Abstract
Oxidative DNA damage has been linked to inflammation, cancer, and aging. Here, we have mapped two types of oxidative DNA damage, oxidized guanines produced by hydrogen peroxide and oxidized thymines created by potassium permanganate, at a single-base resolution. 8-Oxo-guanine occurs strictly dependent on the G/C sequence context and shows a pronounced peak at transcription start sites (TSSs). We determined the trinucleotide sequence pattern of guanine oxidation. This pattern shows high similarity to the cancer-associated single-base substitution signatures SBS18 and SBS36. SBS36 is found in colorectal cancers that carry mutations in MUTYH, encoding a repair enzyme that operates on 8-oxo-guanine mispairs. SBS18 is common in inflammation-associated upper gastrointestinal tract tumors including esophageal and gastric adenocarcinomas. Oxidized thymines induced by permanganate occur with a distinct dinucleotide specificity, 5′T-A/C, and are depleted at the TSS. Our data suggest that two cancer mutational signatures, SBS18 and SBS36, are caused by reactive oxygen species.
Hydrogen peroxide induces a DNA damage pattern with high similarity to two cancer mutational signatures.
INTRODUCTION
Reactive oxygen species (ROS) from internal and external sources represent a threat to genome integrity. Different types of ROS are generated during cellular metabolism or from exposure to chemicals, air pollution, ultraviolet (UV) radiation, or diet (1–6). These ROS include hydrogen peroxide (H2O2) as a common oxidizing molecule present in cells and molecules derived from H2O2 in the presence of transition metals, such as hydroxyl radicals (•OH). Superoxide anion (O2•-) produced by electron leakage from the electron transport chain or by myeloperoxidase during inflammatory processes is converted to H2O2 by superoxide dismutase (7). Another relevant type of ROS is singlet oxygen (1O2) formed by UVA radiation through excitation of endogenous photosensitizers (2). Reactive nitrogen species are produced during inflammatory processes (8). These molecules react with guanine to form 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG). Guanine has the lowest oxidation potential compared to the other DNA bases and is therefore oxidized preferentially. 8-Oxoguanine (8-oxo-G) is the most prevalent DNA base oxidation product, although more than 20 oxidatively damaged DNA bases have been identified (1–3, 5). In addition to direct oxidation of guanine, 8-oxo-dG in DNA may arise from oxidation of the nucleotide pool, i.e., by formation of 8-oxo-dGTP, which is then incorporated into DNA during replication (9). Damage of pyrimidines is a less efficient reaction produced by ROS. One exception is the oxidizing agent potassium permanganate (KMnO4), which readily oxidizes thymine to form mostly thymine glycol (10, 11).
8-oxo-G is repaired primarily by base excision repair initiated by the 8-oxoguanine DNA glycosylase OGG1 (1). When this lesion is not repaired and persists during replication, DNA polymerases may incorporate cytosine or adenine across from 8-oxo-dG. A specialized repair DNA glycosylase, MUTYH (MYH), is present in mammalian cells to remove the mis-incorporated adenine from 8-oxo-G/A mispairs (12). When these repair pathways fail, the most common mutation induced by 8-oxo-G is the G to T transversion (13, 14). Guanine oxidation may contribute to carcinogenesis. MUTYH mutations have been associated with inherited colorectal polyposis and colorectal cancer (15, 16) and possibly other cancers (17). In mouse models, combined deficiency of Myh (encoding MUTYH) and Ogg1 (encoding the OGG1 repair enzyme) predisposes the animals to lung and ovarian tumors and lymphomas. In these mice, 75% of the lung tumors carried G to T mutations in the Kras gene (18).
It has been difficult to unambiguously link ROS and oxidative DNA damage to human cancer. The production of ROS is a well-known process occurring in the setting of inflammation. An estimated 25% of all human cancers have been linked to chronic inflammation, which acts as a tumor-predisposing condition, and many cancer risk factors have inflammation as a common mode of action (19). These malignancies include cancers associated with infections (for example, liver cancers and hepatitis viruses, cervical cancers and head and neck tumors and human papilloma virus, and gastric cancers and Helicobacter pylori) or inflammatory conditions [liver cancers and alcoholic and nonalcoholic steatohepatitis (fatty liver disease), esophageal cancers and Barret’s esophagus, intestinal tumors linked to inflammatory bowel disease, and several others].
As part of the Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium, more than 84 million mutations from whole cancer genomes and exomes were used to establish about 50 single-base substitution (SBS) signatures (20). Some signatures occur in most cancer types and in most individual tumors analyzed. An example is SBS1, with typical C to T mutations at CpG dinucleotides. SBS1 is thought to be derived from spontaneous hydrolytic deamination of 5-methylcytosine at methylated CpG sites in the genome (21). On the other hand, many signatures are quite tumor type specific. For example, lung cancers show a strong enrichment of SBS4, a signature in which G to T mutations are most prevalent. SBS4 is typical for lung cancers in tobacco smokers (22). The patterns of these lung cancer G to T mutations are best correlated with mutations induced by polycyclic aromatic hydrocarbons, exemplified by benzo[a]pyrene (22–26).
G to T signatures have been extracted from a few other specific cancer types, in which they occur along with additional signatures. From the signature collection of the COSMIC database (20), SBS18 has been suggested to be potentially caused by ROS. SBS18 is most common in esophageal adenocarcinomas and in certain stomach and colorectal cancers. Esophageal adenocarcinomas have a clear preinflammatory condition, Barrett’s esophagus. Rare colorectal tumors carry germline or somatic mutations in the oxidative damage repair enzyme MUTYH (16). These mutations, which show a high frequency of G to T events, have been characterized as SBS36 in the COSMIC database (27).
One important goal is to have a clear understanding of the extent and genomic features of the oxidative DNA damage that may cause the mutations. In most cases, DNA damage cannot be read directly by standard DNA polymerase–based sequencing methods. There are several methodologies available for mapping of oxidative DNA damage (28). Using immunoprecipitation of DNA with antibodies against 8-oxo-dG, followed by high-throughput sequencing, it is possible to map 8-oxo-dG in the genome with a resolution of 150 to 250 base pairs (bp) (29, 30), which is analogous to resolution achieved by chromatin immunoprecipitation (ChIP) sequencing. Other affinity-based pulldown methods are 8-oxo-G-sequencing (OG-seq) (31) and apurinic site-sequencing (AP-seq) (32), which achieve similar levels of resolution.
However, it is most desirable to achieve single-base specificity for mapping of oxidized DNA bases. One method developed for this purpose is based on click chemistry (33). This approach involves removing the damage with a repair enzyme and filling the gap with an alkynylated deoxynucleoside, which can then be coupled to a code sequence oligonucleotide. The resulting triazole-linked DNA can be traversed and read by a DNA polymerase. This method has been used to map 8-oxo-dG in the yeast genome (33). Nick-sequencing (nick-seq) has been used for mapping oxidative DNA damage in Escherichia coli (34). 8-oxo-G can be further oxidized and chemically labeled with biotin, which produces a polymerase stop signal (35). However, to our knowledge, there is currently no straightforward enzyme-based method to map oxidized DNA bases genome-wide and at single-base resolution in mammalian cells.
We recently developed the circle-damage-sequencing (CD-seq) method for base level mapping of UV-induced DNA damage in human cells (36). This method relies on the efficient conversion of DNA damage into strand breaks and bidirectional sequencing from the break. Here, we have adopted CD-seq for mapping the sequence distribution of 8-oxo-G and of oxidized thymine in human cells. These damaged bases form with unique sequence specificities. The patterns of 8-oxo-dG resemble two mutational signatures found in cancer genomes.
RESULTS
Mapping of oxidized guanines at base resolution
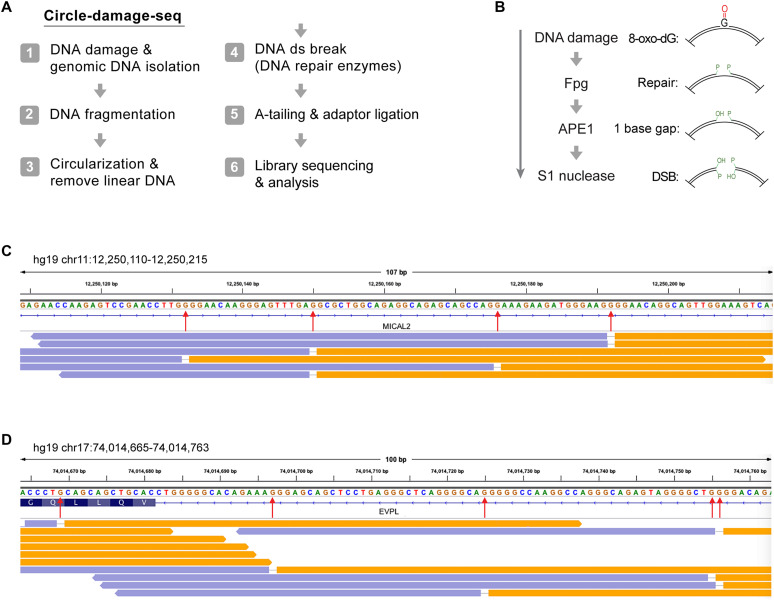
We used hydrogen peroxide as the ROS with physiological relevance and incubated human dermal fibroblasts with this oxidizing agent at concentrations ranging from 5 to 25 mM for 30 min. We isolated DNA from the treated cells and initially verified the formation of oxidized guanines by cleavage of the high–molecular weight DNA with the Fpg DNA glycosylase. Fpg protein efficiently excises 8-oxo-G. It also releases ring-opened formamidopyrimidine (FAPY) DNA adducts (37–39), which may form under reducing conditions through one-electron reduction after free-radical reaction of guanine (1, 40). After incubation of the DNA with Fpg protein, we produced DNA double-strand breaks at the incision sites using single-strand–specific S1 nuclease. We separated the treated DNA molecules on nondenaturing agarose gels (fig. S1). Increasing concentrations of H2O2 produced dose-dependently increasing levels of strand breakage, consistent with formation of 8-oxo-dG in the treated cells at frequencies of about one modification per 10 kb at the highest concentration used and less at the lower concentrations.
