging experiments [13], [14]. Although a few single-cell 3C-based studies have been reported, they generated data at a more moderate resolution and within a relatively limited number of cells. Many studies have therefore preferred to combine data from both imaging and genomics. Recent technological developments in the field of “spatial genomics” have taken this a step further, by directly allowing the positioning of specific DNA sequences within the cell nucleus [15], [16], [17].
The 3D organization of chromosomes in human cells is not random, with distinct types of organization visible at different levels. At the first level, as observed by fluorescence in situ hybridization (FISH), each interphase chromosome occupies its own distinct “chromosome territory” (CT) in the nucleus [18]. The radial positioning of CTs is linked to gene activity, with inactive heterochromatin preferentially located at the periphery of the nucleus and gene-rich euchromatic regions more towards the interior. Indeed, the repositioning of proto-oncogenes within the cell nucleus has been observed in breast cancer cells, confirming the intimate link between global genome organization and transcriptional activity [19].
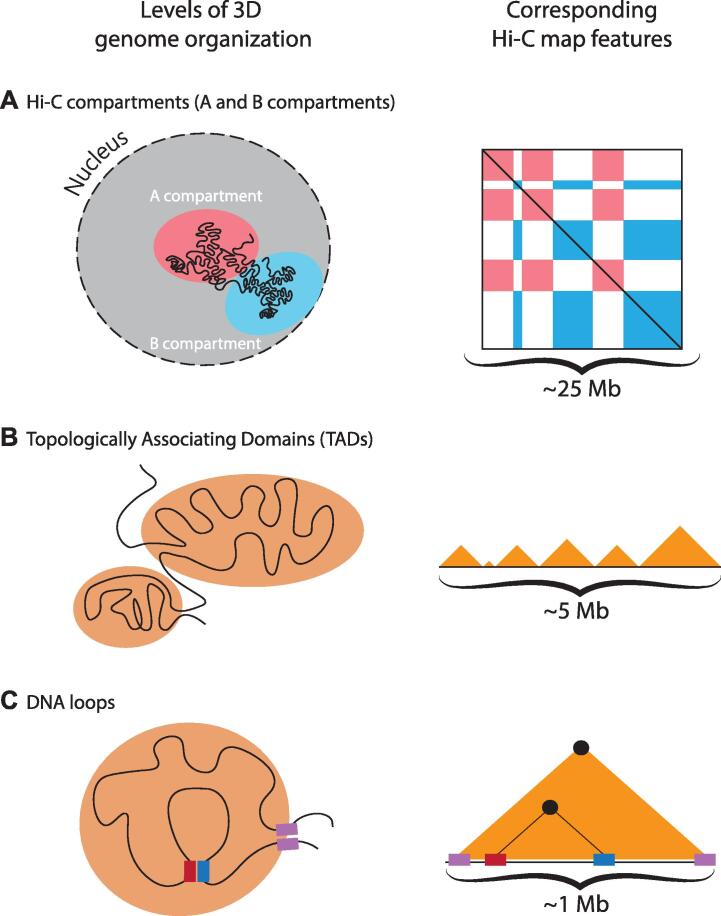
Hi-C has subsequently allowed the identification of multiple scales of 3D genome organization within chromosomes. The first Hi-C study provided interaction maps at a resolution of 1 Megabase (Mb), allowing a first genome-wide view of chromosomal domain positioning in human cells. These Hi-C maps, which are a 2D representation of 3D interactions among all regions within chromosomes, revealed a ‘plaid’ pattern of alternating interactions among domains of several megabases in size (Fig. 1A). At the multi-Mb scale, the human genome is thus spatially separated into two types of Hi-C compartments, with contacts between the same type of domains (homotypic domains) being able to interact over long distance and between chromosomes, and contacts between different types of compartments (heterotypic domains) being depleted. These mutually exclusive compartments have been named A and B compartments, with the A compartment being enriched for gene-rich and actively-transcribed regions and the B compartment being more gene-poor and mostly transcriptionally inactive [12].
Fig. 1.
Different scales of intra-chromosomal 3D genome organization in human cells and their corresponding appearance in Hi-C interaction maps. A: Hi-C compartments (also known as A and B compartments) constitute the largest scale of organization and represent alternating active regions and inactive regions that each preferentially engage in homotypic interactions. B: TADs are sub-Megabase domains embedded within Hi-C compartments. Within a TAD, interactions are enriched over surrounding domains. TADs appear as triangles along the diagonal of the Hi-C map. C: DNA loops represent interactions between two genomic loci, e.g. an enhancer and a promoter (red and blue bars) or the two extremities of a TAD (purple bars). They appear as a punctuated increase of signal (black dots) within or on top of a TAD in a Hi-C map. Below each schematic Hi-C map, approximate length-scales are indicated. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
With further improvements to the resolution of Hi-C experiments, interaction maps revealed the organization of human chromosomes at the sub-Mb scale into discrete domains [20], [21]. These domains, which were named “topologically associating domains” (TADs), are defined as regions where 3D interactions within the domain are considerably enriched as compared to interactions with neighboring TADs (Fig. 1B). Intersection with other types of genomics data have revealed that CREs that regulate the same gene (i.e. promoters and enhancers) co-occupy the same TAD, giving rise to the idea that they constitute insulated regulatory neighborhoods that restrict enhancer-promoter interactions [22], [23], [24].
At the highest resolution, two further types of organization have been detected at length scales that are generally smaller than TADs. Large numbers of DNA loops have been identified, both between CREs (i.e. promoter-promoter, promoter-enhancer and enhancer-enhancer contacts) and among sites bound by the CTCF insulator protein [25], [26], [27], [28]. A schematic illustration of DNA loops is depicted in (Fig. 1C), where they emerge as singular dots within the Hi-C map. In parallel, genomic regions that carry stretches of the same histone modifications can form local aggregates, so-called contact domains. Similar to Hi-C compartments, contact domains that carry the same histone modifications can cluster over long distances and between chromosomes [25].
3. The CTCF insulator protein and 3D genome organization
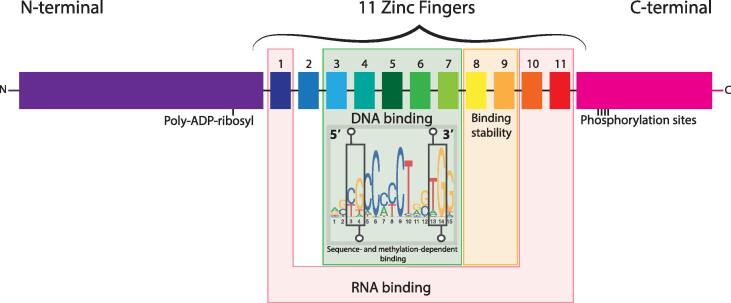
The discovery of the CTCF insulator protein, or CCCTC-binding factor, was first reported by Lobanenkov and colleagues, who detected a strong association of this protein with three regulatory regions flanking the chicken c-myc gene [29]. CTCF is a well-conserved DNA-binding protein that is found in metazoans, with a particularly strong conservation in vertebrates [30]. The many regulatory functions of human CTCF are a direct consequence of the complex structure of this protein, which incorporates 11 distinct zinc finger domains [31] (Fig. 2). These zinc fingers (ZFs), alone or in combination, allow binding to a range of DNA sequences, as well as to RNA and other proteins [32].
Fig. 2.
Domain organization of the CTCF protein. The linear organization of the CTCF protein is indicated on top, including its different functional domains. The reverse complement of the 15 bp core DNA binding motif that is recognized by ZFs 3–7 is indicated below. Lollipops highlight the positions where the presence of a methylated CpG dinucleotide can prevent CTCF binding [46].
CTCF is ubiquitously expressed and several studies have confirmed its essential nature, either in cell survival or cell proliferation (see e.g. [33], [34], [35], [36]). CTCF is thought to be the only insulator protein in somatic human cells. A closely-related paralogue, CTCFL or BORIS (Brother of Regulator of Imprinted Sites) is present in spermatocytes, where it binds a subset of CTCF sites in the genome [37], [38]. Incorrect activation of BORIS has been observed in many different types of cancer, where its competition with CTCF for DNA binding leads to the inappropriate activation of cancer-associated genes [39].
