Abstract
Background
Mosquito-borne viruses pose a serious threat to humans worldwide. There has been an upsurge in the number of mosquito-borne viruses in Europe, mostly belonging to the families Togaviridae, genus Alphavirus (Sindbis, Chikungunya), Flaviviridae (West Nile, Usutu, Dengue), and Peribunyaviridae, genus Orthobunyavirus, California serogroup (Inkoo, Batai, Tahyna). The principal focus of this study was Inkoo (INKV) and Sindbis (SINV) virus circulating in Norway, Sweden, Finland, and some parts of Russia. These viruses are associated with morbidity in humans. However, there is a knowledge gap regarding reservoirs and transmission. Therefore, we aimed to determine the prevalence of INKV and SINV in blood sucking insects and seroprevalence for INKV in semi-domesticated Eurasian tundra reindeer (Rangifer tarandus tarandus) in Norway.
Materials and methods
In total, 213 pools containing about 25 blood sucking insects (BSI) each and 480 reindeer sera were collected in eight Norwegian reindeer summer pasture districts during 2013–2015. The pools were analysed by RT-PCR to detect INKV and by RT-real-time PCR for SINV. Reindeer sera were analysed for INKV-specific IgG by an Indirect Immunofluorescence Assay (n = 480, IIFA) and a Plaque Reduction Neutralization Test (n = 60, PRNT).
Results
Aedes spp. were the most dominant species among the collected BSI. Two of the pools were positive for INKV-RNA by RT-PCR and were confirmed by pyrosequencing. The overall estimated pool prevalence (EPP) of INKV in Norway was 0.04%. None of the analysed pools were positive for SINV. Overall IgG seroprevalence in reindeer was 62% positive for INKV by IIFA. Of the 60 reindeer sera- analysed by PRNT for INKV, 80% were confirmed positive, and there was no cross-reactivity with the closely related Tahyna virus (TAHV) and Snowshoe hare virus (SSHV).
Conclusion
The occurrence and prevalence of INKV in BSI and the high seroprevalence against the virus among semi-domesticated reindeer in Norway indicate that further studies are required for monitoring this virus. SINV was not detected in the BSI in this study, however, human cases of SINV infection are yearly reported from other regions such as Rjukan in south-central Norway. It is therefore essential to monitor both viruses in the human population. Our findings are important to raise awareness regarding the geographical distribution of these mosquito-borne viruses in Northern Europe.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12985-022-01815-0.
Keywords: Arbovirus, Blood sucking insects (BSI), Mosquito, Reindeer, INKV, SINV, Prevalence, Estimated pooled prevalence, IIFA, Seroprevalence
Introduction
Mosquito-borne viruses emerge as a group that poses a serious threat to human health worldwide [1]. There has been an upsurge in the number of mosquito-borne viruses in Europe mostly belonging to the families Togaviridae (Sindbis virus SINV, Chikungunya virus), Flaviviridae (West Nile virus, Dengue virus), and Peribunyaviridae (Inkoo virus INKV, Batai virus BATV, Tahyna virus TAHV) [2, 3].
Surveillance of mosquito-borne viruses and their prevalence in the human and animal populations is highly required, since INKV, SINV, Chikungunya virus and West Nile virus infections often are not reported and remain undiagnosed [2]. Among other mosquito-borne viruses, INKV and SINV are known to be circulating in Norway, Sweden [1, 4, 5], Finland, and some parts of Russia [6–9]. In humans, INKV causes mild fever to fatal encephalitis while SINV causes arthritis and rashes [10, 11].
INKV, a member of the California serogroup together with TAHV and Snowshoe hare virus (SSHV), is an enveloped virus belonging to the genus Orthobunyavirus in the Peribunyaviridae family with a tri-segmented, negative-sense, single-stranded (ss) 12.4-kb (kilo-base pair) RNA genome [12]. Phylogenetically, INKV is closely related to Jamestown Canyon Virus (JCV) found in the US [13, 14]. INKV has been detected previously in adult Aedes (Ae.) communis, Ae. hexodontus and Ae. punctor mosquitoes, and in Ae. communis larvae, although there is a necessity for identification of mosquito vectors capable of transmitting INKV [7, 15–17]. INKV has been found to be circulating in Northern Europe, including Norway [2, 16]. Cattle (Bos taurus), reindeer (Rangifer tarandus), moose (Alces alces), red fox (Vulpes vulpes) and snow hare (Lepus timidus), are considered to be major vertebrate hosts of INKV besides humans [6].
SINV is a member of the Western equine encephalomyelitis virus complex. It is an enveloped, positive sense, 11.7-kb ssRNA virus belonging to the genus Alphavirus in the Togaviridae family. Wild birds are considered as virus reservoir [18]. Antibodies against SINV have been detected in migratory and residential birds [19, 20]. Ornithophilic mosquitoes (mosquitoes that feed on birds) of the genera Culex (Cx.), and Culiseta as well as Ochlerotatus spp., and Aedes spp. are considered to be the major SINV vectors [2]. Experimental infection studies implicate Cx. torrentium as a competent vector for SINV in northern Sweden [21–23]. The vector species of SINV in Sweden prefer lowland forested wetlands and humid forests with deciduous and coniferous trees as habitats [19]. SINV causes human arthritic diseases [24, 25], known by various names in Fennoscandia; “Bærplukkersyken” in Norway [26], Ockelbo disease in Sweden [27], Karelian fever in Russia [28], and Pogosta disease in Finland [6].
Both INKV and SINV have been isolated from diverse species of mosquitoes including Ae. cinereus, Cx. torrentium, Cx. pipiens, Culiseta morsitans, Ae. Communis, and Ae. punctor [8, 29]. Moreover, evidence pointing to vertical transmission of these viruses in vectors has been demonstrated through detection of viral RNA in mosquito larvae [1]. In addition, SINV has been suggested to overwinter in Cx. pipiens mosquitoes [30]. Antibodies against INKV and SINV have been detected in human populations in Sweden and Finland [31–33]. In Russia, chronic neurological disease was reported in patients with antibodies to INKV [34]. Acute human INKV infections in association with encephalitis have been reported in Finland [11].
In Norway, INKV and SINV have not been studied extensively despite findings of viruses in humans and mosquitoes in 1978 and 1992 [7, 35, 36]. There is a lack of knowledge covering the distribution and prevalence of these viruses in BSI vectors, humans, potential reservoir species, and the regions affected. Other California Encephalitis (CE) group viruses now categorized as California serogroup viruses have previously been isolated in Norway, but the methods used to identify them were not specific enough to confirm if it was INKV or related viruses [16]. There are several cases of encephalitis reported to Norwegian Surveillance System for Communicable Diseases (MSIS) with unknown aetiology [26]. These may be associated with INKV or other vector-borne pathogens. SINV cases are not reported to the MSIS but about 15–20 clinical cases have been diagnosed since 1982.