Having confirmed the formation of 8-oxo-dG, we selected the samples treated with 5 and 10 mM H2O2 and processed them for modified base mapping using CD-seq (Fig. 1A) (36). After DNA circularization, the 8-oxo-G bases were released with E. coli Fpg protein, which also has AP lyase activity. We used APE1 to remove the modified sugar residues at the 3′ ends near the single-strand breaks. Using S1 nuclease, we then produced ligatable DNA double-strand breaks at the ring-opened molecules. After adapter ligation, sequencing libraries were prepared and subjected to paired-end sequencing on Illumina flow cells (see Methods). In the CD-seq procedure, the aligned reads show a unique pattern in genome browser views; they appear as divergent reads (due to opening of the rings) with a single-nucleotide gap, whereby the gap position represents the base that was eliminated by the DNA glycosylase enzyme (Fig. 1, B to D). Considering all divergent gapped bases obtained, we observed that guanine was the base in the gaps at a frequency of over 70% of all bases. Adenine or thymine appeared in ~30% of the divergent reads. The presence of adenine may be caused by adenine oxidation or, alternatively, by the presence of abasic sites at adenine positions, which are cleaved by the AP lyase activity inherent to Fpg protein. However, in terms of the number of total divergent reads normalized to the total number of all reads obtained, our untreated control sample had 500 to 700 times fewer divergent reads with single-base gaps, suggesting that most of the damaged sequence positions we mapped were due to the treatment with H2O2 and were not due to a background of such modifications in cells or due to artifactual damage to the DNA during sample processing. The untreated samples contained 54% A or T and 46% G or C bases in the gap. The H2O2-treated cell samples had predominantly G or C in the gap at frequencies of 67 to 77.5%. One technical challenge with mapping oxidative DNA damage is its potential occurrence as background, which could be induced in part by the DNA preparation methods, most notably in the presence of phenol (41). Therefore, care needs to be taken to minimize background oxidation.
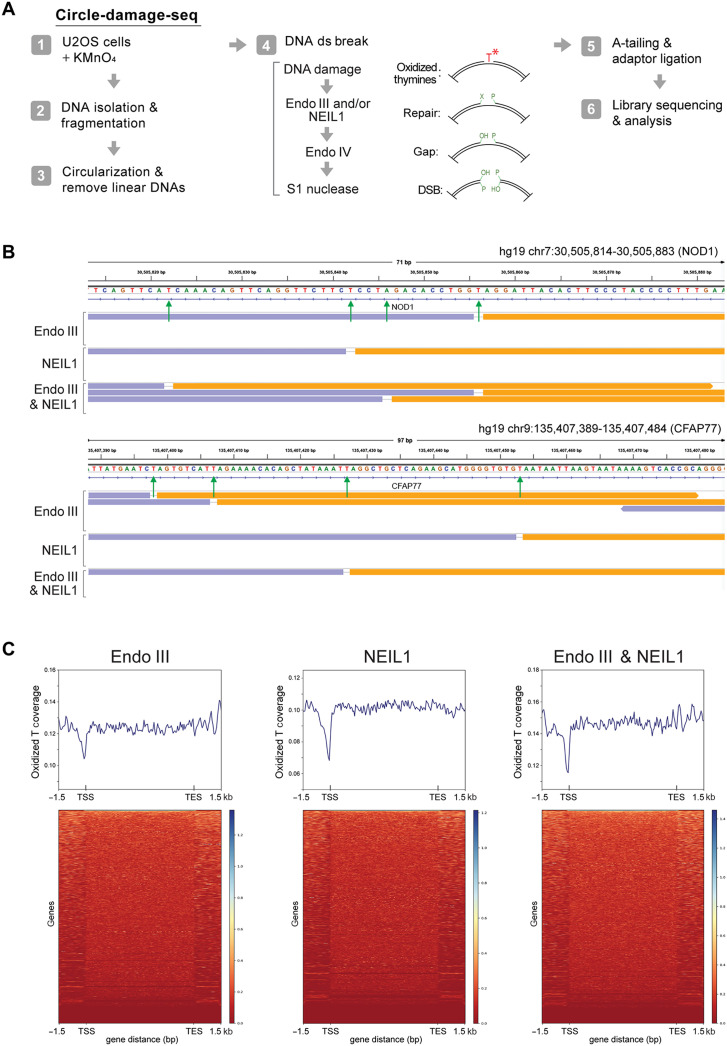
Fig. 1. Mapping of oxidized guanines by CD-seq.
(A) Outline of the CD-seq method. (B) Outline of the enzymatic cleavage procedure to map 8-oxo-dG at single-base resolution. (C) Genome browser view of divergent DNA sequencing read pairs on chromosome 11 at the MICAL2 gene. Orange and lavender segments represent the divergent read pairs. Red arrows indicate single-base gaps matching the excised DNA base. (D) Genome browser view of divergent DNA sequencing read pairs on chromosome 17 at the EVPL gene. ds, double strand; DSB, double-strand break.
Distribution of oxidized guanines along genes
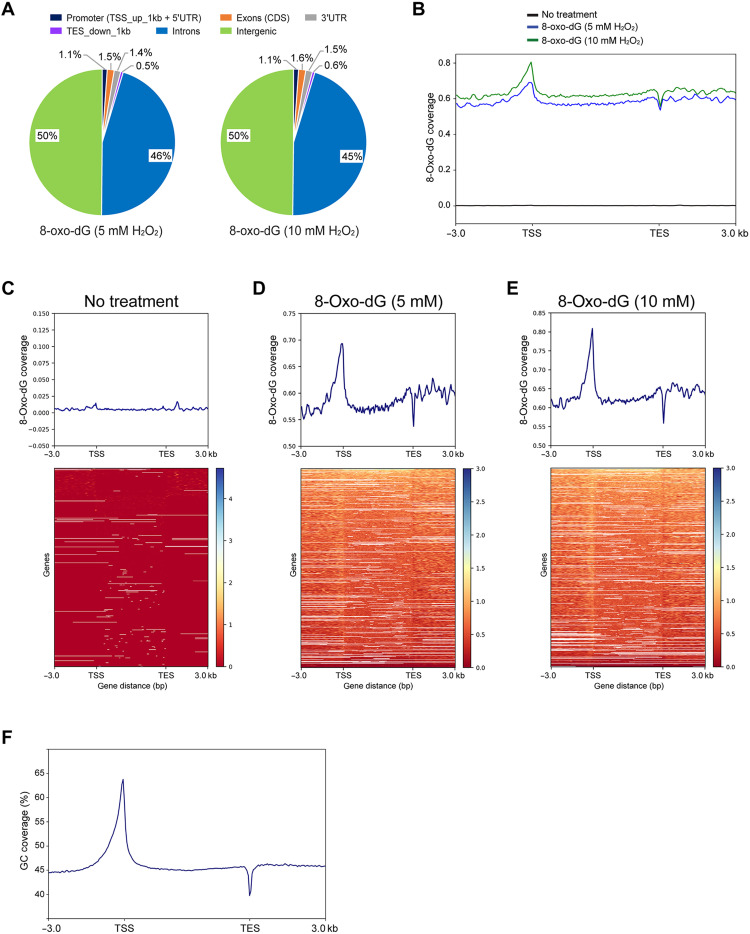
Using 5 million to 6 million divergent single-base gapped reads from cells treated with 5 or 10 mM H2O2 and much fewer reads from nontreated cells (n = ~12,000), the reads were mapped to the hg19 human genome. The oxidized bases were most strongly enriched in intergenic regions and in introns (Fig. 2A). Using gene-level (metagene) analysis, we profiled the oxidized base signals along all genes from −3 kb upstream of transcription start sites (TSSs) and then binned them according to gene length from the TSS to the transcription end sites (TESs) and continuing 3 kb downstream of the TES (Fig. 2B). The H2O2-treated samples showed similar profiles between the different concentrations, which were characterized by strong peaks near the TSS and a dip near the TES (Fig. 2, B to E). This distribution of signal roughly followed the GC-content of the human genome as plotted in Fig. 2F. Sequences near the TSS are generally GC rich and often include CpG islands, and sequences near the TES are AT rich due to the presence of polyadenylation signals and their surrounding sequences (42, 43).
