Abstract
Parkinson’s disease is a progressive neurodegenerative disorder characterized by the intracellular accumulation of insoluble alpha-synuclein aggregates into Lewy bodies and neurites. Increasing evidence indicates that Parkinson’s disease progression results from the spread of pathologic alpha-synuclein through neuronal networks. However, the exact mechanisms underlying the propagation of abnormal proteins in the brain are only partially understood. The objective of this study was first to describe the long-term spatiotemporal distributions of Lewy-related pathology in mice injected with alpha-synuclein preformed fibrils and then to recreate these patterns using a computational model that simulates in silico the spread of pathologic alpha-synuclein.
In this study, 87 2–3-month-old non-transgenic mice were injected with alpha-synuclein preformed fibrils to generate a comprehensive post-mortem dataset representing the long-term spatiotemporal distributions of hyperphosphorylated alpha-synuclein, an established marker of Lewy pathology, across the 426 regions of the Allen Mouse Brain Atlas. The mice were injected into either the caudoputamen, nucleus accumbens or hippocampus, and followed over 24 months with pathologic alpha-synuclein quantified at seven intermediate time points. The pathologic patterns observed at each time point in this high-resolution dataset were then compared to those generated using a Susceptible-Infected-Removed (SIR) computational model, an agent-based model that simulates the spread of pathologic alpha-synuclein for every brain region taking simultaneously into account the effect of regional brain connectivity and Snca gene expression.
Our histopathological findings showed that differentially targeted seeding of pathological alpha-synuclein resulted in unique propagation patterns over 24 months and that most brain regions were permissive to pathology. We found that the SIR model recreated the observed distributions of pathology over 24 months for each injection site. Null models showed that both Snca gene expression and connectivity had a significant influence on model fit.
In sum, our study demonstrates that the combination of normal alpha-synuclein concentration and brain connectomics contributes to making brain regions more vulnerable to the pathological process, providing support for a prion-like spread of pathologic alpha-synuclein. We propose that this rich dataset and the related computational model will help test new hypotheses regarding mechanisms that may alter the spread of pathologic alpha-synuclein in the brain.
Keywords: alpha-synuclein, Parkinson’s disease, synucleinopathy, modelling, Snca
Rahayel et al. inject alpha-synuclein preformed fibrils into the mouse brain and quantify their spread over 24 months. Differentially targeted seeding results in unique propagation patterns, which can be recreated using agent-based computational modelling that considers both regional connectivity and Snca gene expression.
Introduction
Parkinson’s disease is a progressive neurodegenerative condition characterized by the intracellular accumulation of insoluble alpha-synuclein aggregates into Lewy bodies and neurites.1–3 Post-mortem studies suggest that the distribution of Lewy pathology follows a stereotypical caudo-rostral pattern in the brain, characterized by inclusions appearing in the caudal brainstem and olfactory bulb, followed in time by the involvement of the midbrain, limbic areas and neocortex.4,5 This has led to the hypothesis that pathologic alpha-synuclein spreads through an iterative transneuronal process.6,7 Once exogenous pathologic alpha-synuclein enters a recipient cell, it acts as a template that causes natively unfolded alpha-synuclein proteins to misfold.6
Accumulating evidence from a diverse collection of clinical, histological and experimental observations indicates that pathologic alpha-synuclein behaves in a prion-like fashion. First, post-mortem studies demonstrate the possibility of neuron-to-neuron transfer of pathologic alpha-synuclein in patients with Parkinson’s disease.8,9 Second, brain atrophy patterns derived from MRI in Parkinson’s disease appear to be shaped by connectivity, indicating a propagating mechanism.10–12 Third, consistent with the Braak hypothesis, human studies provide evidence of gut-to-brain transfer of pathologic alpha-synuclein.13,14 Finally, synthetic alpha-synuclein preformed fibrils or post-mortem brain lysates from patients with synucleinopathy induce the formation and CNS spread of pathologic alpha-synuclein in the brains of transgenic and wild-type mice, rats and non-human primates, providing direct evidence of pathological templating and transneuronal spread.15–18 Moreover, alpha-synuclein preformed fibrils injected in the gastrointestinal system of mice also propagate to the CNS.19,20 However, despite these findings, the propagation hypothesis is still subject to debate, with some authors arguing that alpha-synuclein is not the causal pathological agent of so-called synucleinopathies but rather a non-specific response to injury, arising from upstream mechanisms and compensatory responses to cellular stress.21–23 To shed light on the pathological nature of alpha-synuclein, a comprehensive assessment of the distributions of pathologic alpha-synuclein after injection is needed to describe and understand the long-term evolution of pathologic alpha-synuclein in the brain.
Computational modelling provides a versatile approach for uncovering the biological factors and processes that determine the spread of misfolded proteins in the brain24 and for informing in a cost-effective way predictions and decisions regarding therapeutic strategies that target alpha-synuclein accumulation. A limited number of studies have applied simulation frameworks to model the spread of pathologic alpha-synuclein in the mouse brain.25,26 For example, when using connectivity to model the spread of alpha-synuclein, the pathology at five coronal levels was replicated for up to 6 months after injection into the caudoputamen (CP).26 Another study also recreated the spread of alpha-synuclein 9 months after injection into the olfactory bulb or the substantia nigra.25 While highly informative, the focus on transneuronal factors may overshadow the contribution of cell-autonomous processes. In contrast, agent-based simulation allows modelling the dynamics at the level of individual proteins using rules about their mutual interactions and their environment. Such a framework may allow testing hypotheses related to the prion-like network transmission of pathology but also the selective vulnerability hypothesis of certain regions or cell types to pathology. We therefore developed an agent-based Susceptible-Infected-Removed (SIR) model,27 which simulates the dynamics of normal and pathologic alpha-synuclein proteins on a realistic brain connectome.12 This specific agent-based model was previously demonstrated to be relevant for understanding the pathological mechanisms underlying the brain atrophy occurring in Parkinson’s disease, recreating the spatial pattern of atrophy with high accuracy and demonstrating that both the local concentration of alpha-synuclein and the brain’s regional connectivity pattern made regions more vulnerable to pathology and more effective at propagating it.12 However, this previous study in patients used tissue atrophy measurements from human MRI data as a proxy measure of alpha-synuclein accumulation in the brain. Therefore, before testing new potential disease modifiers of the spread of alpha-synuclein in humans, it is required that the model be validated using quantification of actual alpha-synuclein pathology, that is, to test whether the model can recreate the spatiotemporal distributions of pathologic alpha-synuclein observed in animals injected with pathology. To date, no investigation has yet applied the SIR model to direct observations of alpha-synuclein spread in animal models.
We generated a comprehensive high-resolution histopathological dataset from non-transgenic mice followed over 24 months after the injection of recombinant alpha-synuclein preformed fibrils into either the striatum, nucleus accumbens or hippocampus. We stained for hyperphosphorylated alpha-synuclein at Ser129, an established marker of Lewy pathology in human disease,28 in 426 regions of the Allen Mouse Brain Atlas. We then tested whether the agent-based SIR model simulated in silico the observed distribution of pathologic alpha-synuclein. We show here that different injection sites lead to distinct neuroanatomical distribution patterns of pathologic alpha-synuclein, which the model could accurately recreate using high-resolution gene expression and structural connectomics data. The model also demonstrated that regional Snca gene expression, connection topology and spatial embedding of the brain significantly shape the spread of alpha-synuclein. Taken together, this study demonstrates that the simulated spread of alpha-synuclein based on gene expression and connectomics recreates the propagation of pathologic alpha-synuclein induced in wild-type mice; this model may therefore represent an elegant tool to test hypotheses to stop, impair or hinder brain spread of pathologic alpha-synuclein.
Materials and methods
Animals
All housing, breeding and procedures were performed according to the National Institutes of Health Guide for the Care and Use of Experimental Animals and approved by the University of Pennsylvania Institutional Animal Care and Use Committee. The injection studies described used 2–3-month-old female C57B16/C3H mice (Stock No.100010; The Jackson Laboratories). Animals were maintained on a 12-h light/dark schedule and provided with food ad libitum.
