Abstract
The COVID-19 pandemic caused by SARS-CoV-2 infections has led to excess deaths worldwide. Neutralizing antibodies (nAbs) against viral spike protein acquired from natural infections or vaccinations contribute to protection against new- and re-infections. Besides neutralization, antibody-mediated cellular cytotoxicity (ADCC) and phagocytosis (ADCP) are also important for viral clearance. However, due to the lack of convenient methods, the ADCC and ADCP responses elicited by viral infections or vaccinations remain to be explored. Here, we developed cell-based assays using target cells stably expressing SARS-CoV-2 spikes and Jurkat-NFAT-CD16a/CD32a effector cells for ADCC/ADCP measurements of monoclonal antibodies and human convalescent COVID-19 plasmas (HCPs). In control samples (n = 190), the specificity was 99.5% (95%CI: 98.4–100%) and 97.4% (95%CI: 95.1–99.6%) for the ADCC and ADCP assays, respectively. Among 87 COVID-19 HCPs, 83 (sensitivity: 95.4%, 95%CI: 91.0–99.8%) and 81 (sensitivity: 93.1%, 95%CI: 87.8–98.4%) showed detectable ADCC (titer range: 7.4–1721.6) and ADCP activities (titer range: 4–523.2). Notably, both ADCC and ADCP antibody titers positively correlated with the nAb titers in HCPs. In summary, we developed new tools for quantitative ADCC and ADCP analysis against SARS-CoV-2, which may facilitate further evaluations of Fc-mediated effector functions in preventing and treating against SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Antibody-mediated cellular cytotoxicity, Antibody-mediated phagocytosis, Cell-based reporter assay
1. Introduction
Coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) still continues and has caused 518 million cases and 6.3 million deaths as of May 13, 2022 (World Health Organization, 2022). The spike protein of SARS-CoV-2 mediates the viral cellular binding and entry via interaction with the ACE2 receptors. SARS-CoV-2 spike proteins contain the furin cleavage site (RRAR motif), which allows S1/S2 cleavage during virus packaging and is required for cell-to-cell virus transmission (Hoffmann et al., 2020). Immune-response against spike proteins, particularly for the neutralization antibodies (nAbs), essentially contribute to the protection acquired from vaccination and natural infection recovery (Baum et al., 2020, Chi et al., 2020, Feng et al., 2021, Gilbert et al., 2022, Khoury et al., 2021). In addition to the neutralization effects, spike-specific antibodies may also eliminate viruses or virus-infected cells via antibody-dependent cellular cytotoxicity (ADCC), phagocytosis (ADCP), or complement-dependent cytotoxicity (CDC) (Ji et al., 2019, Tay et al., 2019, Zohar and Alter, 2020).
So far, several assays for analysis activities of antibodies in neutralizing SARS-CoV-2 have been developed, including authentic virus-based cytopathogenic effect (CPE) inhibition or plaque reduction (PRNT) tests and pseudotyped lentivirus (LV) or vesicular stomatitis virus (VSV) based neutralization tests (Case et al., 2020, Manenti et al., 2020, Xie et al., 2020, Zhang et al., 2022). However, convenient and high-throughput methods for measurements of ADCC and ADCP activities in serum or plasma samples are still limited. The nuclear factor of activated T-cells (NFAT) signaling pathway is essential for Fcγ receptors (FcγRs) activation, which plays a critical role in ADCC and ADCP (Aramburu et al., 1995, Oh-hora and Rao, 2009); the NFAT-reporter systems are commonly used as surrogate ADCC or ADCP effectors. Human FcγRIIa (CD16a) or FcγRIIIa (CD32a)-expressing NFAT reporter cells are widely-used effector cells for ADCC or ADCP measurements for therapeutic monoclonal antibodies (mAbs) (Cao et al., 2020, Cheng et al., 2014, Liu et al., 2021, Parekh et al., 2012). In this study, we generated cells with stable expression of full-length SARS-CoV-2 spikes as target cells and developed NFAT-reporter cell-based assays for measurements of ADCC and ADCP activities of antibodies. The performance of new assays was systematically evaluated in several mAbs and human plasmas.
2. Materials and methods
2.1. Monoclonal antibodies
A total of 10 reported SARS-CoV-2 anti-spike mAbs were tested for ADCC and ADCP activities (Baum et al., 2020, Cerutti et al., 2021, Chi et al., 2020, Pinto et al., 2021, Sauer et al., 2021, Tortorici et al., 2020, Zhang et al., 2021). All mAbs were produced in ExpiCHO cells by transient transfection using a modified EIRBdMie vector (EIRBsMie-derived) containing dual promoters for human IgG light chain and heavy chain. The coding sequences of the variable regions of 4A8, NTD4–8, BD368–2, COVA2–15, S2M11, REGN10933, S2P6 were synthesized in the human IgG1 backbone (Generalbiol, Anhui, China) according to the published literature. The 36H6, 83H7, and 85F7 were developed in our lab, and their variable regions cDNA sequences were obtained via RT-PCR using mRNA isolated from the hybridoma cells, codon-optimized synthesized, and were cloned into the same EIRBdMie vector and human IgG1 backbone. Recombinant expressions of these mAbs were performed in ExpiCHO-S cells using the ExpiFectamine™ CHO transfection kit (Thermo Scientific, A29129) following previously described. Purified mAbs were obtained from cell culture supernatants using MabSelect SuRe (Cytiva). Among these mAbs, 2 (4A8 and NTD4–8) target the N-terminal domain (NTD), 1 (S2P6) recognizes S2 subunit, and the remaining ones are all RBD-targeting mAbs.
2.2. Human samples for assay validations
Human convalescent COVID-19 plasmas (HCPs, n = 87) were collected from recovered COVID-19 patients (one sample for each patient) from the Xinglin Branch of the First Affiliated Hospital of Xiamen University. All involved COVID-19 HCP samples were collected in the convalescent phase (≥14 days from illness onset or diagnosis, range 14–122 days) and showed detectable anti-SARS-CoV-2 RBD antibodies in a double-sandwich immunoassay (Wantai Biological Pharmacy). Among these samples, 70 were collected between 14 and 90 days (early convalescent phase), and 17 were collected between 91 and 122 days (later convalescent phase) since illness or diagnosis onset. For specificity evaluation, 190 stored plasma samples pre-dating the SARS-CoV-2 outbreak (collected in 2019) were used. All samples were heat-inactivated for pseudovirus (PsV) neutralization tests and ADCC/ADCP assays. Informed consent was obtained from each subject. The study was approved by the institutional review board of the School of Public Health of Xiamen University following the Declaration of Helsinki.