A recent study indicated the need of monitoring INKV in vertebrate hosts such as moose and reindeer to identify the possible risk of transmission to humans [29]. The present study aimed to provide new knowledge on the prevalence and distribution of INKV and SINV in blood sucking insects, and to investigate their exposure in semi-domesticated reindeer in Norway. The combined analysis of blood sucking insects and reindeer serum samples collected on the reindeer summer pastures provides an indication of the distribution of these viruses in Norway, which could be addressed further and evaluated as potential causes of human encephalitis in Norway.
Materials and methods
Study area and collection of samples
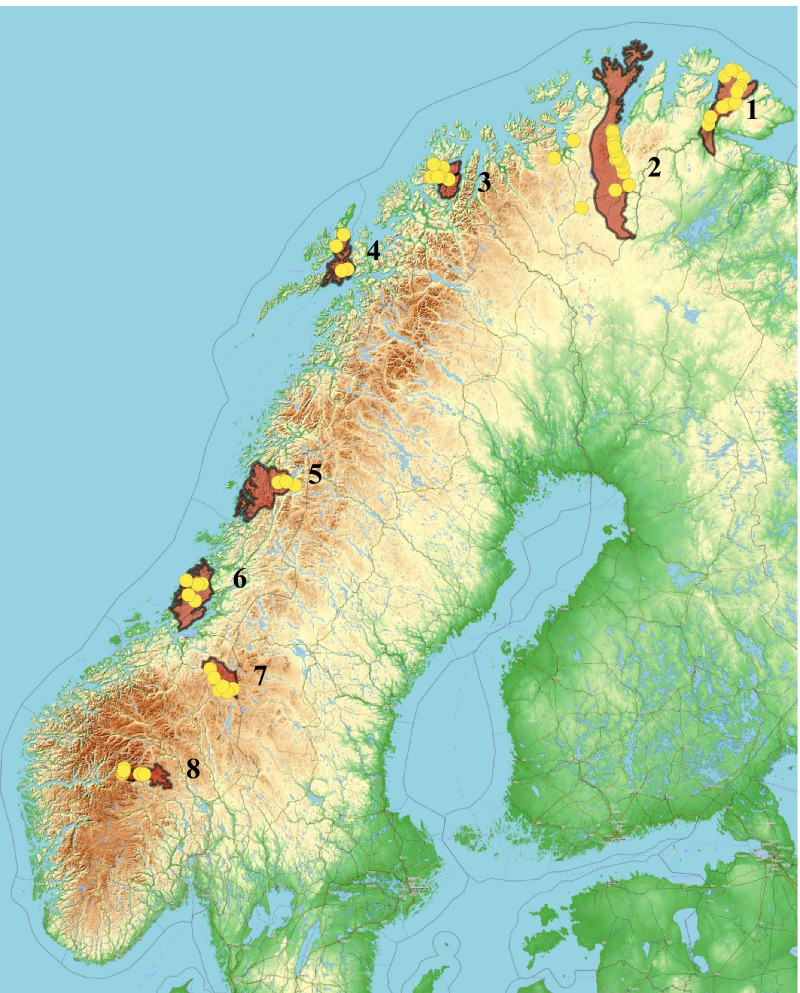
Blood sucking insects
Mosquitoes and midges, hereafter named blood sucking insects (BSI), were collected by The Arctic University of Norway (UiT) as a part of the research project Climate and Reindeer Diseases (CARD), during the summer season between July and August each year from 2013 to 2015. BSI were caught with a mosquito trap (Mosquito Magnet Independence; Woodstream® Corporation, Pennsylvania, USA), using propane gas to produce carbon dioxide (CO2) and a biting insect attractant (Mosquito Magnet Octenol, Woodstream® Corporation). The trap was set to run for two to 24 h depending on the abundance of BSI and weather conditions. The mosquito trap attracts all kinds of BSI and were collected for several research purpose. To cover a variety of different BSI species the sampling was performed at four to 24 multiple sites from each of the eight reindeer summer pasture locations and whenever possible, each site was visited twice during the summer season (Table 1, Fig. 1). One of the main purposes of sampling BSI for this study was to test if there were any mosquito-borne viruses, like INKV and SINV, within these pools. BSI were killed by freezing and stored at − 20 °C from 2 days and up to 2 weeks after capture, until a − 80 °C freezer was available. The BSI were collected in cryotubes (1.8 ml, Thermo Fisher Scientific, Rochester, NY, USA) with appr. 25 insects per tube from each site. One pool from each site were analyzed for detection of INKV and SINV.
Table 1.
An overview of selected summer pastures for semi-domesticated reindeer (Rangifer tarandus tarandus)
| County | Locations | UTM-coordinates* | |
|---|---|---|---|
| Latitude (S–N) | Longitude (E–W) | ||
| Troms and Finnmark | Tana | 35 W 0573349 | 7820819 |
| Troms and Finnmark | Lakselv | 35 W 0441724 | 707695 |
| Troms and Finnmark | Tromsø | 34 W 0429573 | 7738767 |
| Nordland | Lødingen | 33 W 0540824 | 7589369 |
| Nordland | Hattfjelldal | 33 W 0458873 | 7281479 |
| Trøndelag | Fosen | 32 W 0568340 | 7129347 |
| Trøndelag | Røros | 32 V 0619596 | 6966550 |
| Innlandet | Valdres | 32 V 0490781 | 6794290 |
Serum samples were collected during the following winter seasons (November–April) of 2013–2014, 2014–2015, and 2015–2016. Blood sucking insects (BSI) were collected from the reindeer summer pastures during the summer seasons (July–August) each year from 2013–2014, 2014–2015, and 2015–2016
*UTM-coordinates Universal Transverse Mercator coordinates
Fig. 1.
The map illustrates the sampling locations where the blood sucking insects (BSI) and reindeer sera were collected. 1: Tana, 2: Lakselv, 3: Tromsø, 4: Lødingen, 5: Hattfjelldal, 6: Fosen, 7: Røros, and 8: Valdres. The brown areas represent the summer pastures for each of the reindeer herds, whereas the yellow dots indicate the sampling locations for blood sucking insects (BSI) in or in the vicinity of each summer pasture
Reindeer serum samples
Blood samples were collected from 480 semi-domesticated reindeer grazing on eight different summer pastures (Table 1, Fig. 1). Reindeer were sampled about 3–6 months after the BSI sampling, i.e., during the winter seasons (November–April) of 2013–2014, 2014–2015 and 2015–2016, with the exception of the Lødingen herd which was not sampled during the winter 2015–2016. Adult reindeer males were underrepresented compared to adult females due to restricted availability in the herd at the time of sampling. From live animals, blood was obtained by venepuncture of external jugular vein using a venoject needle (Terumo, Leuven, Belgium) and blood collecting tubes (BD Vacutainer®; BD, Plymouth, UK). From slaughtered animals, blood was collected directly in the blood collecting tubes when bled. Blood tubes were centrifuged at 3500 rpm for 10 min to collect the serum. Sera were stored at − 20 °C until analysis [37].
