Abstract
Brachyspira (Serpulina) hyodysenteriae, the etiologic agent of swine dysentery, uses the enzyme NADH oxidase to consume oxygen. To investigate possible roles for NADH oxidase in the growth and virulence of this anaerobic spirochete, mutant strains deficient in oxidase activity were isolated and characterized. The cloned NADH oxidase gene (nox; GenBank accession no. U19610) on plasmid pER218 was inactivated by replacing 321 bp of coding sequence with either a gene for chloramphenicol resistance (cat) or a gene for kanamycin resistance (kan). The resulting plasmids, respectively, pCmΔNOX and pKmΔNOX, were used to transform wild-type B. hyodysenteriae B204 cells and generate the antibiotic-resistant strains Nox-Cm and Nox-Km. PCR and Southern hybridization analyses indicated that the chromosomal wild-type nox genes in these strains had been replaced, through allelic exchange, by the inactivated nox gene containing cat or kan. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblot analysis revealed that both nox mutant cell lysates were missing the 48-kDa Nox protein. Soluble NADH oxidase activity levels in cell lysates of Nox-Cm and Nox-Km were reduced 92 to 96% compared to the activity level in parent strain B204. In an aerotolerance test, cells of both nox mutants were at least 100-fold more sensitive to oxygen exposure than were cells of the wild-type parent strain B204. In swine experimental infections, both nox mutants were less virulent than strain B204 in that fewer animals were colonized by the mutant cells and infected animals displayed mild, transient signs of disease, with no deaths. These results provide evidence that NADH oxidase serves to protect B. hyodysenteriae cells against oxygen toxicity and that the enzyme, in that role, contributes to the pathogenic ability of the spirochete.
Brachyspira (Serpulina) hyodysenteriae cells colonize the oxygen-respiring mucosal tissues of the swine cecum and colon. During the early stages of swine dysentery, cells of this spirochete are visible first along the intestinal epithelium and then among epithelial cells and within goblet cells (7, 20). As the disease progresses, lesions appear in the mucosa at sites of spirochete colonization and host blood passes from underlying capillaries into the intestinal lumen through the lesions (7, 17). For the most part, bacterial characteristics essential for B. hyodysenteriae colonization and pathogenesis have not been thoroughly investigated, although there is evidence that hemolytic activity (15, 48) and bacterial motility and chemotaxis (18, 26, 32) are important contributing factors.
B. hyodysenteriae is an aerotolerant anaerobe. Cells of this spirochete grow beneath a culture atmosphere containing 1% O2–99% N2 and consume substrate amounts of oxygen (46). A major mechanism for oxygen metabolism by B. hyodysenteriae and other Brachyspira species is NADH oxidase, based on the high specific activities of the enzyme in soluble (membrane-free) cell fractions of the spirochetes (40, 43). The purified NADH oxidase of B. hyodysenteriae B204 is a flavin adenine dinucleotide-dependent, monomeric protein with an apparent molecular mass, based on gel migration, of 47 to 48 kDa (45). The enzyme carries out a four-electron reduction of oxygen, yielding water. The gene for the B. hyodysenteriae NADH oxidase has been cloned (47).
NADH oxidase has been viewed as a mechanism by which B. hyodysenteriae cells either contend with oxygen (as an antioxidant defense mechanism) or take advantage of oxygen (as an alternative NADH-regenerating pathway) in their native habitat, the oxygen-respiring tissues of the swine intestinal tract (40, 41). The enzyme may be important in early stages of the disease when cells first populate mucosal tissues or in later stages when oxygen-carrying erythrocytes enter the spirochete habitat and are possibly lysed by the B. hyodysenteriae hemolysin. Additionally, NADH oxidase may protect cells from oxygen exposure during fecal-oral passage between hosts. In any of these roles, NADH oxidase would likely contribute to the virulence of B. hyodysenteriae.
The objectives of this study were threefold: first, to inactivate the NADH oxidase (nox) gene and produce B. hyodysenteriae mutant strains deficient in NADH oxidase activity; second, to use those nox mutants to investigate a possible role for NADH oxidase in the growth and oxygen sensitivity of B. hyodysenteriae; and third, to determine whether the loss of NADH oxidase affects the virulence of this mucosal pathogen for its host species, swine.
MATERIALS AND METHODS
Strains and culture conditions.
B. hyodysenteriae B204 is a virulent strain commonly used in experimental infections of swine in the United States. Cells were routinely cultured, with stirring, in BHIS broth (Difco brain heart infusion broth containing 10% [vol/vol] heat-treated calf serum) beneath an initial culture atmosphere of 1% O2–99% N2 (40, 42). Trypticase soy blood (TSB) agar medium was made by adding defibrinated bovine blood (final concentration, 5% [vol/vol]) to sterile, melted Trypticase soy agar medium containing glucose (BBL, Becton Dickinson, Cockeysville, Md.), and then the medium was poured into petri plates. Agar plates were prepared and stored in an air atmosphere until use. After inoculation, agar plate cultures were incubated in a Coy anaerobe chamber inflated with 5% H2, 10% CO2, and 85% N2.
B. hyodysenteriae Nox-Cm and Nox-Km were derived from strain B204. Cells of these NADH oxidase mutant strains were routinely cultured anaerobically in BHIS broth or on TSB agar plates containing chloramphenicol (final concentration, 10 μg/ml [wt/vol]) or kanamycin (final concentration, 200 to 400 μg/ml [wt/vol]). Cells of the nox mutants used in physiology and virulence experiments were cultured in antibiotic-free media. All antibiotics were purchased from Sigma Chemical Company, St. Louis, Mo.
Strain EPC (stands for electroporation and passage control) was subcultured from B204 cells that had been electroporated without DNA and cultured on medium without antibiotics. Inasmuch as there were no detectable differences between EPC cells and wild-type B204 cells in growth characteristics, genotypic properties, and virulence for swine in separate studies, data is not presented for that strain.
Construction of nox gene mutations.
