Abstract
The rate and extent of bacterial Fe(III) mineral reduction are governed by molecular-scale interactions between the bacterial cell surface and the mineral surface. These interactions are poorly understood. This study examined the role of surface proteins in the adhesion of Shewanella alga BrY to hydrous ferric oxide (HFO). Enzymatic degradation of cell surface polysaccharides had no effect on cell adhesion to HFO. The proteolytic enzymes Streptomyces griseus protease and chymotrypsin inhibited the adhesion of S. alga BrY cells to HFO through catalytic degradation of surface proteins. Trypsin inhibited S. alga BrY adhesion solely through surface-coating effects. Protease and chymotrypsin also mediated desorption of adhered S. alga BrY cells from HFO while trypsin did not mediate cell desorption. Protease removed a single peptide band that represented a protein with an apparent molecular mass of 50 kDa. Chymotrypsin removed two peptide bands that represented proteins with apparent molecular masses of 60 and 31 kDa. These proteins represent putative HFO adhesion molecules. S. alga BrY adhesion was inhibited by up to 46% when cells were cultured at sub-MICs of chloramphenicol, suggesting that protein synthesis is necessary for adhesion. Proteins extracted from the surface of S. alga BrY cells inhibited adhesion to HFO by up to 41%. A number of these proteins bound specifically to HFO, suggesting that a complex system of surface proteins mediates S. alga BrY adhesion to HFO.
Dissimilatory iron-reducing bacteria (DIRB) use ferric iron as a terminal electron acceptor for anaerobic respiration and growth (13, 23, 24, 26, 29). This metabolism greatly influences the geochemistry of anaerobic soils and sediments and may also provide a mechanism for both intrinsic and engineered bioremediation of contaminated environments (25). Although ferric iron is abundant in many nonsulfidogenic anaerobic environments, the estimated solubility product constant of Fe(III) at neutral pH is 10−38, which limits the concentration of soluble Fe(III) to approximately 10−18 M (20). The rate and extent of dissimilatory Fe(III) reduction and the potential for bioremediation by DIRB are thus limited by the bioavailability of Fe(III) (22). A fundamental understanding of the interactions between DIRB and insoluble Fe(III) minerals is requisite to understanding the role of these organisms in geochemical cycling within anaerobic soils and sediments and to the effective application of this metabolism in bioremediation.
While a number of studies have suggested that DIRB cell contact with insoluble Fe(III) minerals is necessary for Fe(III) mineral reduction (1, 5, 6, 15, 21, 27, 28, 42), the mechanisms by which DIRB adhere to Fe(III) minerals are not completely understood. In one study, an Fe(III)-reducing Pseudomonas sp. that attached to and corroded steel oil pipelines was shown to colonize the surface of the steel coupons by means of exopolysaccharide excretion (30). Scanning electron micrographs of Pseudomonas sp. strain 200 growing on hematite or goethite also showed that this organism colonized the Fe(III) minerals by what appeared to be extracellular polymer (1). Although both of these studies suggested that extracellular polymers were involved in DIRB colonization of Fe(III) minerals, neither study examined the initial adhesion of the DIRB cells. A recent study that quantitatively examined the mechanisms of Shewanella alga adhesion to hydrous ferric oxide (HFO) suggested that the initial contact between DIRB and HFO was mediated by hydrophobic interactions (7).
The purpose of this study was to identify the molecules that S. alga BrY uses to adhere to HFO. The results demonstrate that surface proteins play a significant role in this adhesion process.
MATERIALS AND METHODS
Culture conditions and cell preparation.
S. alga BrY is a gram-negative rod that was isolated from anaerobic sediments of the Great Bay estuary, New Hampshire (5, 34). S. alga BrY was grown aerobically in 100 ml of tryptic soy broth (TSB; 30 g/liter; Difco Laboratories, Detroit, Mich.) at 28°C on a rotary shaker at 150 rpm for 15 h. Cells were harvested by centrifugation (5,520 × g, 4°C, 20 min) during the late exponential-early stationary growth phase. Optimal Fe(III) reductase activity is expressed at this stage of growth (14). Cells were washed once in sodium bicarbonate buffer (2.5 g/liter of distilled H2O, pH 7.0) that had been made anaerobic by boiling and cooling under a stream of O2-free gas containing 80% (vol/vol) N2 and 20% (vol/vol) CO2 (2–4). The washed cells were suspended in bicarbonate buffer. In some experiments, cells were washed with and suspended in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (20 mM, pH 7.0) that had been made anaerobic by boiling and cooling under a stream of 100% N2.
Adhesion assay.
The adhesion assay was conducted as described previously (7). The final cell concentration in adhesion assays ranged from 0.1 × 108 to 0.8 × 108 cells ml−1. All adhesion assays were performed in triplicate.
Oxidation and degradation of surface polysaccharides.
Washed S. alga BrY cells suspended in PIPES buffer (final concentration of 2.0 × 109 cells ml−1) were incubated with β-glucuronidase (Sigma Chemical Co., St. Louis, Mo.) (final concentration of 100 U ml−1) for 60 min at 50 rpm and ambient temperature. The negative control was incubated in buffer alone. Cells from each treatment were then harvested by centrifugation (5,520 × g, 4°C, 20 min), washed once in PIPES buffer, and suspended in PIPES buffer. These cells were used to perform adhesion assays as described above.
