Abstract
A laccase from Coprinus cinereus is active at alkaline pH, an essential property for some potential applications. We cloned and sequenced three laccase genes (lcc1, lcc2, and lcc3) from the ink cap basidiomycete C. cinereus. The lcc1 gene contained 7 introns, while both lcc2 and lcc3 contained 13 introns. The predicted mature proteins (Lcc1 to Lcc3) are 58 to 80% identical at the amino acid level. The predicted Lcc1 contains a 23-amino-acid C-terminal extension rich in arginine and lysine, suggesting that C-terminal processing may occur during its biosynthesis. We expressed the Lcc1 protein in Aspergillus oryzae and purified it. The Lcc1 protein as expressed in A. oryzae has an apparent molecular mass of 66 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and absorption maxima at 278 and 614 nm. Based on the N-terminal protein sequence of the laccase, a 4-residue propeptide was processed during the maturation of the enzyme. The dioxygen specificity of the laccase showed an apparent Km of 21 ± 2 μM and a catalytic constant of 200 ± 10 min−1 for O2 with 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) as the reducing substrate at pH 5.5. Lcc1 from A. oryzae may be useful in industrial applications. This is the first report of a basidiomycete laccase whose biosynthesis involves both N-terminal and C-terminal processing.
Laccases are multicopper enzymes (EC 1.10.3.2) that catalyze the oxidation of a variety of phenolic compounds and are widely distributed among plants (37) and fungi (6). In plants, laccase is involved in lignification (37, 39). In fungi, laccases may be involved in many cellular processes, including delignification (17, 30), sporulation (33), pigment production (2, 12, 46), fruiting body formation (33), and plant pathogenesis (10, 19, 36). Proposed industrial applications for laccases include paper processing (8, 42), prevention of wine discoloration (32), detoxification of environmental pollutants (7), oxidation of dye and dye precursors (14), enzymatic conversion of chemical intermediates (1), and production of chemicals from lignin. Before laccase can be used commercially for any of these applications, an inexpensive source must be available.
Most fungi that produce laccases do so at levels that are far too low to be an economical source. Laccase genes have been cloned from Neurospora crassa (20), Cryphonectria parasitica (10), Aspergillus nidulans (2), Agaricus bisporus (41), Coriolus hirsutus (31), Phlebia radiata (44), Coriolus versicolor (25), Trametes versicolor (24, 27, 28, 40), Trametes villosa (54, 55), Rhizoctonia solani (52), Myceliophthora thermophila (5), the ligninolytic basidiomycetes PM1 (13) and CECT 20197 (34), Podospora anserina (18), and Pycnoporus cinnabarinus (16).
The C. hirsutus laccase has been expressed in Saccharomyces cerevisiae (31), and the P. radiata laccase has been expressed in Trichoderma reesei (43). The yields obtained for these laccases were too low to be commercially feasible. Recently, the expression of the T. versicolor Lcc1 laccase in Pichia pastoris was reported (27). The M. thermophila laccase, three of the laccases from R. solani, and one of the laccases from T. villosa have been expressed in Aspergillus oryzae (5, 52, 55). The work on expression in A. oryzae resulted in commercialization of laccase for use by the textile industry in denim processing (29).
Coprinus cinereus is an ink cap basidiomycete that secretes a laccase (4) which has been purified from culture broth and characterized enzymologically (47), and its three-dimensional crystal structure has been determined (15). Our objectives in this study were (i) to clone the gene for the purified C. cinereus laccase, (ii) to express the laccase in A. oryzae to produce enough material for further characterization and applications testing, and (iii) to determine if there was more than one laccase gene that was expressed under the growth conditions under which the laccase had been previously purified. The work to clone and express this laccase was undertaken due to its neutral-to-alkaline pH optimum, which is required for some of the potential industrial applications of laccases (8, 14, 42). Because it has been demonstrated that, unlike other basidiomycetes, C. cinereus contains only a single peroxidase gene (4), we were also interested in whether C. cinereus had a laccase gene family, as do many other basidiomycetes. We found that C. cinereus contains a family of at least three laccase genes and that the previously biochemically characterized C. cinereus laccase can be expressed in A. oryzae.
MATERIALS AND METHODS
Strains.
Plasmid and library construction was done with Escherichia coli Y1090(ZL) (Gibco BRL, Gaithersburg, Md.), E. coli DH10B(ZL) (Gibco BRL), and E. coli DH5α (Stratagene, La Jolla, Calif.). The fungal strains were Coprinus cinereus var. microsporus IFO 8371 (Institute for Fermentation, Osaka, Japan) and A. oryzae HowB104 (5), a pyrG amyA amyB amyC mutant of IFO4177.
RNA isolation.
C. cinereus A3387 was cultivated in FG4 medium (1.5% maltodextrin, 3% soy flour, 0.5% Bacto Peptone, 0.2% PLURONIC L61 [BASF, Mount Olive, N.J.]) at 26°C; the mycelium was harvested after 6 days of growth, frozen in liquid N2, and stored at −80°C. Total RNA was prepared from frozen, powdered mycelia of C. cinereus A3387 by extraction with guanidinium thiocyanate followed by ultracentrifugation through a 5.7 M CsCl cushion (9). Poly(A)+ RNA was isolated by oligo(dT)-cellulose affinity chromatography (3).