Fig. 2. Distribution of 8-oxo-dG along human genes.
(A) Distribution of 8-oxo-dG in different compartments of the human genome. (B) Metagene profiles of 8-oxo-dG distribution along all genes of the hg19 human genome. The signal was plotted from 3 kb upstream of TSSs and then binned over the entire gene length, continuing 3 kb downstream of TESs. The y-axis scale is 0 to 0.8. (C to E) Heatmaps of 8-oxo-dG coverage are sorted from high (top, blue) to low (bottom, red). The signals were mapped and binned in 50-bp windows from 3 kb upstream of the TSS and then normalized relative to gene length over the gene bodies to the TES and 3 kb downstream of the TES. (C) No H2O2 treatment. (D) Treatment of cells with 5 mM H2O2. (E) Treatment of cells with 10 mM H2O2. (F) GC content along hg19 genes.
Relationship of guanine oxidation to gene expression and chromatin features
Using publicly available gene expression data for human fibroblasts, we examined whether 8-oxo-dG formation is correlated with gene expression levels. Figure S2 shows that there is no correlation between transcript levels (FPKM, fragments per kilo base per million mapped reads) and the extent of 8-oxo-dG formation in the region covering 2.5 kb upstream of the TSS.
Using chromatin state descriptions from the ENCODE (The Encyclopedia of DNA Elements) project (ChromHMM), we determined whether higher levels of 8-oxo-G bases accumulate in genomic regions that are associated with specific chromatin features (fig. S3). The highest levels of oxidized guanines were found in intragenic enhancer regions, in bivalent enhancers and TSS regions, and in genomic regions targeted by the Polycomb complex. Regions of heterochromatin and quiescent regions of the genome showed the lowest levels of 8-oxo-dG formation (fig. S3). These data are consistent with higher GC content of specific genomic segments being associated with higher levels of guanine oxidation, with limited contribution of gene expression state or specific chromatin features.
A previous report, using an immunoprecipitation technique, indicated that DNA in the nuclear periphery is more susceptible to H2O2-induced guanine oxidation than DNA in the center of the nucleus (29). The nuclear periphery–located DNA contains lamin-associated domains (LADs), which are regions of heterochromatin (44). We used published LAD data from an earlier study of human fibroblasts (45) and determined the distribution of 8-oxo-dG in LAD regions and in LAD-flanking regions, up to 100 kb from the border regions. 5-Oxo-dG was moderately depleted in LADs and enhanced at the border and flanking regions (fig. S4A). In addition, here, the distribution of the oxidized bases largely followed the GC content of the genome, which is lower in LADs (fig. S4B).
Distribution of oxidized guanines along specific DNA sequences
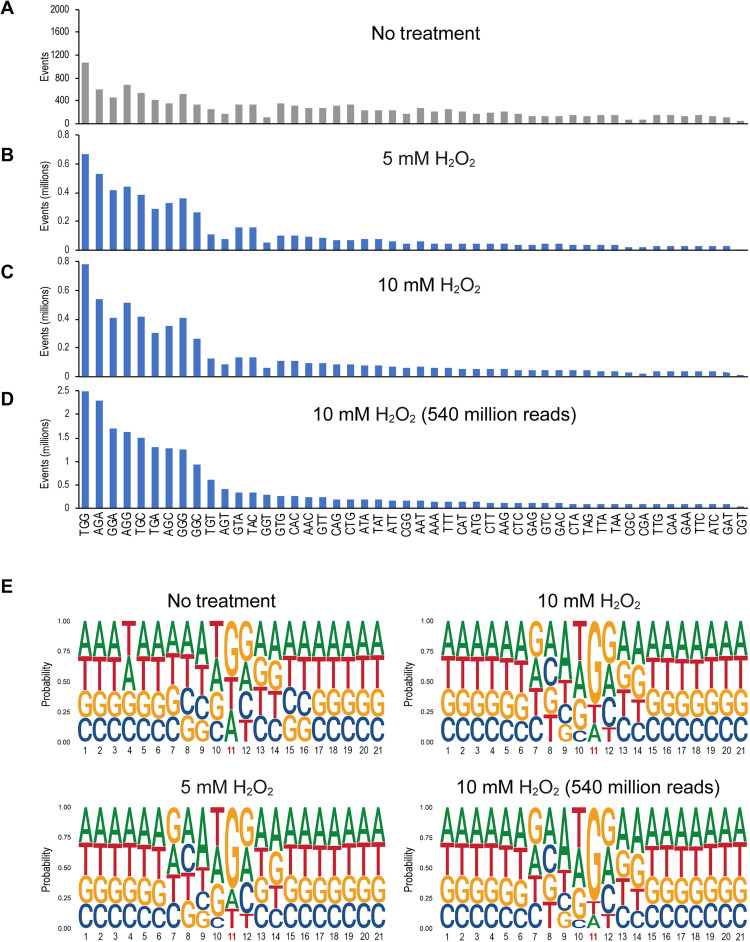
We next analyzed the trinucleotide sequence contexts of guanine oxidation in H2O2-treated human cells. Because the strands are symmetrical, for example, AGC corresponds to GCT on the opposite strand, we considered 48 combinations because we treated A or T in the gap separately. The middle base is displayed as either G, A, or T, and the 5′ and 3′ flanking bases can be any of the four bases, which leads to 48 combinations. For the nontreated control, the distribution of the different trinucleotides was relatively flat, with some enhancement at the sequences 5′TGG, 5′AGG, and 5′GGG (Fig. 3A). However, for the samples treated with 5 or 10 mM H2O2, trinucleotides with a central guanine were much enhanced (Fig. 3, B to D, consider y-axis scales compared to Fig. 3A). Here, we also included a sample derived from a 10 mM H2O2 treatment that was run on the sequencer at 540 million reads (Fig. 3D). The most highly damaged trinucleotide sequence always was 5′TGG. Additional major damage sites were found at 5′AGA, 5′AGG, and 5′GGA. Two neighboring purines with G as the damaged base as well as the sequence 5′TGC characterized the most frequent trinucleotides harboring oxidized guanine in the center. Expanding the analysis to a broader sequence context, we generated logo plots (Fig. 3E). These sequence plots show guanine as the preferred base in position 11 (the base in the gaps of divergent reads). The bases most preferred in the 5′ direction were T, A, and G. Cytosine was clearly underrepresented, largely because 5′CG sequences are depleted in mammalian genomes. The bases most preferred in the 3′ direction of the 8-oxo-G were G, A, and C, with T being strongly underrepresented. Outside of this trinucleotide context, no further sequence-specific enrichment of 8-oxo-G was apparent.
Fig. 3. Sequence context of 8-oxo-dG formation in human fibroblasts treated with H2O2.
(A) Trinucleotide sequence context of read gaps in control cells (no treatment). (B to D) Trinucleotide sequence context of read gaps in cells treated with 5 mM (B) or 10 mM (C and D) H2O2. The read depth was 90 million reads in (C) or 540 million reads in (D). (E) Sequence context of guanine oxidation by using logo plot analysis. Position 11 represents the base in the divergent read gaps. At each base position, the height of each letter represents the relative frequency of that nucleic acid base.
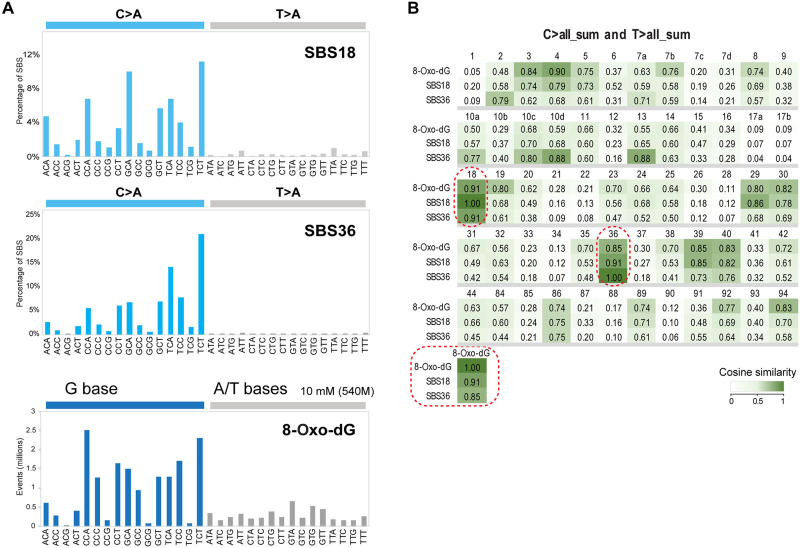
It is of interest to compare this 8-oxo-dG DNA damage signature with the set of mutation signatures collected in the COSMIC cancer mutation database. We used cosine similarity analysis to establish the relationship between our 8-oxo-dG signature (established from 540 million reads) and the various COSMIC signatures. The highest similarities between the 8-oxo-dG signature and COSMIC SBS signatures were found for SBS4, SBS18, SBS36, and SBS39 (Fig. 4). SBS4, which is enriched in G to T mutations, has been explained by tobacco smoke–associated polycyclic aromatic hydrocarbons (25). SBS39, a signature of unknown etiology, has predominantly G to C transversions, in addition to C to A mutations, and would therefore not qualify for a specific 8-oxo-dG signature. Those signatures that are dominated by C to A (or G to T) transversions are of greater relevance.