Understanding the different functions of the CTCF insulator protein is the key to establish the involvement of different levels of 3D genome organization in gene regulation, as this jack-of-all-trades has structural roles in most levels of genome structure. Initially, CTCF was identified as a tumor suppressor gene, as the CTCF locus was frequently overlapping the smallest region of overlap between recurrent heterozygous deletions in both breast and prostate cancers [40]. Since then, different changes in the structure of the protein itself, in its activity or in its patterns of binding to the DNA have been found to influence transcriptional programs in cancer cells as well. A precise characterization of the changes in the structure and function of CTCF is therefore essential to unravel the relationships between reorganization of 3D genome structure and transcriptional deregulation in cancer cells.
3.1. What is known about binding of CTCF in the genome?
Genomics studies have reported several tens of thousands of CTCF binding sites (CBSs) in the human genome, with many constitutive sites bound in all or nearly all cell types and some sites being highly cell type specific [41], [42], [43], [44]. Initial genome-wide characterization of CTCF binding revealed a complex and non-symmetric consensus binding motif, which permits binding of the protein to a wide range of related DNA sequences [44] (Fig. 2). This recognition of diverse sequences is achieved through the combinatorial use of its 11 ZFs, with deletion and structural studies identifying that ZFs 3 to 7 each interact with 3 bases of a 15 bp core binding motif [45], [46]. Upstream and downstream DNA binding motifs have been identified as well, which provide further selectivity of CTCF binding at subsets of CBSs [45], [47]. Further fine tuning may be achieved through the implication of additional ZFs, with ZF3 only being crucial when both the core motif and a downstream motif are bound and ZF8 improving the stability of CTCF binding to the DNA [45], [48].
DNA methylation has been identified as an important determinant for differential DNA binding, with the presence of methylated CpG dinucleotides at two positions in the consensus binding motif directly interfering with CTCF binding [41], [46], [49], [50] (Fig. 2, lollipops). However, changes in DNA methylation alone cannot explain all changes in CTCF binding between cell types, as the majority of differentially bound CTCF motifs do not contain CpG dinucleotides and upon demethylation most CpG dinucleotide-containing motifs remain unbound [41], [51]. Changes in DNA methylation in the region around the CBS can also influence CTCF binding, thereby further extending the effect of this modification [52]. This may be linked to the unique pattern of nucleosome positioning around sites that are bound by CTCF, with differences in remodeling capacity providing a potential explanation for cell-type specific binding at individual sites [53], [54].
The association of CTCF with RNA has been implicated in the reorganization of CTCF occupancy at its DNA binding sites as well, with blocking of transcription leading to a moderate reduction in global CTCF binding levels. Deletion studies have identified RNA binding regions (RBRs) in the protein, which include the ZFs 1, 10 and 11 and a part of the C-terminal domain [55], [56], [57] (Fig. 2). Binding of RNA can promote the oligomerization of CTCF, whereas perturbations in the RBRs can cause a complete loss of binding at subsets of CBSs and the misregulation of hundreds of genes. The interaction of the CTCF RBRs with RNA may thus promote the spatial clustering of CTCF within the nuclear space, which in turn may instruct binding to a subset of sites in the genome and may influence global binding affinities through either direct or more indirect effects as well.
Similarly, post-translational modifications can regulate various aspects of CTCF binding (Fig. 2). In the absence of phosphorylation of either the C-terminal domain or the linkers between certain ZFs, the activity of a large number of genes becomes deregulated [58], [59]. In the case of the C-terminally phosphorylated variant of CTCF, it was found to specifically occupy a subset of CBSs. Phosphorylation therefore appears to further modulate CTCF binding specificity. In contrast, poly(ADP-ribosyl)ation (PARylation) of the N-terminal CTCF domain can inhibit DNA binding [60]. Upon PARylation, CTCF relocalizes to the cytoplasm and binding to most sites is strongly reduced. DNA binding of CTCF can be regulated by other proteins as well, although the reported effects are often limited to few gene loci (CTCF protein partners reviewed in [61]). Finally, CTCF does not only compete for binding to the DNA with other proteins (as is the case for the previously mentioned BORIS protein), but at subsets of sites competition occurs with the binding of the Jpx noncoding RNA as well [62].
3.2. What are the different regulatory functions of CTCF?
CTCF was initially identified as a transcriptional repressor, because of its negative impact on the expression of the chicken c-myc gene [29], [31]. Subsequent studies revealed that CTCF binds so-called insulator elements in the genome, which are short DNA sequences that prevent DNA contacts between promoters and enhancers when positioned in-between these CREs [30]. Due to this conserved function in enhancer blocking, CTCF has become known as an insulator protein. While enhancer blocking mostly results in transcriptional silencing, CTCF occupancy has also been found to activate genes, like the APP (Amyloid Beta Precursor Protein) gene [63]. This capacity thus distinguishes the CTCF insulator protein from more conventional repressors. Here, we discuss how CTCF achieves these different regulatory functions.
3.2.1. TAD insulation
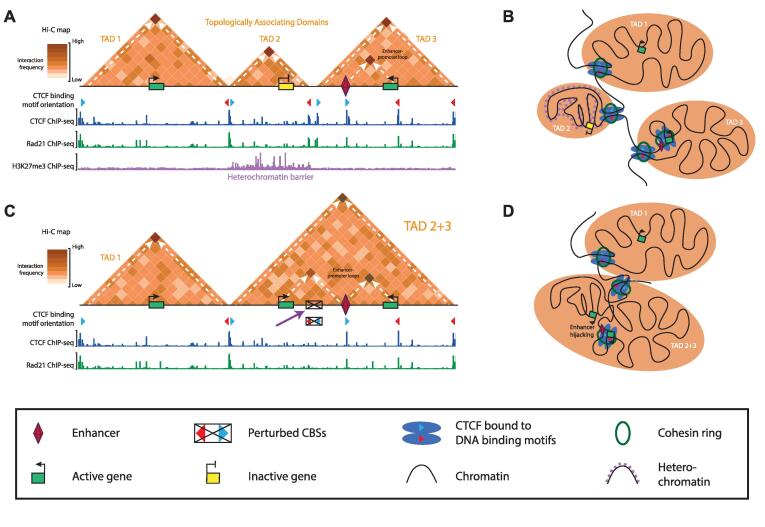
TADs represent discrete domains in the human genome where the insulation between neighboring domains promotes enhancer blocking. Based on this similarity with insulator elements, it may not be surprising that CTCF binding is strongly enriched at the boundaries between TADs [20]. The functional link between insulator elements and TADs provides a further insight into how enhancer blocking is achieved within the 3D space of the nucleus. CTCF binding coincides with an accumulation of the cohesin complex, a ring-shaped protein complex that can entrap DNA [64], [65]. Combinations of biophysical modeling and experimental studies where CTCF, the cohesin complex subunit RAD21 or the cohesin loading factor NIPBL were depleted from the DNA have revealed that TADs are formed through a process of loop extrusion [66], [67], [68], [69], [70]. In this mechanism, the cohesin complex is loaded on the DNA where it extruding function starts to actively create a loop of increasing size. When the complex encounters CTCF bound to the DNA, loop extrusion on this side will be blocked, whereas it continues in the other direction until it encounters CTCF as well. The result of blocking on both sides, particularly when involving multiple copies of the cohesin complex, creates a TAD with enriched contacts within the domain. Interestingly, the efficiency of blocking by the CTCF protein depends on the orientation of binding to the DNA template, as mediated by its non-symmetrical binding motif [25], [71], [72] (Fig. 2, Fig. 3A and B). Indeed, up to 95% of CBSs at the boundaries that surround TADs are in a convergent orientation, which allows the most efficient blocking of loop extrusion [25], [71], [72], [73], [74]. The CTCF-mediated blocking of loop extrusion on both sides of a TAD can bring the CBSs at the boundaries in spatial proximity. Indeed, at the summit of many TADs, an enrichment of signal can be observed that reflects the association between the two boundaries (Fig. 3A). Due to this apparent presence of a DNA loop between these CBSs in Hi-C maps, TADs and other CTCF-mediated structures are also referred to as “loop domains” [25].
Fig. 3.
