SUMMARY
This study investigated the molecular prevalence of Trypanosoma lewisi and T. evansi in wild rodents from Cambodia, Lao PDR and Thailand. Between 2008 and 2012, rodents (and shrews) were trapped in nine locations and 616 of these were tested using three sets of primers: TRYP1 (amplifying ITS1 of ribosomal DNA of all trypanosomes), TBR (amplifying satellite genomic DNA of Trypanozoon parasites) and LEW1 (amplifying ITS1 of ribosomal DNA of T. lewisi). Based on the size of the PCR products using TRYP1, 17% were positive for T. lewisi and 1·0% positive for Trypanozoon. Results were confirmed by sequencing PCR products and by using more specific primers (LEW1 and TBR). The specificity of TRYP1 primers, however, failed as rodent DNA was amplified in some instances, giving unexpected product sizes. Using LEW1 primers, 13·3% of the samples were confirmed positive for T. lewisi, both by PCR and sequencing. In Thailand, T. lewisi was found in Rattus tanezumi, R. exulans and Berylmys; in Lao PDR, in R. tanezumi and R. exulans, and in Cambodia in R. tanezumi, R. exulans and R. norvegicus. Using TBR, 1·3% of the samples tested positive for Trypanozoon by PCR and sequencing; T. evansi is the only species of the Trypanozoon subgenus possibly present in wild Asian rodents. These results confirmed its presence in rodents from Thailand (R. tanezumi), Lao PDR (R. tanezumi, R. nitidus) and Cambodia (R. tanezumi, Niviventer fulvescens, Maxomys surifer). Based on the information related to rodent trapping, it was found that rodent species trapped in and around human dwellings had a higher prevalence of T. lewisi infection. R. tanezumi and R. exulans, two synanthropic species, were mainly found infected in this habitat suggesting a role as a reservoir and thus a potential source of T. lewisi for human infection.
Key words: Animal pathogens, rodent-borne diseases, rodents, Southeast Asia, trypanosomes
INTRODUCTION
Trypanosomosis is a disease of medical and veterinary importance, mainly distributed in tropical areas around the world. The disease is caused by haemoflagellate protozoan parasites transmitted by biting insects [1]. Trypanosomoses cause severe human syndromes such as American Chagas' disease due to Trypanosoma cruzi, or African sleeping sickness due to Trypanosoma brucei spp. (T. b. rhodesiense, East African acute disease, and T. b. gambiense, West and Central African chronic disease). The main pathogenic animal trypanosomes include (i) the complex T. vivax, T. congolense and T. brucei brucei, found in tropical Africa, causing ‘nagana’ in domestic animals (mainly transmitted by tsetse flies), (ii) T. evansi transmitted mechanically (by tabanids and stomoxes) and causing El debab in camels (Africa), mal de caderas in horses in South America, and ‘surra’ in Asia, and (iii) T. equiperdum transmitted venereally in horses [1]. Beside these pathogenic parasites, some Trypanosoma species are considered as non-pathogenic, such as Trypanosoma theileri in bovines, T. melophagium in sheep, T. rangeli in humans and T. lewisi in rats [1].
Recent studies have reported an increasing number of atypical human cases of T. lewisi or T. lewisi-like [2–4] and T. evansi [5, 6] infections, in Malaysia, Sri Lanka, India and Thailand. For this reason, the transmission of these two Trypanosoma species requires investigation. Forty-four Trypanosoma species have been described in 144 rodent species [1, 7]; most of them are from the Stercoraria section.
T. lewisi is almost exclusively found in rodents and belongs to the non-pathogenic subgenus Herpetosoma. It is a cosmopolitan parasite of rats, transmitted by fleas. Rats are most probably infected by metatrypanosomes when ingesting fleas or their excretions. Rattini rodents are the original hosts of T. lewisi including commensal species such as the black rat (Rattus rattus), the Pacific rat (R. exulans) and the Norwegian rat (R. norvegicus), which have disseminated outside their original range following human migration and trade [8]. This has provided the opportunity for T. lewisi to spread worldwide along with its rodent hosts.
The other atypical trypanosome found in humans is T. evansi, from the Salivaria section, subgenus Trypanozoon, which was recently described in India [9], and has also been reported from Egypt [10]. T. evansi can infect a very large range of animals, including livestock, domestic animals and wild hosts [1]. Together with livestock, rodents may act as a reservoir for T. evansi, thus contributing to putting animals and humans at risk of infection. The presence of T. evansi and T. lewisi in rodents has been reported in previous studies [7, 11]. Here we evaluate the role of wild rodents in atypical human infections by animal trypanosomes in relation to the environmental habitat of rodents [12, 13].
There are several diagnostic tests for trypanosomes, which have both advantages and limitations [14–18]; however, internal transcribed spacer 1 (ITS1) sequences have recently been widely used in the diagnosis, characterization and phylogenetic studies of trypanosomes [19–21].