Construction of the cDNA library.
Double-stranded cDNA was synthesized from 5 μg of C. cinereus poly(A)+ RNA as described previously (21, 45), except that an oligo(dT)-NotI anchor primer instead of an oligo(dT)12–18 primer was used in the first-strand reaction. After synthesis the cDNA was treated with mung bean nuclease, blunt ended with T4 DNA polymerase, and ligated to nonpalindromic BstXI adaptors (Invitrogen) with a ca. 50-fold molar excess of the adaptors. The adapted cDNA was digested with NotI, size fractionated for 1.2- to 3.0-kb cDNAs by agarose gel electrophoresis, and ligated into BstXI/NotI-cleaved pYES 2.0 vector (Invitrogen), and the ligation mixture was transformed into electrocompetent E. coli DH10B cells (Gibco BRL). The library consisted of 106 independent clones.
Generation of a cDNA probe for C. cinereus laccase by PCR.
One μg of plasmid DNA from a C. cinereus cDNA library pool was used as a template in a PCR. The reaction mixture contained 500 pmol of primer and 2.5 U of Taq polymerase (Perkin-Elmer, Foster City, Calif.). The primers used were as follows: sense, 5′-ATICAC/TTGGCAC/TGGIC/TTIC/TTI-3′, and antisense, 5′-GGIACCAAA/GA/GAIGTA/GACA/GGTA/GTAICT-3′. I denotes inosine. Thirty cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min were performed. The amplified fragment was subcloned into pCR2.1 (Invitrogen) with the TA cloning kit and sequenced with universal forward and reverse primers (45).
Genomic DNA isolation.
A culture of C. cinereus A3387 was grown in YEG medium (0.5% yeast extract, 2% dextrose) at room temperature (23 to 25°C) at 200 rpm for 4 days. Mycelia were harvested through Miracloth (Calbiochem, La Jolla, Calif.), washed twice with TE, and frozen in liquid nitrogen. DNA was isolated as previously described (52).
Preparation of C. cinereus genomic library.
A genomic library of C. cinereus A3387 was constructed with a λZipLox kit (EcoRI arms) (Gibco BRL). A partial digestion of genomic DNA with Tsp509I (New England Biolabs, Beverly, Mass.) was done at 65°C, and samples were taken after 3, 5, 7, 8, and 9 min of digestion. Fragments were separated on a 1% agarose preparative gel, and DNA fragments of 3 to 8 kb in size were recovered from the gel slices with a Qiaex kit (Qiagen, Chatsworth, Calif.). The size-fractionated DNA was ligated overnight at room temperature to λZipLox EcoRI arms following the protocols provided with the kit. The ligations were packaged into phage with a Gigapak Gold packaging kit (Stratagene).
Probe preparation for library screening.
A digoxigenin (DIG)-labeled probe for nonradioactive screening of the library was prepared by PCR with the C. cinereus lcc1 partial cDNA as a template. The primers used in the reaction were 5′-ACTGCGATGGTCTCCGTGGTC-3′ and 5′-GGGGCCTGGGTTATCGGTGAC-3′. The PCR conditions were 1 cycle of 95°C for 5 min, 50°C for 1 min, and 72°C for 1 min 30 s; 29 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min 30 s; and 1 cycle of 95°C for 30 s, 50°C for 1 min, and 72°C for 3 min. The reaction mixture contained 0.1 μg of C. cinereus lcc1 partial cDNA, 10 μl of 10× PCR buffer (Perkin-Elmer), 5 μl of 10× DIG labeling mix (Boehringer Mannheim, Mannheim, Germany), 75 pmol of each primer, and 0.5 U of Taq DNA polymerase.
32P-labeled probes were prepared with a RadPrime kit (Gibco BRL).
Library screening.
Appropriate dilutions of the λZipLox C. cinereus genomic library were plated with E. coli Y1090 cells on NZY plates (0.5% NaCl, 0.2% MgSO4, 0.5% yeast extract, 1% N-Z-Amine A, pH 7.5) with 0.7% top agarose. Plaques were lifted to Hybond N+ filters (Amersham, Arlington Heights, Ill.) by standard procedures (45). The filters were hybridized in Engler Blue hybridization buffer at 65°C for 1 h or in DIG Easy Hyb (Boehringer Mannheim) at 42°C, and after prehybridization the DIG-labeled probe was added at a final concentration of 3 ng/ml. The filters and probe were hybridized overnight at the same temperature used for prehybridization and were washed under conditions recommended by the manufacturer. The filters were processed to detect hybridized DIG label with the Genius kit (Boehringer Mannheim) and either Lumi-Phos 530 or CDP Star substrate.
For screening with 32P-labeled probes, filter lifts were prehybridized at 65°C in 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 1% sodium dodecyl sulfate (SDS), 0.5% nonfat dry milk, and 200 μg of denatured salmon sperm DNA. Probes were added to a final concentration of 106 cpm/ml, and hybridizations were done overnight at 65°C. The filters were washed once at room temperature for 5 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS and twice at 65°C for 15 min in 0.2× SSC–1% SDS–0.1% sodium pyrophosphate.
DNA sequencing.
Nucleotide sequences were determined by Taq polymerase cycle sequencing with fluorescently labeled nucleotides, and reactions were electrophoresed on an Applied Biosystems automatic DNA sequencer (model 373A, version 2.0.1).