Functions of CTCF in transcriptional regulation and impact of perturbed CTCF binding in cancer cells. A: TAD insulation, enhancer-promoter looping and heterochromatin barrier function through CTCF binding. A schematic Hi-C map with 3 TADs and simulated CTCF, Rad21 (cohesin complex) and H3K27me3 ChIP-seq are depicted. The yellow gene (center TAD) is inactive because of the enhancer-blocking activity of CTCF at the TAD boundary. The green gene on the right is activated by its enhancer though DNA loop formation. B: Schematic chromatin organization of the 3 TADs from panel A, showing the relative position between neighboring TADs, the containment of heterochromatin and the physical proximity between the green gene and its enhancer at loop anchors mediated by CTCF and the cohesin complex. C: Consequence of changes in the DNA sequence or methylation status of a CBS on TAD structure. Perturbation of CTCF binding at the CBSs that separate TAD 2 and TAD 3 (purple arrow) causes a fusion between the domains. This allows the hijacking of the enhancer located in former TAD 3 by the gene previously located in TAD 2. D: Schematic chromatin organization of the TADs from panel C. TAD 2 and 3 have fused, with the disruption of the boundary causing enhancer hijacking and gene activation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2.2. Enhancer-promoter looping
Although CTCF binding is strongly enriched at TAD boundaries, the majority of CBSs are found elsewhere in the genome. Within TADs, the formation of DNA loops between enhancers and their target promoters can also depend on CTCF binding. Importantly, CTCF needs to bind both the promoter and the enhancer to promote the formation of such interactions, which can achieve gene activation across large genomic distances. Thousands of genes are thought to benefit from CTCF-mediated enhancer-promoter interactions, as removal of CTCF leads to both a loss of enhancer-promoter loops and a decrease in activity of subsets of genes [75], [76] (Fig. 3A and B).
Depending on where it binds in the genome, CTCF can thus both prevent the formation of enhancer-promoter interactions, if present at TAD boundaries, or promote enhancer-promoter contacts, if bound at CREs. This paradox confirms the versatility of this jack-of-all-trades in gene regulation and 3D genome structure.
3.2.3. Heterochromatin/euchromatin segregation
Whereas removal of CTCF does not appear to directly affect the presence of A/B compartments in Hi-C interactions maps [66], it does have an impact on the spread of heterochromatin domains across the genome, although most likely in a cohesin-independent manner [77]. CTCF binding is enriched at the boundaries between regions that carry the repressive H3K27me3 histone mark and active histone marks, which can both form intra-TAD contact domains [25], [78]. The role of CTCF at these boundaries is not well established though, as a strong reduction of CTCF levels in the cell does not noticeably change the spread of the H3K27me3 mark [66]. In contrast, upon removal of a single CBS in the mouse genome, a noticeable invasion of an active mark into a H3K27me3-marked domain could be observed [79]. CTCF binding may thus protect the integrity of H3K27me3-marked contact domains, rather than prevent the spreading of this heterochromatin mark.
4. CTCF binding changes and 3D genome reorganization in cancer cells
CTCF is involved in gene regulation through a diverse range of mechanisms, including the formation of multiple layers of 3D genome organization. As a result, its perturbed function may cause misexpression of oncogenes and tumor suppressor genes. In this section, we will focus on the different mechanisms whereby incorrect binding to the genome contributes to the cancerous state. For this purpose, we distinguish three categories of changes: changes in the amount of CTCF protein, changes in CTCF protein function and changes in CTCF binding to the genome.
4.1. Category 1: Changes in CTCF dosage
CTCF is known as a tumor suppressor, with a deletion or inactivation of the gene detected in a range of cancers [40], [80], [81], [82], [83]. Interestingly though, CTCF function is essential for cell proliferation and cell survival in normal cells and particularly during development [33], [34], [35], [36]. The maintenance or promotion of cell proliferation in cancers thus requires additional transformation events. Indeed, both in endometrial cancer and immortalized MEFs, haploinsufficiency of CTCF results in the downregulation of tumor-suppressor genes and the upregulation of estrogen-sensitive genes [34], [84]. Moreover, Ctcf haploinsufficiency in mouse cells causes destabilization of DNA methylation patterns and increased cancer susceptibility [85]. A reduction of CTCF levels may thus further promote cellular transformation and proliferative capacity. Conversely, upregulation of CTCF has also been described in certain cancers. In hepatocellular carcinoma cells, CTCF dosage can be increased, which correlates with poorer prognosis. A reduction of CTCF protein amounts in these cells diminishes cell proliferation and, in mouse models, inhibits tumor progression [86]. In breast cancer, CTCF overexpression has been proposed to provide a proliferative advantage protecting cancer cells from apoptosis [87]. More recently, changes in CTCF dosage have also been linked to the reversible epithelial-mesenchymal transition in breast cancers, where CTCF upregulation favors the mesenchymal phenotype while its downregulation favors the epithelial traits via transcriptional changes [88]. Combined, these observations indicate that correct physiological levels of CTCF are essential for normal cellular function. Depending on the cancer type, and their genomic transformations, either the decrease or increase of CTCF dosage may associate with uncontrolled proliferation.
4.2. Category 2: Changes in CTCF protein function
Besides mutations that completely abrogate the CTCF function (null mutations), other mutations can induce changes to its functionality (missense mutations). A number of studies have characterized recurrent missense mutations in various cancers, which are particularly enriched in the ZFs [81], [89], [90]. These mutations can be further divided into those that perturb the structure of a ZF, and therefore the interaction with the zinc moiety, and those that have an impact on the binding capacity to specific DNA motifs. Deletions of individual ZFs have confirmed the direct effect on CTCF binding affinity, by either changing the specificity or the stability of binding to the DNA [45], [46], [48].
4.3. Category 3: Changes in CTCF binding to the genome
A different mechanism whereby the function of CTCF is changed, is through localized reorganization of DNA binding or through the relative positioning of CTCF at CBSs within regulatory neighborhoods. This reorganization can either be caused by changes to the DNA sequence itself—ranging from substitutions of single base pairs that directly affects its binding to complex rearrangements like translocations and inversions that can perturb TAD structure—or by deregulation of DNA methylation. In the absence of CTCF binding at a TAD boundary, two neighboring TADs can fuse, thereby allowing the formation of inappropriate contacts between promoters and enhancers. Such “enhancer hijacking” can cause the upregulation of oncogenes (Fig. 3C and D). Conversely, loss of CTCF binding at CREs may perturb the formation of enhancers-promoter loops, which may result in loss of gene activity.
4.3.1. Changes to the DNA sequence
Studies in a range of cancers have identified CBSs as mutational hotspots [91], [92], [93], [94], [95], [96]. Sequencing of gastrointestinal cancers from a large number of patients confirmed that such mutations are directly linked to deregulation of neighboring genes [97]. Mutations that influence CTCF binding at CBSs are enriched for changes of purines (A or T) to pyrimidines (C or G) in the DNA binding motif itself or in the adjacent bases [91], [92], [93], [94], [95], [96].
Similarly, mutations within TAD boundaries are also enriched in a variety of cancers, albeit usually with considerable cancer-type specificity for the affected boundaries [98], [99], [100], [101]. A detailed study in T cell acute lymphoblastic leukemia (T-ALL) cells identified micro-deletions near oncogenes that removed CBSs from TAD boundaries [96]. Recreation of these microdeletions in normal cells increased interactions between the neighboring TADs and the activation of oncogenes contained within those domains. These results thus confirm the instructive nature of CTCF binding to prevent inappropriate activation of oncogenes (Fig. 3C and D).
Interestingly, several studies have suggested that CBSs are fragile sites where mutations are enriched also in the absence of (obvious) positive selection [101], [102]. Indeed, CBSs have been detected as cancer-associated hotspots for chromosomal instability and recombination [102]. As a consequence of this chromosomal instability, the position of TAD boundaries in the genome may become reorganized, thereby permitting the fusion between TADs that in normal cells are located far away from each other [103].
Combined, these studies confirm how genetic changes at CBSs and TAD boundaries can cause fusions of TADs, which creates the potential for enhancer hijacking and oncogene activation.
4.3.2. Changes in DNA methylation
A second means by which CTCF binding can be perturbed at specific sites in the genome is through changes in DNA methylation at CBSs. Methylation of CpG dinucleotides within the CTCF binding motif can directly interfere with DNA binding, although most motifs appear to be insensitive to changes in DNA methylation [41], [51]. Mining of data from a large number of human cell types and six types of cancers revealed that a large fraction of changes in CTCF binding could be explained by changes in DNA methylation in the region directly surrounding the CBS [104]. In contrast, in the same six cancer types, few differences in CTCF binding could be assigned to mutations in the CBS.
