Abstract
Purpose
Despite the growing interest in bacteriophage (phage) usage for the prevention, control, and removal of bacterial biofilms, few scientific data exist on phage applications on medical implant surfaces, while none exists on multiple implants. In this study, we aimed to isolate, biophysically characterize and assess phages as potential antibiofilm agents to inhibit and remove multidrug-resistant (MDR) Pseudomonas aeruginosa biofilm on catheter and endotracheal tube surfaces.
Methods
The well-identified stored clinical isolates (n = 7) of MDR P. aeruginosa were obtained from Jimma Medical Center. Specific phages were isolated and characterized based on standard protocols. The phages were tested for their antibiofilm effects in preventing colonization and removing preformed biofilms of MDR P. aeruginosa, following phage coating and treatment of catheter and endotracheal tube segments.
Results
Two P. aeruginosa-specific phages (ΦJHS-PA1139 and ΦSMK-PA1139) were isolated from JMC compound sewage sources. The phages were biophysically characterized as being thermally stable up to 40°C and viable between pH 4.0 and 11.0. The two phages tested against clinical MDR strains of P. aeruginosa showed broad host ranges but not on other tested bacterial species. Both phages reduced MDR bacterial biofilms during the screening step. The phage-coated segments showed 1.2 log10 up to 3.2 log10 inhibition relative to non-coated segments following 6 h coating of segments prior to microbial load exposure. In both phages, 6 h treatment of the segments with 106 PFU/mL yielded 1.0 log10 up to 1.6 log10 reductions for ΦJHS and 1.6 log10 up to 2.4 log10 reductions for ΦSMK.
Conclusion
Our results suggest that phages have great potential to serve the dual purpose as surface coating agents for preventing MDR bacterial colonization in medical implants and as biofilm removal agents in implant-associated infections.
Keywords: bacteriophage, biofilm, multidrug-resistant P. aeruginosa, catheter, endotracheal tube
Introduction
Pseudomonas aeruginosa (PA), a ubiquitous, opportunistic, and notorious biofilm-forming bacteria, causes a wide range of severe life-threatening hospital-acquired infections (HAIs). These are associated with contamination of medical devices, equipment used in hospitals, and other hard or liquid surfaces, which act as reservoirs for biofilm-acquired infections. The ability of PA to remain viable on hospital devices or grow in used dilutions of disinfectants is unmatched, as survival advantage results from its nutritional versatility, unique outer membrane that forms an effective barrier to the passage of antimicrobials, and/or efflux systems.1 As a more frequently transferable bacteria in clinical settings, PA is responsible for high rates of morbidity and mortality due to its resistance to several antibiotics, which is attributed to multidrug efflux pumps, extracellular polymeric substance (EPS) –protected biofilm that makes its outer membrane impermeable to antibiotics, mutation, and acquisition of resistance genes.2
The report from the National Healthcare Safety Network managed by the Centers for Disease Control and Prevention (CDC) in 2020 stated that more than 28,000 PA were isolated from adult HAIs in the United States, representing 8.0% of total pathogens isolated. Adult HAIs reported, include device-associated: central line-associated bloodstream infections (CLABSIs), catheter-associated urinary tract infections (CAUTIs), ventilator-associated pneumonia (VAP); and surgical site infections (SSIs). P. aeruginosa was among the 3 most frequently reported CAUTI and VAP pathogens in device-associated adult HAIs. In CAUTI, PA was reported in 22.6% of long-term acute-care hospitals, 15.4% from inpatient rehabilitation facilities, 13.2% from hospital oncology units, and 12.8% from hospital wards and intensive care units. Likewise, in VAP, PA was reported in 32.6% of long-term acute-care hospitals, 21.8% from hospital wards, and 12.9% from hospital intensive care units.3 In sub-Saharan Africa, inadequate comprehensive data exist on the burden of PA in HAIs due to the lack of resources for a surveillance system. However, studies available in Ethiopia reported 14.3–18.4% isolation rate of PA in HAIs.4,5
On AMR pattern of device-associated HAIs, PA exhibited 26.2% resistance to fluoroquinolones (ciprofloxacin or levofloxacin), 20.7% resistance to carbapenems (imipenem, meropenem, or doripenem), 20.3% resistance to extended-spectrum cephalosporins (cefepime or ceftazidime), 15.0% resistance to piperacillin or piperacillin/tazobactam, and 14.4% resistance to aminoglycosides (amikacin, gentamicin or tobramycin). In SSIs, PA exhibited 11.0%, 9.1%, 10.2%, 7.7%, and 5.8% resistance to the antibiotic classes above, in the same order, respectively. The multidrug-resistant (MDR) pattern, operationalized as resistance to one agent in at least three of the different antibiotic classes above, showed that PA exhibited 14.2% resistance in device-associated HAIs and 4.5% resistance in SSIs.3
The potential threat of MDR PA biofilms in medical implants has resulted in a growing interest in the development of antimicrobial-coated biomaterials. The use of essential oils6 and metal/hydroxyapatite-synthesized nanoparticles7 has demonstrated significant antibacterial activities against implant-related infections. In addition, the application of quaternary ammonium compounds, chlorhexidine, antibiotics, and antimicrobial peptides, has shown a significant decrease in biofilms on implant surfaces.8,9 However, these antimicrobial-coated biomaterials have been implicated in bacterial resistance in some studies.9–11 Novel strategies to prevent device-associated infections by material scientists, biologists, and microbiologists are urgently needed. Hence, new approaches to prevent, control, and remove MDR PA on medical devices and implants should be investigated. For microbiologists, this includes the use of bacteriophages (phages).
Bacteriophages, first identified and characterized in the twentieth century, are bacterial viruses that infect bacterial cells with high specificity.12 Phages are reckoned to be the most bountiful life forms on earth with numbers estimated to be 10 times more than their bacterial hosts.13 Phage therapy was being practiced once globally before the advent of antibiotics. It has been reemerging recently in the world due primarily to the threat posed by the increasing incidence of antibiotic-resistant bacteria, coupled with the scarcity of new antibiotics.14 Similarly, the ineffectiveness of antibacterial drug interventions on biofilms has brought about a steep growth of scientific interest in phages as an alternative remedy in controlling and preventing biofilm formation.15 Indeed, the interaction of phage strategies and biofilms as research titles in scientific publications has jumped up rapidly in the last decade.16
Despite a plethora of studies on phage – biofilm interactions, few reports exist on phage applications on implant surfaces, though promising results were reported recently on new isolated lytic bacteriophages against endotracheal tube (ET)-associated PA biofilms17,18 and E. faecalis cells in biofilms formed on Foley silicone catheters.19 To date, no data has been reported on phage coating of multiple implants to serve the dual purpose of bacterial colonization prevention and removal of preformed biofilms. As one of the most frequently isolated pathogens in CAUTIs and VAP, the burden of PA in catheters and ET cannot be over-emphasized. To the best of our knowledge, this research is one of the few studies on phage therapy in sub-Saharan Africa, particularly against biofilm-associated with medical implants. Therefore, in this study, we aimed to isolate, biophysically characterize and assess phages as potential antibiofilm agents to prevent and remove MDR P. aeruginosa clinical isolate biofilm on catheter and ET surfaces.