2.3. Target cells with stable expression of SARS-CoV-2 spike
The codon-optimized full-length spike cassette (SFLwt) of SARS-CoV-2 (referring to MN908947.3) was synthesized (Generalbiol, Anhui, China). A furin-site mutated spike cassette (SFLfk, changed RRAR to GSAS) was generated via site-directed mutagenesis based on SFLwt. Both SFLwt and SFLfk were cloned into the EIRBsMie vector, which is a modified PiggyBac vector with IRES-driven dual-selection markers of red-fluorescent protein (mRuby3) and Blasticidin S-resistance gene (Blasticidin S deaminase, Bsd). The Expi293F Cells (Thermo Scientific, A14528, 293 F) were co-transfected with the full-length spike expression vector (EIRBsMie-SFLwt or EIRBsMie-SFLfk) and a PiggyBac transposase expression vector (at a ratio of 4:1) using the PEI transfection system (BIOHUB, 78PEI25000–5 G). Then, blasticidin was supplied at 30 μg/mL in the culture medium for selections of cells with successful transfection since 24 h after transfection. Cells were cultured at 37 ℃ in a stackable CO2 (8%) incubator shaker (Kühner AG, SMX1503C). Subsequently, cells that were stably integrated with the transfected cassettes were obtained via continuous blasticidin selection and flow cytometry (FACS) cell sortings (mRuby3-activated).
2.4. Effector cells
The Jurkat-Lucia™ NFAT and Jurkat-Lucia™ NFAT-CD16 (hereafter designated Jurkat-NFAT-hCD16) cells were purchased from InvivoGen. Both cells were derived from the human Jurkat cell line by stable integration of an NFAT-inducible Lucia luciferase reporter (InvivoGen). The former is the parental reporter cell line and the latter having additional expressing of human Fc receptor CD16A (FcγRIIIA; V158 allotype). For the generation of Jurkat-NFAT-hCD32 cells, the parental Jurkat-NFAT cells were stably transduced with a lentiviral vectored human CD32A (FcγRIIA; H131 allotype) expression cassette. The CD32A cDNA is linked with an IRES-mRuby3-P2A-BsR cassette (synthesized by Generalbiol, Anhui, China) and cloned into the pLV-EF1α-MCS-IRES-Bsd vector. The presence of IRES-driven expression of mRuby3 and Bsd enable convenient positive-selections via blasticidin supplement and FACS-sorting. The Jurkat-NFAT-hCD16 and Jurkat-NFAT-hCD32 cells were cultured in SuperCulture serum-free lymphocyte medium (DAKEWEI) with Normocin (10 μg/mL), Blasticidin (10 μg/mL), and Zeocin (10 μg/mL) at 37 ℃ in a stackable CO2 (8%) incubator shaker.
2.5. FACS and western blot analyses
FACS analyses were used to determine the expression levels of human CD16 or CD32 in Jurkat-NFAT-hCD16 or Jurkat-NFAT-hCD32 cells. In brief, cells were incubated with PE/Cyanine7 anti-human CD16 antibody (BioLegend, 302016, for Jurkat-NFAT-CD16 cells) or FITC CD32/FCGR2/Fc gamma RII antibody (Sino Biological, 10374-MM01-F, for Jurkat-NFAT-hCD32) for 30 min at 4 ℃. After twice washes with PBS buffer, stained cells were subjected for FACS analyses using LSRFortessaX-20 (BD Biosciences). The cell lysates of 239 F, SFLwt, and SFLfk (1 ×105 cells) were prepared and subjected to western blots using HRP-conjugated mouse antibodies specific to the S1 subunit and S2 subunit (developed in our lab).
2.6. ADCC and ADCP assays
Antibodies or plasmas were pre-diluted in a reaction medium of IMDM medium containing 10% heat-inactivated FBS (Gibco, heating to 56 ℃ for 30 min). Aliquots (20 μL) of pre-diluted antibodies or plasmas were incubated with of SFLwt or SFLfk target cells (2 ×105 cells in 90 μL reaction medium) in 96-well plates for 1-hour at 37 ℃ in 5% CO2 incubator. Subsequently, Jurkat-NFAT-hCD16 or Jurkat-NFAT-hCD32 effector cells (2 ×105 cells in 90 μL) were added into the mixtures for further incubation at 37 ℃ in 5% CO2 incubator for 18-hour. Finally, 20 μL supernatants from co-cultures were mixed with 50 μL pre-diluted Coelenterazine substrate (Topscience, T6808, 10 mM, 1:4000 diluted with Opti-MEM medium) in a 96-well plate for immediate bioluminescence measurements using a multimode microplate reader (Spark 20 M, Tecan). The luciferase (Luc) induction fold (ADCC or ADCP Luc induction fold) was calculated by the fold change of relative light unit (RLU) values of the test well to control well (mock buffer reaction with target and effector cells).
2.7. Quantitative and statistical analyses
The four-parameter logistic (4-PL) model was used for fitting dose-dependent responses (ADCC or ADCP Luc induction fold) of antibodies or human plasma. The cut-off value for a positive response in ADCC or ADCP assay was defined as the Luc induction fold ≥ 2. For quantitative analyses, the parameter of EC2 (for mAbs) or ED2 (for plasma or serum samples), which were defined as the minimal concentration (EC2) or the maximal dilution fold (ED2), to achieve a positive reaction (Luc induction fold=2), was calculated following the 4-PL fitting equation. The Gaussian function was used for frequency distribution determination. Linear regression models and the Pearson coefficient were used for correlation analyses. All statistical tests and data visualization were conducted using GraphPad Prism (version 9.3.1).