As described below, an antibiotic resistance gene (cat or kan) was inserted into the cloned nox gene and this constructed gene was used to mutagenize B. hyodysenteriae B204 cells through allelic exchange, that is, by replacing the wild-type gene with the constructed defective gene (Fig. 1). Allelic exchange has been used to mutate two B. hyodysenteriae flagellar genes singly (31) or in combination (32) and a gene that confers hemolytic activity when it is cloned into Escherichia coli (48).
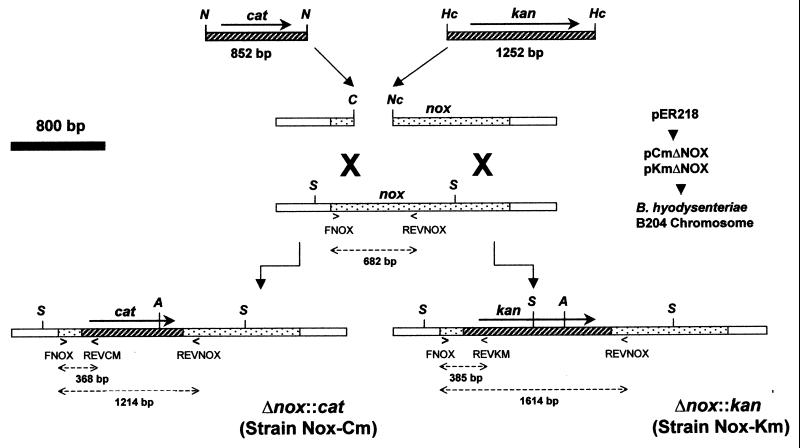
FIG. 1.
Construction and use of plasmids to insertionally inactivate the nox gene of B. hyodysenteriae B204 cells by allelic exchange. The nox gene cloned in plasmid pER218 was inactivated by replacing a 321-bp portion (ClaI-NcoI fragment) of the gene with either a chloramphenicol resistance (cat) or a kanamycin resistance (kan) gene to yield, respectively, plasmids pCmΔNOX and pKmΔNOX. B. hyodysenteriae B204 cells were transformed by electroporation with either pCmΔNOX or pKmΔNOX. Allelic exchange (indicated by large X’s) occurred between the plasmid nox gene and the chromosomal nox gene. B. hyodysenteriae resistant strains in which the mutated nox gene had recombined with the nox chromosomal locus were selected by plating on antibiotic-containing media. Two strains, Nox-Cm and Nox-Km, were chosen for characterization in these investigations. Gene sites complementary to the PCR primers FNOX, REVNOX, REVCM, and REVKM and positions of certain restriction enzyme sites, namely, ClaI (C), HincII (Hc), NlaIV (N), NcoI (Nc), SspI (S), and AseI (A), are indicated.
The B. hyodysenteriae nox gene was subcloned as an EcoRI fragment from plasmid pCRNOX (47) into pUC19 to yield plasmid pER218. An internal 321-bp fragment, corresponding to nucleotide positions 214 to 534 in the nox coding sequence (GenBank accession no. U19610), was removed by digesting the plasmid with the restriction enzymes ClaI and NcoI (Fig. 1). The ends of the plasmid were made blunt by adding missing nucleotides with the Klenow fragment of DNA polymerase.
The deleted 321-bp DNA sequence contained codons for amino acid positions Asp72 to Asn178 of the Nox protein. Inasmuch as this region encodes two conserved domains of NADH oxidase proteins (47), including a domain (Val162 to Phe176) essential for NADH binding (34), the nox gene mutations were constructed to irreversibly inactivate NADH oxidase activity, even if the antibiotic resistance inserts, described below, should spontaneously exit the mutated gene.
The cat gene was obtained following digestion of pER919a (33) with NlaIV. The resulting 852-bp fragment was ligated into the deleted portion of the nox gene to yield plasmid pCmΔNOX. By the same strategy, a kan resistance gene was removed from plasmid pUC4K (Pharmacia, Piscataway, N.J.) after digestion with HincII and the 1,252-bp fragment was ligated into the nox gene to yield plasmid pKmΔNOX. The constructed plasmids were used to transform E. coli JM83 cells by standard electroporation techniques (36). Both plasmid inserts contained promoters and ribosome binding sites for expression of antibiotic resistance. In both pCmΔNOX and pKmΔNOX, the antibiotic resistance genes were oriented so that their sense strands were aligned with the sense strand of nox. The plasmids are unable to replicate within B. hyodysenteriae cells and thus serve as suicide vectors.
Derivation of B. hyodysenteriae nox mutants.
Transformation of B. hyodysenteriae B204 cells by electroporation was based on previously described methods (31). Cultures in the early exponential phase of growth in BHIS broth were harvested by centrifugation and concentrated 75-fold by resuspension in 0.1 ml of 0.5 M sucrose. Cells (approximately 3.5 × 109 CFU in 0.1 ml) were electroporated in chilled cuvettes (0.1-cm gap, 15-kV/cm discharge) containing 500 ng of plasmid DNA. Control electroporation mixes received no DNA. Following electric discharge, cells were immediately inoculated under anaerobic conditions into 0.5 ml of BHIS broth in a 18-mm-diameter culture tube containing a magnetic stirring flea and incubated with mixing at 38°C in a Coy anaerobic chamber. After 7 h of incubation to allow expression of antibiotic resistance, either chloramphenicol (final concentration, 10 μg/ml) or kanamycin (200 μg/ml) was added to the culture broth to select for cells in which the plasmid nox gene had exchanged with the chromosomal nox gene (Fig. 1).
After an additional 12 h of incubation, 0.25 ml of each culture was spread on the surfaces of TSB agar plates containing either no antibiotic (control cultures), kanamycin, or chloramphenicol. The agar plates were incubated in an anaerobic chamber at 38°C. After 10 days, cells from individual colonies (hemolytic zones) were inoculated into BHIS broth containing antibiotic. Strains were purified by subculturing single colonies on agar plates at least twice before use in experiments.
Genotypic analysis of nox mutants.