Alpha-synuclein purification and preformed fibril assembly
Recombinant mouse alpha-synuclein purification was performed as previously described elsewhere.16,29 Briefly, BL21 (DE3) RIL-competent E. coli cells (Agilent Technologies catalogue no. 230245) were transformed with a plasmid containing the mouse alpha-synuclein cDNA (pRK172/mSyn). Single colonies were expanded in Terrific Broth (12 g/l of Bacto-tryptone, 24 g/l of yeast extract 4% (vol/vol) glycerol, 17 mM KH2PO4 and 72 mM K2HPO4) containing ampicillin (Fisher Scientific). Bacterial pellets were resuspended in high salt buffer (750 mM NaCl in 10 mM Tris, pH 7.6), sonicated and boiled. The resulting supernatant was dialysed against 10 mM Tris, pH 7.6, 50 mM NaCl, 1 mM EDTA overnight at 4°C, passed through a 0.22-µm filter and concentrated using Amicon Ultra-15 centrifugal filter units (Millipore no. UFC901008). Proteins were separated over a Superdex 200 column (GE Healthcare) and 1-ml fractions collected. Fractions enriched in alpha-synuclein were identified by SDS–PAGE followed by Coomassie blue staining, dialysed in 10 mM Tris, pH 7.6, 50 mM NaCl, 1 mM EDTA overnight and run on a HiTrapQ HP column (GE Health 645932) over a linear gradient (25 to 1000 mM NaCl). Fractions containing alpha-synuclein were collected and dialysed into DPBS. Protein was filtered through a 0.22-µm filter, concentrated to 5 mg/ml and frozen at −80°C until used. For preparation of preformed fibrils, alpha-synuclein monomer was shaken at 1000 rpm for 7 days at 37°C using a Thermomixer C (Eppendorf). Preparations were aliquoted and validated for conversion to preformed fibrils SDS–PAGE following sedimentation (100 000g for 30 min) and Thioflavin T fluorescence. Remaining aliquots were stored at −80°C until used for stereotaxic injections.
Stereotaxic injection of preformed fibrils
Mice received a single unilateral stereotaxic injection of preformed fibrils targeted to either dorsal striatum, lateral accumbens shell or hippocampus CA1 as previously described.30 For this study, 2–3-month-old female B6C3F1 mice were used. Thawed preformed fibrils were diluted to 2 mg/ml in DPBS, then sonicated using a Biorupter UCD-300 bath sonicator (Diagenode) on high for 10 cycles (30 s on; 30 s off) at constant temperature (10°C). A fresh fibril aliquot was used every 4 h; transmission electron microscopy of unsonicated and sonicated fibrils did not reveal the presence of amorphous aggregates (Supplementary Fig. 1). Mice were injected unilaterally by insertion of a single Hamilton syringe (33 gauge) into the right forebrain targeting either the dorsal striatum (anterior-posterior: +0.2 mm relative to bregma, medial-lateral: +2.0 mm from midline, depth: −3.2 mm from surface of skull), lateral accumbens shell (anterior-posterior: +1.45 mm, medial-lateral: +1.75 mm, depth: −4.4 mm) or hippocampus CA1 region (anterior-posterior: −2.5 mm, medial-lateral: +2.0 mm, depth: −2.4 mm). For each target, 5 µg of preformed fibrils (in 2.5 µl) was injected using a 10-µl syringe (Hamilton) at a rate of 0.4 µl/min. At the post-injection time points indicated, mice were perfused transcardially with heparinized PBS and brains were fixed in 70% ethanol in 150 mM NaCl, pH 7.4. overnight.
Immunohistochemistry
Fixed mouse brains were embedded in paraffin, sectioned at 6 µm and transferred to glass slides for immunohistochemistry. Every 30th section (i.e. representing 180 µm of coronal separation) through the brain was stained for alpha-synuclein phosphorylated at serine 129, an established marker of Lewy pathology.28 Samples were deparaffinized with two sequential 5-min washes in xylenes, followed by 1-min washes in a descending series of ethanols: 100, 100, 95, 80 and 70%. Slides were then incubated in deionized water for 1 min before antigen retrieval as noted. After antigen retrieval, slides were incubated in 5% hydrogen peroxide in methanol to quench endogenous peroxidase activity. Slides were washed for 10 min in running tap water, 5 min in 0.1 M Tris, then blocked in 0.1 M Tris with 2% foetal bovine serum (FBS). Slides were incubated with an antibody against alpha-synuclein phosphorylated at serine 129 (clone 81A)31 at a final concentration of 0.1 µg/ml. Sections were washed in 0.1 M Tris for 5 min, then incubated with biotinylated horse anti-mouse (1:1000; Vector BA2000) biotinylated IgG in 0.1 M Tris containing 2% FBS for 1 h. Secondary antibody was rinsed off with 0.1 M Tris for 5 min, then incubated with avidin-biotin solution (Vector PK-6100) for 1 h. Slides were then washed with 0.1 M Tris and developed with ImmPACT DAB peroxidase substrate (Vector SK-4105) and counterstained briefly with haematoxylin (Fisher 67-650-01). Slides were washed in running tap water for 5 min, dehydrated in ascending ethanol for 1 min each: 70, 80, 95, 100 and 100%, then washed twice in xylenes for 5 min and coverslipped in Cytoseal Mounting Media (Fisher 23-244-256). All slides were digitized using a Lamina Scanner (Perkin Elmer) in brightfield mode.
Pathology assessment and scoring
For each brain, a 1:30 series of pathologic alpha-synuclein sections was scored manually by one of three independent raters blinded to the treatment group using CaseViewer v.2.3 (3DHistech). Pathology was categorized as either neuritic (i.e. process-like) or cell body (i.e. contained within the soma) and scored on a scale of 0 (no pathology) to 3 (highest level of inclusions). The mean pathology score per brain for each CNS region as defined by the Allen Mouse Brain Atlas was calculated from all sections examined. For each target/time point combination, four to seven mice were imaged and analysed. Heat maps were generated using Matrix2png (https://matrix2png.msl.ubc.ca/).32
Agent-based Susceptible-Infected-Removed model
The agent-based model simulated the spread of pathologic alpha-synuclein within an SIR framework.12 Every agent was an alpha-synuclein protein that was either ‘Susceptible’ when normal (soluble alpha-synuclein), ‘Infected’ when abnormal (misfolded alpha-synuclein) or ‘Removed’ when undergoing degradation or spreading to connected regions. Agents were independent and their mobility patterns and life spans depended on rules that guided their interactions. The agent-based SIR model was implemented into four modules: (i) the production of normal alpha-synuclein; (ii) the clearance of normal and misfolded alpha-synuclein; (iii) the misfolding of normal alpha-synuclein (infection transmission); and (iv) the propagation of normal and pathologic alpha-synuclein.
Snca expression
The structure-wise gene expression was obtained from the Allen Mouse Brain Atlas API (http://help.brain-map.org/display/mousebrain/API). The section images containing gene expression values were divided into a 200 × 200-µm grid to generate a low-resolution three-dimensional volume and then mapped to the three-dimensional reference model. Pixel-based gene expression statistics were then summarized within each division and expression energy was chosen as the gene expression measurement. For each experiment, the expression energy of each structure was calculated by unionizing the corresponding grid voxels with the same structure label in the three-dimensional reference atlas. These values were subsequently z-scored and averaged across the associated experiments, giving the final expression map.
Anatomical connectivity
The connectivity data were obtained from the viral tract-tracing experiments available in the Allen Mouse Brain Connectivity Atlas.33 The connectivity matrix consisted of 213 rows representing source regions and 426 columns representing the 213 ipsilateral regions and the 213 contralateral regions. The retrograde spread of agents was simulated using a matrix in which each element represented the tracer-derived connectivity strength of every incoming connection to a region, corresponding to a connectivity matrix with a sparsity of 64%. A sparser connectivity matrix (94% sparsity), which was made of only the connections that were associated with a statistical confidence of P < 0.05 in the original tract-tracing study (i.e. based on the best-fit model generated from a bounded optimization and a subsequent linear regression),33 was also used to test the robustness of our findings.
Model specifications
Production of normal alpha-synuclein
Equation (1) describes the synthesis of susceptible agents in region i, which occurred per unit time with probability αi:
| (1) |
where Φ0,1(·) was the standard normal cumulative distribution function and Snca expressioni was the Snca expression of region i. The increment of susceptible agents in region i at each simulation timestep was αiSiΔt, where Si was the size of region i and Δt was the total time. The main analyses were performed with a Δt of 0.1, but findings were similar for values from 0.001 to 1 (Supplementary Fig. 2).