Extraction of total nucleic acid from BSI
Each of the 213 pools of BSI were transferred to 2 ml tubes containing six steel beads (MP Biomedical Life Science, CA, USA) and 350 μl of RLT® lysis buffer with β-mercaptoethanol in a ratio of 1:10 (RNeasy Mini Kit, QIAGEN Inc., Valencia, CA, USA). They were homogenized at 4 °C by FastPrep®-24 5G instrument (MP Biomedical Life Science, CA, USA) operated at a relative velocity of 4.0 m/s for 60 s. The homogenates were centrifuged at 20,817×g for 5 min to remove BSI debris. The total nucleic acid (TNA) was extracted with MagNA Pure LC 2.0 (software v 1.1.24) instrument and MagNA Pure LC Total Nucleic Acid High-Performance Isolation kit (Roche Diagnostics GmbH, Penzburg, Germany) according to the manufacturer’s protocol. The elution volume of TNA was 60 μl.
Detection of INKV by one-step RT-PCR
To detect INKV, a RT-PCR amplifying the fragment targeting the gene encoding the non-structural protein (nsp1) of the Lövånger strain of INKV (GenBank accession number KX554935) was carried out. TNA extracted from BSI were amplified by RT-PCR in a 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA) using SuperScript™ III One-Step RT-PCR System with Platinum™ Taq DNA polymerase kit (Invitrogen, Carlsbad, CA, USA). The amplification was carried out in a total volume of 25 µl per reaction, containing 20 µl mastermix and 5 µl extracted TNA template. The concentration of each primer and cycling conditions were modified from [1, 13, 38]. The concentrations of the INKV forward and reverse primers were 0.25 μM each. The optimized cycling conditions were: 55 °C for 30 min, 95 °C for 2 min followed by 45 cycles of 95 °C for 15 s, 60.5 °C for 30 s and 72 °C for 1 min, before a final extension at 72 °C for 5 min and 4 °C until the PCR products were further analysed. The PCR products were visualized on E-Gel® 48 Gels (2% agarose) with EtBr according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA) and stored at − 80 °C until further analysis. A tenfold serial endpoint dilution (10–1 to 10–4) of INKV positive controls and RNase free water as negative control were included in each test.
Detection of SINV by one-step RT-real-time PCR
Total nucleic acid from pooled BSI were analysed to detect SINV RNA by probe-based RT-real-time PCR, using qScript™ XLT One-step RT-real-time PCR ToughMix® kit (Quanta Biosciences, Gaithersburg, MD, USA) in a Rotor-gene 6000 (QIAGEN, Hilden, Germany). The primers and probe used for the amplification targeted the non-structural gene (nsp1) from the Ockelbo strain of SINV (GenBank accession number M69205.1) [1, 39]. The optimized cycling conditions for the reaction were: 55 °C for 30 min, 95 °C for 2 min followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 1 min, before a final extension at 72 °C for 5 min. The PCR products were stored at − 80 °C until further analysis.
Included on each plate was SINV RNA positive control, from the Lövånger strain, cultured in the lab, in a tenfold serial endpoint dilution (10–4 to 10–7) along with two RNase free water samples as negative control.
Pyrosequencing
All the PCR positive samples for INKV RNA were further analysed by Pyrosequencing [40], for sequence analysis (SQA) with the BioTage (Pyromark Q24) system (QIAGEN, Hilden, Germany). A tenfold serial endpoint dilution (10–1 to 10–4) of INKV positive controls and RNase free water as negative control were included in each test.
The positive controls were used as a standard to compare the sequences obtained from pyrosequencing of PCR positive samples. Samples with sequence similarity above 70% was regarded as positive in this study. Sequences were then aligned to previously identified INKV positive control and GenBank sequences using the Basic Local Alignment Search Tool (BLAST) provided by National Centre for Biotechnology Information.
Barcoding/speciation of BSI
Barcoding/speciation of BSI (10% of the total pool) was conducted on the same sample tube/pool that was subjected to the RT-PCR and pyrosequencing.
Amplification of the 710-bp fragment of the mitochondrial cytochrome c oxidase subunit I gene (COI) was performed. Briefly, amplification of cDNA from 10% of the total number of pools was performed using Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher Scientific) with universal DNA primers namely; LCO1490 and HCO2198 as described by [41]. For each reaction, 2 μl of template was used together with 10 μl of 2 × Phusion mix, 1.25 μl of both forward and reverse primers (10 pmol), 0.6 μl of DMSO and 4.9 μl of nuclease free water, up to a total reaction volume of 20 μl. Conditions for reactions were 98 °C for 30 s for initial denaturation. Further, amplification was performed using 35 cycles of: 98 °C for 7 s, 50 °C for 15 s and 72 °C for 20 s. Final extension was performed at 72 °C for 7 min. PCR product was analysed by gel electrophoresis using 1.2% agarose in 1 × TAE with GelRed (Biotium Inc. Hayward, CA, US) and later purified with ExoSAP-IT kit (Thermo Fisher Scientific) and sent to Eurofin Genomics (Germany) for Sanger sequencing. Sequences were then aligned to previously identified mosquito species in GenBank using the Basic Local Alignment Search Tool (BLAST) provided by National Centre for Biotechnology Information.
Calculation of prevalence
Two hundred and thirteen pools with approximately 25 BSI in each pool were analysed. The sample size was designed for other research purposes by the CARD project. Estimated pooled prevalence (EPP) of INKV was calculated by the Epitools Epidemiological calculators [42]. The estimated prevalence was assumed to be close to zero with 95% confidence interval (CI) using Method 2 [43]. This method utilizes the frequentists approach assuming a fixed pool size, and perfect (100%) test sensitivity and specificity for the estimation of prevalence and confidence limits [40].
Serological screening of reindeer samples
Indirect immunofluorescence assay (IIFA) for detection of IgG antibody against INKV
Slides were fixed with INKV antigen, a positive fluorescence signal from 40–50% of the cells was required for a positive result. The samples evaluated as borderline had a positive fluorescence signal from less than 40% of the cells [33]. Two INKV positive controls; anti-Tahyna mouse-ascites and INKV positive deer whole blood were included in each run.
Initially, mouse ascites (1:100), deer whole blood, and reindeer serum samples (1:20), were diluted in 1X PBS (Dulbecco’s solution A, pH = 7.4, Norwegian Institute of Public Health, Oslo, Norway).
Twenty μl of the diluted reindeer sera (n = 480) and the controls were analysed on slides and incubated at 37 °C for 1 h, and washed with cold PBS and water. The slides were stained with anti-mouse Alexa Fluor® 488 conjugate (Invitrogen Life Technology, Inc., Carlsbad, CA, USA) (dilution1:30) and anti-deer IgG fluorescein isothiocyanate conjugate (FITC) (KPL, Gaithersburg, MD, USA) (dilution 1:25) for 30 min at 37 °C and washed as described previously. Each slide was air dried, coated with glycerol before examining with a fluorescence microscope (Nikon Eclipse Ci, Nikon Corporation, Tokyo, Japan). The overall seroprevalence of IgG against INKV in reindeer sera was calculated as the percentage of the total number of positives of the total number of samples tested.