Construction of vector for expression in A. oryzae.
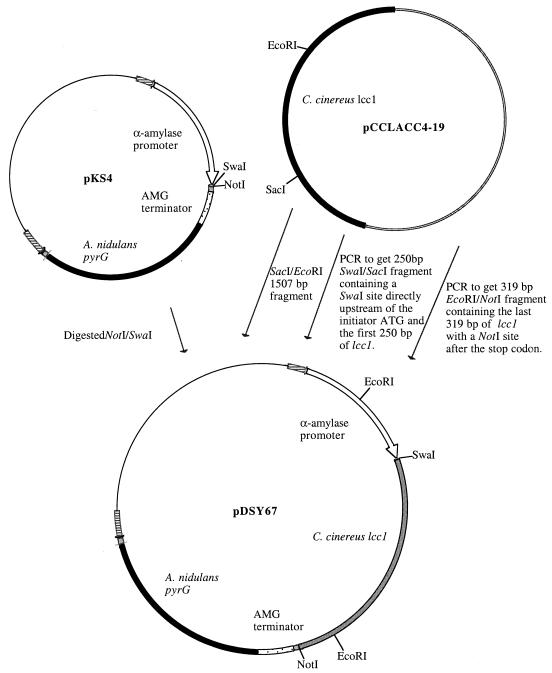
A vector, pDSY67 (Fig. 1), was constructed for the expression of C. cinereus Lcc1 in A. oryzae. The lcc1 gene was cloned into the expression vector, pKS4, that contains the A. oryzae α-amylase promoter (11), glucoamylase terminator (11) and A. nidulans pyrG for selection. The lcc1 gene was inserted as three fragments into pKS4 digested with SwaI/NotI to obtain pDSY67.
FIG. 1.
Construction of expression vector pDSY67 for C. cinereus lcc1.
Transformation of A. oryzae.
An A. oryzae pyrG auxotroph of HowB104 (5) was grown for 18 h in YEG at 34°C, and protoplasts were generated and transformed as described previously (11, 49). The protoplasts were transformed with 10 μg of pDSY67. Transformants were selected on minimal medium plates containing 1 M sucrose.
Screening of laccase transformants.
Primary transformants were screened on minimal medium plates containing 1% glucose and 1 mM 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS). Transformants producing green zones on the ABTS plates were picked and purified by subculturing single conidiospores. The transformants were grown at 37°C in MY51 [(per liter) 30 g of maltose, 2 g of MgSO4, 10 g of KH2PO4, 2 g of K2SO4, 2 g of citric acid, 10 g of yeast extract, 0.5 ml of trace metals, 1 g of urea, 2 g of (NH4)2SO4, pH 6.0].
Materials and instrumentation.
The chemicals used as buffers and substrates were of at least reagent grade. Chromatography was done on a Pharmacia (Piscataway, N.J.) fast protein liquid chromatograph. Spectroscopic assays were conducted on either a spectrophotometer (PC160; Shimadzu, Columbia, Md.) or a microplate reader (Molecular Devices, Menlo Park, Calif.). Amino acid analysis was done on a Hewlett-Packard AminoQuant and an Applied Biosystems 476A protein sequencer.
Enzymatic assay.
ABTS or syringaldazine oxidase activity was determined as previously reported (55). Redox potential was measured with an I2-NaI (0.536 V) couple in 0.9 mM MES (morpholineethanesulfonic acid)-NaOH, pH 5.5, by monitoring the absorbance change of Lcc1 at 600 nm as previously reported (53). The laccase-catalyzed O2 reduction, accompanied by the concomitant oxidation of ABTS, was monitored by a Hansatech (Norfolk, United Kingdom) DW1/AD O2 cell at 20°C with 0.3 ml of 10 mM MES-NaOH (pH 5.5), 1 mM ABTS, and 92 μM laccase. The O2 concentration was controlled by bubbling the solution with an O2-N2 mixture humidified by passing it through water. The initial output voltage changes were used to calculate the initial reaction rate (v). The apparent kinetic parameter, Km, was determined by fitting v and [O2] to v = Vmax × [O2]/(Km + [O2]) with the Prizm program (GraphPad, San Diego, Calif.), and the apparent catalytic constant (kcat) was determined from kcat = Vmax/[laccase]. The [O2] in the air-saturated assay solution was assumed to be 0.28 mM, the same as in plain water. Standard deviation was used to estimate the range of Km and kcat.
Protein purification.
Miracloth-filtered culture supernatant (pH 7.2; 15 mS) was filtered through Whatman no. 2 filter paper and then concentrated and washed on a spiral concentrator (Amicon, Beverly, Mass.) equipped with an S1Y30 membrane (16-fold; 0.8 mS). The broth was frozen overnight at −20°C, thawed the next day, filtered again on Whatman no. 2 paper, and loaded onto a Q-Sepharose XK26 column (120 ml), preequilibrated with 10 mM Tris, pH 7.7, 0.9 mS (buffer A). After the Q-Sepharose column was loaded and washed with buffer A, a linear gradient with buffer B (buffer A plus 2 M NaCl) was applied and the active fractions were eluted around 7% buffer B. These fractions were dialyzed in buffer A and loaded onto a Mono-Q 16/10 (40-ml) column preequilibrated with buffer A.
