Abstract
The coenzyme A (CoA)-acylating aldehyde dehydrogenase (ALDH) catalyzes a key reaction in the acetone- and butanol (solvent)-producing clostridia. It reduces acetyl-CoA and butyryl-CoA to the corresponding aldehydes, which are then reduced by alcohol dehydrogenase (ADH) to form ethanol and 1-butanol. The ALDH of Clostridium beijerinckii NRRL B593 was purified. It had no ADH activity, was NAD(H) specific, and was more active with butyraldehyde than with acetaldehyde. The N-terminal amino acid sequence of the purified ALDH was determined. The open reading frame preceding the ctfA gene (encoding a subunit of the solvent-forming CoA transferase) of C. beijerinckii NRRL B593 was identified as the structural gene (ald) for the ALDH. The ald gene encodes a polypeptide of 468 amino acid residues with a calculated Mr of 51,353. The position of the ald gene in C. beijerinckii NRRL B593 corresponded to that of the aad/adhE gene (encoding an aldehyde-alcohol dehydrogenase) of Clostridium acetobutylicum ATCC 824 and DSM 792. In Southern analyses, a probe derived from the C. acetobutylicum aad/adhE gene did not hybridize to restriction fragments of the genomic DNAs of C. beijerinckii and two other species of solvent-producing clostridia. In contrast, a probe derived from the C. beijerinckii ald gene hybridized to restriction fragments of the genomic DNA of three solvent-producing species but not to those of C. acetobutylicum, indicating a key difference among the solvent-producing clostridia. The amino acid sequence of the ALDH of C. beijerinckii NRRL B593 was most similar (41% identity) to those of the eutE gene products (CoA-acylating ALDHs) of Salmonella typhimurium and Escherichia coli, whereas it was about 26% identical to the ALDH domain of the aldehyde-alcohol dehydrogenases of C. acetobutylicum, E. coli, Lactococcus lactis, and amitochondriate protozoa. The predicted secondary structure of the C. beijerinckii ALDH suggests the presence of an atypical Rossmann fold for NAD+ binding. A comparison of the proposed catalytic pockets of the CoA-dependent and CoA-independent ALDHs identified 6 amino acids that may contribute to interaction with CoA.
Acetone, 1-butanol, and 2-propanol (all solvents) can be produced to commercially important levels by several Clostridium species (10). Industrial production of solvents by fermentation has utilized different clostridia, including Clostridium acetobutylicum for the earlier starch-based Weizmann process and Clostridium beijerinckii and other clostridia for the later molasses-based processes (19, 21). Based on DNA-DNA reassociation and other characteristics, industrial strains of solvent-producing clostridia have been identified as strains of four species: C. acetobutylicum, C. beijerinckii, “Clostridium saccharoperbutylacetonicum,” and a presently unnamed species represented by strains NRRL B643 and NCP 262 (20, 22). Among these four species, 2-propanol production is a trait found only in some strains of C. beijerinckii (7, 20). However, 2-propanol is also produced by strains of Clostridium aurantibutyricum (12).
Most of the enzymes involved in the production of solvents have been purified (5, 9). Two enzymes, acetoacetate:butyrate/acetate coenzyme A (CoA) transferase and acetoacetate decarboxylase, are responsible for the production of acetone, and these two enzymes are conserved in C. acetobutylicum and C. beijerinckii (8, 29, 30, 34, 35, 35a). A primary-secondary alcohol dehydrogenase is responsible for the production of 2-propanol (15, 27). On the other hand, the enzymes that play a role in butanol production are not as well defined because of the presence of multiple primary alcohol dehydrogenases (ADHs) and possibly multiple aldehyde dehydrogenases (ALDHs) in these organisms (6, 9).
ALDH is responsible for the formation of butyraldehyde and acetaldehyde for the production of, respectively, 1-butanol and ethanol. ALDHs and the related dehydrogenases can be differentiated into two types by their dependence on CoA. The CoA-independent ALDHs (EC 1.2.1.3) are present in both prokaryotes and eukaryotes (see reference 28 and the web site cited therein), and they catalyze the irreversible oxidation of the aldehydes to their corresponding acids, such as the oxidation of acetaldehyde to acetic acid by the mitochondrial ALDH (ALDH2 or DHAM) in humans for the clearance of ethanol (39). The CoA-dependent ALDHs (EC 1.2.1.10) are predominantly found in bacteria, and they catalyze the reversible conversion of acyl-CoAs to their corresponding aldehydes (3). The physiological reaction of the CoA-dependent ALDH can be either the formation of acyl-CoAs, such as the conversion of acetaldehyde to acetyl-CoA in the utilization of ethanolamine by Salmonella typhimurium (31), or the formation of aldehydes by alcohol-producing bacteria (5).
For the reduction of butyryl-CoA to butyraldehyde in the solvent-producing clostridia, it was not clear whether the different species use similar ALDHs to catalyze the reaction. Palosaari and Rogers (26) first reported the purification of a CoA-acylating ALDH from Clostridium sp. strain NRRL B643; this was followed by a report detailing the purification of a CoA-acylating ALDH from C. beijerinckii NRRL B592 (37). The ALDHs from these two organisms have similar molecular properties. Different ALDH activities have been measured in cell extracts of C. acetobutylicum (1), but the purification of an ALDH from this species was only recently described (9). C. acetobutylicum also contains an aldehyde-alcohol dehydrogenase, encoded by the adhE (11) or the aad (25) gene. The Aad/AdhE protein has been purified, and it was found to exhibit a high level of ALDH activity (CoA dependent) but a low level of ADH activity (14).
To elucidate the enzymology of butanol formation, we wished to determine whether an aad/adhE-like gene is present in C. beijerinckii and whether similar ALDHs are present in significantly different strains of C. beijerinckii. The genomic DNA of C. beijerinckii NRRL B593 is 73% similar to that of C. beijerinckii NRRL B592, as measured by the DNA-DNA reassociation technique (20), suggesting that the two strains are only distantly related within the species, because a similarity level of 70% has been proposed as the limit for assigning strains to a bacterial species (17). The two strains also have distinct ADHs, which parallels the different capacities of the two strains to produce 2-propanol. In this paper, we report the purification of an ALDH from C. beijerinckii NRRL B593 and compare its properties to those of ALDHs purified from other clostridia. The structural gene for the ALDH of C. beijerinckii NRRL B593 was cloned and sequenced. Results of Southern analyses suggest that similar ALDHs are present in three species of solvent-producing clostridia other than C. acetobutylicum, whereas the aad/adhE gene is present only in C. acetobutylicum. A structural comparison of the CoA-dependent ALDHs with the CoA-independent ALDHs identified several amino acid residues that may play a role in the CoA-dependent reaction.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
Spores of C. beijerinckii NRRL B593 were produced in a potato extract-glucose medium (12) and stored at −70°C. C. beijerinckii cells were grown at 35°C and harvested between 10 and 12 h after inoculation (33). Cell paste was stored in liquid nitrogen until used. Cells for DNA isolation were grown as described previously (20), with the harvested cells being used immediately for DNA isolation. The sources for Clostridium strains were reported previously (20). Plasmids LITMUS 28 and 29 and Escherichia coli DH5α and DH5αF′ were maintained as recommended by New England BioLabs (Beverly, Mass.).