Materials and Methods
Clinical Isolates and Growth Conditions
The clinical bacterial isolates used in this study were obtained from Jimma Medical Center (JMC) microbiology laboratory. Stored MDR PA isolates were recovered from different specimens of patients (Table 1). The isolates were biochemically re-identified as PA strains through different biochemical reactions. From pure cultures grown on Columbia agar (CA; bioMérieux, Marcy l’Etoile, France), bacteria inoculums were checked for their multidrug resistance properties, as depicted in Table 1. Furthermore, pure cultures were prepared and suspended in sterile 0.85% NaCl and kept in the refrigerator at 4°C until use. Inoculums were routinely grown with agitation on nutrient broth (NB; Oxoid, Hampshire, UK) at 37°C for phage isolation tests. Biofilms were grown in tryptic soy broth (TSB; Oxoid) containing 1% glucose at 37°C.
Table 1.
Clinical Bacterial Isolates and Their Antimicrobial Resistance Patterns
| Isolates | Source | Antibiotic Resistance |
|---|---|---|
| PA 1095 | Wound abscess | AMP, AMC, CTX, CXM, TZP, TET |
| PA 1098 | Sputum | AMP, AMC, AMK, CRO, CXM, CHL, SXT, CN, MEM |
| PA 1139 | Wound abscess | AMP, AMC, AMK, CXM, CHL, SXT, CRO, CTX, MEM |
| PA 1280 | Pleural fluid | AMP, AMC, CFZ, CXM, CHL, SXT, TET |
| PA 1321 | Wound abscess | AMP, AMC, CFZ, CAZ, CTX, CXM, CHL, SXT, |
| PA 1329 | Wound abscess | AMP, AMC, CAZ, CRO, CTX, CXM, CHL, SXT, CN, TET, TOB |
| PA 1668 | Urine | AMP, AMK, CRO, CXM, CIP, CN, NIT, NOR |
Abbreviations: PA, Pseudomonas aeruginosa, AMC, amoxicillin-clavulanic acid; AMK, amikacin; AMP, ampicillin; CAZ, ceftazidime; CFZ, cefazolin; CHL, chloramphenicol; CIP, ciprofloxacin; CN, gentamicin; CRO, ceftriaxone; CTX, cefotaxime; CXM, cefuroxime; MEM, meropenem; NIT, nitrofurantoin; NOR, norfloxacin; SXT, sulfamethoxazole-trimethoprim; TET, tetracycline; TOB, tobramycin; TZP, piperacillin-tazobactam.
Isolation of Bacteriophages from Hospital Sewage
Bacteriophage Isolation and Enrichment
Isolation of bacteriophages specific to PA was carried out from hospital sewage sources (JMC, Ethiopia) according to the standard enrichment protocol described earlier20 with some modifications. The clinical PA 1095, 1139, and 1329 strains were chosen at random and used as the host strains for phage isolation. Sewage samples were collected in sterile 500 mL containers from three different areas around the JMC compound and quickly transported to the medical microbiology laboratory of Jimma University for enrichment. Using 50 mL falcon tubes, sewage samples were centrifuged at 10,000×g, 4°C for 15 min, to remove particulate materials. The supernatants were filter-sterilized through 0.45 μm membrane filters and mixed with equal volume (50 mL) of sterile double strength NB containing 2mM MgCl2, alongside 5 mL log phase of grown PA host strains. After an overnight aerobic incubation at 37°C with frequent agitation, the mixtures were centrifuged at 10,000×g, 4°C for 15 mins, filter sterilized through 0.45 μm membrane filters, and enriched for the second time with the same host strain to amplify the filtrate.
Spot Assay
The amplified filtrates obtained above were re-filtered through sterile membranes of pore size 0.45 μm and tested for phage activity following the spot assay as described elsewhere.14 Briefly, 100 μL of PA hosts inoculum was added to 5–7 mL molten soft agar and poured onto the CA plate surface. After solidification, 10 μL of amplified filtrates were spotted and plates were allowed to dry (absorb) at room temperature (RT) for a few minutes and incubated overnight at 37°C. Positive spotted phage activities were purified by successive single plaque isolation until homogenous plaques were obtained according to the standard procedure described previously.21 In a 5 mL broth of fresh log-phase PA host, one plaque from a plate was added and incubated at 37°C under shaking conditions, alongside a control tube without the host strain until complete lysis occurred in the test preparation. Afterward, tubes were centrifuged at 10,000×g, 4°C for 15 minutes. Supernatants were chloroform treated and serially diluted for plaque assay. The procedure was repeated three times to ascertain the purity and activity of isolated phages.
Quantitative Assay of Bacteriophages
The number of phage particles or titers was estimated by the double agar overlay method as described before by counting plaque-forming units per milliliter (PFU/mL).22 Serial dilutions of phage lysates (10 folds) were made in sterile saline magnesium (SM) buffer solution (100 mM NaCl, 25 mM Tris-HCl, 8 mM MgSO4, pH 7.5). Then, 100 μL of phage suspension from each dilution mixed with 100 μL of host inoculum in a 5–7 mL molten soft agar, was quickly poured on the CA plate surface without creating air bubbles. After an overnight incubation at 37°C aerobically, the number of plaques was counted and plates with 20–200 plaques were selected to determine the phage titer from two countable plates. Subsequently, plaques morphology and diameter were determined.22
Determination of Host Ranges of Phages
The host ranges of phages were determined following the standard spot test procedure described earlier14 with undiluted phage stocks, having a predetermined plaque count of 107 or 106 per milliliter of phage lysates. All seven MDR PA isolates as well as ten clinically used American Type Culture Collection (ATCC) strains of PA and other species were used in this experiment, as listed in Table 2. Briefly, 100 µL of each of the bacterium inoculum was mixed with 5–7 mL of molten soft agar and layered over a CA plate. The phage lysates were serially diluted (10-folds) in SM buffer and 10 µL was spotted onto the solidified soft agar with the bacterium. Plates were left undisturbed until the drop became absorbed in RT. After an overnight incubation aerobically at 37°C, bacterial sensitivity to phages was confirmed by the presence of a zone of clearance at the sites of phage application. Positive spotted tests were assayed for plaques to verify the lysis and sensitivity to the phages. The obtained results were differentiated as clear plaques or complete lysis (++), turbid plaque or partial lysis (+), or no plaque or no lysis (–).
Table 2.
Lytic Activity of Phages ΦJHS and ΦSMK Against Tested Bacterial Strains
| Bacterial Strain | Phage Sensitivity | |
|---|---|---|
| ΦJHS | ΦSMK | |
| Pseudomonas aeruginosa 1095 | + | + |
| Pseudomonas aeruginosa 1098 | ++ | ++ |
| Pseudomonas aeruginosa 1139* | ++ | ++ |
| Pseudomonas aeruginosa 1280 | ++ | + |
| Pseudomonas aeruginosa 1321 | + | + |
| Pseudomonas aeruginosa 1329 | ++ | ++ |
| Pseudomonas aeruginosa 1668 | ++ | ++ |
| Pseudomonas aeruginosa ATCC 27853 | ++ | + |
| Escherichia coli ATCC 25922 | – | – |
| Salmonella typhimurium ATCC 13311 | – | – |
| Proteus mirabilis ATCC 35659 | – | – |
| Klebsiella pneumoniae ATCC 700603 | – | – |
| Enterobacter cloacae ATCC 13047 | – | – |
| Acinetobacter baumannii ATCC 19606 | – | – |
| Shigella dysenteriae ATCC 13313 | – | – |
| Staphylococcus aureus ATCC 25923 | – | – |
| Staphylococcus saprophyticus ATCC 15305 | – | – |
Notes: (++), clear plaques or complete lysis; (+), turbid plaque or partial lysis; (–) no plaque or no lysis; (*), host.