Fig. 4. 8-Oxo-dG signature and cancer mutational signatures.
(A) COSMIC signatures SBS18 and SBS36 in comparison to the 8-oxo-dG signature. For the cancer signatures, only the C>A and T>A mutation windows are shown. The 8-oxo-dG DNA damage signature is displayed to reflect the style of cancer mutation signatures. (B) Heatmap showing the cosine similarity scores for 8-oxo-dG mapping at the trinucleotide level and the COSMIC mutational signatures found in the COSMIC v3.2 mutation database. We used the sum of all C to N and T to N mutation windows for comparisons with the 8-oxo-dG trinucleotide signature. We excluded COSMIC signatures that are thought to be sequencing artifacts. Darker green colors indicate higher similarity. Signatures SBS18 and SBS36 are indicated by dotted round rectangles.
SBS18 has been proposed to be linked to ROS-induced DNA damage (COSMIC database). SBS36 is thought to be due to deficiency of the MUTYH DNA glycosylase known to excise adenines from 8-oxo-G/A mispairs, is enriched in colorectal tumors from patients with MUTYH mutations, and therefore should reflect a signature from oxidative DNA damage (27, 46). A similar signature of mutations has been found in Mth1/Ogg1/Mutyh triple knockout mice (47, 48). The cosine similarity between the 8-oxo-dG signature and SBS18 was highest at 0.91, and the cosine similarity between 8-oxo-dG and SBS36 was 0.85 (Fig. 4B). Our analysis suggests that the trinucleotide patterns we established for oxidized guanines are concordant with two G/C to T/A mutational signatures enriched in human cancers.
Analysis of oxidized thymines in permanganate-treated cells
Potassium permanganate oxidizes thymine and produces thymine glycol as the main DNA oxidation product (11). The sequence specificity and genomic preferences for thymine oxidation have remained unknown due to a lack of available methodology.
To analyze permanganate-oxidized thymines, we treated human U2OS cells with potassium permanganate and isolated the DNA. To test the global extent of thymine oxidation in the treated cells, we initially used agarose gel electrophoresis after cleavage of the DNA with two DNA glycosylases, Endo III and NEIL1. These enzymes remove oxidized DNA bases from DNA (49, 50). Whereas Endo III prefers to excise oxidized bases from double-stranded DNA, NEIL1 preferentially operates on single-stranded regions (51), although there likely is overlap in their substrate preference depending on enzyme concentrations and incubation conditions. The enzyme Endo IV was used as an AP endonuclease to remove ligation-blocking lesions from the 3′ ends. Double-strand cleavage was achieved with S1 nuclease digestion. The data show that the combined enzyme treatment produces double-strand breaks in a KMnO4 concentration–dependent manner (fig. S5) and that the treatment with 10 mM KMnO4 creates approximately one lesion per 3 kb of DNA.
Following these initial experiments, we used NEIL1 or Endo III in the CD-seq procedure (Fig. 5A). We also included samples in which both enzymes were used in combination. Figure 5B shows high-resolution genome browser views of the amplified libraries. As expected, we almost exclusively observed thymines in the single-base gaps, consistent with the properties of permanganate oxidation and DNA glycosylase specificity.
Fig. 5. Mapping of oxidized thymines.
(A) Outline of the CD-seq and enzymatic cleavage methods used to map oxidized thymines at single-base resolution. (B) Base resolution view of oxidized thymines along the human NOD1 and CFAP77 genes. Orange and lavender segments represent the divergent read pairs. Green arrows indicate single-base gaps matching the excised oxidized DNA base. (C) Heatmaps and metagene profiles of permanganate-oxidized thymines along human genes. DNA from untreated cells produced very few divergent read pairs, and the data could not be used in similar displays. Different cleavage enzymes, Endo III or NEIL1, or a combination of the two was used.
Distribution of oxidized thymines along genes
We created several million divergent reads with single-base gaps and mapped them to the hg19 genome. We analyzed the damage detected with the different enzyme combinations along genes from −3 kb upstream of TSSs and then binned according to gene length from the TSS to the TES, and 3 kb downstream of the TES (Fig. 5). These profiles show a pronounced reduction of oxidized thymine near TSS regions and a relatively flat profile along other parts of the genes. It is likely that the reduced levels of oxidized Ts near the TSS are a consequence of the GC richness of these sequences.
The levels of oxidized thymines in regions representing upstream gene promoters did not correlate with gene expression levels (fig. S6). We then derived ChromHMM maps for human U2OS cells using published ChIP-sequencing (ChIP-seq) data for histone modifications (fig. S7). We found the strongest enrichment of oxidized thymines at zinc finger genes and at repeats (fig. S7A). The enrichment of thymine glycol in zinc finger genes may be explained, at least in part, by the relative AT richness (GC poorness) of these genes relative to other hg19 genes (fig. S7B). However, with regard to LADs, the distribution of oxidized thymines was uniform along those regions and flanking sequences and was not negatively correlated with GC content of the regions (fig. S8).
Distribution of oxidized thymines along specific DNA sequences
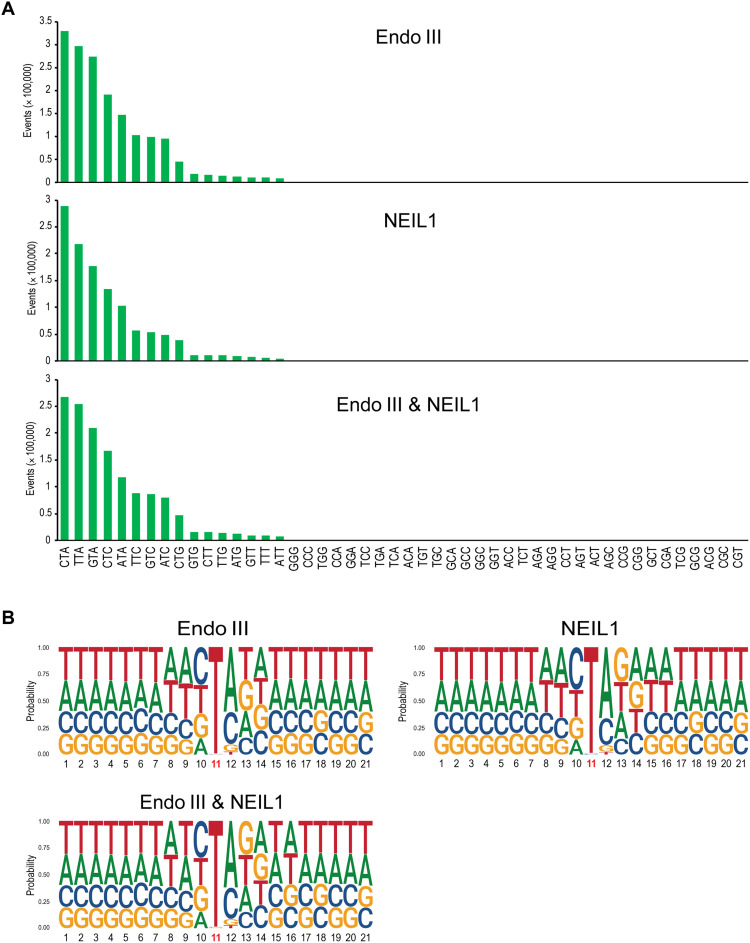
We next analyzed the trinucleotide sequence contexts of thymine oxidation damage in permanganate-treated cells. Because the strands are symmetrical, for example, ATC corresponds to GAT on the opposite strand, we considered 48 combinations. The middle base is displayed as either T, C, or G, and the 5′ and 3′ flanking bases can be any of the four bases, which creates 48 combinations. For nontreated cells, we obtained only 392 divergent reads with single-base gaps. One reason for this very small number of background reads is the low frequency of oxidized pyrimidines in untreated cells. The number of reads was too low to establish a reliable trinucleotide distribution list or gene profiles for nontreated cells. The different enzyme treatments with Endo III, NEIL1, or the combination of the two gave very similar trinucleotide sequence patterns. The preferred sequence contexts of oxidized thymines were 5′CTA and 5′TTA, followed by 5′GTA, 5′CTC, and 5′ATA (Fig. 6A).
Fig. 6. Sequence-level distribution of oxidized thymines in permanganate-treated human cells.
