Abstract
Lactococcus lactis produced more exopolysaccharide (EPS) on glucose than on fructose as the sugar substrate, although the transcription level of the eps gene cluster was independent of the sugar source. A major difference between cells grown on the two substrates was the capacity to produce sugar nucleotides, the EPS precursors. However, the activities of the enzymes required for the synthesis of nucleotide sugars were not changed upon growth on different sugars. The activity of fructosebisphosphatase (FBPase) was by far the lowest of the enzymes involved in precursor formation under all conditions. FBPase catalyzes the conversion of fructose-1,6-diphosphate into fructose-6-phosphate, which is an essential step in the biosynthesis of sugar nucleotides from fructose but not from glucose. By overexpression of the fbp gene, which resulted in increased EPS synthesis on fructose, it was proven that the low activity of FBPase is indeed limiting not only for EPS production but also for growth on fructose as a sugar source.
Lactic acid bacteria are widely used in the food industry, mainly for lactic acid formation but also for the production of minor food components important for structure, flavor, or preservation.
Several lactic acid bacteria are able to produce exopolysaccharides (EPS). These EPS-forming bacteria play a considerable role in the rheology and texture of fermented milks. Ropy starter cultures of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus, for example, are used for yogurt manufacture in order to improve the viscosity and to prevent syneresis and gel fracture. Furthermore, the presence of mucoid Lactococcus lactis subsp. cremoris strains in starter cultures for the production of the Scandinavian ropy sour milks Viili and Långfil is essential for the desired textures of these products (7). Polysaccharides produced by lactic acid bacteria also provide a source of stabilizing, viscosifying, emulsifying, gelling, or water binding agents for use as natural additives in various food products, which may be an alternative to texturizing agents of plant or animal origin (32).
The strain, the culture conditions, and the medium composition influence the amount of microbial EPS that is produced by a certain species. The type of carbon source has a huge influence on EPS productivity and may also affect the composition of EPS. L. delbrueckii subsp. bulgaricus NCFB 2772 produces three times more EPS with glucose than with fructose as a sugar source, and the type of EPS produced by this organism is influenced by the sugar source as well (16). The yields of EPS produced by Lactobacillus casei CG11, Lactobacillus rhamnosus C83, and Streptococcus salivarius subsp. thermophilus are also significantly influenced by the carbon source (8, 10, 11). In a previous report we described how Lactococcus lactis subsp. cremoris NIZO B40 produces about nine times more EPS with glucose than with fructose as a sugar source under acidifying conditions (22).
Biosynthesis of polysaccharides that are produced by lactococci starts with the intracellular formation of EPS precursors, the sugar nucleotides, followed by the formation of a repeating unit on a lipid carrier, which is located in the cytoplasmic membrane. The repeating unit of EPS produced by Lactococcus lactis NIZO B40 is composed of glucose, galactose, rhamnose, and phosphate in a ratio of 2:2:1:1 (31, 33). The sugar nucleotides UDP-glucose, UDP-galactose, and dTDP-rhamnose are the donors of monomers for the biosynthesis of this pentasaccharide unit. The last steps of EPS formation most likely involve transport of the repeating units across the membrane to the outer layer and polymerization of several hundred to several thousand repeating units to form the final EPS (7, 28).
The formation of sugar nucleotides and the use of a lipid carrier are not unique to EPS biosynthesis; both are also involved in the formation of cell wall sugars (30). Enzymes necessary for the other reactions involved in the biosynthesis of EPS by lactococci are specific and their genes are borne by an EPS plasmid. EPS production by strain NIZO B40 is encoded by a 12-kb region containing 14 genes with the order epsRXABCDEFGHIJKL from the 40-kb EPS plasmid called pNZ4000 (33). The eps gene cluster is transcribed from a single promoter upstream of epsR (33). The gene products EpsD, -E, -F, and -G are glycosyltransferases required for synthesis of the EPS backbone (34).
Regulation of EPS production may be possible at all the different steps involved in its biosynthesis. We determined the steps during which the sugar source influences the final EPS yield of Lactococcus lactis subsp. cremoris.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and analysis of growth and product formation.
Bacterial strains and plasmids used in this study are listed in Table 1. Fermentations with Lactococcus lactis were performed in a chemically defined medium (CDM) at 30°C and pH 5.8 as described before (22). For fermentations without pH control, 1.9 g of β-glycerophosphate per liter was added to the medium and the concentration of the sugar source was reduced to 5 g liter−1. For leucine-limited growth in chemostat cultures, the concentration of leucine was reduced to 30 mg liter−1. Escherichia coli was grown in tryptone yeast extract (TY) broth with aeration at 37°C. If appropriate, the media contained chloramphenicol (10 mg liter−1) and erythromycin (5 mg liter−1). Cell growth was determined by measuring the optical density of the culture fluid at 600 nm (OD600). The amount of residual sugars was quantified by high-performance liquid chromatography (HPLC) (35). Organic acids were analyzed by HPLC with a Rezex Organic Acid column (Phenomenex Inc., Torrance, Calif.) at 60°C with 0.6 ml of 5 mM H2SO4 min−1 as the eluent and detection based on a refractive index. The amount of EPS was measured in duplicate by gel permeation chromatography with dextran as the standard as described previously (22). The standard deviation of this method was 2%.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| Lactococcus lactis | ||
| NIZO B40 | Lac+ Eps+ | 33 |
| MG1614 | Rfr Smr, plasmid free | 12 |
| NZ4010 | MG1614 derivative containing pNZ4000 | 33 |
| NZ9000 | MG1363 pepN::nisRK | 20 |
| E. coli MC1061 | 6 | |
| Plasmids | ||
| pNZ4000 | 40-kb plasmid bearing genes for EPS production | 33 |
| pNZ4030 | Emr, 27-kb derivative of pNZ4000 | 33 |
| pNZ4040 | Cmr, plasmid with the marker gene gusA under control of the eps promoter | 33 |
| pNZ8048 | Cmr, lactococcal cloning and expression vector with the nisA promoter upstream of a multiple cloning site | 20 |
| PNZ4150 | pNZ8048 derivative containing the E. coli fbp gene translationally fused to the nisA promoter | This work |
Lac+, lactose-fermenting phenotype; Eps+, EPS-producing phenotype; Rfr, rifampin resistant; Smr, streptomycin resistant; Emr, erythromycin resistant; Cmr, chloramphenicol resistant.