External Factors Stability Tests on Phages
The stability of phages at varying ranges of temperature and pH is important for their ability to act at various physiological or environmental conditions. The stability of phages to different physical and chemical factors, including temperature, pH, and organic solvents was tested according to the protocols described earlier23 with some modifications. All assays were performed in triplicates and plating was by double agar overlay procedure.
Temperature Stability Assay
Phage suspensions were diluted in SM buffer (1:9 dilution) and incubated for 1 h at different temperatures (15, RT, 37, 40, 50, and 90°C). Next, the phage suspension was withdrawn, 10-fold serially diluted in SM buffer, and used for plating. After an overnight incubation at 37°C, the percentage of viable phages able to lyse the host bacterial cells was estimated. Initial plaque count of phages kept at 4°C was taken as control.
pH Stability Assay
Phage suspensions (100 µL) were diluted in SM buffer (900 µL) of different pH values (2.0, 4.0, 7.0, 10.0 and 12.0) with 1 M HCl or 1 M NaOH and incubated for 1 h at 37°C. Next, 10-fold serial dilutions were prepared in SM buffer and used for plating. After an overnight incubation at 37°C, the percentage of viable phages able to lyse host bacterial cells was calculated. Plaque count of phages incubated in SM buffer of pH 7.0 was taken as control.
Organic Solvents Tolerance
The stability of phage particles was tested against three different organic solvents: ethanol, acetone, and chloroform. A stock solution of phage lysate was added to chloroform, acetone, 96%, and 48% ethanol. The mixture was incubated for 1.5 h at RT (chloroform, acetone, and ethanol). Next, 10-fold serial dilutions in SM buffer were prepared and used for plating. After an overnight incubation at 37°C, the percentage of viable phages able to lyse host bacterial cells was estimated. Phage particles incubated in SM buffer under the conditions described above were used as controls.
Biofilm Formation Assay
A quantitative assessment of MDR PA biofilm formation was performed based on the method described earlier24 with some modifications. Briefly, the concentrations of overnight cultures of PA were adjusted to that of the turbidity of a 0.5 McFarland standard. The suspensions were then diluted to 100-fold, containing approximately 106 colony forming units (CFU/mL) in a TSB medium supplemented with 1% glucose. 200 µL of these were seeded into a sterile flat-bottomed 96-well polystyrene microplate (Greiner Bio-one CELLSTAR). PA static biofilms were grown at 37°C for 24 h without renewal of media. After incubation, the non-adherent cells were removed from the wells and washed twice with 200 µL of sterile phosphate-buffered saline (PBS), pH 7.4. Biofilm was fixed with methanol for 15 min, and it was removed, air-dried, and stained with 220 µL of 0.1% crystal violet for 15 min at RT. Again, the wells were washed twice with PBS to remove excess stain and allowed to air-dry. The stained biofilms were solubilized with 220 µL of 96% ethanol for 15 min and optical density (O.D) of eluted stain was measured with a microtiter plate reader (Elisys Uno Human) at a wavelength of 630 nm. For quantitative assays, experiments were performed in triplicate wells. Sterile TSB medium in the wells left without the bacteria was used as the negative control. The cut-off O.D for biofilm formation and its strength were calculated and defined as three standard deviations above the mean O.D of the negative control.24
Screening of Lytic Activity of Phages on Biofilms
To test the lytic activity of phages in mature biofilms, static biofilms of MDR PA were cultured as described above. After incubation, biofilms were washed in PBS, pH 7.4, to remove planktonic cells, and then proceeded to phage treatment. Two hundred microliters of phage lysates were added to each well to a final titer of 106 PFU per well, agitated at 120 rpm for 1 h, and incubated for 6 h at 37°C. Control biofilms of MDR PA were treated with sterile TSB in place of the phage. Next, the mixture was removed and washed twice with 200 µL of PBS. The plate was fixed with methanol, and phage treated and untreated biofilms proceeded with crystal violet staining and measurement of O.D as described above. All assays were conducted in triplicate wells.
Assessment of Phage Activity on Biofilm Formation in Catheters and Endotracheal Tubes
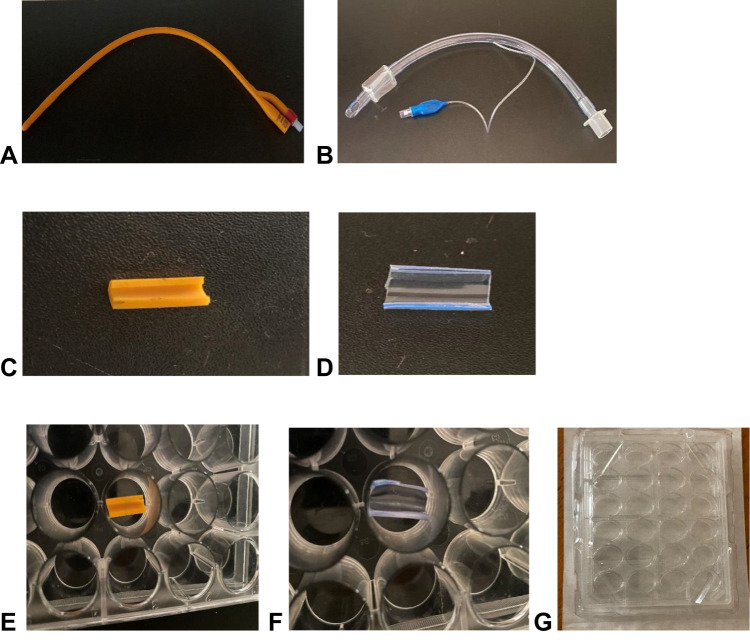
PA 1098 and 1668 strains, isolated from sputum and urine, respectively, were selected for this experiment. The two PA strains were also adequately lysed by the isolated virulent bacteriophages. Sterile silicone Foley balloon catheters (Ramsons Int, Noida, India) and Endotracheal Tubes (Henso Ltd., Hangzhou, China) were prepared before experimentation, as described earlier19 with some modifications. Briefly, with the use of sterile scissors, catheters and ETs were cut into 15-mm and 12-mm-long segments, respectively, followed by cutting in half lengthwise to expose the interior surfaces of the tubes (Figure 1A–D). The segments were soaked in 70% ethanol followed by UV light irradiation for 2 h. The segments were then placed in sterile flat-bottomed 24-well polystyrene culture plates (Becton Dickinson Labware, NJ, USA) for pre- and post-treatment with phage lysates (Figure 1E–G).
Figure 1.
Medical implants used for biofilm experiment. (A) Foley balloon catheter. (B) Endotracheal tube. (C) 15-mm long catheter tube cut in half. (D) 12-mm long endotracheal tube cut in half. (E) Catheter segment and (F) endotracheal tube segment placed in (G) sterile 24-well culture plate.
Pre-Treatment Experiments
1 mL of phage lysates containing 106 PFU/mL were added to the sterile segments, agitated at 120 rpm for 1 h, and incubated for 6 h at 37°C to allow phage adsorption on the catheter and ET surfaces. Afterward, the suspension was removed and segments were washed with PBS, pH 7.4, to remove non-adhered phages. Control segments were covered with a sterile TSB medium. Phage-coated and non-coated catheter and ET segments were covered with 1 mL of prepared overnight cultures of MDR PA in TSB, containing approximately 106 bacterial cells as described above. Plates were incubated at 37°C for 24 h with static-non-renewal conditions for biofilm formation assessment.