Proteolytic enzyme experiments.
Protease from Streptomyces griseus (type XI), chymotrypsin (type II), and trypsin (type III-S) were obtained from Sigma Chemical Co. The effect of these proteolytic enzymes on S. alga BrY adhesion to HFO was examined in three separate experiments. The first experiment examined the effect of cell surface proteolysis on adhesion to HFO. Washed S. alga BrY cells suspended in PIPES buffer (final concentration of 2.0 × 109 cells ml−1) were incubated with protease, chymotrypsin, and trypsin (final concentrations of 250 μg ml−1) or protease and chymotrypsin (final concentration of 250 μg ml−1 for each enzyme) for 60 min at 50 rpm and ambient temperature. The negative control was incubated in buffer alone. Cells from each treatment were then harvested by centrifugation (5,520 × g, 4°C, 20 min), washed once in PIPES buffer, and suspended in PIPES buffer. These cells were used to perform adhesion assays as described above. The second experiment examined adhesion to HFO in the presence of proteolytic enzymes. Native enzymes or enzymes boiled for 1 h were added to the tubes of adhesion buffer at final concentrations of 0, 5, 25, 50, 100, and 250 μg ml−1. These tubes were mixed and then used for adhesion assays as described above. The third experiment examined the ability of the different proteolytic enzymes to remove previously adhered cells. Cells were added to tubes of the adhesion assay buffer and incubated for 1 h at 150 rpm and ambient temperature to allow complete adhesion. The native or boiled enzymes were then added to tubes of the adhesion assay buffer (final concentration, 250 μg ml−1). The number of unadhered cells was determined in subsamples from each tube at intervals during a 1-h incubation. The Student t test was used to determine whether the adherence inhibition curves for native and heat-denatured enzyme treatments were statistically different. Differences were considered significant when P was <0.05. Cell lysis induced by proteolytic enzyme treatment was determined by incubating washed cells with protease, chymotrypsin, or trypsin (final concentration of 2.5 mg ml−1) at a cell density of 4 × 1010 to 5 × 1010 cells ml−1 for 90 min. Samples were collected for direct counts as described above. Lysis was determined by comparing counts of cells treated with proteolytic enzymes to counts of cells treated only with buffer.
Chloramphenicol experiments.
Serial twofold dilutions of chloramphenicol (3.125- to 25-μg ml−1 final concentration) in TSB were inoculated with log-phase bacteria to provide a final cell density of 107 cells ml−1 and incubated at 28°C and 150 rpm for 18 h. The lowest concentration of chloramphenicol that completely inhibited growth, as determined by measuring the optical density of each culture at 600 nm, was defined as the MIC (44).
A 15-h, exponential-growth-phase culture of S. alga BrY was used to inoculate test tubes with 5.0 ml of TSB containing chloramphenicol at final concentrations of 1/32, 1/16, and 1/8 MIC (0.195, 0.391, and 0.781 μg ml−1, respectively). Controls contained no chloramphenicol and chloramphenicol at the MIC (6.25 μg ml−1). The final cell density in each culture was 1.73 × 107 cells ml−1. The cultures were incubated at 28°C and 150 rpm for 18 h, after which the optical density at 600 nm of each culture was determined. Washed cells from each treatment were used in adhesion assays as described above.
Extraction of surface proteins.
A cation-exchange method was used to obtain the cell surface extract (12). Dowex (50 by 8, 20/50 mesh, sodium form) (18 g) that had been prewashed for 1 h in sodium bicarbonate buffer was added to 60 ml of cell suspension (optical density of 2.0 at 600 nm) and stirred on ice for 30 min at 300 rpm. Dowex beads were then removed by centrifugation (614 × g, 4°C, 15 min), and cells were removed by a subsequent centrifugation (15,300 × g, 4°C, 15 min). The resulting supernatant was centrifuged again (15,300 × g, 4°C, 30 min). The supernatant resulting from this second centrifugation was lyophilized and suspended in 3 ml of dH2O. This constituted the cell surface extract.
Aliquots of the cell surface extract were added to tubes of HFO buffer to provide final protein concentrations of 0, 4.84, 12.4, 24.2, 36.3, and 48.4 μg ml−1. A series of negative control tubes contained the same concentrations of bovine serum albumin. All tubes were incubated for 30 min at 150 rpm and ambient temperature. S. alga BrY cells were added to the tubes, which were then incubated for 15 min at 150 rpm and ambient temperature. The number of adhered and unadhered cells in each tube was determined after the incubation period, as described above.
SDS-PAGE analyses.
Aliquots (50 μl) of cell suspensions from the first proteolysis experiment were heated at 95°C for 10 min in 50 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (Bio-Rad, Hercules, Calif.). Approximately 2 × 107 cells (25 μl) were loaded into each lane of a 12.5% polyacrylamide slab gel, and SDS-PAGE was conducted according to the methods of Laemmli (19). Bio-Rad low-molecular-weight standards (Mrs, 14,400, 21,500, 31,000, 42,700, 66,200, and 97,400) were used as standards. Estimates of molecular size were made by comparing the relative mobilities of the unknown proteins to the mobilities of the proteins of known molecular mass on SDS-PAGE gels.