More mechanistic insights into the underlying causes of DNA methylation changes have been obtained from gliomas and gastrointestinal stromal tumors (GISTs). In both types of cancers, the inactivation of specific proteins prevents the removal of DNA methylation from the DNA, which results in global hypermethylation of the genome [105], [106]. The resulting hypermethylation subsequently reduces CTCF binding at subsets of CBSs, which in both cases causes the fusion of an oncogene-containing TAD. In gliomas, this permits the PDGFRA oncogene to hijack the FIP1L1 enhancer [105], whereas in GISTs the FGF4 and KIT oncogenes establish inappropriate contacts with super-enhancers in fused TADs as well [106]. Similar to genetic changes at CBSs, DNA methylation changes can thus create TAD fusions, thereby permitting enhancer hijacking and oncogene activation. Curiously, the global hypermethylation that is at the root of both these cancer types is responsible for the inappropriate CTCF-mediated activation of a single oncogene. Despite the global reorganization of 3D genome organization in these cancer cells, the true transformative event may thus ultimately be limited to enhancer hijacking by a single “master” oncogene.
The effect of perturbed DNA methylation may extend beyond CBSs themselves though, as changes in chromatin accessibility due to DNA methylation changes can influence CTCF binding as well [52]. Exploration of a large number of human cancers detected frequent gains and losses of CTCF binding at promoters, which positively correlated with gene activity (i.e a gain of CTCF binding was associated with gene upregulation and a loss of binding with downregulation) and negatively correlated with DNA methylation changes in the 300 bp surrounding the CBS at the promoter [104]. The effect of DNA methylation changes in cancer cells is therefore not restricted to TAD reorganization, but can directly influence the formation of promoter-enhancer loops as well.
5. Non-canonical functions of CTCF in transcriptional regulation and genome (in)stability
Besides its direct role in 3D genome organization and transcriptional regulation, CTCF has been implicated in a number of non-canonical processes which may impact cancer cells as well. Here, we briefly discuss recent insights into CTCF function.
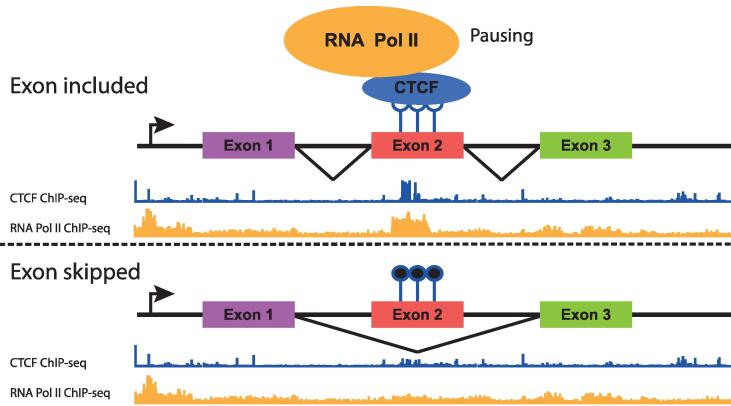
Alternative splicing (AS) represents an additional level of gene regulation, albeit at the level of co– or post-transcriptional tuning of transcriptional output. AS generates differentially spliced mRNA molecules through the selective incorporation of exons and introns. Alterations in AS can lead to oncogenesis, as different isoforms of a protein can have opposing functions, as illustrated by alternative isoforms that promote or inhibit apoptosis [107], [108]. CTCF functions as a direct and indirect regulator of AS through various mechanisms that act at a genomic, epigenomic or co-transcriptional level (reviewed in [109]). Notable examples include the formation of CTCF-mediated DNA loops between promoters and specific exons, which promotes their inclusion in the transcript [110] and CTCF-mediated pausing of RNA Polymerase II that promotes the inclusion of exons that are otherwise ignored due to the presence of weak splicing sites [111] (Fig. 4). More recently, CTCF haploinsufficiency has been linked to increased intron retention at selected genes [112]. The functional implication in AS thus provides a further means whereby CTCF can modulate the abundance of specific mRNA or transcripts or isoforms.
Fig. 4.
The non-canonical function of CTCF in alternative splicing. CTCF-mediated alternative splicing occurs in a methylation-dependent manner. When exon 2 is unmethylated, CTCF can bind and will pause RNA Pol II, leading to the inclusion of the exon. When exon 2 is methylated, CTCF will not bind and the exon will be skipped.
Another non-canonical regulatory function for CTCF has been proposed at super-enhancers (SEs). SEs are a subset of CREs that can confer their particularly strong activating influence through the formation of phase-separated condensates where factors dedicated to transcription are enriched [113]. During carcinogenesis, cancer cells acquire new SEs near oncogenes [114]. In a recent study, the involvement of CTCF in this process was determined. Whereas CTCF itself did not form phase-separated condensates, its depletion perturbed the observable clustering of factors associated with SEs. CTCF binding to the DNA, and possibly the resulting 3D genome organization, may thus have an instructive role in the formation of phase-separated condensates at these strong CREs [115].
CTCF has also been implicated in the activation of the MYC oncogene through a non-canonical mechanism. In colon cancer cells, the binding of CTCF to a distal MYC SE promotes an interaction of the SE with the nucleoporin AHCTF1. In turn, the SE positions the active allele of the MYC oncogene near the nuclear pores, which improves the nuclear export of its transcripts and induces its transcriptional upregulation [116].
Finally, CTCF is involved in DNA double strand break (DSB) repair through various pathways as well (reviewed in [61], [117]). During DSB repair, the 53BP1 and MDC1 proteins and the γH2AX histone modification (ser-139 phosphorylation of the histone variant H2AX) spread over large genomic intervals that overlap the TADs that existed prior to the formation of the DSB [118], [119]. Indeed, upon the formation of a DSB, the ATM kinase that is recruited to the break will associate with the loop extrusion machinery to rapidly deposit the ser-139 phosphorylation mark within the surrounding TAD. In turn, this allows the rapid recruitment of repair proteins and the chromatin remodeling associated with this process [119]. Considering the essential function of CTCF at TAD boundaries, it is therefore directly involved in determining the spread of DSB repair domains. Additionally, CTCF has been proposed as a regulator of Homologous Recombination DSB Repair through the recruitment of the BRCA2 protein at CBSs [120]. This regulation requires the PARylation of CTCF, which is a posttranslational modification that is frequently lost in breast cancer cells [121], [122]. These two examples thus show that the impact of modified CTCF function or binding to the DNA expand beyond transcriptional regulation and can directly influence the outcome of DNA DSB repair in cancer cells as well.
6. How to identify changes in CTCF binding and 3D genome organization?
In this last section, we will provide guidelines on how to identify changes to the genome-wide binding of CTCF in cancer cells and how to link these changes to the reorganization of 3D genome structure. Particularly, we will focus on the data analysis of genome-wide assays for protein-DNA binding and 3D genome organization.
6.1. Identification of differential CTCF binding
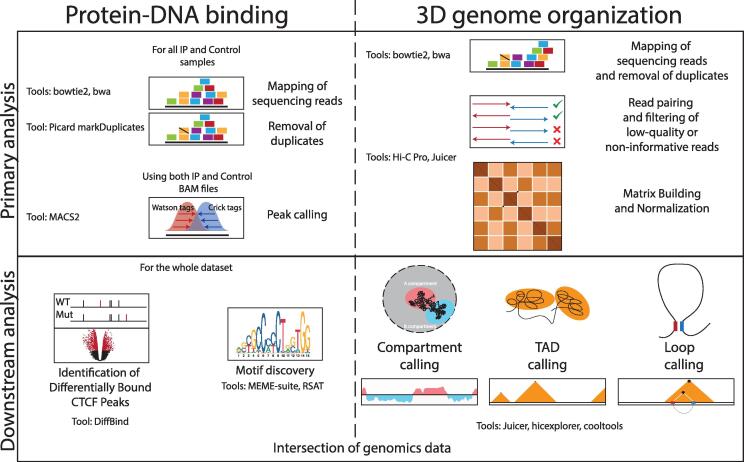
In order to identify differential CBSs on a genome-wide scale, genomics data for CTCF binding should be generated or obtained for different conditions (e.g. ChIP-seq or CUT&Tag data from cancer cells and matching healthy control cells). An outline of the experimental assays to obtain such data is provided in [123]. Data to control for sequencing biases (e.g. from input material without enrichment or from material after enrichment using a control antibody) should be included as well. Particularly in cancer cells, these controls will help to correct for sequencing biases due to copy number variations (CNVs). To improve the reliability of identified CBS, the experiments in both the cancer cells and their matching controls should be replicated.