Cytopathic effect neutralization test (CPE-NT) from reindeer sera
Fifty-five reindeer sera samples evaluated as borderline from the IIFA analysis were further verified by the CPE-neutralization test at the Department of Virology, University of Helsinki as previously described [33].
Plaque reduction neutralization test (PRNT)
Reindeer sera (n = 60) with strongly positive (25), borderline (25), or negative (10) result by IIFA were confirmed with a PRNT to determine whether the IgG antibodies were specific for INKV, TAHV, or SSHV as previously described [32]. A 50% plaque reduction neutralization titer (PRNT50) was calculated as the reciprocal of the highest serum dilution based on 50% or greater reduction in the plaque counts. Samples with titre < 20 was considered negative and titres ≥ 20 were defined as INKV seropositive.
Statistical method for calculation of seroprevalence
All statistical analyses were performed by the program STATA Software v. 16 (Stata Corporation, College Station, TX, USA). The statistical difference between infection rate within each location were analysed by χ2 for each year. In addition, the interaction between infection, location, and year was analysed by logistic regression. In all analyses statistical significance was indicated by p < 0.05.
Result
Identification of blood sucking insects
Two-hundred and thirteen pools of BSI were visually identified with regards to species [44]. Besides Culicidae (mosquitoes), the BSI samples also contained Simuliidae (black flies), Culicoides (biting midges). Aedes spp. was the most common identified mosquito species with very few numbers of Culex spp. Of the mosquitoes identified were Ae. excrucians, Ae. punctor/hexodontus, Ae. communis, Ae. nigripes, Ae. impiger, Ae. cinereus, Ae. diantaeus, Ae. pionips, and Culiseta spp. that were most frequently observed from all different locations during the sampling period. Ae. punctor and/or hexodontus, Ae. diantaeus and/or Ae. impiger were the dominating species of Aedes mosquitoes in the BSI pools from the two INKV positive locations, Fosen and Røros. Similarly, Culiseta spp. could be identified from one sample from each of the two locations. Ae. excrucians and Ae. cinereus were only recorded in Røros.
In order to confirm the results from the visual identification barcoding analysis was performed. These results from randomly selected BSI samples (10%) indicated most of the identified pools as arthropods in addition to mosquitoes. Many of the pools contained mosquito species but other contained midges. The barcoding method only amplifies the most common, or best fit, not all insect species in the pools. Eight pools of BSI from Tromsø indicated that four of the pools contained a majority of midges Prosimulium hirtipes, two contained a majority of mosquitoes, Ae. communis, while the remaining two had a majority of fungus gnats Marcocera spp. and biting midges Culicoides chioptera. Similarly, eight pools from Lødingen demonstrated six pools of mosquitoes, four with Ae. punctor and/or Ae. hexodontus, two of Ae. communis, and two containing a majority of midges, P. hirtipes. One pool each from Fosen and Røros indicated a majority of the two midge's species, Simulium monticola and P. hirtipes, respectively.
Screening for INKV by one-step RT-PCR
Two out of 213 analysed pools of 25 BSI were positive for INKV by RT-PCR. The results from these two positive pools were further verified as INKV by pyrosequencing (Table 2). One of the positive pools had 88% and the other had 94% sequence similarity to the INKV positive controls (10–1 to 10–4). The two positive pools were from Fosen and Røros from 2014. From this, the overall EPP was calculated to be 0.04% (95% CI) by the Epitools Ausvet calculator [42], with a site EPP of 0.5%.
Table 2.
Analysis of blood sucking insects (BSI) for detection of INKV from eight different reindeer pastures in Norway by RT-PCR and pyrosequencing
| Locations | Number of pools of BSI analysed | Number of pools positive for INKV by RT-PCR | Number of pools positive by pyrosequencing | ||
|---|---|---|---|---|---|
| 2013–2014 | 2014–2015 | 2015–2016 | |||
| Tana | 5 | 9 | 8 | 0 | 0 |
| Lakselv | 12 | 26 | 18 | 0 | 0 |
| Tromsø | 1 | 8 | 7 | 0 | 0 |
| Lødingen | 8 | 8 | 8 | 0 | 0 |
| Hattfjelldal | 8 | 8 | 8 | 0 | 0 |
| Fosen | 8 | 8 | 8 | 1* | 1 |
| Røros | 8 | 8 | 8 | 1* | 1 |
| Valdres | 8 | 8 | 7 | 0 | 0 |
| Total = 213 | 2 | 2 | |||
Bold indicates the positive samples and they were collected in 2014
* = the two positive pools were collected in 2014
Screening for SINV by one-step RT-real-time PCR
None of the analysed pools of BSI were positive for SINV on RT-real-time PCR.
IgG against INKV in reindeer sera by indirect immunofluorescence assay
Sera from 480 reindeer, representing eight different reindeer pastures in Norway were analysed for detection of IgG antibodies against INKV by indirect immunofluorescence assay (IIFA). In total, 296 (62%) serum samples tested positive with INKV-IIFA.
IgG antibodies against INKV were detected in reindeer from all the eight different reindeer pastures of Norway (Fig. 2, Additional file 1: Table S1).
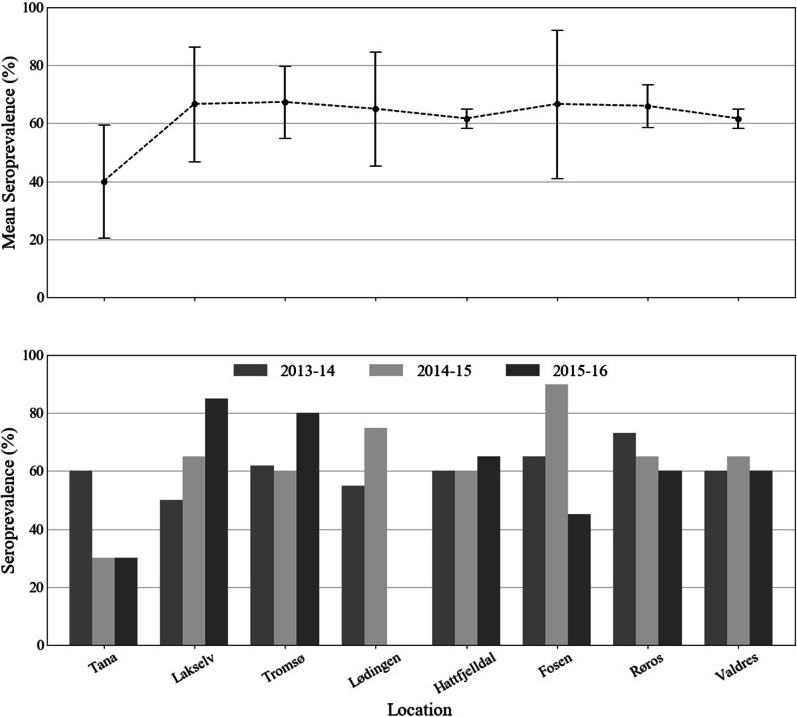
Fig. 2.