Protein determination.
Protein was determined by the Bradford dye binding assay (2), with bovine gamma globulin as a standard.
Enzyme assays.
ALDH was routinely assayed in the nonphysiological direction under Ar (37). Bovine serum albumin (1 mg/ml) was included when dilute ALDH samples were assayed. ADH activity with butyraldehyde was measured in either 50 mM potassium 2-(N-morpholino)ethanesulfonic acid (MES) buffer at pH 6 with 0.1 mM NADH or in 50 mM Tris-chloride buffer at pH 7.5 with 0.1 mM NADPH. Butyraldehyde (5.5 mM) was diluted 10-fold in methanol before use (38).
Purification of ALDH.
Cells were suspended in anaerobic 50 mM potassium HEPES buffer (1 g of cells per 3 ml of buffer), pH 8, containing lysozyme (2 mg/ml), DNase I (0.1 mg/ml), and dithiothreitol (DTT; 1 mM). Other steps were as described elsewhere (37). The crude extract was immediately mixed with anaerobic glycerol to give a final glycerol concentration of 20% (vol/vol). Purification of the ALDH was performed by a previously published procedure (37). Fractions with the highest purity were eluted from a Cibacron Blue 3GA-agarose column with 3 mM NAD+ in 50 mM Tris-acetate buffer, pH 7, also containing 20% (vol/vol) glycerol, 5 mM DTT, and 0.2 mM KCl. Purified ALDH was stored as frozen droplets in liquid nitrogen.
Determination of purity and the subunit and native Mrs.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed on slab gels (23) to determine the purity and the subunit Mr. Protein bands were stained with Coomassie brilliant blue. Mr standards used were bovine serum albumin (66,000), ovalbumin (45,000), glyceraldehyde-3-phosphate dehydrogenase (36,000), and carbonic anhydrase (29,000). The native Mr of purified ALDH was determined by gel filtration on a Sephacryl S-300 column (2.5 by 50 cm), which was eluted with Tris-acetate buffer (50 mM, pH 7.0) containing 0.1 M KCl, 5 mM DTT, and 20% (vol/vol) glycerol. Mr standards used were thyroglobulin (669,000), β-amylase (200,000), yeast ADH (150,000), conalbumin (77,000), and ribonuclease A (13,700).
Determination of coenzyme specificity.
The ALDH activity in the crude extract and in the purified enzyme sample was measured with 2 mM NAD(P)+ within the pH range of 6.5 to 9.5. Potassium 3-(N-morpholino)-propanesulfonic acid (MOPS) buffer (50 mM) was used at pH 6.5 and 7.5, whereas potassium 2-(N-cyclohexylamino)ethanesulfonate) (CHES) buffer (50 mM) was used at pH 6.5, 7.5, 8.5, and 9.5. The ALDH activity was also measured with 20 mM of NAD(P)+ at pH 8.6 in potassium CHES buffer. Other conditions were as for the routine assay.
The search for other ALDHs in crude extracts.
Crude extracts were fractionated by using either a Sephacryl S-300 column or a DE-52 column under the conditions described above. The fractions were assayed for acetaldehyde- and butyraldehyde-linked activities, using NAD+ as the coenzyme. The ratios of the butyraldehyde-linked activity to the acetaldehyde-linked activity of the active fractions and the patterns of elution of the ALDH from these columns were examined to determine the presence of different ALDHs.
Determination of the N-terminal amino acid sequence.
The purified ALDH was desalted in a Centricon 30 ultrafiltration unit (Millipore, Bedford, Mass.) and collected on the polyvinylidene difluoride membrane of a ProSpin cartridge (Perkin-Elmer Applied Biosystems, Foster City, Calif.). The N-terminal amino acid sequence was determined with an Applied Biosystems model 477A sequencer at the Protein Sequencing Facility of Virginia Tech.
Isolation of DNA from solvent-producing clostridia.
DNA was isolated by a variation of the Marmur procedure (18, 20). Cells of Clostridium sp. strain NCP 262 were converted to protoplasts before lysis (40).
DNA cloning and sequencing.
Plasmid DNA was purified by using a QIAprep Spin Plasmid Kit in accordance with the procedure recommended by the manufacturer (Qiagen, Valencia, Calif.). The genes encoding the acetoacetate:butyrate/acetate CoA transferase of C. beijerinckii NRRL B593 were cloned first. The probe for the CoA transferase genes was a 700-bp DNA fragment that was generated by PCR based on the N-terminal amino acid sequences of the two subunits (8). Automated DNA sequencing was performed with a DuPont Genesis 2000 sequencer, using single-stranded DNA as the template. The ctfA and ctfB genes for the small and large subunits, respectively, of the CoA transferase of C. beijerinckii NRRL B593 were detected on a 4.5-kb BglII fragment, which was cloned in plasmids pJT293 and pJT295 (data not shown) in opposite orientations (34, 35a). Analysis of the DNA sequence preceding the ctfA gene revealed an open reading frame (ORF), which has been identified as the structural gene (ald) for the ALDH. A 2.7-kb XbaI-EcoRI fragment containing the ald gene and part of the ctfA gene was subcloned into LITMUS 29, resulting in pJT308.
Analysis of DNA sequences.
Both the MacVector (Genetics Computer Group, Madison, Wis.) and the Lasergene (DNASTAR, Madison, Wis.) software packages were used for the management of DNA sequences. The BLAST programs, provided by the National Center for Biotechnology Information, were used in searching databases for related amino acid sequences.
Southern analyses of genomic DNA of solvent-producing clostridia.
PCR was used to amplify a region of the ald gene of C. beijerinckii NRRL B593 and a region of the adhE gene of C. acetobutylicum DSM 792 to be used as probes. The primer pair 5′-CATGAATAAAGACACACTAATAC and 5′-CAATAGTGAAAGTTGTAAATC amplified a 1,334-bp fragment that encompassed the first 444 amino acids of the C. beijerinckii NRRL B593 ALDH. The primer pair 5′-ATAAAGTCCGTGAAGTGATT and 5′-AGTACCCACATTAGCTTTGC amplified a 836-bp fragment that encompassed amino acid residues 278 to 556 of the C. acetobutylicum aldehyde-alcohol dehydrogenase.