Clearance of normal and pathologic alpha-synuclein
The clearance of susceptible and infected agents from region i occurred per unit time with probability βi (i.e. clearance rate). The cleared proportion of agents within time step Δt corresponded to 1 – e−βΔt given the probability limδτ→0(1 − βδτ)Δt/δτ = e−βΔt that an agent was still active after total time Δt. Therefore, the higher the clearance rate, the lower the probability of an agent to be still active after Δt; similarly, the longer the total time Δt, the lower the probability of an agent to be still active.
Misfolding of normal alpha-synuclein: infection transmission
Equation (2) describes the probability γi of susceptible agents from region i to become infected per unit time:
| (2) |
where Mi was the population of infected agents in region i, was the baseline likelihood that an infected agent induced the misfolding of susceptible agents in region i, set as the reciprocal of the size of the region (1/Si) and was the probability that a susceptible agent did not get infected by any of the Mi infected agents. Therefore, the larger the population of infected agents in a region, the higher the transmission rate and the lower the probability of not being infected; similarly, the larger the size of a region, the lower the transmission rate and the higher the probability of not being infected. Similar to the previous module, the probability that a susceptible agent remained susceptible after a total time Δt was given by , such that the proportion of susceptible agents that became infected after a total time Δt was . The longer the total time Δt, the lower the probability of an agent of being still susceptible.
To determine the baseline density of susceptible agents in every region, the population of susceptible agents Ni was incremented with equation (3):
| (3) |
Once the system reached its stable point, the pathogenic spread and update of the population of susceptible (Ni) and infected agents (Mi) was based on equations (4) and (5), respectively:
| (4) |
| (5) |
Propagation of normal and pathologic alpha-synuclein
The probabilities of susceptible and infected agents in region i to either remain in region i or to spread to other regions via fibre tracts were determined by equations (6) and (7), respectively, according to a multinomial distribution per unit time:
| (6) |
| (7) |
where wij was the directed connection weight between region i and region j. The probability ρi that an agent remained in region i was set to 0.01, but peak fits remained robust at the lowest spreading rates (Supplementary Fig. 2). The stronger the connection between two regions among the total set of projections from the region, the higher the probability of an agent to spread.
Equations (8) and (9) describe the binary probabilities that susceptible and infected agents located in an edge exited the edge per unit time:
| (8) |
| (9) |
where lij was the edge length linking regions i and j. The population of susceptible and infected agents in the edge between regions i and j were denoted by Nij and Mij, respectively. The shorter the length of the connection between two regions, the higher the probability of an agent to spread to the target region.
The increments of Ni and Mi in region i after a total time Δt were described by equations (10) and (11):
| (10) |
| (11) |
whereas the increment of Nij and Mij were described by equations (12) and (13):
| (12) |
| (13) |
Model validation
The spread of pathologic alpha-synuclein was run for 30 000 iterations and done separately with CP, ACB or CA1 as seed region. For every region, the regional number of infected agents was simulated at each timestep and compared with the alpha-synuclein total pathology scores (i.e. the sum of the average soma and neurite pathology scores) observed at each post-injection time point using Spearman’s rank correlation coefficients. The peak fit was considered as the maximal correlation coefficient among the 30 000 coefficients surviving the Bonferroni-corrected threshold of P < 1.67 × 10−6. The peak fits were investigated in each hemisphere separately. Correlations with observed somal and neuritic pathology were also investigated separately.
Snca null models
The impact of Snca expression on the spread was tested by first comparing, for each injection site and post-injection time point, the original peak fit observed in the 426 regions (i.e. empirical peak fit) with the average peak fit derived from 500 null models in which Snca expression was randomized (i.e. null peak fit). The peak fits were compared using Mann–Whitney U exact tests; the fit was considered disrupted when the empirical peak fit was significantly higher than the null peak fit based on an unbiased Monte Carlo estimate of P < 0.05 (10 000 permutations). Snca expression was also replaced by the expression of genes associated with other pathological entities, namely App, Mapt or Nf1, respectively, coding for proteins involved in amyloidopathies, tauopathies and neurofibromatosis,34–36 to assess how this led to a disrupted peak fit between patterns. Fit disruption was also assessed by replacing Snca expression by homogeneous values ranging from 0.1 to 0.9.
Connectome null models
To assess the impact of the connectome’s topology and/or geometry on pathologic alpha-synuclein spread, the empirical peak fit was compared to the average null peak fit derived from sets of 500 rewired or repositioned null models, separately. Rewired null models were generated by swapping edge pairs using the Maslov–Sneppen algorithm as part of the Brain Connectivity Toolbox (https://sites.google.com/site/bctnet/),37,38 with 100 rewirings per edge. This rewired the network’s connectivity while preserving the original degree sequence and density. Repositioned null models were generated by shuffling randomly the physical position of regions inside the connectivity and distance matrices, disrupting spatial autocorrelation and preserving the original degree sequence and connectivity profile. These null networks were generated using the sparse connectivity matrices (94% sparsity), which included only the connections associated with a P < 0.05 in the original tract-tracing study.33
Assessment of regional vulnerability
To understand how gene expression and connectivity associated with regional resilience to pathology, the Snca expression and incoming connectivity strength of regions in which pathologic alpha-synuclein was overestimated by the model were compared to those of regions showing pathology. In addition, every brain region was assigned to one of 13 divisions from the Allen Reference Atlas Ontology (i.e. isocortex, olfactory areas, hippocampal formation, cortical subplate, striatum, pallidum, thalamus, hypothalamus, midbrain, pons, medulla, cerebellar cortex, cerebellar nuclei) to understand the spatial distribution of regions with overestimated pathology.
Data availability
The pathological datasets and the Python code for running the agent-based SIR model in the mouse brain are available at https://github.com/srahayel.
Results
Spatiotemporal evolution of pathologic alpha-synuclein following stereotaxic seeding
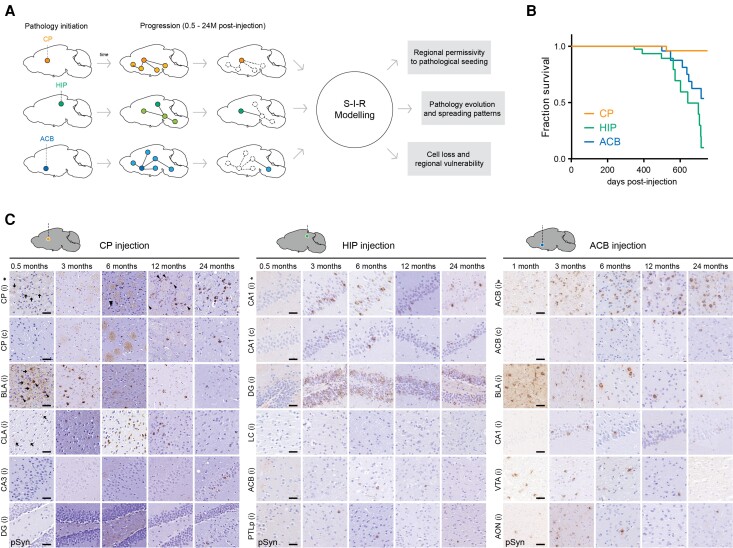
To investigate the spread of pathologic alpha-synuclein across different parts of the connectome, we seeded pathology in three distinct regions by stereotaxically targeting the same dosage of mouse preformed fibrils to the CP, anterior nucleus accumbens (ACB) or hippocampal CA1 region (HIP) (Fig. 1A), respectively, representing components of nigrostriatal, limbic and cognitive circuits that are affected in Parkinson’s disease.39 Although all CP-injected mice survived up to 24 months, mice that received preformed fibril injection in either the ACB or HIP showed a clear reduction in survival. After injections in these two regions, approximately 50% of animals died before 24 months, developing a rapidly progressive hypokinetic phenotype starting at 347 (HIP) and 499 (ACB) days after injection on average (Fig. 1B). Thus, pathologic alpha-synuclein originating within differing circuits leads to distinct survival times.
Figure 1.
