SUMMARY
The influence of an immunosuppressive cytokine, interleukin-10 (IL-10), on the outcome of hepatitis C virus (HCV) infection has been increasingly reported recently. A number of polymorphisms appear to control the level of IL-10 production. Among them, −592C/A, −819C/T and −1082G/A in the IL-10 gene are three most studied single nucleotide polymorphisms. To provide a more definitive conclusion about their association with the risk of HCV infection, a meta-analysis was performed by combining and summarizing a total of 17 studies. A biological justification for the choice of genetic model was provided. The results indicated no significant association between these IL-10 polymorphisms and the susceptibility to HCV infection [–592C/A: odds ratio (OR) 0·99, 95% confidence interval (CI) 0·78–1·25; –819C/T: OR 0·90, 95% CI 0·69–1·18; –1082G/A: OR 1·34, 95% CI 0·90–2·00]. However, this analysis did not account for the possible risk modifications by other factors, such as ethnicity and virus persistence. Therefore, the effects of ethnicity and virus persistence were investigated using Bayesian meta-regression and subgroup analysis. Finally, an extended case-control association study was conducted in a Chinese population involving 1140 subjects. Both serum level and genotype data of IL-10 −1082G/A were determined. As a result, a low prevalence of G allele was observed. Significantly higher IL-10 production was observed in HCV patients, especially patients with the GG genotype.
Key words: Association study, HCV, IL-10, meta-analysis, polymorphism
INTRODUCTION
Hepatitis C virus (HCV) is a leading cause of liver disease worldwide [1]. About 80% of infected patients fail to clear the virus and develop chronic infection [2]. Most of these patients may further progress to liver cirrhosis and eventually hepatocellular carcinoma (HCC). To date, HCV infection has been attributed to several molecular mechanisms concerning transcription, translation and secretion pathways [3, 4]. Most of these proposed mechanisms have implicated that cytokines may play an important role in the regulation of immune responses against viral infection [5–7].
Interleukin-10 (IL-10) is one of the cytokines involved in HCV infection. It is located on chromosome 1 and encodes five exons (5·1 kb) [8]. The IL-10 level differs widely between individuals probably due to its polymorphisms in the promoter region of the IL-10 gene, which includes three frequent point mutations −592C/A (rs1800872), −819C/T (rs1800871) and −1082G/A (rs1800896) [9]. Although −592C, −819C and −1082G alleles have been previously associated with higher IL-10 production [10], the population-based studies [11] investigating the association between IL-10 polymorphisms and risk of HCV infection have yielded conflicting results [12–25]. Some of the studies show a significant association [17, 18, 20], while others do not [6, 14]. In the presence of these inconsistent results, a new meta-analysis, conducted by compiling all available data and organizing them into a coherent summary, may provide a more definitive conclusion about the association between these IL-10 polymorphisms and the susceptibility to HCV.
The fact that some studies did not find the effect of IL-10 polymorphisms on the risk of HCV infection also implies an involvement of other risk factors. These factors might modify the outcome of meta-analysis. For example, ethnic difference in the examined populations could lead to a differential distribution of IL-10 promoter genotypes, resulting in a different response to HCV infection [26]. This type of heterogeneity can be explored by the inclusion of covariates at study level [27], such as meta-regression and subgroup analysis [28]. In subgroup analysis, the studies are stratified into homogeneous subgroups and meta-analysis is then conducted in each subgroup. Compared to subgroup analysis, meta-regression can accommodate study-level factors directly [29]. The study is the unit of analysis and the outcomes are effect size [i.e. odds ratios (ORs)]. Hence the effect size outcomes are correlated with risk factors measured at study level.
This report aims to determine the association of IL-10 − 592C/A, −819C/T and −1082G/A polymorphisms with the risk of HCV infection. The possible risk modifications by ethnicity and HCV persistence were investigated using both Bayesian meta-regression and subgroup analysis. Finally, an extended case-control study was conducted in a Chinese population involving 1140 subjects for the further assessment of the association evaluated here.
METHODS
Literature search
The systematic survey of eligible studies for meta-analysis was performed using electronic database engines, including PubMed, the Cochrane Database for Systematic Reviews and EMBASE. The language of the reviewed articles was limited to English. The key words used for search were ‘hepatitis C’ or ‘HCV’, ‘interleukin-10’ or ‘IL-10’, ‘polymorphism’ and combinations thereof. The abstract of each citation was screened in order to select studies that explicitly focused on the association of IL-10 −592C/A, −819C/T and −1082G/A polymorphisms with the risk of HCV infection. Articles satisfying these criteria were further evaluated by two independent reviewers to determine if the inclusion criteria, listed below, were met. If so, the full article text was obtained. In addition, each reference cited in the obtained articles was reviewed in order to identify additional work not indexed by the database engines provided above. Efforts were also made to contact authors for missing data.
Inclusion/exclusion criteria
Inclusion criteria were (1) an original article, (2) a population-based association study of IL-10 polymorphisms at positions −592, −819 and −1082 with HCV infection being the outcome, and (3) a human study. Studies were excluded if they: (1) contained overlapped data, (2) were an animal study, (3) were without healthy controls, or (4) contained incomplete data and failed to respond to a data request. If more than one paper with the same data was found, the one that contained original data was reviewed.
Statistical analysis
Relevant data, including details of genotypes and frequencies of IL-10 at three positions −592, −819 and −1082, were first extracted. The effect size for each study was the OR. As both dominant and recessive modes have been employed in the selected studies, a biological justification for the choice of genetic model was required [30]. The procedure to capture the inheritance mode in association studies has been described previously [12, 28]. Using −592C/A polymorphism as an example here, λ (the ratio of log ORCA and log ORCC) was determined; the pairwise estimates of the OR of CA vs. AA (ORCA) and the OR of CC vs. AA (ORCC) were performed by conducting two separate meta-analyses [30]. Values of λ equal to 0 and 1 correspond to the recessive and dominant genetic model, respectively. Afterwards, the OR in the full meta-analysis was estimated using the selected inheritance mode.