After high-throughput sequencing of the different samples, significant CBS will first be identified in the individual data sets. After validation of the quality of sequencing quality, the sequencing reads should be aligned to the reference genome (e.g. available from https://www.ncbi.nlm.nih.gov/genome/guide/human/) (Fig. 5). Widely-used mapping algorithms for short-read sequencing data are bowtie2 [124] and BWA [125]. Each of the resulting data files (BAM format) contains the genome-wide alignments of the sequencing reads for one individual sample. As most sequencing library preparation protocols include a PCR-amplification step, duplicate alignments should be removed from these individual files. Tools for such filtering are included in the Picard suite (https://broadinstitute.github.io/picard) (Fig. 5). Significant CBSs in each data set can subsequently be identified by using peak calling algorithms (e.g. MACS2; [126]). In such an analysis, data on CTCF binding is compared to controls to identify regions where signal is significantly enriched only in the CTCF binding data set. Reproducible lists of CBSs can subsequently be obtained by filtering for regions that are identified in all or multiple replicates.
Fig. 5.
Overview of computational strategies for the analysis of protein-DNA binding and 3D genome organization in cancer cells. The left panel shows an outline for the computational analysis of protein-DNA binding data from the mapping of raw sequencing reads to the identification of genomic features like differential peaks and binding motifs. Commonly used tools are indicated for each step. The right panel shows an outline for the computational analysis of 3D genome organization data from the mapping of raw sequencing reads to the identification of genomic features like A/B compartments, TAD boundaries and DNA loops. The outcomes from both analyses can be intersected to identify correlations between protein binding (e.g. CTCF) and 3D genome organization.
To identify differentially bound CBSs between conditions, it is preferred to perform a dedicated differential binding analysis. Such analyses can be further refined using corresponding control samples. Moreover, to compensate for differences in immunoprecipitation efficiency between samples, signal can be normalized by spiking-in a DNA reference of known concentration. Practically, prior to immunoprecipitation each spiked-in sample is supplemented with a fixed amount of control chromatin from a different species. Importantly, CTCF from this control species should be recognized by the same antibody as used for recognition of the human protein. This control material, added as a small percentage relative to the sample of interest (typically in the order of 5%), will act as an external reference to permit quantitative normalization for the differential analysis. Indeed, the observed CTCF enrichment in the control material can be used to adjust the enrichment efficiencies between different conditions, thereby allowing to distinguish between experimental bias and significant biological difference [127]. One option for differential CBS calling is to use the DiffBind package after independent peak calling using the MACS2 algorithm [126], [128]. In a first step, DiffBind can perform different normalizations, including spike-in normalization. Next, two approaches for differential analysis that were originally developed for the identification of differentially expressed genes: DESeq2 and edgeR [129], [130] can be used in Diffbind(Fig. 5).
A consensus binding motif, either from all CBSs or from differentially bound CBSs can be identified by extracting enriched sequences within the population of CBSs. Available tools for such analysis are the MEME suite [131] or the RSAT software [132] (Fig. 5). Such analysis can be particularly useful to detect sequence biases due to ZF mutations or other modifications that change CTCF binding specificity in the genome.
6.2. Identification of differential 3D genome organization
To identify changes in 3D genome organization at a genome-wide scale, Hi-C or Micro-C data should be generated or obtained for the different conditions. Procedures for high resolution in-situ Hi-C and Micro-C are outlined in [133]. Alternatively, genome-wide 3C assays can be used that incorporate a chromatin immunoprecipitation step to enrich for CTCF bound regions in the genome. Examples of such assays include ChIA-PET and Hi-ChIP [134], [135].
After high-throughput sequencing of the different samples and validation of sequencing quality, paired-end sequencing reads can be aligned to the reference genome using widely used mapping tools like bowtie2 or BWA [124], [125] (Fig. 5). After removal of duplicate read pairs, a further filtering step is required to remove read pairs that are the result of artifacts that are specific to the Hi-C assay (i.e. read pairs within the same restriction fragment or from self-ligated restriction fragments) (Fig. 5). The remaining pairs of interactions are subsequently used to create a non-normalized (“raw”) Hi-C data file, which is a matrix that contains the number of contacts between all sites in the genome. An overview of available packages for Hi-C specific filtering, matrix building, file generation and analysis of 3D genome organization is provided in Table 1 (see also [136], [137], [138], [139], [140], [141]).
Table 1.
Comparison of Hi-C analysis pipelines. Asterisks indicate the inclusion of tools with comparable output. The cooler file format allows interoperability between pipelines.
 |
* For the HOMER toolbox, citation number is not restricted to Hi-C related tools.
* The Juicer pipeline allows the identification of contact domains, not TAD boundaries.
* The HOMER toolbox does not provide a “Distance vs Counts” but a “Distal-To-Local” tool to analyze chromatin compaction.
Current protocols for high-resolution Hi-C and Micro-C use enzymes that can fragment the human genome in millions of different fragments that can potentially all interact among each other. Obtaining high coverage information about 3D organization therefore requires a sequencing depth that is both expensive and highly data dense. For this reason, in most experiments, data is sequenced to a lower degree of coverage followed by binning into genomic intervals of consistent size. Appropriate bin size can be determined post-hoc using the criteria defined in [25]: at least 80% of bins within the contact matrix should be covered by at least 1,000 reads. This bin size represents the resolution of the Hi-C matrix, i.e. the size of the genomic interval that represents a single pixel within the Hi-C map.
Due to the large numbers of possible genome-wide interactions, particularly in ultra-high-resolution interactions matrices, the data management of Hi-C matrices can become limited (an interaction matrix of the human genome at 1 kb resolution contains over 9.6 * 1012 fields). As most interactions in the human genome are intra-chromosomal and restricted to a few Mbs in distance, a large fraction of bins in the Hi-C matrix will have a value of 0. More computationally efficient file formats have been developed that store only informative bins, with efficiency further improved by storing in a binary manner. Among the most used and adapted interaction matrix formats is the Cooler format [139]. Cool files can be stored in both binary and computationally-efficient formats. These files can be used as input in the “cooltools ecosystem”, a suite of command line tools and python3 libraries for the analysis of 3D genome organization [139]. The hicConvertFormat tool from the hicexplorer toolbox permits efficient conversion of various other file formats into the Cooler format [138].
After this conversion, the computationally efficient interaction matrix must be normalized to correct for experimental biases that stem from variation in GC content, mappability and restriction fragment length (Fig. 5). This normalization is needed to make sure that each locus has a similar visibility in the matrix despite such biases. The most frequently used approaches for normalization are the Iterative Correction and Eigenvector Decomposition (ICE) approach [142] and the Knight Ruiz (KR) [143] matrix balancing algorithm, which ultimately generate a smoother and more accurate interaction matrix. These approaches are not suited for cancer genomes that contain structural variation though, as their normalization assumes a normal genome (i.e. absence of CNVs, translocations and inversions). Instead, a CNV-aware normalization can be performed, after segmenting the genome into blocks of similar coverage. A normalization similar to the above-mentioned approaches is applied to each CNV-block in the genome, separately, to remove the visibility bias CNVs create. Two recent examples for CNV-aware normalization are the NeoLoopFinder toolbox [144] and the HiNT tool [145].
Normalized interaction matrices can be used to identify different layers of 3D genome organization. Hi-C compartments (i.e. A/B compartments; Fig. 1A) can be distinguished using eigenvector decomposition in almost all Hi-C analysis packages (Table 1). Boundaries between TADs (Fig. 1B) can be called using an “Insulation score” that identifies local minima of interactions between domains in the Hi-C matrix. Using these boundaries, TADs themselves can subsequently be called as well. Most Hi-C analysis packages can identify TADs or similar domains using insulation scores or comparable approaches (Table 1). DNA loops, which emerge as punctuated local enrichments of signal in the interaction matrices, can also be called by most Hi-C analysis pipelines (Table 1). However, this finer-scale 3D organization level requires a Hi-C matrix at a high resolution of 2.5 – 10 kb. Hi-C interaction matrices can be visualized and interactively explored using the HiGlass tool [146], which allows the highlighting of identified 3D genome features and the addition of external data as well, at different resolutions from a single mcool file (multi-resolution cool files).