Protein sequencing and NH2-terminal deblocking.
Laccase was reduced, S-carboxymethylated, and digested by a lysyl-specific protease or chymotrypsin, and the resulting peptides were purified by high-pressure liquid chromatography and sequenced. NH2-terminal deblocking was done with pyroglutamate aminopeptidase (Sigma, St. Louis, Mo.), acylase I (Sigma), and acylamino acid peptidase (Boehringer Mannheim) in accordance with the manufacturers’ instructions. For pyroglutamate aminopeptidase treatment, a laccase sample (2 mg/ml) was incubated at 4°C for 16 h with the peptidase (horse liver) in a 0.1 M Na-phosphate (pH 8) solution containing 10 mM EDTA, 5% glycerol, and 0.7 mM dithiothreitol, with or without 1 M urea or 0.5 M guanidine-HCl. For acylamino acid peptidase treatment, a laccase sample (2 mg/ml) was incubated at 37°C for 20 h with the peptidase (Boehringer Mannheim) (0.4 mg/ml) in a 0.2 M NH4HCO3 (pH 7.8) solution containing 1 mM EDTA, 1 mM 2-mercaptoethanol, and 0.01% SDS, 0.08 M guanidine-HCl, or 0.7 M urea. For acylase I treatment, a laccase sample (2 mg/ml) was incubated with the acylase (porcine kidney; 0.6 mg/ml) in 0.1 M Na-phosphate, pH 7, at 37°C for 22 h. The treated laccase was blotted onto a polyvinylidene difluoride membrane and sequenced. Under these conditions, the native laccase yielded an open NH2 terminus whereas the recombinant laccase remained blocked at its NH2 terminus.
Nucleotide sequence accession numbers.
The nucleotide sequences in GenBank for the C. cinereus laccase genes are AF118267, AF118268, and AF118269 for lcc1, lcc2, and lcc3, respectively.
RESULTS
Isolation of partial laccase cDNA clones.
Two oligonucleotide primers corresponding to the conserved amino acid regions IHWHGL and PWFLHCHID found in fungal laccases were used to amplify cDNA sequences from a C. cinereus library. A PCR product of the expected size, 1.2 kb, was obtained, and the nucleotide sequences of seven subclones were determined. All seven clones encoded amino acids with identity to fungal laccases, and the seven clones represent at least three laccases: the products of C. cinereus lcc1, C. cinereus lcc2, and C. cinereus lcc3. We compared the deduced protein sequences of lcc1, lcc2, and lcc3 with peptide sequences from the previously purified protein and determined that this protein was encoded by lcc1.
Isolation and characterization of genomic clones.
We probed the genomic library with a DIG-labeled fragment of lcc1 and identified nine clones, two of which appeared to be identical. Four of the eight clones carried two fragments unique to lcc1. The nucleotide sequence of one clone containing the complete Lcc1 open reading frame was determined on both strands. The deduced amino acid sequence of this clone matches the NH2-terminal sequence of the purified protein, although the predicted signal peptide cleavage site (51) is between A18 and Q19 while the NH2-terminal sequence begins 4 residues downstream at S23. The open reading frame of the lcc1 gene is interrupted by seven introns ranging in size from 54 to 77 bp. The positions of introns 3, 4, and 5 were confirmed from the partial cDNA. The positions of the other four introns were identified based on the consensus sequences found at the 5′ and 3′ splice sites of fungal introns (22) and by homology of the deduced protein to other laccases. The deduced protein contains three potential N-linked glycosylation sites (AsnXThr/Ser), and the predicted size of the mature protein after removal of the signal peptide is 521 amino acids. Lcc1 has the highest percent identity (58%) to the laccase from the unidentified basidiomycete PM1. Alignments of Lcc1 with other laccases suggest that Lcc1 may have either a COOH-terminal extension or a COOH-terminal peptide that is removed by processing (Fig. 2).
FIG. 2.
Partial alignment of the deduced amino acid sequence of the C. cinereus Lcc1 laccase and other known laccase amino acid sequences. The sequences were aligned with the Clustal algorithm (DNASTAR, Madison, Wis.). The numbers refer to the amino acid sequence. The region in boldface is the peptide that may be removed during the biosynthesis of Lcc1. C. cin lcc1, lcc2, and lcc3, C. cinereus Lcc1, Lcc2, and Lcc3, respectively; T. vil lcc1 and lcc2, T. villosa Lcc1 (GenBank accession no. L49376) and Lcc2 (L49377), respectively; T. ver. lcc1, T. versicolor Lcc1 (X84683); C. hir and P. rad, C. hirsutus (M60560; J05562) and P. radiata (X52134), respectively; P. cin and PM1, P. cinnabarinus (AF025481) and the basidiomycete PM1 (Z12156) laccases, respectively; A. bis, N. cra, and M. ther, A. bisporus Lcc1 (L10664), N. crassa laccase (M18333; M18334), and M. thermophila Lcc1 (T10922), respectively; R. sol lcc1 and lcc4, R. solani Lcc1 (Z54275) and Lcc4 (Z54277), respectively.
Two lcc3 clones were identified in the genomic library. The lcc3 gene contains 13 introns and codes for a precursor protein of 517 amino acids. There is one potential N-glycosylation site, and mature Lcc3 is predicted to be 501 amino acids long.