To confirm that B. hyodysenteriae nox mutant strains had either a cat or kan insertion in their nox genes, genomic DNAs were analyzed by PCR amplification and Southern hybridization. For PCR analysis, bacterial cells were harvested by centrifugation; lysed by resuspending them in buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.05% Tween 20, and 0.05% Triton X-100; and digested with proteinase K (100 μg/ml) at 60°C for 1 h. The suspension was then heated at 95°C for 10 min to inactivate proteinase K, and a sample equivalent to 60 μl (107 cells) of the original culture was used in the PCRs. PCR amplification was performed with VENT DNA polymerase supplemented with 6 mM MgSO4 in accordance with recommendations of the manufacturer (New England Biolabs, Beverly, Mass.). Amplification was at 95°C for 3 min, followed by 30 cycles of denaturation (95°C, 1 min), annealing (50°C, 1.5 min), and extension (72°C, 2 min), and then by a final 72°C extension for 7 min. Primers based on the sequence of the nox gene included the forward primer FNOX, 5′-ATGAAAGTTATTGTAATAGG-3′, which corresponds to nucleotide positions 1 to 20 of the coding sequence (CDS) (47), and the reverse primer REVNOX, 5′-CACCTTCAAATTTCTTAAC-3′, which corresponds to base positions 681 to 663. Reverse primers, based on the antibiotic resistance genes (31), were REVCM, 5′-GATTAAATATCTCTTTTCTCTTCC-3′ (positions 55 to 32 of the cat CDS), and REVKM, 5′-CGCGGCCTCGAGCAAGACG-3′ (positions 41 to 23 of the kan CDS). PCR products were detected and their sizes were determined after horizontal electrophoresis on 1% agarose gels in 0.5× Tris-borate-EDTA buffer (36), staining with ethidium bromide, and UV transillumination.
For Southern hybridization analysis, DNAs were prepared from small-volume (7-ml) cultures of B. hyodysenteriae strains by a scaled-down version of the Marmur technique (24), except that lysozyme was not needed to lyse bacteria. Genomic DNAs were digested by using the restriction enzyme SspI, AseI, or EcoRV according to the instructions of the supplier (Gibco-BRL). DNA fragments were separated by electrophoresis on a 1% agarose gel (100 V, 2 h) in 0.5× Tris-borate-EDTA buffer. Conditions and reagents for blotting DNA onto nylon membranes, for radiolabelling the oligonucleotide nox probe, and for hybridization have been reported previously (43). The oligonucleotide probe for the nox gene was the same as the FNOX primer used for PCR amplification.
NADH oxidase assays.
Spirochete cells in the exponential phase of growth (3 × 108 to 5 × 108 cells/ml, direct counts) were harvested by centrifugation from 700 ml of BHIS broth (approximately 1.5 g [wet weight] of cells), washed once in 150 ml of 0.05 M sodium phosphate buffer (pH 7.0), and resuspended at 1 g (wet weight) of cells per ml of PBCF buffer (45). This suspension on ice was sonicated with two 25-s bursts with a Kontes KT40 Micro Ultrasonic Cell Disrupter (setting 25) with 1 min of cooling between bursts. This method resulted in greater than 99% cell lysis, as determined by microscopy. NADH oxidase assay conditions, control assays, and methods for calculating enzyme-specific activities in cell lysates have been described previously (45). Soluble oxidase activity refers to the activity in the supernate fractions (S1 fractions) after cell lysates had been ultracentrifuged at 147,000 × g at 5°C for 2 h in order to remove unbroken cells and cell membranes (45).
Proteins in S1 fractions of B. hyodysenteriae cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western immunoblot techniques used to detect expression of the spirochete NADH oxidase in recombinant E. coli cells (47). Swine antiserum D40, raised against partially purified NADH oxidase from B. hyodysenteriae cells, was the source of polyclonal antibodies. Twenty milliliters of a 1/250 dilution of the antiserum was placed in a glass test tube containing membrane filter strips on which mutant strain S1 fractions had been spotted (15 μg of protein of each strain). The antiserum was gently mixed for 6 h at room temperature, diluted one-fourth, and then used as the primary antibody source to identify proteins present in wild-type cell lysates and absent from mutant cell lysates.
Oxygen sensitivity assay.
To evaluate oxygen sensitivities of wild-type and mutant strains, cells were cultured overnight in BHIS broth (exponential growth phase; 1 × 108 to 3 × 108 CFU/ml). In a Coy anaerobic chamber, cultures were serially diluted 10-fold to the 10−6 dilution in anaerobic, basal (no serum added) BHI broth. A 2-μl sample of the original culture and of each dilution was spotted in a clockwise pattern onto the surfaces of each of seven TSB agar plates. The plates had been stored in the Coy chamber at room temperature for 12 h prior to inoculation. After the liquid sample had been absorbed into the agar (approximately 10 min), the plates were inverted and removed from the chamber. Zero-time exposure plates were immediately returned to the chamber and incubated at 38°C. Other plates were exposed to laboratory air at 38°C for periods of 2, 4, 6, 8, 10, or 12 h before incubation in the anaerobic chamber. After 72 h of incubation, plates were examined to determine the highest dilution (lowest cell density) at which growth, as judged by hemolysis, was visible.
Animal challenge experiments.
The virulence of the nox mutant strains was evaluated independently in two experiments at two sites (Ames, Iowa, and Kalamazoo, Mich.). Crossbred postweaning piglets, both male and female, were randomly assigned to groups housed in separate rooms. Animals were 5 to 7 weeks old and weighed 10 to 15 kg at the time of challenge. Swine were fed antibiotic-free starter ration ad lib. Animals appeared healthy and were free of hemolytic E. coli, Salmonella, and B. hyodysenteriae based on culture analysis of rectal swabs taken prior to inoculation.
Animals were fasted for 24 h before and 2 h after inoculation. Animals were inoculated once by intragastric gavage with 100 ml of culture. All bacterial cultures were in the exponential growth phase and contained 2 × 108 to 7 × 108 viable cells (CFU) per ml of medium.
Blood in feces was assessed visually or by an occult blood test (44). Shedding of B. hyodysenteriae cells in feces was monitored by direct microscopy and by culturing rectal swab samples on TSB agar plates containing spectinomycin (39). In addition to spectinomycin, either chloramphenicol (10 μg/ml) or kanamycin (200 μg/ml) was added to plates for recovery of the mutant strains.