Seroprevalence (%) of INKV (IgG) in reindeer sera determined by IIFA from Tana, Lakselv, Tromsø, Lødingen, Hattfjelldal, Fosen, Røros, and Valdres from the winter seasons of 2013–2014, 2014–2015 and 2015–2016, indicated by bar graphs (bottom). Above each bar graph, the dots indicate the mean seroprevalence and the whiskers represent the confidence interval (95% CI) of each site throughout the year (2013–2014, 2014–2015, 2015–2016) (top)
The IIFA results illustrated a high average seroprevalence of 67% in Lakselv, Tromsø, and Fosen while the lowest average seroprevalence of 40% was in Tana. There was no statistically significant difference in seroprevalence among the eight different reindeer pastures for the season 2013–2014 (50–73%). However, there was a significant difference between the locations both in 2014–2015 (p = 0.011) and 2015–2016 (p = 0.006) (χ2 test, between eight locations, STATA). Fosen 2014–2015 had the highest seroprevalence reaching up to 90% with an OR of 6.0 (95% CI [1.1–33.3]) p > 0.04, while Tana (2014–2015) had the lowest seroprevalence of 30% with an OR of 0.2 (95% CI [0.1–0.9]) p > 0.03 (logistic regression analysis, STATA). Similarly, for the winter season 2015–2016, 30% of the reindeer samples from Tana were seropositive and the highest seroprevalence was detected in Lakselv (85%).
Of the seropositives, 37% (175/480) of the reindeer were female, 23% (112/480) were male, and 2% (9/480) of unknown gender (Table 3). The seroprevalence of INKV among the calves (< 1 year of age) indicate recent exposure to the virus (previous summer). The highest number of seropositive were among adult females, 36% (107/296) (Table 3).
Table 3.
Seropositivity (%) of IgG antibodies against INKV reindeer sera determined by IIFA for the winter seasons of 2013–2014, 2014–2015, and 2015–2016, based on age (Adult = > 1 year, Calves = < 1 year) and sex presented as seropositive/tested (%)
| Seropositive (n)* | % of total (480) | % of total seropositive (296) | |
|---|---|---|---|
| Male adults | 28 | 6 | 9 |
| Male calves | 84 | 18 | 28 |
| Female adults | 107 | 22 | 36 |
| Female calves | 68 | 14 | 23 |
| Unknown adults | 0 | 0 | 0 |
| Unknown calves | 9 | 2 | 3 |
| Total | 296 | 62 | 100 |
*n = Number
Cytopathic effect neutralization test (CPE-NT) from reindeer sera
Of the 55 borderline reindeer sera samples from IIFA, 42 (76%) of the samples were confirmed to be negative and had a titre ˂ 20 while 13 (24%) of the remaining samples were positive on neutralization test.
Plaque reduction neutralization test (PRNT)
Sixty samples of the 480 reindeer sera with strongly positive (25), borderline (25), or negative (10) on IIFA were analyzed by PRNT. Of these 48 (80%) were confirmed seropositive and had neutralizing antibodies against INKV with titre ≥ 20, six samples (10%) were seronegative with titre ≤ 10 and six samples (10%) had no neutralization. Among the seropositives 39% (19/48) had titre ≥ 20–40, 52% (25/48) had titre ≥ 80–160, 8% (4/48) had titre ≥ 320. None of the samples showed any cross-reactivity with TAHV and SSHV.
Discussion
The frequent movement of humans and animals across countries might contribute to the spread of viruses and vectors. Currently, there is no INKV surveillance in Norway. As part of the surveillance analysis of viral infections that may cause encephalitis, an IgG INKV IIFA was performed. We analysed reindeer sera as sentinel animals for the surveillance of possible circulation of INKV-infection in Norway. IIFA can be used in INKV monitoring programs to test antibodies in reindeer as well as for diagnosis of INKV infection in other animal species and humans.
Reindeer have previously been suggested to serve as amplification hosts for INKV [6]. The virus is mostly transmitted by Ae. communis mosquitoes feeding on large mammals, such as cattle, reindeer, and moose which show high INKV antibody prevalence [6]. Detection of INKV in mosquitoes and anti-INKV specific antibodies in reindeer are good indications that infected mosquitoes are circulating in the area posing a risk of transmission of INKV to humans in Norway. Interestingly, in our study, 62% of the reindeer investigated showed specific antibodies against INKV. This correlates well with previous studies on INKV seropositive from Finland in adult moose, reindeer and sheep with 64%, 89% and 7–75%, respectively [7, 45, 46]. In our study there was highest seroprevalence among the adult females and calves (Table 3). The seroprevalence of INKV among the calves (< 1 year of age) indicate recent exposure to the virus from the environment or by active immunization via mothers' milk [47].
IIFA for INKV is known to cross-react with other California serogroups of viruses such as TAHV and SSHV in Finland [33, 46]. Nevertheless, none of the reindeer sera analysed with the INKV neutralization test in this study cross-reacted with TAHV or SSHV. As IgG antibodies can be detected for several months, and up to years, or even a lifetime, after exposure to the virus [33] detection of INKV IgG among reindeer may reflect an old infection. However, it is likely that the seropositive reindeer found in Røros and Fosen may have acquired INKV the previous summer as the detection of the virus in the BSI confirmed the circulation of INKV in these regions. The lack of detection of INKV in the BSI pools at the other sites may be due to low viral load. Another possibility is lacking or low numbers of the actual vector species in the analysed BSI pool. It cannot be ruled out that the antibodies detected in reindeer sera could have been acquired from infections gained outside Norway since some reindeer cross the borders to Sweden, Finland, or Russia [48]. However, the cross-border herding is rather limited most places, and reindeer can be regarded as good sentinels for the local ecosystems, feeding blood sucking insects through the summer season. To get information about acute infections in reindeer, a test to detect IgM antibodies in their sera could have been performed. However, this was not possible due to lack of conjugates and positive controls. It is therefore essential to study IgM antibodies to INKV as a sign of recent infection among the human and animal population during mosquito seasons. High seropositivity of INKV in human populations has been indicated by recent studies in Sweden and Finland [11, 32]. A study in Norway demonstrated mean IgG prevalence against CE viruses of 20% among the militaries without any clinical symptoms [16]. It could be interesting to study if the exposure of reindeer to INKV have any impact on reindeer health in future studies.