Genomic DNA of the following strains were used in Southern analyses to determine the distribution of the ald gene and the aad/adhE gene among solvent-producing clostridia: C. acetobutylicum ATCC 824, DSM 792, and NRRL B528; C. beijerinckii VPI 5481, NCIMB 8052, NCIMB 6444, NRRL B592, NRRL B593, NCP 193, and ATCC 39057; an unnamed Clostridium species represented by strains NCP 262 and NRRL B643; and “C. saccharoperbutylacetonicum” N1-4. The ECL direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech, Piscataway, N.J.) was used under low-stringency conditions, with 1 M NaCl in the hybridization buffer and 0.3 M NaCl and 0.03 M sodium citrate (2× SSC) in the posthybridization wash buffer. Other conditions were as recommended by the manufacturer.
Nucleotide sequence accession number.
The sequence of the 2,697-bp insert in pJT308 has been deposited in the GenBank nucleotide sequence database under accession no. AF132754.
RESULTS AND DISCUSSION
Purification of ALDH.
The ALDH of C. beijerinckii NRRL B593 was purified 95-fold, with a 10% yield, from extracts of cells at the solvent-producing stage of growth, during which the level of ALDH activity is elevated (38). Throughout the purification, the buffer contained 20% (vol/vol) glycerol and 5 mM DTT; omission of DTT from the buffer resulted in a substantial loss of activity during ultrafiltration (data not shown).
The purified ALDH reached a specific activity of 3.8 U per mg of protein (1 U is defined as the amount of enzyme resulting in the production of 1 μmol of NADH per min with butyraldehyde as the substrate). In comparison, the purified ALDH from C. beijerinckii NRRL B592 had an activity of 2 U per mg of protein (37). Like the ALDHs of C. beijerinckii NRRL B592 (37) and Clostridium sp. strain NRRL B643 (26), the ALDH of C. beijerinckii NRRL B593 had no ADH activity. The purified ALDH gave a single band (data not shown) when examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and it corresponded to a subunit Mr of 57,000. The calculated Mr based on the deduced amino acid sequence (see below) was 51,353. The Mr of the native ALDH was 90,500 (data not shown), suggesting that the ALDH is a homodimer. The ALDHs of C. beijerinckii NRRL B592 (37) and Clostridium sp. strain NRRL B643 (26) had subunit Mrs of 55,000 and 56,000, respectively, and native Mrs of 100,000 and 115,000, respectively. The rat class 3 ALDH, for which the X-ray crystal structure has been solved, is a homodimer (24).
Coenzyme specificity of the ALDH.
Within the pH range of 6.5 to 9.5 and with 2 mM NAD+ as the coenzyme, the ALDH of C. beijerinckii NRRL B593 showed increasing activity with increasing pH (data not shown). Except at pH 8.5, the ALDH showed no detectable activity when 2 mM NADP+ was used as the coenzyme. At pH 8.5, the activity with NADP+ was about 5% of the activity with NAD+. No activity was detected when either coenzyme was used at a concentration of 20 mM. The ALDHs of C. beijerinckii NRRL B592 (37) and Clostridium sp. strain NRRL B643 (26) were active with either NAD(H) or NADP(H) as the coenzyme. However, the Vmax/Km values indicate that NAD(H) is a more effective coenzyme than NADP(H) for the ALDHs of C. beijerinckii NRRL B592 and Clostridium sp. strain NRRL B643.
The N-terminal amino acid sequence.
The N-terminal amino acid sequence of the purified ALDH of C. beijerinckii NRRL B593 was MNKD(T/N)LIPT(T/N)K(D/N)LKL(K/V)TNVENI(N/V)L. The sequence agreed with that deduced from the cloned gene (see below). For positions 5, 10, 12, 16, and 23 (in parentheses), two amino acids were detected at each position, and the amino acids in boldface were the predicted ones. We previously determined that the N-terminal amino acid sequence of the purified ALDH of C. beijerinckii NRRL B592 is MNKDTLIPTTKDLKVKTNGENINLK(N/D)YKD (36), which is very similar to the sequence of the C. beijerinckii NRRL B593 ALDH, and there was one uncertainty at position 26. It is unclear what caused the ambiguity at the positions described above.
The search for additional ALDHs of C. beijerinckii NRRL B593.
Multiple ALDHs with different substrate specificities are commonly observed in an organism, and the completed microbial genome sequences further corroborate the general presence of distinct ALDH-like genes in each organism (28, 39). Because more than one ALDH is present in C. acetobutylicum (1, 9), we examined extracts of C. beijerinckii NRRL B593 to determine if more than one ALDH is produced by this organism. We compared the ratios of the butyraldehyde-linked and the acetaldehyde-linked ALDH activities (the B/A ratio) in the crude extract and in ALDH samples at different stages of purification, using NAD+ as the coenzyme (there was little NADP+-linked ALDH activity in this organism).
At all stages of purification, the acetaldehyde-linked ALDH activity was less than 10% of the butyraldehyde-linked activity. The measurement of acetaldehyde-linked ALDH activity in the crude extract was prone to error because of the intrinsically low level of activity and because of the presence in the crude extract of a relatively high-level endogenous NAD+-reducing activity. This interfering NAD+-reducing activity was not dependent on CoA or DTT, which were normally present in the assay mixture, but it was dependent on a low-molecular-weight substance(s) that could be completely removed by passing the crude extract through an anaerobic Sephadex G-25 column (data not shown). When measuring the ALDH activity in dilute samples of purified ALDH, addition of bovine serum albumin to the assay mixture at a 1-mg/ml final concentration was necessary for an accurate activity measurement.
When the acetaldehyde-linked activity was properly measured, a B/A ratio of about 8 was observed for the ALDH of C. beijerinckii NRRL B593 at different stages of purification. During the purification, only one ALDH activity peak was eluted from each type of chromatographic column used (data not shown). Because the butyraldehyde-linked activity was accountable throughout the purification, the purified ALDH should be the predominant, if not the only, ALDH present in solvent-producing cells of C. beijerinckii NRRL B593. The result did not exclude, however, the possible presence in C. beijerinckii of other types of ALDH that may be expressed under different growth conditions.
The nucleotide sequence of the ald gene and its deduced amino acid sequence.