(A) Trinucleotide sequence specificity. Forty-eight trinucleotide contexts were analyzed, but the gapped reads occurred chiefly at trinucleotides with central thymine. Different enzymes (Endo III and NEIL1 in combination and alone) were used to excise the oxidized bases. (B) Sequence context of thymine oxidation by permanganate using logo plot analysis. Position 11 represents the permanganate-oxidized thymine base. At each base position, the height of each letter represents the relative frequency of that nucleic acid base. Note the enrichment of A or C 3′ to the thymine.
A sequence logo plot analysis (Fig. 6B) revealed T as the overwhelming base in position 11, the gapped position of divergent reads. The position 5′ to the damaged T showed a minor preference for pyrimidines. However, the position 3′ to the oxidized T showed interesting characteristics, the common presence of A followed by C and much fewer occurrences of G or T. The data reveal a heretofore unknown DNA sequence specificity of permanganate-induced thymine oxidation.
DISCUSSION
We applied the CD-seq method for comprehensive mapping of two types of oxidative DNA damage. CD-seq is applicable for analysis of any type of DNA damage or DNA modifications for which a specific cleavage enzyme is available. In our study, we used DNA glycosylases of the base excision repair pathway. These enzymes may have a narrow or broader selectivity for the damaged bases. In this case, Fpg protein is known to excise 8-oxo-G from DNA but also acts on other oxidized bases, preferentially purines, such as FAPY-G or oxidized adenines. The DNA damaging agent we used, H2O2, is known to induce 8-oxo-G as a major modified DNA base. Therefore, the signals we mapped are expected to be mostly 8-oxo-G. For permanganate-induced DNA damage, we used E. coli Endo III, which preferentially recognizes oxidized pyrimidines. We also used the NEIL1 protein, which has a broader specificity and can remove oxidized purines and oxidized pyrimidines. The DNA damaging agent KMnO4 oxidizes thymine, preferentially leading to the formation of thymine glycol (10, 11). Notably, the sequence patterns we obtained with Endo III and NEIL1 cleavage were almost identical and consisted of almost 100% thymine, which suggests that the procedure mapped thymine oxidation products. Our work revealed a previously unrecognized sequence specificity of permanganate oxidation, 5′TA/C.
Oxidized guanines obtained after treatment of cells with H2O2 formed with a defined sequence pattern in human cells. This oxidizing agent is present endogenously in many cell types and tissues. H2O2 is generated as a product of white blood cells that are active during immune responses and under inflammatory conditions. In the presence of transition metal ions, hydroxyl radical and other oxidizing short-lived intermediates are formed, which damage guanine preferentially. We found that guanine oxidation by H2O2 is chiefly related to the GC content of the genome with GC-rich regions, such as TSSs, acquiring the highest levels of 8-oxo-dG. This result is partially consistent with previous results using DNA immunoprecipitation, in which the authors observed the highest levels of 8-oxo-dG just downstream of the TSS (52). However, Poetsch et al. (32) reported a substantially decreased AP and 8-oxo-dG signal using their AP-seq method near TSS regions. This contrasts with our observations, but neither one of these two earlier publications used H2O2 for treatment of the cells. The base oxidation patterns were not noticeably dependent on gene expression state and chromatin environments except that such environments have differential GC content.
Oxidative DNA damage has long been suspected to have a causative role in cancer, aging, and other diseases such as neurodegeneration. However, definitive proof of this connection has been difficult to obtain. Regimens of diets rich in antioxidants are thought to prevent cancer and promote longevity. Studies in mouse models defective in DNA repair pathways that deal with oxidative DNA damage are perhaps the best available evidence to support the oxidative damage and cancer relationship. These studies, for example, with Ogg1 knockout mice, often do not lead to a major cancer phenotype, presumably because of effective antioxidant defense systems and redundant DNA repair capacities (53, 54). However, mice double-deficient in OGG1, which removes 8-oxo-G from 8-oxo-G/C base pairs, and deficient in MUTYH, which removes adenine from 8-oxo-G/A mispairs, are clearly cancer prone (18).
For human cancers, a direct relationship between oxidative DNA damage and cancer has been even more difficult to obtain. Although inflammation is a cancer-predisposing condition, which has been suspected to contribute to a large fraction of human tumors, the inflammatory process induces many other changes in addition to direct DNA damage. For example, inflammation is contributing to altered signaling pathways, altered immune responses, changes in gene expression, and epigenetic changes such as DNA hypermethylation (55).
There has been great technical difficulty in precisely measuring endogenous oxidative DNA damage in tissues. The methods used are often confounded by artifactual damage introduced during DNA isolation or sample processing (41, 56, 57). Therefore, it has been challenging to determine DNA damage in human tissues including precancerous lesions. With further improvements and effective precautions to prevent in vitro damage to DNA (such as inclusion of antioxidants during DNA isolation), the CD-seq method may eventually achieve high enough sensitivity to measure oxidized DNA bases in human tissue specimens. In a previous study, Kucab et al. (25) used cell cloning and mutation sequencing as an end point to measure mutagenesis induced by H2O2 but did not observe clear mutation patterns. This approach requires DNA replication, which necessitates using a rather low concentration of hydrogen peroxide. To produce sufficient DNA damage, in our hands required millimolar concentrations of H2O2, which may not permit DNA replication to occur due to cell cycle arrest.
Human cancer genome sequencing efforts have provided catalogs of cancer-specific mutational patterns for dozens of human tumor types. Several of these mutational patterns give an indication for what the premutagenic DNA damaging processes might be. For example, sunlight-induced skin cancers carry a clear mutation signature (referred to as SBS7a and SBS7b in the current COSMIC database). This signature consists chiefly of C to T mutations at dipyrimidine sequences, for example, at 5′TC. Using CD-seq, we recently showed that this signature is highly similar to the distribution of UVB irradiation–induced cyclobutane pyrimidine dimers that have undergone cytosine deamination (36). For oxidative DNA damage, the signatures should be dominated by G to T (or C to A) transversions, which are known mutations induced by 8-oxo-dG (13, 14). There are several COSMIC signatures that have these required features. One is SBS4, which is smoking associated and has been linked to tobacco-related polycyclic aromatic hydrocarbons (22, 25, 58). SBS10c, SBS10d, SBS14, and SBS20, all enriched in G to T mutations, have been linked to DNA polymerase mutations. SBS24 is linked to aflatoxin exposure, and SBS29 is found in tobacco chewers.
Another G to T mutation signature is SBS18, which is of unproven etiology. When we compared our 8-oxo-dG signature with SBS18, we obtained a strong cosine similarity value of 0.91, which was higher than that with any other signature (Fig. 4). We point out that mutational signatures are not solely the product of a one-to-one relationship between DNA damage and mutations. The mutations are shaped by additional factors that are currently unknown such as a potential sequence-specific repair of 8-oxo-G (46) or a sequence-dependent bypass of the oxidized guanines by DNA polymerases. Nonetheless, SBS18 is so similar to the 8-oxo-dG signature that they are likely directly related. Because repair of 8-oxo-G follows rapid kinetics with most damage removed within minutes (59, 60), our measurements most likely reflect an equilibrium between ongoing H2O2-induced damage formation and repair of this damage in cells. We propose that this equilibrium is perhaps the best way of reflecting mutational patterns because it is the equilibrium (unrepaired) DNA damage that will translate into mutations. Knockout of OGG1 in human untreated induced pluripotent stem cells also produced mutations resembling SBS18, albeit at lower similarity levels (46). It is worth noting that SBS18 is most enriched and prevalent in gastrointestinal cancers that have a strong inflammatory component, such as esophageal adenocarcinoma (linked to Barret’s esophagus), gastric adenocarcinoma (linked to gastritis), and a few colorectal cancers (fig. S9). Colonic crypts from patients with inflammatory bowel disease show an increase of SBS18 mutations (61). Inflammation is strongly associated with ROS (5, 6).
SBS36 is also defined by G to T/C to A transversions (Fig. 4A) and is quantitatively similar to SBS18 (cosine = 0.91 between the two signatures). This signature is found in colorectal cancers of patients that have biallelic germline mutations or somatic mutations in the MUTYH gene (27). Lack of MUTYH activity will eliminate an important correction pathway that would otherwise remove mispaired adenine opposite 8-oxo-G. When this adenine is not removed, mutations at 8-oxo-dG will be created and fixed more readily. For these reasons, SBS36 will be a good indicator of 8-oxo-dG–induced mutations. Comparison of our 8-oxo-dG trinucleotide distribution data with SBS36 indicated a cosine similarity value of 0.85, which is among the highest observed for all available COSMIC signatures. Another type of tumor in which a ROS-mediated mutagenesis pathway seems to be at work is neuroblastoma with copy number loss of OGG1 or MUTYH as recently reported (62).