Subsequent identification of sites where changes in CBSs correlate with 3D genome reorganization can be done by intersection of genomics features. Juicer can identify CTCF motifs at loop anchors, Cooltools can calculate the enrichment of CTCF binding at TAD boundaries and HOMER can annotate loops with CTCF binding peaks. Currently, we are not aware of dedicated tools for systematic differential analysis of combined CTCF binding and 3D organization. The intersection of both types of data therefore requires the development of tools dedicated to this particular research question.
7. Summary and outlook
Cancer cells are characterized by changes in gene expression programs, which are intimately linked to changes in their 3D genome organization. The CTCF insulator protein has emerged as a jack-of-all-trades in the formation of the different levels of 3D genome organization. The functional impact of CTCF can be perturbed through various mechanisms in cancer cells, which include changes in the cellular dosage of the CTCF protein, changes to the CTCF protein that have an impact on its function or changes to CBSs in the genome. These later changes can both be caused by changes to the DNA sequence (mutations) or by changes in DNA methylation. CTCF binding perturbations cause different types of 3D genome reorganization, with particularly the fusion of TADs causing the activation of oncogenes. Combined with its non-canonical functions with relevance to cancer, CTCF has thus established itself as a jack-of-all-trades that is misguided in various ways in cancer cells. To identify links between the reorganization of CTCF binding and 3D genome organization, we have provided a description of computational strategies and tools in the last section of the manuscript.
Despite these expanding insights into the function of CTCF, many questions relevant to the emergence and progression of cancers remain. One major question is what determines the specificity of CBS, resulting in potential loss or gain in cancer cells. DNA methylation patterns are often profoundly reorganized in cancer cells, which may interfere with CTCF binding. A large fraction of dynamic CBS do not contain CpG dinucleotides though, suggesting that other mechanisms must be involved in the regulation of CTCF binding to the DNA as well [41], [51]. Another open question is how the formation of TADs, which are relatively weakly insulated domains, can prevent inappropriate contacts between enhancers and oncogenes in neighboring domains [20], [147]. Besides TAD structure, the loop extrusion machinery itself may thus be important for bringing CREs together in the nuclear space [147], [148]. The functional importance of CTCF binding at the level of TAD organization may therefore expand towards the physical separation of loop extrusion between neighboring domains. Finally, it remains to be determined how the different and often contradictory changes in CTCF function can lead to cancers in different cellular contexts. Here, it will be of particular interest to determine how CTCF dysfunction, either locally or globally, is mechanistically involved in the wide spectrum of different cancer cell types. A particularly interesting angle will be to distinguish between inappropriate activation of a single “master” oncogene, as appears to be the outcome of the global epigenetic deregulation in gliomas and GISTs [105], [106], the active involvement of more global patterns of transcriptional deregulation, or the contribution of CTCF through its non-canonical functions. Further mechanistic characterization will help to position the various perturbations of CTCF function and DNA binding within the larger context of gene deregulation, which can ultimately open up new opportunities for the development of tailored treatments and therapies in different cancers.
CRediT authorship contribution statement
Julie Segueni: Writing – original draft, Writing – review & editing, Visualization. Daan Noordermeer: Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We apologize to our colleagues whose work was excluded due to space constraints. We thank Benoit Moindrot and Francois Charon for critical reading of the manuscript and the members of the Noordermeer lab for useful discussion. We thank Wolfgang Heymes for help with compiling the figures. This work has been supported by funds from PlanCancer (19CS145-00), the Agence Nationale de la Recherche (ANR-18-CE12-0022-02 and ANR-21-CE12-0034-01) and the Fondation Bettencourt Schueller to D.N. J.S. is supported by a PhD grant from the Université Paris-Saclay (Ecole Doctorale Structure et dynamique des systèmes vivants). The funders had no influence on the design of the manuscript or the decision to publish.
Footnotes
This manuscript should go in the 10th anniversary celebration issue of the Computational and Structural Biotechnology Journal.
References
- 1.Bradner J.E., Hnisz D., Young R.A. Transcriptional Addiction in Cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You J.S., Jones P.A. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarthi B.V.S.K., Nepal S., Varambally S. Genomic and Epigenomic Alterations in Cancer. Am J Pathol. 2016;186:1724–1735. doi: 10.1016/j.ajpath.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Y., Shan G., Xue J., Chen C., Zhang C. Defining the multivalent functions of CTCF from chromatin state and three-dimensional chromatin interactions. Nucleic Acids Res. 2016;44:6200–6212. doi: 10.1093/nar/gkw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braccioli L., de Wit E. CTCF: a Swiss-army knife for genome organization and transcription regulation. Essays Biochem. 2019;63:157–165. doi: 10.1042/EBC20180069. [DOI] [PubMed] [Google Scholar]
- 6.Wu Q., Liu P., Wang L. Many facades of CTCF unified by its coding for three-dimensional genome architecture. J Genet Genomics. 2020;47:407–424. doi: 10.1016/j.jgg.2020.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Merkenschlager M., Nora E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 8.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 9.Davies J.O., Oudelaar A.M., Higgs D.R., Hughes J.R. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods. 2017;14:125–134. doi: 10.1038/nmeth.4146. [DOI] [PubMed] [Google Scholar]
- 10.Sati S., Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2017;126:33–44. doi: 10.1007/s00412-016-0593-6. [DOI] [PubMed] [Google Scholar]
- 11.Chang L.H., Noordermeer D. Of Dots and Stripes: The Morse Code of Micro-C Reveals the Ultrastructure of Transcriptional and Architectural Mammalian 3D Genome Organization. Mol Cell. 2020;78:376–378. doi: 10.1016/j.molcel.2020.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giorgetti L., Heard E. Closing the loop: 3C versus DNA FISH. Genome Biol. 2016;17:215. doi: 10.1186/s13059-016-1081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fudenberg G., Imakaev M. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nat Methods. 2017;14:673–678. doi: 10.1038/nmeth.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne A.C., Chiang Z.D., Reginato P.L., Mangiameli S.M., Murray E.M., Yao C.-C., et al. In situ genome sequencing resolves DNA sequence and structure in intact biological samples. Science. 2021;371:eaay3446. doi: 10.1126/science.aay3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takei Y., Yun J., Zheng S., Ollikainen N., Pierson N., White J., et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature. 2021;590:344–350. doi: 10.1038/s41586-020-03126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao T., Chiang Z.D., Morriss J.W., LaFave L.M., Murray E.M., Del Priore I., et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 2022;601:85–91. doi: 10.1038/s41586-021-04217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 19.Meaburn K.J., Gudla P.R., Khan S., Lockett S.J., Misteli T. Disease-specific gene repositioning in breast cancer. J Cell Biol. 2009;187:801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nora E.P., Dekker J., Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? BioEssays. 2013;35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh T.-H.-S., Cattoglio C., Slobodyanyuk E., Hansen A.S., Rando O.J., Tjian R., et al. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol Cell. 2020;78:539–553.e8. doi: 10.1016/j.molcel.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krietenstein N., Abraham S., Venev S.V., Abdennur N., Gibcus J., Hsieh T.-H.-S., et al. Ultrastructural Details of Mammalian Chromosome Architecture. Mol Cell. 2020;78:554–565.e7. doi: 10.1016/j.molcel.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobanenkov V.V., Nicolas R.H., Adler V.V., Paterson H., Klenova E.M., Polotskaja A.V., et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5’-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 30.Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 31.Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612-7624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chernukhin I.V., Shamsuddin S., Robinson A.F., Carne A.F., Paul A., El-Kady A.I., et al. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor. CTCF J Biol Chem. 2000;275:29915–29921. doi: 10.1074/jbc.M001538200. [DOI] [PubMed] [Google Scholar]