Preparation of cell extracts.
Bacteria were harvested by centrifugation (16,000 × g, 30 min, 4°C) at an OD600 of 1 to 1.5, washed twice with 0.85% NaCl, and suspended in 20 mM phosphate buffer (pH 6.5) containing 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol (25). The bacteria were disrupted ultrasonically (20 kHz) at 0°C for 36 cycles of 5 s (XL2020 sonicator; Heat Systems New York, N.Y.). Cell debris was removed by centrifugation (13,000 × g, 10 min, 4°C). The protein content of the cell extract was determined by the method of Bradford (5). For the assays of the phosphoenolpyruvate (PEP)-glucose-phosphotransferase system (PTS), the PEP-fructose-PTS, UDP-glucose pyrophosphorylase, and dTDP-glucose pyrophosphorylase, cell debris was not removed because these enzymes are probably linked to the cell membranes.
Enzyme assays.
Enzyme assays were performed at 30°C in a total volume of 1 ml with freshly prepared cell extracts. The formation or consumption of NAD(P)H was determined by measuring the change in the absorbance at 340 nm. Values are the means of results from at least two independent duplicate measurements. The blank contained the reaction buffer, the cofactors, and the substrate but lacked the cell extract.
The PEP-PTS uptake systems for glucose and fructose were assayed with a mixture containing 50 mM KPO4 buffer (pH 6.8), 5 mM MgCl2, 5 mM PEP, 0.5 mM NADH, 4 U of lactate dehydrogenase, and cell extract. The reaction was started by adding 1 mM glucose or fructose (15).
The reaction mixtures for 1- and 6-phosphofructokinase (EC 2.7.1.56 and EC 2.7.1.11) contained 50 mM Tris-HCl buffer (pH 7.5), 5 mM MgCl2, 50 mM KCl, 1.25 mM ATP, 0.15 mM NADH, 4.5 U of aldolase, 18 U of triose-phosphate isomerase, 6.2 U of glycerol-3-phosphate dehydrogenase, and cell extract. Addition of 5 mM fructose-1-phosphate or fructose-6-phosphate started the reactions (15).
The reaction mixture for the α-phosphoglucomutase (EC 2.7.5.1) assay contained 50 mM triethanolamine buffer (pH 7.2), 5 mM MgCl2, 0.4 mM NADP+, 50 μM glucose-1,6-diphosphate, 4 U of glucose-6-phosphate dehydrogenase, and cell extract. The reaction was started by the addition of 1.4 mM α-glucose-1-phosphate (27).
The phosphoglucose isomerase (EC 5.3.1.9) reverse-reaction mixture contained 50 mM potassium phosphate buffer (pH 6.8), 5 mM MgCl2, 0.4 mM NADP+, 4 U of glucose-6-phosphate dehydrogenase, and cell extract. The reaction was started by adding 5 mM fructose-6-phosphate (15).
The UDP-galactose-4-epimerase (EC 5.1.3.2) activity was assayed with a mixture of 50 mM Tris-HCl buffer (pH 8.5), 5 mM MgCl2, 0.5 mM NAD, 0.015 U of UDP-glucose dehydrogenase, and cell extract. The reaction was started by the addition of 0.2 mM UDP-galactose.
The activity of the dTDP-rhamnose biosynthetic enzyme system was assayed in a reaction mixture containing 50 mM Tris-HCl buffer (pH 8.0), 0.5 mM NADPH, and cell extract. Addition of 0.3 mM TDP-glucose started the reaction (15).
The UDP-glucose pyrophosphorylase (EC 2.7.7.9) reverse reaction mixture contained 50 mM Tris-HCl buffer (pH 7.8), 14 mM MgCl2, 0.3 mM NADP+, 0.1 mM UDP-glucose, 2.1 U of α-phosphoglucomutase, 4 U of glucose-6-phosphate dehydrogenase, and cell extract. The reaction was started by adding 4 mM inorganic pyrophosphate (4).
The reaction mixture of the dTDP-glucose pyrophosphorylase (EC 2.7.7.24) reverse-reaction assay contained 50 mM Tris-HCl buffer (pH 7.8), 8 mM MgCl2, 0.3 mM NADP+, 0.1 mM TDP-glucose, 2.1 U of α-phosphoglucomutase, 4 U of glucose-6-phosphate dehydrogenase, and cell extract. The reaction was started by the addition of 4.7 mM inorganic pyrophosphate (4).