Animals were monitored daily for signs of disease and weighed at least every three days. At the recommendation of attending veterinary staff, animals with severe, chronic dysentery (manifested as a 20 to 30%, or greater, loss in body weight) were euthanized and necropsied. For euthanasia, sodium pentobarbital (Sleepaway, T-61; Ft. Dodge Laboratories) at a dosage of 1 ml/4.5 kg of body weight was given intravenously, followed by exsanguination in accordance with National Animal Disease Center Animal Care and Use Committee guidelines. Experiments were terminated 3 and 4 weeks after inoculation, at which time surviving animals were euthanized and necropsied. Necropsy evaluations included visual examination of cecal and colonic tissues for inflammation and gross lesions typical of swine dysentery. At necropsy, tissue samples were fixed in formalin for later histopathological examination by light microscopy (7).
RESULTS
Isolation of B. hyodysenteriae nox mutant strains.
Constructed plasmids pCmΔNOX and pKmΔNOX were used to genetically transform B. hyodysenteriae B204 cells, with selection for either chloramphenicol or kanamycin resistance (Fig. 1). Based on PCR analysis, each of several colonies (eight Cmr and two Kmr colonies) contained DNA with either cat or kan inserted into the nox gene. One strain of each resistance phenotype, designated Nox-Cm and Nox-Km, respectively, was selected for further study.
Genotypic analysis of nox mutant strains.
Extrachromosomal DNA was not detected after gel electrophoresis and ethidium bromide staining of genomic DNA prepared from either strain Nox-Cm or strain Nox-Km. DNA from each strain was analyzed by PCR with the primers FNOX and REVNOX, which are complementary to the B. hyodysenteriae nox gene sequence upstream and downstream of the insertion sites of the antibiotic resistance genes (Fig. 1). The estimated sizes of the amplification products were 0.7 kbp for strain B204, 1.6 kbp for strain Nox-Km, and 1.2 kbp for strain Nox-Cm. These amplicon sizes are consistent with insertion of either the kan (1,252-bp) or the cat (852-bp) gene into the nox gene after removal of the 321-bp ClaI-NcoI nox fragment. DNAs from strains Nox-Cm and Nox-Km also yielded, as expected, products approximately 0.38 kb in size after amplification with the primer FNOX and either the reverse primer REVCM or REVKM. These results indicated that antibiotic resistance genes were present within nox genes in the mutant strains.
The sizes of DNA restriction fragments hybridizing with a nox probe (Fig. 2) confirmed that gene exchange had occurred between the introduced plasmid and chromosomal DNA in the mutant strains. Only a single hybridizing fragment was detected for each digest. DNA from wild-type strain B204 contained a 1.2-kb SspI fragment that hybridized with the nox probe (Fig. 2). Strain Nox-Cm DNA gave a 1.8-kb hybridizing fragment, consistent with the absence of an SspI site from the cat gene (Fig. 1). Nox-Km DNA contained a hybridizing SspI fragment (0.9 kb) that was smaller than that of the wild-type DNA, as predicted from the internal SspI site in the kan gene (Fig. 1). The sizes of the hybridizing fragments from the mutant strain DNAs cut with AseI or EcoRV were also consistent with the insertion of the constructed nox deletions into the wild-type nox gene (Fig. 1 and 2).
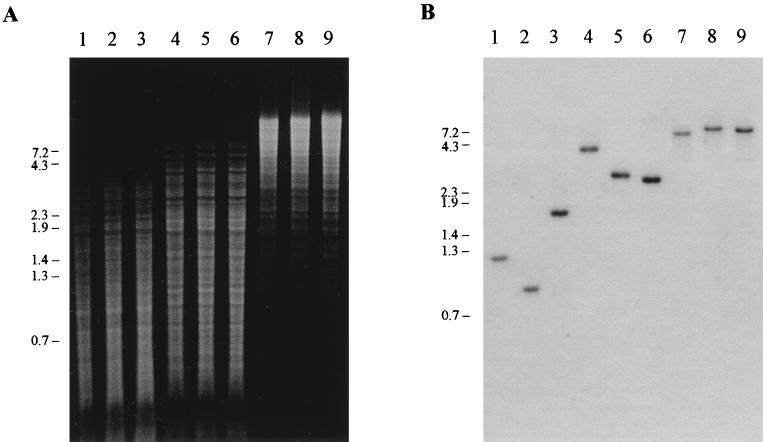
FIG. 2.
Hybridization analysis of B. hyodysenteriae strains B204, Nox-Km, and Nox-Cm. (A) Electrophoresis of DNA fragments of genomic DNAs digested with SspI (lanes 1 to 3), AseI (lanes 4 to 6), or EcoRV (lanes 7 to 9). Genomic DNAs were extracted from strains B204 (lanes 1, 4, and 7), Nox-Km (lanes 2, 5, and 8), and Nox-Cm (lanes 3, 6, and 9). (B) Southern hybridization blot of the gel shown in panel A. DNA fragments were hybridized with a 32P-labelled nox probe. Based on this and other hybridizations, estimated sizes (in kilobase pairs) of hybridizing fragments for B204, Nox-Km, and Nox-Cm were, respectively, 1.2, 0.9, and 1.8 (SspI); 4.0, 3.0, and 2.8 (AseI); and 7.2, 8.1, and 7.7 (EcoRV). The positions (in kilobase pairs) of DNA fragment size markers are indicated along the left sides of the panels.
Both PCR and Southern hybridization evidence suggested that, in strains Nox-Cm and Nox-Km, the wild-type nox genes had been replaced with genes inactivated by insertional mutagenesis. This allelic exchange was associated with a double-crossover recombination event within the nox gene or close to the ends of the gene. A similar allelic exchange has been observed for B. hyodysenteriae fla genes (31).
NADH oxidase activities.
Soluble oxidase activities of strains Nox-Cm and Nox-Km were, respectively, 0.4 and 0.2 μmol of NADH oxidized/min/mg of cell protein. These values were 8 and 4% of wild-type activity (4.7 μmol of NADH oxidized/min/mg of cell protein), respectively. In (control) enzyme assays under nitrogen, the mutant strains’ NADH-oxidizing abilities were reduced by 90%, confirming that oxygen was essential for the low-level activity.
