Abstract
The emerging technology of brain organoids deriving from human pluripotent stem cells provides unprecedented opportunities to study human brain development and associated disorders. Various brain organoid protocols have been developed that can recapitulate some key features of cell type diversity, cytoarchitectural organization, developmental processes, functions, and pathologies of the developing human brain. In this review, we focus on patterning of human stem cell-derived brain organoids. We start with an overview of general procedures to generate brain organoids. We then highlight some recently developed brain organoid protocols and chemical cues involved in modeling development of specific human brain regions, subregions, and multiple regions together. We also discuss limitations and potential future improvements of human brain organoid technology.
Introduction
Human brain organoids are neural tissues differentiated from pluripotent stem cells (PSCs) with self-organized three-dimensional (3D) structures that recapitulate some key characteristics of the cell type diversity, cytoarchitectural organization, and developmental trajectories of the embryonic human brain [1,2]. Brain organoid technology overcomes many limitations of conventional approaches, such as 2D neural cultures, and provides platforms for investigation of human brain development and disorders. Human brain organoids have recently been widely used in modeling human neural development (e.g. differentiation [3–5], migration [6] and evolution [7]), neurological and psychiatric disorders (e.g. schizophrenia [8], lissencephaly [9], hypoxia [10], viral infections [4,11–14]), brain cancers [15], drug screening and testing [16,17], AAV capsid selection for gene therapy [18], and have been transplanted into the rodent brain [19–21]. In this review, we first describe common procedures for brain organoid generation, and then focus on patterning of brain region- or subregion-specific organoids. We also highlight emerging technologies for engineering organoids consisting of multiple CNS regions in continuum. Finally, we discuss current limitations and prospects for future improvements.
Generation of brain organoids from human pluripotent stem cells
Generation of brain organoids starts from undifferentiated human embryonic stem cells (hESCs) or induced pluripotent stem cells (hiPSCs) cultured either on mouse-derived feeder cells or under feeder-free conditions (Figure 1). Notably, protocols are usually different between feeder and feeder-free conditions. Second, 3D embryoid bodies (EBs) are formed through the re-aggregation of single-PSC suspensions in micro-wells, such as Aggrewell [22], V/U-bottom wells, or 3D printed wells [23], or through the self-aggregation of PSCs colonies in low-adherence plates [3,4]. Additionally, engineered materials, such as microfilaments [24] and microfluidic chips [25], have been used. The third step is to differentiate EBs into neural progenitors. Brain organoid protocols are generally divided into unguided and guided categories based on this step. The unguided protocol takes advantage of the property that EBs differentiate preferentially toward neuroectoderm through intrinsic signals simply by the absence of exogenous growth factors in the culture media. The derived cerebral organoids contain tissues resembling multiple brain regions in an unpredictable fashion, including the cerebral cortex, ventral telencephalon, choroid plexus and retina, and sometimes non-neural cells [26]. The guided protocol is based on timed modulation of key morphogen-related signaling pathways, which generates organoids possessing specific brain region components with higher reproducibility [3,4]. For both protocols, EBs are sometimes embedded in the extracellular matrix (ECM), such as Matrigel, to support morphogen gradients and tissue growth, and promote organization in organoids. Brain organoids are further cultured to promote neural differentiation and maturation under various conditions, such as the slicing method [27], the air–liquid interface method [28,29], and they can be transplanted into animals [19–21]. Organoids of different kinds can also be fused together as “assembloids” to investigate their interactions [6,30]. For organoid characterization, cellular and molecular identities are usually investigated with histological, immunocytochemical, and omics methods [31], including single-cell/bulk RNA-sequencing and ATAC-sequencing [32], whereas physiological functions and synaptic connections can be analyzed by calcium imaging [33], electrophysiology recording, and rabies viral tracing.
Figure 1. Procedures for generation of brain organoids from human pluripotent stem cells.