The fructose-1,6-bisphosphatase (EC 3.1.3.11) (FBPase) assay mixture contained 50 mM glycylglycine buffer (pH 8.5), 5 mM MgCl2, 0.4 mM NADP+, 4 U of glucose-6-phosphate dehydrogenase, 3.5 U of phosphoglucose isomerase, and cell extract. The reaction was started by adding 5 mM fructose-1,6-diphosphate (15).
Sugar nucleotide analysis.
Cell extracts were prepared as described above. Immediately after preparation of the cell extracts, the enzymes were separated from the sugar nucleotides and other small water-soluble components by means of centrifugal filtration (5,000 × g, 2°C) with filter units with a nominal molecular weight limit of 10,000 (Ultrafree-MC; Millipore, Bedford, Mass.). The concentration of sugar nucleotides in the filtrates was measured by HPLC according to the method described by Harding et al. (18), with a detection limit of 0.5 μmol liter−1. The results are the average determinations of results with bacteria harvested during three independent fermentations.
Isolation of cell wall sugars and characterizations of EPS and cell wall sugars.
The isolation of cell wall sugars is based on a method described by Gopal and Reilly (13). The bacteria were grown in CDM with either 6% glucose or fructose at 30°C and pH 5.8. The bacteria were harvested at an OD600 of about 1.5. Lysed-cell extracts of the cultures were prepared as described above. After ultrasonic treatment, whole cells were removed by centrifugation (3,000 × g, 10 min, 4°C) and the supernatant was centrifuged (20,000 × g, 20 min, 4°C) to harvest the cell walls. The crude cell wall fraction was suspended in buffer containing 140 μg of RNase and 100 μg of DNase per ml and incubated for 90 min at 37°C. The cell walls were collected by centrifugation (20,000 × g, 20 min, 4°C). The obtained pellet was resuspended in buffer with 2% sodium dodecyl sulfate (SDS) and incubated at 70°C for 1 h. After centrifugation (20,000 × g, 20 min, 4°C), the pellet was washed three times with distilled water to remove SDS and freeze-dried, which resulted in the purified cell wall fraction. Isolated EPS or cell walls were hydrolyzed in 4 mol of HCl liter−1 for 30 min at 100°C. Samples were dried under vacuum and dissolved in distilled water. The monomeric sugar composition after hydrolysis was determined by HPLC (35).
Activity of the eps promoter.
Lactococcus lactis MG1614 harboring plasmid pNZ4040 and MG1614 harboring both pNZ4040 and the EPS plasmid pNZ4030 were used to determine the activity of the promoter of the eps operon (Table 1). Plasmid pNZ4040 contains the eps promoter fused to the promoterless gusA reporter gene, which encodes β-glucuronidase (33). The activity of β-glucuronidase was determined in an assay with 950 μl of GUS buffer (50 mM NaHPO4 [pH 7.0], 10 mM β-mercaptoethanol, 1 mM EDTA, 0.1% Triton X-100) and 40 μl of cell extract. The reaction was started by adding 10 μl of 100 mM para-nitro-β-d-phenyl-glucuronic acid. The increase in the A405 was measured at 37°C (26). The β-glucuronidase activity was measured with cell extracts of MG1614 harboring pNZ4040 or of MG1614 harboring pNZ4040 and pNZ4030 to make it possible to distinguish between regulation by chromosomally encoded or plasmid-borne factors.
Controlled overexpression of the fbp gene.
The E. coli fbp gene was amplified by PCR with chromosomal DNA from E. coli MC1061 as a template and the primers 5′-CATGCCATGGCAAAAACGTTAGGTGAATTTATTGTCG-3′ and 5′-CTAGTCTAGATTACGCGTCCGGGAACTC-3′. Primer design was based on sequence data of the E. coli fbp gene (GenBank accession no. P09200 [17]), and the primers introduced flanking NcoI and XbaI restriction sites (underlined). The fbp gene was translationally fused to the nisA promoter by cloning the NcoI- and XbaI-digested PCR product in NcoI- and XbaI-digested pNZ8048, yielding pNZ4150. Plasmid pNZ4150 was transformed into Lactococcus lactis NZ9000 by electroporation with E. coli as an intermediate host. This resulted in a nisin-controlled expression system for fbp (20). The EPS plasmid pNZ4030 was transformed into Lactococcus lactis NZ9000 harboring pNZ4150 by means of electroporation.
Lactococcus lactis NZ9000 containing pNZ4150 (and pNZ4030) was grown in CDM with a 0.5% sugar source at 30°C until an OD600 of 0.1 was reached and induced with various levels of lactococcal nisin A (0 to 1 ng ml−1), resulting in different levels of expression of the fbp gene.
RESULTS
Influence of the sugar source on EPS production.
In a previous study we showed that the natural-EPS-producing strain Lactococcus lactis subsp. cremoris NIZO B40 produces more EPS with glucose than with fructose as the source of sugar (22). Here we studied the regulation of EPS production by the carbon source by comparison of EPS-producing and non-EPS-producing cells with isogenic backgrounds, i.e., from strains NZ4010 and MG1614, respectively. Strain NZ4010 was constructed by conjugal transfer of the EPS plasmid pNZ4000 of strain NIZO B40 to the EPS− strain MG1614 (12), resulting in an EPS+ phenotype (33). First, EPS production of strains NZ4010 and NIZO B40 grown on glucose and fructose was determined with pH-controlled batch cultures. The amounts of EPS produced by both strains were considerably lower with fructose than with glucose as the source of sugar. Growing on glucose, the transconjugant produced less EPS than the wild-type strain (Table 2).