Targeted initiation of alpha-synuclein pathology. (A) Schematic outlining the current study. Alpha-synuclein preformed fibrils were targeted to one of three independent CNS regions in non-transgenic mice to initiate pathology in distinct connectomes. (B) Survival curves of CP-, HIP- and ACB-injected mice. Reduced survival was observed in HIP- and ACB-injected mice, while the CP cohort remained unaffected (P < 0.0011 and P < 0.0174 versus CP, respectively; log-rank/Mantel–Cox test). (C) Representative images of brain sections immunostained for pathologic alpha-synuclein at various time points after preformed fibril injection. Examples of neuritic (arrows) and soma pathology (arrowheads) within the indicated regions are shown. Scale bars = 50 µm. Asterisk indicates the injection site; i = ipsilateral hemisphere to injection; c = contralateral hemisphere to injection. ACB = lateral accumbens; AON = anterior olfactory nucleus; BLA = basolateral amygdala; CA = cornu ammonis; CLA = claustrum; CP = caudoputamen; DG = dentate gyrus; HIP = hippocampus; LC = locus coeruleus; PTLp = posterior parietal association areas; pSyn = phosphorylated alpha-synuclein; VTA = ventral tegmental area.
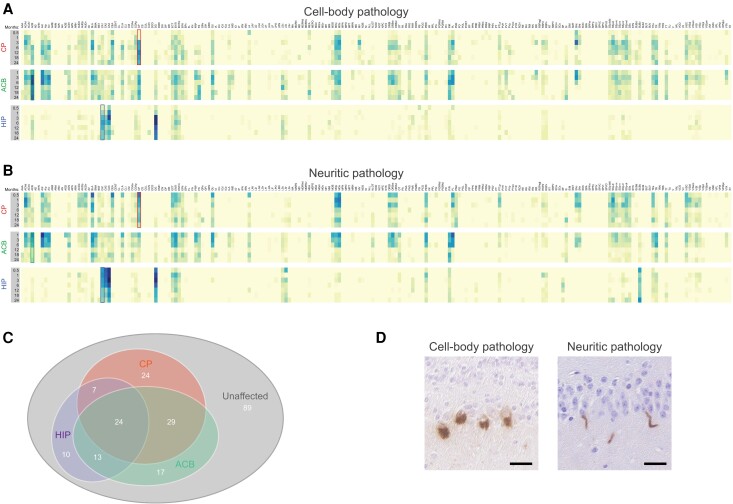
To define the evolution of pathologic alpha-synuclein, we examined the mesoscale distribution of inclusions from 0.5 up to 24 months post-preformed fibril injection (Fig. 1C and Supplementary material). The spatiotemporal pathology maps (Fig. 2A and B) revealed unique and highly dynamic global patterns of pathologic alpha-synuclein. Of the 213 ipsilateral regions from the Allen Mouse Brain Atlas, pathology was detectable in 124 sites (58%) (Fig. 2C). Affected regions were characterized by the appearance of neuritic pathology (i.e. within neuronal processes; Fig. 2D) followed by the establishment of inclusions in neuronal soma as previously observed in vitro and in vivo.40,42 The proportion of regions that developed detectable pathology at least one time point after preformed fibril injection ranged from 25% following CA1 injection to 39% for injections into the ACB or CP (Fig. 2C).
Figure 2.
Spatiotemporal distribution of pathologic alpha-synuclein following single preformed fibril-inoculation. Heatmaps illustrating the evolution of cell body (A) and neuritic (B) pathology following a single unilateral preformed fibril injection into CP, ACB or HIP. Analysed regions representing 213 areas from the Allen Mouse Brain Atlas are listed on the x-axis in alphabetical order. Data represent mean pathology scores from the hemisphere ipsilateral to the injection site (n = 4–7 mice per time point). The ipsilateral and contralateral hemisphere data along with names and abbreviations are available in the Supplementary material. Boxes in the heatmaps indicate the site of fibril injection for CP (red), ACB (green) and HIP (blue). (C) Venn diagram showing unique and common regions affected by pathologic alpha-synuclein following inoculation into CP, ACB or HIP. (D) Examples of intraneuronal (cell body) and neuritic inclusions following preformed fibril injection in the olfactory bulb and hippocampus, respectively. Scale bars = 20 µm.
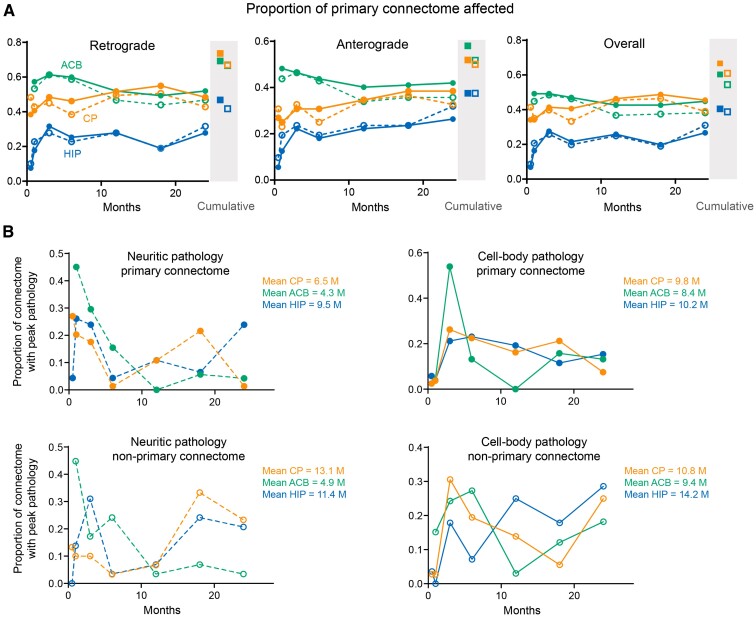
To discern the effect of direct preformed fibril exposure on regional development of pathology, regions were classified based on their connectivity to the injection site. Among sites that shared direct afferent or efferent projections with the injection site, the proportion of affected regions increased to 67% for CP, 60% for ACB and 41% for HIP, indicating that areas with monosynaptic connections, and hence higher exposure to pathologic alpha-synuclein, are more susceptible. Directly connected regions were infiltrated (i.e. defined as the first time point with detectable inclusions) over an extended period after preformed fibril injection, although the overall connectome involvement remained steady after 1 month post-injection (Fig. 3A). Pathologic alpha-synuclein was also found outside the injection site’s primary connectome, particularly in the CP and HIP datasets where the pathology in the second- and third-order connected regions became greater as the post-injection duration increased (Supplementary Figs 3–5 for the high-resolution layouts). Consistent with pathological spread occurring via neuronal networks, the mean time to achieve peak pathology (i.e. the post-injection time point at which a brain region had the highest amount of pathologic alpha-synuclein) was lower for regions directly connected with the seeds compared to regions not directly connected with the seeds, although there was considerable variability between regions (Fig. 3B). In addition, both neuritic and somal pathology underwent a reduction over time in multiple regions (e.g. basolateral amygdala and piriform cortex), suggesting the removal of pathologic alpha-synuclein and/or the cells in which it was contained. To determine whether cell loss occurred in such regions, cell nuclei were quantified in selected regions at 24 MPI following PFF injection into either CP, ACB or HIP (Supplementary Fig. 6). Whereas inoculation into the CP and ACB resulted in significantly reduced nuclei in the basolateral amygdala ipsilateral to injection, HIP injected mice showed decreased nuclei in the dentate gyrus. We failed to detect changes within the piriform cortex possibly due to a smaller proportion of neurons developing pathology following injection into either CP or ACB.
Figure 3.
Infiltration of primary connectome after preformed fibril-inoculation into different injection sites. (A) Graphs show the proportion of regions affected by either cell body (filled circles) or neuritic (open circles) pathology within the retrograde or anterograde primary connectomes associated with the indicated injection sites at various time points after preformed fibril exposure. Regions with anterograde and/or retrograde connectivity are represented in the overall primary connectome (right panel). Closed and open squares represent the cumulative proportion of connectome involvement over the 24-month duration of the study. (B) Distribution of pathology-affected regions based on the time after preformed fibril injection when peak pathology was detected. Peak pathology was defined as the post-injection time point at which a brain region had the highest amount of pathologic alpha-synuclein. Non-primary areas represent afflicted regions outside of the injection site’s primary anterograde and retrograde connectome. Data represent means from 4–7 mice per time point and are based on values in Fig. 2 and Supplementary material.
There was also significant overlap between the injection sites with regards to the regions affected by pathology (e.g. entorhinal, ectorhinal and perirhinal cortices), particularly between the CP and ACB cohorts. Of the 124 regions with pathology, 59% (73/124) were affected in at least two cohorts with different injection sites while 19% (24/124) were affected in all three sites (Fig. 2C). Despite differences in the time after preformed fibril injection required for pathology to accumulate within these areas and differences in pathologic alpha-synuclein load, such overlaps point to a subset of regions permissive to alpha-synuclein misfolding, transmission and accumulation. In contrast, pathology was absent from 89 regions across all three cohorts, primarily in the thalamus and cerebellum, identifying areas that are relatively resistant to pathology (Supplementary material). Interestingly, pathology within the CP and ACB increased at a slower rate than most other affected regions, despite being initial sites of injection.