Post-Treatment Experiments
Catheter and ET segments were covered with 1 mL of MDR PA cultures in TSB, containing approximately 106 bacterial cells, and incubated for 96 h at 37°C for biofilm formation, with the renewal of half the volume of media every 24 h to mimic in vivo contamination conditions. Afterward, the medium with planktonic bacterial cells was aspirated from each well-containing segment and washed twice with PBS, pH 7.4. Segments were then treated with 1 mL for each of 102, 104, and 106 PFU/mL titer or 1 mL of sterile TSB (control), agitated for 1 h at 120 rpm, and incubated for 6 h at 37°C.
Recovery and Determination of Surface-Attached Bacteria Cells
After the required incubation time, the liquid contents of the wells were removed, and segments were washed twice with PBS, and aseptically transferred to Eppendorf tubes containing 1 mL of 0.85% NaCl. The tubes were vortexed at a maximum speed for 60 s to detach cells from the segments. Ten-fold serial dilutions were prepared in 0.85% NaCl, and 100 µL of each dilution was spread onto CA plates. After an overnight incubation at 37°C, the number of viable recovered PA cells was estimated based on counted colonies and expressed as log10CFU/mL.
Data Analysis
All experimental data were analyzed as mean ± standard deviation (SD) using GraphPad Prism, version 9.0.0 for Windows (GraphPad Software, San Diego, CA, USA). Statistically significant differences between the mean values of experimental samples and controls were performed using an unpaired t-test followed by Bonferroni-Dunn multiple comparisons test and were marked with asterisks when p < 0.05 (⁎), p < 0.01 (⁎⁎), or p < 0.001 (⁎⁎⁎).
Results
Isolation of Phages Specific Against Pseudomonas aeruginosa
Two PA bacteriophages were isolated from samples of hospital sewage after testing for the presence of phages that may infect MDR PA clinical strains. The phages were named ΦJHS-PA1139 and ΦSMK-PA1139 (hereafter, called ΦJHS and ΦSMK, respectively) based on the source of the sewage (Jimma Hospital Sewage; sewage from Surgical, Medical, and Kitchen block) and the host strain used for propagation (PA 1139). Wastewater samples were collected from JMC, Ethiopia, in June 2021.
Plaques Morphology
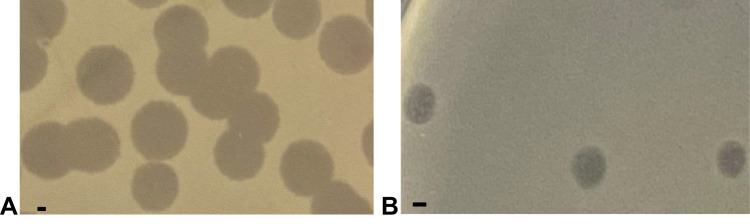
Phages ΦJHS and ΦSMK were propagated using the host strain, MDR PA 1139, from clinical specimens. Subjecting the phage lysates to further analysis revealed that both phages formed clear plaques on the lawn of the host and produced complete lysis in moderate titers (1.0–6.0 х107 PFU/mL) for ΦJHS and (1.0–5.0 × 106 PFU/mL) for ΦSMK. Plaques of ΦJHS (Figure 2A) and ΦSMK (Figure 2B) had an average diameter of about 5 mm and 2 mm, respectively, on the lawn of the PA 1139 host. Such plaque morphology indicated that these viruses are lytic bacteriophages.
Figure 2.
Plaques formed by bacteriophages (A) ΦJHS and (B) ΦSMK on the lawn of PA 1139 strain using the double agar overlay method. The bar corresponds to 1 mm.
Host Ranges
The host ranges of phages ΦJHS and ΦSMK were tested with clinical PA strains as well as strains of other clinically relevant Gram-negative and Gram-positive species. Both phages exhibited a broad host range against tested PA clinical strains from different patients. The proportion of clear complete lysis was 75% (6/8) for ΦJHS and 50% (4/8) for ΦSMK. To verify the positive spotted results, further spot tests in serial dilutions were performed to obtain plaques that were assayed in all cases. Compared to the host, the plaque sizes did not vary among the sensitive strains tested. No cross-sensitivity to non-PA strains, such as E. coli ATCC 25922, A. baumannii ATCC 19606, and S. aureus ATCC 25923 was detected as appeared in Table 2.
Stability of Phages ΦJHS and ΦSMK to External Factors
The stability of ΦJHS and ΦSMK to various physical and chemical factors, including different temperatures, pH conditions, and organic solvents, were tested. The virions of both phages appeared relatively stable for various temperatures and pH conditions, though the virions could not withstand extreme conditions (pH of 2.0 and 12.0 and temperature of 90°C). The stability of organic solvents varied depending on the nature of the tested solution. Notwithstandingly, the virions of both phages could not survive under acetone and 96% ethanol (Table 3).
Table 3.
Stability of Phages ΦJHS and ΦSMK to External Physical and Chemical Factors
| External Factors | Time and Conditions of Incubation | Percentage Viability of Phages ± SD | |
|---|---|---|---|
| ΦJHS | ΦSMK | ||
| Temperature | |||
| 4°C* | 1 h | 100 ± 0.0 | 100 ± 0.0 |
| 15°C | 1 h | 100 ± 7.5 | 100 ± 4.4 |
| RT | 1 h | 100 ± 6.5 | 100 ± 3.2 |
| 37°C | 1 h | 100 ± 6.1 | 100 ± 6.5 |
| 40°C | 1 h | 100 ± 3.1 | 100 ± 5.5 |
| 50°C | 1 h | 76.9 ± 0.6 | 55.4 ± 2.5 |
| 90°C | 1 h | 0.0 ± 0.0 | 0.0 ± 0.0 |
| pH | |||
| pH 2.0 | 1 h; 37°C | 0.0 ± 0.0 | 0.0 ± 0.0 |
| pH 4.0 | 1 h; 37°C | 38.0 ± 2.0 | 29.8 ± 0.6 |
| pH 7.0* | 1 h; 37°C | 100.0 ± 0.0 | 100.0 ± 0.0 |
| pH 10.0 | 1 h; 37°C | 78.9 ± 3.1 | 68.1 ± 1.1 |
| pH 12.0 | 1 h; 37°C | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Organic solvent | |||
| SM buffer* | 1.5 h; RT | 100 ± 0.0 | 100 ± 0.0 |
| Chloroform | 1.5 h; RT | 82.0 ± 2.5 | 70.8 ± 1.5 |
| Acetone | 1.5 h; RT | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 96% ethanol | 1.5 h; RT | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 48% ethanol | 1.5 h; RT | 90.0 ± 4.9 | 57.0 ± 2.1 |
Note: (*), controls.