TABLE 2.
EPS production by Lactococcus lactis strains in CDM with either 6% glucose or fructose as the sugar source at 30°C and pH 5.8 and concentrations of sugar nucleotides in cell extracts of these strains
| Strain | Sugar source | EPS (mg liter−1) | Mean concn (μmol g of protein−1) ± SD (n = 3) ofa:
|
|
|---|---|---|---|---|
| UDP-glucose | UDP-galactose | |||
| NIZO B40 | Glucose | 460 | 29.9 ± 6.4 | 9.6 ± 2.2 |
| Fructose | 65 | ND | ND | |
| NZ4010 | Glucose | 310 | 4.9 ± 2.9 | 2.2 ± 0.5 |
| Fructose | 85 | 0.8 ± 0.7 | 0.4 ± 0.3 | |
| MG1614 | Glucose | 14.2 ± 1.9 | 4.5 ± 0.7 | |
| Fructose | 1.8 ± 0.6 | 0.6 ± 0.1 | ||
ND, not detected (values were below the detection limit).
Growth of the three strains was only slightly lower with fructose as the source of sugar. The growth phase, during which most EPS is produced, was also influenced by the sugar source. During growth on glucose most of the EPS was produced during the exponential growth phase, while during growth on fructose about 60% of the EPS was produced in the stationary phase (not shown).
Influence of the sugar source on the levels of expression of eps genes.
Van Kranenburg et al. (33) showed that all the eps genes are under the control of the eps promoter; hence, the activity of this promoter is a measure for the transcription levels of the eps genes. Plasmid pNZ4040 contains the gusA reporter gene, which encodes β-glucuronidase, under the control of the eps promoter. The activity of the eps promoter was determined by measuring the β-glucuronidase activity with cell extracts of strain MG1614 harboring pNZ4040 (and pNZ4030) grown in CDM with either glucose or fructose as the source of sugar. The activity of β-glucuronidase was about 98 nmol mg of protein−1 min−1 for both strains grown on glucose as well as fructose, which means that the activity of eps promoter does not depend on these sugar sources. From these results it can be concluded that the transcription level of the eps genes is not regulated by the source of sugar.
Influence of the sugar source on the concentration of sugar nucleotides.
Lactococcus lactis NIZO B40 and NZ4010 produce EPS composed of glucose, galactose, rhamnose, and phosphate (31). For the biosynthesis of this EPS, the activated sugar monomers UDP-glucose, UDP-galactose, and dTDP-rhamnose are necessary (Fig. 1). As glucose, galactose, and rhamnose are components of the cell walls of lactococci, these sugar nucleotides are also necessary for cell wall synthesis and hence for growth. The intracellular concentrations of UDP-glucose and UDP-galactose were much lower for fructose- than for glucose-grown cells (Table 2). The amounts of sugar nucleotides present in cells grown on glucose were higher for strain MG1614 than for strain NZ4010 (Table 2). All the sugar nucleotides that were found in MG1614 but not in NZ4010 were probably used for the biosynthesis of EPS. After the growth on fructose the sugar nucleotide levels were only slightly lower in the EPS producer. This may mean that when the cells grow on fructose, only just enough sugar nucleotides are produced to fulfil the need for cell wall biosynthesis. The affinity of the eps genes for the sugar nucleotides is apparently not high enough to be able to produce EPS when the concentration of these activated sugars is as low as that measured in the fructose-grown cells. EPS production on fructose took place mainly during the stationary phase, when there is no need of sugar nucleotides for growth. Cells harvested during the stationary phase contained a much higher concentration of UDP-glucose than that of cells harvested during the exponential growth phase when fructose was the substrate (not shown).
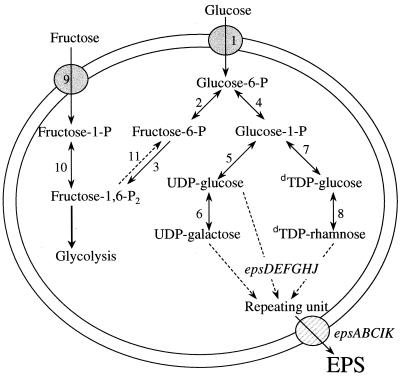
FIG. 1.
Schematic representation of the metabolism of an EPS-producing Lactococcus cell grown on glucose and fructose. 1, Mannose PEP PTS; 2, phosphoglucose isomerase; 3, 6-phosphofructokinase; 4, α-phosphoglucomutase; 5, UDP-glucose pyrophosphorylase; 6, UDP-galactose-4-epimerase; 7, deoxy-TDP (dTDP)-glucose pyrophosphorylase; 8, dTDP-rhamnose biosynthetic enzyme system; 9, fructose PEP PTS; 10, 1-phosphofructokinase; 11, FBPase. Genes borne by plasmid pNZ4000 are involved in the formation of the repeating unit (epsDEFGHJ) and in export and polymerization (epsABCIK) (33).
Activities of enzymes involved in biosynthesis of sugar nucleotides.
