Abstract
The performance of artificial intelligence (AI) for brain MRI can improve if enough data are made available. Generative adversarial networks (GANs) showed a lot of potential to generate synthetic MRI data that can capture the distribution of real MRI. Besides, GANs are also popular for segmentation, noise removal, and super-resolution of brain MRI images. This scoping review aims to explore how GANs methods are being used on brain MRI data, as reported in the literature. The review describes the different applications of GANs for brain MRI, presents the most commonly used GANs architectures, and summarizes the publicly available brain MRI datasets for advancing the research and development of GANs-based approaches. This review followed the guidelines of PRISMA-ScR to perform the study search and selection. The search was conducted on five popular scientific databases. The screening and selection of studies were performed by two independent reviewers, followed by validation by a third reviewer. Finally, the data were synthesized using a narrative approach. This review included 139 studies out of 789 search results. The most common use case of GANs was the synthesis of brain MRI images for data augmentation. GANs were also used to segment brain tumors and translate healthy images to diseased images or CT to MRI and vice versa. The included studies showed that GANs could enhance the performance of AI methods used on brain MRI imaging data. However, more efforts are needed to transform the GANs-based methods in clinical applications.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13244-022-01237-0.
Keywords: Artificial intelligence, Data augmentation, Generative adversarial networks, Magnetic resonance imaging, Medical imaging
Key points
This article aims to provide a comprehensive review on the applications of generative adversarial networks (GANs) in brain MRI.
The specific focus of this education review is on brain MRI.
It covers a large number of studies on GANs in brain MRI and the most recently published studies on brain MRI.
Introduction
Magnetic resonance imaging (MRI) is a widely used medical imaging technology. MRI is non-intrusive and considered safe for humans. MRI can generate different modalities of an image and can provide valuable insights into a specific disease. The frequent sequences of MRI are T1-weighted and T2-weighted scans [1, 2]. The major difference between MRI and other medical imaging technologies is that MRI is free from using X-ray radiography. The radiologists use MRI to analyze brain tissue and diagnose brain-related diseases such as brain tumors (i.e., the abnormal and uncontrolled growth of brain cells). This process requires trained radiologists, and the accuracy is heavily dependent on the expertise of the radiologists and the quality of MRI data acquisition [1, 2].
Computer-aided diagnosis (CAD) can aid in the process of MRI analysis. Recently, there has been a significant increase in interest in developing artificial intelligence and deep learning-based methods for CAD. However, deep learning methods rely on training using large medical imaging data. Generative adversarial networks (GANs) have the potential to generate new samples of the data and represent the distribution of the real data. GANs are particular types of deep learning models formed of two neural networks, namely the generator and the discriminator. The generator generates new samples, while the discriminator attempts to classify the images as real or synthetic. The adversarial training effectively improves the overall training of the model. While GANs methods were initially popular for generating synthetic data in the medical imaging domain, they have also been used for other applications such as super-resolution, segmentation, and diagnosis.
This study performed a scoping review to find out the role of GANs-based methods in brain MRI. While many reviews have been performed on the use of GANs in medical imaging and GANs in MRI [1–3], their scope is too broad. For example, the review in [1] covers a broad range of MRI and does not focus on brain MRI only. Similarly, the review in [2] covers many different deep learning techniques and does not limit the discussion to GANs-based methods only. The review in [3] covers the discussion on GANs for all types of medical imaging data. Table 1 provides a comparison of our work with previous reviews. The growing number of studies on the use of GANs in brain MRI demands a dedicated review. In this regard, this review presents a review of how GANs-based methods were used to address many challenges in advancing the performance of AI for brain MRI data. More specifically, it summarizes the applications of GANs-based methods in brain MRI such as synthesis of brain MRI, segmentation of brain tumor, and super-resolution of brain MRI. Furthermore, it also highlights the different evaluation metrics such as structural similarity index measure (SSIM) and the peak signal-to-noise ratio (PSNR) used in the literature for evaluation of the performance of GANs. The following research questions related to the role of GANs-based method in brain MRI were considered for this review.
What were the typical applications of GANs proposed for brain MRI?
Which architectures of GANs are most commonly applied for brain MRI?
What was the purpose of using GAN in brain MRI?
What were the most commonly used datasets for brain MRI?
How many datasets were publicly accessible?
What evaluation matrices were used for the validation of the model?
Table 1.
Comparison with previous reviews
| Previous review | Year | Scope and coverage | Comparative contribution of our review |
|---|---|---|---|
| An overview of deep learning in medical imaging focusing on MRI [1] | 2019 |
(1) It did not focus on GANs but rather covered many different deep learning methods (2) It did not focus on just brain MRI but rather focused on different MRI (3) It did not cover many recent studies as there has been an exponential rise in GANs-based methods for brain MRI during the last 2 years |
(1) Our review is focused on GANs (2) Our review is focused on brain MRI (3) Our review covers many recent studies, published in 2020 and 2021 |
| Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI [2] | 2020 |
(1) It did not focus on GANs but rather covered a broad range of deep learning methods (2) It did not cover applications for brain MRI such as synthesis of brain MRI data, translation of brain MRI data, and deep learning for noise removal from brain MRI, etc. |
(1) Our review is focused on GANs (2) Our review covers all the possible applications for brain MRI |
| Generative adversarial network in medical imaging: A review [3] | 2019 |
(1) It did not focus on brain MRI but rather covered all modalities of medical imaging (2) It did not cover many recent studies published in 2020 and 2021, as there has been an exponential rise in studies for brain MRI during the last 2 years |
(1) Our review is focused on brain MRI (2) Our review covers many recent studies, published in 2020 and 2021 |
The study will be helpful for researchers and professionals in the medical imaging and healthcare domain who are considering using GANs methods to diagnose and predict the brain tumors from the MRI images. The review also lists publicly available brain MRI datasets that will be helpful for AI researchers to develop advanced research methods.
Methods
We performed a literature search in famous databases and conducted a scoping review as per the guidelines of the PRISMA-ScR (Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews) [4]. Additional file 1: Table S1 provides the adherence to the PRISMA-ScR checklist. The following methods were used for the search and the study selection.
Search strategy
Search sources
This review searched five different databases for relevant literature, namely PubMed, Scopus, IEEE Xplore, ACM Digital Library, and Google Scholar. We note here that MEDLINE is covered in PubMed. The search was performed between September 20 and 22, 2021. For the search outcomes of Google Scholar, only the first 100 results were considered, as, beyond the first 100 entries, the search results were quickly losing match and relevancy to the topic of the review. In addition to the search on the five databases, we also screened the reference lists of the included studies to find additional relevant studies.
Search terms
We defined the search terms from the available literature and by referring to the experts in the fields. The search terms were selected based on the intervention (e.g., deep learning, generative adversarial networks (GANs)), the target anatomy (brain), and the target data modality (e.g., MRI, fMRI, sMRI). The search strings used in this study are provided in Additional file 1: Table S2.
Search eligibility criteria
We focused on GANs-based approaches used for brain MRI data. We considered studies published in English from January 2015 to September 2021. Studies for all applications of GANs were included, such as segmentation, synthesis, noise removal, and super-resolution of brain MRI. We included studies that used GANs for brain MRI data and excluded studies that used other deep learning methods (such as convolutional neural networks or recurrent neural networks) but did not use GANs. Similarly, we excluded studies that used GANs for non-image data or image data of modalities other than MRI (such as ultrasound, X-ray, or computed tomography (CT)). We also excluded studies that used GANs for MRI data other than the brain.
We included peer-reviewed articles and conference proceedings and excluded preprints, commentaries, short reviews, editorials, and abstracts. Similarly, we excluded studies that presented a survey of GANs methods. No restrictions were imposed on the country of publication, comparators, and outcomes of the GANs methods.
Study selection
Two reviewers, namely authors AJ and OT, independently reviewed the titles and abstracts of the studies identified in the search and made initial flagging for inclusion and exclusion. The flagging was then verified by a third reviewer (HA). The studies that passed the title and abstract screening were shortlisted for the full-text reading phase to perform study selection. Any disagreement between the reviewers (AJ and OT) was investigated and resolved through discussion and consensus. The Cohen’s kappa score [5] was calculated to measure the agreement between the two reviewers.
Data extraction
We prepared a purpose-built form for data extraction. Additional file 1: Table S4 shows the data extraction form. The entries for the form were pilot-tested using ten relevant studies to extract the data accurately. Two reviewers (MB and FA) independently performed the data extraction according to the data extraction form. The data were extracted for the applications of the studies, the purpose of using GAN, the type of GAN, features of the dataset, and the evaluation mechanism of the GANs-based methods. Any disagreement between the two reviewers was resolved through discussion and consensus.