The recent cases of atypical infection of humans by Trypanosoma species in Southeast Asia casts doubt on the identification of rodent reservoirs and the role of habitats in their infection. In particular, on-going land use changes may affect not only the transmission of Trypanosoma in rodents but also the risk of transmission to humans. The investigation of rodent infection in different habitats at different locations in Southeast Asia may make it possible to evaluate the particular effect of habitat, taking into account the influence of locality and season.
The aims of this study were (1) to provide a more intensive survey of the presence of T. lewisi and T. evansi in murid rodents from Southeast Asia using molecular investigation, and (2) to determine the type of environmental habitat that may explain the infection of rodents.
MATERIAL AND METHODS
Rodent sampling and identification
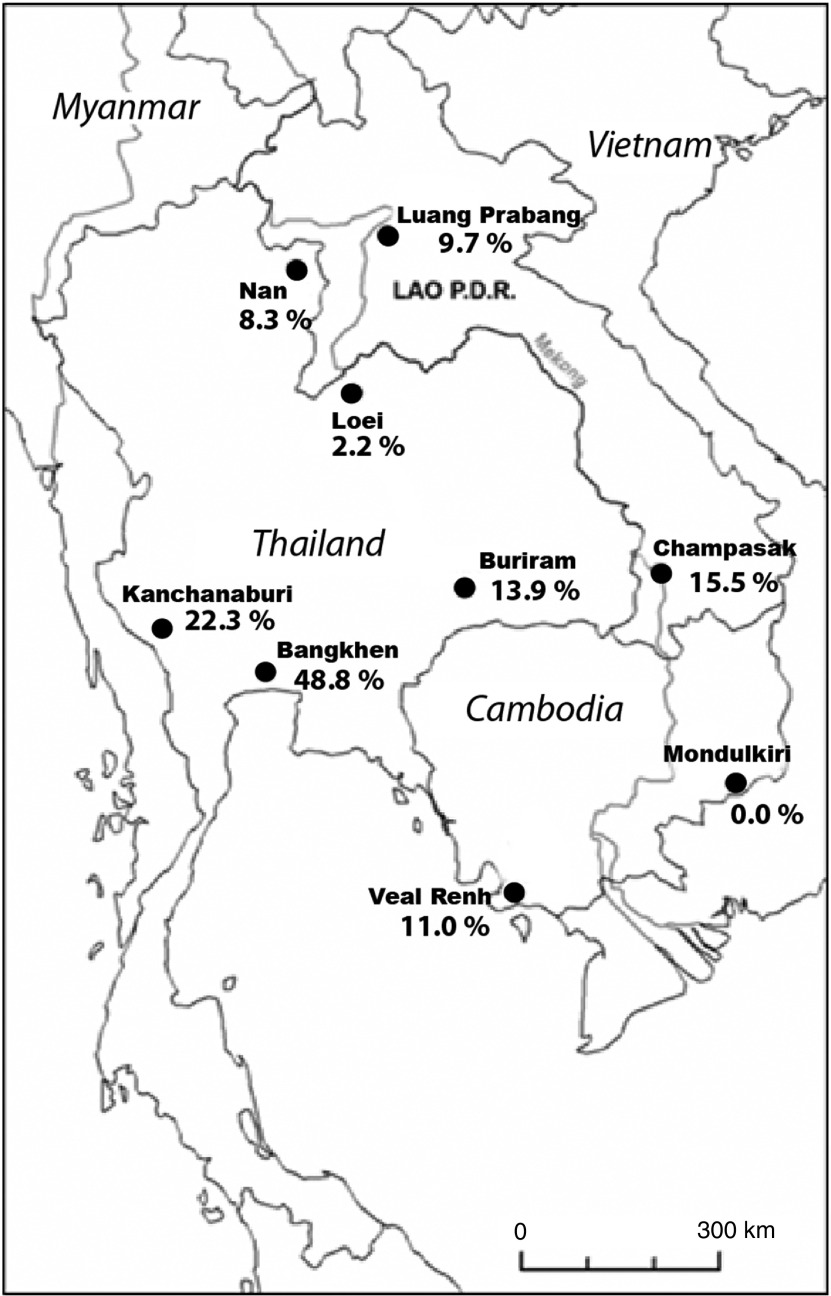
Eight different sites were chosen in three countries (Thailand, Cambodia, Lao PDR), as offering a representative overview of the various ecosystems that are affected by land-use changes. These sampling sites were part of the CERoPath project (www.ceropath.org). Four main habitats were investigated in each locality. At each site, 30 lines of 10 traps were installed over four nights targeting three different habitats, i.e. forests, non-flooded lands (agricultural lands such as orchards, dry rice field, cassava field or non-cultivated land such as bush) and flooded-irrigated agricultural lands (i.e. paddy rice fields), for a total of 1200 night-traps. Villages and isolated houses, which corresponded to a fourth habitat category, i.e. human settlement, were also sampled using cage-traps distributed to residents. Pictures, a habitat description and coordinates of the trap lines are available in the ‘Research/study’ areas and ‘Research/protocols’ sections of the Ceropath project website (www.ceropath.org) (see online Supplementary material). In addition, a ninth site situated in an urban environment, the Bangkhen campus of the University of Kasetsart, was investigated using traps for 2 days (Fig. 1).