A difference in the substrate fluxes into the direction of sugar nucleotides in fructose- and glucose-grown bacteria may be caused by a difference in the activities of the enzymes involved in their biosynthesis or by a difference in the initial levels of sugar metabolism.
The activities of enzymes involved in the biosynthesis of the EPS precursors and the initial metabolism of glucose and fructose were not influenced by the ability of cells to produce EPS (Table 3). During growth on fructose, the activity of 1-phosphofructokinase was significantly increased for both strains compared to that during growth on glucose. As the bacteria need only this enzyme for growth on fructose (Fig. 1), it is presumably induced by the presence of fructose. All the other enzymes were not significantly influenced by the source of sugar (Table 3).
TABLE 3.
Activities of enzymes involved in initial sugar metabolism of glucose and fructose and biosynthesis of sugar nucleotides in glucose- and fructose-grown cultures of Lactococcus lactis NZ4010 and MG1614
| Enzyme | Mean activity (nmol mg of protein−1 min−1 ± SD (n = ≥4) in indicated strain grown on:
|
|||
|---|---|---|---|---|
| Glucose
|
Fructose
|
|||
| NZ4010 | MG1614 | NZ4010 | MG1614 | |
| Mannose PTS | 29 ± 13 | 21 ± 5 | 21 ± 5 | 28 ± 8 |
| Phosphoglucose isomerasea | 4,318 ± 349 | 4,085 ± 357 | 3,415 ± 289 | 3,148 ± 544 |
| 6-Phosphofructokinase | 173 ± 44 | 189 ± 105 | 205 ± 37 | 210 ± 44 |
| α-Phosphoglucomutasea | 345 ± 41 | 295 ± 37 | 295 ± 7 | 290 ± 40 |
| UDP-glucose pyrophosphorylasea | 5.5 ± 2.2 | 4.2 ± 1.3 | 5.3 ± 1.9 | 4.4 ± 0.8 |
| UDP-galactose-4-epimerasea | 182 ± 36 | 143 ± 35 | 182 ± 7 | 190 ± 18 |
| TDP-glucose pyrophosphorylasea | 34 ± 9 | 29 ± 2 | 27 ± 6 | 22 ± 9 |
| TDP-rhamnose biosynthetic system | 16 ± 3 | 14 ± 6 | 14 ± 1 | 15 ± 1 |
| Fructose PTS | 29 ± 9 | 30 ± 14 | 22 ± 6 | 23 ± 9 |
| 1-Phosphofructokinase | 248 ± 93 | 246 ± 54 | 658 ± 86 | 628 ± 134 |
| FBPase | 3.1 ± 0.8 | 3.5 ± 0.6 | 1.9 ± 0.2 | 2.2 ± 0.6 |
The reversed reaction was used for measuring the activities.
Striking is the fact that the activity of FBPase was considerably lower than the activities of other enzymes involved in precursor formation and seemed to be even somewhat lower in fructose-grown cultures (Table 3). This enzyme is needed for the biosynthesis of sugar nucleotides when the bacteria grow on fructose but not when glucose is used as the sugar source (Fig. 1).
Overexpression of fbp.
To verify if the low activity of FBPase is the bottleneck for EPS production with fructose as the sugar source, the activity of this enzyme was increased by overexpression of the fbp gene, for which the nisin-controlled expression system was used (9, 20). The fbp gene from E. coli was fused to the nisA promoter in pNZ8048 (pNZ4150) and transformed into strain NZ9000. This strain contains the nisR and nisK genes, which are necessary for sensing nisin and subsequent activation of the nisA promoter (20), so the presence of nisin A in the medium resulted in transcription of the fbp gene. An SDS-polyacrylamide gel of cell extracts of strain NZ9000 harboring pNZ4150 showed the appearance of a protein band when the bacteria were induced with nisin (not shown). This protein band had a molecular mass of 36.8 kDa, which corresponds well with that of the E. coli FBPase (17). The intensity of the FBPase band as well as the FBPase activity increased when the concentration of nisin was increased, which proved that the expressed protein was functional.
The EPS plasmid pNZ4030 was transformed into strain NZ9000 harboring pNZ4150. This new strain was grown in CDM with 0.5% fructose at 30°C under acidifying conditions. The concentration of EPS in the broth was measured at the end of the experiment. Compared to the level of EPS production in strain NZ9000 harboring pNZ4030 and the control plasmid pNZ8048, a fivefold increase in EPS production per ml was obtained with an induction level of 1 ng of nisin liter−1 (Table 4). Not only the EPS concentration but also the growth rate and the final OD increased when the concentration of nisin in the medium was raised (Table 4). Apparently, the activity of FBPase was not only limiting for EPS production but also limiting for growth of this organism with fructose as the source of sugar. Comparable experiments with medium containing glucose as the source of sugar did not result in a change in either growth or EPS production (data not shown).
TABLE 4.