MDR Pseudomonas aeruginosa Biofilm Formation
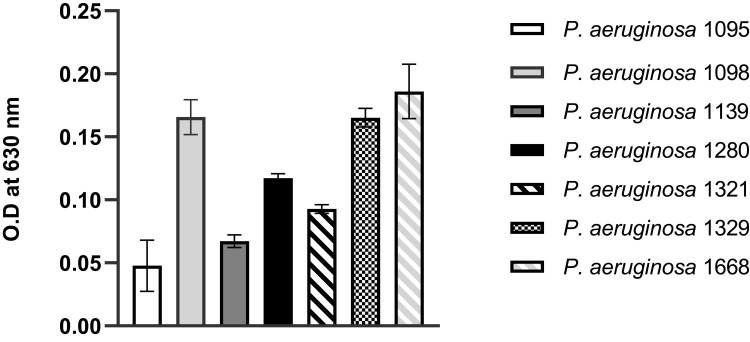
Biofilms of MDR PA were cultured for 24 h at static non-renewal conditions in 96-well culture plates and stained with crystal violet. The intensity of the color generated by the biofilms with crystal violet (O.D measured at 630 nm) is a direct indication of the biomass formed. In general, all the MDR PA clinical isolates obtained from JMC were biofilm formers. Categorically, except for PA 1095 and PA 1139 strains that exhibited weak and moderate biofilms, respectively, all isolates were strong biofilm formers (Figure 3).
Figure 3.
Biofilms formed by MDR PA clinical isolates cultured for 24 h at static non-renewal condition, as analyzed by crystal violet staining procedure and shown as O.D values measured at a wavelength of 630 nm. All assays were performed in triplicates. The values presented are mean ± SD from two readings of triplicate experiments (n = 6).
Screening of Phage Effect on MDR Pseudomonas aeruginosa Biofilms
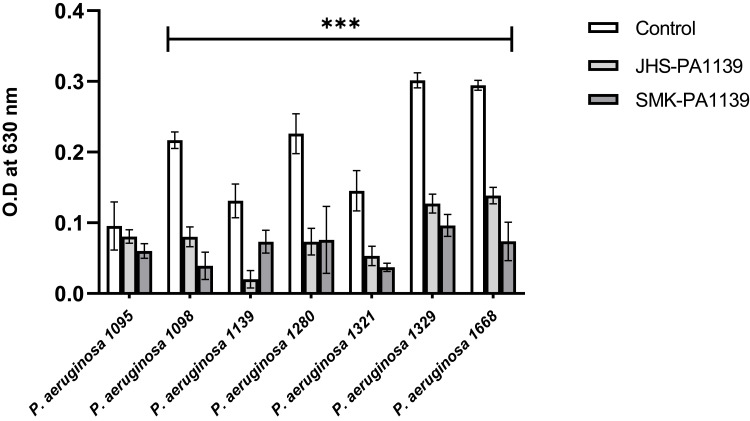
The lytic activity of phages ΦJHS and ΦSMK on biofilms formed by clinical isolates of MDR PA was assessed. Twenty-four-hour-old biofilms obtained under static non-renewal conditions were treated with the phages for 6 h. After incubation with the phages, biofilms were stained with crystal violet and the biomasses were determined by O.D measurement at 630 nm. As depicted in Figure 4, upon biofilm treatment with phages ΦJHS and ΦSMK, except for PA 1095, biomasses of MDR PA 1098, 1139, 1280, 1321, 1329, and 1668 decreased significantly (p < 0.001, n = 6).
Figure 4.
Lytic activity of bacteriophages on biofilms formed by MDR PA clinical isolates after 6 h treatment with phages ΦJHS-PA1139 and ΦSMK-PA1139, as analyzed by crystal violet staining procedure and shown as O.D values measured at a wavelength of 630 nm. The values presented are mean ± SD from two readings of triplicate experiments (n= 6). Statistically significant differences between control and analyzed samples are marked with asterisks (p < 0.001 (⁎⁎⁎) in the multiple unpaired t-tests).
Effect of Phage Coating on Biofilm Formation on Catheters and Endotracheal Tubes
The coating effect of phages ΦJHS and ΦSMK against MDR PA biofilm formation on catheters and ETs was assessed. Catheter and ET segments were first coated with ΦJHS and ΦSMK in a final titer of 106 PFU/mL for 6 h incubation before biofilm formation under static non-renewal conditions. Following 24 h incubation at 37°C, the number of viable surface-attached bacterial cells was estimated.
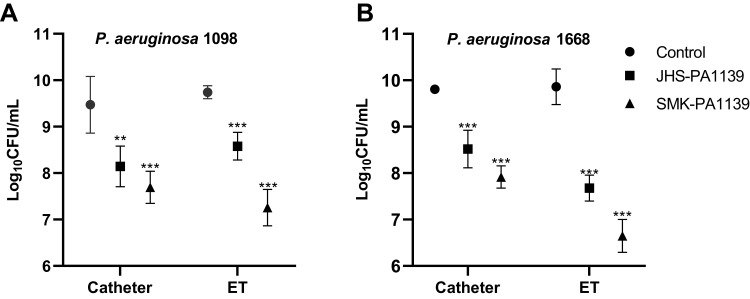
Comparing the CFU values, microbial load on non-coated catheter and ET segments were similar among both strains. However, different microbial growth patterns were observed on phage-coated segments. Phage-coated segments showed statistically significant log10 inhibition of microbial load compared to the controls. Phage ΦJHS achieved 1.3 log10 inhibition on catheter segments in both strains, while phage ΦSMK achieved 1.8 log10 inhibition. The activities of the phages were, however, different and dominant in ET segments. Phage ΦJHS exhibited a 1.2 log10 inhibition in PA 1098 strain and a 2.2 log10 (> 99%) inhibition in the PA 1668 strain. In the same order, phage ΦSMK achieved 2.5 log10 inhibition and 3.2 log10 (> 99.9%) inhibition, respectively (Figure 5).
Figure 5.
Microbial load inhibition of MDR PA 1098 (A) and 1668 (B) biofilm formation on non-coated and phage-coated catheter and ET segments, quantified as log10CFU/mL. The values presented are mean ± SD from two counts of triplicate experiments (n= 6). Statistically significant differences between control and analyzed samples are marked with asterisks (p < 0.01 (⁎⁎), p < 0.001 (⁎⁎⁎) in the multiple unpaired t-tests).
Effect of Phage Treatment of Preformed Biofilms on Catheter and Endotracheal Tubes
The treatment effects of phages ΦJHS and ΦSMK on MDR PA biofilms formed on catheters and ETs were assessed. In an in vitro model, MDR PA biofilms were formed on catheter and ET segments for 96 h with the renewal of media to mimic in vivo contamination conditions, followed by treatment with phages ΦJHS and ΦSMK at different titers of 102, 104, and 106 PFU/mL. Following 6 h incubation at 37°C, the number of viable surface-attached bacterial cells was estimated.