The Susceptible-Infected-Removed model replicates pathologic alpha-synuclein distribution
The agent-based SIR model was used to simulate in silico the spread of pathologic alpha-synuclein. Every alpha-synuclein protein in a region is an agent that can belong to one of three compartments: ‘susceptible’ when soluble (normal alpha-synuclein), ‘infected’ when misfolded or ‘removed’ when metabolized or after travelling to another region. High-resolution Snca gene expression and whole-brain connectivity data were used to model the agents’ synthesis rate and mobility pattern. The simulation was performed over 30 000 iterations during which agents spread and transited between compartments based on predefined rules guiding the interactions of agents with each other and with their local environment. To benchmark the model against its empirical analogue, the simulated number of infected agents in every region at each iteration was compared to the amount of phosphorylated alpha-synuclein observed in every region at each post-injection time point.
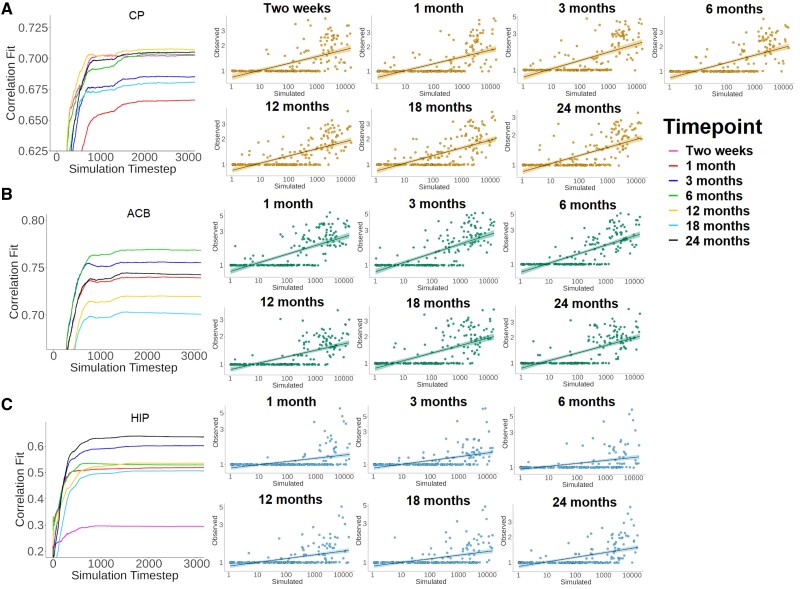
The ipsilateral comparisons between regional pathologic alpha-synuclein observed in the mice and the simulated regional number of infected agents revealed significant correlations for virtually every injection site and post-injection time point (CP: 0.67−0.71, ACB: 0.70−0.77, HIP: 0.51−0.64) (Table 1 and Fig. 4). The model fit gradually increased with the number of iterations, with a stable point reached at around the 1000th timestep, where the peak fit was generally found. Only the pathology at 0.5 months after HIP injection was not recreated, likely owing to the low number of regions showing pathology (8.5%). Contralateral comparisons yielded similar although slightly lower fits (Supplementary Table 1). Although the model does not attempt to simulate somal and neuritic pathology separately, the simulated regional number of infected agents recreated both somal and neuritic distributions in mice for virtually every injection site and time point (Supplementary Table 2). Again, somal pathology at the 0.5-month post-injection time point into the HIP was not predicted by the model. Similar findings were generally found contralaterally (Supplementary Table 2). Also, using matrices consisting of either retrograde or anterograde connections yielded similar findings (results shown here are for retrograde connectivity). These findings demonstrate that the agent-based SIR model recreates the distribution of pathologic alpha-synuclein.
Table 1.
Peak correlation fits between simulated and observed pathologic alpha-synuclein
| Post-injection time point | CP | ACB | HIP | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Peak fit | P-value | 95% CI | Peak fit | P-value | 95% CI | Peak fit | P-value | 95% CI | |
| Two weeks | 0.703 | 4.6 × 10−33 | 0.64–0.76 | – | – | – | ns | ns | ns |
| 1 MPI | 0.668 | 7.7 × 10−29 | 0.59–0.74 | 0.740 | 3.1 × 10−38 | 0.67–0.79 | 0.521 | 3.3 × 10−16 | 0.43–0.60 |
| 3 MPI | 0.686 | 5.2 × 10−31 | 0.60–0.76 | 0.756 | 1.0 × 10−40 | 0.69–0.80 | 0.603 | 1.6 × 10−22 | 0.51–0.68 |
| 6 MPI | 0.705 | 2.9 × 10−33 | 0.63–0.76 | 0.769 | 6.2 × 10−43 | 0.72–0.81 | 0.535 | 3.4 × 10−17 | 0.44–0.61 |
| 12 MPI | 0.708 | 1.0 × 10−33 | 0.62–0.77 | 0.720 | 2.2 × 10−35 | 0.65–0.77 | 0.537 | 2.6 × 10−17 | 0.43–0.62 |
| 18 MPI | 0.683 | 1.2 × 10−30 | 0.60–0.75 | 0.703 | 4.0 × 10−33 | 0.64–0.76 | 0.507 | 2.5 × 10−15 | 0.41–0.60 |
| 24 MPI | 0.707 | 1.5 × 10−33 | 0.63–0.77 | 0.745 | 6.9 × 10−39 | 0.68–0.79 | 0.639 | 7.9 × 10−26 | 0.55–0.70 |
The peak correlation fit represents the maximal Spearman’s rank correlation coefficient in the ipsilateral hemisphere between the simulated number of infected agents and the observed total pathologic alpha-synuclein score. Note that observed pathology 2 weeks after injection was quantified only for the CP and HIP datasets. CI = confidence interval; MPI = month post-injection; ns = not significant.
Figure 4.
Fit between the simulated and observed pathologic alpha-synuclein. Graphs (left) show the progression of the model fit between simulated and observed pathology when seeding from the CP (A), ACB (B) or CA1 field (C). Only the first 3000 simulation steps are presented since the model reached its stable state. Plots (right) show the distribution of regional pathology at the peak model fit. Data are presented using logarithmic axes and shaded areas around the regression lines are the 95% confidence intervals. HIP = CA1 field.
Snca expression shapes the spread of pathologic alpha-synuclein
One cell-autonomous factor that may render cells more vulnerable is Snca gene expression.43,44 In our model, Snca expression determines the synthesis rate of alpha-synuclein agents. We tested whether Snca expression shaped the spread of pathology by comparing the peak fits to those obtained in sets of null models in which gene expression was randomized. For every injection site and post-injection time point, randomization led to a substantial fit disruption between modelled and measured pathologic alpha-synuclein patterns (Fig. 5A and Supplementary Table 3). In other words, the distribution of pathology simulated by the model correlated with the distribution of pathology observed by immunohistochemistry; however, when randomizing the expression of Snca used by the model, the simulated distribution of pathology no longer correlated with the observed distribution of pathology, suggesting that the spatial pattern of Snca expression is needed to model pathology spread.
Figure 5.
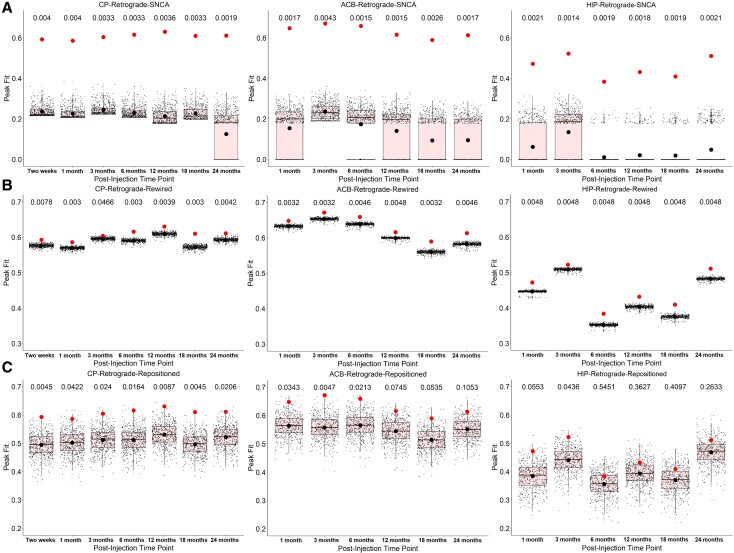
Snca expression and connection topology shape the spread of pathologic alpha-synuclein. Null models were generated by either randomizing (A) Snca expression, (B) the connection topology (‘rewired’) or (C) the physical position of regions (‘repositioned’) of the retrograde network. The red dots represent the original peak fits, and the black dots represent the average null peak fits. Unbiased Monte Carlo estimates of the exact P-values are reported above the box plot. HIP = CA1 field.