In conclusion, we present a method based on CD-seq, which can be used to map various types of oxidatively damaged DNA bases at single-base resolution in the human genome. We define a sequence-specific pattern for oxidized guanines induced by H2O2 and of oxidized thymine induced by permanganate. The 8-oxo-dG trinucleotide distribution pattern closely resembled cancer mutation signatures SBS18 and SBS36, suggesting the involvement of guanine oxidation in specific types of human cancers of the gastrointestinal tract.
METHODS
Cell culture
Human male skin fibroblast cells [human dermal fibroblast (HDF); American Type Culture Collection (ATCC), PCS-201-012] with no genetic modification and human osteosarcoma cells (U2OS; ATCC, HTB-96) were cultured in the Fibroblast Growth Kit (ATCC, catalog no. PCS-201-041) supplemented with 2% fetal bovine serum (FBS) and in Dulbecco’s modified Eagle’s medium/high glucose supplemented with 10% FBS, respectively, at 37°C in a 5% CO2 standard incubator.
Oxidative DNA damage and genomic DNA isolation
To induce 8-oxo-dGs, HDF cells at 80 to 90% confluence on 10-cm culture plates were washed with 1× phosphate-buffered saline (PBS) and treated in PBS with H2O2 solution diluted to final concentrations of 5, 10, and 25 mM for 30 min at 37°C in a 5% CO2 incubator. After treatment, the cells were immediately trypsinized and pelleted, and then genomic DNA was isolated using the Quick-DNA Miniprep Plus Kit (Zymo Research, Irvine, CA) according to the manufacturer’s instruction manual. To evaluate 8-oxo-dG damage in the isolated genomic DNAs, the DNAs were treated with Fpg and APE1 [New England Biolabs (NEB), Ipswich, MA] followed by S1 nuclease (Thermo Fisher Scientific) treatment, and then damage-specific DNA cleavage events were observed on 1% agarose gels.
To induce oxidized thymines, 1 × 107 U2OS cells were washed with 1× PBS and treated with 10 or 40 mM KMnO4 for 70 s at 37°C in a solution with the following components: 15 mM tris-HCl (pH 7.5), 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, and 300 mM sucrose. The reaction was quenched by adding β-mercaptoethanol to a final concentration of 700 mM and EDTA to a final concentration of 50 mM. SDS was added to 1%, and the DNA was purified by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. The quenching reaction and the proteinase K digestion were carried out at 37°C for 30 min and 4 hours, respectively. DNA base damage was evaluated by treatment with Endo III (NEB) and/or NEIL1 (OriGene, Rockville, MD) DNA glycosylases, followed by Endo IV and S1 treatment, and then observed on a 1% agarose gel.
CD-seq mapping of 8-oxo-dGs
CD-seq library preparations were performed as previously described (36) with slight modifications to generate damage-specific DNA double-strand breaks. Briefly, 2 μg of the genomic DNAs prepared from H2O2-treated cells was sheared to an average length of 300 bp by sonication with a Covaris E220 sonicator (Covaris, Woburn, MA) and end-repaired to prepare blunt-ended DNA with ribonuclease H, T4 DNA polymerase, and T4 polynucleotide kinase (NEB). To circularize DNA fragments, 1 μg of the blunt-ended DNA was incubated with T4 DNA ligase (NEB) in a final reaction volume of 200 μl by overnight incubation at 16°C. Then, to remove noncircularized linear DNA from the circularized DNA pool, the ligase-treated DNA was incubated with Plasmid-Safe ATP-Dependent DNase (Lucigen, Middleton, WI).
To generate DNA double-strand breaks at 8-oxo-dG sites, the circularized DNA was incubated in 1× NEBuffer 1 reaction buffer with 1 μl (8 U/μl) of Fpg protein (NEB) in a final volume of 40 μl for 1 hour at 37°C. The DNA was cleaned up with 72 μl (1.8×) of AMPure XP beads (Beckman Coulter, Indianapolis, IN) and eluted in 35 μl of 10 mM tris-HCl (pH 8.0). To remove the modified sugar residues at the 3′ ends near the single-strand breaks, the DNA from above was incubated in 1× NEBuffer 4 reaction buffer with 1 μl (10 U/μl) of APE1 (NEB) for 20 min at 37°C. The DNA was cleaned up and cleaved on the opposite strand of the nicked positions using 1 μl (5 U/μl) of single strand–specific S1 nuclease (Thermo Fisher Scientific) for 4 min at room temperature. Then, the reaction was stopped by adding 2 μl of 0.5 M EDTA and 1 μl of 1 M tris-HCl (pH 8.0) to the reaction mixture, and DNA was further incubated for 10 min at 70°C. The DNA sample was cleaned up with 72 μl (1.8×) of AMPure XP beads and eluted in 48 μl of 10 mM tris-HCl (pH 8.0).
The double-strand cleaved DNA was A-tailed with Klenow fragment exo- (NEB) and subsequently ligated with 50 nM (final concentration) of T-overhang NEBNext hairpin adaptors (NEB): 5′-phos-GATCGGAAGAGCACACGTCTGAACTCCAGTCdUACACTCTTTCCTACACGACGCTCTTCCGATC*T-3′ and with 50 nM of C-overhang adaptors synthesized by Integrated DNA Technologies (Coralville, IA): 5′-phos-GATCGGAAGAGCACACGTCTGAACTCCAGTCdUACACTCTTTCCTACACGACGCTCTTCCGATC*C-3′ (*; phosphorothioate linkage), and this reaction was followed by incubation with USER enzyme (NEB). The polymerase chain reaction (PCR)–amplified CD-seq library was sequenced as 150-bp paired-end sequencing runs on an Illumina Hiseq2500 platform to obtain between 100 million and 540 million reads.
CD-seq mapping of oxidized Ts
To generate double-strand breaks at oxidized thymine sites, the circularized DNAs prepared from KMnO4-treated cells were initially incubated in 1× reaction buffer [20 mM tris-HCl (pH 8.0), 1 mM EDTA, 50 mM NaCl, and bovine serum albumin (0.1 mg/ml)] with 0.5 μl (10 U/μl) of Endo III (NEB) and/or 1 μl (0.24 μg/μl) of NEIL1 (OriGene) in a final volume of 20 μl for 30 min at 37°C. Then, the DNAs were cleaned up and treated with Endo IV (NEB) to remove ligation-blocking lesions from the 3′ ends, followed by S1 nuclease digestion to achieve double-strand cleavage. The cleaved DNAs were subjected to A-tailing, and this was followed by adaptor ligation with 50 nM (final concentration) of T-overhang NEBNext hairpin adaptors (NEB). The adaptor-ligated DNAs were treated with USER enzyme and amplified by PCR. The CD-seq library was sequenced as 150-bp paired-end reads on an Illumina Hiseq2500 platform to obtain 50 million reads.
Data analysis
Identification of oxidized DNA bases
To identify the positions of oxidized DNA bases, fastq files containing the CD-seq reads were processed using the pipeline as previously described (36). Briefly, all reads were trimmed from the 3′ end to 2 × 80 nucleotide length after removal of sequencing adapters and low-quality reads (phred < 20) using Trim_Galore (https://bioinformatics.babraham.ac.uk/projects/trim_galore/). BWA-MEM (version 0.7.17) was then used to align the reads to the human reference genome (hg19) and to remove duplicate alignments with the “removeDups” SAMBLASTER (version 0.1.26) option. Only divergently aligned pairs of reads with a single-nucleotide gap between reads were retained by selecting SAM records with TLEN of 3 and saved as bam files for downstream analyses. TLEN = 3 represents divergently aligned reads with one nucleotide between mates. In addition, chromosome name, start position, stop position, and inferred strand (+/−) for all damaged base loci were written to a BED file as processed data, and this information has been deposited into the Gene Expression Omnibus (GEO) database under accession number GSE184820.
Visualization of read alignments using the IGV viewer
To view aligned reads on the Integrative Genomics Viewer (IGV) downloaded from the Broad Institute (http://software.broadinstitute.org/software/igv/), the TLEN = 3 filtered bam files were sorted by genomic coordinates and indexed using SAMtools (version 1.11). The bam files were loaded on IGV viewer (version 2.8.9), and the tracks were displayed across the hg19 human genome with a “view as pairs” option to show pairs together with a line joining the ends.
Metagene profiles and genomic region analysis
To analyze the distribution of oxidized DNA damage along the hg19 human reference genes, the TLEN = 3 filtered bam files were processed on the Galaxy web platform (https://usegalaxy.org/). The bam files were converted to bigwig files using bamCoverage at 20-bp resolution, and computeMatrix and plotHeatmap programs in deepTools were then used to assess the oxidized bases over genic regions that included 3 kb upstream of the TSS and 3 kb downstream of the TES with average coverage calculated in nonoverlapping 50-nucleotide bins and normalized to gene length. For analysis of GC content of the hg19 human genome, we downloaded the hg19.gc5Base.wigVarStep.gz file from the UCSC Genome Browser database and converted it to bigwig file format. Then, the mean GC content distribution was analyzed using the same process as for metagene profiles above. The range of GC content was scaled to 35 to 70% GC content. The TLEN = 3 filtered bam files were also processed using the “Read Distribution” program in RseQC package provided on the Galaxy web platform to analyze the distribution of oxidized DNA bases over genomic features of the hg19 human reference genes.