Data synthesis
After the extraction of the data from the included studies, we synthesized the data using a narrative approach. First, we classified the included studies in terms of their applications, such as synthesis (data augmentation), diagnosis (e.g., tumor detection), prognosis, and super-resolution. We also classified the studies based on the purpose of using GANs, such as synthesis, noise removal, and translation. Based on dataset types, we organized the data into two broad categories: studies that used publicly available datasets and studies that used privately collected MRI data. We also summarized the studies based on the size of the dataset, the evaluation mechanisms, and the reporting of external validation. We performed and managed the data synthesis using MS Excel.
Results
Search and study selection results
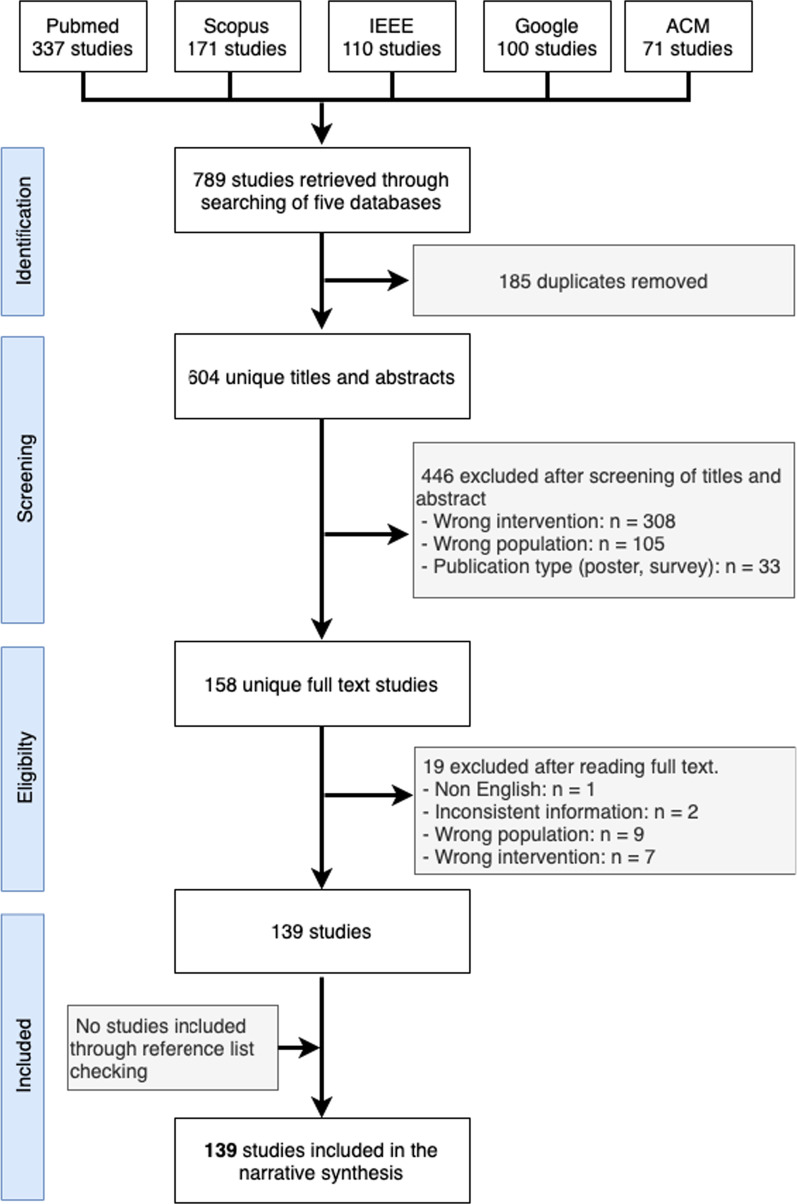
We retrieved 789 studies as a search result. We removed 185 duplicates. We then did the screening of the titles and abstracts of the remaining studies. As a result of title and abstract screening, we excluded 446 studies following the criteria defined in the protocol. We then performed the full-text reading of the remaining 158 studies. Among these, we removed 19 studies that did not fulfill the criteria of inclusion. Finally, we were left with 139 studies for inclusion in this survey. See Fig. 1 for the flowchart of the study selection process. No additional studies were identified by forward-and-backward reference checking. The Cohen’s kappa score was 86.3% for the title and abstract screening, which shows a good agreement between the reviewers. The Cohen’s kappa score was 84.7% for the full-text reading phase, which shows a good agreement between the reviewers. Additional file 1: Table S3 shows the matrix for the calculation of the Cohen’s kappa score.
Fig. 1.
The PRISMA-ScR flowchart for the selection of the included studies
Demographics of the included studies
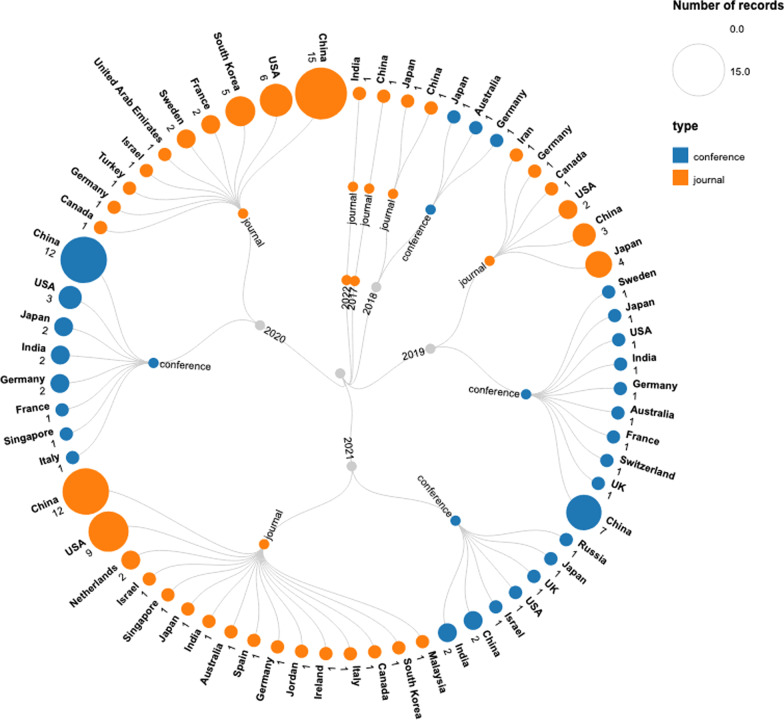
Among the included studies, 87 were peer-reviewed journal articles and 52 were conference publications. More than two-thirds of the studies (n = 104) were published in the last 2 years, i.e., 2020 and 2021. In comparison, only five studies were published in 2018 and only one study was published in 2017. A total of 27 countries contributed to the studies. Around one-third of the studies (n = 53) were published in China. The only two other countries that published more than ten studies were the USA (n = 21) and Japan (n = 12). Table 2 summarizes the demographics of the included studies. Figure 2 shows a visualization of the year-wise and country-wise distribution of the included studies.
Table 2.
Demographics of the included studies
| Number of studies | |
|---|---|
| Year | |
| Year | |
| 2022 | 1 |
| 2021 | 44 |
| 2020 | 60 |
| 2019 | 28 |
| 2018 | 5 |
| 2017 | 1 |
| Countries | |
| Country | |
| China | 53 |
| USA | 22 |
| Japan | 11 |
| Germany | 7 |
| India | 7 |
| South Korea | 6 |
| France | 4 |
| Sweden | 3 |
| Israel | 3 |
| Canada | 3 |
| Australia | 2 |
| UK | 2 |
| Singapore | 2 |
| The Netherlands | 2 |
| Italy | 2 |
| United Arab Emirates | 1 |
| Turkey | 1 |
| Switzerland | 1 |
| Spain | 1 |
| Russia | 1 |
| Malaysia | 1 |
| Jordan | 1 |
| Ireland | 1 |
| Iran | 1 |
| Malaysia | 1 |
| Type of publication | |
| Venue | |
| Conference | 52 |
| Journal | 87 |
Fig. 2.
Year-wise and country-wise distribution of the included studies. The numbers at the terminal node show the number of publications in each country
Applications of GANs in brain MRI
GANs have been used for many applications of brain MRI data. The included studies used GAN-based methods as a sub-module of their deep learning frameworks for different applications, as shown in Table 2. The majority of the included studies targeted applications, namely the generation of synthetic data (n = 43), the segmentation of area of interest in brain MRI (n = 32), and the diagnosis of neurological diseases (n = 22). Other common applications of the studies were super-resolution to improve the quality of the images as reported in ten studies and reconstruction of high-quality images (which can be considered a sub-category of super-resolution) reported in 13 studies. Few studies also reported applications such as noise removal (n = 5), prognosis (n = 4), and image registration (n = 2). Only one study reported the generation of 3D synthetic volumes of MRI data (see Fig. 3).
Fig. 3.