A χ2 test was preformed to determine if observed frequencies of genotypes conformed to Hardy–Weinberg (HW) expectations [31]. Studies that deviated from the HW equilibrium were excluded from meta-analysis. The heterogeneity of the group of ORs was assessed using H statistics and the I2 metric, which suggest the presence of heterogeneity when H ⩾ 1·2 and I2 ⩾ 25% [32]. Publication bias was assessed using a funnel plot. Cumulative meta-analysis was also performed to examine whether the magnitude of effect changed markedly with sample size. Then the ORs were pooled, and 95% confidence intervals (CIs) were calculated using meta-analysis [33, 34]. In the meta-analysis, the Mantel–Haenszel and DerSimonian–Laird methods were employed in fixed-effects and random-effects models, respectively. Furthermore, sensitivity analyses were conducted to evaluate the robustness of our results. We removed each study individually to evaluate its effect on the pooled OR [35]. All parameters of the model were estimated by the ‘metan’ package within Stata software v. 10.1 [36]. The significance of the pooled OR was determined by Z test, and a 95% CI was calculated.
Bayesian meta-regression was performed to determine the impact of ethnicity on the association of IL-10 −592C/A, −819C/T, −1082G/A polymorphisms with the risk of HCV infection [37]. The procedure has been described previously [28]. Briefly, log ORs of selected studies were regressed on an intercept and covariates (i.e. ethnicity). The significance of effect was identified by the value and CI of the corresponding regression coefficient (βethnicity). The regression process was performed using Markov Chain Monte Carlo (MCMC) and Gibbs sampling algorithms through Bayesian computation software WinBUGS. The prior distribution for standard deviation σ was given as a uniform distribution with a mean of 0 and a variance of 100 to reflect the non-informative situation of effect size. Ten thousand iterations were dismissed as burn-in, and the subsequent 100 000 iterations were used for parameter estimation. Satisfactory convergence of the simulated Markov chains to the target posterior distributions was assessed using visual inspection of posterior traces and running chains with different initial values of the model parameters. Next, subgroup meta-analysis was performed by stratifying the existing studies into subgroups according to ethnicity and virus persistence, and conducting quantitative pooling separately for each subgroup.
An association study in a Chinese population involving 1140 subjects
New subjects
This study was approved by the institutional review board of Nanjing Medical University, Nanjing, China. Chronic HCV patients (n = 586) were consecutively recruited between January 2007 and March 2011 at the First Affiliated Hospital of Nanjing Medical University, Nanjing, China [328 men, 258 women; mean age (range) 47·9 (15–75) years]. They were biologically unrelated ethnic Han Chinese from Nanjing City and surrounding regions in Jiangsu Province, China. Diagnosis in all patients was confirmed by both biochemical and molecular assays, including detection of anti-HCV antibody using a third-generation commercial enzyme immunoassay (Chiron Corporation, USA) and detection of HCV RNA in serum using an Amplicor HCV assay (Roche Diagnostics, Germany). Patients presenting with other causes of chronic hepatitis (i.e. alcoholic or autoimmune hepatitis), testing positive for other hepatotropic viral antigens, or treated with antiviral drugs before liver biopsy or echography were excluded. A total of 554 healthy control subjects tested negative for HCV infection were selected randomly from blood donors from the same region during the same time period as the cases were recruited [302 men, 252 women; mean age (range) 51·7 (16–79) years]. Informed consent was obtained from case and control subjects.
DNA extraction and genotyping
Peripheral blood (5 ml) was collected in EDTA tubes (BD Biosciences, Germany). Genomic DNA was extracted and purified from whole-blood lymphocytes using a blood DNA kit (Qiagen, USA) according to the manufacturer's instructions. Genotyping of IL-10 polymorphism at position –1082 was performed by an allele-specific PCR (AS-PCR) assay. The PCR primer sequences were as follows: (i) primer A (sense): 5′-ACTACTAAGGCTTCTTTGGGAA-3′; (ii) primer G (sense): 5′-CTACTAAGGCTTCTTTGGGAG-3′ and (iii) generic primer (antisense): 5′-CAGTGCCAACTGAGAATTTGG-3′ [38]. The PCR was performed in a 20-μl volume of reaction mixture containing 10× PCR buffer (500 mm KCl, 100 mm Tris–HCl, 0·8% Nonidet P40; Fermentas, USA), 2 mm MgCl2, 0·8 mm dNTPs, 0·5 μm specific primer, 50 ng template DNA and 0·5 U Taq polymerase (Fermentas). The PCR cycle conditions were 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 68 °C for 60 s with a final extension step at 72 °C for 10 min. The amplified AS-PCR products were identified by gel electrophoresis on 2% agarose gels stained with ethidium bromide and visualized with ultraviolet light.
Measurement of serum IL-10
Serum samples were subjected to an enzyme-linked immunosorbent assay (ELISA). The assay was performed in the First Affiliated Hospital of Nanjing Medical University using an IL-10 ELISA kit (R&D systems, USA) on a model 680 Microplate Reader (Bio-Rad, USA).
Statistical analysis
The association between IL-10 –1082G/A polymorphism and susceptibility to HCV in this Chinese population was analysed using χ2 test. ORs and the corresponding 95% CIs were calculated. Student's t test was used to test differences in the means of IL-10 serum levels between groups.
RESULTS
Characteristics of studies
Seventeen case-control studies met the inclusion criteria, of which 14 studies/subgroups reported data for polymorphism meta-analysis. Three studies were excluded because they lacked either a control group or genotype data [24, 26, 39]. All the selected studies presented original data on independent samples. Of these, 10, 9 and 14 studies reported data concerning IL-10 polymorphisms of −592C/A, −819C/T and −1082G/A, respectively. The numbers of patients and controls are listed in Table 1.
Table 1.