None of the four positive lcc2 clones contained the complete open reading frame. The largest clone was missing the last 300 bp of the gene, but subsequent screening identified a clone containing this sequence. The lcc2 gene contains 13 introns and codes for a precursor protein of 517 amino acids. There is one potential N-glycosylation site, and the predicted mature protein is 499 amino acids.
Neither Lcc2 nor Lcc3 contains the 23-amino-acid COOH-terminal extension present in Lcc1 (Fig. 2). Lcc1 has 59 and 58% amino acid identity with Lcc2 and Lcc3, respectively. Lcc2 and Lcc3 are similar (80%) to one another. The percent amino acid identity with other fungal laccases ranges from 63% for Lcc2 and the basidiomycete PM1 laccase (13) to 18% for Lcc3 and the A. nidulans laccase (2).
The positions of the 13 introns in lcc2 and lcc3 are strictly conserved. The introns actually interrupt the coding sequence at the same codons. No significant regions of similarity are found when the lcc1, lcc2, and lcc3 promoter regions are compared.
Heterologous expression of lcc1 in A. oryzae.
We transformed A. oryzae HowB425 with the expression vector pDSY67, and more than 90% of the transformants were positive for laccase activity. Transformants cultured in shake flasks of MY51 at 34°C for 3 days produced from 8.0 to 135 mg of laccase per liter.
Purification and characterization of recombinant C. cinereus laccase.
During purification the active fractions passed through the Mono-Q column and showed apparent homogeneity on SDS-polyacrylamide gel electrophoresis. An overall 64-fold purification and a percent recovery of 23 were achieved. Purified recombinant Lcc1 had absorbance maxima at 278 and 614 nm and a redox potential of 0.54 ± 0.05 V at pH 5.5. With syringaldazine as the substrate, Lcc1 had optimum activity at pH 6 to 7. Lysyl-C and chymotryptic digestions yielded 13 internal peptides that matched the deduced protein sequence from the gene. Both the wild-type and the recombinant Lcc1 have a blocked NH2-terminal amino group. Treatment of the native Lcc1 with acylamino acid peptidase and acylase I led to two identical open NH2-terminal sequences. Treatment with the acylamino acid peptidase suggested an acylated amino acid residue prior to S27, but treatment with acylase I suggested an acylation of S27 as the cause for the blocked NH2 terminus. Comparison of the predicted signal sequence processing site with the NH2 terminus suggested that a 4-amino-acid-residue propeptide (QIVN-) was cleaved during the maturation of both the recombinant and native Lcc1 proteins.
At pH 5.5 and 20°C, O2 showed a Km of 21 ± 2 μM and a kcat of 200 ± 10 min−1 for both the native and recombinant Lcc1 with 1 mM ABTS as the reducing substrate.
DISCUSSION
C. cinereus, like several other basidiomycetes, contains a laccase gene family of at least three members. Laccase gene families have been reported in T. villosa (54, 55), R. solani (52), A. bisporus (41), the lignin-degrading basidiomycete CECT 20197 (34), and T. versicolor (40). The physiological importance of these gene families is unknown, but differential expression of the members of the families was observed in A. bisporus (48), T. villosa (55), R. solani (52), and the lignin-degrading basidiomycete CECT 20197 (35). In our study, no quantitative analysis of the expression of the three laccase genes was done to demonstrate differential expression. We do not know if the C. cinereus laccase genes are differentially expressed. We would like to systematically delete the genes in C. cinereus as a step towards understanding why this fungus contains multiple laccase genes.
Recently, laccases were purified from the Coprinaceae members Coprinus friesii, Panaeolus sphinctrinus, and Panaeolus papilionaceus (23). The NH2-terminal sequences of these laccases were identical and differ from the predicted NH2-terminal sequences of the three C. cinereus laccases. For example, Lcc2 differs from that of C. friesii at 6 of the 20 amino-terminal residues.
There are three potential N-linked glycosylation sites in the Lcc1 protein. The analysis of the crystal structure of the heterologously produced Lcc1 protein confirms that one of the three potential sites for N-linked glycosylation, N343, is glycosylated. The crystal structure also suggested O glycosylation, although this was not confirmed (15). In addition to glycosylation, mature C. cinereus Lcc1 laccase requires at least three processing steps (signal peptide removal in the endoplasmic reticulum, propeptide cleavage, and removal of its COOH-terminal extension). Comparison of the NH2 terminus predicted after signal sequence cleavage to the NH2 terminus determined for both the native and recombinant proteins indicates that a 4-residue propeptide (QIVN-) is removed during the maturation of Lcc1. The fact that the NH2 terminus of the recombinant laccase is identical to that of the native protein demonstrates that A. oryzae contains the activity required for propeptide removal. Alignment of the deduced amino acid sequence of Lcc1 with those of other fungal laccases predicts a 23-amino-acid COOH-terminal extension similar in length to the extensions of the P. radiata, M. thermophila, and N. crassa laccases (Fig. 2). This extension is rich in arginine and lysine, and in the crystal structure of the recombinant Lcc1 no electron density is observed for the last 13 predicted COOH-terminal residues (Fig. 2) (15). Therefore, the extension of Lcc1 may be cleaved during its synthesis or secretion in A. oryzae. It is not known if the COOH-terminal extension of P. radiata is removed during its processing.