3. Results
3.1. Characterizations of cells for ADCC and ADCP assays
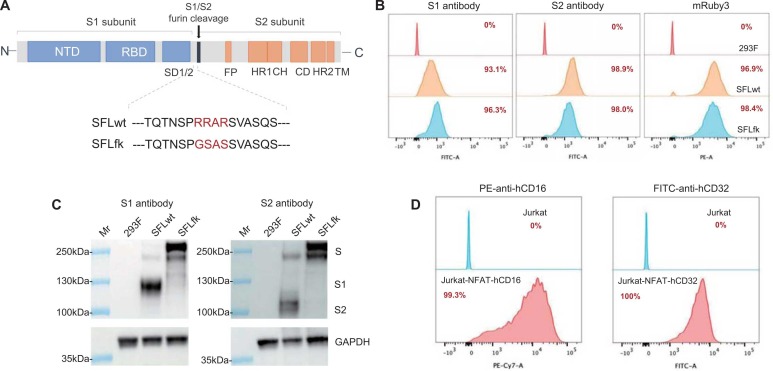
As the presence of the furin site, the SARS-CoV-2 spike may be cleaved by endogenous furin-like enzymes and result in the disassociation of S1 and S2 subunits. We designed two constructs, the SFLfk and SFLwt ( Fig. 1 A), to generate cell lines expressing SARS-CoV-2 spike proteins as target cells. Both constructs contain full-length (aa 1–1273) SARS-CoV-2 spike protein-expressing cassettes. Being different from the SFLwt, the furin site was mutated in the SFLfk. Two cell lines expressing spike proteins were developed via stable transfections of the SFLwt and SFLwt into 293 F cells. FACS analyses (Fig. 1B), for either staining with S1 or S2 antibodies, demonstrated the successful expressions of spike proteins on the two cell lines. Both cell lines presented an over 95% positive rate, as evidenced by the integrated mRuby3 expression and S2 antibody staining (Fig. 1B). Moreover, western blots revealed furin-cleaved spike derivates (furin-cleaved S1 and S2 subunits) were presented in SFLwt cell lines but not in SFLfk cell lines. In contrast, the intact spike protein levels appeared to be higher in SFLfk than that in SFLwt cell lines. These results supported the two cell lines could be used as target cells in ADCC or ADCP assays. For effectors, two Jurkat-derived NFAT reporter cells, Jurkat-NFAT-hCD16 and Jurkat-NFAT-hCD32, were used for ADCC and ADCP assays, respectively. The expressions of hCD16 or hCD32 receptors were evidenced by FACS analyses (Fig. 1D).
Fig. 1.
Characterizations of the target and effector cells for ADCC and ADCP assays against SARS-CoV-2 spike protein, (A). Schematics of constructs for generations of SFLwt and SFLfk cells. The SFLwt contains a codon-optimized gene encoding a full-length SARS-CoV-2 spike (aa 1–1273, identical to the spike protein derived from MN908947.3). The only difference between the SFLwt and SFLfk was the furin site is mutated (RRAR changed into GSAS). (B). Flow cytometry analyses of Expi293F (293 F) cells with stable expressions of SFLwt or SFLfk by antibodies specific to the S1 subunit (left panel), S2 subunit (middle panel), or mRuby3 selection marker (right panel). (C). Western blots for SARS-CoV-2 spike proteins in cell lysates of SFLwt and SFLwt cells. S, intact spike proteins; S1, furin-cleaved S1 subunit; S2, furin-cleaved S2 subunit. (D). Flow cytometry analyses for expressions of hCD16 and hCD32 in Jurkat-NFAT-hCD16 (left panel) and Jurkat-NFAT-hCD32 (right panel) effector cells, respectively.
3.2. Optimizations of experimental conditions for ADCC and ADCP assays
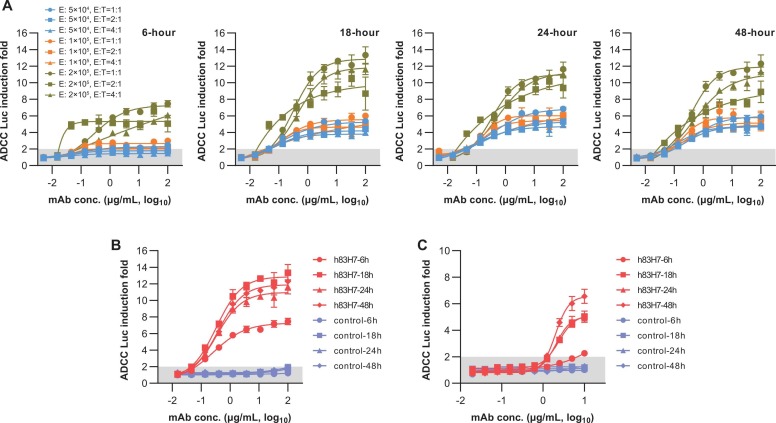
To explore the optimized experimental conditions, we used an RBD-targeting mAb h83H7 as a proof-concept positive control to test the effector cell density, the E:T (cell number ratio of effectors to targets) ratio, and the reaction time. The ADCC Luc fold inductions of 3-fold serial dilutions of h83H7 were tested with Jurkat-NFAT-hCD16 at various cell densities (5 ×104 cells/well, 1 ×105 cells/well, or 2 ×105 cells/well) in combination with SFLfk cells at different E:T ratios (1:1, 2:1, or 4:1). As shown in Fig. 2 A, Jurkat-NFAT-hCD16 using at 2 × 105 cells/well with the same density of target cell (1:1) resulted in higher Luc induction folds. At this condition, the h83H7 mAb generated a dose-dependent response with an upper plateau of 10–14 Luc induction folds when detected at 18-hour or later (Fig. 2B). In contrast, little or even no response was observed for an irrelevant control mAb (an HBsAg specific humanized mAb). In the same condition, h83H7 also exhibited dose-dependent ADCP activity in the Jurkat-NFAT-hCD32/SFLfk reporter system, though the effects appeared to be weaker than that in the ADCC test (Fig. 1 C). To shorten the turnaround time, bioluminescence detections in further experiments of this study were performed at 18-hour after co-cultures of the effector and target cells.
Fig. 2.
Optimizations of the experimental conditions for the ADCC and ADCP assay. (A) Comparisons of the antibody-induced bioluminescence increase using SFLfk (as target) and Jurkat-NFAT-hCD16 (as ADCC effector) cells at various experimental conditions. The 83H7 mAb in chimeric with human IgG1 Fc (h83H7) was used for ADCC assay in a dose-dependent manner. The Y-axis represents the bioluminescence increase (RLU fold changes to RLU of the mAb-free well) in the presence of antibodies. The Jurkat-NFAT-hCD16 were used at 5 × 104 (blue), 1 × 105 (orange), or 2 × 105 (yellow) cells/well in different E: T ratios. E, effector cell; E: T, the cell number ratio of effector and target cells; Bioluminescence (RLU) measurements were performed at 6, 18, 24, or 48 h post incubating cells and antibodies. (B) Dose-dependent response of the h83H7 and a control mAb in ADCC assay using Jurkat-NFAT-hCD16 effector at 2 × 105 cell/well with the same density of SFLfk. The data of h83H7 are identical with that in the corresponding conditions shown in (A). (C) Dose-dependent response of the h83H7 and a control mAb in ADCP assay using Jurkat-NFAT-hCD32 effector at 2 × 105 cell/well with the same density of SFLfk. Data in (A-C) are plotted as mean ± SD of 3 technical replicates. Dark shadows in (A-C) indicate a < 2-fold RLU increase.