The ald gene was located on a 2,697-bp XbaI-EcoRI fragment (GenBank accession no. AF132754). The coding region was 1,404 bp long (nucleotides 882 to 2288), and it was preceded by a putative ribosome-binding site (GGAG) at −13 to −10 bp from the start codon. The predicted ALDH polypeptide was 468 amino acid residues in length, with a calculated Mr of 51,353 and a predicted pI of 5.631. The N-terminal 24 amino acid residues matched the sequence determined from the purified ALDH.
There was no ORF in the 881-bp region upstream from the ald gene. The ORF 90 bp downstream from the ald gene was the beginning of the ctfA gene, which encodes the small subunit of the acetoacetate:butyrate/acetate CoA transferase (8, 34). The ctfA gene was preceded by a putative ribosome-binding site (GGAG) at −11 to −8 bp from the start codon, and there was no potential transcription termination site between the ald and ctfA genes. Northern analysis of RNA isolated from solvent-producing cells of C. beijerinckii NRRL B593 revealed a ca. 4-kb band that hybridized to the ald and ctfA genes, but the transcription start site for the mRNA has not yet been accurately located (data not shown). In C. beijerinckii NCIMB 8052, the mRNA for the corresponding region was also approximately 4 kb in length (4), suggesting that the organization of the solvent production genes is conserved in these two strains of C. beijerinckii.
Southern analysis of solvent-producing clostridia for the presence of DNA sequences related to the ald gene of C. beijerinckii or the aad/adhE gene of C. acetobutylicum.
The ALDH of C. beijerinckii is distinct from Aad/AdhE of C. acetobutylicum. To see if the different species of the solvent-producing clostridia possess one or both of the genes encoding the two proteins, we generated two probes representing the two genes. The aad/adhE probe encompassed amino acid residues 278 to 556 to include regions that are highly conserved between the adhE (aad) genes of C. acetobutylicum and E. coli (25). In Aad/AdhE, amino acid residue 400 is roughly the end of the ALDH domain and amino acid residue 450 is roughly the start of the ADH domain.
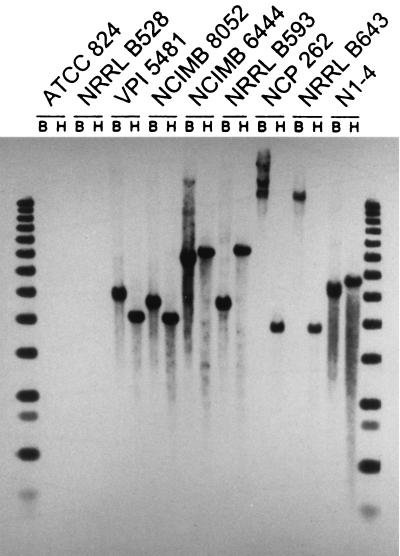
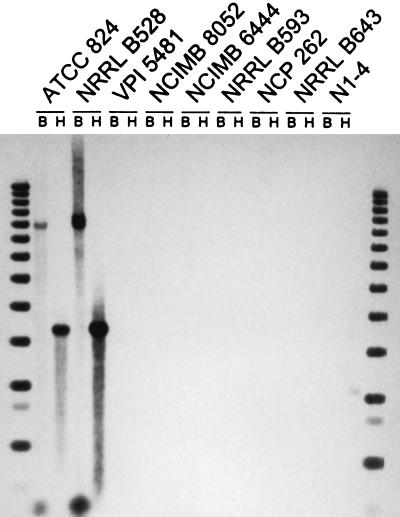
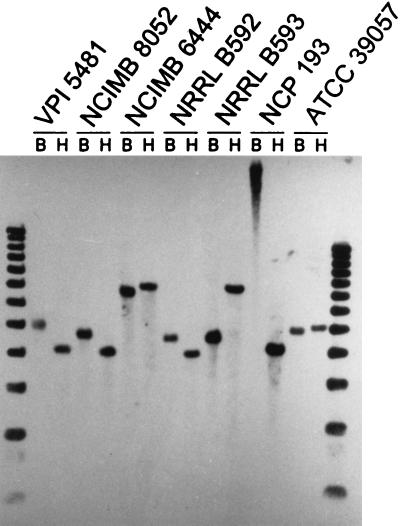
The results of the Southern analysis showed that an ald-like DNA sequence was not present in C. acetobutylicum but was present in the other three species of solvent-producing clostridia (Fig. 1). In contrast, an aad/adhE-like sequence was present in C. acetobutylicum but not in the other three species of solvent-producing clostridia (Fig. 2). In addition, an ald-like sequence was present in all seven strains of C. beijerinckii examined, although the size of the ald-hybridizing fragment differed among the strains (Fig. 3).
FIG. 1.
Southern hybridization analysis of four species of solvent-producing clostridia for the presence of DNA sequences related to the ald gene of C. beijerinckii NRRL B593. Lanes B and H contained, respectively, BglII- and HindIII-digested genomic DNA of the following strains: C. acetobutylicum ATCC 824 (type strain) and NRRL B528; C. beijerinckii VPI 5481 (type strain), NCIMB 8052, NCIMB 6444, and NRRL B593; two strains (NCP 262 and NRRL B643) belonging to an unnamed species; and “C. saccharoperbutylacetonicum” N1-4. The border lanes contained 1-kb ladders.
FIG. 2.
Southern hybridization analysis of four species of solvent-producing clostridia for the presence of DNA sequences related to the aad/adhE gene of C. acetobutylicum. Strains tested were those listed in the legend to Fig. 1. Lanes B and H contained, respectively, BglII- and HindIII-digested genomic DNA of each strain. The border lanes contained 1-kb ladders.
FIG. 3.
Southern hybridization analysis of strains of C. beijerinckii for the presence of DNA sequences related to the ald gene of C. beijerinckii NRRL B593. Lanes B and H contained, respectively, BglII- and HindIII-digested genomic DNA of the indicated strains of C. beijerinckii. The border lanes contained 1-kb ladders.
C. acetobutylicum ATCC 824 and NRRL B528 represent the two distinct groups of this species as differentiated by the DNA-DNA reassociation method (20). The two strains showed the same hybridization pattern when tested with the aad/adhE probe, each displaying an 8-kb BglII fragment and a 3.5-kb HindIII fragment (Fig. 2). The 3.5-kb HindIII fragment is expected from C. acetobutylicum ATCC 824, based on the reported nucleotide sequence (25). Therefore, although the two strains can be differentiated by the DNA-DNA reassociation technique, which measures the relatedness of the DNA sequence throughout the genome, the aad/adhE region is conserved in the two strains.