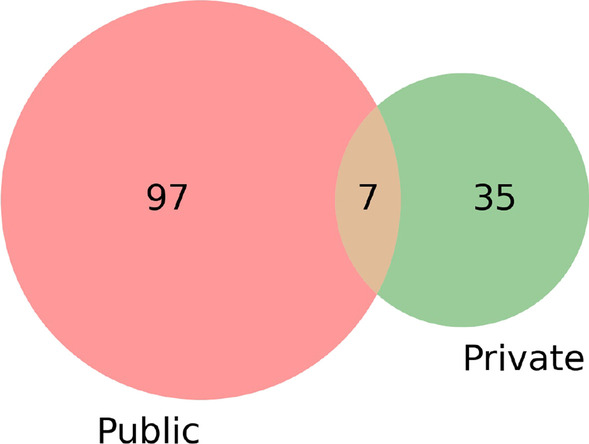
Venn diagram for the number of studies using public vs. privately collected datasets. Some of the studies (n = 7) reported using both public and private datasets
The included studies used GANs for many different applications, namely synthesis (generation of synthetic data), segmentation (generation of the segmentation mask), diagnosis, and translation of data from one modality to another (e.g., translation from CT to MRI and vice versa, or translation form normal MRI to infected MRI). Almost one-third of the studies (n = 45) reported the use of GANs for the synthesis of data. Around one-sixth of the studies (n = 26) reported GANs to perform segmentation. Other popular use cases of GANs were diagnosis reported in 16 studies, reconstruction reported in 15 studies, and translation reported in 12 studies. The reconstruction may also be regarded as a particular case of image synthesis. Only a few studies reported use cases of GANs for other applications, such as super-resolution reported in seven studies, noise removal reported in five studies, prediction reported in five studies, and prognosis reported in four studies. Table 3 provides a summary of the use cases of GANs.
Table 3.
Applications of the use of GANs in brain MRI
| Applications of studies | No. of studies | Reference of the study |
|---|---|---|
| Applications of studies | ||
| Synthesis | 43 | [6–37, 39–49] |
| Segmentation | 32 | [50–81] |
| Diagnosis | 22 | [82–103] |
| Reconstruction | 13 | [119–131] |
| Super-resolution | 10 | [104–112, 118] |
| Prediction | 7 | [132–138] |
| Noise removal | 5 | [113–117] |
| Prognosis | 4 | [139–142] |
| Image registration | 2 | [143, 144] |
| 3D synthesis | 1 | [38] |
| Purpose of using GANs | ||
| Synthesis | 45 | [6–35, 53–59, 96–100, 120, 132, 133] |
| Segmentation | 26 | [60–81, 101, 102, 104, 104] |
| Diagnosis | 16 | [50–52, 82–93, 106] |
| Reconstruction | 15 | [12–123, 125–131] |
| Translation | 12 | [37, 41–49, 95, 118, 143] |
| Super-resolution | 7 | [38, 107–112] |
| Noise removal | 5 | [113–117] |
| Prediction | 5 | [134–138] |
| Prognosis | 4 | [139–142] |
| Features extraction | 1 | [39] |
| Translation | 1 | [37, 41–45, 47–49, 95, 118, 143] |
| Anomaly detection | 1 | [94] |
| Image registration | 1 | [144] |
Types of GANs methods
While there are many different types of GANs usually named based on their architectures, there is a tendency to assign a new name to every GAN even if the fundamental changes in the architecture are not significant. This review found that the most common types of GANs used were the cycleGAN used by 12 studies [15, 17, 48, 51, 55–57, 65, 66, 79, 84, 110, 133] followed by conditional GAN used by 8 studies [53, 54, 71, 72, 101, 112, 118, 119], and Wasserstein GAN used by 7 studies [13, 14, 19, 39, 116, 131, 132]. Other types of GANs reported in more than one study were deep convolutional GAN, reported in three studies [20, 93, 140], unified GAN [21, 49] reported in two studies, and Pix2Pix GAN, reported in two studies [32, 133].
Types of datasets
Most of the studies (n = 97) reported the use of public datasets for brain MRI for the training of GAN models. Thirty-five studies reported the use of privately collected data. A few studies (n = 7) reported using both public and privately collected data. This review identified many different datasets used in the included studies. Table 4 provides a list of publicly available datasets and the access link. In the included studies, the most commonly used dataset was the Alzheimer’s Disease Neuroimaging Initiative dataset reported in 16 studies (also see Table 4). The BRaTs 2018 dataset was reported in eight studies, while the use of the IXI dataset of MR images from three different hospitals in London was reported in seven studies. The accumulative number of studies using the various versions of the BRaTs dataset was 20.
Table 4.
Publicly available datasets used in the included studies. Sorting is done on the basis of the number of studies using the dataset
The names of the dataset are assigned only for identification purposes and do not follow any specific convention
Evaluation procedure
The number of patients was reported in some studies, while other studies reported the number of images. The maximum number of patients for whom the data were used was 2175 [92]. Two studies reported the use of more than 100,000 thousand images [23, 106], and one study reported the use of more than 10,000 images. In 25 studies, the number of images used was between 1000 and 10,000. In 33 studies, the number of images used was between 100 and 1000. Other studies either used less than 100 images or did not include information on the number of images. In the included studies, 38 reported splitting the data into independent training and test sets, while 17 reported splitting the data into training, validation, and test sets. Many other studies used the k-fold cross-validation method for evaluation; for example, twofold cross-validation was reported in three studies and sevenfold cross-validation was reported in two studies (see Table 5). External evaluation by human experts was reported in seven studies only.
Table 5.
Evaluation mechanisms used in different studies
| Evaluation mechanism | Number of studies | IDs of studies |
|---|---|---|
| Train, validate, test split | 17 | [6, 16, 17, 22, 37, 58, 59, 65, 76, 81, 89, 97–99, 106, 121, 126] |
| Training, test split | 38 | [10, 11, 13, 14, 20, 24, 33, 35, 36, 40, 45, 47, 50, 52, 53, 57, 66, 68, 69, 77, 87, 92, 98, 100, 103, 104, 107, 108, 110, 112, 115–117, 122, 125, 127, 128, 130] |
| Twofold cross-validation | 3 | [9, 75, 114] |
| Threefold cross-validation | 2 | [134, 137] |
| Fourfold cross-validation | 2 | [56, 70] |
| Fivefold cross-validation method | 12 | [7, 8, 21, 25, 29, 41, 46, 62, 90, 113, 120, 129] |
| Sevenfold cross-validation | 2 | [79, 139] |
| Tenfold cross-validation | 6 | [42, 80, 84, 95, 96, 101] |
| External | 7 | [31, 32, 43, 45, 48, 118, 135] |
The different metrics used for the evaluation of the quality of the generated images using GANs were SSIM (n = 53 studies), PSNR (n = 49 studies), and FID (n = 8 studies). Other metrics for evaluation of performance for diagnosis, segmentation, or classification were Dice score used in 31 studies, mean absolute error used in 16 studies, and mean square error used in 16 studies. Table 6 summarizes the different evaluation metrics used in the studies.
Table 6.
Most popular evaluation metrics used in different studies
| Evaluation metric | Number of studies | IDs of studies |
|---|---|---|
| SSIM | 53 | [7–12, 15, 16, 18, 21, 25, 27, 36, 40, 42, 43, 45, 47, 48, 55, 56, 58, 62, 66, 69, 72, 85, 86, 103–110, 112, 113, 115–117, 120–123, 125–131] |
| PSNR | 49 | [7–11, 15–17, 21, 25, 36, 38, 42, 43, 45–48, 53, 55, 56, 58, 62, 66, 72, 85, 86, 97, 104–110, 112, 113, 115–118, 120, 121, 123, 124, 128, 129, 131] |
| DSC | 31 | [9, 20, 29, 45, 50–56, 59–61, 68, 72–77, 79–81, 102, 105, 114, 125, 136, 142–144] |
| Accuracy | 22 | [6, 13, 14, 19, 34, 35, 37, 39, 64, 83, 84, 89, 90, 92, 93, 95, 96, 98, 122, 132, 135, 139] |
| MAE | 16 | [7, 17, 21, 23, 29, 42, 46, 53, 58, 69, 85, 100, 115, 120, 129, 134] |
| MSE | 16 | [11, 16, 40, 45, 48, 58, 72, 117, 118, 122, 123, 128, 130, 131, 142] |
| Sensitivity | 11 | [75, 76, 81, 84, 92, 95, 96, 98, 99, 142] |
| Precision | 9 | [19, 26, 54, 64, 75, 132, 135, 138, 139] |
| Recall | 9 | [19, 26, 39, 54, 64, 132, 135, 138, 139] |
| F1 score | 8 | [19, 24, 64, 92, 93, 135, 138, 139] |
| FID | 8 | [21, 22, 42, 59, 109, 130] |
| Specificity | 8 | [68, 76, 84, 92, 95, 98, 142] |
The numbers do not sum up as many studies used more than one evaluation metric, while some studies lack details on evaluation metrics
SSIM structural similarity index measure, PSNR peak signal-to-noise ratio, DSC Dice similarity coefficient, MAE mean absolute error, MSE mean square error, FID Frechet inception distance
Focal diseases in the studies
We also identify the diseases that were the focus of the included studies. In the included studies, 44 studies reported their methods for addressing challenges related to brain tumors, such as tumor segmentation, tumor classification, or tumor growth prediction. Similarly, 20 studies reported the use of their methods for diagnosis, prognosis, or analysis of neurodegenerative disorders, for example, Alzheimer's disease, autism spectrum disorder (ASD), multiple sclerosis, and Parkinson’s disease. The remaining 75 studies did not focus on a particular disease. A summary of the disease-based categorization of the studies is given in Table 7.