Fig. 1.
Geographical locations of trapping sites with percentage of rodents infected by Trypanosoma lewisi, the most prevalent trypanosome species, based on PCR diagnosis (see Table 1 for raw data).
Rodents were identified on the basis of their morphology or using species-specific primers and/or barcoding assignment [22, 23]. Complete data for animals used as a reference for barcoding assignment are available in the ‘Barcoding Tool/RodentSEA’ section of the Ceropath project website (http://www.ceropath.org/).
Rodents were euthanized and dissected to collect organs including kidneys according to CERoPath protocols [24] (www.ceropath.org), which respect animal care, health security for field parasitologists and quality data handling.
Microscopic observation of trypanosomes
When it was possible to perform this examination on site, a small drop (4–5 μl) of freshly citrated blood was deposited between slide and slide cover for immediate direct microscopic observation at ×400 magnification. If trypanosomes were observed, a thin blood smear was made for Giemsa staining and microscopic observation (×1000 in oil immersion) [25]. Pre-identification of T. lewisi-like and T. evansi was based on the morphology and morphometry of the parasites [1].
Molecular identification of trypanosomes
The polymerase chain reaction (PCR) technique, which detects parasite nucleic acids, is a very specific and sensitive method that can detect 1–20 trypanosomes/ml of blood [18]. This property makes PCR a very useful method for the pre-patent period and chronic phase of the disease. ITS1 sequences have recently been widely used in the diagnosis, characterization and phylogenetic studies of trypanosomes [19–21].
Rodent blood was deposited in sodium citrate in a microtube and centrifuged at 9000 g. Plasma was collected in a microtube for further serological tests. The buffy coat was collected in a volume of ∼100 μl and frozen for further DNA preparation.
In the laboratory, buffy-coat samples were thawed and DNA extracted with the phenol-chloroform technique [26].
Trypanosoma species identification was based on microscopic observation of the morphology and morphometry of the parasites, and the results obtained by PCR, followed by sequencing and sequence analysis. Three sets of primers were used systematically, i.e. TRYP1, TBR and LEW1 primers [7, 27–29] (see Supplementary material).
Using TRYP1 primers, a 520 base-pair (bp) and a 620 bp amplified product were characteristic for Trypanozoon and T. lewisi, respectively. Other sized products might be obtained and would require sequencing for species identification.
Using TBR primers only, a 168 bp product is expected and is characteristic of a parasite belonging to the Trypanozoon subgenus. Based on host specificity (T. equiperdum is only infective to Equidae) and geographical distribution (T. brucei ssp. are only present in Africa) [1], T. evansi is the only Trypanozoon species possibly present in wild Asian rodents.
Using LEW1 primers, a 220 bp product is expected with T. lewisi DNA; however (although never described so far but theoretically possible from the in silico data), similar or closely sized products can be obtained with very closely related parasites such as T. rabinowitschae, T. grosi, T. blanchardi and T. otospermophili [29]. Interpretation of the results was based on the size of the PCR product estimated by comparison with the molecular marker GeneRuler®100Plus (Fermentas™, USA), and positive controls including T. evansi and T. lewisi DNA.
DNA sequencing
DNA sequencing was performed in some cases to confirm the specificity of the products obtained. PCR products were excised from agarose gels and purified using gel purification kits according to the manufacturer's instructions (GeneJET®, Fermentas). Sequencing was carried out by Ward Medic Ltd (Thailand; 1st Base Distributor), using an automated sequencer (Applied Biosystems 3730xl DNA Analyzer, USA), and the DNA dye primer sequencing kits (ABI Big Dye version 3.1, USA). Data were analysed using Applied Biosystems Sequencing Analysis software v. 5.2.0. The sequences obtained were blasted with sequences available on Genbank (www.ncbi.nlm.nih.gov).
Statistical analyses
We performed generalized linear models (GLM), using a binomial distribution of individual host infection and logit function, to identify the likely variables that might explain the infection of rodents by T. lewisi in R software [30]. Selection of the best model was based on Akaike's Information Criterion (AIC) using host species, habitat and locality as independent variables. A GLM analysis on the infection of rodents by T. evansi could not performed due to the small number of infected rodents (see Table 1). These localities represented a variety of habitats in relation to human pressures and land usage. Habitats were ranked as: (1) forests and mature plantations, (2) non-flooded lands or fields (shrubby wasteland, young plantations, orchards), (3) rain-fed and irrigated lowland paddy rice fields (cultivated floodplain), (4) households (in villages or city), which corresponded to an increasing gradient of human-dominated habitats. Analyses were performed with and without the urbanized site of Bangkhen.