SDS-PAGE analysis was also used to determine the binding of proteins in the cell surface extract to either whole S. alga BrY cells or HFO. Aliquots (0.5 ml) of washed cell suspension and HFO buffer were centrifuged (16,060 × g, 5 min, and ambient temperature). The supernatants were removed, and each pellet was suspended in 0.5 ml of cell surface extract and incubated for 10 min at 150 rpm and ambient temperature. Proteins in the cell surface extract that bound to either S. alga BrY whole cells or HFO were removed by centrifugation (16,060 × g, 5 min, and ambient temperature). The resulting supernatants, containing cell surface proteins that did not bind to cells or HFO, were analyzed by SDS-PAGE as described above. A sample of the original cell surface extract was also analyzed as a reference. Each lane of the gel was loaded with 1.5 μg of protein. Protein concentrations were determined by using the Micro Protein assay (Pierce Chemical Co., Rockford, Ill.), with bovine serum albumin as a standard.
RESULTS
Degradation of surface polysaccharides.
Previous research demonstrated that the surface of S. alga cells contains carbohydrates composed of uronic acids and hexose (7). These results provided a rationale for using β-glucuronidase to examine the role of carbohydrates in adhesion to HFO. β-Glucuronidase treatment of S. alga BrY cells did not result in cell lysis. Following an adhesion assay, the number of unadhered cells treated with β-glucuronidase ([2.43 ± 0.10] × 107) was not significantly different than the number of unadhered cells that were treated only with buffer ([2.85 ± 0.36] × 107) (P = 0.179). β-Glucuronidase treatment inhibited (7.73 ± 0.29)% of the total cells used in the assay from adhering. Buffer-treated cells were inhibited by (7.0 ± 0.90)%.
Influence of proteolytic enzymes on cell adhesion.
A variety of proteolytic enzymes were used to determine if proteins mediated S. alga BrY adhesion to HFO. The effects of surface proteolysis on cell adhesion are shown in Table 1. The numbers of cells that did not adhere to HFO increased 2.6- and 2.9-fold relative to the untreated control after cells were treated with protease and chymotrypsin, respectively. Surface proteolysis with trypsin had no effect on cell adhesion relative to the untreated control. The number of unadhered cells increased 4.6-fold when cells were treated with both protease and chymotrypsin simultaneously. Although surface proteolysis with protease and/or chymotrypsin inhibited adhesion relative to the untreated control, the percentage of unadhered cells for each of these treatments was below 8% of the total number of cells used in each adhesion assay. Controls for cell lysis by each of the proteolytic enzymes indicated no significant decrease in cell concentration after 1 h of incubation with final concentrations of each enzyme that were 10 times higher than those used in this experiment (data not shown).
TABLE 1.
Effect of surface proteolysis on S. alga BrY adhesion to HFOa
| Treatment | No. of unadhered cells (107) | Unadhered cells (% total cells added) |
|---|---|---|
| No enzyme | 3.32 ± 0.52 | 1.66 ± 0.26 |
| Protease | 8.65 ± 0.70 | 4.33 ± 0.36 |
| Chymotrypsin | 9.68 ± 0.41 | 4.85 ± 0.22 |
| Trypsin | 2.89 ± 0.49 | 1.45 ± 0.25 |
| Protease and chymotrypsin | 15.4 ± 3.96 | 7.71 ± 1.50 |
Values are shown as means ± standard deviations from the means (n = 3).
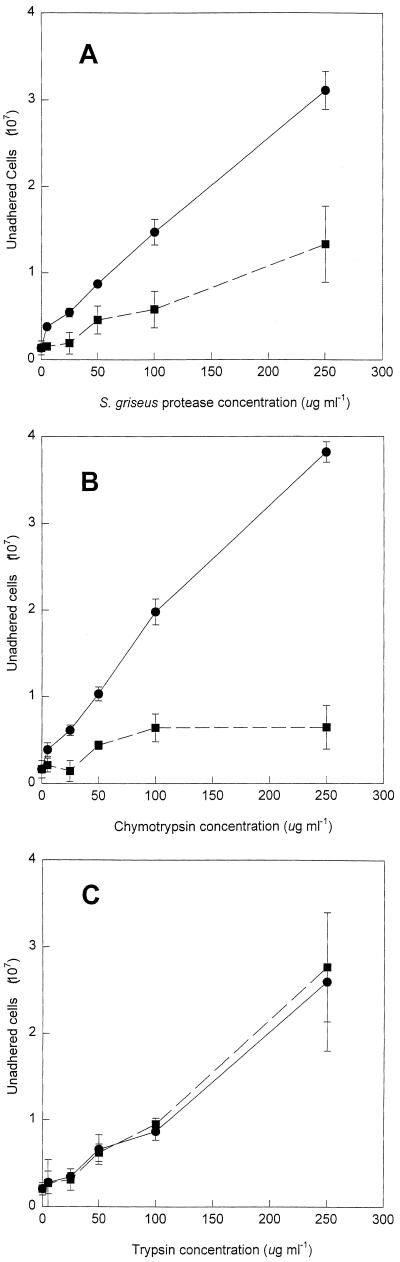
Preincubating HFO with protease from S. griseus inhibited the adhesion of S. alga BrY cells (Fig. 1A). A linear relationship between the protease concentration and the number of unadhered cells was observed (r2 = 0.996). Since proteins are known to coat surfaces and change interfacial energies (31), the effect of heat-denatured protease on S. alga BrY adhesion was also examined. A significantly greater number of cells failed to adhere in native enzyme treatments than in heat-denatured enzyme treatments (P = 0.0009). The highest concentration of native protease inhibited (3.52 ± 0.25)% of the total cells used in the assay from adhering. Chymotrypsin also inhibited S. alga BrY adhesion to HFO (Fig. 1B). A linear relationship between the chymotrypsin concentration and the number of unadhered cells was observed (r2 = 0.991). A significantly greater number of cells failed to adhere in native chymotrypsin treatments than in heat-denatured treatments (P = 0.003). The highest concentration of native chymotrypsin inhibited (4.31 ± 1.31)% of the total cells used in the assay from adhering. While trypsin inhibited S. alga adhesion and there was a linear relationship between the number of unadhered cells and the trypsin concentration (Fig. 1C, r2 = 0.982), there was not a significant difference between native trypsin treatments and heat-denatured treatments (P = 0.885).