3.3. Evaluations for the ADCC and ADCP activities of representative mAbs
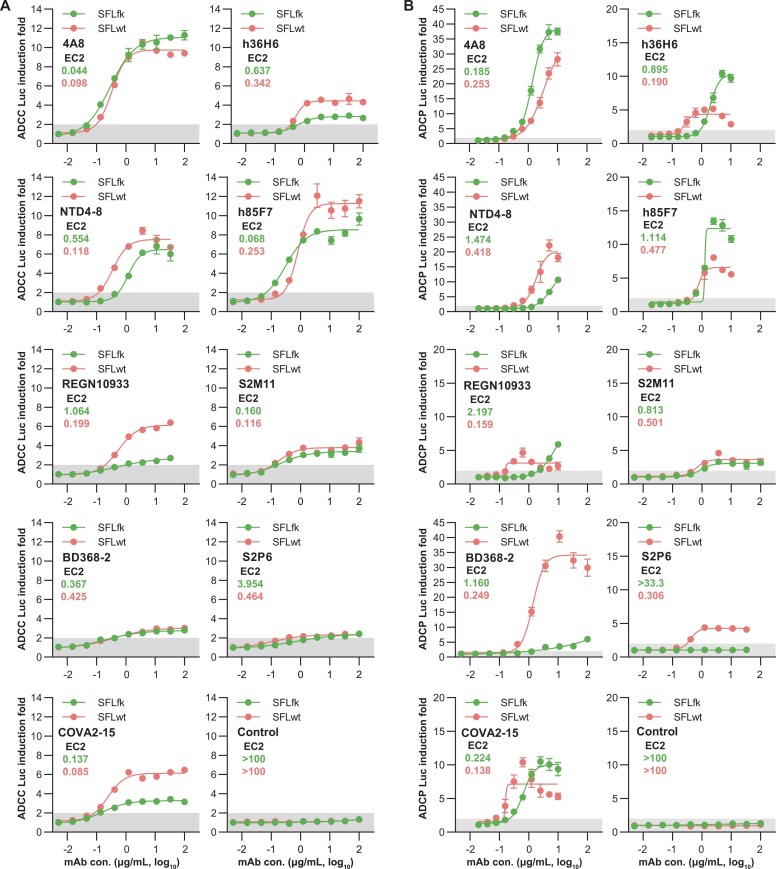
Besides the h83H7, additional nine mAbs were further tested for their activities in ADCC and ADCP assays. Among these mAbs, the 4 A8 and NTD4–8 are NTD-specific mAbs. The S2P6 recognizes the stem helix in the C-terminus (aa 1148–1152) of the S2 subunit. The remaining mAbs, including REGN10933, BD368–2, COVA2–15, h36H6, h85F7, and S2M11, are all RBD-targeting mAbs. All of these mAbs showed neutralization activities in previous studies. In ADCC and ADCP tests, most mAbs presented detectable and dose-dependent activities targeting either the SFLwt or SFLfk cells ( Fig. 3). To quantitative assessments of their potencies, the minimal concentrations to achieve 2 × Luc induction fold (EC2) of these mAbs were calculated using the 4-PL fitting curves. Overall, the EC2 values of these mAbs inversely correlated with their maximal Luc induction folds (values of the upper plateau in fitting curves) in either ADCC (R2 =0.448, p = 0.002) or ADCP (R2 =0.230, p = 0.044) assays (Fig. S1). However, some prozone effects were noted, particularly for the ADCP assay (Fig. 3B). Therefore, we considered the EC2 as the preferred parameter for quantitative comparisons of the potencies of mAbs in the two assays. In ADCC tests, the 4A8, BD238–2, COVA2–15, h36H6, h85F7, and S2M11 showed comparable potencies (<4-fold difference) in targeting the SFLwt and SFLfk cells. However, the NTD4–8, S2P6, and REGN10933 mAbs resulted in markedly higher activities when using the SFLwt as the target cells. In ADCP tests, the 4A8, NTD4–8, COVA2–15, h85F7, and S2M11 showed similar potencies (<4-fold difference) in targeting the SFLwt and SFLfk cells. However, the h36H6, BD368–2, S2P6, and REGN10933 mAbs showed higher activities (>4-fold difference) when using the SFLwt as the target cells. These data suggested the potential differences of some mAbs in mediating FcγR activation when targeting the furin-cleaved and uncleaved SARS-CoV-2 spikes, which may be attributed to their epitope difference. As the SFLwt cell line expressed the natural form of the SARS-CoV-2 spike and presented a better performance for reporter assays in mAbs tests, we selected it as the target cell in subsequent studies.
Fig. 3.
Dose-dependent activities of representative mAbs against SARS-CoV-2 spike in ADCC (A) and ADCP (B) assays. A total of 9 SARS-CoV-2 spike mAbs and 1 control mAb were tested for their ADCC and ADCP activities in a dose-dependent manner. All mAbs were produced in ExpiCHO cells in the same human-IgG1 backbone. The 4A8 and NTD4–8 are NTD-mAbs, the S2P6 is an S2-mAb, and the remaining RBD-targeting mAbs. The Jurkat-NFAT-hCD16 (A) or Jurkat-NFAT-hCD32 (B) were used at 2 × 105 cell/well and effectors combined with SFLwt (red) or SFLfk (green) target cells at the same density. Data are plotted as mean ± SD of 3 technical replicates. Dark shadows indicate a < 2-fold RLU increase. Con., concentration. The EC2 values (the minimal concentration to achieve 2 × Luc induction fold) are shown to represent the potencies of mAbs in ADCC or ADCP assays for targeting SFLwt (red) or SFLfk (green) cells.