Table 1.
List and number of host species infected by Trypanosoma species in localities from Thailand, Lao PDR and Cambodia, using TRYP1, LEW and TBR primers (see Material and methods section)
| Country | Locality | Rodent species | Number tested | Trypanosome positive using TRYP1 (%) | T. lewisi positive using LEW (%) | T. evansi positive using TBR (%) |
|---|---|---|---|---|---|---|
| Thailand | Buriram | Ban. indica | 1 | 0 (0) | 0 (0) | 0 (0) |
| Ban. savilei | 11 | 4 (36·4) | 2 (18·2) | 0 (0) | ||
| M. cervicolor | 4 | 1 (25·0) | 1 (25·0) | 0 (0) | ||
| R. exulans | 21 | 3 (14·3) | 3 (14·3) | 0 (0) | ||
| R. tanezumi | 10 | 1 (10) | 1 (10·0) | 0 (0) | ||
| Kanchanaburi | Ban. savilei | 33 | 12 (36·4) | 8 (24·2) | 1 (3·0) | |
| Max. surifer | 8 | 1 (12·5) | 0 (0) | 0 (0) | ||
| M. caroli | 1 | 0 (0) | 0 (0) | 0 (0) | ||
| M. cervicolor | 4 | 0 (0) | 0 (0) | 0 (0) | ||
| M. cookii | 10 | 0 (0) | 0 (0) | 0 (0) | ||
| Mus sp. | 5 | 1 (20·0) | 0 (0) | 0 (0) | ||
| R. andamanensis | 1 | 0 (0) | 0 (0) | 0 (0) | ||
| R. exulans | 9 | 3 (33·3) | 3 (33·3) | 0 (0) | ||
| R. tanezumi | 13 | 4 (30·8) | 3 (23·1) | 0 (0) | ||
| Loei | Ban. indica | 2 | 0 (0) | 0 (0) | 0 (0) | |
| Ban. savilei | 2 | 0 (0) | 0 (0) | 0 (0) | ||
| Ber. berdmorei | 2 | 0 (0) | 0 (0) | 0 (0) | ||
| N. fulvescens | 4 | 0 (0) | 0 (0) | 0 (0) | ||
| R. exulans | 13 | 0 (0) | 1 (7·7) | 0 (0) | ||
| R. losea | 25 | 0 (0) | 0 (0) | 0 (0) | ||
| Nan | Ban. indica | 13 | 0 (0) | 0 (0) | 0 (0) | |
| Ber. berdmorei | 3 | 1 (33·3) | 1 (33·3) | 0 (0) | ||
| Ber. bowersi | 1 | 1 (100) | 0 (0) | 0 (0) | ||
| M. cervicolor | 3 | 0 (0) | 0 (0) | 0 (0) | ||
| M. cookii | 5 | 0 (0) | 0 (0) | 0 (0) | ||
| R. exulans | 15 | 2 (13·3) | 2 (13·3) | 0 (0) | ||
| R. tanezumi | 9 | 2 (22·2) | 0 (0) | 0 (0) | ||
| Bangkhen | R. tanezumi | 41 | 14 (34·1) | 20 (48·8) | 3 (7·3) | |
| Lao PDR | Champasak | Ban. indica | 1 | 0 (0) | 0 (0) | 0 (0) |
| Ban. savilei | 3 | 0 (0) | 0 (0) | 0 (0) | ||
| Ber. berdmorei | 1 | 1 (100) | 1 (100) | 0 (0) | ||
| Max. surifer | 3 | 0 (0) | 0 (0) | 0 (0) | ||
| N. fulvescens | 1 | 0 (0) | 0 (0) | 0 (0) | ||
| R. exulans | 38 | 10 (26·3) | 10 (26·3) | 3 (7·9) | ||
| R. losea | 6 | 0 (0) | 0 (0) | 0 (0) | ||
| R. tanezumi | 6 | 2 (33·3) | 1 (16·7) | 0 (0) | ||
| Luang Prabang | Ban. indica | 2 | 0 (0) | 0 (0) | 0 (0) | |
| B. berdmorei | 1 | 0 (0) | 0 (0) | 0 (0) | ||
| B. bowersi | 1 | 1 (100) | 0 (0) | 0 (0) | ||
| M. caroli | 7 | 0 (0) | 0 (0) | 0 (0) | ||
| M. cookii | 27 | 7 (25·9) | 0 (0) | 0 (0) | ||
| R. andamanensis | 2 | 0 (0) | 0 (0) | 0 (0) | ||
| R. nitidus | 8 | 1 (12·5) | 0 (0) | 0 (0) | ||
| R. tanezumi | 38 | 10 (26·3) | 6 (15·8) | 1 (2·6) | ||
| Cambodia | Mondulkiri | Ban. savilei | 10 | 1 (10) | 0 (0) | 0 (0) |
| Ber. berdmorei | 1 | 0 (0) | 0 (0) | 0 (0) | ||
| Max. surifer | 3 | 0 (0) | 0 (0) | 0 (0) | ||
| R. exulans | 5 | 0 (0) | 0 (0) | 0 (0) | ||
| R. tanezumi | 4 | 0 (0) | 0 (0) | 0 (0) | ||
| Veal Renh | Ber. berdmorei | 3 | 0 (0) | 0 (0) | 0 (0) | |
| Max. surifer | 41 | 1 (2·4) | 1 (2·4) | 0 (0) | ||
| R. argentiventer | 15 | 0 (0) | 0 (0) | 0 (0) | ||
| R. exulans | 37 | 8 (21·6) | 6 (16·2) | 0 (0) | ||
| R. norvegicus | 17 | 2 (11·8) | 2 (11·8) | 0 (0) | ||
| R. tanezumi | 37 | 2 (5·4) | 2 (5·4) | 0 (0) | ||
| S. murinus | 29 | 8 (27·6) | 8 (27·6) | 0 (0) | ||
| Total | 616 | 104 (16·9) | 82 (13·3) | 8 (1·3) |
RESULTS
Rodent identification