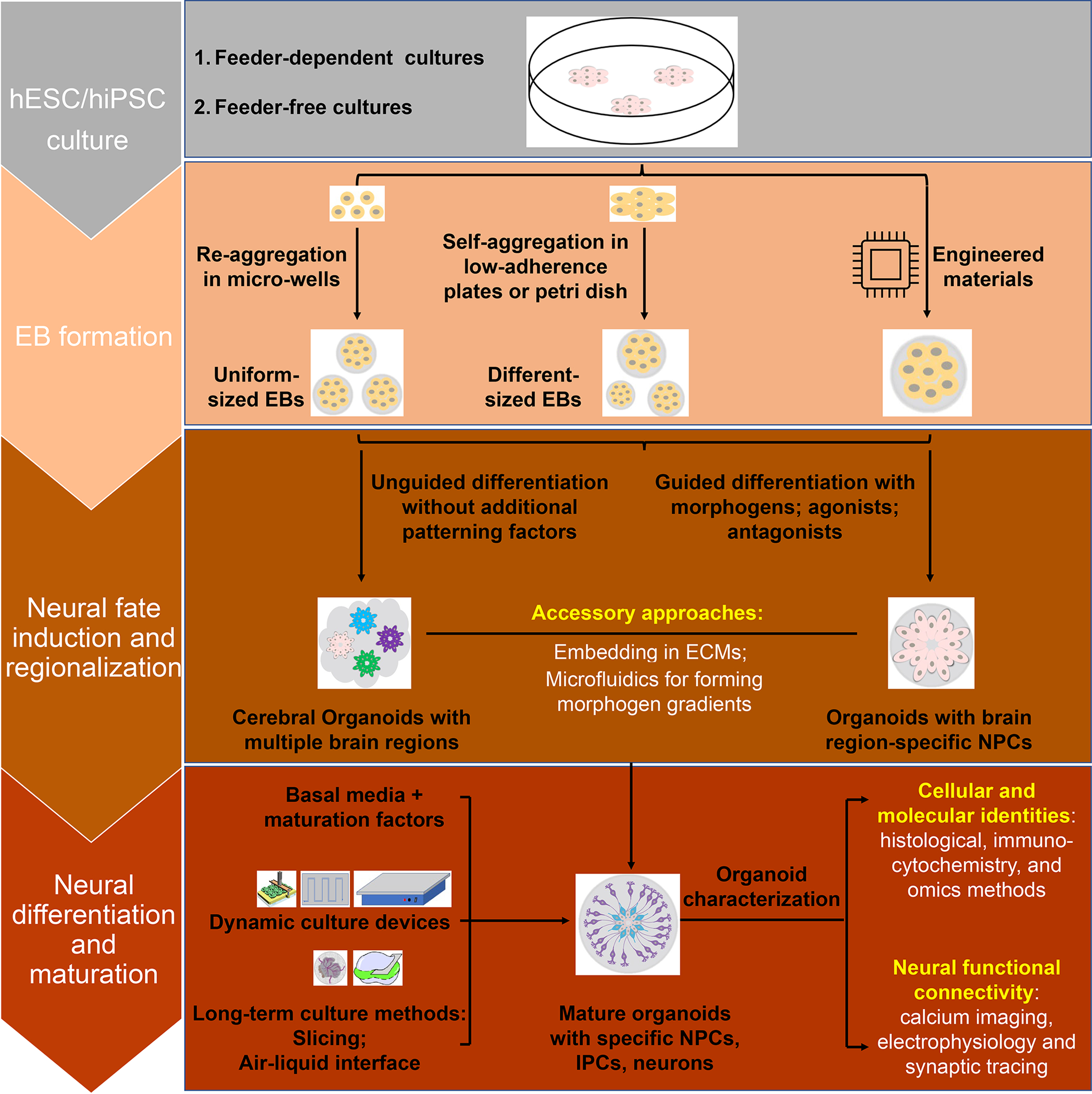
Embryoid bodies (EBs) are formed from hESCs or hiPSCs via re-aggregation, self-aggregation or engineered approaches. EBs can undergo unguided or guided differentiation with specific morphogens or related agonists/antagonists for patterning into organoids with brain region-specific neural progenitors (NPCs). Organoids then go through a long-term culture in basal culture media (such as, Neurobasal or DMEM/F-12) with additional factors for promoting maturation (such as, ascorbic acids, cAMP, NT3, BDNF and GDNF). During this process, dynamic culture devices (e.g. orbital shaker, spinning bioreactor, and microfluidics), and different approaches (e.g. slicing method, or culturing at air-liquid interface) can be used to enhance the diffusion of oxygen, nutrition, and metabolites of organoids and prevent cell death. Organoids are analyzed using different approaches to identify their cellular populations, molecular signatures, neural functions, and network connections.
Patterning of brain region-specific organoids
Significant progress has been made in generating different brain region-specific organoids by fine turning of distinct morphogens mimicking a similar differentiation program as the developing embryonic central nervous system (CNS) (Figure 2). In general, the “dual-SMAD inhibition” by simultaneously inhibiting the bone morphogenic proteins (BMPs) and TGFβ pathways is used for patterning the neuroectoderm fate. During normal brain development, retinoic acid (RA), WNTs, and FGFs cause early caudalization, while their inhibition promotes rostral differentiation. With appropriate gradients, the neuroectoderm further forms the neural tube and develops along the rostral-caudal axis into the prosencephalon (or forebrain), mesencephalon (or midbrain), rhombencephalon (or hindbrain), and spinal cord. During this process, Sonic hedgehog (SHH) is critical for ventral region patterning, while BMP and WNTs are important for dorsal fate patterning.