3.4. Measurements of ADCC and ADCP activities in human convalescent plasmas
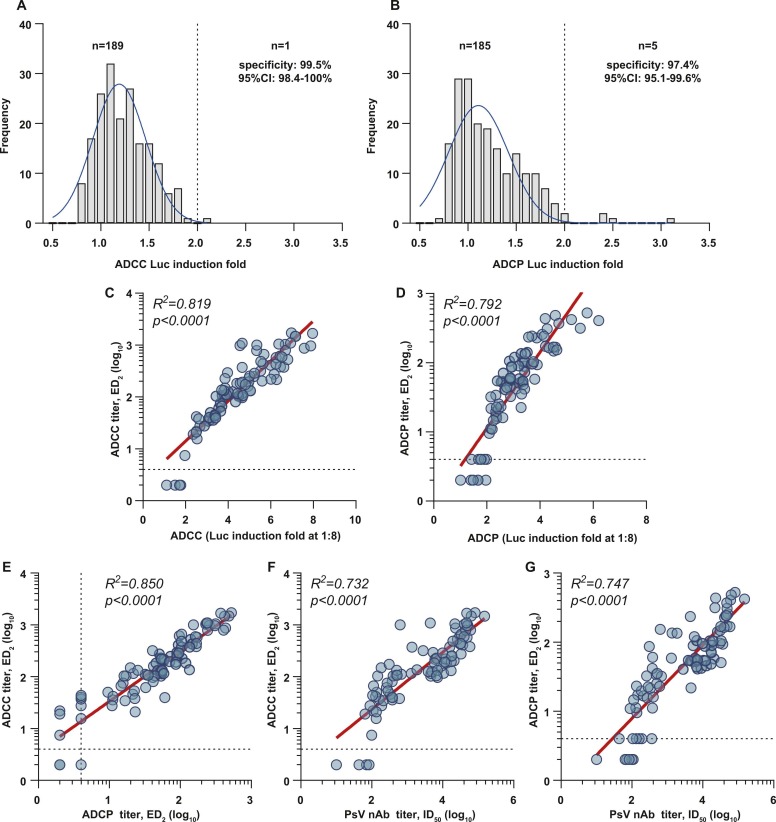
We further investigated the performance of two assays in detecting ADCC and ADCP activities mediated by polyclonal antibodies acquired from natural infections. First, we evaluated the specificities of the assays using 190 plasma samples collected from healthy individuals in the pre-pandemic era. As shown in Fig. 4A, 1 and 5 samples were positive (resulted in >2 Luc induction fold, at 1:4 dilution) in ADCC and ADCP tests, respectively. The specificity was 99.5% (95%CI: 98.4–100%) for the ADCC assay and was 97.4% (95%CI: 95.1–99.6%) for the ADCP assay. Of all 87 COVID-19 HCPs we tested, 83 and 81 samples showed positive reactions in ADCC and ADCP tests (at 1:4 dilution). The sensitivities were 95.4% (95%CI: 91.0–99.8%) and 93.1% (95%CI: 87.8–98.4%) for ADCC and ADCP assays, respectively. Detailedly, the sensitivities of the ADCC and ADCP assays were 97.1% (68/70) and 95.7% (67/70) among samples collected in the early convalescent phase; and were 88.2% (15/17) and 82.4% (14/17) among samples collected in the later convalescent phase.
Fig. 4.
Quantitative measurements of ADCC and ADCP activities of human plasma samples. Distributions of bioluminescence inductions of 190 samples (at 1:4 dilution) from pre-COVID-19 healthy populations in ADCC (A) and ADCP (B) assays. (C) Correlation analyses of the ADCC (C) or ADCP (D) titers (ED2, maximum dilution fold to achieve ≥2-fold induction determined by the 4-PL fitting curve) and bioluminescence induction folds (at 1:8 dilution) in detecting 87 HCP samples from COVID-19 patients. (E) Comparison of the ADCC and ADCP titers (ED2) of 87 HCPs. Correlations between the titers (Y-axis) of ADCC (F) or ADCP (G) of 87 HCPs with their neutralizing antibody titers (X-axis).
We further assessed the ADCC and ADCP antibody titers (ED2) of these HCPs via serial dilution measurements of each sample (Fig. S2, from 1:8–1:17496). These HCPs presented ADCC antibody titers ranging from 7.4 to 1721.6 (for 83 ADCC positive samples) and ADCP titers ranging from 4 to 523.2 (for 81 ADCP positive samples). Overall, the ADCC (Fig. 4 C, R2 =0.819) and ADCP (Fig. 4D, R2 =0.792) antibody titers of these HCPs positively correlated with their Luc induction fold measure at 1:8 dilution. Moreover, the antibody titers determined in the ADCC, ADCP, and PsV neutralization assays were significantly correlated (Fig. 4E to G).
4. Discussions
Antibodies against SARS-CoV-2 spike acquired from natural infections or vaccine immunization play antiviral roles through multiple functions, including the Fab-mediated neutralization and the Fc-mediated immune opsonization. Most previous studies focused on the neutralization functions of spike antibodies, which are essentially required for protection against COVID-19. Beyond neutralization, in vivo therapeutic studies revealed the Fc-mediated effector functions are also critical for viral control, as evidenced that the LALA Fc loss-function mutations significantly diminished the efficacity of spike antibodies to protect animals from lethal SARS-CoV-2 challenges (Ullah et al., 2021, Winkler et al., 2021). By contrast, the GAALIE mutated S309 mAb (VIR-7832) with enhanced Fc-effector function showed improved clearance of SARS-CoV-2 infected cells linking to boost T-cell immunity (Cathcart et al., 2022). Several studies revealed the presence of ADCC and ADCP activities of anti-spike antibodies in plasmas of COVID-19 patients (Adeniji et al., 2021, Natarajan et al., 2021, Tso et al., 2021), and inadequate ADCC or ADCP response appeared to be associated with severe disease and mortality (Bahnan et al., 2021, Brunet-Ratnasingham et al., 2021, Díez et al., 2021, Yu et al., 2021). Moreover, several ADCC assays for SARS-CoV-2 antibodies were established based on the determinations of targeted cell loss using NK cell lines or PBMC (Beaudoin-Bussières et al., 2021, Chen et al., 2021, Garvin et al., 2021). However, due to the lack of robust assays for quantitative assessments, current understandings regarding antibodies' ADCC and ADCP activities remain limited, particularly for their role in linkage with COVID-19 disease and vaccine protection efficacy.
The classical method for accurate ADCC quantification is the chromium-51 (51Cr) release assay, requiring specialized equipment and radioactive materials (Brier et al., 1975, Nelson et al., 2001). Besides, several non-radioactive methods were also developed based on the measurements of cell deaths of the target cells via released lactate dehydrogenase or fluorescent stains with various dyes (calcein-AM, BCECF, CSFE, or EuTPA) (Alrubayyi et al., 2018, Broussas et al., 2013, Gillissen et al., 2016, Gómez-Román et al., 2006, Kolber et al., 1988, Neri et al., 2001, Patel and Boyd, 1995, Roden et al., 1999). The pH-sensitive dyes, which preferentially accumulate in acidic organelles such as phagosomes and present pH-dependent fluorescence (Kamen et al., 2019, Miksa et al., 2009), are commonly used in ADCP assays. However, these methods require flow cytometers or high-resolution microscopes that limit their usage. The NFAT reporter system is an alternative approach to characterize the Fc-effector functions of antibodies via measurements on NFAT pathway activation, which is the downstream action of Fc engagement of Fc gamma receptors in both ADCC and ADCP processes. Here, we developed robust and convenient ADCC and ADCP assays using well-characterized target cells that stably expressed SARS-CoV-2 spike and Jurkat-NFAT-CD16a/CD32a effector cells. We also establish methods and indexes for quantitatively analyzing the potencies of mAbs (Fig. 3) as well as the ADCC and ADCP antibody titers in human plasmas (Fig. 4). As the targets for assays, the SFLwt cells exhibited better performance than SFLfk in some tested mAbs though there was no marked difference noted in spike expressing levels for the two cells. It is possible that partial furin cleavage of the spike trimer may facilitate the engagement of IgG-Fc and Fc receptors. On the other hand, although all test mAbs were expressed with the same human-IgG1 backbone, their activities in ADCC and ADCP assays appeared to be distinctly varied, possibly attributed to the affinity difference or epitope diversity.