Concentrations of EPS and activity of FBPase of strain NZ9000 carrying pNZ4030 and pNZ4150 or pNZ8048 incubated in CDM with 0.5% fructose at 30°C and induced with different levels of nisin at an OD600 of 0.1
| Strain | Nisin (ng ml−1) | EPS (mg liter−1) | OD600 | EPS (mg OD unit−2) | μa (h−1) | FBPase (nmol mg of protein−1 min−1) |
|---|---|---|---|---|---|---|
| NZ9000 + pNZ4030 + pNZ8048 | 0 | 1.4 | 1.045 | 1.37 | 0.22 | 6.2 |
| NZ9000 + pNZ4030 + pNZ8048 | 1 | 1.6 | 1.115 | 1.37 | 0.27 | 6.3 |
| NZ9000 + pNZ4030 + pNZ4150 | 0 | 1.3 | 1.07 | 1.21 | 0.23 | 6.7 |
| NZ9000 + pNZ4030 + pNZ4150 | 0.001 | 1.5 | 1.12 | 1.34 | 0.27 | 11.4 |
| NZ9000 + pNZ4030 + pNZ4150 | 0.01 | 2.4 | 1.305 | 1.84 | 0.47 | 78 |
| NZ9000 + pNZ4030 + pNZ4150 | 0.1 | 5.4 | 1.63 | 3.31 | 0.60 | 303 |
| NZ9000 + pNZ4030 + pNZ4150 | 1 | 8.5 | 1.72 | 4.94 | 0.74 | 628 |
μ, maximum growth rate after induction. The initial growth rate for both strains was 0.47 h−1.
Induction of bacteria grown in medium with fructose resulted in an increase in the growth rate as well as the EPS yield (Table 4). To exclude the influence of the growth rate on EPS production, strain NZ9000 harboring pNZ4030 and pNZ4150 was grown in a continuous culture under leucine limitation with fructose as the source of sugar at a dilution rate of 0.2 h−1 with and without induction. During steady state of these cultures, the EPS concentration was about two times higher in the induced culture than in the uninduced culture. The activities of FBPase at that time were 7.5 and 105 nmol mg of protein−1 min−1 in, respectively, the uninduced and the induced cultures.
Influence of the sugar source on EPS and cell wall composition.
The carbon source did not influence the sugar compositions of the cell wall polysaccharides of strains MG1614 and NZ4010 or the composition of EPS produced by NZ4010 (Table 5). Preliminary results indicate that the amount of cell wall sugars is independent of the sugar source, meaning that sugar nucleotides are used preferentially for the formation of cell wall sugars.
TABLE 5.
Sugar compositions of the polysaccharides of the cell walls of glucose- and fructose-grown Lactococcus lactis MG1614 and NZ4010 and of EPS produced by strain NZ4010
| Strain | Sugar source | mol% rhamnose | mol% galactose | mol% glucose |
|---|---|---|---|---|
| MG1614 | Glucose | 55.4 | 15.4 | 29.2 |
| Fructose | 55.1 | 20.7 | 24.1 | |
| NZ4010 | Glucose | 51.0 | 17.8 | 31.3 |
| Fructose | 58.0 | 17.2 | 24.8 | |
| EPS | Glucose | 22.5 | 29.5 | 48.0 |
| Fructose | 21.2 | 30.6 | 48.2 |
DISCUSSION
It was shown that the EPS production by Lactococcus lactis subsp. cremoris NIZO B40 is far more efficient with glucose than with fructose as the source of sugar. In this paper we describe our investigation of the possible influence of the sugar source during the different steps involved in the production of EPS by lactococci.
Enzymes leading to EPS formation can roughly be divided into four groups: enzymes responsible for the initial metabolism of a carbohydrate, enzymes involved in sugar nucleotide synthesis and interconversion, glycosyltransferases that form the repeating unit attached to the glycosyl carrier lipid, and translocases and polymerases that form the polymer. Possibilities exist for exerting control over polysaccharide synthesis at any of these four levels, and mutants lacking enzymes of any group fail to synthesize EPS (29). For Lactococcus lactis NIZO B40, the genes encoding the enzymes of the third and fourth groups are borne by a plasmid and are all under the control of the eps promoter. The activity of this promoter was shown to be independent of the source of sugar, meaning that the transcription level of the eps gene is not regulated by the source of sugar. Apparently, the sugar source does not exert a specific control over EPS production by Lactococcus lactis but influences the polymer yield by influencing the fist, unspecific steps involved in EPS biosynthesis.
The second group of enzymes has been shown to control EPS synthesis in several organisms. Grobben et al. (15) found that EPS production by L. delbrueckii subsp. bulgaricus NCFB 2772 is lower with fructose than with glucose as the carbon source. The activity of UDP-glucose pyrophosphorylase was higher in glucose-grown cells than in fructose-grown cells of this strain. Others also found a correlation between the activities of EPS precursor-forming enzymes and the amount of EPS produced by Sphingomonas paucimobilis GS1 (1), Azotobacter vinelandii (19), Pseudomonas aeruginosa (21), and E. coli (14).
With EPS production by strain NIZO B40, no relationship between the activities of precursor-forming enzymes and the amounts of EPS produced on glucose and fructose was found, as was also the case for EPS produced by Pseudomonas sp. strain NCIB 11264 (36) and Enterobacter aerogenes (23). All the enzymes necessary for the formation of EPS precursors in strain NIZO B40 are also needed for the formation of cell wall sugars. The genes for these enzymes are household genes and not located on the EPS plasmid (33). Although the activities of enzymes involved in the biosynthesis of EPS precursors were not influenced by the source of sugar, the levels of these sugar nucleotides were much lower in fructose-grown than in glucose-grown Lactococcus lactis. Apparently, during growth on fructose, the metabolic flux in the direction of sugar nucleotides is less than during growth on glucose.