The distribution and prevalence of INKV in mosquitoes in Norway is largely unknown. However, INKV is circulating in Northern Europe including Norway [2, 16]. Similarly, other viruses belonging to the CE group have been isolated from five different Aedes spp. in Norway, but the methods were not specific enough to identify and confirm the findings as INKV or if they represented other related CE group viruses. Those viruses were isolated from mosquitoes collected from Trandum and Øyern (Akershus), and Masi (Troms and Finnmark) in 1975 and Sjusjøen (Innlandet), Trysil (Innlandet) and Trandum (Akershus) in 1976 [7, 16].
In the present study, INKV were detected in two out of 213 pools of BSI collected in Fosen and Røros in the summer of 2014–2015, supporting previous findings indicating that INKV may be circulating in Norway [7, 16]. Although our results show that INKV was present at these two sites in 2014–2015, this does not signify that the virus was not present in these areas before 2014–2015, since these locations were not investigated previously. The low prevalence of the virus from these two regions may indicate that the virus might have been carried to these areas by animals returning from areas with higher prevalence of the virus. The overall EPP of INKV in Norway was calculated to be 0.04% [42]. With this low prevalence it is unlikely that a single pool contains more than one infected insect, however we cannot exclude the possibility of several, which would affect the calculation of the true virus prevalence in the population of BSI [49]. For true knowledge on the virus prevalence in an area, repeated samplings of mosquitoes over several years are necessary, or a confirmation of circulation of the virus in human and animal sera within the same location. The samples containing virus were relatively few in this study. By visual inspection and barcoding of these two pools we suggest that Aedes spp. are the biological vector of INKV in Norway, since INKV has been detected previously in adult Aedes (Ae.) communis, Ae. hexodontus and Ae. punctor mosquitoes and in Ae. communis larvae [7, 15–17]. The prevalence of INKV in the BSI population can possibly be affected by transovarial transmission of the virus, from infected mothers to the BSI progeny [1].
Many mosquitoes collected in this study were identified as Aedes spp. with lower numbers of Culex spp. SINV has mostly been isolated from Culex mosquitoes in studies from Sweden, and Cx. torrentium has been identified as the main vector of SINV in Scandinavia [22]. Nevertheless, the SINV/SINV-like virus have previously been isolated from Aedes in Norway [28, 35]. The first SINV strain isolated from mosquitoes in Norway (Vemork 1/92) was collected in 1992 in Rjukan (Vestfold and Telemark), Norway [35, 36]. The virus is found to be geographically limited in Norway with most of the cases of human SINV-infections being reported from Rjukan [26], which is situated south of the southernmost collection sites, Valdres, in our study. Since there were low numbers of Culex. spp. in the present study we cannot rule out the presence of SINV in the studied areas. Also, the presence of migratory birds could affect incidence of SINV. However, SINV could not be detected from the collected BSI pools by RT-real-time PCR in the present study. Increasing the sample size of BSI or pure mosquito pools might have increased the possibility to detect positives.
The findings in this study are important for public awareness about geographical distribution of the INKV. This knowledge may help to take necessary preventive measures against INKV infections in the future. In further studies it would be interesting to screen mosquitoes and the human population to determine the seroprevalence of INKV during the mosquito season, as well as whole genome sequencing of isolated INKV to identify which strains are circulating in Norway.
Conclusion
There are knowledge gaps regarding the occurrence, distribution, and prevalence of the mosquito-borne viruses, INKV and SINV in Norway. The last study on these viruses was in 1992, reporting circulation of these viruses in the country. Our study is important to establish a baseline and provide new knowledge on the prevalence and distribution of INKV in BSI and reindeer. Moreover, it is important to monitor these viruses in case of an outbreak, and to avoid undiagnosed and unreported human cases. This study indicates that there is a low prevalence of INKV in BSI in Norway. The high seroprevalence of INKV in reindeer may indicate a risk of virus transmission from BSI to humans in these regions. Reindeer are one of the major hosts for INKV-infections and serve as an important sentinel animal for circulation of INKV. The occurrence and prevalence of INKV in vectors, animals, and the human population in Norway and other countries in Northern Europe requires further studies and monitoring.
Supplementary Information
Additional file 1: Table S1. Distribution of IgG antibodies against INKV reindeer sera determined by IIFA for the winter seasons of 2013–2014, 2014–2015 and 2015–2016 with number of total positive/number total tested, seropositivity rate %, standard deviation (SD) and confidence interval (95% CI).
Acknowledgements
Katrine Mørk Paulsen for technical support and proofreading, and Preben Skrede Ottesen for books and advice on mosquito literatures in Norway. Bjørn Heine Strand for statistical analysis, “STATA”. Prof. Göran Bucht for designing the Inkoo virus (INKV) primers. All the staff at Department of Virology, Norwegian Institute of Public Health for technical support. We thank reindeer herders for their patience and assistance during the sampling of reindeer.
Abbreviations
- INKV
Inkoo virus
- SINV
Sindbis virus
- TAHV
Tahyna virus
- BATV
Batai virus
- SSHV
Snowshoe hare virus
- CE
California encephalitis
- RT-PCR
Reverse transcriptase PCR
- EPP
Estimated pool prevalence
- IIFA
Indirect immunofluorescence assay
- CPE-NT
Cytopathic effect neutralization test
- PRNT
Plaque reduction neutralization test
Author contributions
MT, JSR, KÅ, IHN collected blood sucking insects and reindeer sera samples, RS prepared TNA samples, did qPCR, prepared samples for pyrosequencing, IIFA, PRNT analysis, data analysis, interpretation and contributed to the writing, ÅKA designed and initiated the study, supervision, data analysis, interpretation, critically revising the manuscript and proofreading, RV contributed by supervision, writing, revising the manuscript, ENDNOTE revision and proofreading, RM analysed the collected insects for species identification, ME, CA, OWL provided INKV and SINV positive controls for qPCR and optimized qPCR conditions, ME and CA provided a platform including; laboratory space, reagents, equipment and Standard Operating Procedure (SOP) for conducting the PRNT in their laboratory, OWL performed barcoding/speciation of blood sucking insects, also contributed to PRNT assay, and proofreading of the manuscript, OV for providing contacts, facilities and reagents for analysis of slides, revising the manuscript, NP prepared the slides, provided positive controls and evaluated the results for the IIFA test, performed CPE-NT on borderline samples from IIFA and proofreading of the manuscript, WJ for supervision, critically revising the manuscript and proofreading, AS and KSE as part of project planning, technical supervision, and revising the manuscript. All authors revised and approved the final manuscript.
Funding
This study was funded by the Norwegian Ministry of Health and Care Services, Barents region project B1710 “Emerging infections. Capacity building on vector-borne infections in the Barents region, Norway and Russia”. Sample collecting was funded by grants from the Norwegian Reindeer Development Fund (RUF) and by the Fram Center flagship “Klimaeffekter på økosystemer, landskap, lokalsamfunn og urfolk”, grant nr. 362256. Sample analysis was funded by grants from Insamlingsstiftelsen, Medical faculty, Umeå University, Sweden, and through a regional agreement between Umeå University and Västerbotten County Council (ALF).