The two assays developed in our study can also be used in detecting the antibody titers of ADCC and ADCP in human plasmas. Our data demonstrated the specificities of the assays in SARS-CoV-2 native populations (Fig. 4 A). Most of the HCP samples from recovered COVID-19 patients presented detectable antibodies with ADCC and ADCP activities, showing titers correlating with that determined in neutralization tests (Fig. 4). As the nAb level is the well-documented immune marker inversely associated with COVID-19 risk and directly associated with vaccine efficacy (Garcia-Beltran et al., 2021, Khoury et al., 2021), it is reasonable to assume the ADCC and/or ADCP antibody titers may also serve as surrogates of anti-SARS-CoV-2 immunity. However, further studies using the two assays to explore the significance and clinical value in vaccinated populations are certainly required.
In summary, our study developed cell-based reporter assays to evaluate the ADCC and ADCP activities of the mAbs and human blood samples against the SARS-CoV-2 spike. The good performance of the two assays as documented by our data supports their further applications both in basic and clinical studies.
CRediT authorship contribution statement
Yuting Hong: Investigation, Methodology, Data curation, Formal analysis, Writing – original draft. Huilin Guo: Investigation, Methodology, Data curation, Validation. Min Wei: Investigation, Methodology. Yali Zhang: Methodology, Validation. Mujin Fang: Visualization. Tong Cheng: Writing – review & editing. Zhiyong Li: Resources. Shengxiang Ge: Formal analysis, Supervision. Xiangyang Yao: Resources, Conceptualization, Writing – review & editing, Funding acquisition. Quan Yuan: Conceptualization, Project administration, Writing – review & editing, Funding acquisition. Ningshao Xia: Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This project was supported by grants of the Science and Technology Major Project (special for COVID-19 prevention and control) of Fujian province (2021Y0103) and Xiamen city (3502Z2021YJ013) in China.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2022.114564.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available on request.
References
- Adeniji O.S., Giron L.B., Purwar M., Zilberstein N.F., Kulkarni A.J., Shaikh M.W., Balk R.A., Moy J.N., Forsyth C.B., Liu Q., Dweep H., Kossenkov A., Weiner D.B., Keshavarzian A., Landay A., Abdel-Mohsen M. COVID-19 severity is associated with differential antibody fc-mediated innate immune functions. mBio. 2021;12 doi: 10.1128/mBio.00281-21. e00281-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrubayyi A., Schuetz A., Lal K.G., Jongrakthaitae S., Paolino K.M., Ake J.A., Robb M.L., de Souza M.S., Michael N.L., Paquin-Proulx D., Eller M.A. A flow cytometry based assay that simultaneously measures cytotoxicity and monocyte mediated antibody dependent effector activity. J. Immunol. Methods. 2018;462:74–82. doi: 10.1016/j.jim.2018.08.012. [DOI] [PubMed] [Google Scholar]
- Aramburu J., Azzoni L., Rao A., Perussia B. Activation and expression of the nuclear factors of activated T cells, NFATp and NFATc, in human natural killer cells: regulation upon CD16 ligand binding. J. Exp. Med. 1995;182:801–810. doi: 10.1084/jem.182.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahnan W., Wrighton S., Sundwall M., Bläckberg A., Larsson O., Höglund U., Khakzad H., Godzwon M., Walle M., Elder E., Strand A.S., Happonen L., André O., Ahnlide J.K., Hellmark T., Wendel-Hansen V., Wallin R.P., Malmstöm J., Malmström L., Ohlin M., Rasmussen M., Nordenfelt P. Spike-dependent opsonization indicates both dose-dependent inhibition of phagocytosis and that non-neutralizing antibodies can confer protection to SARS-CoV-2. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.808932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., Atwal G.S., Oyejide A., Goez-Gazi Y., Dutton J., Clemmons E., Staples H.M., Bartley C., Klaffke B., Alfson K., Gazi M., Gonzalez O., Dick E., Jr., Carrion R., Jr., Pessaint L., Porto M., Cook A., Brown R., Ali V., Greenhouse J., Taylor T., Andersen H., Lewis M.G., Stahl N., Murphy A.J., Yancopoulos G.D., Kyratsous C.A. Vol. 370. 2020. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters; pp. 1110–1115. (Science). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G., Richard J., Prévost J., Goyette G., Finzi A. A new flow cytometry assay to measure antibody-dependent cellular cytotoxicity against SARS-CoV-2 Spike-expressing cells. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier A.M., Chess L., Schlossman S.F. Human antibody-dependent cellular cytotoxicity. Isolation and identification of a subpopulation of peripheral blood lymphocytes which kill antibody-coated autologous target cells. J. Clin. Invest. 1975;56:1580–1586. doi: 10.1172/JCI108240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussas M., Broyer L., Goetsch L. Evaluation of antibody-dependent cell cytotoxicity using lactate dehydrogenase (LDH) measurement. Methods Mol. Biol. 2013;988:305–317. doi: 10.1007/978-1-62703-327-5_19. [DOI] [PubMed] [Google Scholar]
- Brunet-Ratnasingham E., Anand S.P., Gantner P., Dyachenko A., Moquin-Beaudry G., Brassard N., Beaudoin-Bussières G., Pagliuzza A., Gasser R., Benlarbi M., Point F., Prévost J., Laumaea A., Niessl J., Nayrac M., Sannier G., Orban C., Messier-Peet M., Butler-Laporte G., Morrison D.R., Zhou S., Nakanishi T., Boutin M., Descôteaux-Dinelle J., Gendron-Lepage G., Goyette G., Bourassa C., Medjahed H., Laurent L., Rébillard R.M., Richard J., Dubé M., Fromentin R., Arbour N., Prat A., Larochelle C., Durand M., Richards J.B., Chassé M., Tétreault M., Chomont N., Finzi A., Kaufmann D.E. Integrated immunovirological profiling validates plasma SARS-CoV-2 RNA as an early predictor of COVID-19 mortality. Sci. Adv. 2021;7:eabj5629. doi: 10.1126/sciadv.abj5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wang L., Yu C., Wang K., Wang W., Yan J., Li Y., Yang Y., Wang X., Wang J. Development of an antibody-dependent cellular cytotoxicity reporter assay for measuring anti-Middle East respiratory syndrome antibody bioactivity. Sci. Rep. 2020;10:16615. doi: 10.1038/s41598-020-73960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., Ilagan M.X.G., Tartell M.A., Amarasinghe G., Henderson J.P., Miersch S., Ustav M., Sidhu S., Virgin H.W., Wang D., Ding S., Corti D., Theel E.S., Fremont D.H., Diamond M.S., Whelan S.P.J. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485. doi: 10.1016/j.chom.2020.06.021. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart, A.L., Havenar-Daughton, C., Lempp, F.A., Ma, D., Schmid, M.A., Agostini, M.L., Guarino, B., Di iulio, J., Rosen, L.E., Tucker, H., Dillen, J., Subramanian, S., Sloan, B., Bianchi, S., Pinto, D., Saliba, C., Culap, K., Wojcechowskyj, J.A., Noack, J., Zhou, J., Kaiser, H., Chase, A., Montiel-Ruiz, M., Dellota, E., Park, A., Spreafico, R., Sahakyan, A., Lauron, E.J., Czudnochowski, N., Cameroni, E., Ledoux, S., Werts, A., Colas, C., Soriaga, L., Telenti, A., Purcell, L.A., Hwang, S., Snell, G., Virgin, H.W., Corti, D. and Hebner, C.M., 2022. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv, 2021.03.09.434607.