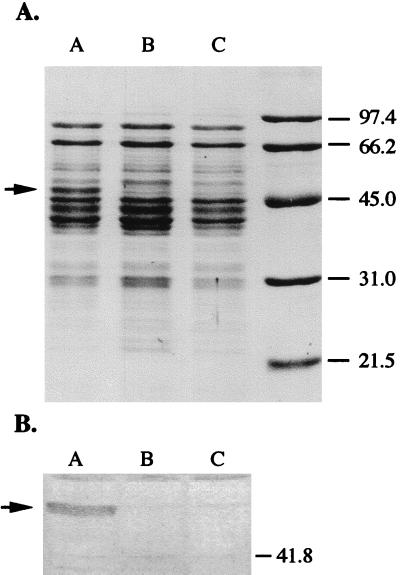
The only detectable difference in the electrophoretic profiles of proteins from soluble cell fractions of wild-type strain B204 and the mutant strains was a protein band with an estimated molecular mass of 48 kDa (Fig. 3A). A similarly migrating protein was previously identified as the B. hyodysenteriae NADH oxidase (45, 47). The protein reacted with antibodies raised against Nox and was missing from both mutant strains (Fig. 3B). The loss of 92 to 96% of the oxidase activity and the disappearance of the 48-kDa protein from soluble cell fractions supported the DNA-based findings that the nox gene in each mutant strain had been inactivated.
FIG. 3.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of supernate fractions of B. hyodysenteriae cell lysates after ultracentrifugation. Each lane contained 7 to 10 μg of protein. Proteins were stained with Coomassie blue. (B) Western immunoblot of proteins depicted in panel A with antibodies raised against B. hyodysenteriae B204 NADH oxidase. Lanes A, wild-type parent strain B204; lanes B, strain Nox-Cm; lanes C, strain Nox-Km; lane D, molecular mass markers of protein standards (in kilodaltons). Arrows indicate the positions of the protein previously identified as NADH oxidase (45).
There were no detectable growth differences (cell yields, population doubling times) between the mutant strains and wild-type strain B204 in broth in sealed culture tubes containing initial atmospheres of 1 or 2% oxygen (data not presented).
Oxygen sensitivities of nox mutant strains.
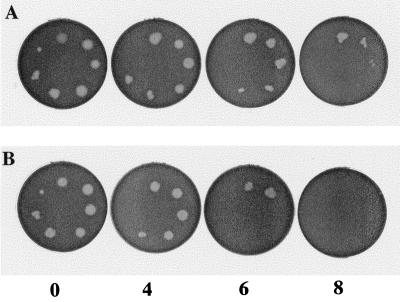
In a test for oxygen sensitivity (Fig. 4), there were no differences in levels of growth at any cell density between nox-deficient bacteria and wild-type bacteria after 0 and 2 h of exposure to air (Table 1). Differences in survival became noticeable after 4 h of exposure (Table 1). After 6- and 8-h exposures, nox mutant strains survived at cell densities that were 100- to 10,000-fold higher than those of strain B204. Wild-type cells at high cell densities (100 to 10−3 dilutions) survived over 10 h of exposure, whereas mutant cells at every dilution were killed between 6 and 8 h of exposure. Based on these results, cells of the Nox-Cm and Nox-Km strains were 100- to 10,000-fold more sensitive to oxygen exposure than were cells of the wild-type parent strain.
FIG. 4.
Oxygen sensitivity assay for B. hyodysenteriae strains. Results with B. hyodysenteriae B204 (wild type) (A) and B. hyodysenteriae Nox-Km (nox mutant) (B) are shown. Two-microliter samples of BHIS broth cultures (approximately 2 × 108 CFU/ml) and dilutions of the culture (10−1 to 10−6) were spotted in a clockwise pattern on TSB agar plates. Undiluted culture is at the top (12 o’clock position). Numbers at the bottom of the figure indicate hours of exposure to laboratory air before plates were incubated anaerobically. B. hyodysenteriae growth appears as hemolytic zones in the blood agar.
TABLE 1.
Oxygen sensitivities of the B. hyodysenteriae wild-type strain B204 and the nox mutant strains Nox-Cm and Nox-Kma
| Exposure time (h) | Highest culture dilution yielding detectable growthb
|
||
|---|---|---|---|
| B204 (nox+) | Nox-Cm (nox mutant) | Nox-Km (nox mutant) | |
| 0 | 10−5 | 10−5 | 10−6 |
| 2 | 10−6 | 10−5 | 10−5 |
| 4 | 10−5 | 10−4 | 10−3 |
| 6 | 10−4 | 10−1 | 10−2 |
| 8 | 10−4 | NG | NG |
| 10 | 10−2 | NG | NG |
| 12 | NG | NG | NG |
Under anaerobic conditions, spirochete cultures were serially diluted 10-fold and the dilutions were spotted on TSB agar plates (Fig. 4). Each plate was exposed to oxygen (laboratory air) at 38°C for 0 to 12 h as indicated and then incubated anaerobically at 38°C. Two cultures of each strain were tested in each of three experiments.
From experiment to experiment, the cell dilution at which growth was inhibited could vary by 1 dilution. For this reason, median dilution values are reported. In any single experiment, the dilution at which growth of a mutant strain was inhibited was always 100- to 10,000-fold greater than the dilution of the wild type that was inhibited. NG, no detectable growth of cells at any dilution.
Swine experimental infections with nox mutant strains.
Combined results of two independent experiments in which swine were inoculated intragastrically with cultures of strain B204, Nox-Km, or Nox-Cm are given in Table 2. Every animal displaying signs of swine dysentery was later confirmed to have had the disease based on necropsy evaluation, histopathological detection of dysentery-like lesions, or both (data not presented). Of the animals inoculated with strain B204, 12 of 14 developed bloody diarrhea typical of swine dysentery within 3 to 12 days after inoculation. Four animals either died or became severely ill and were euthanized.
TABLE 2.