FIG. 1.
Effect of pretreating HFO with various concentrations of native (●) or boiled (■) S. griseus protease (A), chymotrypsin (B), and trypsin (C) on S. alga BrY cell adhesion. Error bars represent the standard deviations from the means (n = 3).
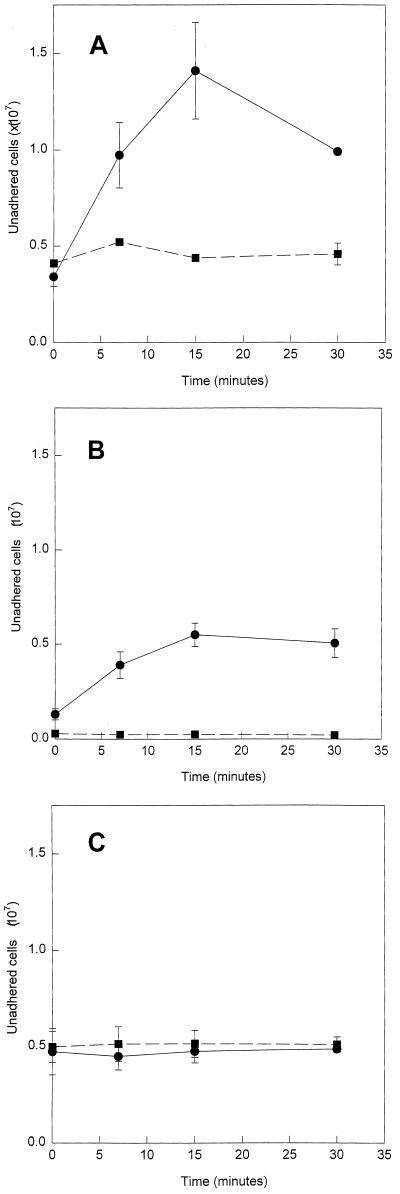
S. griseus protease removed S. alga BrY cells that were adhered to HFO (Fig. 2A). The number of unadhered cells increased with the time of exposure of the adhered cells to native protease but not heat-denatured protease. There was a significant difference between the number of unadhered cells treated with native protease and the number treated with denatured protease (P = 0.002). After 15 min of exposure to native protease, (10.2 ± 1.85)% of the total number of previously adhered cells were unadhered. Chymotrypsin also removed S. alga BrY cells that were adhered to HFO (Fig. 2B). The number of unadhered cells increased significantly with the time of exposure of the adhered cells to native chymotrypsin relative to exposure to heat-denatured chymotrypsin (P = 0.00002). After 15 min of exposure to native chymotrypsin, (3.96 ± 0.43)% of the total number of cells previously adhered were unadhered. Trypsin removed (3.49 ± 0.10)% of the total cells adhered to the HFO. However, the number of unadhered cells did not increase with increased time of exposure to native trypsin, and there was no significant difference between the number of unadhered cells in native trypsin treatment and the number in heat-denatured trypsin treatment (P = 0.1).
FIG. 2.
Kinetics of detachment of S. alga BrY cells from HFO by native (●) or boiled (■) S. griseus protease (A), chymotrypsin (B), and trypsin (C). Error bars represent the standard deviations from the means (n = 3).
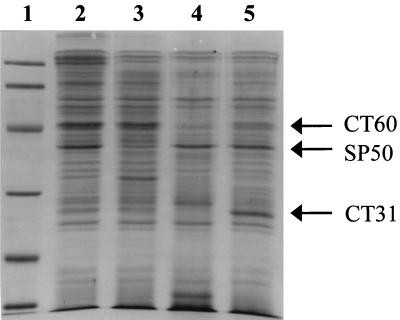
SDS-PAGE analyses of cells exposed to proteolytic enzymes showed that S. griseus protease removed a single peptide band representing a protein with an apparent molecular mass of 50 kDa (Fig. 3, lane 3; arrow labeled SP50), relative to the untreated control (Fig. 3, lane 2). Chymotrypsin treatment resulted in the loss of two peptide bands representing proteins with apparent molecular masses of 60 and 31 kDa (Fig. 3, lane 4; arrows labeled CT60 and CT31, respectively). Peptide bands that were removed by trypsin treatment (Fig. 3, lane 5) as well as bands removed by both trypsin and S. griseus protease or chymotrypsin were not considered to be involved in HFO adhesion.