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., Huang Y., Katsamba P.S., Liu L., Nair M.S., Rawi R., Olia A.S., Wang P., Zhang B., Chuang G.Y., Ho D.D., Sheng Z., Kwong P.D., Shapiro L. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833. doi: 10.1016/j.chom.2021.03.005. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Rostad C.A., Anderson L.J., Sun H.Y., Lapp S.A., Stephens K., Hussaini L., Gibson T., Rouphael N., Anderson E.J. The development and kinetics of functional antibody-dependent cell-mediated cytotoxicity (ADCC) to SARS-CoV-2 spike protein. Virology. 2021;559:1–9. doi: 10.1016/j.virol.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Garvin D., Paguio A., Moravec R., Engel L., Fan F., Surowy T. Development of a robust reporter-based ADCC assay with frozen, thaw-and-use cells to measure Fc effector function of therapeutic antibodies. J. Immunol. Methods. 2014;414:69–81. doi: 10.1016/j.jim.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez J.M., Romero C., Cruz M., Vandeberg P., Merritt W.K., Pradenas E., Trinité B., Blanco J., Clotet B., Willis T., Gajardo R. Anti-SARS-CoV-2 hyperimmune globulin demonstrates potent neutralization and antibody-dependent cellular cytotoxicity and phagocytosis through N and S proteins. J. Infect. Dis. 2021;225(6):938–946. doi: 10.1093/infdis/jiab540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., Humphries H.E., Jepson B., Kelly E.J., Plested E., Shoemaker K., Thomas K.M., Vekemans J., Villafana T.L., Lambe T., Pollard A.J., Voysey M. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., Murali M.R., Alter G., Charles R.C., Dighe A., Branda J.A., Lennerz J.K., Lingwood D., Schmidt A.G., Iafrate A.J., Balazs A.B. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488. doi: 10.1016/j.cell.2020.12.015. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin D., Stecha P., Gilden J., Wang J., Grailer J., Hartnett J., Fan F., Cong M., Cheng Z.J. Determining ADCC activity of antibody-based therapeutic molecules using two bioluminescent reporter-based bioassays. Curr. Protoc. 2021;1 doi: 10.1002/cpz1.296. [DOI] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., Castellino F., Flach B., Lin B.C., O'Connell S., McDanal C., Eaton A., Sarzotti-Kelsoe M., Lu Y., Yu C., Borate B., van der Laan L.W.P., Hejazi N.S., Huynh C., Miller J., El Sahly H.M., Baden L.R., Baron M., De La Cruz L., Gay C., Kalams S., Kelley C.F., Andrasik M.P., Kublin J.G., Corey L., Neuzil K.M., Carpp L.N., Pajon R., Follmann D., Donis R.O., Koup R.A. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen M.A., Yasuda E., de Jong G., Levie S.E., Go D., Spits H., van Helden P.M., Hazenberg M.D. The modified FACS calcein AM retention assay: a high throughput flow cytometer based method to measure cytotoxicity. J. Immunol. Methods. 2016;434:16–23. doi: 10.1016/j.jim.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Gómez-Román V.R., Florese R.H., Patterson L.J., Peng B., Venzon D., Aldrich K., Robert-Guroff M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Lang J., Ning B., Qi F., Wang H., Zhang Y., Zhao R., Yang X., Zhang L., Li W., Shi X., Qin Z., Zhao Y., Nie G. Enhanced Natural Killer Cell Immunotherapy by Rationally Assembling Fc Fragments of Antibodies onto Tumor Membranes. Adv. Mater. 2019;31 doi: 10.1002/adma.201804395. [DOI] [PubMed] [Google Scholar]
- Kamen L., Myneni S., Langsdorf C., Kho E., Ordonia B., Thakurta T., Zheng K., Song A., Chung S. A novel method for determining antibody-dependent cellular phagocytosis. J. Immunol. Methods. 2019;468:55–60. doi: 10.1016/j.jim.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Kolber M.A., Quinones R.R., Gress R.E., Henkart P.A. Measurement of cytotoxicity by target cell release and retention of the fluorescent dye bis-carboxyethyl-carboxyfluorescein (BCECF) J. Immunol. Methods. 1988;108:255–264. doi: 10.1016/0022-1759(88)90427-9. [DOI] [PubMed] [Google Scholar]
- Liu C., Yu C., Yang Y., Huang J., Yu X., Duan M., Wang L., Wang J. Development of a novel reporter gene assay to evaluate antibody-dependent cellular phagocytosis for anti-CD20 therapeutic antibodies. Int Immunopharmacol. 2021;100 doi: 10.1016/j.intimp.2021.108112. [DOI] [PubMed] [Google Scholar]
- Manenti A., Maggetti M., Casa E., Martinuzzi D., Torelli A., Trombetta C.M., Marchi S., Montomoli E. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J. Med Virol. 2020;92:2096–2104. doi: 10.1002/jmv.25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksa M., Komura H., Wu R., Shah K.G., Wang P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods. 2009;342:71–77. doi: 10.1016/j.jim.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan H., Crowley A.R., Butler S.E., Xu S., Weiner J.A., Bloch E.M., Littlefield K., Wieland-Alter W., Connor R.I., Wright P.F., Benner S.E., Bonny T.S., Laeyendecker O., Sullivan D., Shoham S., Quinn T.C., Larman H.B., Casadevall A., Pekosz A., Redd A.D., Tobian A.A.R., Ackerman M.E. Markers of polyfunctional SARS-CoV-2 antibodies in convalescent plasma. mBio. 2021;12 doi: 10.1128/mBio.00765-21. e00765-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.L., Kurman, C.C. and Serbousek, D.E., 2001. 51Cr release assay of antibody-dependent cell-mediated cytotoxicity (ADCC). Curr Protoc Immunol Chapter 7, Unit 7.27. [DOI] [PubMed]