Table 7.
Focal diseases in the studies
| Focal disease | Number of studies (n) | IDs of studies |
|---|---|---|
| Brain tumor | 44 | [5, 10, 20], [22], [25, 32, 35, 37, 44, 50–58, 61–64, 66–69, 71, 74–78, 83, 89, 90, 93, 98–101, 107, 133, 136, 142] |
| Neurodegenerative disorders | 20 | [19, 26, 31, 33, 39, 45, 84, 85, 87, 88, 91, 92, 94–97, 132, 137, 139, 140] |
| None | 75 | [7–9, 11–18, 21, 23, 24, 27–29, 31, 34, 36, 38, 40–43, 46–49, 59, 60, 65, 70, 72, 73, 79–82, 86, 102–106, 108–131, 134, 135, 138, 141, 143, 144] |
Discussion
Principal results
In this study, we conducted a scoping review of the use of GANs in brain MRI data. We found that most of the studies were published in the years 2020 and 2021, while very few (only six) were published in 2016 and 2017 combined. This is not surprising as the interest in using GANs for medical imaging in general and brain MRI, in particular, gained momentum only recently. More than one-third of the studies were published in China (n = 53). The second-largest number of studies were published in the USA (n = 21), although less than half of those published in China. In comparison, only seven studies were published in India and Germany each. The rest of the countries published less than five studies each.
In almost one-third of the studies, the main application of using GANs was the synthesis/generation of data to achieve data augmentation. However, many studies also used GANs for the segmentation of tissues of interest, for example, the segmentation of tumors in brain MRI. Another popular use case of GAN was translating images from one modality to another or translating from normal to cancerous images. Furthermore, GANs can enhance the quality of images and hence were used for super-resolution of images as reported in seven studies and noise removal as reported in five studies. Less common use cases of GANs on brain MRI data were prognosis and image registration reported only in 4 studies and 1 study, respectively. While GANs are more popular for data synthesis, addressing a particular clinical disease is usually not the focus of using GANs. Nevertheless, some studies have demonstrated the effectiveness of GANs by demonstrating the use of the generated data to improve the diagnosis or prognosis of different diseases.
The term synthesis in this review is used in a broader sense and covers the synthesis of brain MRI sequences as well as the synthesis of missing sequences from existing sequences. The synthesized data were then used to enhance the diagnosis, such as detecting Alzheimer’s disease or segmentation of brain tumors.
The cycleGAN architecture that uses two GANs for generating synthetic data was the most popular choice of architecture in the included studies. Other popular choices were the Wasserstein GAN and the deep convolutional GAN. For many studies, the fundamental changes in the architecture were only minor, or the details on the changes introduced were insufficient; it is beyond the scope of this review to analyze all the architectures.
While testing the models on individual test sets or using k-fold cross-validation methods was reported in most of the studies, external validation of the performance is still limited and should be encouraged in future work.
Research and practical implications
The majority of the included studies reported results on publicly available datasets. Among these, the BRaTS dataset and the Alzheimer’s Disease Neuroimaging Initiative dataset were the most popular datasets among the researchers. Since these datasets can be accessed publicly, it would be of great help to provide the associated computer code/software for the results reported in the included studies. This would encourage other researchers to reproduce the results and build upon the existing models/methods. However, some studies reported results on privately collected data. Hence, the opportunity for external validation of the claims made in the research studies or building upon those results is limited.
We did not find any framework implemented on mobile devices in the included studies. The computational requirements of GANs and the memory resources for MRI data can be the possible reasons for the limited transformation of these models to mobile devices. It is only hoped that future research might enable the implementation of these methods on mobile devices.
No studies were found on the transformation of these methods into clinical applications, which shows that their acceptance for clinical use is still limited. Many studies claim the value of their methods for use in clinical tasks; however, they lack reporting of testing for clinical purposes.
GANs were initially popular for generating synthetic image data similar to the original data. However, the perception of realistic-looking is subjective. Furthermore, though some quantitative measures such as peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM) are reported in many included studies, these metrics are principally borrowed from the computer vision literature. Hence, how efficiently these metrics quantify the complex physiological information within the MRI images data is not well understood. Thus, there is a dire need to develop uniform methods to evaluate the performance of GANs on how well they capture complex features within MRI image data.
As used in many of the studies, the publicly available data for MRI are primarily from developed economies. However, there is a lack of medical imaging data from developing economies. Hence, computer models for diagnosis trained on such data may not necessarily generalize well for a population of different geoeconomics characteristics due to a lack of representation in the data. Including MRI data from diverse locations is needed and will help develop better AI methods for clinical applications such as diagnosis, prognosis, and tumor detection in brain MRI.
Strengths and limitations
Strengths
While many reviews have been published on the applications of GANs in medical imaging, to the best of our knowledge, this is the first review on the applications of GANs in brain MRI images. This review includes all the studies that used GANs for brain MRI; hence, this is the most comprehensive review on the topic. This review helps the readers to know the potential of the GANs for the synthesis of brain MRI data and the potential to improve the diagnosis and segmentation of brain tumors within brain MRI. Unlike reviews as [1–3] that covered a broad scope of different deep learning methods, this review focuses specifically on the applications of GANs in brain MRI.
In this review, we followed the scientific review guidelines of the PRISMA-ScR [4]. In addition, we covered the major databases in health sciences, engineering, and technology fields to identify as many as possible published studies. Hence, the number of studies included in this review is high. We devised a strategy to avoid bias in study selection by employing two independent reviewers for study selection and data extraction and a third reviewer to validate the screening and the data extraction. This review provides a comprehensive list of the publicly accessible datasets for brain MRI. Hence, it can be considered a rich resource for the readers to identify valuable datasets of brain MRI.
Limitations
In this review, we included studies from five major databases. So, some studies might have been left out if they were not covered in the included databases. In addition, due to practical limitations, the review only consists of studies published in English. Hence, relevant studies published in other languages might have been left out. This review lists the studies into major applications such as synthesis, segmentation, diagnosis, super-resolution, and noise removal. The definition of some applications may overlap partly with others; for example, super-resolution may be considered as a sub-category of synthesis, and the categorization of super-resolution studies as synthesis studies will then increase the number of the studies in the synthesis category. However, we believe that the categorization in this review will better reflect the notion of the applications. We did not perform validation and assessment of the claims on the diagnosis of a brain tumor or the quality of the synthesized MRI data, as this was beyond the scope of this review.
Conclusion
In this scoping review, we included 139 studies that reported the use of GANs for brain MRI data. We identified the most common applications of GANs. We also identified the most commonly used datasets publicly available for brain MRI. We also summarized the most common architectures of GANs and the evaluation metrics that are widely adopted to evaluate the performance of GANs in brain MRI. It will be most rewarding if these studies find their way into clinical transformations. To achieve this, we remark that encouraging the availability of the software and codes for these studies will facilitate the reproducibility of the results. Eventually, more research progress will be possible. In addition, the need to bridge the gap between the computer scientists and clinicians is widely felt as the input and feedback of clinicians and radiologists is vital for the research outcomes to find their way into clinical uses. Similarly, there is a need to follow standardized comparison protocols for the different architectures of GANs used for brain MRI.
Supplementary Information
Additional file1: Table S1 PRISMA-ScR Checklist. Table S2 Search strategy. Table S3 Interrater agreement matrices for study selection steps. Table S4 Data extraction form. Appendix S5 Characteristics of the studies.
Acknowledgements
The publication of this article was funded by Qatar National Library.
Abbreviations
- ACM
Association for Computing Machinery
- BRaTs
Brain tumor segmentation
- CAD
Computer-aided diagnosis
- CT
Computed tomography
- DSC
Dice similarity coefficient
- FID
Fréchet inception distance
- fMRI
Functional magnetic resonance imaging
- GAN
Generative adversarial networks
- IEEE
Institute of Electrical and Electronic Engineers
- IXI
Information extraction from images
- MAE
Mean absolute error
- MRI
Magnetic resonance imaging
- MSE
Mean square error
- PRISMA-ScR
Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews
- PSNR
Peak signal-to-noise ratio
- sMRI
Structural magnetic resonance imaging
- SSIM
Structural similarity index measure
Author contributions
HA and ZS contributed to conceptualization. MB, FA, and UZ curated the data. AA and OM helped in methodology. HA administered the project. ZS supervised the study. MB and HA contributed to writing—original draft. HA and ZS performed writing—review and editing. All authors read and approved the final manuscript.