Laccases from the ascomycetes Myceliophthora (5), Neurospora (20), and Podospora (18) are processed at both the NH2 and COOH termini. Processing at the NH2 terminus removes propeptides of about 20 amino acids, which are larger than the 4-residue propeptide removed from C. cinereus Lcc1. The COOH-terminal processing site in ascomycetes (Asp-Ser-Gly-Leu↓Arg/Lys) is conserved, but it is not found in the COOH terminus of C. cinereus Lcc1, suggesting that its processing is distinct.
Among various laccases, Lcc1 has a “low redox potential (E°)” (53). Based on protein sequence, Lcc1 has only 24% identity to the low-E° M. thermophila laccase and 56, 56, and 33% homology to the high-E° T. villosa Lcc1, T. versicolor laccase-1, and R. solani Lcc4, respectively. Thus, the microenvironment at the Cu sites in Lcc1 could be quite different from those in other laccases. However, the E° and kcat for Lcc1 fit well to the linear correlation between ΔE° = E° (laccase) − E° (substrate) and log kcat observed for the other laccases, supporting the hypothesis that the ΔE° (or the thermodynamic driving force) dominates the rate-limiting step of the catalysis, the electron transfer from the substrate to the type 1 Cu in laccase (53). The Km (O2) of 21 μM is close to the values reported for other laccases (26, 38, 50), suggesting a conserved O2-binding domain in this enzyme family.
The cloned lcc1, lcc2, and lcc3 genes have 7, 13, and 13 introns, respectively. The 3′ consensus splice sites (C/TAG) are found in all of the introns; however, the 5′ splice sites of many of the introns do not strictly match the consensus (GTANGT). The variant bases of the 5′ intron splice sites are at positions 3 and 6, with the most common being a C at position 6. These variant bases are similar to those in the introns of T. villosa laccase genes (54, 55). The positions of the 13 introns in lcc2 and lcc3 are identical, and the predicted Lcc2 and Lcc3 proteins have the highest identity (80%).
The Lcc2 and Lcc3 proteins were not isolated from the extracellular broth of C. cinereus or expressed in Aspergillus. Therefore, no direct protein characterization confirming they have laccase activity exists. However, the deduced protein sequences have a high degree of identity with other laccases. In addition, there are two highly conserved regions near the COOH termini of all fungal laccases important for the coordination of the four copper ions that form a redox center (13). These regions, HPI/FHLHGHT/EF and PWFILHCHIDWHLVLGL, are conserved within Lcc2 and Lcc3. In these regions all of the histidines and cysteines believed to be critical for coordination of the copper ions are strictly conserved (Fig. 3). In addition, the HWH and HSH motifs important for copper coordination which are found near the NH2 termini of all laccases are conserved in Lcc2 and Lcc3 (Fig. 3), and the size (517 amino acids) of the predicted Lcc2 and Lcc3 proteins is similar to that of other fungal laccases.
FIG. 3.
Alignment of Lcc1, Lcc2, and Lcc3 amino acid sequences with other known laccase amino acid sequences at the four putative copper binding regions. The sequences were aligned with the Clustal algorithm. Conserved histidines and cysteines are boxed. The numbers refer to the amino acid sequence. The dashes are gaps in the alignment.
The yield of Lcc1 from A. oryzae transformants grown in shake flasks was modest (8 to 135 mg per liter) but better than yields reported for heterologous expression of the M. thermophila laccase (5). Furthermore, the yields in A. oryzae were at least 20-fold higher than those obtained from fermentations of C. cinereus. The higher yield obtained in A. oryzae compared to C. cinereus wild-type fermentations is probably due to the use of the highly expressed α-amylase promoter in the expression vector pDSY67. The expression of Lcc1 in A. oryzae should benefit from the years of experience with industrial scale-up, strain improvement and process development for other enzyme products produced in Aspergillus (11). Recombinant Lcc1 from A. oryzae will be used to test this enzyme for industrial applications.
ACKNOWLEDGMENTS
We thank Randy Berka, Joel Cherry, and Alan Klotz for critically reading the manuscript.