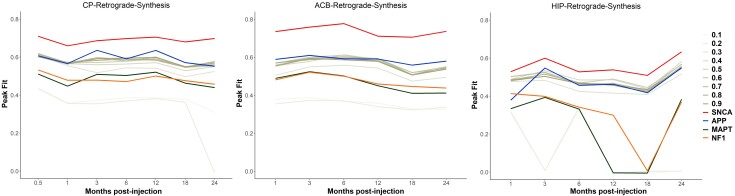
In addition, when replacing the Snca expression by the expression of genes involved in other pathological entities, namely App, Mapt or Nf1, coding for proteins involved in amyloidopathies, tauopathies and neurofibromatosis, respectively,34–36 the peak fits were also lower (Fig. 6 and Supplementary Table 4). Lower peak fits were also observed when replacing the real Snca expression with homogenous values from 0.1 to 0.9 (Fig. 6 and Supplementary Table 4). These findings demonstrate that regional Snca expression is involved in the spread of pathologic alpha-synuclein.
Figure 6.
The effect of synthesis rate on the spread of pathologic alpha-synuclein. Scatterplots showing the original peak fits obtained using Snca expression as the synthesis rate (red lines) versus the peak fits obtained when replacing Snca expression by either the expression of control genes, namely App, Mapt or Nf1 (blue, green, and orange lines), or by homogeneous synthesis rates (tones of grey). HIP = CA1 field.
Connectome topology shapes the spread of pathologic alpha-synuclein
There is some evidence that progression of brain pathology in Parkinson’s disease is mediated by synaptic connectivity, as demonstrated by alpha-synuclein preformed fibril injection in mice15,45 or when recreating MRI-derived atrophy patterns of Parkinson’s disease in humans.10,12 To investigate the role of connectivity in shaping pathologic alpha-synuclein spread, we randomized the connectome’s topology or geometry in two separate ways, either through rewired null networks by swapping pairs of edges or, since connectivity is part of an embedded system constrained by physical laws,46 through repositioned null networks by swapping the physical positions of the nodes. We found that rewired null networks always led to lower peak fits between simulated and true pathology patterns (Fig. 5B and Supplementary Table 3), whereas repositioned null networks disrupted the fit at every time point for the CP, at the earlier time points for the ACB, and at none of the time points for HIP (Fig. 5C and Supplementary Table 3). It is notable that the disruption was more modest when randomizing the connectome than Snca expression. Nonetheless, these findings indicate that the connectome is involved in the pathologic alpha-synuclein spread.
Resistant regions have weaker afferent projections and lower gene expression
Despite simulating the spread in vivo, the model overestimated pathologic alpha-synuclein in some regions. These resistant nodes were in the thalamus, with over 30 of its 35 regions showing overestimation of pathology, followed by the pallidum, hypothalamus, midbrain and pons (Supplementary Table 5). For the regions that were predicted to develop pathology, we compared the characteristics of those that remained resistant with those that developed pathology, and observed lower average incoming connectivity (3.81 versus 14.03, P < 0.001 for the CP; 3.41 versus 10.36, P = 0.001 for the ACB; 4.67 versus 14.10, P = 0.005 for the HIP) and Snca expression (4.22 versus 7.96, P < 0.001 for the CP; 3.50 versus 9.60, P < 0.001 for the ACB; 5.25 versus 13.19, P < 0.001 for the HIP) in resistant regions (Supplementary Table 6), which is consistent with the model’s premises. Note however that the model may fail to account for a threshold effect that might exist in vivo for the accumulation of pathologic alpha-synuclein. Also, regions may be resistant to pathologic alpha-synuclein accumulation for other unknown reasons.
Discussion
Previous studies have provided support for the prion-like spread of pathologic alpha-synuclein; however, the alpha-synuclein propagation model remains a matter of debate in the field. In this study, we generated a comprehensive histopathological dataset from non-transgenic mice followed over 24 months after injection of recombinant alpha-synuclein preformed fibrils into the CP, ACB or HIP and used the agent-based SIR model to recreate in silico the observed neuroanatomical distributions over time. Our observations demonstrate that pathologic alpha-synuclein initiated in different regions leads to distinct patterns of pathology in mice, which can be recreated in silico using our model. Because the pathology was observed in wild-type mice after injection of alpha-synuclein fibrils, the most plausible explanation is that misfolded alpha-synuclein is pathogenic. In addition, by modelling the spread based on Snca expression and connectomics, we demonstrate that these factors are sufficient, and therefore central, for the spread of pathology in animals.
First, introducing pathologic alpha-synuclein at different initiation sites generates distinct patterns of pathology. This recapitulates numerous findings on the propagation of pathologic alpha-synuclein15,17,47–52 and agrees with the distribution patterns of pathology that differ according to the injection site.15,45,47,48,53 In this study, seeding pathologic alpha-synuclein from the ACB or the HIP led to a strikingly reduced survival rate compared to the CP, reminiscent of the clinical heterogeneity observed in human synucleinopathies. The mechanisms underlying this are unclear, but different pathologic alpha-synuclein conformations have been shown to target distinct regions and cell types in animal models.54–57 Nonetheless, our study provides a more complex picture whereby divergent clinical outcomes can be achieved by inoculating an identical dose of the same pathologic alpha-synuclein preparation into different sites. Compared to previous studies that used a more limited number of regions, we generated a comprehensive quantification of pathologic alpha-synuclein sampled over the 426 regions of the Allen Mouse Brain Atlas. Also, whereas previous models quantified the progression of pathology over 18 months and two to three intermediary time points, we studied mice from 2 weeks to 24 months post-injection, with six to seven intermediate time points. While different experimental protocols were used in previous studies, we used the same initiating dosage of preformed fibrils for every cohort, ensuring a better comparability of findings.
Second, the agent-based SIR model recreates in silico the neuroanatomical distributions of pathologic alpha-synuclein in mice. Previous computational models of alpha-synuclein spread in the mouse brain generally relied on connectivity or spatial proximity.25,26 Although important, the spread of pathology does not solely conform to the connectivity map and cell-autonomous factors also shape propagation.45 Here, both gene expression and connectivity data were inserted simultaneously into an agent-based framework to model the dynamics occurring at the level of the individual alpha-synuclein agent. Agent-based SIR models allow the identification of factors that contribute to the dynamics of complex epidemics upon networks and have proven successful in infectious disease epidemiology.27,58 The prion-like hypothesis of neurodegenerative disease suggests that abnormal proteins have the properties of transmissibility and propagation, making SIR models appealing. The validation of our SIR model in preformed fibril-injected mice now further strengthens its validity as a tool to investigate synucleinopathies.
Third, some regions are more prone to pathology due to local cell-autonomous factors.43,44 One such factor is Snca gene expression, which may predispose a cell to increased accumulation of alpha-synuclein. We tested this hypothesis by showing that randomizing Snca expression reduced the correlation between observed and simulated pathology by 60–70%. The contribution of Snca expression is in line with previous studies showing that some cell types appear more vulnerable to showing pathologic alpha-synuclein.40,45 In fact, although the factors underlying selective vulnerability remain to be elucidated, the kinetics of pathology formation appear largely determined by the availability of intraneuronal alpha-synuclein.40 This is supported by primary cultures showing that a higher alpha-synuclein expression level associates with a higher propensity to seed exogenous alpha-synuclein fibrils.59 Also, the early knockdown of alpha-synuclein through antisense-oligonucleotide treatment induces a significant reduction in pathological burden in the presence of alpha-synuclein preformed fibrils and an increase in neuronal viability.40,41 In a previous computational model, the pathology patterns were recreated better when the connectivity-based diffusion of alpha-synuclein was weighted by Snca expression.26 The impact of Snca expression on the development of pathologic alpha-synuclein in humans is further supported by the dose-dependent association of Snca gene multiplication with early-onset Parkinson’s disease.60–64 In post-mortem brains with neocortical Lewy body disease, the expression of physiological alpha-synuclein also appeared lower in regions without pathology.65 Moreover, the previous use of the agent-based SIR model to recreate the atrophy seen in patients with Parkinson’s disease supported that Snca expression significantly shaped the spread.12 It is notable that both pathologic alpha-synuclein accumulation, as modelled here, and brain atrophy, as modelled in Zheng et al.12 are replicated by the SIR model and appear to depend on Snca expression. This suggests a close relationship between pathologic alpha-synuclein accumulation and neurodegeneration, although we did not directly test this association here.