Funding
The publication of this article was funded by Qatar National Library.
Availability of data and material
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/30/2022
A Correction to this paper has been published: 10.1186/s13244-022-01268-7
Contributor Information
Hazrat Ali, Email: haali2@hbku.edu.qa.
Zubair Shah, Email: zshah@hbku.edu.qa.
References
- 1.Lundervold AS, Lundervold A (2019) Arvid lundervold, an overview of deep learning in medical imaging focusing on MRI. Z Med Phys 29(2):102–127. 10.1016/j.zemedi.2018.11.002 [DOI] [PubMed]
- 2.Zeng C, Lin Gu, Liu Z, Zhao S. Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI. Front Neuroinform. 2020;14:55. doi: 10.3389/fninf.2020.610967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 4.Tricco AC, Lillie E, Zarin W, O'Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169(7):467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 5.Higgins J, Deeks J (2008) Chapter 7: selecting studies and collecting data. In: Cochrane handbook for systematic reviews of interventions. Wiley, Hoboken, NJ
- 6.Han C, et al. Combining noise-to-image and image-to-image gans: brain mr image augmentation for tumor detection. IEEE Access. 2019;7:156966–156977. doi: 10.1109/ACCESS.2019.2947606. [DOI] [Google Scholar]
- 7.Dai X, et al. Multimodal MRI synthesis using unified generative adversarial networks. Med Phys. 2020;47(12):6343–6354. doi: 10.1002/mp.14539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma A, Hamarneh G. Missing MRI pulse sequence synthesis using multi-modal generative adversarial network. IEEE Trans Med Imaging. 2020;39(4):1170–1183. doi: 10.1109/TMI.2019.2945521. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Zheng F, Cong R, Huang W, Scott MR, Shao L. MCMT-GAN: multi-task coherent modality transferable GAN for 3D Brain image synthesis. IEEE Trans Image Process. 2020;29:8187–8198. doi: 10.1109/TIP.2020.3011557. [DOI] [PubMed] [Google Scholar]
- 10.Xin B, Hu Y, Zheng Y, Liao H (2020) Multi-modality generative adversarial networks with tumor consistency loss for brain MR image synthesis. In: 2020 IEEE 17th international symposium on biomedical imaging (ISBI), pp 1803–1807
- 11.Liu X, Yu A, Wei X, Pan Z, Tang J. Multimodal MR image synthesis using gradient prior and adversarial learning. IEEE J Sel Top Signal Process. 2020;14(6):1176–1188. doi: 10.1109/JSTSP.2020.3013418. [DOI] [Google Scholar]
- 12.Chong CK, Ho ETW. Synthesis of 3D MRI brain images with shape and texture generative adversarial deep neural networks. IEEE Access. 2021;9:64747–64760. doi: 10.1109/ACCESS.2021.3075608. [DOI] [Google Scholar]
- 13.Yang X, Lin Y, Wang Z, Li X, Cheng K-T. Bi-modality medical image synthesis using semi-supervised sequential generative adversarial networks. IEEE J Biomed Health Inform. 2020;24(3):855–865. doi: 10.1109/JBHI.2019.2922986. [DOI] [PubMed] [Google Scholar]
- 14.Han C et al (2018) GAN-based synthetic brain MR image generation. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), pp 734–738
- 15.Qu Y, Deng C, Su W, Wang Y, Lu Y, Chen Z (2020) Multimodal brain MRI translation focused on lesions. In: Proceedings of the 2020 12th international conference on machine learning and computing, pp 352–359
- 16.Alogna E, Giacomello E, Loiacono D (2020) Brain magnetic resonance imaging generation using generative adversarial networks. In: 2020 IEEE symposium series on computational intelligence (SSCI), pp 2528–2535
- 17.Li Y et al (2020) Synthesize CT from paired MRI of the same patient with patch-based generative adversarial network. In: Progress in biomedical optics and imaging—proceedings of SPIE, vol 11314
- 18.Gu Y, Peng Y, Li H (2020) AIDS brain MRIs synthesis via generative adversarial networks based on attention-encoder. In: 2020 IEEE 6th international conference on computer and communications (ICCC), pp 629–633
- 19.Rejusha TR, KS VK (2021) Artificial MRI image generation using deep convolutional GAN and its comparison with other augmentation methods. In: 2021 international conference on communication, Control and Information Sciences (ICCISc), vol 1, pp 1–6
- 20.Rezaei M et al (2020) “Generative synthetic adversarial network for internal bias correction and handling class imbalance problem in medical image diagnosis. In: Progress in biomedical optics and imaging—proceedings of SPIE, vol 11314
- 21.Sohan K, Yousuf MA (2020) 3D bone shape reconstruction from 2D X-ray images using MED generative adversarial network. In: 2020 2nd international conference on advanced information and communication technology (ICAICT), pp 53–58. 10.1109/ICAICT51780.2020.9333477
- 22.Dikici E, Bigelow M, White RD, Erdal BS, Prevedello LM. Constrained generative adversarial network ensembles for sharable synthetic medical images. J Med Imaging. 2021;8(2):024004. doi: 10.1117/1.JMI.8.2.024004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Tahan H, Mohsenzadeh Y. Reconstructing feedback representations in the ventral visual pathway with a generative adversarial autoencoder. PLoS Comput Biol. 2021;17(3):e1008775. doi: 10.1371/journal.pcbi.1008775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, Du C, Wang S, Wang H, He H. Multi-subject data augmentation for target subject semantic decoding with deep multi-view adversarial learning. Inf Sci. 2021;547:1025–1044. doi: 10.1016/j.ins.2020.09.012. [DOI] [Google Scholar]
- 25.Ma B, et al. MRI image synthesis with dual discriminator adversarial learning and difficulty-aware attention mechanism for hippocampal subfields segmentation. Comput Med Imaging Gr. 2020;86:101800. doi: 10.1016/j.compmedimag.2020.101800. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Cao P, Dou L, Yang J, Zhao D (2020) An auto-encoding generative adversarial networks for generating brain network. In: The fourth international symposium on image computing and digital medicine, pp 14–18
- 27.Segato V, Corbetta MD, Marzo LP, Momi ED. Data augmentation of 3D brain environment using deep convolutional refined auto-encoding alpha GAN. IEEE Trans Med Robot Bionics. 2021;3(1):269–272. doi: 10.1109/TMRB.2020.3045230. [DOI] [Google Scholar]
- 28.Yang H, Qian P, Fan C. an indirect multimodal image registration and completion method guided by image synthesis. Comput Math Methods Med. 2020;2020:2684851. doi: 10.1155/2020/2684851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazemifar S, et al. MRI-only brain radiotherapy: assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother Oncol. 2019;136:56–63. doi: 10.1016/j.radonc.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 30.Barile B, Marzullo A, Stamile C, Durand-Dubief F, Sappey-Marinier D. Data augmentation using generative adversarial neural networks on brain structural connectivity in multiple sclerosis. Comp Methods Prog Biomed. 2021;206:106113. doi: 10.1016/j.cmpb.2021.106113. [DOI] [PubMed] [Google Scholar]
- 31.Hirte AU, Platscher M, Joyce T, Heit JJ, Tranvinh E, Federau C. Realistic generation of diffusion-weighted magnetic resonance brain images with deep generative models. Magn Reson Imaging. 2021;81:60–66. doi: 10.1016/j.mri.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Jw S, et al. Synthetic generation of DSC-MRI-derived relative CBV maps from DCE MRI of brain tumors. Magn Reson Med. 2021;85(1):469–479. doi: 10.1002/mrm.28432. [DOI] [PubMed] [Google Scholar]
- 33.Finck T, et al. Deep-learning generated synthetic double inversion recovery images improve multiple sclerosis lesion detection. Invest Radiol. 2020;55(5):318–323. doi: 10.1097/RLI.0000000000000640. [DOI] [PubMed] [Google Scholar]
- 34.Kazuhiro K, et al. Generative adversarial networks for the creation of realistic artificial brain magnetic resonance images. Tomography. 2018;4(4):159–163. doi: 10.18383/j.tom.2018.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deepak S, Ameer PM (2020) MSG-GAN based synthesis of brain MRI with meningioma for data augmentation. In: 2020 IEEE international conference on electronics, computing and communication technologies (CONECCT), pp 1–6
- 36.Li G, Lv J, Wang C. A modified generative adversarial network using spatial and channel-wise attention for CS-MRI reconstruction. IEEE Access. 2021;9:83185–83198. doi: 10.1109/ACCESS.2021.3086839. [DOI] [Google Scholar]