When tested with the ald probe, BglII- or HindIII-digested DNAs from representative strains of C. beijerinckii, “C. saccharoperbutylacetonicum,” and an unnamed solvent-producing species (represented by strains NCP 262 and NRRL B643) each gave a strong hybridizing band, except for strain NCP 262, which gave two additional high-molecular-weight bands compared with BglII-digested DNA from strain NRRL B643 (Fig. 1). The size of the hybridizing fragment differed among the three species. When DNAs from seven strains of C. beijerinckii were tested with the ald probe, four strains (VPI 5481 [type strain], NCIMB 8052, NRRL B592, and NCP 193) gave a 4-kb HindIII fragment, but the size of the hybridizing BglII fragment differed among the strains (Fig. 3).
Alignment of the amino acid sequences of CoA-dependent and CoA-independent ALDHs.
ALDHs and related proteins form an extended family whose members share a conserved tertiary structure, but they do not exhibit a high degree of sequence identity (13, 28). A BLASTP search of the nonredundant sequence databases through the National Center for Biotechnology Information identified 232 amino acid sequences (with an E value [i.e., the number of times one expects to get a match, based on chance] of less than 1) related to the ALDH of C. beijerinckii NRRL B593. Table 1 shows the relatedness between the ALDH of C. beijerinckii NRRL B593 and selected proteins representing a broad range of organisms. In the conserved region, the level of identity ranged from 44 to 17%, and the aligned proteins included CoA-dependent and CoA-independent ALDHs, semialdehyde dehydrogenases, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenases, and crystallins. The length of the conserved region ranged from about 165 to 460 amino acid residues, which may reflect the different substrate and coenzyme specificities of the ALDHs.
TABLE 1.
Relatedness of the C. beijerinckii ALDH and selected members of the extended ALDH family
| Organism and proteina | Conserved region
|
% Identity based on the full length of C. beijerinckii NRRL B593 ALDH | Accession no. | ||
|---|---|---|---|---|---|
| Positionsb | E valuec | % Identityd | |||
| Giant octopus Ω-crystallin | 2–423, 8–449 | 0.047 | 18.7 (86/460) | 18.4 | L06902 |
| S. typhimurium EutE | 35–466, 31–460 | 1 × 10−101 | 44.3 (192/433) | 41.0 | AAA80209 |
| Entamoeba histolytica Adh2 | 36–431, 9–415 | 1 × 10−45 | 29.8 (122/410) | 26.1 | Q24803 |
| G. intestinalis AdhE | 36–454, 22–448 | 4 × 10−36 | 28.3 (123/435) | 26.3 | U93353 |
| C. kluyveri SSDH | 39–417, 6–382 | 5 × 10−42 | 29.5 (114/387) | 24.4 | P38947 |
| C. acetobutylicum Aad/AdhE | 50–439, 19–409 | 3 × 10−41 | 29.9 (120/402) | 25.6 | P33744 |
| E. coli AdhE | 58–433, 26–406 | 4 × 10−48 | 31.4 (122/389) | 26.1 | P17547 |
| L. lactis AdhE | 58–433, 47–431 | 9 × 10−39 | 28.8 (111/386) | 23.7 | CAA04467 |
| Rat class 3 ALDH | 128–466, 99–429 | 6 × 10−7 | 19.5 (69/354) | 14.7 | P11883 |
Abbreviations: Aad, AdhE, and Adh2, aldehyde-alcohol dehydrogenases; EutE, acetaldehyde dehydrogenase encoded by the eutE gene; SSDH, succinate-semialdehyde dehydrogenase.
The first range in each pair corresponds to the C. beijerinckii NRRL B593 ALDH; the second corresponds to the protein listed in the left column on that line.
The E value is the number of times one expects to see such a match (or a better one) merely by chance.
The percent identity is based on the total number of positions used in the alignment, including the introduced gaps.
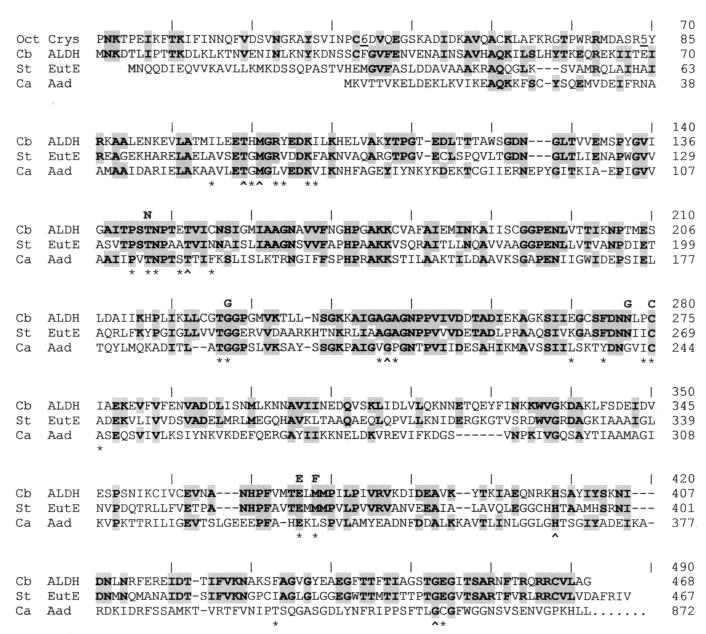
The C. beijerinckii NRRL B593 ALDH was most highly related to the CoA-dependent ALDHs, and the conserved region spanned nearly the entire length of the polypeptide. Figure 4 illustrates the degree of similarity among three CoA-dependent ALDHs. Based on the location and length of the conserved region, the relationship between the C. beijerinckii NRRL B593 ALDH and the other ALDHs falls into several classes. (i) The C. beijerinckii NRRL B593 ALDH was most similar to the eutE-encoded acetaldehyde dehydrogenases of S. typhimurium (accession no. AAA80209) and E. coli (P77445), yielding E values of 10−101 and 10−100, respectively, for the region between residues 35 and 468 (the C terminus). (ii) Between residues 40 and 435, the C. beijerinckii NRRL B593 ALDH was related to the ALDH domains of the aldehyde-alcohol dehydrogenases of E. coli (P17547), Entamoeba histolytica (Q24803 and S53319), C. acetobutylicum (P33744 and A49346), Lactococcus lactis (CAA04467), and Giardia intestinalis (U93353) and the succinate-semialdehyde dehydrogenase of Clostridium kluyveri (P38947), with E values of 4 × 10−48 to 4 × 10−3. (iii) Between residue 60 and the C terminus, the C. beijerinckii NRRL B593 ALDH was related to the ALDHs of Caenorhabditis elegans (AAB66022), mouse DHA4 (P47740), rat DHA4 (P30839), and human ALDH7 (P43353), with E values of 1 × 10−14 to 9 × 10−8. (iv) Between residue 130 and the C terminus, the C. beijerinckii NRRL B593 ALDH was related to human ALDH8 (P48448) and ALDH10 (P51648), rat ALDH3 (P11883), and maize RF2 (U43082), with E values of 10−7 to 10−5. (v) Between residues 130 and 295, the C. beijerinckii NRRL B593 ALDH was related to ALDHs of a wide range of organisms, with E values of 2 × 10−8 to 1 × 10−4. The region encompassed by residues 130 and 295 of the C. beijerinckii NRRL B593 ALDH is hence the most conserved region among ALDHs, and it corresponds to the NAD-binding domain of ALDHs.