Five hundred and eighty-seven rodents were identified at species level: Bandicota (Ban.) indica (19), Ban. savilei (59), Berylmys (Ber.) berdmorei (11), Ber. bowersi (2), Mus caroli (8), M. cervicolor (11), M. cookii (42), Maxomys (Max.) surifer (55), Niviventer fulvescens (5), Rattus andamanensis (3), R. argentiventer (15), R. exulans (138), R. losea (31), R. nitidus (8), R. norvegicus (17), R. tanezumi (101), R. tanezumi clade 3 (a mitochondrial lineage, see [31]) (57), with five individuals identified as Mus sp. and 29 shrews identified as Suncus murinus.
Microscopic identification of trypanosomes
One hundred and twenty-five fresh blood samples were examined by microscope when the examination was possible near the trapping site, e.g. in the two locations of Kanchanaburi and Bangkhen. In total, 22 (17·6%) were found to be positive, with 5/84 (6%) in Kanchanaburi, and 17/41 (41·5%) in Bangkhen. When high parasitaemia was observed (16/22 cases), parasites could easily be found on Giemsa-stained blood smears. All the parasites observed exhibited a morphology and morphometry compatible with T. lewisi or T. lewisi-like.
Molecular identification of trypanosomes
Using TRYP1 primers, 89 individuals were found to be positive, with either a 520 bp product characterizing Trypanozoon or a 620 bp product characterizing T. lewisi, which were confirmed using LEW1 and TBR primers. Other sized products were found, potentially corresponding to other Trypanosoma species, in samples from Ban. savilei (Cambodia, Thailand), Ber. bowersi (Lao PDR, Thailand), Max. surifer (Thailand), M. cookii (Lao PDR), Mus sp. (Thailand), R. exulans (Cambodia), R. nitidus (Lao PDR) and R. tanezumi (Cambodia, Lao PDR, Thailand). PCR products extracted from the gel (950, 900, 750, 720 bp) underwent sequencing and BLAST; however, they proved to be either rodent or non-specific products. Other visible PCR products were of insufficient quantity for sequencing.
Using LEW1 primers, 79 individuals were found to be positive with a 220 bp product characterizing T. lewisi (Table 1). The rodent species were Ban. savilei (Thailand), Ber. berdmorei (Thailand, Lao PDR), Max. surifer (Cambodia), M. cervicolor (Thailand), R. exulans (Lao PDR, Cambodia, Thailand), R. tanezumi (Lao PDR, Cambodia, Thailand) and the shrew S. murinus (Cambodia). In one sample from R. tanezumi, exhibiting a T. lewisi-like parasite by microscopic observation, a 210 bp product was obtained, but of insufficient quantity for sequencing. Interestingly, this sample was negative using TRYP1 primers. Further studies are necessary for parasite identification.
Using TBR primers, eight individuals were found to be positive with a 168 bp product characterizing the Trypanozoon subgenus (Table 1). These individuals belonged to Ban. savilei (in Thailand), R. tanezumi (Lao PDR, Thailand) and R. exulans (Lao PDR).
Rodent infection
Table 1 shows the numbers of Trypanosoma infections in each rodent species for each locality. Of the 616 rodents and shrews investigated, 82 (13·3%) were found infected by T. lewisi and eight (1·3%) by T. evansi. No mixed infections were observed. The prevalence values for rodent infection were 9·5% by T. lewisi and 0% by T. evansi in Cambodia, 12·4% by T. lewisi and 2·8% by T. evansi in Lao PDR, 16·7% by T. lewisi and 1·5% by T. evansi in Thailand. The highest prevalence of T. lewisi infection detected with LEW1 primers was observed in Bangkhen, with 48·8%.