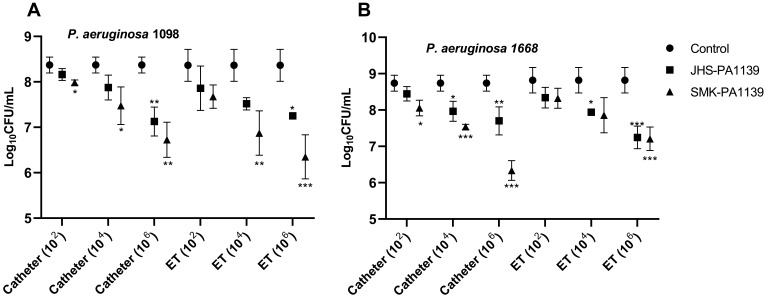
Comparing the CFU values, treatment with lower titers resulted in a slight reduction in microbial load. At a titer of 102 PFU/mL, a statistically significant reduction in microbial load was achieved with phage ΦSMK with 0.4 log10 (p = 0.03, n = 6) (Figure 6A) and 0.7 log10 (p = 0.02, n = 6) (Figure 6B) reductions on catheter segments relative to control experiment (phage-untreated segments). Both phages were, however, effective at titers of 104 PFU/mL with statistically significant log10 reduction values. With the application of this titer, phage ΦJHS was only efficient at reducing the number of viable cells with 0.8 log10 and 0.9 log10 reductions on catheter and ET segments colonized by PA 1668 strain, respectively. Treatment with phage ΦSMK, however, resulted in 0.9 log10 and 1.2 log10 (> 90%) reductions on catheter segments colonized by PA 1098 and PA 1668, respectively, and 1.5 log10 reductions on ET segments colonized by PA 1098 strain. The most efficient log10 reductions were achieved when phages were applied at titers of 106 PFU/mL relative to the control experiment. Phage ΦSMK achieved the highest reduction and was therefore most efficient with 2.4 log10 (> 99%) reduction (p < 0.001, n = 6) (Figure 6B). With regard to phage ΦJHS, a 1.6 log10 reduction was observed as the highest log10 reduction value in comparison to control. The results presented indicate that phages ΦJHS and ΦSMK were efficient in reducing the number of viable MDR PA cells in biofilms formed on catheters and endotracheal tubes.
Figure 6.
Microbial load reduction with 6 h phage treatment of MDR PA 1098 (A) and 1668 (B) biofilms formed on catheter and ET segments, quantified as log10CFU/mL. The values presented are mean ± SD from two counts of triplicate experiments (n = 6). Statistically significant differences between control and analyzed samples are marked with asterisks (p < 0.05 (⁎), p < 0.01 (⁎⁎), p < 0.001 (⁎⁎⁎) in the multiple unpaired t-tests).
Discussion
As natural enemies of bacteria, bacteriophages can be found wherever bacteria lives, spanning from the ocean and freshwater environments to terrestrial and complex environments, with an estimate of at least ten phages existing for each bacterial/archaeal cell.25 In our study, two PA bacteriophages (ΦJHS and ΦSMK) were isolated from hospital sewage in Jimma, Ethiopia, and their biophysical characteristics are described. Moreover, the antibiofilm potential of the phages in preventing colonization and removing preformed biofilm of MDR P. aeruginosa on Foley catheter and ET surfaces is described in our study. In light of the search for alternative approaches to combat biofilm-forming PA and its associated infections in implants, the use of these bacterial viruses that are bountiful on earth and as such economical necessitates our study.
The isolated phages exhibited similarities in their plaque morphologies by forming clear plaques on the lawn of the host strain, although significant differences in plaques size were noted. Depending on the life cycle, phages exist as lytic – by lysing the host cell following genome replication, or lysogenic – by persisting within the host genome as prophage. The formation of clear lytic plaques on the lawn of the host indicated that specific lytic bacteriophages were isolated, with the ability to replicate and rapidly destroy the MDR PA host. Such a feature confers therapeutic potentials in lytic phages as candidates for phage therapy. With phages known to be specific to their hosts and in features, the significant difference in plaque size indicated the existence of distinct bacteriophages, yet infecting the same host bacteria.
Both ΦJHS and ΦSMK phages demonstrated broad lytic spectra on clinical MDR PA strains, as all PA strains tested were lysed by both phages. Infection of seven MDR PA strains indicated the potential of the isolated phages to be developed as candidates for clinical phage therapy or as biocontrol agents on abiotic surfaces. Furthermore, the isolated phages have the potential to be formulated as a phage cocktail to target a single bacterial species (Pseudomonas aeruginosa). Such a cocktail that targets only a single rather than multiple bacterial species is described as generally emphasizing the spectrum of phage activity breadth in its design, rather than necessarily emphasizing the spectrum of phage activity depth.26
Physiological factors, such as pH and temperature, play important roles in bacteria-phage interactions. In the present study, the isolated phages were thermally stable at 15–40°C and displayed maximum viability at pH 7.0. These observations, which are in line with reports from previous studies,17,27 suggest that the phages could be stored at RT, particularly in resource-limited settings. From the observations also, it is suggestive that the phages could be applied, with minimal denaturation, on surfaces of inanimate objects and that their infectivity could be maintained in human conditions, as physiologic systems do not change abruptly.28 The phages tested for organic solvent stability showed that they were not resistant to chloroform, an organic solvent traditionally used in bacteriophage isolation. The results showed that 1.5 h exposure of both phages to chloroform yielded 70–82% of viable phage virions for infection, suggestive that the phages could be lipid enveloped membrane-containing bacteriophages, which are relatively underrepresented among identified phage isolates.29,30 This could also explain the lower phage titers (106–107 PFU/mL) obtained when compared to other studies (≥109 PFU/mL),17,28 as chloroform was continuously used throughout the phage purification processes in the present study. Nonetheless, chloroform reduction of the viability of non-membrane-containing bacteriophages has been reported in some studies.23,27 Without molecular data on the phage genomes and electron micrographs of the phage virions, one could speculate that the high similarity in the response of both phages to the external factors could result from their indistinct structures by belonging to the same viral family.
Assessment of the biofilm-producing capacity of the clinical MDR PA isolates showed that all isolates were biofilm producers, with 71.4%/14.3%/14.3% as strong/moderate/weak biofilm producers. Considering investigations on the association between biofilm formation and multidrug resistance, it has been reported that biofilm formation is significantly higher in MDR PA clinical isolates, due primarily to the presence of biofilm-related genes in these isolates.31 Moreover, the production of strong and moderate biofilms has been reported to be higher in carbapenem-resistant PA than carbapenem-susceptible PA in clinical isolates.32 Besides drug resistance, PA biofilm formation has been reported to be associated in particular with pyocyanin (blue-green lipid-soluble pigment) expression.33,34 On the contrary, no correlation between biofilm formation and drug resistance–or other virulence factors–has been reported,34 suggestive of PA as a notorious biofilm producer and its continuous involvement in persistent chronic infections. Variation in the source of the isolates highlights the enormous burden of PA in the clinical settings, particularly in device-associated infections, which must be dealt with.