FIG. 3.
Coomassie brilliant blue R-250-stained SDS-PAGE analysis showing effect of surface proteolysis on S. alga BrY cells. Lanes: 1, molecular mass standards (molecular weights, 97,400, 66,200, 45,000, 31,000, 21,500, and 14,400, top to bottom, respectively); 2, buffer-treated control; 3, cells treated with S. griseus protease; 4, cells treated with chymotrypsin; 5, cells treated with trypsin.
Influence of chloramphenicol on cell adhesion.
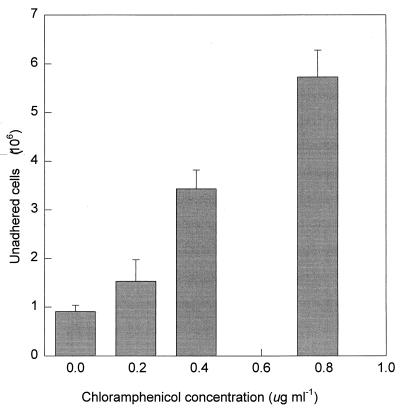
The MIC of chloramphenicol for S. alga BrY was 6.25 μg ml−1. The optical densities of 15-h cultures grown with 0, 1/8, 1/16, and 1/32 MIC of chloramphenicol were 2.47, 2.12, 2.47, and 2.46, respectively. There was no difference in cell morphology between any of these cultures, as determined by phase-contrast microscopy. Cells grown in the presence of a sub-MIC of chloramphenicol were inhibited in their ability to adhere to HFO (Fig. 4). A linear relationship between the concentration of chloramphenicol and the number of unadhered cells existed (r2 = 0.98). The highest sub-MIC of chloramphenicol inhibited (46 ± 5)% of the total cells used in the assay from adhering.
FIG. 4.
HFO adhesion of S. alga BrY cells grown in the presence of various sub-MICs of chloramphenicol. Error bars represent the standard deviations from the means (n = 3).
Influence of cell surface proteins on adhesion.
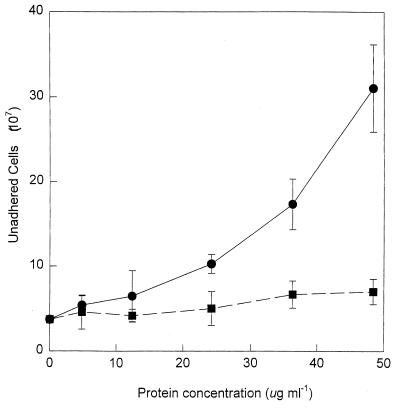
Proteins extracted from the surface of S. alga BrY cells inhibited the adhesion of cells to HFO (Fig. 5). A linear relationship between the amount of cell surface extract added to HFO and the number of unadhered cells existed (r2 = 0.90). The highest concentration of cell surface extract inhibited (41 ± 7)% of the total cells used in the assay from adhering.
FIG. 5.
The influence of preincubating HFO with various concentrations of cell surface extract (●) or bovine serum albumin (■) on S. alga BrY cell adhesion. Error bars represent the standard deviations from the means (n = 3).
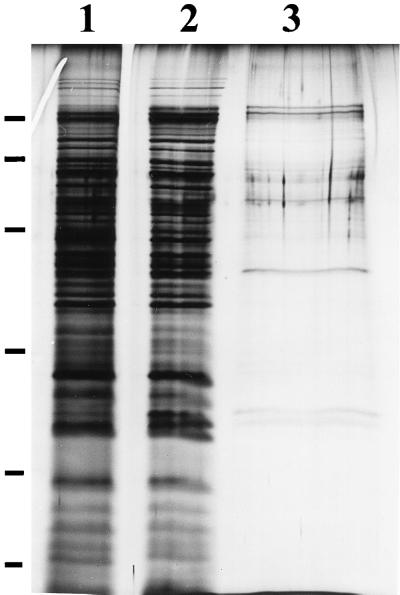
Inhibition by the cell surface extract could have been due to proteins binding to either S. alga BrY cells or HFO in the experiments described above. The proteins in the cell surface extract that did not bind to cells or HFO were thus examined by SDS-PAGE. None of the approximately 38 proteins found in the cell surface extract (Fig. 6, lane 1) bound to S. alga cells (Fig. 6, lane 2). However, all but approximately 10 of the proteins in the cell surface extract bound to HFO (Fig. 6, lane 3). Interaction of the extract proteins with HFO typically caused a widening of the SDS-PAGE lane containing those proteins (Fig. 6, lane 3). This resulted in a decrease in the band intensities within this lane after silver staining.
FIG. 6.
Silver-stained SDS-PAGE analysis of S. alga BrY cell surface extract proteins (lane 1), cell surface extract proteins that did not bind to whole cells (lane 2), and cell surface extract proteins that did not bind to HFO (lane 3). The bars on the left represent molecular mass standards (molecular weights, 97,400, 66,200, 45,000, 31,000, 21,500, and 14,400, top to bottom, respectively).