We used AIC to compare logistic regression models to explain individual rodent infection (Table 2). We performed this analysis without the locality of Bangkhen for which rodents were only investigated in urban habitat (as there are no other types of habitat in the city). Of the rodent species found infected, a fitted logistic regression of rodent infection by T. lewisi showed the lack of influence of season and rodent species and their characteristics such as sex and age, but showed the influence of locality and habitat (Table 2). Including the site of Bangkhen did not change the statistical results.
Table 2.
Comparison of models used to test the effect of several independent variables (locality, habitat, season, sex, age and species of rodents) on individual rodent infection (GLM with logit function) in eight sites in Thailand, Lao PDR and Cambodia. Models are ranked from the least to the most supported according to corrected AIC
| Model ranks | AIC |
|---|---|
| Locality + habitat | 322·9 |
| Locality + habitat + season | 324·6 |
| Locality + habitat + season + sex | 326·5 |
| Species | 327·5 |
| Species + season | 329·0 |
| Species + season + age | 330·8 |
| Locality + habitat + season + sex + age | 330·1 |
| Species + season + age + sex | 333·3 |
| Locality + habitat + season + sex + age + species | 335·6 |
| Locality + habitat + locality*habitat | 342·7 |
| Locality + habitat + season + sex + age + species + locality*species | 7589·1 |
GLM, Generalized Linear Model; AIC, Akaike's Information Criterion.
The rodents from a settlement habitat showed higher prevalence of infection than those from other habitats (Table 3). Although there was a significant effect of locality, only rodents from Loie showed higher prevalence of trypanosome infection than the rodents from Kanchanaburi (Table 3). Including Bangkhen again did not change the statistical patterns, although rodents from this site exhibited higher trypanosome infection than rodents of the other rural localities: Buriram, Loei, Kanchanaburi Nan (in Thailand), Pakse (Lao PDR) and Veal Renh (Cambodia) (not shown).
Table 3.
General Linear Model of rodent infection by Trypanosoma lewisi with binomial distribution and logit link function (Log-Likelihood Type 1 Test) at 8 sites in Thailand, Lao PDR and Cambodia. Selection of this best model using the AIC criterion (see Table 2). Analysis of Deviance Table (Type II tests), with locality and habitat as explanatory variables
| Explanatory variables | Slope estimate (P value) | Log ratio χ2 | Degree of freedom | Probability (>χ2) |
|---|---|---|---|---|
| Locality, Loei vs. | ||||
| Buriram | 1·61 (0·15) | |||
| Kanchanaburi | 2·53 (0·02) | |||
| Nan | 0·97 (0·41) | |||
| Luang Prabang | 1·86 (0·13) | |||
| Pakse | 1·63 (0·174) | |||
| Mondulkiri | −13·98 (0·98) | |||
| Veal Renh | 1·28 (0·22) | |||
| 18·11 | 7 | 0·01 | ||
| Habitat, forest vs. | ||||
| Rain-fed lands | 0·47 (0·63) | |||
| Non-flooded lands | 0·89 (0·27) | |||
| Settlement | 2·01 (<0·001) | |||
| 19·55 | 3 | 0·0002 |
There was no interaction effect of locality and habitat on rodent infection, and also no clear geographical pattern of infection (Fig. 1). An increase in rodent infections was observed from forest to settlement (Table 3). The results of the statistical model were similar when the urban site of Bangkhen was added (results not shown). Results suggest that rodent infection by T. lewisi increased with the increase in urbanization.
DISCUSSION
Direct microscopic observation of parasites in fresh blood and Giemsa-stained blood smears was only possible for T. lewisi infections, most likely due to low parasitaemia in the case of T. evansi infections, which could only be demonstrated by PCR with TBR primers. Sequencing of PCR products obtained with TRYP1 primers demonstrated the species specificity of the tests when 620 bp (T. lewisi) or 520 bp (T. evansi) were observed. Most of the other sized products sequenced proved to be rodent DNA or non-specific products, as previously reported by Milocco et al. [7].
Using LEW1 primers, with the exception of one 210 bp product obtained from a R. tanezumi sample, all the PCR products obtained were 220 bp, which were shown to be specific for T. lewisi by sequencing and BLAST. The 210 bp product observed in one sample found to be positive by microscopic observation was unfortunately of insufficient quantity for sequencing. Further studies will be necessary to identify this ‘T. lewisi-like’ parasite.