In Lactococcus most metabolizable sugars are transported via the PEP PTS. Glucose is transported via the mannose PTS, which has a very low affinity for fructose. During translocation of sugars via this system, the sugars are phosphorylated at C-6. Uptake of fructose is realized mainly via the fructose PTS, resulting in fructose-1-phosphate (2). When fructose is transported via the fructose PTS, the combined actions of 1-phosphofructokinase and FBPase are required in order to form essential biomass precursors. These enzymes are not involved in the formation of biomass precursors from glucose (3). The activity of 1-phosphofructokinase was shown to be significantly higher in fructose-grown cultures (Table 3), but the activity of FBPase was very low on both substrates. The low activity of this enzyme may be responsible for the reduced production of sugar nucleotides on fructose and hence a decreased EPS production. For Lactobacillus bulgaricus NCFB 2772, it was also suggested that the reduced EPS production on fructose could be caused by a more complex pathway involved in the synthesis of EPS precursors, although the levels of sugar nucleotides in this strain were only 1.5 times higher for glucose-grown cultures (15).
FBPase may also be involved in a 6-phosphofructokinase–FBPase-catalyzed ATP-consuming futile cycle in lactococci (24). In our continuous cultures under leucine limitation we found indeed that the concentration of lactic acid was somewhat higher and the concentration of fructose somewhat lower in the induced culture than in the uninduced culture, although the biomass concentrations were equal in these cultures (not shown).
In summary, overexpression of FBPase resulted in increased EPS production on fructose as the growth substrate. It can be concluded that the activity of this enzyme limits the amount of EPS produced by wild-type Lactococcus lactis subsp. cremoris on fructose. Fructose is not a common sugar source for the dairy industry, but FBPase is required for production of biomass and EPS precursors from galactose, if it is phosphorylated at C-6 during transport via the galactose or lactose PTS (3). Furthermore, these results are also of importance when Lactococcus lactis is used as a cell factory for the production of EPS from sucrose or other inexpensive bulk materials containing fructose or galactose.
ACKNOWLEDGMENTS
This work was supported by the Ministry of Economic Affairs, the Ministry of Education, Culture and Science, and the Ministry of Agriculture, Nature Management and Fishery within the framework of an industrially relevant research program of the Netherlands Association of Biotechnology Centres in The Netherlands (ABON).
We thank Richard van Kranenburg for providing several strains and plasmids that were used for this study and for critically reading the manuscript.
REFERENCES
- 1.Ashtaputre A A, Shah A K. Studies on the exopolysaccharide from Shingomonas paucimobilis-GS1: nutritional requirement and precursor-forming enzymes. Curr Microbiol. 1995;31:234–238. [Google Scholar]
- 2.Benthin S, Nielsen J, Villadsen J. Transport of sugars via two anomer-specific sites on mannose-phosphotransferase system in Lactococcus cremoris: in vivo study of mechanism, kinetics, and adaptation. Biotechnol Bioeng. 1993;42:440–448. doi: 10.1002/bit.260420406. [DOI] [PubMed] [Google Scholar]
- 3.Benthin S, Nielsen J, Villadsen J. Two uptake systems for fructose in Lactococcus lactis subsp. cremoris FD1 produce glycolytic and gluconeogenic fructose phosphates and induce oscillations in growth and lactic acid formation. Appl Environ Microbiol. 1993;59:3206–3211. doi: 10.1128/aem.59.10.3206-3211.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein R L. Control aspects of uridine 5′-diphosphate glucose and thymidine 5′-diphosphate glucose synthesis by microbial enzymes. J Biol Chem. 1965;240:391–397. [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 6.Cassadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 7.Cerning J. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol Rev. 1990;87:113–130. doi: 10.1111/j.1574-6968.1990.tb04883.x. [DOI] [PubMed] [Google Scholar]
- 8.Cerning J, Renard C M G C, Thibault J F, Bouillanne C, Landon M, Desmazeaud M, Topisirovic L. Carbon source requirements for exopolysaccharide production by Lactobacillus casei CG11 and partial structure analysis of the polymer. Appl Environ Microbiol. 1994;60:3914–3919. doi: 10.1128/aem.60.11.3914-3919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Ruyter P G G A, Kuipers O P, De Vos W M. Controlled gene expression systems for Lactococcus lactis using the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamar L, Blondeau K, Simonet J-M. Physiological approach to extracellular polysaccharide production by Lactobacillus rhamnosus strain C83. J Appl Microbiol. 1997;83:281–287. [Google Scholar]
- 11.Gancel F, Novel G. Exopolysaccharide production by Streptococcus salivarius ssp. thermophilus cultures. 1. Conditions of production. J Dairy Sci. 1994;77:685–688. [Google Scholar]
- 12.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopal P K, Reilly K I. Molecular architecture of the lactococcal cell surface as it relates to important industrial properties. Int Dairy J. 1995;5:1095–1111. [Google Scholar]