- Neri S., Mariani E., Meneghetti A., Cattini L., Facchini A. Calcein-acetyoxymethyl cytotoxicity assay: standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin. Diagn. Lab Immunol. 2001;8:1131–1135. doi: 10.1128/CDLI.8.6.1131-1135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-hora M., Rao A. The calcium/NFAT pathway: role in development and function of regulatory T cells. Microbes Infect. 2009;11:612–619. doi: 10.1016/j.micinf.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh B.S., Berger E., Sibley S., Cahya S., Xiao L., LaCerte M.A., Vaillancourt P., Wooden S., Gately D. Development and validation of an antibody-dependent cell-mediated cytotoxicity-reporter gene assay. MAbs. 2012;4:310–318. doi: 10.4161/mabs.19873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.K., Boyd P.N. An improved assay for antibody dependent cellular cytotoxicity based on time resolved fluorometry. J. Immunol. Methods. 1995;184:29–38. doi: 10.1016/0022-1759(95)00071-h. [DOI] [PubMed] [Google Scholar]
- Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., Rosen L.E., di Iulio J., Jerak J., Kaiser H., Islam S., Jaconi S., Sprugasci N., Culap K., Abdelnabi R., Foo C., Coelmont L., Bartha I., Bianchi S., Silacci-Fregni C., Bassi J., Marzi R., Vetti E., Cassotta A., Ceschi A., Ferrari P., Cippà P.E., Giannini O., Ceruti S., Garzoni C., Riva A., Benigni F., Cameroni E., Piccoli L., Pizzuto M.S., Smithey M., Hong D., Telenti A., Lempp F.A., Neyts J., Havenar-Daughton C., Lanzavecchia A., Sallusto F., Snell G., Virgin H.W., Beltramello M., Corti D., Veesler D. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden M.M., Lee K.H., Panelli M.C., Marincola F.M. A novel cytolysis assay using fluorescent labeling and quantitative fluorescent scanning technology. J. Immunol. Methods. 1999;226:29–41. doi: 10.1016/s0022-1759(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Sauer M.M., Tortorici M.A., Park Y.J., Walls A.C., Homad L., Acton O.J., Bowen J.E., Wang C., Xiong X., de van der Schueren W., Quispe J., Hoffstrom B.G., Bosch B.J., McGuire A.T., Veesler D. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Wiehe K., Pollara J. Antibody-dependent cellular phagocytosis in antiviral immune responses. Front Immunol. 2019;10:332. doi: 10.3389/fimmu.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., De Marco A., Guarino B., Bianchi S., Lauron E.J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J.A., Case J.B., Chen R.E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S.C., Schmid M.A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M.S., Neyts J., Diamond M.S., Virgin H.W., Snell G., Corti D., Fink K., Veesler D. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso F.Y., Lidenge S.J., Poppe L.K., Peña P.B., Privatt S.R., Bennett S.J., Ngowi J.R., Mwaiselage J., Belshan M., Siedlik J.A., Raine M.A., Ochoa J.B., Garcia-Diaz J., Nossaman B., Buckner L., Roberts W.M., Dean M.J., Ochoa A.C., West J.T., Wood C. Presence of antibody-dependent cellular cytotoxicity (ADCC) against SARS-CoV-2 in COVID-19 plasma. PLoS One. 2021;16 doi: 10.1371/journal.pone.0247640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., Gasser R., Fink C., Chen Y., Tauzin A., Goyette G., Bourassa C., Medjahed H., Mack M., Chung K., Wilen C.B., Dekaban G.A., Dikeakos J.D., Bruce E.A., Kaufmann D.E., Stamatatos L., McGuire A.T., Richard J., Pazgier M., Bjorkman P.J., Mothes W., Finzi A., Kumar P., Uchil P.D. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158. doi: 10.1016/j.immuni.2021.08.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2022. Corona virus disease (COVID-19) pandemic. https://covid19.who.int; WHO, Geneva, Switzerland. [Internet] Accessed 13 May 2022.
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., Case J.B., Sutton R.E., Carnahan R.H., Darling T.L., Boon A.C.M., Mack M., Head R.D., Ross T.M., Crowe J.E., Jr., Diamond M.S. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820. doi: 10.1016/j.cell.2021.02.026. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., Tseng C.K., Makino S., Menachery V.D., Bilello J.P., Shi P.Y. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang M., Zhang X., Li S., Lu Q., Zeng H., Hou H., Li H., Zhang M., Jiang F., Wu J., Ding R., Zhou Z., Liu M., Si W., Zhu T., Li H., Ma J., Gu Y., She G., Li X., Zhang Y., Peng K., Huang W., Liu W., Wang Y. Antibody-dependent cellular cytotoxicity response to SARS-CoV-2 in COVID-19 patients. Signal Transduct. Target Ther. 2021;6:346. doi: 10.1038/s41392-021-00759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang S., Wu Y., Hou W., Yuan L., Shen C., Wang J., Ye J., Zheng Q., Ma J., Xu J., Wei M., Li Z., Nian S., Xiong H., Zhang L., Shi Y., Fu B., Cao J., Yang C., Li Z., Yang T., Liu L., Yu H., Hu J., Ge S., Chen Y., Zhang T., Zhang J., Cheng T., Yuan Q., Xia N. Virus-free and live-cell visualizing SARS-CoV-2 cell entry for studies of neutralizing antibodies and compound inhibitors. Small Methods. 2021;5 doi: 10.1002/smtd.202001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei M., Wu Y., Wang J., Hong Y., Huang Y., Yuan L., Ma J., Wang K., Wang S., Shi Y., Wang Z., Guo H., Xiao J., Yang C., Ye J., Chen J., Liu Y., Fu B., Lan M., Gong P., Huang Z., Su Y., Chen Y., Zhang T., Zhang J., Zhu H., Yu H., Yuan Q., Cheng T., Guan Y., Xia N. Cross-species tropism and antigenic landscapes of circulating SARS-CoV-2 variants. Cell Rep. 2022 doi: 10.1016/j.celrep.2022.110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.