REFERENCES
- 1.Agematu H, Kominato K, Shibamoto N, Yoshika T, Nishida H, Okamoto R, Shin T, Mura S. Transformation of 7-(4-hydroxyphenylacetamido)cephalosporanic acid into a new cephalosporin antibiotic, 7-[1-oxaspiro (2,5)octa-6-oxo-4,7-diene-2-carboxamido]cephalosporonic acid, by laccase. Biosci Biotechnol Biochem. 1993;57:1387–1388. [Google Scholar]
- 2.Aramayo R, Timberlake W E. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 1990;18:3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aviv H, Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baunsgaard L, Dalbøge H, Houen G, Rasmussen E M, Welinder K G. Amino acid sequence of Coprinus macrohizus peroxidase and cDNA sequence encoding Coprinus cinereus peroxidase. Eur J Biochem. 1993;213:605–611. doi: 10.1111/j.1432-1033.1993.tb17800.x. [DOI] [PubMed] [Google Scholar]
- 5.Berka R M, Schneider P, Golightly E J, Brown S T, Madden M, Brown K M, Halkier T, Mondorf K, Xu F. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl Environ Microbiol. 1997;63:3151–3157. doi: 10.1128/aem.63.8.3151-3157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollag J-M, Leonowicz A. Comparative studies of extracellular fungal laccases. Appl Environ Microbiol. 1984;48:849–854. doi: 10.1128/aem.48.4.849-854.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollag J-M. Decontaminating soil with enzymes. Environ Sci Technol. 1992;26:1876–1881. [Google Scholar]
- 8.Bourbonnais R, Paice M G, Reid I D, Lanthier P, Yaguchi M. Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol. 1995;61:1876–1880. doi: 10.1128/aem.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chirgwin J M, Przybyla A F, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 10.Choi G H, Larson T G, Nuss D L. Molecular analysis of the laccase gene from the chestnut blight fungus and selective suppression of its expression in an isogenic hypovirulent strain. Mol Plant-Microbe Interact. 1992;5:119–128. doi: 10.1094/mpmi-5-119. [DOI] [PubMed] [Google Scholar]
- 11.Christensen T, Wøldike H, Boel E, Mortensen S B, Hjortshøj K, Thim L, Hansen M T. High level expression of recombinant genes in Aspergillus oryzae. Biotechnology. 1988;6:1419–1422. [Google Scholar]
- 12.Clutterbuck A. Absence of laccase from yellow-spored mutants of Aspergillus nidulans. J Gen Microbiol. 1972;70:423–435. doi: 10.1099/00221287-70-3-423. [DOI] [PubMed] [Google Scholar]
- 13.Coll P M, Tabernero C, Santamaria R, Perez P. Characterization and structural analysis of the laccase I gene from the newly isolated ligninolytic basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:4129–4135. doi: 10.1128/aem.59.12.4129-4135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damhus, T., and P. Schneider. March 1996. International Patent Application WO96/06930.
- 15.Ducros V, Brzozowski A M, Wilson K S, Brown S H, Østergaard P, Schneider P, Yaver D S, Pedersen A H, Davies G J. Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nat Struct Biol. 1998;5:310–316. doi: 10.1038/nsb0498-310. [DOI] [PubMed] [Google Scholar]
- 16.Eggert C, LaFatette P R, Temp U, Eriksson K-E L, Dean J F D. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl Environ Microbiol. 1998;64:1766–1772. doi: 10.1128/aem.64.5.1766-1772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eggert C, Temp U, Eriksson K-E L. Laccase is essential for lignin degradation by the white-rot fungus Pycnoporus cinnabarinus. FEBS Lett. 1997;407:89–92. doi: 10.1016/s0014-5793(97)00301-3. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Larrea J, Stahl U. Isolation and characterization of a laccase gene from Podospora anserina. Mol Gen Genet. 1996;252:539–551. doi: 10.1007/BF02172400. [DOI] [PubMed] [Google Scholar]
- 19.Geiger J P, Nicole M, Nandris D, Rio B. Root rot diseases of Hevea brasiliensis. I. Physiological and biochemical aspects of root aggression. Eur J For Pathol. 1986;16:22–37. [Google Scholar]
- 20.Germann U A, Müller G, Hunziker P E, Lerch K. Characterization of two allelic forms of Neurospora crassa laccase. J Biol Chem. 1988;263:885–896. [PubMed] [Google Scholar]
- 21.Gubler U, Hoffman B J. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 22.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 23.Heinzkill M, Bech L, Halkier T, Schneider P, Anke T. Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae) Appl Environ Microbiol. 1998;64:1601–1606. doi: 10.1128/aem.64.5.1601-1606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunolstein C V, Valenti P, Visca P, Antonini G, Nicolini L, Orsi N. Production of laccases A and B by a mutant strain of Trametes versicolor. J Gen Appl Microbiol. 1986;32:185–191. [Google Scholar]
- 25.Iimura Y, Takenouchi K, Nakamura M, Kawai S, Katayama Y, Morohoshi N. Cloning and sequence analysis of laccase genes and its use for an expression vector in Coriolus versicolor. 1992. Presented at the Proceedings of the Fifth International Conference on Biotechnology in the Pulp and Paper Industry, Kyoto, Japan. [Google Scholar]
- 26.Iwasaki H, Matsubara T, Mori T. A fungal laccase, its properties and reconstitution from its protein and copper. J Biochem. 1967;61:814–816. doi: 10.1093/oxfordjournals.jbchem.a128618. [DOI] [PubMed] [Google Scholar]
- 27.Jönsson L, Saloheimo M, Penttil M. Laccase from the white-rot fungus Trametes versicolor: cDNA cloning of lcc1 and expression in Pichia pastoris. Curr Genet. 1997;32:425–430. doi: 10.1007/s002940050298. [DOI] [PubMed] [Google Scholar]