Characteristics of case-control studies included in the full meta-analysis
| Study (first-named author) | Country | −592 | −819 | −1082 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype for cases | Genotype for controls | Genotype for cases | Genotype for controls | Genotype for cases | Genotype for controls | ||||||||
| CC + CA | AA | CC + CA | AA | CC + CT | TT | CC + CT | TT | GG | GG + AA | GG | GA + AA | ||
| Afzal [23] | Pakistan | – | – | – | – | 73 | 16 | 84 | 15 | 15 | 74 | 3* | 96 |
| Dogra [25] | India | – | – | – | – | – | – | – | – | 38 | 32 | 42* | 28 |
| Gao [21] | China | 35 | 20 | 40 | 35 | – | – | – | – | 2 | 53 | 1* | 73 |
| Chuang [12] | Taiwan | 70 | 73 | 69 | 65 | 71 | 72 | 69 | 65 | 0 | 143 | 0* | 133 |
| Bouzgarrou [13] | Tunisia | – | – | – | – | – | – | – | – | 19 | 81 | 12 | 91 |
| Abbas [14] | Pakistan | 87 | 12 | 57 | 5 | 87 | 12 | 57 | 5 | 17 | 82 | 9 | 53 |
| Pereira [6] | Brazil | 110 | 18 | 84 | 10 | 110 | 18 | 84 | 10 | 17 | 111 | 13 | 81 |
| Chen [15] | Taiwan | 36 | 36 | 87 | 93 | 36 | 36 | 86 | 94 | 0 | 72 | 0* | 180 |
| Falleti [22] | Italy | 21 | 29 | 35 | 61 | – | – | – | – | 8 | 42 | 25 | 71 |
| Persico [16] | Ireland | 218 | 10 | 104 | 6 | 218 | 10 | 104 | 6 | 74 | 154 | 18 | 92 |
| Mangia [20] | Italy | 246 | 24 | 136 | 9 | 246 | 24 | 136 | 9 | 40 | 230 | 23 | 122 |
| Zein [19] | USA | 29 | 1 | 33 | 3 | 29 | 1 | 33 | 3 | 12 | 18 | 6 | 30 |
| Egypt | 20 | 2 | 41 | 3 | 20 | 2 | 41 | 3 | 5 | 17 | 14 | 30 | |
| Lio [18] | Italy | – | – | – | – | – | – | – | – | 27 | 33 | 34* | 101 |
| Vidigal [17] | Ireland | 69 | 9 | 33 | 3 | 69 | 9 | 33 | 3 | 27 | 51 | 6 | 30 |
The distribution of genotypes in controls was not consistent with Hardy–Weinberg equilibrium law (P < 0·0001).
Six studies were excluded from the meta-analysis assessing the association between IL-10 −1082G/A polymorphism and the susceptibility to HCV because the observed distribution of genotypes in controls was inconsistent with HW equilibrium [12, 15, 18, 21, 23, 25]. All control samples of −592C/A and −819C/T polymorphisms were in HW proportions. The results of homogeneity analysis suggested no statistically significant heterogeneity across the studies of IL-10 polymorphisms at positions −592 (H = 1·00, I2 = 0%, P = 0·758) and −819 (H = 1·00, I2 = 0%, P = 0·932), whereas a high degree of heterogeneity was found in −1082G/A polymorphism (H = 1·56, I2 = 53·3%, P = 0·029). In addition, no clear evidence for publication bias was observed in the inversed funnel plots of the polymorphisms evaluated here. The resulting P values were 0·19, 0·53 and 0·70, respectively. The cumulative meta-analyses did not show that sample size had a significant impact on the pooled ORs (see Supplementary Fig. S1). Sensitivity analyses excluding one study at a time for studies on the association of IL-10 polymorphisms with the risk of HCV infection confirmed the direction and magnitude of statistical significance of the overall effect, even after the exclusion of the largest study (see Supplementary Fig. S2).