DISCUSSION
Although cell adhesion is assumed to be a crucial step in the enzymatic reduction of insoluble ferric iron minerals, the mechanisms of adhesion are still obscure. This study presents four independent lines of evidence that indicate that proteins mediate the adhesion of the DIRB S. alga BrY to HFO. Cell surface polysaccharide degradation had no effect on cell adhesion. Three separate experiments demonstrated the inhibitory effect of proteolytic enzymes on cell adhesion to HFO. Cells grown in the presence of sub-MICs of an antibiotic that inhibits protein synthesis showed a decreased ability to adhere to HFO. Finally, proteins extracted from the cell surface bound specifically to HFO and competitively inhibited cell adhesion.
Previous work in this laboratory provided evidence against polysaccharide-mediated adhesion of S. alga BrY to HFO (7). A mutant that overproduced exopolysaccharide showed a reduced ability to adhere to HFO relative to the wild-type S. alga strain BrY. It was postulated that this exopolysaccharide sterically hindered the interaction of hydrophobic adhesive proteins with HFO, resulting in weak adhesion of the mutant strain. The experiment using β-glucuronidase to examine the effect of enzymatic cell surface polysaccharide degradation on S. alga BrY adhesion to HFO supported these previous findings and suggested that the initial adhesion of S. alga BrY to HFO is independent of cell surface polysaccharide.
A number of studies have employed proteolytic enzymes to examine the role of proteins in bacterial adhesion to a variety of surfaces (8, 9, 11, 16, 17, 33, 38, 39, 41, 43). The surface proteolysis experiments in this study demonstrated that preincubating S. alga BrY cells with S. griseus protease and chymotrypsin, but not trypsin, inhibited adhesion. SDS-PAGE analysis of these cells demonstrated that proteolytic digestion with protease removed a single 50-kDa protein (SP50) from the S. alga BrY cell surface, while digestion with chymotrypsin removed proteins of 60 kDa (CT60) and 31 kDa (CT31). The inhibitory effects of protease and chymotrypsin on adhesion and the digestion of specific cell surface proteins by these enzymes suggest that SP50, CT60, and CT31 represent putative HFO adhesion molecules.
Preincubation of HFO with each proteolytic enzyme also supported the role of these proteins in HFO adhesion. The significant difference in adhesion between native and heat-denatured S. griseus protease and chymotrypsin suggested that the primary effect of these enzymes on adhesion was catalytic degradation of adhesion proteins on the cell surface rather than surface-coating effects. Preincubation of HFO with trypsin, on the other hand, resulted in inhibition solely from surface-coating effects, since there was no significant difference between native and heat-denatured trypsin treatments. Similar surface-coating effects have been observed in a study examining the adhesion of Vibrio proteolytica to hydrophilic and hydrophobic surfaces (31).
S. alga BrY cells display a high affinity for HFO, as nearly 100% of the cells adhere immediately during HFO adhesion assays and remain adhered indefinitely despite constant physical agitation (7). Given the strong affinity of these cells for HFO, the ability of the proteolytic enzymes S. griseus protease and chymotrypsin to remove cells from HFO substantiates the role of SP50, CT60, and CT31 in S. alga BrY adhesion to HFO.
Although the results of three separate proteolysis experiments each supported the role of SP50, CT60, and CT31 in S. alga BrY adhesion to HFO, proteolysis was not particularly efficient at inhibiting cell adhesion. Previous studies have shown that the efficiency of proteolytic enzymes in inhibiting protein-mediated bacterial adhesion to different surfaces is varied (8, 9, 16, 32, 33, 38). Proteolytic experiments with S. alga BrY typically inhibited cell adhesion to HFO by less than 10%. A series of experiments were performed to explore the possibility that adhesion is mediated by a complement of proteins including but not limited to the SP50, CT60, and CT31 putative adhesion molecules.
Concentrations of antibiotics below the MIC may induce changes in the surface properties of bacteria, and a number of studies have investigated the influence of sub-MICs of antibiotics on bacterial adhesion (18, 40, 44). Chloramphenicol is a peptidal transferase inhibitor that binds to the 50S ribosomal subunit (31) and inhibits protein-mediated adhesion in a number of bacterial strains (10, 31, 35–37). Inhibition of S. alga BrY protein synthesis resulted in adhesion inhibition of a much larger proportion (46%) of S. alga cells than in the proteolytic enzyme digestion experiments. These results support the hypothesis that an array of proteins may play a role in S. alga BrY adhesion to HFO.
Inhibition of adhesion by preincubating HFO with the S. alga BrY cell surface extract suggested that one or more HFO binding proteins associated with the cell surface were involved in adhesion. The inability of the control, bovine serum albumin, to inhibit adhesion suggested that proteins in the cell surface extract specifically and competitively inhibited adhesion by up to 41%. SDS-PAGE experiments demonstrated that 28 of the 38 proteins in the extract bound specifically to HFO. The cell surface extract included peptide bands with apparent molecular masses of 50, 60, and 31 kDa. Both the 60- and 31-kDa peptides bound to HFO, while the 50-kDa peptide did not.
The results of this study suggest that cell adhesion to HFO is a complex process. This study has identified a proteinaceous, HFO-adhesion system composed of multiple proteins, including but not limited to SP50, CT60, and CT31, that serve both iron binding and as yet unidentified functions.