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable due to studies on bloodsucking insects and blood samples from semi-domesticated animals.
Consent for Publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tingström O, Wesula Lwande O, Näslund J, Spyckerelle I, Engdahl C, Von Schoenberg P, Ahlm C, Evander M, Bucht G. Detection of Sindbis and Inkoo virus RNA in genetically typed mosquito larvae sampled in Northern Sweden. Vector Borne Zoonotic Dis. 2016;16(7):461–467. doi: 10.1089/vbz.2016.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubálek Z. Mosquito-borne viruses in Europe. Parasitol Res. 2008;103(1):29–43. doi: 10.1007/s00436-008-1064-7. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Gao X, Liang G. Newly recognized mosquito-associated viruses in mainland China, in the last two decades. Virol J. 2011;8:1. doi: 10.1186/1743-422X-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergqvist J, Forsman O, Larsson P, Näslund J, Lilja T, Engdahl C, Lindström A, Gylfe Å, Ahlm C, Evander M, Bucht G. Detection and isolation of Sindbis virus from mosquitoes captured during an outbreak in Sweden, 2013. Vector Borne Zoonotic Dis. 2015;15(2):133–140. doi: 10.1089/vbz.2014.1717. [DOI] [PubMed] [Google Scholar]
- 5.Gylfe Å, Ribers Å, Forsman O, Bucht G, Alenius GM, Wållberg-Jonsson S, Ahlm C, Evander M. Mosquitoborne Sindbis virus infection and long-term illness. Emerg Infect Dis. 2018;24(6):1141. doi: 10.3201/eid2406.170892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brummer-Korvenkontio MA. Arboviruses in Finland. V. Serological survey of antibodies against Inkoo virus in human, cow, reindeer, and wildlife sera. Am J Trop Med Hygiene. 1973;22(5):654–661. doi: 10.4269/ajtmh.1973.22.654. [DOI] [PubMed] [Google Scholar]
- 7.Traavik T, Mehl R, Wiger R. California encephalitis group viruses isolated from mosquitoes collected in Southern and Arctic Norway. Acta Path Microbiol Scand B Microbiol. 1978;86(1–6):335–342. doi: 10.1111/j.1699-0463.1978.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 8.Francy DB, Jaenson T, Lundström JO, Schildt E-B, Espmark A, Henriksson B, Niklasson B. Ecologic studies of mosquitoes and birds as hosts of Ockelbo virus in Sweden and isolation of Inkoo and Batai viruses from mosquitoes. Am J Trop Med Hygiene. 1989;41(3):355–363. doi: 10.4269/ajtmh.1989.41.355. [DOI] [PubMed] [Google Scholar]
- 9.Butenko AM, Vladimirtseva EA, Lvov SD, Calisher CH, Karabatsos N. California serogroup viruses from mosquitoes collected in the USSR. Am J Trop Med Hygiene. 1991;45(3):366–370. doi: 10.4269/ajtmh.1991.45.366. [DOI] [PubMed] [Google Scholar]
- 10.Kurkela S, Manni T, Myllynen J, Vaheri A, Vapalahti O. Clinical and laboratory manifestations of Sindbis virus infection: prospective study, Finland, 2002–2003. J Infect Dis. 2005;191(11):1820–1829. doi: 10.1086/430007. [DOI] [PubMed] [Google Scholar]
- 11.Putkuri N, Kantele A, Levanov L, Kivistö I, Brummer-Korvenkontio M, Vaheri A, Vapalahti O. Acute human Inkoo and Chatanga virus infections, Finland. Emerg Infect Dis. 2016;22(5):810. doi: 10.3201/eid2205.151015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ICTV. 2016. Retrieved 27 Oct 10. https://second.wiki/wiki/inkoo-virus.
- 13.Vapalahti O, Plyusnin A, Cheng Y, Manni T, Brummer-Korvenkontio M, Vaheri A. Inkoo and Tahyna, the European California serogroup bunyaviruses: sequence and phylogeny of the S RNA segment. J Gen Virol. 1996;77(8):1769–1774. doi: 10.1099/0022-1317-77-8-1769. [DOI] [PubMed] [Google Scholar]
- 14.Campbell WP, Huang C. Sequence comparisons of medium RNA segment among 15 California serogroup viruses. Virus Res. 1999;61(2):137–144. doi: 10.1016/S0168-1702(99)00033-7. [DOI] [PubMed] [Google Scholar]
- 15.Brummer-Korvenkontio M. Arboviruses of the California complex and the Bunyamwera group. Bratislava: Publ. House of Slov at Acad. Sci; 1969. p. 412. [Google Scholar]
- 16.Traavik T, Mehl R, Wiger R. Mosquito-borne arboviruses in Norway: further isolations and detection of antibodies to California encephalitis viruses in human, sheep and wildlife sera. J Hygiene. 1985;94(1):111–122. doi: 10.1017/S0022172400061180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lwande OW, Bucht G, Ahlm C, Ahlm K, Näslund J, Evander M. Mosquito-borne Inkoo virus in northern Sweden-isolation and whole genome sequencing. Virol J. 2017;14(1):1–9. doi: 10.1186/s12985-017-0725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krauss H, Weber A, Appel M, Enders B, Isenberg HD, Schiefer HG, Slenczka W, Graeventiz A, Zahner H. Zoonoses: infectious diseases transmissible from animals to humans. 3. Washington, DC: ASM Press; 2003. [Google Scholar]
- 19.Lundström JO, Lindstrom KM, Olsen B, Dufva R, Krakower DS. Prevalence of Sindbis virus neutralizing antibodies among Swedish passerines indicates that thrushes are the main amplifying hosts. J Med Entomol. 2001;38(2):289–297. doi: 10.1603/0022-2585-38.2.289. [DOI] [PubMed] [Google Scholar]
- 20.Kurkela S, Rätti O, Huhtamo E, Uzcátegui NY, Nuorti JP, Laakkonen J, Manni T, Helle P, Vaheri A, Vapalahti O. Sindbis virus infection in resident birds, migratory birds, and humans, Finland. Emerg Infect Dis. 2008;14(1):41. doi: 10.3201/eid1401.070510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundström JO, Niklasson B, Francy DB. Swedish Culex torrentium and Cx. pipiens (Diptera: Culicidae) as experimental vectors of Ockelbo virus. J Med Entomol. 1990;27(4):561–563. doi: 10.1093/jmedent/27.4.561. [DOI] [PubMed] [Google Scholar]
- 22.Hesson JC, Verner-Carlsson J, Larsson A, Ahmed R, Lundkvist Å, Lundström JO. Culex torrentium mosquito role as major enzootic vector defined by rate of Sindbis virus infection, Sweden, 2009. Emerg Infect Dis. 2015;21(5):875. doi: 10.3201/eid2105.141577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lwande OW, Näslund J, Lundmark E, Ahlm K, Ahlm C, Bucht G, Evander M. Experimental infection and transmission competence of Sindbis virus in Culex torrentium and Culex pipiens mosquitoes from Northern Sweden. Vector Borne Zoonotic Dis. 2019;19(2):128–133. doi: 10.1089/vbz.2018.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurkela S, Manni T, Vaheri A, Vapalahti O. Causative agent of Pogosta disease isolated from blood and skin lesions. Emerg Infect Dis. 2004;10(5):889–894. doi: 10.3201/eid1005.030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhrbier A, Jaffar-Bandjee MC, Gasque P. Arthritogenic alphaviruses—an overview. Nat Rev Rheumatol. 2012;8(7):420–429. doi: 10.1038/nrrheum.2012.64. [DOI] [PubMed] [Google Scholar]
- 26.Norwegian Institute of Public Health. Bærplukkersyke - veileder for helsepersonell. Retrieved 7 Nov 2017. 2013. https://www.fhi.no/nettpub/smittevernveilederen/sykdommer-a-a/barplukkersyke---veileder-for-helse/.