Figure 2. Patterning of brain region-specific organoids.
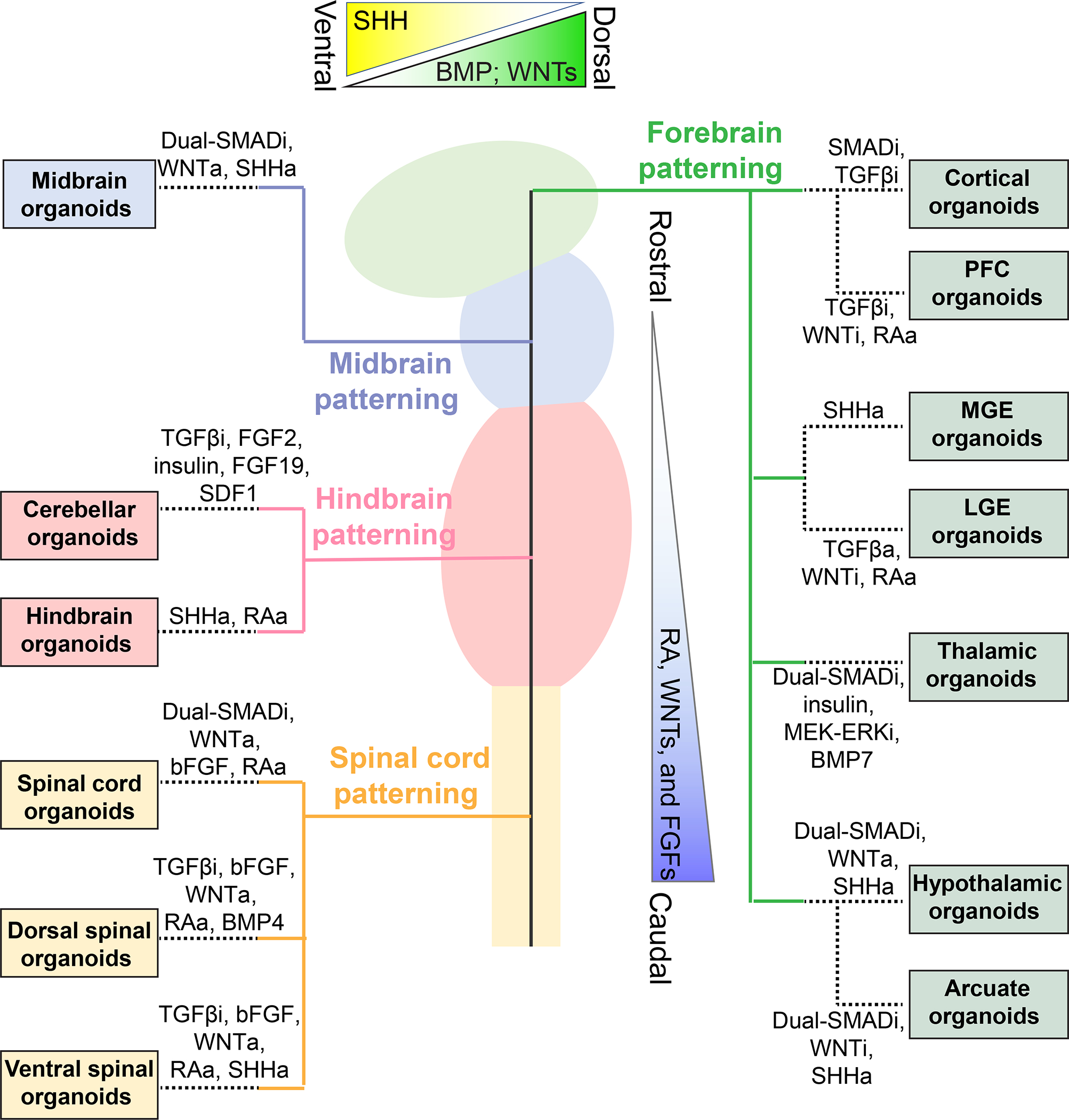
The ex vivo brain organoids are patterned following similar differentiation programs of distinct brain regions as in vivo. The developing neural tube is generally patterned into rostral-caudal regions by low to high gradients of RA, WNTs, and FGFs, and into dorsal-ventral regions by the opposite gradients of BMP/WNTs and SHH. Shown are examples of patterning of different brain region specific organoids by manipulating specific signaling pathways. SMADi: SMAD inhibitors including BMPi or TGFβi; BMPi: BMP inhibitors including Noggin, LDN-193189, or Dorsomorphin; TGFβi: TGFβ inhibitors including SB-431542 or A-83; WNTi: WNT inhibitors including IWR-1, IWP-2, or XAV-939; Notchi: Notch inhibitor DAPT; MEK-ERKi: MEK-ERK inhibitor PD0325901; WNTa: WNT activators including WNT3A or CHIR 99021; SHHa: SHH activators including recombined SHH, purmorphamine, or SAG; TGFβa: TGFβ activator Activin A; RAa: RA activators including RA, SR11237 or Vitamin A.