ACKNOWLEDGMENTS
This research was partly supported by a summer faculty fellowship from the Graduate School of the University of New Hampshire.
I thank J. D. Coates for critically reviewing the manuscript.
REFERENCES
- 1.Arnold R G, DiChristina T J, Hoffman M R. Reductive dissolution of Fe(III) oxides by Pseudomonas sp. 200. Biotechnol Bioeng. 1988;32:1081–1096. doi: 10.1002/bit.260320902. [DOI] [PubMed] [Google Scholar]
- 2.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balch W E, Wolfe R S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant M P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 5.Caccavo F, Jr, Blakemore R P, Lovley D R. A hydrogen-oxidizing, Fe(III)-reducing microorganism from the Great Bay estuary, New Hampshire. Appl Environ Microbiol. 1992;58:3211–3216. doi: 10.1128/aem.58.10.3211-3216.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caccavo F, Jr, Frølund B, Kloeke F V, Nielsen P H. Deflocculation of activated sludge by the dissimilatory Fe(III)-reducing bacterium Shewanella alga BrY. Appl Environ Microbiol. 1996;62:1487–1490. doi: 10.1128/aem.62.4.1487-1490.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caccavo F, Jr, Schamberger P C, Keiding K, Nielsen P H. Role of hydrophobicity in adhesion of the dissimilatory Fe(III)-reducing bacterium Shewanella alga to amorphous Fe(III) oxide. Appl Environ Microbiol. 1997;63:3837–3843. doi: 10.1128/aem.63.10.3837-3843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coconnier M, Klaenhammer T R, Kernes S, Bernet M, Servin A L. Protein-mediated adhesion of Lactobacillus acidophilus BG2FO4 on human enterocyte and mucus-secreting cell lines in culture. Appl Environ Microbiol. 1992;58:2034–2039. doi: 10.1128/aem.58.6.2034-2039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway P L, Kjelleberg S. Protein-mediated adhesion of Lactobacillus fermentum strain 737 to mouse stomach squamous epithelium. J Gen Microbiol. 1989;135:1175–1186. doi: 10.1099/00221287-135-5-1175. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher M. The question of passive versus active attachment mechanisms in non-specific bacterial adhesion. In: Berkeley R C W, Lynch J M, Melling J, Rutter P R, Vincent B, editors. Microbial adhesion to surfaces. Chichester, England: Ellis Horwood, Publisher, Ltd.; 1980. pp. 197–210. [Google Scholar]
- 11.Flint S H, Brooks J D, Bremer P J. The influence of cell surface properties of thermophilic streptococci on attachment to stainless steel. J Appl Microbiol. 1997;83:508–517. doi: 10.1046/j.1365-2672.1997.00264.x. [DOI] [PubMed] [Google Scholar]
- 12.Frolund B, Palmgren R, Keiding K, Nielsen P H. Extraction of extracellular polymers from activated sludge using a cation exchange resin. Water Res. 1996;30:1749–1758. [Google Scholar]
- 13.Ghiorse W C. Microbial reduction of manganese and iron. In: Zehnder J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons, Inc.; 1988. pp. 305–331. [Google Scholar]
- 14.Gorby Y A, Bolton H., Jr . Abstracts of the 93rd General Meeting of the American Society for Microbiology 1993. Washington, D.C.: American Society for Microbiology; 1993. Effects of O2 on metal-reductase activity and cytochrome content in a facultative Fe(III)-reducing bacterium, abstr. Q-124; p. 37. [Google Scholar]
- 15.Grantham M C, Dove P M, DiChristina T J. Microbially catalyzed dissolution of iron and aluminium oxyhydroxide mineral surface coatings. Geochim Cosmochim Acta. 1997;61:4467–4477. [Google Scholar]
- 16.Greene J D, Klaenhammer T R. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl Environ Microbiol. 1994;60:4487–4494. doi: 10.1128/aem.60.12.4487-4494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogt A H, Dankert J, Hulstaert C E, Feijen J. Cell surface characteristics of coagulase-negative staphylococci and their adherence to fluorinated poly(ethylenepropylene) Infect Immun. 1986;51:294–301. doi: 10.1128/iai.51.1.294-301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klainer A S, Perkins R L. Surface manifestations of antibiotic-induced alterations in protein synthesis in bacterial cells. Antimicrob Agents Chemother. 1972;1:164–170. doi: 10.1128/aac.1.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lindsay W L, Schwab A P. The chemistry of iron in soils and its availability to plants. J Plant Nutr. 1982;5:821–840. [Google Scholar]
- 21.Little B J, Wagner P A, Lewandowski Z. The role of biomineralization in microbially influenced corrosion. In: Banfield J F, Nealson K H, editors. Geomicrobiology: interactions between microbes and minerals. Washington, D.C.: Mineralogical Society of America; 1997. pp. 123–159. [Google Scholar]
- 22.Lovley D R. Organic matter mineralization with the reduction of ferric iron: a review. Geomicrobiol J. 1987;5:375–399. [Google Scholar]