FIG. 4.
Alignment of the amino acid sequences of three CoA-dependent ALDHs and the N-terminal region of an ALDH-related protein. Markers above the sequences indicate the positions used in the alignment of the C. beijerinckii NRRL B593 ALDH. The sources and abbreviations are as follows: Oct Crys, Ω-crystallin of the giant octopus (41); Cb ALDH, ALDH of C. beijerinckii NRRL B593 (accession no. AF132754); St EutE, the eutE-encoded ALDH of S. typhimurium (AAA80209); Ca Aad, the ALDH domain encoded by the aad gene of C. acetobutylicum ATCC 824 (25). Amino acid residues that are shared by the C. beijerinckii NRRL B593 ALDH and at least one of the other sequences are highlighted. Positions of amino acids that line the proposed catalytic pocket of the rat ALDH3 are indicated by an asterisk or a caret (the latter indicates a residue that is conserved in nine CoA-dependent ALDHs but not in CoA-independent ALDHs at these positions). An underlined number in a sequence indicates the number of amino acids omitted at that position to save space. Dashes in the sequence indicate the gaps that were introduced to refine the alignment. Boldfaced letters above the sequences indicate amino acid residues conserved in previously aligned ALDHs with demonstrated dehydrogenase activity (28).
There exists an interesting relationship between the terminal region of the C. beijerinckii NRRL B593 ALDH and those of some moderately related proteins. The N-terminal region of the C. beijerinckii NRRL B593 ALDH was most related to the corresponding region of the giant octopus Ω-crystallin (Fig. 4), an ALDH-related structural protein found in cephalopod (squid and octopus) lenses (41). This relatedness is intriguing considering the evolutionary distance between the two organisms. The C-terminal region of the C. beijerinckii NRRL B593 ALDH, on the other hand, was highly related to those of ALDHs from Salmonella, Escherichia, Mycobacterium, maize, rat, mouse, cattle, and humans, but not to the corresponding region in the Aad/AdhE proteins of bacteria and protozoa (Fig. 4 and data not shown). The terminal regions of the ALDHs may be especially informative for the deduction of the evolutionary pathway for the ALDHs.
Although ALDHs share few invariant amino acid residues, their tertiary structures are conserved, including an atypical Rossmann fold for NAD binding, as seen in their X-ray crystal structures (16, 24, 32). When 145 full-length ALDH-related sequences were aligned recently (28), only 6 amino acid residues (shown above the sequences in Fig. 4) were invariant among sequences with demonstrated dehydrogenase activity. This recent alignment (28) did not, however, include the CoA-dependent bacterial and protozoal ALDHs covered in this study. When the CoA-dependent ALDHs were included, only 3 amino acid residues were invariant: Gly at alignment position 227, Cys at position 280, and Glu at position 376. Two of these residues (Gly and Glu) are for NAD binding, and the other (Cys) is involved in substrate binding (28).
Both CoA and NAD contain an ADP moiety. Whether the binding of CoA and NAD involves separate or overlapping regions of the ALDH remains to be elucidated, but the result of the activation experiment (37) suggests that the binding of CoA and NAD involves a shared region that probably recognizes the ADP moiety of CoA and NAD. It can be speculated that in CoA-dependent ALDHs, some of the amino acids that line the proposed catalytic pocket (24) recognize and interact with CoA and are characteristic of these ALDHs. A comparison of these positions between nine CoA-dependent ALDHs and the CoA-independent ALDHs identified the six residues (marked by carets under the sequences in Fig. 4) Thr-89, Met-91, Thr-148, Gly-242, His-398, and Gly-449 as being characteristic of the CoA-dependent ALDHs. Among these, Thr-148, Gly-242, His-398, and Gly-449 were invariant in all nine CoA-dependent ALDHs examined. Thr-89 was replaced by Ser in E. coli AdhE, whereas Met-91 was replaced by Arg in AdhE of Entamoeba histolytica and L. lactis. These residues may be further analyzed for a possible role in the enzyme’s interaction with CoA.
The predicted secondary structure and coenzyme-binding sites of the ALDH of C. beijerinckii NRRL B593.
The secondary-structure prediction for the C. beijerinckii NRRL B593 ALDH was performed by the Chou-Fasman method and the Robson-Garnier method as implemented by MacVector (Genetics Computer Group). The predicted secondary structure of the C. beijerinckii NRRL B593 ALDH was compared with the X-ray crystal structures of a dimeric ALDH (24) and two tetrameric ALDHs (16, 32) to allow a prediction of the coenzyme-binding sites in the former. Figure 5 shows the consensus prediction for an α-helix and a β-sheet in the NAD-binding domain. Like the other ALDHs, the C. beijerinckii NRRL B593 ALDH also lacked a GXGXXG motif in an atypical Rossmann fold for NAD binding.
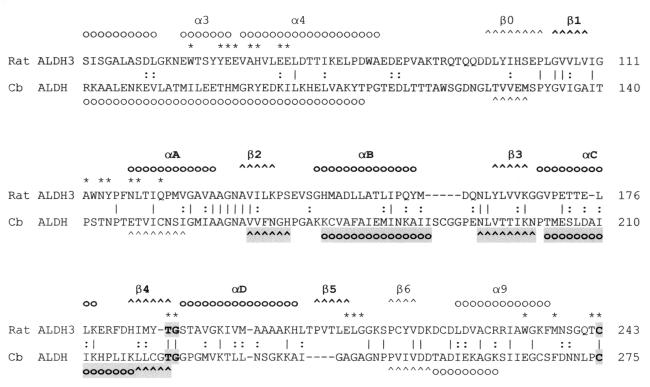
FIG. 5.