Rodents showed higher infection by T. lewisi than by T. evansi. This was as expected because (i) T. lewisi is a well tolerated parasite of rats, (ii) cyclical transmission by fleas is an efficient means of transmission compared to mechanical transmission (T. evansi), and (iii) fleas are common and abundant vectors in rats. On the other hand, a low prevalence of T. evansi was expected because it is mechanically transmitted and highly pathogenic for rodents [32, 33], which may limit its transmission in these animals. Moreover, Stomoxys spp. and tabanids, the major vectors of T. evansi, do not usually feed on rodents, thus other means of transmission such as contamination due to eating on a recently dead animal may lead to occasional infections of rodents by T. evansi.
In this study our results show trypanosome infections are more frequent in rodents from a domestic environment, and that Rattus and Bandicota species are therefore possible sources of animal and human exposure to these parasites.
Interestingly, a significant effect of habitat on T. lewisi infection was observed with an increase in human-dominated habitat (i.e. from forests to settlement). Stercorarian trypanosome infections were more frequent in rodents from a domestic environment. Although more investigations are needed, particularly on rodent ecology and habitat preferences, this suggests that human settlements represent potential risks for trypanosome infection in both humans and domestic animals. Synanthropic rodent species such as R. tanezumi and R. exulans trapped in human settlement habitats may be potential reservoirs for these parasites.
Our results challenge the study of Jittapalapong et al. [11], in which rodents trapped in a forest habitat in Thailand were more infected by stercorarian trypanosomes than rodents trapped in other habitats, including human settlements. However, the main difference between the two studies lies in the standardization of rodent sampling in the present study [24], which allowed the statistical investigation of the effects of habitat on rodent infection with minimum bias. The results presented here suggest that the increase in urbanization in rural habitats (linked to an increase in habitat fragmentation), which culminates in cities, may favour the spread of T. lewisi and increase risks of transmission to humans.
Supplementary Material
Supplementary information supplied by authors.
ACKNOWLEDGEMENTS
This study was funded by the French ANR Biodiversity grant ANR 07 BDIV 012 CERoPath project ‘Community Ecology of Rodents and their Pathogens in a Changing Environment’ (www.ceropath.org), the French ANR CP&ES grant ANR 11 CPEL 002 BiodivHealthSEA (Local Impacts and Perceptions of Global Changes: Biodiversity, Health and Zoonoses in Southeast Asia) (www.biodivhealthsea.org), and by the Thailand International Cooperation Agency (TICA). Special thanks go to all participants in field work for their great help.
We are grateful for the support of the network for coordination of information and research on Atypical Human Infection by Animal Trypanosomes (NAHIAT) created in May 2011 and coordinated by IRD and CIRAD with the support of FAO, OIE, WHO [contacts: Dr Philippe Truc (philippe.truc@ird.fr), and Dr Marc Desquesnes (marc.desquesnes@cirad.fr)].
We thank two anonymous referees and the Editor for their helpful comments.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268813002161.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Hoare CA. The Trypanosomes of Mammals. A Zoological Monograph. Oxford: Blackwell Scientific Publications, 1972, pp. 749. [Google Scholar]
- 2.Shrivastava KK, Shrivastava GP. Two cases of Trypanosoma (Herpetosoma) species infection of man in India. Transactions of Royal Society of Tropical Medicine and Hygiene 1974; 68: 143–144. [DOI] [PubMed] [Google Scholar]
- 3.Howie S, et al. A Gambian infant with fever and an unexpected blood film. PLoS Medicine 2006; 3: e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarataphan N, et al. Diagnosis of a Trypanosoma lewisi-like (Herpetosoma) infection in a sick infant from Thailand. Journal of Medical Microbiology 2007; 56: 1118–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joshi PP, et al. Treatment and follow-up of the first case of human trypanosomiasis caused by Trypanosoma evansi in India. Transactions of Royal Society of Tropical Medicine and Hygiene 2006; 100: 989–991. [DOI] [PubMed] [Google Scholar]
- 6.Shegokar VR, et al. Short report: human trypanosomiasis caused by Trypanosoma evansi in a village in India: preliminary serologic survey of the local population. American Journal of Tropical Medicine and Hygiene 2006; 75: 869–870. [PubMed] [Google Scholar]
- 7.Milocco C, et al. Molecular demonstration of Trypanosoma evansi and Trypanosoma lewisi DNA in wild rodents from Cambodia, Lao PDR and Thailand. Transboundary and Emerging Diseases 2012; 60: 17–26. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DE, Reeder DAM. Mammal Species of the World: A Taxonomic and Geographic Reference. Baltimore: Johns Hopkins University Press, 2005, pp. 2142. [Google Scholar]