The choice of genetic model
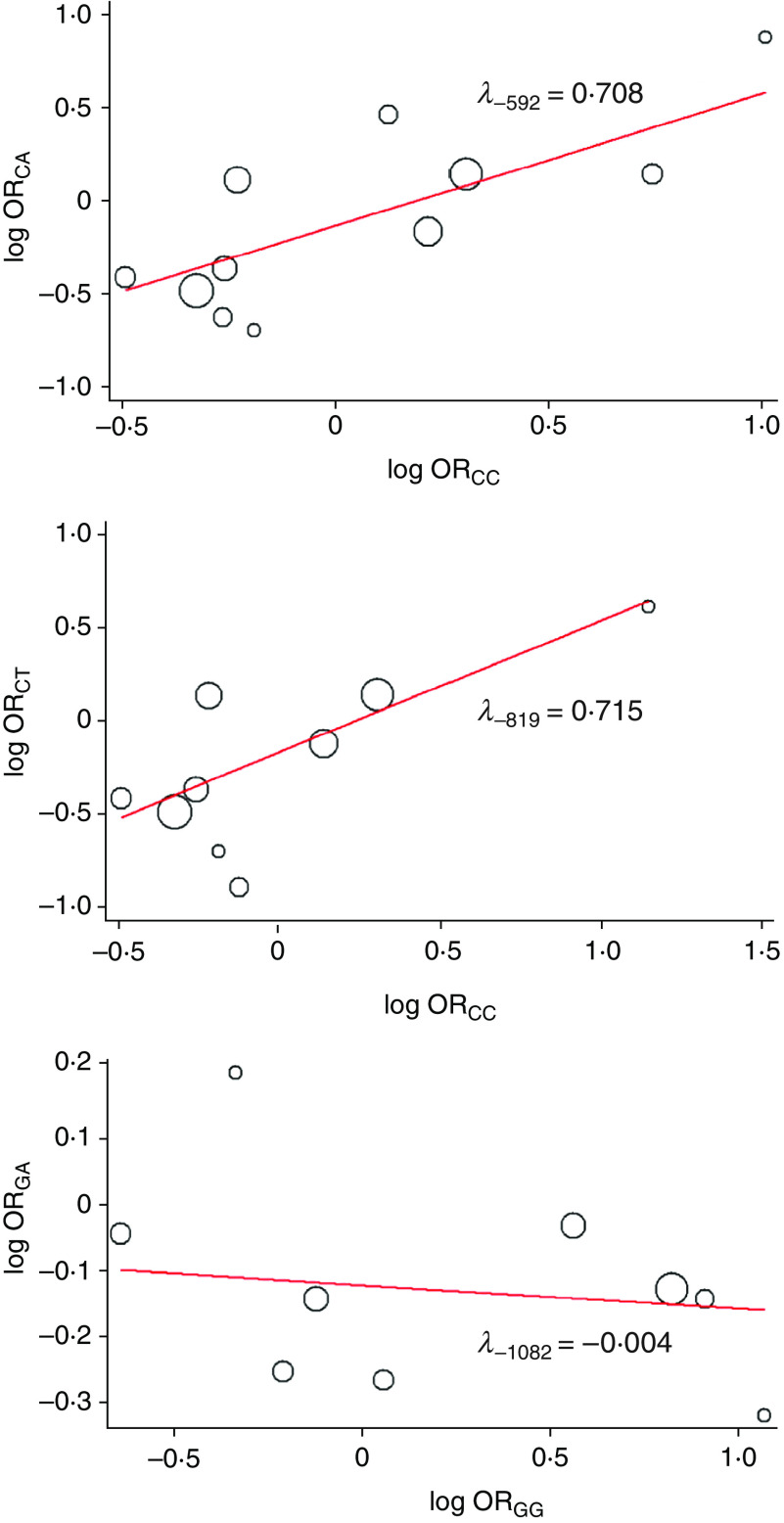
The plots of log ORCAvs. log ORCC (−592C/A), log ORCTvs. log ORCC (−819C/T) and log ORGAvs. log ORGG (−1082G/A) are shown in Figure 1, in which the slope represents λ. The resulting values of λ for −592C/A and −819C/T polymorphisms were 0·708 ± 0·014 and 0·715 ± 0·017, respectively, suggesting dominant models. In addition, a recessive model was pronounced in the −1082G/A polymorphism, λ = −0·004 ± 0·005. Therefore, full meta-analyses were performed using dominant inheritance mode for polymorphisms at positions −592 (CC + CA vs. AA) and −819 (CC + CT vs. TT), and recessive mode for −1082 (GG vs. GA + AA). Notably, the plots in Figure 1 can help to check the consistency of λ across studies and identify outlier studies. Ideally, all studies would be expected to lie along a straight line with slope λ in the absence of heterogeneity in the genetic model. Here no significant outliers or influential studies were visually identified. To further elucidate the advantages of biological justification, study-specific estimates of λ and bootstrapped 95% CIs are depicted in the Supplementary material (Supplementary Fig. S3), which can help assess whether any departures from linearity in Figure 1 can be explained by sampling error [30]. The findings revealed that the selected genetic models were consistent across studies. Otherwise, it may be better to perform joint pairwise comparisons using a general bivariate meta-analysis model [40].
Fig. 1.
[colour online]. Plots of log odds ratio (OR)CA against log ORCC (−592C/A), log ORCT against log ORCC (−819C/T) and log ORGA against log ORGG (−1082G/A) for IL-10 polymorphisms and risk of HCV infection. The slope of the solid line represents λ and size of circular symbols denotes the weight of study.
Meta-analysis of the association between IL-10 polymorphisms and the risk of HCV infection
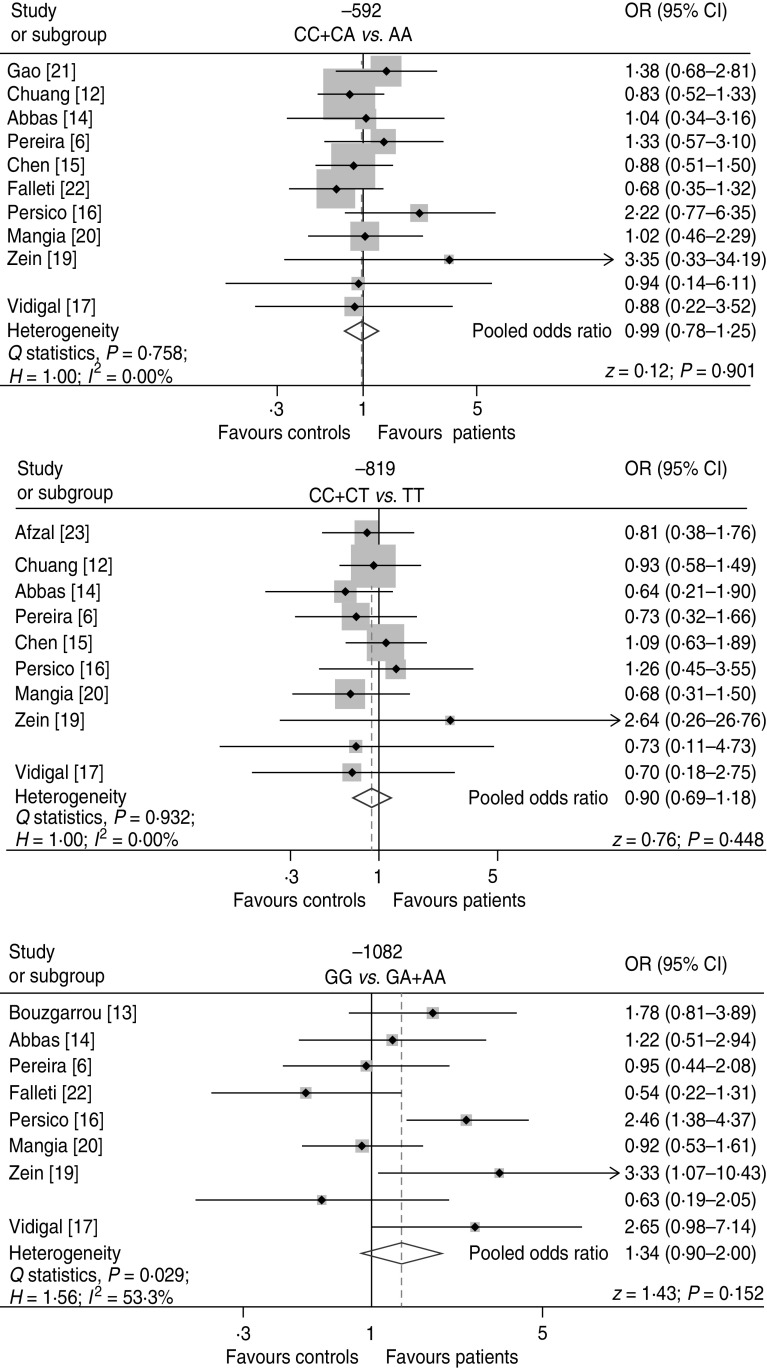
Fixed-effects models were employed for the full meta-analysis of the relationship between IL-10 −592C/A and −819C/T polymorphisms and the susceptibility to HCV. The pooled ORs were 0·99 (95% CI 0·78–1·25) and 0·90 (95% CI 0·69–1·18), respectively (Fig. 2). In the case of −1082 G/A polymorphism, a random-effects model was used due to the presence of heterogeneity. The pooled OR and 95% CI were 1·34 and 0·90–2·00, respectively (Fig. 2). Overall, no significant association was found between the risk of HCV infection and the IL-10 polymorphisms investigated here.