Alignment of the region of the amino acid sequence that forms the Rossmann fold in the class 3 ALDH of rat (Rat ALDH3) with the corresponding region of the ALDH of C. beijerinckii NRRL B593 (Cb ALDH). Positions of amino acids that are identical in the two sequences are linked by vertical lines; those occupied by similar amino acids are linked by colons. The predicted secondary structure of the C. beijerinckii NRRL B593 ALDH is compared with the X-ray crystal structure of the rat ALDH3 (24). The α-helices and β-sheets are marked, respectively, by circles and carets either above or below the amino acid sequence. The secondary structures in the NAD-binding domains of the two ALDHs that are apparently conserved are highlighted below the C. beijerinckii ALDH sequence. Amino acid residues that line the proposed catalytic pocket of the rat ALDH3 are marked by asterisks above the sequence. Dashes in the sequence indicate the gaps that were introduced to refine alignment.
Although the amino acid sequences of the C. beijerinckii NRRL B593 ALDH and rat ALDH3 exhibit only a low level of similarity, several regions of α-helices and β-sheets are conserved between the two ALDHs. The β-α-β-α-β motif lying between residues 160 and 230 of the C. beijerinckii NRRL B593 ALDH (highlighted in Fig. 5) closely resembles the β2-αB-β3-αC-β4 motif of rat ALDH3. The invariant Gly-223 at the end of β4 plays a crucial role in NAD binding (24). This region of the C. beijerinckii NRRL B593 ALDH is probably also a crucial part of the NAD-binding domain, although it does not seem to have the β1-αA and αD-β5 flanking regions present in rat ALDH3. We also subjected the amino acid sequences of rat ALDH3 (24) and cod liver betaine ALDH (16) to secondary-structure prediction with MacVector and compared the results with the X-ray crystallographic data. Not surprisingly, αA, αD, and β5 of rat ALDH3 were not predicted. With the cod liver ALDH, structures corresponding to αA, αB, αC, β4, and β5 of rat ALDH3 were not predicted. Therefore, the β-α motif of the NAD-binding site of the C. beijerinckii NRRL B593 ALDH may resemble that of rat ALDH3 to a greater extent than was predicted by the method used in this study.
Between residues 240 and 330, the α-helices and β-sheets also resemble the β6-α9-β7-α10-α11-β8 region of rat ALDH3, and this region contains the invariant Cys-275, which is the catalytic thiol for the ALDHs. During the BLASTP search, the region bounded by residues 130 and 295 of the C. beijerinckii NRRL B593 ALDH aligned with the largest number of ALDH-related proteins. The conservation between the CoA-dependent C. beijerinckii NRRL B593 ALDH and the CoA-independent rat ALDH3 of the secondary structure in this region suggests that the domain surrounding the catalytic thiol and the NAD-binding site is preserved among all ALDHs.
Concluding remarks.
We have determined that a CoA-acylating ALDH is the predominant ALDH in solvent-producing cells of C. beijerinckii. Results of Southern analyses showed that the ald gene encoding this ALDH is present in strains of C. beijerinckii and two other species of solvent-producing clostridia but not in C. acetobutylicum. In contrast, the aad/adhE gene, encoding an aldehyde-alcohol dehydrogenase, was detected only in C. acetobutylicum. The reversible CoA-acylating ALDH is a key enzyme in bacterial alcohol production, but little is known about its structure and function because most of the current knowledge about the structure of ALDH was based on the CoA-independent ALDHs, which irreversibly oxidize aldehydes to the corresponding acids. To further enhance the properties of ALDHs for industrial alcohol production, future research may be directed toward the elucidation of the structural elements that determine the substrate and coenzyme specificities as well as the catalytic efficiency of the CoA-acylating ALDH.
ACKNOWLEDGMENTS
This study was supported by U.S. Department of Energy grants DE-FG05-85-ER13368 and DE-FG02-97-ER20276 and by the CSREES, U.S. Department of Agriculture, under project 6122000.
REFERENCES
- 1.Bertram J, Kuhn A, Dürre P. Tn916-induced mutants of Clostridium acetobutylicumdefective in regulation of solvent formation. Arch Microbiol. 1990;153:373–377. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Burton R M, Stadtman E R. The oxidation of acetaldehyde to acetyl coenzyme A. J Biol Chem. 1953;20:873–890. [PubMed] [Google Scholar]
- 4.Chen C-K, Blaschek H P. Effect of acetate on molecular and physiological aspects of Clostridium beijerinckiiNCIMB 8052 solvent production and strain degeneration. Appl Environ Microbiol. 1999;65:499–505. doi: 10.1128/aem.65.2.499-505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J-S. Properties of acid- and solvent-forming enzymes of clostridia. In: Woods D R, editor. The clostridia and biotechnology. Stoneham, Mass: Butterworth-Heinemann; 1993. pp. 51–76. [PubMed] [Google Scholar]
- 6.Chen J-S. Alcohol dehydrogenase: multiplicity and relatedness in the solvent-producing clostridia. FEMS Microbiol Rev. 1995;17:263–273. doi: 10.1111/j.1574-6976.1995.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen J-S, Hiu S F. Acetone-butanol-isopropanol production by Clostridium beijerinckii (synonym, Clostridium butylicum) Biotechnol Lett. 1986;8:371–376. [Google Scholar]
- 8.Colby G D. CoA-transferase and 3-hydroxybutyryl-CoA dehydrogenases: acetoacetyl-CoA-reacting enzymes from Clostridium beijerinckii NRRL B593. Ph.D. thesis. Blacksburg: Virginia Polytechnic Institute and State University; 1993. [Google Scholar]
- 9.Dürre P. New insights and novel developments in clostridial acetone/butanol/isopropanol fermentation. Appl Microbiol Biotechnol. 1998;49:639–648. [Google Scholar]
- 10.Dürre P, Bahl H. Microbial production of acetone/butanol/isopropanol. In: Roehr M, editor. Products of primary metabolism. 2nd ed. Vol. 6. Weinheim, Germany: VCH Publishers; 1996. pp. 230–268. [Google Scholar]