- 28.Jönsson L, Sjöström K, Häggström I, Nyman P O. Characterization of a laccase gene from the white-rot fungus Trametes versicolor and structural features of basidiomycete laccases. Biochim Biophys Acta. 1995;1251:210–215. doi: 10.1016/0167-4838(95)00104-3. [DOI] [PubMed] [Google Scholar]
- 29.Kierulff J V, Pedersen A H. Providing a bleached look to dyed fabric, e.g., denim, by contact with an aq. phenol oxidising enzyme system and an enhancing agent, e.g. a syringate. International Patent Application WO96/12845. May 1996. [Google Scholar]
- 30.Kirk T K, Farrell R L. Enzymic “combustion”: the microbial degradation of lignin. Annu Rev Biochem. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 31.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 32.Lante A, Crapisi A, Pasini G, Zamorani A, Spettoli P. Immobilized laccase for must and wine processing. Enzyme Eng. 1992;11:558–562. [Google Scholar]
- 33.Leatham G, Stahman M A. Studies on the laccase of Lentinus edodes: specificity, localization and association with the development of fruiting bodies. J Gen Microbiol. 1981;125:147–157. [Google Scholar]
- 34.Mansur M, Suárez T, Fernández-Larrea J B, Brizuela M A, González A E. Identification of a laccase gene family in the new lignin-degrading basidiomycete CECT20197. Appl Environ Microbiol. 1997;63:2637–2646. doi: 10.1128/aem.63.7.2637-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansur M, Suárez T, González A E. Differential gene expression in the laccase gene family from basidiomycete I-62 (CECT 20197) Appl Environ Microbiol. 1998;64:771–774. doi: 10.1128/aem.64.2.771-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marbach I, Harel E, Mayer A M. Pectin, a second inducer for laccase production by Botrytis cinerea. Phytochemistry. 1985;24:2559–2561. [Google Scholar]
- 37.Mayer A M. Polyphenol oxidases in plants: recent progress. Phytochemistry. 1987;26:11–20. [Google Scholar]
- 38.Naki A, Varfolomeyev S D. Mechanism of the inhibition of laccase activity from Polyporus versicolor by halide-ions. Biokhimiia. 1981;46:1694–1702. [PubMed] [Google Scholar]
- 39.O’Malley D M, Whetten R, Bao W, Chen C, Sederoff R R. The role of laccase in ligninification. Plant J. 1993;4:751–757. [Google Scholar]
- 40.Ong E, Brent W, Pollock R, Smith M. Cloning and sequence analysis of two laccase complementary DNAs from the ligninolytic basidiomycete Trametes versicolor. Gene. 1997;196:113–119. doi: 10.1016/s0378-1119(97)00215-1. [DOI] [PubMed] [Google Scholar]
- 41.Perry C R, Smith M, Britnell C H, Wood D A, Thurston C F. Identification of two laccase genes in the cultivated mushroom Agaricus bisporus. J Gen Microbiol. 1993;139:1209–1218. doi: 10.1099/00221287-139-6-1209. [DOI] [PubMed] [Google Scholar]
- 42.Reid I D. Biological pulping in paper manufacture. Trends Biotechnol. 1991;9:262–265. [Google Scholar]
- 43.Saloheimo M, Leena M, Paavola N. Heterologous production of a ligninolytic enzyme: expression of the Phlebia radiata laccase gene in Trichoderma reesei. Biotechnol. 1991;9:987–990. [Google Scholar]
- 44.Saloheimo M, Niku-Paavola M L, Knowles J K C. Isolation and structural analysis of the laccase gene from the lignin-degrading fungus Phlebia radiata. J Gen Microbiol. 1991;137:1537–1544. doi: 10.1099/00221287-137-7-1537. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sannia G, Giardina P, Luna M, Rossi M, Buonocore V. Laccase from Pleurotus ostreatus. Biotechnol Lett. 1986;8:797–800. [Google Scholar]
- 47.Schneider, P., M. B. Caspersen, K. Mondorf, T. Halkier, L. K. Skov, P. R. Østergaard, K. M. Brown, S. H. Brown, and F. Xu. Characterization of a Coprinus cinereus laccase. Enzyme Microbiol. Technol., in press.
- 48.Smith M, Shnyreva A, Wood D A, Thurston C F. Tandem organization and highly disparate expression of the two laccase genes lcc1 and lcc2 in the cultivated mushroom Agaricus bisporus. Microbiology. 1998;144:1063–1069. doi: 10.1099/00221287-144-4-1063. [DOI] [PubMed] [Google Scholar]
- 49.Tilburn J T, Scazzocchio C, Taylor G G, Zabicky-Zissman J H, Lockington R A, Davies R W. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:29–37. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- 50.Varfolomeyev S D, Naki A, Yaropolov A I, Berezin I V. Kinetics and mechanism of catalytic reduction of molecular oxygen in the presence of laccase. Biokhimiia. 1985;50:1411–1420. [Google Scholar]
- 51.von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 52.Wahleithner J A, Xu F, Brown K M, Brown S H, Golightly E J, Halkier T, Kauppinen S, Pederson A, Schneider P. The identification and characterization of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr Genet. 1996;29:395–403. doi: 10.1007/BF02208621. [DOI] [PubMed] [Google Scholar]
- 53.Xu F, Shin W, Brown S H, Wahleithner J A, Sundaram U M, Solomon E L. A study of a series of fungal laccases and bilirubin oxidase that exhibits significant differences in redox potential, substrate specificity, and stability. Biochim Biophys Acta. 1996;1292:303–311. doi: 10.1016/0167-4838(95)00210-3. [DOI] [PubMed] [Google Scholar]
- 54.Yaver D S, Golightly E J. Cloning and characterization of three laccase genes from the white-rot basidiomycete Trametes villosa: genomic organization of the laccase gene family. Gene. 1996;181:95–102. doi: 10.1016/s0378-1119(96)00480-5. [DOI] [PubMed] [Google Scholar]
- 55.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalbøge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]