Fig. 2.
Association between IL-10 polymorphisms and risk of HCV in full meta-analysis. The pooled odds ratio (OR) and 95% confidence interval (CI) were generated using either a fixed-effects model (Mantel–Haenszel method) or a random-effects model (DerSimonian–Laird method). Studies are ordered by publication year.
It should be noted that the above conclusion cannot prevent an assumption of possible risk modifications by other factors, and also that the selected studies contained data on several ethnic populations, including primarily Caucasians and Asians. Thus, subgroup meta-analysis was performed to evaluate the effect of polymorphisms in these two ethnic populations, by conducting quantitative pooling individually. A forest plot assessing the ethnicity effect is shown in Figure 3. All the 95% CIs included the null value 1. For studies on the association of −1082G/A polymorphism, the interaction appeared a little stronger in Caucasians (P = 0·193). Further, considering the high heterogeneity described previously, a binomial logistic regression analysis using a Bayesian model [41] with ethnic status (Caucasians = 1, Asians = 0) as the dependent variable and the distribution of genotypes as an independent variable was performed to illustrate whether ethnicity was a risk modification factor. As a result, no clear evidence for the modification effect of ethnicity was observed (β−1082 = − 0·274 ± 0·636) when HCV patients were compared to healthy controls.
Fig. 3.
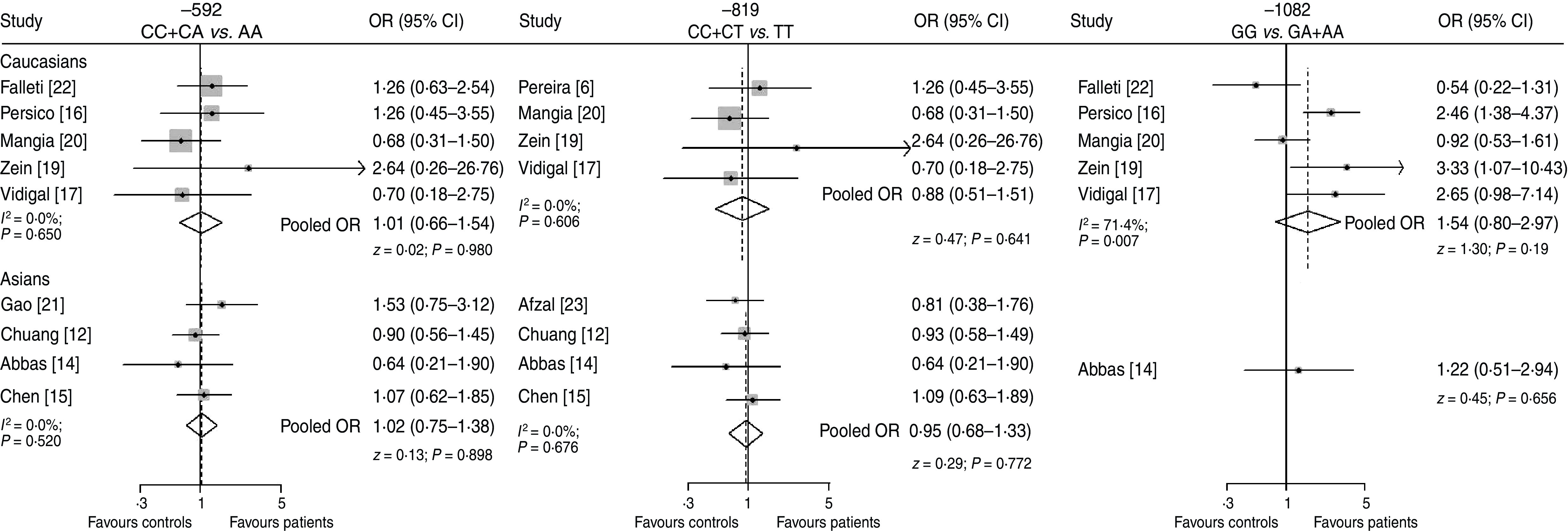
IL-10 polymorphisms and risk of HCV in (a) Caucasian and (b) Asian populations. Studies are ordered by publication year. OR, Odds ratio; CI, confidence interval.
Moreover, HCV infection can become chronic in most infected individuals. Only a few infected people spontaneously clear the virus. To date, HCV persistence has also been related to the production of inappropriate levels of IL-10 [17]. Therefore, the studies on patients with chronic HCV were grouped and another meta-analysis was performed in comparison with healthy controls. Under the premise of chronic HCV, no significant associations were further observed for these polymorphisms (data not shown).
Chinese association study
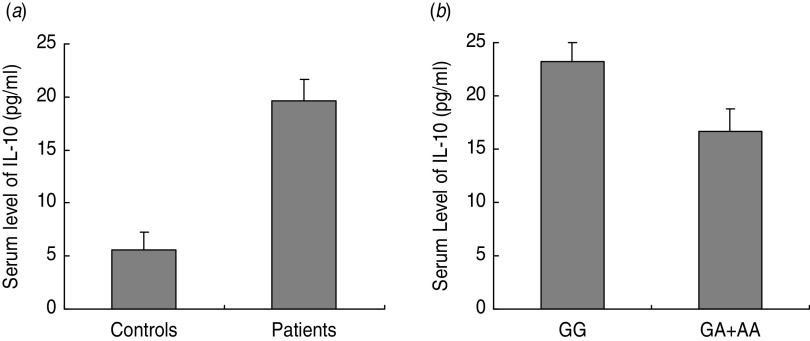
The demographic data of the study is presented in Table 2. Representative genotyping data are shown in Supplementary Fig. S4. The size of the AS-PCR product was 258 bp. The genotype and allele frequencies of IL-10 –1082G/A were 79·2% for AA genotype, 16·9% for GA genotype, 3·9% for GG genotype, 87·5% for A allele and 12·5% for G allele in controls and 71·9% for AA genotype, 15·6% for GA genotype, 12·5% for GG genotype, 79·7% for A allele and 20·3% for G allele in cases. No statistically significant difference was observed between chronic HCV patients and healthy controls for IL-10 − 1082G/A polymorphism (OR 2·64, 95% CI 0·690–10·1, P = 0·156). Notably, the G allele was found less frequently in this Chinese population (i.e. 12·5%) than in Caucasians (36·1–57·0%), while approaching its prevalence in Taiwanese (3·4% and 7·8%). Furthermore, the ELISA results indicated that the serum level of IL-10 was significantly higher in HCV patients than in controls (P < 0·0001, Fig. 4 a). In particular, a significantly higher serum IL-10 was observed in patients with the GG genotype (P < 0·0001, Fig. 4 b).