Screening the efficacy of the phages in reducing adherent biofilms of MDR PA under static conditions, significant reductions (p < 0.001) were observed in all the strains except for one, after 6 h of phage treatment. Such reductions in biomass were reported in a recent study with four mono phages infecting each of MDR PA, S. aureus, K. pneumoniae, and E. coli biofilms in static, dynamic with medium renewal, and dynamic with nonrenewal of media conditions.35 To maximize the lytic activity of the phages in a resource-limited setting, we employed combinatory dynamic and static conditions in the phage treatment of the biofilms in our study. The experimental setup was first agitated to aid in the spread of the phages to adsorb onto the entire span of the biofilm and then incubated statically to induce the release of progeny viruses to attach to the neighboring bacteria within the biofilm. Although phage ΦJHS presented a dominant advantage over phage ΦSMK in infecting the MDR PA strains, phage ΦSMK presented dominance in reducing the biofilms formed by these strains. Our screening results showed that one MDR PA isolate did not show a statistically significant reduction in biofilm at the evaluated time and could be attributed to the inevitable development of phage resistance. A recent in vitro study reported the appearance of phage-resistant mutants, which sheds more light on the fact that the development of phage resistance by bacteria occurs more frequently in vitro.19
To assess the phages’ potential in inhibiting biofilm formation, catheter and ET segments were coated with the isolated phages by physical adsorption.18,36 The results of our study indicated that physical adsorption promoted phage immobilization on the segments, as evidenced by the statistically significant differences between phage-coated segments and non-coated segments. The results showed that phage coating of catheters and ET inhibited bacteria colonization on these devices. Inhibitory activities of the isolated phages on catheter segments remained similar in the two MDR PA 1098 and 1668 strains tested. These MDR strains were selected based on their source of isolation (sputum and urine), which are in parallel with the medical implants used in this study. In addition, compared to the phage-isolation host strain (PA 1139), which formed moderate biofilm, PA 1098 and 1668 formed strong biofilms, making them bacterial strains of choice for in vitro biofilm study. With both MDR strains, phage ΦSMK was the most efficient with >99.9% inhibition of microbial load compared to phage ΦJHS with >99% inhibition of microbial load. The high log10 inhibition values achieved in ET segments could suggest that phages adsorb more effectively on ET than catheters. Contrary to this finding, reduced phage adsorption on ET was reported recently.18 With limited literature on phage immobilization on polyvinyl chloride ET surfaces, the increased phage adsorption on ET segments in this study could be attributed to the use of segments other than the whole tube in the physical adsorption process. Another possible explanation could be the combinatory dynamic-static conditions used in this study.
On the other hand, our study further assessed the potential of using bacteriophages as destructive agents in preformed biofilms on medical implants. Our results showed that 6 h phage treatment efficiently reduced the number of viable bacterial cells, providing further evidence for their possible use in preventing bacterial colonization of medical implants and eradication of preformed biofilm on the implants. The interactions showed that efficient reductions were observed with phage titers of 106 PFU/mL. Differences in efficiency were noted between the isolated phages against the two MDR PA strains as well as between the different phage titers used. Though efficient, slight reductions were observed with the use of lower phage titers, which are in agreement with a previous study19 that reported an efficient decrease in viable E. faecalis cells in biofilms formed on catheters when low phage titers were applied. With 2.4 log10 reductions, corresponding to >99% removal, phage ΦSMK achieved the highest log10 reduction value, further confirming its dominance over phage ΦJHS. Our results further showed that increasing phage titer increased log10 reduction value, indicating that the greatest significance of ≥3 log10 could be achieved with phage titers above or equal to 108 PFU/mL.
Our results clearly show that monophages alone can be used to either prevent PA colonization of medical implants or reduce the number of preformed biofilms on these implants. Besides using mono phages, positive results have been reported on the use of phage cocktails to prevent ET colonization18 by this bacterium. Outside the medical scope, effective use of mono phages or phage cocktails as biocontrol agents of PA in water37 and phage combination with disinfectants to remove plastic-surface associated PA38 have been recently reported. These studies highlight the urgent need to investigate new strategies to prevent, control, or remove PA biofilms from surfaces. Our study has demonstrated that phages could serve dual purposes in coating surfaces to prevent bacterial colonization and reduce bacterial bioburden on multiple implant surfaces. Most research, however, has been conducted on the latter, at the expense of the former, although the results of our study clearly indicate that phages are more effective at inhibiting bacterial colonization, than removing preformed biofilms. Although some issues remain in phage immobilization, future research must turn more attention to the use of phages as preventive agents rather than removal agents. As a limitation, our study relied on the use of only phenotypic techniques during phage characterization, biofilm formation, and assessment of phage antibiofilm potentials. We, therefore, did not have micrograph data (electron or confocal) and molecular data to complement our results generated from a resource-limited setting.
Conclusion
The results of our study suggest that phages have great potential for the development of phage-coated catheters and ETs and phage-biofilm removal from these implants. The encouraging results, obtained with the inhibition of MDR PA biofilm formation on catheter and ET surfaces, emphasize the potential of using phages for dual purposes of bacterial colonization prevention and bacterial biofilm removal. However, the use of monophages alone is not enough to completely inhibit bacterial colonization or remove biofilms, suggesting that monophages alone may be used as complementary strategies rather than alternative strategies in bacterial colonization prevention and biofilm removal. Furthermore, the results of our study depict that phages serve better as preventive agents than removal agents and, therefore, would require more effort to enhance phage-coating and immobilization methods.
Acknowledgments
We would like to express our sincere thanks to Jimma University Medical Microbiology laboratory for giving us the space to carry out this study. We are also grateful to Dr. Adane Mihret, senior scientist at the Armauer Hansen Research Institute (AHRI), Addis Ababa, Ethiopia, for his generosity in providing us with the culture plates for this study.
Funding Statement
This study was supported by Jimma University, Ethiopia.
Data Sharing Statement
The data sets generated and/or analyzed for this study are available from the corresponding authors on reasonable request.
Ethics Approval
Ethical clearance and approval were obtained from the Institutional Review Board (IRB) of Jimma University Institute of Health with reference number JHRPGN/166/2021. The clinical bacterial isolates and their antimicrobial susceptibility test results were obtained without patient names to maintain patient confidentiality.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; agreed on the journal to which the article will be submitted to; reviewed and agreed on the article in its current version; gave final approval of the version to be published; and agree to take responsibility and be accountable for the contents of the article.
Disclosure
The authors report no conflicts of interest in relation to this work and declare that the study was carried out in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Centers for Disease Control and Prevention. Disinfection of healthcare equipment. Available from: https://www.cdc.gov/infectioncontrol/guidelines/disinfection/healthcare-equipment.html. Accessed October 20, 2020.