Swine experimental infections with B. hyodysenteriae strains B204, Nox-Km, and Nox-Cm
| Inoculum | No. of animals | No. of animal deathsa | Detection of blood in feces
|
Recovery of B. hyodysenteriae from feces
|
||
|---|---|---|---|---|---|---|
| No. of positive animals | Avg duration/positive animal (days) | No. of positive animals | Avg duration/positive animal (days) | |||
| Sterile medium | 7 | 1 | 0 | 0 | ||
| B204 (wild type) | 14 | 4 | 12 | 8.2b | 12 | 9.9b |
| Nox-Km (nox mutant) | 14 | 0 | 1 | 2.0 | 5 | 2.6 |
| Nox-Cm (nox mutant) | 7 | 0 | 3 | 2.0 | 3 | 3.8 |
Includes natural deaths and deaths due to euthanasia. In the control (sterile medium) group, one animal died unexpectedly with symptoms of a systemic infection of unknown etiology. In the B204 group, one animal died of dysentery and three animals with severe dysentery were euthanized.
Animals that died or were euthanized are not included in these determinations. Three animals inoculated with B. hyodysenteriae B204 remained positive for blood in feces and were shedding B. hyodysenteriae cells at the end of the experiment.
In contrast to the results with strain B204, clinical signs of dysentery in animals inoculated with nox mutants were not as common, were less severe, and were transient. There were no animal deaths due to either mutant strain. Only 1 of 14 animals inoculated with strain Nox-Km and 3 of 7 animals inoculated with strain Nox-Cm had detectable blood in fecal samples over a 2-day (average) period. Fewer animals inoculated with the nox mutants shed detectable B. hyodysenteriae in their feces, and the duration of shedding was shorter than for animals inoculated with wild-type cells (Table 2). These results indicate the nox mutant cells were less virulent than wild-type cells and had a reduced capacity to colonize their animal hosts, that is, a reduced ability to both establish and maintain populations in the swine intestinal tract.
DISCUSSION
NADH oxidase activity has been reported for cells of obligately anaerobic bacteria (1, 2, 5, 6, 21, 23, 43), archaebacteria (8, 25), lactic acid bacteria (4, 10, 13, 19, 37), mycoplasma (30), microaerophilic bacteria (38), and protozoa lacking mitochondria (3, 50). NADH oxidase activity is displayed by several biochemically distinct enzymes which directly reduce molecular oxygen with electrons derived from NADH-H+. The broad distribution of this activity suggests that NADH oxidase is a common adaptation by which microorganisms lacking a cytochrome-mediated reduction of oxygen are able to contend with or take advantage of oxygen in their environments.
Several roles for NADH oxidase in the growth and survival of microorganisms have been proposed. NADH oxidase provides certain lactic acid bacteria with an alternative NADH-H+-oxidizing mechanism, resulting in more rapid growth, higher growth yields, and an ability to grow on substrates more chemically reduced than glucose, e.g., mannitol (9, 11, 22, 28). By using NADH oxidase as an O2-scavenging mechanism, anaerobes such as Peptostreptococcus anaerobius, Clostridium spp., and Selenomonas ruminantium are thought to gain some measure of aerotolerance by protecting cell components and intracellular redox reactions from inactivation due to oxygen and oxygen radicals (6, 14, 29, 35). In several anaerobic species, NADH oxidase activities increase in response to elevated oxygen exposure (1, 12, 27, 29, 35). Strain differences in the levels of oxygen tolerance of Streptococcus mutans correlated with the NADH oxidase activities in the strains (12). Although these observations are consistent with the hypothesis that NADH oxidase protects anaerobic cells from oxygen exposure, direct evidence is limited.
To investigate the influence of NADH oxidase on the oxygen sensitivity and virulence of B. hyodysenteriae, the NADH oxidase gene of strain B204 was inactivated by insertional mutagenesis. This approach led to the isolation of two strains, Nox-Cm and Nox-Km, each with a mutated nox gene (Fig. 2), a corresponding diminution of over 90% of soluble NADH oxidase activity, and loss of the Nox protein (Fig. 3).
Based on the viability of cells on agar plates incubated in laboratory air for various times, both B. hyodysenteriae nox mutant strains were 100- to 10,000-fold more sensitive to oxygen exposure than were cells of the wild-type strain B204 (Table 1). These findings are direct evidence that NADH oxidase plays a role in protecting B. hyodysenteriae cells from the lethal effects of oxygen. The spirochete has additional oxidative stress defenses, NADH peroxidase, superoxide dismutase, and catalase (16, 40), and appears well equipped for contending with oxygen in its natural environment.
We hypothesized that oxygen-metabolizing enzymes are important adaptations enabling B. hyodysenteriae cells to establish and persist among the O2-respiring tissues of the swine intestinal tract (45). In this capacity, NADH oxidase might contribute to the colonizing ability and virulence of this anaerobic spirochete. The behavior of the nox mutant strains in animal challenge experiments supports this hypothesis inasmuch as both Nox-Km and Nox-Cm were less virulent for swine than was the parent, wild-type strain B204 (Table 2).
We are aware of two alternative explanations for the decreased virulence of the nox mutants. These explanations are unrelated to increased oxygen sensitivity. First, it is possible that the presence or expression of antibiotic resistance genes somehow reduces B. hyodysenteriae virulence. This explanation seems unlikely since both mutant strains, with different antibiotic resistance genes, exhibited similar in vitro and in vivo characteristics. Furthermore, we are unaware of examples of other pathogenic bacteria whose virulence is diminished by the expression of chloramphenicol or kanamycin resistance. Second, mutations in the nox gene may cause “polar effects,” affecting transcription of virulence genes downstream from nox. A strong (ΔG = −25.5 kcal/mol) DNA inverted repeat sequence, commonly associated with transcription termination, lies immediately downstream of the B. hyodysenteriae nox gene (47). The existence of this hairpin loop makes a polar transcription effect appear less likely but does not rule it out. Unfortunately, genetic techniques, such as rescue complementation of the mutated gene, to completely rule out these alternative explanations are currently unavailable for B. hyodysenteriae.
Although both nox mutants exhibited reduced virulence for swine by comparison to that of the wild-type parent strain, they were not avirulent. Fewer animals were colonized, and infected animals exhibited mild disease symptoms, namely, short duration of bloody feces and no animal deaths. It seems worthwhile to determine whether or not the transient dysentery associated with the mutant strains provides protective immunity against challenge with the wild-type strain. If immunity develops and the incidence of animals colonized by the nox mutant cells can be increased, the nox mutant stains might find practical use as rationally attenuated, live vaccine strains.