Table 2.
Demographic data of HCV patients and controls
| Data | HCV patients (n = 586) | Controls (n = 554) |
|---|---|---|
| Age (yr) | 47·9 ± 15·3 | 51·7 ± 16·1 |
| Body mass index | 23·1 ± 4·2 | 23·7 ± 4·1 |
| Anti-HCV/HCV RNA | Positive | Negative |
| Gender | ||
| Male | 328 | 302 |
| Female | 258 | 252 |
| IL-10 –1082 | ||
| GG | 73 | 22 |
| GA | 92 | 94 |
| AA | 421 | 438 |
Fig. 4.
Serum levels of IL-10 in (a) HCV patients and controls, and (b) HCV patients with GG and GA + AA genotypes.
DISCUSSION
Genetic association studies commonly have three genotype groups for a bi-allelic polymorphism, one of which is thought to be associated with a disease. In practice, three groups are often reduced to two by assuming a specific genetic model, such as a dominant or recessive model. However, a biological justification for the choice of generic model is rarely available [30, 31]. Lack of coherence or transparency is a common issue in choosing a model of meta-analysis. In this report, different genetic models were found in the selected epidemiological studies. As mentioned previously, inappropriate specification of inheritance mode could increase the risk of false negatives. For example, the association between IL-10 − 1082G/A polymorphism and the risk of HCV infection was more significant using a recessive model (P = 0·0018) than a dominant one (P = 0·0742) in the study by Persico et al. [16]. Therefore, we used a linear regression of pairwise estimates (two log ORs), weighted by study to determine the most appropriate genetic mode [40]. The results indicated dominant inheritance in IL-10 − 592C/A and −819C/T polymorphisms and recessive inheritance in –1082G/A. Furthermore, the presence of both dominant and recessive inheritances in the IL-10 gene suggested a complex interaction between genotype and protein expression. Therefore, the genetic contribution of single genes towards a complex disease is unlikely to act in a simple Mendelian fashion. To confirm dominantly and recessively inherited polymorphisms, additional phenotype–genotype studies need to be performed.
No overall significant association was observed between the IL-10 polymorphisms and the risk of HCV infection using meta-analysis. However, the effect of polymorphisms on the susceptibility to HCV may be modified by other risk factors such as ethnicity. This hypothesis has been recently suggested by the fact that African patients composed 53% of all HCV-positive patients while constituting only slightly less than one third of HCV-negative patients [42]. In addition, significantly higher IL-10 production was generated in vitro by peripheral blood mononuclear cells (PBMCs) isolated from healthy Caucasian subjects compared to first nation subjects [43]. One possible explanation can be the different amounts of IL-10 produced in populations, which was probably results from differential distribution of the cytokine polymorphisms [44]. It has been reported that PBMCs obtained from individuals presenting IL-10 –1082G/G genotype produced twofold greater quantities of IL-10 compared to individuals exhibiting GA or AA genotypes at the same position [45]. Moreover, Asians had a significantly higher proportion of genotypes that resulted in low IL-10 production [13]. Nevertheless, the significance of either subgroup analysis or Bayesian meta-regression in this study was not enough to accept or reject the hypothesis suggested above. Therefore, to further examine whether IL-10 –1082G/A is associated with the risk of HCV infection, an extended case-control study was conducted in a Chinese population. Despite no significant association being found, a low prevalence of G allele was observed. Furthermore, significantly higher IL-10 production was observed in HCV patients and especially patients with the GG genotype, which were also observed previously [13, 17].
Surprisingly, no essential role of IL-10 polymorphisms was found in the persistence of HCV. On the contrary, the incidence of chronicity has been associated with the ability of proteins to modulate immune responses upon various occasions. For example, enhanced production of IL-10 from antigen-presenting cells in the early stages of infection can alter the development of protective T-cell responses [46], contributing to impaired IFN-γ production and proliferation of HCV-specific T cells [47, 48]. However, it should be noted that some of those patients with chronic HCV infection may progress to liver fibrosis, cirrhosis and eventually HCC [49]. Thus, the association between the genetic polymorphisms and chronic HCV may depend on the severity/progress of HCV. Case-control studies have shown that women carrying IL-10 − 1082G/G genotype were associated with a higher risk of HCV persistence but a lower risk of progress to cirrhosis [50]. This finding was also supported by the protective role of IL-10 against progressive fibrosis in chronic infection [51]. The variability of outcomes in the progress of HCV precludes making a more definitive statement on the association evaluated here. The effort in grading the severity of HCV could help elucidate the role of IL-10 polymorphisms in HCV progress. Notably, other suspected risk factors for the susceptibility to HCV (e.g. age, sex and body mass index) also deserve extensive investigation.
It should be noted that the risk of bias of individual studies such as selection bias could be a potential issue in meta-analysis [52]. In this meta-analysis, sampling bias as one of the types of selection biases might cause some members of the population to be less likely to be included than others, resulting in that all participants are not equally balanced or objectively represented. Since we cannot obtain more information about the individual studies in meta-analysis, we are incertain whether there are other selection biases existing in those individual studies, e.g. if the selection process does not aim to include a patient spectrum similar to the population in which the test will be used in practice, if the populations are systematically different between comparison groups within a study (e.g. important baseline imbalances), if the studies apply inclusion/exclusion criteria uniformly to all comparison groups, etc. Furthermore, selection bias may be present in the process of meta-analysis due to the inability to identify and include all conducted and relevant studies [53]. Such selection bias can cause exaggerated or even false-positive gene-disease associations. Failure to include all relevant studies is largely caused by selective publication of studies with certain results, and the inability to identify studies published in languages other than English. All these biases could lead to meta-analysis an incomplete set of the evidence and produce summary results potentially biased.