- 33.Moore J.M., Rabaia N.A., Smith L.E., Fagerlie S., Gurley K., Loukinov D., et al. Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE. 2012;7:e34915. doi: 10.1371/journal.pone.0034915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey C.G., Metierre C., Feng Y., Baidya K., Filippova G.N., Loukinov D.I., et al. CTCF Expression is Essential for Somatic Cell Viability and Protection Against Cancer. Int J Mol Sci. 2018;19:3832. doi: 10.3390/ijms19123832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath H., de Almeida C.R., Sleutels F., Dingjan G., van de Nobelen S., Jonkers I., et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedoriw A.M., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 37.Loukinov D.I., Pugacheva E., Vatolin S., Pack S.D., Moon H., Chernukhin I., et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugacheva E.M., Rivero-Hinojosa S., Espinoza C.A., Méndez-Catalá C.F., Kang S., Suzuki T., et al. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015;16:161. doi: 10.1186/s13059-015-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debaugny R.E., Skok J.A. CTCF and CTCFL in cancer. Curr Opin Genet Dev. 2020;61:44–52. doi: 10.1016/j.gde.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filippova G.N., Lindblom A., Meincke L.J., Klenova E.M., Neiman P.E., Collins S.J., et al. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 41.Wang H., Maurano M.T., Qu H., Varley K.E., Gertz J., Pauli F., et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen H., Tian Y., Shu W., Bo X., Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS ONE. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakahashi H., Kieffer Kwon K.-R., Resch W., Vian L., Dose M., Stavreva D., et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hashimoto H., Wang D., Horton J.R., Zhang X., Corces V.G., Cheng X. Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA. Mol Cell. 2017;66:711–720.e3. doi: 10.1016/j.molcel.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang H., Zhu Q., Jussila A., Han Y., Bintu B., Kern C., et al. CTCF mediates dosage- and sequence-context-dependent transcriptional insulation by forming local chromatin domains. Nat Genet. 2021;53:1064–1074. doi: 10.1038/s41588-021-00863-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soochit W., Sleutels F., Stik G., Bartkuhn M., Basu S., Hernandez S.C., et al. CTCF chromatin residence time controls three-dimensional genome organization, gene expression and DNA methylation in pluripotent cells. Nat Cell Biol. 2021;23:881–893. doi: 10.1038/s41556-021-00722-w. [DOI] [PubMed] [Google Scholar]
- 49.Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 50.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 51.Maurano M.T., Wang H., John S., Shafer A., Canfield T., Lee K., et al. Role of DNA Methylation in Modulating Transcription Factor Occupancy. Cell Rep. 2015;12:1184–1195. doi: 10.1016/j.celrep.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 52.Wiehle L., Thorn G.J., Raddatz G., Clarkson C.T., Rippe K., Lyko F., et al. DNA (de)methylation in embryonic stem cells controls CTCF-dependent chromatin boundaries. Genome Res. 2019;29:750–761. doi: 10.1101/gr.239707.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Y., Sinha M., Peterson C.L., Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barisic D., Stadler M.B., Iurlaro M., Schübeler D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature. 2019;569:136–140. doi: 10.1038/s41586-019-1115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saldaña-Meyer R., González-Buendía E., Guerrero G., Narendra V., Bonasio R., Recillas-Targa F., et al. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014;28:723–734. doi: 10.1101/gad.236869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen A.S., Hsieh T.-H.-S., Cattoglio C., Pustova I., Saldaña-Meyer R., Reinberg D., et al. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol Cell. 2019;76:395–411.e13. doi: 10.1016/j.molcel.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saldaña-Meyer R., Rodriguez-Hernaez J., Escobar T., Nishana M., Jácome-López K., Nora E.P., et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol Cell. 2019;76:412–422.e5. doi: 10.1016/j.molcel.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Rosario B.C., Kriz A.J., Del Rosario A.M., Anselmo A., Fry C.J., White F.M., et al. Exploration of CTCF post-translation modifications uncovers Serine-224 phosphorylation by PLK1 at pericentric regions during the G2/M transition. ELife. 2019;8:e42341. doi: 10.7554/eLife.42341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo H., Yu Q., Liu Y., Tang M., Liang M., Zhang D., et al. LATS kinase-mediated CTCF phosphorylation and selective loss of genomic binding. Science. Advances. 2020;6:eaaw4651. doi: 10.1126/sciadv.aaw4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavlaki I., Docquier F., Chernukhin I., Kita G., Gretton S., Clarkson C.T., et al. Poly(ADP-ribosyl)ation associated changes in CTCF-chromatin binding and gene expression in breast cells. Biochim Biophys Acta Gene Regul Mech. 2018;1861:718–730. doi: 10.1016/j.bbagrm.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang M.A., Lee J.-S. A Newly Assigned Role of CTCF in Cellular Response to Broken DNAs. Biomolecules. 2021;11:363. doi: 10.3390/biom11030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh H.J., Aguilar R., Kesner B., Lee H.-G., Kriz A.J., Chu H.-P., et al. Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell. 2021;184:6157–6173.e24. doi: 10.1016/j.cell.2021.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vostrov A.A., Quitschke W.W. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- 64.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 65.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 66.Nora E.P., Goloborodko A., Valton A.-L., Gibcus J.H., Uebersohn A., Abdennur N., et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanborn A.L., Rao S.S.P., Huang S.-C., Durand N.C., Huntley M.H., Jewett A.I., et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rao S.S.P., Huang S.-C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.-R., et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nora E.P., Caccianini L., Fudenberg G., So K., Kameswaran V., Nagle A., et al. Molecular basis of CTCF binding polarity in genome folding. Nat Commun. 2020;11:5612. doi: 10.1038/s41467-020-19283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y., Haarhuis J.H.I., Sedeño Cacciatore Á., Oldenkamp R., van Ruiten M.S., Willems L., et al. The structural basis for cohesin-CTCF-anchored loops. Nature. 2020;578:472–476. doi: 10.1038/s41586-019-1910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Wit E., Vos E.S.M., Holwerda S.J.B., Valdes-Quezada C., Verstegen M.J.A.M., Teunissen H., et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 74.Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kubo N., Ishii H., Xiong X., Bianco S., Meitinger F., Hu R., et al. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat Struct Mol Biol. 2021;28:152–161. doi: 10.1038/s41594-020-00539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim Y.J., Cecchini K.R., Kim T.H. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cuddapah S., Jothi R., Schones D.E., Roh T.-Y., Cui K., Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fiorentino F.P., Giordano A. The tumor suppressor role of CTCF. J Cell Physiol. 2012;227:479–492. doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- 81.Tiffen J.C., Bailey C.G., Marshall A.D., Metierre C., Feng Y., Wang Q., et al. The cancer-testis antigen BORIS phenocopies the tumor suppressor CTCF in normal and neoplastic cells. Int J Cancer. 2013;133:1603–1613. doi: 10.1002/ijc.28184. [DOI] [PubMed] [Google Scholar]
- 82.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oreskovic E, Wheeler EC, Mengwasser KE, Fujimura E, Martin TD, Tothova Z, et al. Genetic analysis of cancer drivers reveals cohesin and CTCF as suppressors of PD-L1. Proc Natl Acad Sci U S A 2022;119:e2120540119. https://doi.org/10.1073/pnas.2120540119. [DOI] [PMC free article] [PubMed]
- 84.Marshall A.D., Bailey C.G., Champ K., Vellozzi M., O’Young P., Metierre C., et al. CTCF genetic alterations in endometrial carcinoma are pro-tumorigenic. Oncogene. 2017;36:4100–4110. doi: 10.1038/onc.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kemp C.J., Moore J.M., Moser R., Bernard B., Teater M., Smith L.E., et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang B., Zhang Y., Zou X., Chan A.W., Zhang R., Lee T.-K.-W., et al. The CCCTC-binding factor (CTCF)-forkhead box protein M1 axis regulates tumour growth and metastasis in hepatocellular carcinoma. J Pathol. 2017;243:418–430. doi: 10.1002/path.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Docquier F., Farrar D., D’Arcy V., Chernukhin I., Robinson A.F., Loukinov D., et al. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 2005;65:5112–5122. doi: 10.1158/0008-5472.CAN-03-3498. [DOI] [PubMed] [Google Scholar]