ACKNOWLEDGMENTS
We thank Sam Humphrey, Ger Bos, and Robert A. Rzepkowski for excellent technical support in this study. Evelyn Nystrom and Vijay Sharma provided comprehensive manuscript reviews, for which we are grateful.
REFERENCES
- 1.Abdollahi H, Wimpenny J W T. Effects of oxygen on the growth of Desulfovibrio desulfuricans. J Gen Microbiol. 1990;136:1025–1030. [Google Scholar]
- 2.Bentzen G, Larsen H. Oxygen activation and defence against oxygen toxicity in a psychrophilic bacteriodaceae. Arch Microbiol. 1989;151:95–100. doi: 10.1007/BF00414420. [DOI] [PubMed] [Google Scholar]
- 3.Brown D M, Upcroft J A, Upcroft P. A H2O-producing NADH oxidase from the protozoan parasite Giardia duodenalis. Eur J Biochem. 1996;241:155–161. doi: 10.1111/j.1432-1033.1996.0155t.x. [DOI] [PubMed] [Google Scholar]
- 4.Condon S. Responses of lactic acid bacteria to oxygen. FEMS Microbiol Rev. 1987;46:269–280. [Google Scholar]
- 5.Cox R P, Marling N. High-affinity oxygen uptake by Bifidobacterium bifidum. Antonie Leeuwenhoek. 1992;62:291–297. doi: 10.1007/BF00572597. [DOI] [PubMed] [Google Scholar]
- 6.Dolin M I. Oxidation of reduced diphosphopyridine nucleotide by Clostridium perfringens. I. Relation of peroxide to the over-all reaction. J Bacteriol. 1959;77:383–392. doi: 10.1128/jb.77.4.383-392.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glock R D, Harris D L, Kluge J P. Localization of spirochetes with the structural characteristics of Treponema hyodysenteriae in the lesions of swine dysentery. Infect Immun. 1974;9:167–178. doi: 10.1128/iai.9.1.167-178.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes C M, Teixeira M. The NADH oxidase from the thermoacidophilic archaea Acidianus ambivalens: isolation and physicochemical characterisation. Biochem Biophys Res Commun. 1998;243:412–415. doi: 10.1006/bbrc.1998.8111. [DOI] [PubMed] [Google Scholar]
- 9.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 10.Gotz F, Sedewitz B, Elstner E F. Oxygen utilization by Lactobacillus plantarum. I. Oxygen consuming reactions. Arch Microbiol. 1980;125:209–214. doi: 10.1007/BF00446878. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi M. The effect of oxygen on the growth and mannitol fermentation of Streptococcus mutans. J Gen Microbiol. 1984;130:1819–1826. doi: 10.1099/00221287-130-7-1819. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi M. Reduced nicotinamide adenine dinucleotide oxidase involvement in defense against oxygen toxicity of Streptococcus mutans. Oral Microbiol Immunol. 1992;7:309–314. doi: 10.1111/j.1399-302x.1992.tb00594.x. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Koga T, Kamio Y. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J Gen Microbiol. 1993;139:2343–2351. doi: 10.1099/00221287-139-10-2343. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino E, Frolander F, Carlsson J. Oxygen and the metabolism of Peptostreptococcus anaerobius VPI4330-1. J Gen Microbiol. 1978;107:235–248. [Google Scholar]
- 15.Hyatt D R, ter Huurne A A H M, van der Zeijst B A M, Joens L A. Reduced virulence of Serpulina hyodysenteriae hemolysin-negative mutants in pigs and their potential to protect pigs against challenge with a virulent strain. Infect Immun. 1994;62:2244–2248. doi: 10.1128/iai.62.6.2244-2248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen N S, Stanton T B. Abstracts of the 93rd General Meeting of the American Society for Microbiology. 1993. Production of a hydrogen peroxide-inducible catalase activity by Serpulina hyodysenteriae, abstr. D-175; p. 126. Washington, D.C. [Google Scholar]
- 17.Kennedy G A, Strafuss A C. Scanning electron microscopy. Vol. 2. Chicago, Ill: IIT Research Institute; 1977. Scanning electron microscopy of the lesions of swine dysentery; pp. 283–290. [Google Scholar]
- 18.Kennedy M J, Yancey R J., Jr Motility and chemotaxis in Serpulina hyodysenteriae. Vet Microbiol. 1996;49:21–30. doi: 10.1016/0378-1135(95)00174-3. [DOI] [PubMed] [Google Scholar]
- 19.Koike K, Kobayashi T, Ito S, Saitoh M. Purification and characterization of NADH oxidase from a strain of Leuconostoc mesenteroides. J Biochem. 1985;97:1279–1288. doi: 10.1093/oxfordjournals.jbchem.a135179. [DOI] [PubMed] [Google Scholar]
- 20.Kubo M, Nakagawa M, Kashiwazaki M, Konno S. Pathological observation on experimental swine dysentery. Natl Inst Anim Health Q. 1979;19:83–90. [PubMed] [Google Scholar]
- 21.Le Gall J, Xavier A V. Anaerobes’ response to oxygen: the sulfate-reducing bacteria. Anaerobe. 1996;2:1–9. doi: 10.1006/anae.1996.0001. [DOI] [PubMed] [Google Scholar]
- 22.Lucey C A, Condon S. Active role of oxygen and NADH oxidase in growth and energy metabolism of Leuconostoc. J Gen Microbiol. 1986;132:1789–1796. [Google Scholar]
- 23.Maeda K, Truscott K, Liu X L, Scopes R K. A thermostable NADH oxidase from anaerobic extreme thermophiles. Biochem J. 1992;284:551–555. doi: 10.1042/bj2840551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 25.Masullo M, Raimo G, Russo A D, Bocchini V, Bannister J V. Purification and characterization of NADH oxidase from the archaea Sulfolobus acidocaldarius and Sulfolobus solfataricus. Biotechnol Appl Biochem. 1996;23:47–54. [PubMed] [Google Scholar]
- 26.Milner J A, Sellwood R. Chemotactic response to mucin by Serpulina hyodysenteriae and other porcine spirochetes: potential role in intestinal colonization. Infect Immun. 1994;62:4095–4099. doi: 10.1128/iai.62.9.4095-4099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy M G, Condon S. Correlation of oxygen utilization and hydrogen peroxide accumulation with oxygen induced enzymes in Lactobacillus plantarum cultures. Arch Microbiol. 1984;138:44–48. doi: 10.1007/BF00425405. [DOI] [PubMed] [Google Scholar]