- 11.Fischer R J, Helms J, Dürre P. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol. 1993;175:6959–6969. doi: 10.1128/jb.175.21.6959-6969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George H A, Johnson J L, Moore W E C, Holdeman L V, Chen J-S. Acetone, isopropanol, and butanol production by Clostridium beijerinckii (syn. Clostridium butylicum) and Clostridium aurantibutyricum. Appl Environ Microbiol. 1983;45:1160–1163. doi: 10.1128/aem.45.3.1160-1163.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hempel J, Nicholas H, Lindahl R. Aldehyde dehydrogenase: widespread structural and functional diversity within a shared framework. Protein Sci. 1993;2:1890–1900. doi: 10.1002/pro.5560021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ismaiel A A, Chen J-S. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C.: American Society for Microbiology; 1998. Purification of the aldehyde-alcohol dehydrogenase encoded by the aad gene from Clostridium acetobutylicum ATCC 824, abstr. O-40; p. 400. [Google Scholar]
- 15.Ismaiel A A, Zhu C-X, Colby G D, Chen J-S. Purification and characterization of a primary-secondary alcohol dehydrogenase from two strains of Clostridium beijerinckii. J Bacteriol. 1993;175:5097–5105. doi: 10.1128/jb.175.16.5097-5105.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson K, El-Ahmad M, Ramaswamy S, Hjelmqvist L, Jornvall H, Eklund H. Structure of betaine aldehyde dehydrogenase at 2.1Å resolution. Protein Sci. 1998;7:2106–2117. doi: 10.1002/pro.5560071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson J L. Bacterial classification. III. Nucleic acids in bacterial classification. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. pp. 8–11. [Google Scholar]
- 18.Johnson J L. Similarity analysis of DNAs. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 655–682. [Google Scholar]
- 19.Johnson J L, Chen J-S. Taxonomic relationships among strains of Clostridium acetobutylicumand other phenotypically similar organisms. FEMS Microbiol Rev. 1995;17:233–240. [Google Scholar]
- 20.Johnson J L, Toth J, Santiwatanakul S, Chen J-S. Cultures of “Clostridium acetobutylicum” from various collections comprise Clostridium acetobutylicum, Clostridium beijerinckii, and two other distinct types based on DNA-DNA reassociation. Int J Syst Bacteriol. 1997;47:420–424. doi: 10.1099/00207713-47-2-420. [DOI] [PubMed] [Google Scholar]
- 21.Jones D T, Keis S. Origins and relationships of industrial solvent-producing clostridial strains. FEMS Microbiol Rev. 1995;17:223–232. [Google Scholar]
- 22.Keis S, Bennett C F, Ward V K, Jones D T. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int J Syst Bacteriol. 1995;45:693–705. doi: 10.1099/00207713-45-4-693. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z-J, Sun Y-J, Rose J, Chung Y-J, Hsiao C-D, Chang W-R, Kuo I, Perozich J, Lindahl R, Hempel J, Wang B-C. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 25.Nair R V, Bennett G N, Papoutsakis E T. Molecular characterization of an alcohol/aldehyde dehydrogenase gene of Clostridium acetobutylicumATCC 824. J Bacteriol. 1994;176:871–885. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palosaari N R, Rogers P. Purification and properties of the inducible coenzyme A-linked butyraldehyde dehydrogenase from Clostridium acetobutylicum. J Bacteriol. 1988;170:2971–2976. doi: 10.1128/jb.170.7.2971-2976.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peretz M, Bogin O, Tel-Or S, Cohen A, Li G, Chen J-S, Burstein Y. Molecular cloning, nucleotide sequencing, and expression of genes encoding alcohol dehydrogenase from the thermophile Thermoanaerobacter brockii and the mesophile Clostridium beijerinckii. Anaerobe. 1997;3:259–270. doi: 10.1006/anae.1997.0083. [DOI] [PubMed] [Google Scholar]
- 28.Perozich J, Nicholas H, Wang B-C, Lindahl R, Hempel J. Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 1999;8:137–146. doi: 10.1110/ps.8.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen D J, Bennett G N. Purification of acetoacetate decarboxylase from Clostridium acetobutylicum ATCC 824 and cloning of the acetoacetate decarboxylase gene in Escherichia coli. Appl Environ Microbiol. 1990;56:3491–3498. doi: 10.1128/aem.56.11.3491-3498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen D J, Cary J W, Vanderleyden J, Bennett G N. Sequence and arrangement of genes encoding enzymes of the acetone-production pathway of Clostridium acetobutylicumATCC 824. Gene. 1993;123:93–97. doi: 10.1016/0378-1119(93)90545-e. [DOI] [PubMed] [Google Scholar]
- 31.Roof D M, Roth J R. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol. 1992;174:6634–6643. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinmetz C G, Xie P, Weiner H, Hurley T D. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 33.Thompson D K, Chen J-S. Purification and properties of an acetoacetyl coenzyme A-reacting phosphotransbutyrylase from Clostridium beijerinckii (“Clostridium butylicum”) NRRL B593. Appl Environ Microbiol. 1990;56:607–613. doi: 10.1128/aem.56.3.607-613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toth J, Chen J-S. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C.: American Society for Microbiology; 1997. Cloning and sequence analysis of the ctfA and ctfB genes encoding the acetoacetate:butyrate/acetate coenzyme A-transferase from Clostridium beijerinckii NRRL B593, abstr. H-8; p. 286. [Google Scholar]
- 35.Toth J, Chen J-S. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C.: American Society for Microbiology; 1998. Organization of the acetone-butanol production genes in Clostridium beijerinckii NRRL B593, abstr. O-39; p. 399. [Google Scholar]
- 35a.Toth, J., G. D. Colby, and J.-S. Chen. Unpublished data.
- 36.Yan R-T. Enzymology of butanol formation in Clostridium beijerinckii NRRL B592. Ph.D. thesis. Blacksburg: Virginia Polytechnic Institute and State University; 1991. [Google Scholar]
- 37.Yan R-T, Chen J-S. Coenzyme A-acylating aldehyde dehydrogenase from Clostridium beijerinckiiNRRL B592. Appl Environ Microbiol. 1990;56:2591–2599. doi: 10.1128/aem.56.9.2591-2599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan R-T, Zhu C-X, Golemboski C, Chen J-S. Expression of solvent-forming enzymes and onset of solvent production in batch cultures of Clostridium beijerinckii (“Clostridium butylicum”) Appl Environ Microbiol. 1988;54:642–648. doi: 10.1128/aem.54.3.642-648.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida A, Rzhetsky A, Hsu L C, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- 40.Zappe H, Jones D T, Woods D R. Cloning and expression of Clostridium acetobutylicum endoglucanase, cellobiase and amino acid biosynthesis genes in Escherichia coli. J Gen Microbiol. 1986;132:1367–1372. doi: 10.1099/00221287-132-5-1367. [DOI] [PubMed] [Google Scholar]
- 41.Zinovieva R D, Tomarev S I, Piatigorsky J. Aldehyde dehydrogenase derived Ω-crystallins of squid and octopus. J Biol Chem. 1993;268:11449–11455. [PubMed] [Google Scholar]