Trinucleotide sequence profiles and logo plots
Sequences for the trinucleotides representing the 5′ and 3′ flanking bases and the gapped base in the middle were identified and counted using BEDtools (63). For 8-oxo-dGs, when there is a cytosine as the gapped nucleotide, the damage would have occurred on the (−) strand; thus, reverse complement trinucleotides with G or C in the middle were combined. For oxidized thymine, an adenine as the gapped nucleotide would indicate that the damage would have occurred on the (−) strand; thus, reverse complement trinucleotide sequences with T or A in the middle were combined, and the 48 possible trinucleotide combinations were used for trinucleotide sequence profiles. Sequence logo plots for nucleotide frequencies surrounding damaged bases (−10 to +10 positions relative to the oxidized base) were drawn using the ggseqlogo package in R (64).
Cosine similarity analysis
SBS mutational signatures with trinucleotide frequencies (version 3.2) were obtained in numerical form from the COSMIC database (http://cancer.sanger.ac.uk/cosmic/signatures/SBS/). We excluded 18 of the COSMIC signatures that are thought to be sequencing artifacts. To directly compare the two datasets, cosine similarity analyses were performed using the R (v. 4.0.2) package “coop” (https://R-project.org/). Because the mutated base of SBS signatures is represented by the pyrimidine of the base pair in the middle position, reverse complement trinucleotide sequences from CD-seq were combined and represented by the pyrimidine in the middle base. In addition, we used the sum of all C to N and T to N mutation windows for comparisons with the CD-seq trinucleotide sequence signature.
Chromatin state analysis
To determine the relative enrichment of oxidative DNA damage along defined chromatin states, ChromHMM (65) was used to perform genome annotations and to compute the fold enrichment scores of each state for oxidized damage mapping data. Genomic segmentation files for the chromatin states (core 18-state model) based on epigenomic data for human dermis fibroblasts were obtained from the Roadmap Epigenomics Project (http://roadmapepigenomics.org) (ENCODE accession no. ENCSR071BVW), and the scores showing the relative enrichment of 8-oxo-dG were calculated using the “OverlapEnrichment” option in ChromHMM with default parameters.
To establish (“learn”) the chromatin state model for U2OS cells, the “LearnModel” option in ChromHMM with 200-bp bin size was used using histone modification ChIP-seq data obtained from GEO database accession no. GSE141081 for H3K4me1, H3K4me3, and H3K27ac, and from accession no. GSE31755 for H3K9me3 and H3K36me3. After annotation of 14 chromatin states using the state emissions data and the TSS neighborhood data generated by the ChromHMM LearnModel, the fold enrichment scores for oxidized thymine along defined chromatin states were obtained as described above.
Lamin-associated domains
To determine the distribution of oxidized bases over LAD regions, previously defined LAD data (n = 1302 LADs) for human fibroblasts were obtained from published sources (45) and U2OS LAD data (n = 1046 LADs) were downloaded from the GEO database (GES87831) (66). The ComputeMatrix program in deepTools was used to calculate the coverages of oxidized DNA bases over LADs and 100-kb regions flanking LADs in nonoverlapping 1-kb bins. The plotProfile program in deepTools was used to plot the average coverages computed by computeMatrix.
Distribution of oxidized bases and gene expression profiles
Transcript levels (FPKM, fragments per kilo base per million mapped reads) determined by RNA-seq for human dermis fibroblasts and for U2OS cells were obtained from the GEO database accession numbers GSE78610 and GES139190, respectively. The transcript data were converted to bed file format and sorted in descending order with FPKM value for downstream analysis. The datasets were grouped into detectable (FPKM > 0) and nondetectable (FPKM = 0) transcripts. To determine the coverages of oxidized bases in extended promoter regions (within 2.5 kb upstream of the TSS) of detectable transcripts, the ComputeMatrix program in deepTools was used to obtain matrices containing scores for coverage of oxidized bases over promoter regions of each transcript. Scatter plots showing the correlation between oxidized DNA bases and gene expression levels were drawn using scores computed by computeMatrix and corresponding FPKM values.
Acknowledgments
Funding: This work was supported by NIH grant CA228089 to G.P.P. and by the Van Andel Institute.
Author contributions: S.-G.J., P.E.S., and G.P.P. conceptualized and designed the project. S.-G.J., Y.M., and J.J. performed experiments. S.-G.J., P.E.S., and G.P.P. performed data analysis. G.P.P. and S.-G.J. wrote the manuscript. All authors commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The code for data analysis in this project is archived at DOI: 10.5281/zenodo.6458592. Sequencing data have been deposited into the GEO database (accession number GSE184820).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
REFERENCES AND NOTES
- 1.Boiteux S., Coste F., Castaing B., Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 107, 179–201 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Cadet J., Davies K. J. A., Medeiros M. H., Di Mascio P., Wagner J. R., Formation and repair of oxidatively generated damage in cellular DNA. Free Radic. Biol. Med. 107, 13–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans M. D., Dizdaroglu M., Cooke M. S., Oxidative DNA damage and disease: Induction, repair and significance. Mutat. Res. 567, 1–61 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Giorgio M., Dellino G. I., Gambino V., Roda N., Pelicci P. G., On the epigenetic role of guanosine oxidation. Redox Biol. 29, 101398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y., Cui Y., Niedernhofer L. J., Wang Y., Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem. Res. Toxicol. 29, 2008–2039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonkar P., Dedon P. C., Reactive species and DNA damage in chronic inflammation: Reconciling chemical mechanisms and biological fates. Int. J. Cancer 128, 1999–2009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn J. M., Melov S., SOD2 in mitochondrial dysfunction and neurodegeneration. Free Radic. Biol. Med. 62, 4–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niles J. C., Wishnok J. S., Tannenbaum S. R., Peroxynitrite-induced oxidation and nitration products of guanine and 8-oxoguanine: Structures and mechanisms of product formation. Nitric Oxide 14, 109–121 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Maki H., Sekiguchi M., MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355, 273–275 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Frenkel K., Goldstein M. S., Duker N. J., Teebor G. W., Identification of the cis-thymine glycol moiety in oxidized deoxyribonucleic acid. Biochemistry 20, 750–754 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Rouet P., Essigmann J. M., Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res. 45, 6113–6118 (1985). [PubMed] [Google Scholar]
- 12.David S. S., O’Shea V. L., Kundu S., Base-excision repair of oxidative DNA damage. Nature 447, 941–950 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibutani S., Takeshita M., Grollman A. P., Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxo-dG. Nature 349, 431–434 (1991). [DOI] [PubMed] [Google Scholar]
- 14.Wood M. L., Dizdaroglu M., Gajewski E., Essigmann J. M., Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry 29, 7024–7032 (1990). [DOI] [PubMed] [Google Scholar]
- 15.Mazzei F., Viel A., Bignami M., Role of MUTYH in human cancer. Mutat. Res. 743-744, 33–43 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Al-Tassan N., Chmiel N. H., Maynard J., Fleming N., Livingston A. L., Williams G. T., Hodges A. K., Davies D. R., David S. S., Sampson J. R., Cheadle J. P., Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 30, 227–232 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Thibodeau M. L., Zhao E. Y., Reisle C., Ch’ng C., Wong H. L., Shen Y., Jones M. R., Lim H. J., Young S., Cremin C., Pleasance E., Zhang W., Holt R., Eirew P., Karasinska J., Kalloger S. E., Taylor G., Majounie E., Bonakdar M., Zong Z., Bleile D., Chiu R., Birol I., Gelmon K., Lohrisch C., Mungall K. L., Mungall A. J., Moore R., Ma Y. P., Fok A., Yip S., Karsan A., Huntsman D., Schaeffer D. F., Laskin J., Marra M. A., Renouf D. J., Jones S. J. M., Schrader K. A., Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harb. Mol. Case Stud. 5, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D. E., Lindahl T., McIlhatton M., Fishel R., Miller J. H., Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 64, 3096–3102 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Todoric J., Antonucci L., Karin M., Targeting inflammation in cancer prevention and therapy. Cancer Prev. Res. 9, 895–905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov L. B., Kim J., Haradhvala N. J., Huang M. N., Tian Ng A. W., Wu Y., Boot A., Covington K. R., Gordenin D. A., Bergstrom E. N., Islam S. M. A., Lopez-Bigas N., Klimczak L. J., McPherson J. R., Morganella S., Sabarinathan R., Wheeler D. A., Mustonen V.; PCAWG Mutational Signatures Working Group, Getz G., Rozen S. G., Stratton M. R., Consortium P. C. A. W. G., The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeifer G. P., Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 301, 259–281 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Alexandrov L. B., Ju Y. S., Haase K., Van Loo P., Martincorena I., Nik-Zainal S., Totoki Y., Fujimoto A., Nakagawa H., Shibata T., Campbell P. J., Vineis P., Phillips D. H., Stratton M. R., Mutational signatures associated with tobacco smoking in human cancer. Science 354, 618–622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denissenko M. F., Pao A., Tang