- 88.Johnson K.S., Hussein S., Chakraborty P., Muruganantham A., Mikhail S., Gonzalez G., et al. CTCF Expression and Dynamic Motif Accessibility Modulates Epithelial-Mesenchymal Gene Expression. Cancers (Basel) 2022;14:209. doi: 10.3390/cancers14010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marshall A.D., Bailey C.G., Rasko J.E.J. CTCF and BORIS in genome regulation and cancer. Curr Opin Genet Dev. 2014;24:8–15. doi: 10.1016/j.gde.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 90.Bailey C.G., Gupta S., Metierre C., Amarasekera P.M.S., O’Young P., Kyaw W., et al. Structure-function relationships explain CTCF zinc finger mutation phenotypes in cancer. Cell Mol Life Sci. 2021;78:7519–7536. doi: 10.1007/s00018-021-03946-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katainen R., Dave K., Pitkänen E., Palin K., Kivioja T., Välimäki N., et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 92.Kaiser V.B., Taylor M.S., Semple C.A. Mutational Biases Drive Elevated Rates of Substitution at Regulatory Sites across Cancer Types. PLoS Genet. 2016;12:e1006207. doi: 10.1371/journal.pgen.1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ji X., Dadon D.B., Powell B.E., Fan Z.P., Borges-Rivera D., Shachar S., et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y., et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 95.Umer H.M., Cavalli M., Dabrowski M.J., Diamanti K., Kruczyk M., Pan G., et al. A Significant Regulatory Mutation Burden at a High-Affinity Position of the CTCF Motif in Gastrointestinal Cancers. Hum Mutat. 2016;37:904–913. doi: 10.1002/humu.23014. [DOI] [PubMed] [Google Scholar]
- 96.Hnisz D., Weintraub A.S., Day D.S., Valton A.L., Bak R.O., Li C.H., et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo Y.A., Chang M.M., Huang W., Ooi W.F., Xing M., Tan P., et al. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat Commun. 2018;9:1520. doi: 10.1038/s41467-018-03828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Akdemir K.C., Le V.T., Kim J.M., Killcoyne S., King D.A., Lin Y.-P., et al. Somatic mutation distributions in cancer genomes vary with three-dimensional chromatin structure. Nat Genet. 2020;52:1178–1188. doi: 10.1038/s41588-020-0708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jablonski K.P., Carron L., Mozziconacci J., Forné T., Hütt M.-T., Lesne A. Contribution of 3D genome topological domains to genetic risk of cancers: a genome-wide computational study. Hum Genomics. 2022;16:2. doi: 10.1186/s40246-022-00375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nieboer M.M., Nguyen L., de Ridder J. Predicting pathogenic non-coding SVs disrupting the 3D genome in 1646 whole cancer genomes using multiple instance learning. Sci Rep. 2021;11:14411. doi: 10.1038/s41598-021-93917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu EM, Martinez-Fundichely A, Diaz BJ, Aronson B, Cuykendall T, MacKay M, et al. Identification of Cancer Drivers at CTCF Insulators in 1,962 Whole Genomes. Cell Syst 2019;8:446-455.e8. https://doi.org/10.1016/j.cels.2019.04.001. [DOI] [PMC free article] [PubMed]
- 102.Kaiser V.B., Semple C.A. Chromatin loop anchors are associated with genome instability in cancer and recombination hotspots in the germline. Genome Biol. 2018;19:101. doi: 10.1186/s13059-018-1483-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dixon J.R., Xu J., Dileep V., Zhan Y., Song F., Le V.T., et al. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet. 2018;50:1388–1398. doi: 10.1038/s41588-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fang C., Wang Z., Han C., Safgren S.L., Helmin K.A., Adelman E.R., et al. Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation. Genome Biol. 2020;21:247. doi: 10.1186/s13059-020-02152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Flavahan W.A., Drier Y., Johnstone S.E., Hemming M.L., Tarjan D.R., Hegazi E., et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature. 2019;575:229–233. doi: 10.1038/s41586-019-1668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schwerk C., Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 108.David C.J., Manley J.L. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alharbi A.B., Schmitz U., Bailey C.G., Rasko J.E.J. CTCF as a regulator of alternative splicing: new tricks for an old player. Nucleic Acids Res. 2021;49:7825–7838. doi: 10.1093/nar/gkab520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruiz-Velasco M., Kumar M., Lai M.C., Bhat P., Solis-Pinson A.B., Reyes A., et al. CTCF-Mediated Chromatin Loops between Promoter and Gene Body Regulate Alternative Splicing across Individuals. Cell Syst. 2017;5:628–637.e6. doi: 10.1016/j.cels.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 111.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alharbi A.B., Schmitz U., Marshall A.D., Vanichkina D., Nagarajah R., Vellozzi M., et al. Ctcf haploinsufficiency mediates intron retention in a tissue-specific manner. RNA Biol. 2021;18:93–103. doi: 10.1080/15476286.2020.1796052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee R., Kang M.-K., Kim Y.-J., Yang B., Shim H., Kim S., et al. CTCF-mediated chromatin looping provides a topological framework for the formation of phase-separated transcriptional condensates. Nucleic Acids Res. 2022;50:207–226. doi: 10.1093/nar/gkab1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chachoua I., Tzelepis I., Dai H., Lim J.P., Lewandowska-Ronnegren A., Casagrande F.B., et al. Canonical WNT signaling-dependent gating of MYC requires a noncanonical CTCF function at a distal binding site. Nat Commun. 2022;13:204. doi: 10.1038/s41467-021-27868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tanwar V.S., Jose C.C., Cuddapah S. Role of CTCF in DNA damage response. Mutat Res – Rev Mut Res. 2019;780:61–68. doi: 10.1016/j.mrrev.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Collins P.L., Purman C., Porter S.I., Nganga V., Saini A., Hayer K.E., et al. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner. Nat Commun. 2020;11:3158. doi: 10.1038/s41467-020-16926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arnould C., Rocher V., Finoux A.L., Clouaire T., Li K., Zhou F., et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature. 2021;590:660–665. doi: 10.1038/s41586-021-03193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hilmi K., Jangal M., Marques M., Zhao T., Saad A., Zhang C., et al. CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair. Sci Adv. 2017;3:e1601898. doi: 10.1126/sciadv.1601898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Docquier F., Kita G.-X., Farrar D., Jat P., O’Hare M., Chernukhin I., et al. Decreased poly(ADP-ribosyl)ation of CTCF, a transcription factor, is associated with breast cancer phenotype and cell proliferation. Clin Cancer Res. 2009;15:5762–5771. doi: 10.1158/1078-0432.CCR-09-0329. [DOI] [PubMed] [Google Scholar]
- 122.Witcher M., Emerson B.M. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaya-Okur H.S., Janssens D.H., Henikoff J.G., Ahmad K., Henikoff S. Efficient low-cost chromatin profiling with CUT&Tag. Nat Protoc. 2020;15:3264–3283. doi: 10.1038/s41596-020-0373-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bonhoure N., Bounova G., Bernasconi D., Praz V., Lammers F., Canella D., et al. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res. 2014;24:1157–1168. doi: 10.1101/gr.168260.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thomas-Chollier M., Sand O., Turatsinze J.-V., Janky R., Defrance M., Vervisch E., et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Akgol O.B., Yang L., Abraham S., Venev S.V., Krietenstein N., Parsi K.M., et al. Systematic evaluation of chromosome conformation capture assays. Nat Methods. 2021;18:1046–1055. doi: 10.1038/s41592-021-01248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.-J., Vert J.-P., et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Durand N.C., Shamim M.S., Machol I., Rao S.S.P., Huntley M.H., Lander E.S., et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wolff J., Bhardwaj V., Nothjunge S., Richard G., Renschler G., Gilsbach R., et al. Galaxy HiCExplorer: a web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 2018;46:W11–W16. doi: 10.1093/nar/gky504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Abdennur N., Mirny L.A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics. 2020;36:311–316. doi: 10.1093/bioinformatics/btz540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Serra F., Baù D., Goodstadt M., Castillo D., Filion G.J., Marti-Renom M.A. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol. 2017;13:e1005665. doi: 10.1371/journal.pcbi.1005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Knight P.A., Ruiz D. A fast algorithm for matrix balancing. IMA Journal of Numerical Analysis. 2013;33:1029–1047. doi: 10.1093/imanum/drs019. [DOI] [Google Scholar]
- 144.Wang X., Xu J., Zhang B., Hou Y., Song F., Lyu H., et al. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nat Methods. 2021;18:661–668. doi: 10.1038/s41592-021-01164-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang S., Lee S., Chu C., Jain D., Kerpedjiev P., Nelson G.M., et al. HiNT: a computational method for detecting copy number variations and translocations from Hi-C data. Genome Biol. 2020;21:73. doi: 10.1186/s13059-020-01986-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kerpedjiev P., Abdennur N., Lekschas F., McCallum C., Dinkla K., Strobelt H., et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19:125. doi: 10.1186/s13059-018-1486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chang L.-H., Ghosh S., Noordermeer D. TADs and Their Borders: Free Movement or Building a Wall? J Mol Biol. 2020;432:643–652. doi: 10.1016/j.jmb.2019.11.025. [DOI] [PubMed] [Google Scholar]
- 148.Vian L., Pękowska A., Rao S.S.P., Kieffer-Kwon K.-R., Jung S., Baranello L., et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell. 2018;175:292–294. doi: 10.1016/j.cell.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]