Fourth, the brain’s connectivity pattern and spatial embedding also shape the spread of pathologic alpha-synuclein. This is in line with the transneuronal spread of alpha-synuclein15,25,26,47,48 and the spatiotemporal distribution of Lewy pathology observed at post-mortem examination in Parkinson’s disease.4,5,66 The randomization of spatial properties disrupted the spread, suggesting that both topology and geometry constrain the pathologic alpha-synuclein propagation. However, the decrease in fit was around 10−20% (i.e. lower than for Snca expression), highlighting that brain connectivity is not the sole contributor to pathologic alpha-synuclein distribution and that region-intrinsic factors are also involved in the resilience of some regions to pathological spread. In line with this, regions that were predicted to show pathology by the model, but that had less pathology in vivo (i.e. thalamus, pallidum, hypothalamus, midbrain, pons), had lower incoming connectivity and Snca expression compared to regions that had pathology in vivo. This may suggest the existence of a threshold effect below which alpha-synuclein arrival in a region does not lead to Lewy pathology, although other protective regional factors may also be involved in this resilience. As mentioned, the agent-based SIR model used here was previously demonstrated to accurately recreate the atrophy pattern measured with MRI in people with Parkinson’s disease.12 In humans, the simulated outcome was tissue loss, while in the mouse model used here, it was the accumulation of pathologic alpha-synuclein. In both cases, connectivity and alpha-synuclein concentration were shown to shape the pattern of disease. This concordance between mouse and human experiments supports the hypothesis that pathologic alpha-synuclein causes tissue loss in Parkinson’s disease. With this model, new potential mechanisms other than Snca expression and connectomics can eventually be tested; in this study, we focused on Snca expression and connectomics because these factors were previously reported to influence the spread and because the immediate goal of this study was to validate the agent-based model using comprehensive pathological datasets. In future studies, the agent-based model can be used to generate hypotheses computationally that will be tested empirically in the laboratory setting to better understand the pathological mechanisms underlying synucleinopathies.
One limitation of this study is that the model used mesoscale gene expression and connectivity information. Undoubtedly, the resilience of some brain regions to alpha-synuclein spread may be due to factors related to cell subpopulations inside each region.40,45 Once information about cell type distribution becomes available, this could be incorporated in the model. Another limitation is that we did not model cell or synaptic loss, despite signs of neuron loss in regions such as the basolateral amygdala and dentate gyrus. The increment in the number of infected agents therefore occurred without accounting for local changes that may interfere with the spread. However, although some studies reported neuron loss following preformed fibril-induced pathologic alpha-synuclein, this has not been universally observed and more knowledge is needed before implementing this process into the model.15,26,40 Finally, several other factors influence pathologic alpha-synuclein spread, such as alpha-synuclein strains67 and comorbid pathologies.68 These could benefit from being modelled to understand the mechanisms leading to different pathological and clinical trajectories, although the level of detail embedded in an agent-based model, if too complex, may lead to over-fitting.69
In sum, alpha-synuclein preformed fibrils inoculated in different brain regions lead to unique patterns of pathology that can be recreated with the agent-based SIR model. This model represents a powerful tool to understand the mechanisms underlying pathologic alpha-synuclein spread and test hypotheses about therapeutic mechanisms that may combat neurodegeneration in synucleinopathies.
Supplementary Material
Acknowledgements
We thank Jacob W. Vogel for providing comments on the manuscript.
Glossary
Abbreviations
- ACB
nucleus accumbens
- CP
caudoputamen
- HIP
hippocampus
- SIR
Susceptible-Infected-Removed
Funding
This work was supported by National Institutes of Health grant NS088322 to K.C.L. S.R. was supported by a fellowship from the Fonds de recherche du Québec - Santé (FRQS). H.Y. was supported by a fellowship from Yonsei University. A.D. declares funding from the Canadian Institutes of Health Research (CIHR), Michael J. Fox Foundation, The W. Garfield Weston Foundation and Healthy Brain for Healthy Lives initiative. The authors have no conflicts of interest to declare that are relevant to the content of this article.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Dickson DW, Braak H, Duda JE, et al. Neuropathological assessment of Parkinson’s disease: Refining the diagnostic criteria. Lancet Neurol. 2009;8(12):1150–1157. [DOI] [PubMed] [Google Scholar]
- 2. Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. [DOI] [PubMed] [Google Scholar]
- 3. Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9(1):13–24. [DOI] [PubMed] [Google Scholar]
- 4. Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24(2):197–211. [DOI] [PubMed] [Google Scholar]
- 5. Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318(1):121–134. [DOI] [PubMed] [Google Scholar]
- 6. Brundin P, Melki R. Prying into the prion hypothesis for Parkinson’s disease. J Neurosci. 2017;37(41):9808–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jucker M, Walker LC. Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci. 2018;21(10):1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson’s disease. Nat Med. 2008;14(5):504–506. [DOI] [PubMed] [Google Scholar]
- 9. Li JY, Englund E, Holton JL, et al. Lewy bodies in grafted neurons in subjects with Parkinson’s disease suggest host-to-graft disease propagation. Nat Med. 2008;14(5):501–503. [DOI] [PubMed] [Google Scholar]
- 10. Yau Y, Zeighami Y, Baker TE, et al. Network connectivity determines cortical thinning in early Parkinson’s disease progression. Nat Commun. 2018;9(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeighami Y, Ulla M, Iturria-Medina Y, et al. Network structure of brain atrophy in de novo Parkinson’s disease. Elife. 2015;4:e08440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng YQ, Zhang Y, Yau Y, et al. Local vulnerability and global connectivity jointly shape neurodegenerative disease propagation. PLoS Biol. 2019;17(11):e3000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu B, Fang F, Pedersen NL, et al. Vagotomy and Parkinson disease: A Swedish register-based matched-cohort study. Neurology. 2017;88(21):1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Svensson E, Horvath-Puho E, Thomsen RW, et al. Vagotomy and subsequent risk of Parkinson’s disease. Ann Neurol. 2015;78(4):522–529. [DOI] [PubMed] [Google Scholar]
- 15. Luk KC, Kehm V, Carroll J, et al. Pathological alpha-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338(6109):949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luk KC, Kehm VM, Zhang B, O’Brien P, Trojanowski JQ, Lee VM. Intracerebral inoculation of pathological alpha-synuclein initiates a rapidly progressive neurodegenerative alpha-synucleinopathy in mice. J Exp Med. 2012;209(5):975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masuda-Suzukake M, Nonaka T, Hosokawa M, et al. Prion-like spreading of pathological alpha-synuclein in brain. Brain. 2013;136(Pt 4):1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watts JC, Giles K, Oehler A, et al. Transmission of multiple system atrophy prions to transgenic mice. Proc Natl Acad Sci USA. 2013;110(48):19555–19560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uemura N, Yagi H, Uemura MT, Hatanaka Y, Yamakado H, Takahashi R. Inoculation of alpha-synuclein preformed fibrils into the mouse gastrointestinal tract induces Lewy body-like aggregates in the brainstem via the vagus nerve. Mol Neurodegener. 2018;13(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S, Kwon SH, Kam TI, et al. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron. 2019;103(4):627–641.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fearon C, Lang AE, Espay AJ. Reply to: ‘The logic and pitfalls of Parkinson’s as brain- versus body-first subtypes’. Mov Disord. 2021;36(3):786–787. [DOI] [PubMed] [Google Scholar]
- 22. Fearon C, Lang AE, Espay AJ. The logic and pitfalls of Parkinson’s disease as ‘brain-first’ versus ‘body-first’ subtypes. Mov Disord. 2021;36(3):594–598. [DOI] [PubMed] [Google Scholar]