M., Pfeifer G. P., Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274, 430–432 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Pfeifer G. P., Denissenko M. F., Olivier M., Tretyakova N., Hecht S. S., Hainaut P., Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21, 7435–7451 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Kucab J. E., Zou X., Morganella S., Joel M., Nanda A. S., Nagy E., Gomez C., Degasperi A., Harris R., Jackson S. P., Arlt V. M., Phillips D. H., Nik-Zainal S., A compendium of mutational signatures of environmental agents. Cell 177, 821–836 e816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nik-Zainal S., Kucab J. E., Morganella S., Glodzik D., Alexandrov L. B., Arlt V. M., Weninger A., Hollstein M., Stratton M. R., Phillips D. H., The genome as a record of environmental exposure. Mutagenesis 30, 763–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viel A., Bruselles A., Meccia E., Fornasarig M., Quaia M., Canzonieri V., Policicchio E., Urso E. D., Agostini M., Genuardi M., Lucci-Cordisco E., Venesio T., Martayan A., Diodoro M. G., Sanchez-Mete L., Stigliano V., Mazzei F., Grasso F., Giuliani A., Baiocchi M., Maestro R., Giannini G., Tartaglia M., Alexandrov L. B., Bignami M., A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine 20, 39–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mingard C., Wu J., McKeague M., Sturla S. J., Next-generation DNA damage sequencing. Chem. Soc. Rev. 49, 7354–7377 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Yoshihara M., Jiang L., Akatsuka S., Suyama M., Toyokuni S., Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 21, 603–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amente S., Di Palo G., Scala G., Castrignano T., Gorini F., Cocozza S., Moresano A., Pucci P., Ma B., Stepanov I., Lania L., Pelicci P. G., Dellino G. I., Majello B., Genome-wide mapping of 8-oxo-7,8-dihydro-2′-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells. Nucleic Acids Res. 47, 221–236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y., Fleming A. M., Burrows C. J., Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-seq. J. Am. Chem. Soc. 139, 2569–2572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poetsch A. R., Boulton S. J., Luscombe N. M., Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol. 19, 215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J., McKeague M., Sturla S. J., Nucleotide-resolution genome-wide mapping of oxidative DNA damage by click-code-seq. J. Am. Chem. Soc. 140, 9783–9787 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Cao B., Wu X., Zhou J., Wu H., Liu L., Zhang Q., DeMott M. S., Gu C., Wang L., You D., Dedon P. C., Nick-seq for single-nucleotide resolution genomic maps of DNA modifications and damage. Nucleic Acids Res. 48, 6715–6725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An J., Yin M., Yin J., Wu S., Selby C. P., Yang Y., Sancar A., Xu G. L., Qian M., Hu J., Genome-wide analysis of 8-oxo-7,8-dihydro-2′-deoxyguanosine at single-nucleotide resolution unveils reduced occurrence of oxidative damage at G-quadruplex sites. Nucleic Acids Res. 49, 12252–12267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S. G., Pettinga D., Johnson J., Li P., Pfeifer G. P., The major mechanism of melanoma mutations is based on deamination of cytosine in pyrimidine dimers as determined by circle-damage-sequencing. Sci. Adv. 7, eabi6508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boiteux S., O’Connor T. R., Lederer F., Gouyette A., Laval J., Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 265, 3916–3922 (1990). [PubMed] [Google Scholar]
- 38.Chetsanga C. J., Lindahl T., Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 6, 3673–3684 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tchou J., Kasai H., Shibutani S., Chung M. H., Laval J., Grollman A. P., Nishimura S., 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U.S.A. 88, 4690–4694 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalam M. A., Haraguchi K., Chandani S., Loechler E. L., Moriya M., Greenberg M. M., Basu A. K., Genetic effects of oxidative DNA damages: Comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 34, 2305–2315 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helbock H. J., Beckman K. B., Shigenaga M. K., Walter P. B., Woodall A. A., Yeo H. C., Ames B. N., DNA oxidation matters: The HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc. Natl. Acad. Sci. U.S.A. 95, 288–293 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian B., Hu J., Zhang H., Lutz C. S., A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Jung, G. P. Pfeifer, CpG islands, in Brenner’s Encyclopedia of Genetics (Second Edition) (Elsevier, 2013), pp. 205–207. [Google Scholar]
- 44.Hoskins V. E., Smith K., Reddy K. L., The shifting shape of genomes: Dynamics of heterochromatin interactions at the nuclear lamina. Curr. Opin. Genet. Dev. 67, 163–173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., van Steensel B., Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Zou X., Koh G. C. C., Nanda A. S., Degasperi A., Urgo K., Roumeliotis T. I., Agu C. A., Badja C., Momen S., Young J., Amarante T. D., Side L., Brice G., Perez-Alonso V., Rueda D., Gomez C., Bushell W., Harris R., Choudhary J. S.; Genomics England Research Consortium, Jiricny J., Skarne W. C., Nik-Zainal S., A systematic CRISPR screen defines mutational mechanisms underpinning signatures caused by replication errors and endogenous DNA damage. Nat. Cancer 2, 643–657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilati C., Shinde J., Alexandrov L. B., Assie G., Andre T., Helias-Rodzewicz Z., Ducoudray R., Le Corre D., Zucman-Rossi J., Emile J. F., Bertherat J., Letouze E., Laurent-Puig P., Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J. Pathol. 242, 10–15 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Ohno M., Sakumi K., Fukumura R., Furuichi M., Iwasaki Y., Hokama M., Ikemura T., Tsuzuki T., Gondo Y., Nakabeppu Y., 8-Oxoguanine causes spontaneous de novo germline mutations in mice. Sci. Rep. 4, 4689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eide L., Bjoras M., Pirovano M., Alseth I., Berdal K. G., Seeberg E., Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 93, 10735–10740 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hazra T. K., Izumi T., Boldogh I., Imhoff B., Kow Y. W., Jaruga P., Dizdaroglu M., Mitra S., Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 3523–3528 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dou H., Mitra S., Hazra T. K., Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Gorini F., Scala G., Di Palo G., Dellino G. I., Cocozza S., Pelicci P. G., Lania L., Majello B., Amente S., The genomic landscape of 8-oxodG reveals enrichment at specific inherently fragile promoters. Nucleic Acids Res. 48, 4309–4324 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D. E., Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U.S.A. 96, 13300–13305 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sampath H., Oxidative DNA damage in disease—Insights gained from base excision repair glycosylase-deficient mouse models. Environ. Mol. Mutagen. 55, 689–703 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Todoric J., Karin M., The fire within: Cell-autonomous mechanisms in inflammation-driven cancer. Cancer Cell 35, 714–720 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Dizdaroglu M., Jaruga P., Birincioglu M., Rodriguez H., Free radical-induced damage to DNA: Mechanisms and measurement. Free Radic. Biol. Med. 32, 1102–1115 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Ravanat J. L., Douki T., Duez P., Gremaud E., Herbert K., Hofer T., Lasserre L., Saint-Pierre C., Favier A., Cadet J., Cellular background level of 8-oxo-7,8-dihydro-2′-deoxyguanosine: An isotope based method to evaluate artefactual oxidation of DNA during its extraction and subsequent work-up. Carcinogenesis 23, 1911–1918 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Pfeifer G. P., How tobacco smoke changes the (epi)genome. Science 354, 549–550 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Lan L., Nakajima S., Oohata Y., Takao M., Okano S., Masutani M., Wilson S. H., Yasui A., In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 13738–13743 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parlanti E., D’Errico M., Degan P., Calcagnile A., Zijno A., van der Pluijm I., van der Horst G. T., Biard D. S., Dogliotti E., The cross talk between pathways in the repair of 8-oxo-7,8-dihydroguanine in mouse and human cells. Free Radic. Biol. Med. 53, 2171–2177 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Olafsson S., McIntyre R. E., Coorens T., Butler T., Jung H., Robinson P. S., Lee-Six H., Sanders M. A., Arestang K., Dawson C., Tripathi M., Strongili K., Hooks Y., Stratton M. R., Parkes M., Martincorena I., Raine T., Campbell P. J., Anderson C. A., Somatic evolution in non-neoplastic IBD-affected colon. Cell 182, 672–684.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Boogaard M. L., Oka R., Hakkert A., Schild L., Ebus M. E., van Gerven M. R., Zwijnenburg D. A., Molenaar P., Hoyng L. L., Dolman M. E. M., Essing A. H. W., Koopmans B., Helleday T., Drost J., van Boxtel R., Versteeg R., Koster J., Molenaar J. J., Defects in 8-oxo-guanine repair pathway cause high frequency of C > A substitutions in neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 118, e2007898118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan A. R., BEDTools: The Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–11.12.34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagih O., ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Ernst J., Kellis M., Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ibarra A., Benner C., Tyagi S., Cool J., Hetzer M. W., Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30, 2253–2258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9