- 9.Joshi PP, et al. Human trypanosomiasis caused by Trypanosoma evansi in India: the first case report. American Journal of Tropical Medicine and Hygiene 2005; 73: 491–495. [PubMed] [Google Scholar]
- 10.Haridy FM, et al. Trypanosoma evansi in dromedary camel: with a case report of zoonosis in greater Cairo, Egypt. Journal of the Egyptian Society of Parasitology 2011; 41: 65–76. [PubMed] [Google Scholar]
- 11.Jittapalapong S, et al. Molecular detection of divergent trypanosomes among rodents of Thailand. Infection Genetics and Evolution 2008; 8: 445–449. [DOI] [PubMed] [Google Scholar]
- 12.Herbreteau V, et al. Rodent-borne diseases in Thailand: targeting rodent carriers and risky habitats. Infection Ecology and Epidemiology 2012; 2: 18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordes F, et al. The diversity of microparasites of rodents: a comparative analysis that helps in identifying rodent-borne rich habitats in Southeast Asia. Infection Ecology and Epidemiology 2013; 3: 20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo PTK. The heamatocrit centrifuge technique for diagnosis of African trypanosomiasis. Acta Tropica 1970; 27: 384–386. [PubMed] [Google Scholar]
- 15.Milleliri JM, Tirandibaye HN, Nan-Madjoum B. The focus of human African trypanosomiasis in Moissala (Chad): prospective study of 16 villages using the direct card agglutination test (Testryp CATT) and the ion exchange mini-column (mAECT). Médecine Tropicale 1989; 49: 253–258. [PubMed] [Google Scholar]
- 16.Bajyana-Songa E, Hamers R. A card agglutination test (CATT) for veterinary use based on an early vat RoTat 1/2 of Trypanosoma evansi. Annales de la Société Belge de Médecine Tropicale 1988; 68: 233–240. [PubMed] [Google Scholar]
- 17.Desquesnes M, Bosseno MF, Breniere SF. Detection of Chagas infections using Trypanosoma evansi crude antigen demonstrates high cross-reactions with Trypanosoma cruzi. Infection Genetics and Evolution 2007; 7: 457–462. [DOI] [PubMed] [Google Scholar]
- 18.Desquesnes M, Dávila AMR. Applications of PCR-based tools for detection and identification of animal trypanosomes; a review and perspectives. Veterinary Parasitology 2002; 109: 213–231. [DOI] [PubMed] [Google Scholar]
- 19.Desquesnes M, et al. Detection and identification of Trypanosoma of African livestock through a single PCR based on internal transcribed spacer 1 of rDNA. International Journal for Parasitology 2001; 31: 610–614. [DOI] [PubMed] [Google Scholar]
- 20.Njiru ZK, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitology Research 2005; 95: 186–192. [DOI] [PubMed] [Google Scholar]
- 21.Rodrigues AC, et al. Phylogeny of Trypanosoma (Megatrypanum) theileri and related trypanosomes reveals lineages of isolates associated with artiodactyl hosts diverging on SSU and ITS ribosomal sequences. Parasitology 2006; 132: 215–224. [DOI] [PubMed] [Google Scholar]
- 22.Chaval Y, et al. A multi-approach survey as the most reliable tool to accurately assess biodiversity: the example of Thai murine rodents. Kasetart Journal (Natural Sciences) 2010; 44: 590–603. [Google Scholar]
- 23.Pagès M, et al. Revisiting the taxonomy of the Rattini tribe: a phylogeny-based delimitation of species boundaries. BMC Evolutionary Biology 2010; 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbreteau V, et al. Protocols for Field and Laboratory Rodent Studies. Bangkok: Kasetsart University Press, 2011, pp. 46. [Google Scholar]
- 25.Desquesnes M. Livestock Trypanosomoses and their Vectors in Latin America. Paris: CIRAD-EMVT Office International des Epizooties, 2004, pp. 174. [Google Scholar]
- 26.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual, 3rd edn. New York: Cold Spring Harbor Laboratory Press, 2001, pp. 2028. [Google Scholar]
- 27.Masiga DK, et al. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. International Journal of Parasitology 1992; 22: 909–918. [DOI] [PubMed] [Google Scholar]
- 28.Desquesnes M, Ravel S, Cuny G. PCR identification of Trypanosoma lewisi, a common parasite of laboratory rats. Kinetoplastid Biology and Disease, 2002; 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desquesnes M, et al. Specific primers for PCR amplification of the ITS1 (ribosomal DNA) of Trypanosoma lewisi. Infection Genetics and Evolution 2011; 11: 1361–1367. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: a language and environment for statistical computing, 2008. (http://cran.r-project.org).
- 31.Pagès M, et al. Cytonuclear discordance among Southeast Asian black rats (Rattus rattus complex). Molecular Ecology 2013; 22: 1019–1034. [DOI] [PubMed] [Google Scholar]
- 32.Singla N, Singla LD, Parshad VR. Potential of Trypanosoma evansi as a biocide of rodent pests. In: Singleton GR, et al. eds. Rats, Mice and People: Rodent Biology and Management. Canberra: Australian Centre for International Agricultural Research, 2003, pp. 43–46. [Google Scholar]
- 33.Singla LD, et al. Rodents as reservoirs of parasites in India. Integrative Zoology 2008; 3: 21–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information supplied by authors.
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268813002161.
click here to view supplementary material