- 23. Espay AJ, Vizcarra JA, Marsili L, et al. Revisiting protein aggregation as pathogenic in sporadic Parkinson and Alzheimer diseases. Neurology. 2019;92(7):329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raj A, Powell F. Models of network spread and network degeneration in brain disorders. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3(9):788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mezias C, Rey N, Brundin P, Raj A. Neural connectivity predicts spreading of alpha-synuclein pathology in fibril-injected mouse models: involvement of retrograde and anterograde axonal propagation. Neurobiol Dis. 2020;134:104623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henderson MX, Cornblath EJ, Darwich A, et al. Spread of alpha-synuclein pathology through the brain connectome is modulated by selective vulnerability and predicted by network analysis. Nat Neurosci. 2019;22(8):1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newman MEJ. Networks. 2nd edn. Oxford University Press; 2018:xi, 780 pages. [Google Scholar]
- 28. Fujiwara H, Hasegawa M, Dohmae N, et al. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4(2):160–164. [DOI] [PubMed] [Google Scholar]
- 29. Volpicelli-Daley LA, Luk KC, Lee VM. Addition of exogenous alpha-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous alpha-synuclein to Lewy body and Lewy neurite-like aggregates. Nat Protoc. 2014;9(9):2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang B, Kehm V, Gathagan R, et al. Stereotaxic targeting of alpha-synuclein pathology in mouse brain using preformed fibrils. Methods Mol Biol. 2019;1948:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waxman EA, Giasson BI. Characterization of kinases involved in the phosphorylation of aggregated alpha-synuclein. J Neurosci Res. 2011;89(2):231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pavlidis P, Noble WS. Matrix2png: A utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. [DOI] [PubMed] [Google Scholar]
- 33. Oh SW, Harris JA, Ng L, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508(7495): 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283(44):29615–29619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strang KH, Golde TE, Giasson BI. MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab Invest. 2019;99(7):912–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rasmussen SA, Friedman JM. NF1 gene and neurofibromatosis 1. Am J Epidemiol. 2000;151(1):33–40. [DOI] [PubMed] [Google Scholar]
- 37. Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;52(3):1059–1069. [DOI] [PubMed] [Google Scholar]
- 38. Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296(5569):910–913. [DOI] [PubMed] [Google Scholar]
- 39. Attems JToledo JB, Walker L, et al. Neuropathological consensus criteria for the evaluation of Lewy pathology in post-mortem brains: A multi-centre study. Acta Neuropathol. 2021;141(2):159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luna E, Decker SC, Riddle DM, et al. Differential alpha-synuclein expression contributes to selective vulnerability of hippocampal neuron subpopulations to fibril-induced toxicity. Acta Neuropathol. 2018;135(6):855–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cole Tracy A, Zhao Hien, Collier Timothy J., et al. α-Synuclein antisense oligonucleotides as a disease-modifying therapy for Parkinson’s disease. JCI Insight 2021;6. ( 10.1172/jci.insight.135633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Volpicelli-Daley LA, Luk KC, Patel TP, et al. Exogenous alpha-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron. 2011;72(1):57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gonzalez-Rodriguez P, Zampese E, Surmeier DJ. Selective neuronal vulnerability in Parkinson’s disease. Prog Brain Res. 2020;252:61–89. [DOI] [PubMed] [Google Scholar]
- 44. Surmeier DJ, Obeso JA, Halliday GM. Selective neuronal vulnerability in Parkinson disease. Nat Rev Neurosci. 2017;18(2):101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henrich MT, Geibl FF, Lakshminarasimhan H, et al. Determinants of seeding and spreading of alpha-synuclein pathology in the brain. Sci Adv. 2020;6(46):eabc2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stiso J, Bassett DS. Spatial embedding imposes constraints on neuronal network architectures. Trends Cogn Sci. 2018;22(12):1127–1142. [DOI] [PubMed] [Google Scholar]
- 47. Rey NL, George S, Steiner JA, et al. Spread of aggregates after olfactory bulb injection of alpha-synuclein fibrils is associated with early neuronal loss and is reduced long term. Acta Neuropathol. 2018;135(1):65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rey NL, Steiner JA, Maroof N, et al. Widespread transneuronal propagation of alpha-synucleinopathy triggered in olfactory bulb mimics prodromal Parkinson’s disease. J Exp Med. 2016;213(9):1759–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Recasens A, Dehay B, Bove J, et al. Lewy body extracts from Parkinson disease brains trigger alpha-synuclein pathology and neurodegeneration in mice and monkeys. Ann Neurol. 2014;75(3):351–362. [DOI] [PubMed] [Google Scholar]
- 50. Peelaerts W, Bousset L, Van der Perren A, et al. alpha-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature. 2015;522(7556): 340–344. [DOI] [PubMed] [Google Scholar]
- 51. Paumier KL, Luk KC, Manfredsson FP, et al. Intrastriatal injection of pre-formed mouse alpha-synuclein fibrils into rats triggers alpha-synuclein pathology and bilateral nigrostriatal degeneration. Neurobiol Dis. 2015;82:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mougenot AL, Nicot S, Bencsik A, et al. Prion-like acceleration of a synucleinopathy in a transgenic mouse model. Neurobiol Aging. 2012;33(9):2225–2228. [DOI] [PubMed] [Google Scholar]
- 53. Shen Y, Yu WB, Shen B, et al. Propagated alpha-synucleinopathy recapitulates REM sleep behaviour disorder followed by parkinsonian phenotypes in mice. Brain. 2020;143(11):3374–3392. [DOI] [PubMed] [Google Scholar]
- 54. Lau A, So RWL, Lau HHC, et al. alpha-Synuclein strains target distinct brain regions and cell types. Nat Neurosci. 2020;23(1):21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo JL, Covell DJ, Daniels JP, et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell. 2013;154(1):103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Candelise N, Schmitz M, Llorens F, et al. Seeding variability of different alpha synuclein strains in synucleinopathies. Ann Neurol. 2019;85(5):691–703. [DOI] [PubMed] [Google Scholar]
- 57. Bousset L, Pieri L, Ruiz-Arlandis G, et al. Structural and functional characterization of two alpha-synuclein strains. Nat Commun. 2013;4:2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brockmann D, Helbing D. The hidden geometry of complex, network-driven contagion phenomena. Science. 2013;342(6164):1337–1342. [DOI] [PubMed] [Google Scholar]
- 59. Courte J, Bousset L, Boxberg YV, Villard C, Melki R, Peyrin JM. The expression level of alpha-synuclein in different neuronal populations is the primary determinant of its prion-like seeding. Sci Rep. 2020;10(1):4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chartier-Harlin MC, Kachergus J, Roumier C, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364(9440):1167–1169. [DOI] [PubMed] [Google Scholar]
- 61. Ibanez P, Bonnet AM, Debarges B, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364(9440):1169–1171. [DOI] [PubMed] [Google Scholar]
- 62. Konno T, Ross OA, Puschmann A, Dickson DW, Wszolek ZK. Autosomal dominant Parkinson’s disease caused by SNCA duplications. Parkinsonism Relat Disord. 2016;22(Suppl 1):S1–S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Muenter MD, Forno LS, Hornykiewicz O, et al. Hereditary form of parkinsonism–dementia. Ann Neurol. 1998;43(6):768–781. [DOI] [PubMed] [Google Scholar]
- 64. Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646): 841. [DOI] [PubMed] [Google Scholar]
- 65. Erskine D, Patterson L, Alexandris A, et al. Regional levels of physiological alpha-synuclein are directly associated with Lewy body pathology. Acta Neuropathol. 2018;135(1):153–154. [DOI] [PubMed] [Google Scholar]
- 66. Adler CH, Beach TG, Zhang N, et al. unified staging system for lewy body disorders: clinicopathologic correlations and comparison to braak staging. J Neuropathol Exp Neurol. 2019;78(10):891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rey NL, Bousset L, George S, et al. alpha-Synuclein conformational strains spread, seed and target neuronal cells differentially after injection into the olfactory bulb. Acta Neuropathol Commun. 2019;7(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bassil F, Brown HJ, Pattabhiraman S, et al. Amyloid-beta (Aβ) plaques promote seeding and spreading of alpha-synuclein and tau in a mouse model of lewy body disorders with Aβ pathology. Neuron. 2020;105(2):260–275.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaul H, Ventikos Y. Investigating biocomplexity through the agent-based paradigm. Brief Bioinform. 2015;16(1):137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pathological datasets and the Python code for running the agent-based SIR model in the mouse brain are available at https://github.com/srahayel.