- 14.Grant W D, Sutherland I W, Wilkinson J F. Control of colanic acid synthesis. J Bacteriol. 1970;103:89–96. doi: 10.1128/jb.103.1.89-96.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grobben G J, Smith M R, Sikkema J, De Bont J A M. Influence of fructose and glucose on the production of exopolysaccharides and the activities of enzymes involved in the sugar metabolism and the synthesis of sugar nucleotides in Lactobacillus delbrueckii subsp. bulgaricus NCFB 2772. Appl Microbiol Biotechnol. 1996;46:279–284. [Google Scholar]
- 16.Grobben G J, Van Casteren W H M, Schols H A, Oosterveld A, Sala G, Smith M R, Sikkema J, De Bont J A M. Analysis of the exopolysaccharides produced by Lactobacillus delbrueckii subsp. bulgaricus NCFB 2772 grown in continuous culture on glucose and on fructose. Appl Microbiol Biotechnol. 1997;48:516–521. [Google Scholar]
- 17.Hamilton W D O, Harrison D A, Dyer T A. Sequence of the Escherichia coli fructose-1,6-bisphosphatase gene. Nucleic Acids Res. 1988;16:8707. doi: 10.1093/nar/16.17.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding N E, Raffo S, Raimondi A, Cleary J M, Ielpi L. Identification, genetic and biochemical analysis of genes involved in synthesis of sugar nucleotide precursors of xanthan gum. J Gen Microbiol. 1993;139:447–457. doi: 10.1099/00221287-139-3-447. [DOI] [PubMed] [Google Scholar]
- 19.Horan N J, Jarman T R, Dawes E A. Effects of carbon source and inorganic phosphate concentration on the production of alginic acid by a mutant of Azotobacter vinelandii and on the enzymes involved in its biosynthesis. J Gen Microbiol. 1981;127:185–191. [Google Scholar]
- 20.Kuipers O P, De Ruyter P G G A, Kleerebezem M, De Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 21.Leitão J H, Correia I S. Growth-phase-dependent alginate synthesis, activity of biosynthetic enzymes and transcription of alginate genes in Pseudomonas aeruginosa. Arch Microbiol. 1995;163:217–222. doi: 10.1007/BF00305356. [DOI] [PubMed] [Google Scholar]
- 22.Looijesteijn P J, Hugenholtz J. Uncoupling of growth and exopolysaccharide production by Lactococcus lactis subsp. cremoris NIZO B40 and optimisation of its exopolysaccharide synthesis. J Biosci Bioeng. 1999;88:159–163. doi: 10.1016/s1389-1723(99)80198-4. [DOI] [PubMed] [Google Scholar]
- 23.Noval M, Sutherland I W. The production of enzymes involved in exopolysaccharide synthesis in Klebsiella aerogenes types 1 and 8. Eur J Biochem. 1973;35:209–215. doi: 10.1111/j.1432-1033.1973.tb02827.x. [DOI] [PubMed] [Google Scholar]
- 24.Otto R. Uncoupling of growth and acid production in Streptococcus cremoris. Arch Microbiol. 1984;140:225–230. [Google Scholar]
- 25.Petit C, Grill J P, Maazouzi N, Marczak R. Regulation of polysaccharide formation by Streptococcus thermophilus in batch and fed-batch cultures. Appl Microbiol Biotechnol. 1991;36:216–221. [Google Scholar]
- 26.Platteeuw C, Simons G, De Vos W M. Use of Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian N, Stanley G A, Hahn-Hägerdal B, Rådström P. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J Bacteriol. 1994;176:5304–5311. doi: 10.1128/jb.176.17.5304-5311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikkema J, Oba T. Extracellular polysaccharides of lactic acid bacteria. Snow Brand R&D Rep. 1998;107:1–31. [Google Scholar]
- 29.Sutherland I W. Bacterial exopolysaccharides. Adv Microb Physiol. 1972;8:143–213. doi: 10.1016/s0065-2911(08)60190-3. [DOI] [PubMed] [Google Scholar]
- 30.Sutherland I W. Biosynthesis of microbial exopolysaccharides. Adv Microb Physiol. 1982;23:79–150. doi: 10.1016/s0065-2911(08)60336-7. [DOI] [PubMed] [Google Scholar]
- 31.Van Casteren W H M, Dijkema C, Schols H A, Beldman G, Voragen A G J. Characterisation and modification of the exopolysaccharide produced by Lactococcus lactis subsp. cremoris B40. Carbohydr Polym. 1998;37:123–130. [Google Scholar]
- 32.Van den Berg D J C, Robijn G W, Janssen A C, Giuseppin M L F, Vreeker R, Kamerling J P, Vliegenthart J F G, Ledeboer A M, Verrips C T. Production of a novel extracellular polysaccharide by Lactobacillus sake 0-1 and characterization of the polysaccharide. Appl Environ Microbiol. 1995;61:2840–2844. doi: 10.1128/aem.61.8.2840-2844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Kranenburg R, Marugg J D, Van Swam I I, Willem J, De Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 34.Van Kranenburg R, Van Swam I I, Marugg J D, Kleerebezem M, De Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactis NIZO B40: functional analysis of the glycosyltransferase genes involved in synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Riel J, Olieman C. Selectivity control in the anion-exchange chromatographic determination of saccharides in dairy products using pulsed amperometric detection. Carbohydr Res. 1991;215:39–46. [Google Scholar]
- 36.Williams A G, Wimpenny J W T. Extracellular polysaccharide biosynthesis by Pseudomonas NCIB 11264. Studies on precursor-forming enzymes and factors affecting exopolysaccharide production by washed suspensions. J Gen Microbiol. 1980;116:133–141. doi: 10.1099/00221287-116-1-133. [DOI] [PubMed] [Google Scholar]