As mentioned previously, a number of cytokines are involved in HCV infection. Among the cytokines identified, T-cell cytokines have been highly associated with the host immune response to HCV infection. IL-10 is an anti-inflammatory T-helper cytokine (Th2), while it can also be produced by regulatory T (Treg) cells [54]. The balance between the T cell-mediated immunities has been proven to be a determinant in many viral or non-viral infections [15]. On the other hand, the imbalance in immune-mediated diseases is commonly mediated by a variety of genetic polymorphisms in the genes. As HCV infection is an interaction between multiple genetic events, effects due to linkage disequilibrium (LD) between genotype variants cannot be excluded. In this study, some of these were considered in meta-analysis using a Bayesian hierarchical model. As a result, in the presence of strong LD between IL-10 –592C/A, –819C/T and –1082G/A polymorphisms, no more evidence for their clear association with the risk of HCV infection was found (data not shown). It is noteworthy that the interactions of these single nucleotide polymorphisms (SNPs) with other genetic variants were not taken into account here. To further model the effects of more SNPs in a region as well as gene × gene interactions, highly computationally efficient methods will be needed, in addition to the amount of previous information available on haplotype frequencies. Therefore, large whole-genome association studies (GWAS) are eagerly awaited to resolve this issue in the near future.
CONCLUSIONS
The present analysis provides increased support for clarifying the overall association of IL-10 − 592C/A, −819C/T and −1082G/A polymorphisms with the risk of HCV infection. Moreover, Bayesian meta-regression and subgroup analysis have demonstrated their attractiveness for exploration of the possible impacts of ethnicity and HCV persistence on the association evaluated here. Finally, an extended case-control study conducted in a Chinese population showed a low prevalence of G allele, and significantly higher IL-10 levels in HCV patients, especially patients with GG genotype.
Supplementary Material
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812002154.
click here to view supplementary material
ACKNOWLEDGEMENTS
National Natural Science Fund (21175071), Research Fund for the Doctoral Programme of Higher Education of China (20093234120010), the project sponsored by SRF for ROCS, SEM (39) and Jiangsu Six-type Top Talents programme (D) to Dr Chen are gratefully acknowledged. The authors thank American Journal Experts for reading the article.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Dai CY, et al. Tumor necrosis factor- alpha promoter polymorphism at position-308 predicts response to combination therapy in hepatitis C virus infection. Journal of Infectious Diseases 2006; 193: 98–101. [DOI] [PubMed] [Google Scholar]
- 2.Lauer GM, Walker BD. Hepatitis C virus infection. New England Journal of Medicine 2001; 345: 41–52. [DOI] [PubMed] [Google Scholar]
- 3.Bidwell J, et al. Cytokine gene polymorphism in human disease: on-line databases, supplement 1. Genes and Immunity 2001; 2: 61–70. [DOI] [PubMed] [Google Scholar]
- 4.Bidwell J, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes and Immunity 1999; 1: 3–19. [DOI] [PubMed] [Google Scholar]
- 5.Jeng JE, et al. Tumor necrosis factor-alpha 308.2 polymorphism is associated with advanced hepatic fibrosis and higher risk for hepatocellular carcinoma. Neoplasia 2007; 9: 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pereira FA, et al. Association of TGF-beta1 codon 25 (G915C) polymorphism with hepatitis C virus infection. Journal of Medical Virology 2008; 80: 58–64. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Pei J. An assessment of a TNF polymorphic marker for the risk of HCV infection: meta-analysis and a new clinical study design. Infection, Genetics and Evolution 2009; 9: 1356–1363. [DOI] [PubMed] [Google Scholar]
- 8.Orhan I, et al. Seminal plasma cytokine levels in the diagnosis of chronic pelvic pain syndrome. International Journal of Urology 2001; 8: 495–499. [DOI] [PubMed] [Google Scholar]
- 9.Hochreiter WW, et al. Evaluation of the cytokines interleukin 8 and epithelial neutrophil activating peptide 78 as indicators of inflammation in prostatic secretions. Urology 2000; 56: 1025–1029. [DOI] [PubMed] [Google Scholar]
- 10.Serdar A, et al. Determination of IL-10 levels in syndrome patients. Advances in Molecular Biology 2008; 2: 87–91. [Google Scholar]
- 11.Kreutzer R. MCS: the status of population-based research. International Journal of Hygiene and Environmental Health 2002; 205: 411–414. [DOI] [PubMed] [Google Scholar]
- 12.Chuang JY, et al. IL-10 promoter gene polymorphisms and sustained response to combination therapy in Taiwanese chronic hepatitis C patients. Digestive and Liver Disease 2009; 41: 424–430. [DOI] [PubMed] [Google Scholar]
- 13.Bouzgarrou N, et al. Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis C severity. Human Immunology 2009; 70: 230–236. [DOI] [PubMed] [Google Scholar]
- 14.Abbas Z, Moatter T. Interleukin (IL) 1beta and IL-10 gene polymorphism in chronic hepatitis C patients with normal or elevated alanine aminotransferase levels. Journal of the Pakistan Medical Association 2003; 53: 59–62. [PubMed] [Google Scholar]
- 15.Chen TY, et al. Impact of serum levels and gene polymorphism of cytokines on chronic hepatitis C infection. Translational Research 2007; 150: 116–121. [DOI] [PubMed] [Google Scholar]
- 16.Persico M, et al. Interleukin-10 -1082 GG polymorphism influences the occurrence and the clinical characteristics of hepatitis C virus infection. Journal of Hepatology 2006; 45: 779–785. [DOI] [PubMed] [Google Scholar]
- 17.Vidigal PG, Germer JJ, Zein NN. Polymorphisms in the interleukin-10, tumor necrosis factor-alpha, and transforming growth factor-beta1 genes in chronic hepatitis C patients treated with interferon and ribavirin. Journal of Hepatology 2002; 36: 271–277. [DOI] [PubMed] [Google Scholar]
- 18.Lio D, et al. IL-10 and TNF-alpha polymorphisms and the recovery from HCV infection. Human Immunology 2003; 64: 674–680. [DOI] [PubMed] [Google Scholar]
- 19.Zein NN, et al. Ethnic differences in polymorphisms of tumor necrosis factor-alpha, interleukin-10, and transforming growth factor-beta1 genes in patients with chronic hepatitis C virus infection. American Journal of Tropical Medicine and Hygiene 2004; 70: 434–437. [PubMed] [Google Scholar]