- 37.Ge C, Gu IY, Jakola AS Yang J (2019) Cross-modality augmentation of brain Mr images using a novel pairwise generative adversarial network for enhanced glioma classification. In: 2019 IEEE international conference on image processing (ICIP), pp 559–563
- 38.Hongtao Z, Shinomiya Y, Yoshida S (2020) 3D brain MRI reconstruction based on 2D super-resolution technology. In: 2020 IEEE international conference on systems, man, and cybernetics (SMC), pp 18–23
- 39.Zhang X, Yang Y, Wang H, Ning S, Wang H (2019) Deep neural networks with broad views for parkinson’s disease screening. In: 2019 IEEE international conference on bioinformatics and biomedicine (BIBM) pp 1018–1022
- 40.Qiao K, Chen J, Wang L, Zhang C, Tong L, Yan B. BigGAN-based bayesian reconstruction of natural images from human brain activity. Neuroscience. 2020;444:92–105. doi: 10.1016/j.neuroscience.2020.07.040. [DOI] [PubMed] [Google Scholar]
- 41.Koike Y, et al. Feasibility of synthetic computed tomography generated with an adversarial network for multi-sequence magnetic resonance-based brain radiotherapy. J Radiat Res. 2019;61(1):92–103. doi: 10.1093/jrr/rrz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu S, Lei B, Wang S, Wang Y, Feng Z, Shen Y. Bidirectional mapping generative adversarial networks for brain MR to PET synthesis. IEEE Trans Med Imaging. 2021;41:145–157. doi: 10.1109/TMI.2021.3107013. [DOI] [PubMed] [Google Scholar]
- 43.Abu-Srhan A, Almallahi I, Abushariah MA, Mahafza W. Al-Kadi, paired-unpaired unsupervised attention guided GAN with transfer learning for bidirectional brain MR-CT synthesis. Comput Biol Med. 2021;136:104763. doi: 10.1016/j.compbiomed.2021.104763. [DOI] [PubMed] [Google Scholar]
- 44.Conte GM, et al. Generative adversarial networks to synthesize missing T1 and FLAIR MRI sequences for use in a multisequence brain tumor segmentation model. Radiology. 2021;299(2):313–323. doi: 10.1148/radiol.2021203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Rosa F, Yu T, Barquero G, Thiran JP, Granziera C, Cuadra MB. MPRAGE to MP2RAGE UNI translation via generative adversarial network improves the automatic tissue and lesion segmentation in multiple sclerosis patients. Comput Biol Med. 2021;132:104297. doi: 10.1016/j.compbiomed.2021.104297. [DOI] [PubMed] [Google Scholar]
- 46.Tang B, et al. Dosimetric evaluation of synthetic CT image generated using a neural network for MR-only brain radiotherapy. J Appl Clin Med Phys. 2021;22(3):55–62. doi: 10.1002/acm2.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu Y, Zheng Q (2021) A transfer deep generative adversarial network model to synthetic brain CT generation from MR images. Wirel Commun Mobile Comput 2021:1–10
- 48.Cheng D, Qiu N, Zhao F, Mao Y, Li C. Research on the modality transfer method of brain imaging based on generative adversarial network. Front Neurosci. 2021;15:655019. doi: 10.3389/fnins.2021.655019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei Y et al (2020) Multi-modality MRI arbitrary transformation using unified generative adversarial networks. In: Progress in biomedical optics and imaging—proceedings of SPIE, vol 11313
- 50.Chen H, Qin Z, Ding Y, Lan T (2019) Brain tumor segmentation with generative adversarial nets. In: 2019 2nd international conference on artificial intelligence and big data (ICAIBD), pp 301–305
- 51.Tokuoka Y, Suzuki S, Sugawara Y (2019) An inductive transfer learning approach using cycle-consistent adversarial domain adaptation with application to brain tumor segmentation. In: Proceedings of the 2019 6th international conference on biomedical and bioinformatics engineering, pp 44–48
- 52.Asma-Ull H, Yun ID, Han D. Data efficient segmentation of various 3d medical images using guided generative adversarial networks. IEEE Access. 2020;8:102022–102031. doi: 10.1109/ACCESS.2020.2998735. [DOI] [Google Scholar]
- 53.Yu B, Zhou L, Wang L, Fripp J, Bourgeat P (2018) 3D cGAN based cross-modality MR image synthesis for brain tumor segmentation. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), pp 626–630
- 54.Wu W, Lu Y, Mane R, Guan C (2020) Deep learning for neuroimaging segmentation with a novel data augmentation strategy. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC), pp 1516–1519 [DOI] [PubMed]
- 55.Hamghalam M, Wang T, Lei B. High tissue contrast image synthesis via multistage attention-GAN: application to segmenting brain MR scans. Neural Netw. 2020;132:43–52. doi: 10.1016/j.neunet.2020.08.014. [DOI] [PubMed] [Google Scholar]
- 56.Hamghalam M, Lei B, Wang T (2020) High tissue contrast MRI synthesis using multi-stage attention-GAN for glioma segmentation. In: AAAI—AAAI conference on artificial intelligence, pp 4067–4074
- 57.Lee H, Jo J, Lim H, Lee S (2020) Study on optimal generative network for synthesizing brain tumor-segmented MR images. Mathematical Problems in Engineering, 2020
- 58.Carver EN, Dai Z, Liang E, Snyder J, Wen N. Improvement of multiparametric MR image segmentation by augmenting the data with generative adversarial networks for glioma patients. Front Comput Neurosci. 2021;14:107. doi: 10.3389/fncom.2020.495075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kossen T, et al. Synthesizing anonymized and labeled TOF-MRA patches for brain vessel segmentation using generative adversarial networks. Comput Biol Med. 2021;131:104254. doi: 10.1016/j.compbiomed.2021.104254. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Yang X, Cheng K, Li Y, Liu Z, Shi Y (2020) Efficient 3D neural networks with support vector machine for hippocampus segmentation. In: 2020 international conference on artificial intelligence and computer engineering (ICAICE), pp 337–341
- 61.Jang J, Lee HH, Park JA, Kim H. Unsupervised anomaly detection using generative adversarial networks in 1H-MRS of the brain. J Magn Reson. 2021;325:106936. doi: 10.1016/j.jmr.2021.106936. [DOI] [PubMed] [Google Scholar]
- 62.Hamghalam M, Wang T, Qin J, Lei B (2020) Transforming intensity distribution of brain lesions via conditional gans for segmentation. In: 2020 IEEE 17th international symposium on biomedical imaging (ISBI), pp 1–4
- 63.Xi N (2019) Semi-supervised attentive mutual-info generative adversarial network for brain tumor segmentation. In: 2019 international conference on image and vision computing New Zealand (IVCNZ), pp 1–7
- 64.Thirumagal E, Saruladha K (2020) Design of FCSE-GAN for dissection of brain tumour in MRI. In: 2020 international conference on smart technologies in computing, electrical and electronics (ICSTCEE), pp 1–6
- 65.Özbey M, Çukur T (2020) T1-weighted contrast-enhanced synthesis for multi-contrast MRI segmentation. In: 28th signal processing and communications applications conference (SIU), pp 1–4. 10.1109/SIU49456.2020.9302109
- 66.Pradhan N, Dhaka VS, Rani G, et al. Transforming view of medical images using deep learning. Neural Comput Appl. 2020;32:15043–15054. doi: 10.1007/s00521-020-04857-z. [DOI] [Google Scholar]
- 67.Zhuang P, Chapman B, Li R, Koyejo S (2019) Synthetic power analyses: empirical evaluation and application to cognitive neuroimaging. In: 2019 53rd asilomar conference on signals, systems, and computers, pp 1192–1196
- 68.Das J, Patel R, Pankajakshan V (2019) Brain tumor segmentation using discriminator loss. In: 2019 National conference on communications (NCC), pp 1–6
- 69.Bernal J, Valverde S, Kushibar K, Cabezas M, Oliver A, Llado X. Generating longitudinal atrophy evaluation datasets on brain magnetic resonance images using convolutional neural networks and segmentation priors. Neuroinformatics. 2021;19(3):477–492. doi: 10.1007/s12021-020-09499-z. [DOI] [PubMed] [Google Scholar]
- 70.Tang Z, Liu X, Li Y, Yap P, Shen D. Multi-atlas brain parcellation using squeeze-and-excitation fully convolutional networks. IEEE Trans Image Process. 2020;29:6864–6872. doi: 10.1109/TIP.2020.2994445. [DOI] [Google Scholar]
- 71.Rezaei M, Yang H, Harmuth K, Meinel C (2019) Conditional generative adversarial refinement networks for unbalanced medical image semantic segmentation. In: 2019 IEEE winter conference on applications of computer vision (WACV), pp 1836–1845
- 72.Tao L, Fisher J, Anaya E, Li X, Levin CS. Pseudo CT image synthesis and bone segmentation from MR images using adversarial networks with residual blocks for mr-based attenuation correction of brain PET data. IEEE Trans Radiat Plasma Med Sci. 2021;5(2):193–201. doi: 10.1109/TRPMS.2020.2989073. [DOI] [Google Scholar]