- 27.Skogh M, Espmark Å. Ockelbo disease: epidemic arthritis-exanthema syndrome in Sweden caused by Sindbis-virus like agent. Lancet. 1982;1:795–796. doi: 10.1016/S0140-6736(82)91834-7. [DOI] [PubMed] [Google Scholar]
- 28.L’vov DK, Skvortsova TM, Gromashevskii VL, Berezina LK, Iakovlev VI. Isolation of the causative agent of Karelian fever from Aedes sp. mosquitoes. Vopr Virusol. 1985;30(3):311–313. [PubMed] [Google Scholar]
- 29.Wesula Lwande O, Bucht G, Ahlm C, Evander M. Inkoo virus: a common but unrecognized mosquito-borne virus in northern Europe. Infect Dis Hub. 2017.
- 30.Bergman A, Dahl E, Lundkvist Å, Hesson JC. Sindbis virus infection in non-blood-fed hibernating Culex pipiens mosquitoes in Sweden. Viruses. 2020;12(12):1441. doi: 10.3390/v12121441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahlm C, Eliasson M, Vapalahti O, Evander M. Seroprevalence of Sindbis virus and associated risk factors in northern Sweden. Epidemiol Infect. 2014;142(7):1559–1565. doi: 10.1017/S0950268813002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evander M, Putkuri N, Eliasson M, Lwande OW, Vapalahti O, Ahlm C. Seroprevalence and risk factors of Inkoo virus in Northern Sweden. Am J Trop Med Hygiene. 2016;94(5):1103–1106. doi: 10.4269/ajtmh.15-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putkuri N, Vaheri A, Vapalahti O. Prevalence and protein specificity of human antibodies to Inkoo virus infection. Clin Vaccine Immunol. 2007;14(12):1555–1562. doi: 10.1128/CVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demikhov V, Chaĭtsev V. Neurologic characteristics of diseases caused by Inkoo and Tahyna viruses. Vopr Virusol. 1995;40(1):21–25. [PubMed] [Google Scholar]
- 35.Norder H, Lundström JO, Kozuch O, Magnius LO. Genetic relatedness of Sindbis virus strains from Europe, Middle East, and Africa. Virology. 1996;222(2):440–445. doi: 10.1006/viro.1996.0441. [DOI] [PubMed] [Google Scholar]
- 36.Storm N, Weyer J, Markotter W, Nel LH, Paweska JT, Leman PA, Kemp A. Phylogeny of Sindbis virus isolates from South Africa. S Afr J Epidemiol Infect. 2013;28(4):207–214. [Google Scholar]
- 37.Tryland M, Sánchez Romano J, Nymo IH, Breines EM, Murguzur FJA, Kjenstad OC, Li H, Cunha CW. A screening for virus infections in eight herds of semi-domesticated Eurasian Tundra Reindeer (Rangifer tarandus tarandus) in Norway, 2013–2018. Front Vet Sci. 2021;8:707787. doi: 10.3389/fvets.2021.707787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C, Shope RE, Spargo B, Campbell WP. The S RNA genomic sequences of Inkoo, San Angelo, Serra do Navio, South River and Tahyna bunyaviruses. J Gen Virol. 1996;77(8):1761–1768. doi: 10.1099/0022-1317-77-8-1761. [DOI] [PubMed] [Google Scholar]
- 39.Sane J, Kurkela S, Levanov L, Nikkari S, Vaheri A, Vapalahti O. Development and evaluation of a real-time RT-PCR assay for Sindbis virus detection. J Virol Methods. 2012;179(1):185–188. doi: 10.1016/j.jviromet.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Andreassen A, Jore S, Cuber P, Dudman S, Tengs T, Isaksen K, Hygen HO, Viljugrein H, Anestad G, Ottesen P, Vainio K. Prevalence of tick borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Parasites Vectors. 2012;5(1):1–2. doi: 10.1186/1756-3305-5-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 42.Sergeant ES. Epitools epidemiological calculators. Bruce: AusVet Pty Ltd; 2018. [Google Scholar]
- 43.Cowling DW, Gardner IA, Johnson WO. Comparison of methods for estimation of individual-level prevalence based on pooled samples. Prev Vet Med. 1999;9(39):211–225. doi: 10.1016/S0167-5877(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 44.Stojanovich CJ, Scott HG. Illustrated key to the mosquitoes of Fennoscandia, Finland, Sweden, Denmark, Norway, CJ Stojanovich; 1995. p. 1–132.
- 45.Medlock JM, Snow KR, Leach S. Possible ecology and epidemiology of medically important mosquito-borne arboviruses in Great Britain. Epidemiol infect. 2007;135(3):466–482. doi: 10.1017/S0950268806007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brummer-Korvenkontio M. Bunyamwera arbovirus supergroup in Finland. A study on Inkoo and Batai viruses. Comment Biol. 1974;76:1–52. [Google Scholar]
- 47.Castro N, Gómez-González LA, Earley B, Argüello A. Use of clinic refractometer at farm as a tool to estimate the IgG content in goat colostrum. J Appl Anim Res. 2018;46(1):1505–1508. doi: 10.1080/09712119.2018.1546585. [DOI] [Google Scholar]
- 48.Kirchner S. Cross-border forms of animal use by indigenous peoples. In: Peters A, editor. Studies in global animal law. Berlin: Springer; 2020. p. 57. [Google Scholar]
- 49.Ebert TA, Brlansky R, Rogers M. Reexamining the pooled sampling approach for estimating prevalence of infected insect vectors. Ann Entomol Soc Am. 2010;103(6):827–837. doi: 10.1603/AN09158. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Distribution of IgG antibodies against INKV reindeer sera determined by IIFA for the winter seasons of 2013–2014, 2014–2015 and 2015–2016 with number of total positive/number total tested, seropositivity rate %, standard deviation (SD) and confidence interval (95% CI).
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.