- 28.Nuraida L, Grigolava I, Owens J D, Campbell-Platt G. Oxygen and pyruvate as external electron acceptors for Leuconostoc spp. J Appl Bacteriol. 1992;72:517–522. [Google Scholar]
- 29.O’Brien R W, Morris J G. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J Gen Microbiol. 1971;68:307–318. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- 30.Pollack J D, Williams M V, McElhaney R N. The comparative metabolism of the mollicutes (mycoplasmas): the utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit Rev Microbiol. 1997;23:269–354. doi: 10.3109/10408419709115140. [DOI] [PubMed] [Google Scholar]
- 31.Rosey E L, Kennedy M J, Petrella D K, Ulrich R G, Yancey R J., Jr Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J Bacteriol. 1995;177:5959–5970. doi: 10.1128/jb.177.20.5959-5970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosey E L, Kennedy M J, Yancey R J., Jr Dual flaA1 flaB1 mutant of Serpulina hyodysenteriae expressing periplasmic flagella is severely attenuated in a murine model of swine dysentery. Infect Immun. 1996;64:4154–4162. doi: 10.1128/iai.64.10.4154-4162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosey E L, Oskouian B, Stewart G C. Lactose metabolism by Staphylococcus aureus: characterization of lacABCD, the structural genes of the tagatose 6-phosphate pathway. J Bacteriol. 1991;173:5992–5998. doi: 10.1128/jb.173.19.5992-5998.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross R P, Claiborne A. Molecular cloning and analysis of the gene encoding the NADH oxidase from Streptococcus faecalis 10C1: comparison with NADH peroxidase and the flavoprotein disulfide reductases. J Mol Biol. 1992;227:658–671. doi: 10.1016/0022-2836(92)90215-6. [DOI] [PubMed] [Google Scholar]
- 35.Samah O A, Wimpenny J W T. Some effects of oxygen on the physiology of Selenomonas ruminantium WPL 151/1 grown in continuous culture. J Gen Microbiol. 1982;128:355–360. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schmidt H-L, Stocklein W, Danzer J, Kirch T, Limbach B. Isolation and properties of an H2O-forming NADH oxidase from Streptococcus faecalis. Eur J Biochem. 1986;156:149–155. doi: 10.1111/j.1432-1033.1986.tb09560.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith M A, Edwards D I. Oxygen scavenging, NADH oxidase and metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1997;39:347–353. doi: 10.1093/jac/39.3.347. [DOI] [PubMed] [Google Scholar]
- 39.Songer J G, Kinyon J M, Harris D L. Selective medium for isolation of Treponema hyodysenteriae. J Clin Microbiol. 1976;4:57–60. doi: 10.1128/jcm.4.1.57-60.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanton T B. Glucose metabolism and NADH recycling by Treponema hyodysenteriae, the agent of swine dysentery. Appl Environ Microbiol. 1989;55:2365–2371. doi: 10.1128/aem.55.9.2365-2371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanton T B. Physiology of ruminal and intestinal spirochaetes. In: Hampson D J, Stanton T B, editors. Intestinal spirochaetes in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1997. pp. 7–45. [Google Scholar]
- 42.Stanton T B, Cornell C P. Erythrocytes as a source of essential lipids for Treponema hyodysenteriae. Infect Immun. 1987;55:304–308. doi: 10.1128/iai.55.2.304-308.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanton T B, Hanzelka B L, Jensen N S. Survey of intestinal spirochaetes for NADH oxidase by gene probe and by enzyme assay. Microb Ecol Health Dis. 1995;8:93–100. [Google Scholar]
- 44.Stanton T B, Jensen N S. Monitoring experimental swine dysentery: rectal swab blood test and Serpulina (Treponema) hyodysenteriae detection. Vet Microbiol. 1993;34:389–396. doi: 10.1016/0378-1135(93)90064-e. [DOI] [PubMed] [Google Scholar]
- 45.Stanton T B, Jensen N S. Purification and characterization of NADH oxidase from Serpulina (Treponema) hyodysenteriae. J Bacteriol. 1993;175:2980–2987. doi: 10.1128/jb.175.10.2980-2987.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanton T B, Lebo D F. Treponema hyodysenteriae growth under various culture conditions. Vet Microbiol. 1988;18:177–190. doi: 10.1016/0378-1135(88)90063-6. [DOI] [PubMed] [Google Scholar]
- 47.Stanton, T. B., and R. Sellwood. Cloning and characteristics of a gene encoding NADH oxidase, a major mechanism for oxygen metabolism by the anaerobic spirochete Brachyspira (Serpulina) hyodysenteriae. Anaerobe, in press.
- 48.ter Huurne A A H M, van Houten M, Muir S, Kusters J G, van der Zeijst B A M, Gaastra W. Inactivation of a Serpula (Treponema) hyodysenteriae hemolysin gene by homologous recombination: importance of this hemolysin in pathogenesis of S. hyodysenteriae in mice. FEMS Microbiol Lett. 1992;92:109–114. doi: 10.1016/0378-1097(92)90550-8. [DOI] [PubMed] [Google Scholar]
- 49.Trott D J, Stanton T B, Jensen N S, Duhamel G E, Johnson J L, Hampson D J. Serpulina pilosicoli sp. nov.: the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol. 1996;46:206–215. doi: 10.1099/00207713-46-1-206. [DOI] [PubMed] [Google Scholar]
- 50.Williams A G, Coleman G S. The rumen protozoa. In: Hobson P N, Stewart C S, editors. The rumen microbial ecosystem. 2nd ed. London, United Kingdom: Blackie Academic & Professional; 1997. pp. 73–139. [Google Scholar]