- 23.Lovley D R. Dissimilatory Fe(III) and Mn(VI) reduction. Microbiol Rev. 1991;55:259–287. doi: 10.1128/mr.55.2.259-287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley D R. Dissimilatory metal reduction. Annu Rev Microbiol. 1993;47:263–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 25.Lovley D R. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J Ind Microbiol. 1995;14:85–93. doi: 10.1007/BF01569889. [DOI] [PubMed] [Google Scholar]
- 26.Lovley D R. Microbial reduction of iron, manganese, and other metals. Adv Agron. 1995;54:176–217. [Google Scholar]
- 27.Lovley D R, Phillips E J P. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurice P, Forsythe J, Hersman L, Sposito G. Application of atomic-force microscopy to studies of microbial interactions with hydrous Fe(III)-oxides. Chem Geol. 1996;132:33–43. [Google Scholar]
- 29.Nealson K H, Saffarini D. Iron and manganese in anaerobic respiration: environmental significance, physiology and regulation. Annu Rev Microbiol. 1994;48:311–343. doi: 10.1146/annurev.mi.48.100194.001523. [DOI] [PubMed] [Google Scholar]
- 30.Obuekwe C O, Westlake D W S, Cook F D, Costerton J W. Surface changes in mild steel coupons from the action of corrosion-causing bacteria. Appl Environ Microbiol. 1981;41:766–774. doi: 10.1128/aem.41.3.766-774.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul J H. Effects of antimetabolites on the adhesion of an estuarine Vibrio sp. to polystyrene. Appl Environ Microbiol. 1984;48:924–929. doi: 10.1128/aem.48.5.924-929.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul J H, Jeffrey W H. Evidence for separate adhesion mechanisms for hydrophilic and hydrophobic surfaces in Vibrio proteolytica. Appl Environ Microbiol. 1985;50:431–437. doi: 10.1128/aem.50.2.431-437.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruzzo C, Crippa A, Bertone S, Pane L, Carli A. Attachment of Vibrio alginolyticus to chitin mediated by chitin-binding proteins. Microbiology. 1996;142:2181–2186. doi: 10.1099/13500872-142-8-2181. [DOI] [PubMed] [Google Scholar]
- 34.Rossello-Mora R, Caccavo F, Jr, Springer N, Spring S, Osterlehner K, Shuler D, Ludwig W, Amann R, Schleifer K H. Isolation and taxonomic characterization of a halotolerant facultatively iron-reducing bacterium. Syst Appl Microbiol. 1994;17:569–573. [Google Scholar]
- 35.Sandberg T, Stenqvist K, Svanborg-Eden C. Effects of subminimal inhibitory concentrations of ampicillin, chloramphenicol, and nitrofurantoin on the attachment of Escherichia coli to human uroepithelial cells in vitro. Rev Infect Dis. 1979;1:838–844. doi: 10.1093/clinids/1.5.838. [DOI] [PubMed] [Google Scholar]
- 36.Scaglione F, Demartini G, Dugnani S, Ferrara F, Maccarinelli G, Cocuzza C, Fraschini F. Effect of antibiotics on Bordetella pertussis adhering activity: hypothesis regarding mechanism of action. Chemotherapy. 1994;40:215–220. doi: 10.1159/000239195. [DOI] [PubMed] [Google Scholar]
- 37.Scheld W M, Zak O, Vosbeck K, Sande M A. Effect of subinhibitory antibiotic concentrations on streptococcal adhesion in vitro and the development of endocarditis in rabbits. J Clin Investig. 1981;68:1381–1384. doi: 10.1172/JCI110388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit G, Kijne J W, Lugtenberg B J J. Roles of flagella, lipopolysaccharide, and a Ca2+-dependent cell surface protein in attachment of Rhizobium leguminosarum biovar viciae to pea root hair tips. J Bacteriol. 1989;171:569–572. doi: 10.1128/jb.171.1.569-572.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit G J, Logman T J J, Boerrigter M E T I, Kijne J W, Lugtenberg B J J. Purification and partial characterization of the Rhizobium leguminosarum biovar viciae Ca2+-dependent adhesin, which mediates the first step in attachment of cells of the family Rhizobiaceae to plant root hair tips. J Bacteriol. 1989;171:4054–4062. doi: 10.1128/jb.171.7.4054-4062.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sud I J, Feingold D S. Detection of agents that alter the bacterial cell surface. Antimicrob Agents Chemother. 1975;8:34–37. doi: 10.1128/aac.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timmerman C P, Fleer A, Besnier J M, De Graaf L, Cremers F, Verhoef J. Characterization of a proteinaceous adhesin of Staphylococcus epidermidis which mediates attachment to polystyrene. Infect Immun. 1991;59:4187–4192. doi: 10.1128/iai.59.11.4187-4192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tugel J B, Hines M E, Jones G E. Microbial iron reduction by enrichment cultures isolated from estuarine sediments. Appl Environ Microbiol. 1986;52:1167–1172. doi: 10.1128/aem.52.5.1167-1172.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veenstra G J C, Cremers F F M, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vosbeck K, Handschin H, Menge E. Effects of subminimal inhibitory concentrations of antibiotics on adhesiveness of Escherichia coli in vitro. Rev Infect Dis. 1979;1:845–851. doi: 10.1093/clinids/1.5.845. [DOI] [PubMed] [Google Scholar]