- 73.Mahapatra D, Ge Z (2019) Training data independent image registration with gans using transfer learning and segmentation information. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019)0, pp 709–713
- 74.Hou Y, Li T, Zhang Q, Yu H, Ge H (2021) Brain tumor segmentation based on knowledge distillation and adversarial training. In: 2021 international joint conference on neural networks (IJCNN), pp 1–7
- 75.Wu X, Bi L, Fulham M, Feng DD, Zhou L, Kim J. Unsupervised brain tumor segmentation using a symmetric-driven adversarial network. Neurocomputing. 2021;455:242–254. doi: 10.1016/j.neucom.2021.05.073. [DOI] [Google Scholar]
- 76.Cheng G, Ji H, He L. Correcting and reweighting false label masks in brain tumor segmentation. Med Phys. 2021;48(1):169–177. doi: 10.1002/mp.14480. [DOI] [PubMed] [Google Scholar]
- 77.Yuan W, Wei J, Wang J, Ma Q, Tasdizen T. Unified generative adversarial networks for multimodal segmentation from unpaired 3D medical images. Med Image Anal. 2020;64:101731. doi: 10.1016/j.media.2020.101731. [DOI] [PubMed] [Google Scholar]
- 78.Liu J, Yin P, Wang X, Yang W, Cheng K (2019) Glioma subregions segmentation with a discriminative adversarial regularized 3D unet. In: ACM international conference proceeding series, pp 269–273
- 79.Liu Y, et al. A 3D fully convolutional neural network with top-down attention-guided refinement for accurate and robust automatic segmentation of amygdala and its subnuclei. Front Neurosci. 2020;14:260. doi: 10.3389/fnins.2020.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi Y, Cheng K, Liu Z. Hippocampal subfields segmentation in brain MR images using generative adversarial networks. Biomed Eng Online. 2019;18(1):5. doi: 10.1186/s12938-019-0623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tong N, Gou S, Yang S, Cao M, Sheng K. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med Phys. 2019;46(6):2669–2682. doi: 10.1002/mp.13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang J, Lu W, Zhang W. Fusion of brain PET and MRI images using tissue-aware conditional generative adversarial network with joint loss. IEEE Access. 2020;8:6368–6378. doi: 10.1109/ACCESS.2019.2963741. [DOI] [Google Scholar]
- 83.Ge C, Gu IY, Jakola AS, Yang J. Enlarged training dataset by pairwise gans for molecular-based brain tumor classification. IEEE Access. 2020;8:22560–22570. doi: 10.1109/ACCESS.2020.2969805. [DOI] [Google Scholar]
- 84.Lin W (2020) Synthesizing missing data using 3D reversible GAN for alzheimer's disease. In: Proceedings of the 2020 international symposium on artificial intelligence in medical sciences, pp 208–213
- 85.Pan Y, Liu M, Lian C, Xia Y, Shen D. Spatially-constrained fisher representation for brain disease identification with incomplete multi-modal neuroimages. IEEE Trans Med Imaging. 2020;39(9):2965–2975. doi: 10.1109/TMI.2020.2983085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen T, Song X, Wang C. Preserving-texture generative adversarial networks for fast multi-weighted MRI. IEEE Access. 2018;6:71048–71059. doi: 10.1109/ACCESS.2018.2877932. [DOI] [Google Scholar]
- 87.Yu W, Lei B, Ng MK, Cheung AC, Shen Y, Wang S (2021) Tensorizing GAN with high-order pooling for alzheimer’s disease assessment. IEEE Trans Neural Netw Learn Syst [DOI] [PubMed]
- 88.Zhou X, et al. Enhancing magnetic resonance imaging-driven Alzheimer’s disease classification performance using generative adversarial learning. Alzheimer’s Res Ther. 2021;13(1):60. doi: 10.1186/s13195-021-00797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yerukalareddy DR, Pavlovskiy E (2021) Brain tumor classification based on mr images using GAN as a pre-trained model. In: 2021 IEEE ural-siberian conference on computational technologies in cognitive science, genomics and biomedicine (CSGB), pp 380–384
- 90.Ghassemi N, Shoeibi A, Rouhani M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Signal Process Control. 2020;57:101678. doi: 10.1016/j.bspc.2019.101678. [DOI] [Google Scholar]
- 91.Budianto T, Nakai T, Imoto K, Takimoto T, Haruki K (2020) Dual-encoder bidirectional generative adversarial networks for anomaly detection. In: 2020 19th IEEE international conference on machine learning and applications (ICMLA), pp 693–700
- 92.Gao X, Shi F, Shen D, Liu M (2021) Task-induced pyramid and attention GAN for multimodal brain image imputation and classification in alzheimers disease. IEEE J Biomed Health Inform [DOI] [PubMed]
- 93.Sandhiya B, Priyatharshini R, Ramya B, Monish S, Raja GRS (2021) Reconstruction, identification and classification of brain tumor using gan and faster regional-CNN. In: 2021 3rd international conference on signal processing and communication (ICPSC), pp 238–242
- 94.Han C, et al. MADGAN: unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinform. 2021;22:31. doi: 10.1186/s12859-020-03936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin W, et al. Bidirectional mapping of brain MRI and PET with 3D reversible GAN for the diagnosis of alzheimer’s disease. Front Neurosci. 2021;15:646013. doi: 10.3389/fnins.2021.646013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McKenna MC, Murad A, Huynh W, Lope J, Bede P. Differential diagnosis of frontotemporal dementia, alzheimer’s disease, and normal aging using a multi-scale multi-type feature generative adversarial deep neural network on structural magnetic resonance images. Front Neurosci. 2020;14:853. doi: 10.3389/fnins.2020.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaur S, Aggarwal H, Rani R. Diagnosis of parkinson’s disease using deep CNN with transfer learning and data augmentation. Multimed Tools Appl. 2021;80(7):10113–10139. doi: 10.1007/s11042-020-10114-1. [DOI] [Google Scholar]
- 98.Ge C, Gu IY, Jakola AS, Yang J. Deep semi-supervised learning for brain tumor classification. BMC Med Imaging. 2020;20(1):87. doi: 10.1186/s12880-020-00485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han C et al (2019) Learning more with less: Conditional PGGAN-based data augmentation for brain metastases detection using highly-rough annotation on MR images. In: The conference on information and knowledge management, pp 119–127
- 100.Kazemifar S, et al. Dosimetric evaluation of synthetic CT generated with GANs for MRI-only proton therapy treatment planning of brain tumors. J Appl Clin Med Phys. 2020;21(5):76–86. doi: 10.1002/acm2.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rezaei M, Yang H, Meinel C (2018) Generative adversarial framework for learning multiple clinical tasks. In: 2018 digital image computing: techniques and applications (DICTA), pp 1–8
- 102.Nguyen B, Feldman A, Bethapudi S, Jennings A, Willcocks CG (2021) Unsupervised region-based anomaly detection in brain MRI with adversarial image inpainting. In: 2021 IEEE 18th international symposium on biomedical imaging (ISBI), pp 1127–1131
- 103.Datta S, Dandapat S, Deka B. A deep framework for enhancement of diagnostic information in CSMRI reconstruction. Biomed Signal Process Control. 2022;71:103117. doi: 10.1016/j.bspc.2021.103117. [DOI] [Google Scholar]
- 104.Pham C-H et al (2019) Simultaneous super-resolution and segmentation using a generative adversarial network: application to neonatal brain MRI. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), pp 991–994
- 105.Delannoy Q, et al. SegSRGAN: super-resolution and segmentation using generative adversarial networks—Application to neonatal brain MRI. Comput Biol Med. 2020;120:103755. doi: 10.1016/j.compbiomed.2020.103755. [DOI] [PubMed] [Google Scholar]
- 106.Lv J, Zhu J, Yang G. Which GAN A comparative study of generative adversarial network-based fast MRI reconstruction. Philos Trans A Math Phys Eng Sci. 2021;379(2200):20200203. doi: 10.1098/rsta.2020.0203. [DOI] [PubMed] [Google Scholar]
- 107.Bourbonne V, et al. Dosimetric validation of a GAN-based pseudo-CT generation for MRI-only stereotactic brain radiotherapy. Cancers. 2021;13(5):1082. doi: 10.3390/cancers13051082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang H, Shinomiya Y, Yoshida S. 3D MRI reconstruction based on 2D generative adversarial network super-resolution. Sensors (Basel) 2021;21(9):2978. doi: 10.3390/s21092978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu J, Tan C, Yang J, Yang G, Lio P. Arbitrary scale super-resolution for medical images. Int J Neural Syst. 2021;31(10):2150037. doi: 10.1142/S0129065721500374. [DOI] [PubMed] [Google Scholar]