- 2.Brindhadevi K, LewisOscar F, Mylonakis E, Shanmugam S, Verma TN, Pugazhendhi A. Biofilm and quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020;96:49–57. doi: 10.1016/j.procbio.2020.06.001 [DOI] [Google Scholar]
- 3.Weiner-Lastinger LM, Abner S, Edwards JR, et al. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect Control Hosp Epidemiol. 2020;41(1):1–18. doi: 10.1017/ice.2019.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yallew WW, Takele AK, Yehuala FM. Point prevalence of hospital-acquired infections in two teaching hospitals of Amhara region in Ethiopia. Drug Healthc Patient Saf. 2016;8:71–76. doi: 10.2147/DHPS.S107344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endalafer N, Gebre-Selassie S, Kotiso B. Nosocomial bacterial infections in a tertiary hospital in Ethiopia. J Infect Prev. 2011;12(1):38–43. doi: 10.1177/1757177410376680 [DOI] [Google Scholar]
- 6.Amorese V, Donadu MG, Usai D, et al. In vitro activity of essential oils against Pseudomonas aeruginosa isolated from infected hip implants. J Infect Dev Ctries. 2018;12(11):996–1001. doi: 10.3855/jidc.10988 [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Wang W, Han J, et al. Synthesis of silver- and strontium-substituted hydroxyapatite with combined osteogenic and antibacterial activities. Biol Trace Elem Res. 2022;200(2):931–942. doi: 10.1007/s12011-021-02697-z [DOI] [PubMed] [Google Scholar]
- 8.Kazemzadeh-Narbat M, Cheng H, Chabok R, et al. Strategies for antimicrobial peptide coatings on medical devices: A review and regulatory science perspective. Crit Rev Biotechnol. 2020;41(1):94–120. doi: 10.1080/07388551.2020.1828810 [DOI] [PubMed] [Google Scholar]
- 9.Sun D, Shahzad MB, Li M, Wang G, Xu D. Antimicrobial materials with medical applications. Mater Technol. 2014;30(sup6):B90–95. doi: 10.1179/1753555714Y.0000000239 [DOI] [Google Scholar]
- 10.Cieplik F, Jakubovics NS, Buchalla W, Maisch T, Hellwig E, Al-Ahmad A. Resistance toward chlorhexidine in oral bacteria – is there cause for concern? Front Microbiol. 2019;10:587. doi: 10.3389/fmicb.2019.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Update. 2016;26:43–57. doi: 10.1016/j.drup.2016.04.002 [DOI] [PubMed] [Google Scholar]
- 12.Endersen L, O’Mahony J, Hill C, Ross RP, McAuliffe O, Coffey A. Phage therapy in the food industry. Annu Rev Food Sci Technol. 2014;5(1):327–349. doi: 10.1146/annurev-food-030713-092415 [DOI] [PubMed] [Google Scholar]
- 13.Batinovic S, Wassef F, Knowler SA, et al. Bacteriophages in natural and artificial environments. Pathogens. 2019;8(3):100. doi: 10.3390/pathogens8030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerveny KE, DePaola A, Duckworth DH, Gulig PA. Phage therapy of local and systemic disease caused by Vibrio vulnificus in iron-dextran-treated mice. Infect Immun. 2002;70(11):6251–6262. doi: 10.1128/IAI.70.11.6251-6262.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Z, Lin H, Ji X, et al. Therapeutic applications of lytic phages in human medicine. Microb Pathog. 2020;142:104048. doi: 10.1016/j.micpath.2020.104048 [DOI] [PubMed] [Google Scholar]
- 16.Amankwah S, Abdusemed K, Kassa T. Bacterial biofilm destruction: A focused review on the recent use of phage-based strategies with other antibiofilm agents. Nanotechnol Sci Appl. 2021;14:161–177. doi: 10.2147/NSA.S325594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira VC, Bim FL, Monteiro RM, et al. Identification and characterization of new bacteriophages to control multidrug-resistant Pseudomonas aeruginosa biofilm on endotracheal tubes. Front Microbiol. 2020;11:580779. doi: 10.3389/fmicb.2020.580779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira VC, Macedo AP, Melo LDR, et al. Bacteriophage cocktail-mediated inhibition of Pseudomonas aeruginosa biofilm on endotracheal tube surface. Antibiotics. 2021;10(1):78. doi: 10.3390/antibiotics10010078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Topka-Bielecka G, Nejman-Faleńczyk B, Bloch S, et al. Phage-bacteria interactions in potential applications of bacteriophage vB_EfaS-271 against Enterococcus faecalis. Viruses. 2021;13(2):318. doi: 10.3390/v13020318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van TR, Kropinski AM. Bacteriophage enrichment from water and soil. In: Clokie MRJ, Kropinski AM editors. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Humana Press; Vol. 501, 2009:15–21. doi: 10.1007/978-1-60327-164-6 [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 22.Azeredo J, Sillankorva S, Pires DP. Pseudomonas bacteriophage isolation and production. In: Filloux A, Ramos J-L editors. Pseudomonas Methods and Protocols. Springer; Vol. 1149, 2014:23–32. doi: 10.1007/978-1-4939-0473-0 [DOI] [PubMed] [Google Scholar]
- 23.Jurczak-Kurek A, Gąsior T, Nejman-Faleńczyk B, et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep. 2016;6(1):34338. doi: 10.1038/srep34338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudina I, Gizachew Z, Woyessa D, Tefera TK. Isolation of bacteriophage and assessment of its activity against biofilms of uropathogenic Escherichia coli in Jimma town, South-western Ethiopia. Am J Curr Microbiol. 2018;6(1):52–66. [Google Scholar]
- 25.Clokie MRJ, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45. doi: 10.4161/bact.1.1.14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abedon ST, Danis-Wlodarczyk KM, Wozniak DJ. Phage cocktail development for bacteriophage Therapy: Toward improving spectrum of activity breadth and depth. Pharmaceuticals. 2021;14(10):1019. doi: 10.3390/ph14101019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topka-Bielecka G, Bloch S, Nejman-Faleńczyk B, et al. Characterization of the bacteriophage vB_EfaS-271 infecting Enterococcus faecalis. Int J Mol Sci. 2020;21(17):6345. doi: 10.3390/ijms21176345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y, Chen P, Lin Z, Wang T. Characterization of two Pseudomonas aeruginosa viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2. Viruses. 2019;11(4):318. doi: 10.3390/v11040318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mäntynen S, Sundberg L-R, Oksanen H, Poranen M. Half a century of research on membrane-containing bacteriophages: bringing new concepts to modern virology. Viruses. 2019;11(1):76. doi: 10.3390/v11010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei H, Cheng RH, Berriman J, et al. Three-dimensional structure of the enveloped bacteriophage Φ12: An incomplete T = 13 lattice is superposed on an enclosed T = 1 shell. PLoS One. 2009;4(9):e6850. doi: 10.1371/journal.pone.0006850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdulhaq N, Nawaz Z, Zahoor MA, Siddique AB. Association of biofilm formation with multidrug resistance in clinical isolates of Pseudomonas aeruginosa. EXCLI J. 2020;19:201–208. doi: 10.17179/excli2019-2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidari R, Farajzadeh Sheikh A, Hashemzadeh M, Farshadzadeh Z, Salmanzadeh S, Saki M. Antibiotic resistance, biofilm production ability and genetic diversity of carbapenem-resistant Pseudomonas aeruginosa strains isolated from nosocomial infections in southwestern Iran. Mol Biol Rep. 2022;(0123456789). doi: 10.1007/s11033-022-07225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thees AV, Pietrosimone KM, Melchiorre CK, et al. PmtA regulates pyocyanin expression and biofilm formation in Pseudomonas aeruginosa. Front Microbiol. 2021;12:789765. doi: 10.3389/fmicb.2021.789765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gajdács M, Baráth Z, Kárpáti K, et al. No correlation between biofilm formation, virulence factors, and antibiotic resistance in Pseudomonas aeruginosa: Results from a laboratory-based in vitro study. Antibiotics. 2021;10(9):1–16. doi: 10.3390/antibiotics10091134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pallavali RR, Degati VL, Narala VR, Velpula KK, Yenugu S, Durbaka VRP. Lytic bacteriophages against bacterial biofilms formed by multidrug-resistant Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus isolated from burn wounds. PHAGE. 2021;2(3):120–130. doi: 10.1089/phage.2021.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melo LDR, Veiga P, Cerca N, et al. Development of a phage cocktail to control Proteus mirabilis catheter-associated urinary tract infections. Front Microbiol. 2016;7:1024. doi: 10.3389/fmicb.2016.01024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kauppinen A, Siponen S, Pitkänen T, et al. Phage biocontrol of Pseudomonas aeruginosa in water. Viruses. 2021;13(5):928. doi: 10.3390/v13050928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stachler E, Kull A, Julian TR. Bacteriophage treatment before chemical disinfection can enhance removal of plastic-surface-associated Pseudomonas aeruginosa. Appl Environ Microbiol. 2021;87:20. doi: 10.1128/AEM.00980-21 [DOI] [PMC free article] [PubMed] [Google Scholar]