- 20.Mangia A, et al. IL-10 haplotypes as possible predictors of spontaneous clearance of HCV infection. Cytokine 2004; 25: 103–109. [DOI] [PubMed] [Google Scholar]
- 21.Gao QJ, et al. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World Journal of Gastroenterology 2009; 15: 5610–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falleti E, et al. Genetic polymorphisms of inflammatory cytokines and liver fibrosis progression due to recurrent hepatitis C. Journal of Interferon and Cytokine Research 2007; 27: 239–246. [DOI] [PubMed] [Google Scholar]
- 23.Afzal MS, et al. Analysis of interleukin-10 gene polymorphisms and hepatitis C susceptibility in Pakistan. Journal of Infection in Developing Countries; 5: 473–479. [DOI] [PubMed] [Google Scholar]
- 24.Ishida C, et al. Functional gene polymorphisms of interleukin-10 are associated with liver disease progression in Japanese patients with hepatitis C virus infection. Internal Medicine; 50: 659–666. [DOI] [PubMed] [Google Scholar]
- 25.Dogra G, et al. Polymorphism of tumor necrosis factor-alpha and interleukin-10 gene promoter region in chronic hepatitis C virus patients and their effect on pegylated interferon-alpha therapy response. Human Immunology; 72: 935–939. [DOI] [PubMed] [Google Scholar]
- 26.Kusumoto K, et al. Interleukin-10 or tumor necrosis factor-alpha polymorphisms and the natural course of hepatitis C virus infection in a hyperendemic area of Japan. Cytokine 2006; 34: 24–31. [DOI] [PubMed] [Google Scholar]
- 27.Baker WL, et al. Understanding heterogeneity in meta-analysis: the role of meta-regression. International Journal of Clinical Practice 2009; 63: 1426–1434. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Pei J. An assessment of a TNF polymorphic marker for the risk of HCV infection: meta-analysis and a new clinical study design. Infection, Genetics and Evolution 2009; 9: 1356–1363. [DOI] [PubMed] [Google Scholar]
- 29.Schmid CH, et al. Meta-regression detected associations between heterogeneous treatment effects and study-level, but not patient-level, factors. Journal of Clinical Epidemiology 2004; 57: 683–697. [DOI] [PubMed] [Google Scholar]
- 30.Minelli C, et al. The choice of a genetic model in the meta-analysis of molecular association studies. International Journal of Epidemiology 2005; 34: 1319–1328. [DOI] [PubMed] [Google Scholar]
- 31.Trikalinos TA, et al. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. American Journal of Epidemiology 2006; 163: 300–309. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis JP, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. International Journal of Epidemiology 2008; 37: 120–132. [DOI] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials 1986; 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 34.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute 1959; 22: 719–748. [PubMed] [Google Scholar]
- 35.Copas J, Shi JQ. Meta-analysis, funnel plots and sensitivity analysis. Biostatistics 2000; 1: 247–262. [DOI] [PubMed] [Google Scholar]
- 36.Boston RC, Sumner AE. Stata: a statistical analysis system for examining biomedical data. Advances in Experimental Medicine and Biology 2003; 537: 353–369. [DOI] [PubMed] [Google Scholar]
- 37.Thompson SG, Higgins JPT. How should meta-regression analyses be undertaken and interpreted? Statistics in Medicine 2002; 21: 1559–1573. [DOI] [PubMed] [Google Scholar]
- 38.Perrey C, et al. ARMS-PCR methodologies to determine IL-10, TNF-alpha, TNF-beta and TGF-beta 1 gene polymorphisms. Transplant Immunology 1999; 7: 127–128. [DOI] [PubMed] [Google Scholar]
- 39.Knapp S, et al. Interleukin-10 promoter polymorphisms and the outcome of hepatitis C virus infection. Immunogenetics 2003; 55: 362–369. [DOI] [PubMed] [Google Scholar]
- 40.Nam IS, Mengersen K, Garthwaite P. Multivariate meta-analysis. Statistics in Medicine 2003; 22: 2309–2333. [DOI] [PubMed] [Google Scholar]
- 41.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. Journal of the American Statistical Association 1993; 88: 9–25. [Google Scholar]
- 42.Kalantar-Zadeh K, et al. Hepatitis C virus and death risk in hemodialysis patients. Journal of the American Society of Nephrology 2007; 18: 1584–1593. [DOI] [PubMed] [Google Scholar]
- 43.Aborsangaya KB, et al. Impact of aboriginal ethnicity on HCV core-induced IL-10 synthesis: interaction with IL-10 gene polymorphisms. Hepatology 2007; 45: 623–630. [DOI] [PubMed] [Google Scholar]
- 44.Zabaleta J, et al. Ethnic differences in cytokine gene polymorphisms: potential implications for cancer development. Cancer Immunology, Immunotherapy 2008; 57: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner DM, et al. An investigation of polymorphism in the interleukin-10 gene promoter. European Journal of Immunogenetics 1997; 24: 1–8. [DOI] [PubMed] [Google Scholar]
- 46.Dolganiuc A, et al. Hepatitis C virus core and nonstructural protein 3 proteins induce pro- and anti-inflammatory cytokines and inhibit dendritic cell differentiation. Journal of Immunology 2003; 170: 5615–5624. [DOI] [PubMed] [Google Scholar]
- 47.Thimme R, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. Journal of Experimental Medicine 2001; 194: 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lechner F, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. Journal of Experimental Medicine 2000; 191: 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene 2006; 25: 3834–3847. [DOI] [PubMed] [Google Scholar]
- 50.Paladino N, et al. Gender susceptibility to chronic hepatitis C virus infection associated with interleukin 10 promoter polymorphism. Journal of Virology 2006; 80: 9144–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaplan DE, et al. Peripheral virus-specific T-cell interleukin-10 responses develop early in acute hepatitis C infection and become dominant in chronic hepatitis. Journal of Hepatology 2008; 48: 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viswanathan M, et al. Assessing the risk of bias of individual studies in systematic reviews of health care interventions, 2008. [PubMed]
- 53.Tang JL. Selection bias in meta-analyses of gene-disease associations. PLoS Medicine 2005; 2: e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unutmaz D, Pulendran B. The gut feeling of Treg cells: IL-10 is the silver lining during colitis. Nature Immunology 2009; 10: 1141–1143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
Supplementary information supplied by authors.
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268812002154.
click here to view supplementary material