- 110.Huang Y et al (2020) Super-resolution and inpainting with degraded and upgraded generative adversarial networks. vol 2021, pp 645–651
- 111.Han S, Carass A, Schär M, Calabresi PA, Prince JL (2021) Slice profile estimation from 2D MRI acquisition using generative adversarial networks. In: 2021 IEEE 18th international symposium on biomedical imaging (ISBI), pp 145–149
- 112.Gu Y, et al. MedSRGAN: medical images super-resolution using generative adversarial networks. Multimed Tools Appl. 2020;79(29):21815–21840. doi: 10.1007/s11042-020-08980-w. [DOI] [Google Scholar]
- 113.Goldfryd T,Gordon S, Raviv TR (2021) Deep semi-supervised bias field correction of Mr images. In: 2021 IEEE 18th international symposium on biomedical imaging (ISBI), pp 1836–1840
- 114.Arabi H, Zeng G, Zheng G, Zaidi H. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. Eur J Nucl Med Mol Imaging. 2019;46(13):2746–2759. doi: 10.1007/s00259-019-04380-x. [DOI] [PubMed] [Google Scholar]
- 115.Johnson PM, Drangova M. Conditional generative adversarial network for 3D rigid-body motion correction in MRI. Magn Reson Med. 2019;82(3):901–910. doi: 10.1002/mrm.27772. [DOI] [PubMed] [Google Scholar]
- 116.Ran M, et al. Denoising of 3D magnetic resonance images using a residual encoder-decoder Wasserstein generative adversarial network. Med Image Anal. 2019;55:165–180. doi: 10.1016/j.media.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 117.Armanious K, Kumar V, Abdulatif S, Hepp T, Gatidis S, Yang B (2020) ipA-MedGAN: inpainting of arbitrary regions in medical imaging. In: 2020 IEEE international conference on image processing (ICIP), pp 3005–3009
- 118.Hagiwara A. Improving the quality of synthetic FLAIR images with deep learning using a conditional generative adversarial network for pixel-by-pixel image translation. AJNR Am J Neuroradiol. 2019;40(2):224–230. doi: 10.3174/ajnr.A5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang H, Wei ZX, Zhou JQ, Tian J (2020) Reconstructing the perceived faces from brain signals without large number of training samples*. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC), pp 1108–1111 [DOI] [PubMed]
- 120.Emami H, Dong M, Glide-Hurst CK (2020) Attention-guided generative adversarial network to address atypical anatomy in synthetic CT generation. In: 2020 IEEE 21st international conference on information reuse and integration for data science (IRI), pp 188–193 [DOI] [PMC free article] [PubMed]
- 121.Wang K, Tao J, Zhu J, Ye Z, Qiu B, Xu J (2019) Compressed sensing MRI reconstruction using generative adversarial network with enhanced antagonism. In: 2019 12th international conference on intelligent computation technology and automation (ICICTA), pp 282–285
- 122.Mozafari M, Reddy L, VanRullen R (2020) reconstructing natural scenes from fMRI patterns using BigBiGAN. In: 2020 international joint conference on neural networks (IJCNN), pp 1–8
- 123.Dar SUH, Yurt M, Shahdloo M, Ildız ME, Tınaz B, Çukur T. Prior-guided image reconstruction for accelerated multi-contrast MRI via generative adversarial networks. IEEE J Sel Top Signal Process. 2020;14(6):1072–1087. doi: 10.1109/JSTSP.2020.3001737. [DOI] [Google Scholar]
- 124.Li Z, Tian Q, et al. High-fidelity fast volumetric brain MRI using synergistic wave-controlled aliasing in parallel imaging and a hybrid denoising generative adversarial network (HDnGAN) Med Phys. 2022;49(2):1000–1014. doi: 10.1002/mp.15427. [DOI] [PubMed] [Google Scholar]
- 125.Shaul R, David I, Shitrit O, Raviv TR. Subsampled brain MRI reconstruction by generative adversarial neural networks. Med Image Anal. 2020;65:101747. doi: 10.1016/j.media.2020.101747. [DOI] [PubMed] [Google Scholar]
- 126.Ren Z, et al. Reconstructing seen image from brain activity by visually-guided cognitive representation and adversarial learning. Neuroimage. 2021;228:117602. doi: 10.1016/j.neuroimage.2020.117602. [DOI] [PubMed] [Google Scholar]
- 127.Shen G, Dwivedi K, Majima K, Horikawa T, Kamitani Y. End-to-end deep image reconstruction from human brain activity. Front Comput Neurosci. 2019;13:21. doi: 10.3389/fncom.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lv J, et al. Transfer learning enhanced generative adversarial networks for multi-channel MRI reconstruction. Comput Biol Med. 2021;134:104504. doi: 10.1016/j.compbiomed.2021.104504. [DOI] [PubMed] [Google Scholar]
- 129.Gu J, Li Z, Wang Y, Yang H, Qiao Z, Yu J. Deep generative adversarial networks for thin-section infant MR image reconstruction. IEEE Access. 2019;7:68290–68304. doi: 10.1109/ACCESS.2019.2918926. [DOI] [Google Scholar]
- 130.Do WJ, Seo S, Han Y, Ye JC, Choi SH, Park SH. Reconstruction of multicontrast MR images through deep learning. Med Phys. 2020;47(3):983–997. doi: 10.1002/mp.14006. [DOI] [PubMed] [Google Scholar]
- 131.Chen Y, Jakary A, Avadiappan S, Hess CP, Lupo JM. QSMGAN: improved quantitative susceptibility mapping using 3D generative adversarial networks with increased receptive field. Neuroimage. 2020;207:116389. doi: 10.1016/j.neuroimage.2019.116389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wegmayr V, Hörold M, Buhmann JM (2019) Generative aging of brain MRI for early prediction of MCI-AD conversion. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), pp 1042–1046
- 133.Ali MB, et al. Domain mapping and deep learning from multiple MRI clinical datasets for prediction of molecular subtypes in low grade gliomas. Brain Sci. 2020;10(7):463. doi: 10.3390/brainsci10070463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bessadok A, Mahjoub MA, Rekik I. Brain multigraph prediction using topology-aware adversarial graph neural network. Med Image Anal. 2021;72:102090. doi: 10.1016/j.media.2021.102090. [DOI] [PubMed] [Google Scholar]
- 135.Ji J, Liu J, Han L, Wang F. Estimating effective connectivity by recurrent generative adversarial networks. IEEE Trans Med Imaging. 2021;40:3326–3336. doi: 10.1109/TMI.2021.3083984. [DOI] [PubMed] [Google Scholar]
- 136.Elazab A et al (2020) Glioma growth prediction via generative adversarial learning from multi-time points magnetic resonance images. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC), pp 1750–1753 [DOI] [PubMed]
- 137.Wei W, et al. Predicting PET-derived myelin content from multisequence MRI for individual longitudinal analysis in multiple sclerosis. Neuroimage. 2020;223:117308. doi: 10.1016/j.neuroimage.2020.117308. [DOI] [PubMed] [Google Scholar]
- 138.Liu J, Ji J, Xun G, Yao L, Huai M, Zhang A (2020) EC-GAN: inferring brain effective connectivity via generative adversarial networks. In: AAAI—AAAI conference on artificial intelligence, pp 4852–4859
- 139.Zhao Y, Ma B, Jiang P, Zeng D, Wang X, Li S. Prediction of alzheimer’s disease progression with multi-information generative adversarial network. IEEE J Biomed Health Inform. 2021;25(3):711–719. doi: 10.1109/JBHI.2020.3006925. [DOI] [PubMed] [Google Scholar]
- 140.Roychowdhury S, Roychowdhury S (2020) A modular framework to predict alzheimer’s disease progression using conditional generative adversarial networks. In: 2020 international joint conference on neural networks (IJCNN), pp 1–8
- 141.Rachmadi MF, C Valdés-Hernández MD, Makin S, Wardlaw JM, Komura T (2019) Predicting the evolution of white matter Hyperintensities in brain MRI using generative adversarial networks and irregularity map. In: International conference on medical image computing and computer-assisted intervention, pp 146–154
- 142.Elazab A, et al. GP-GAN: brain tumor growth prediction using stacked 3D generative adversarial networks from longitudinal MR images. Neural Netw. 2020;132:321–332. doi: 10.1016/j.neunet.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 143.Mahapatra D, Ge Z. Training data independent image registration using generative adversarial networks and domain adaptation. Pattern Recognit. 2020;100:107109. doi: 10.1016/j.patcog.2019.107109. [DOI] [Google Scholar]
- 144.Zheng Y et al (2021) SymReg-GAN: symmetric image registration with generative adversarial networks. IEEE transactions on pattern analysis and machine intelligence [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file1: Table S1 PRISMA-ScR Checklist. Table S2 Search strategy. Table S3 Interrater agreement matrices for study selection steps. Table S4 Data extraction form. Appendix S5 Characteristics of the studies.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.