Forebrain organoids
The forebrain is further segregated into the telencephalon containing the cerebral cortex and basal ganglia and the diencephalon containing retina, thalamus, and hypothalamus. Several protocols have been developed to pattern brain organoids specifically representing some of these regions (Figure 2).
The cortical organoid is sometimes termed as dorsal forebrain organoid as the cerebral cortex is a major component of the forebrain. The first reported cortical organoid protocol [34] used TGFβ and WNT inhibitors for telencephalic fate patterning and produced tissues with self-organized multilayered ventricular and neuronal zones that expressed markers for radial glia cells and neurons of different cortical layers (CTIP2, TBR1, and SATB2). The Pasca group [3] used dual-SMAD inhibition and then FGF2/EGF for expanding ventricular progenitors without the exogenous ECM. Organoids generated using this approach contain astrocytes and neurons with synapses and appeared to be consistent from multiple batches and cell lines [22]. The Ming group [4,35] used dual-SMAD inhibition, followed by continuous TGFβ inhibition and WNT activation, then embedding EBs in Matrigel to promote expansion of ventricular structures and cortical-specific progenitors. After long-term culturing with a spinning bioreactor or the slicing method, they observed well-organized six-layer structures identified by expression of REELIN, CUX1, BRN2, SATB2, CTIP2, or TBR1 markers, as well as a prominent progenitor layer containing human enriched outer radial cells. One limitation of all these approaches is the presence of multiple ventricular structures, which could compromise their applications. Recent studies reported methods to generate a single neural tube by geometric constraint [36], or manual dissection of a single neural rosette [37], although it remains challenging to reliably maintain the single-ventricular structure over the long-term.
The ventral forebrain comprises the medial (MGE) and lateral (LGE) ganglionic eminence, which is patterned by high SHH and low WNT activity, respectively. Several protocols generated MGE organoids by activating the SHH pathway [6,34,38,39]. RNA-sequencing and immunostaining identified the presence of MGE-specific neural progenitors and diverse interneurons. These organoids were fused with cortical organoids to model interneuron migration, cross-region interactions, and brain disorders, such as Timothy syndrome. The striatum mainly originates from the LGE. Striatal organoids were patterned with TGFβ activation (Activin A), WNT inhibition (IWP-2), and RA activation (SR11237), leading to expression of LGE-specific and striatal medium spiny neuron markers [40]. Fused cortico-striatal assembloids exhibited unidirectional synaptic connections from excitatory cortical neurons to striatal GABAergic medium spiny neurons and were used to model Phelan-McDermid syndrome.
Thalamus and hypothalamus are developed from caudal and rostral diencephalon, respectively. Thalamic organoids were generated using dual-SMAD inhibitors and insulin for early caudal neural fate induction, then a MEK-ERK inhibitor (PD0325901) for antagonizing excessive caudalization and BMP7 for thalamic fate patterning [41]. Thalamic identities, including specific neural and progenitor cell populations, were validated by scRNA-sequencing. Furthermore, fused thalamic-cortical organoids exhibited reciprocal axonal projections. Hypothalamic organoids were generated by exposing hiPSC-derived neuroectoderm to high SHH and WNT signaling [4], leading to expression of hypothalamic neural progenitor and peptidergic neuronal markers.
Midbrain and hindbrain organoids
The substantia nigra, a dopaminergic neuron-enriched region in the midbrain, plays key roles in Parkinson’s disease. At least seven midbrain organoid protocols were developed by six research groups [4,42–47] (Figure 2). Neuroectodermal EBs were first induced by dual-SMAD inhibition together with WNT activation. Some protocols embed EBs in Matrigel to promote tissue growth and structural organization [43–45,47]. To pattern EBs toward the midbrain floor plate, SHH activation and FGF8 treatment were commonly used [4,42,43,47], although some protocols showed that FGF8 is not essential [44–46]. They all reported the presence of dopaminergic neurons with TH or DAT markers and dopamine synthesis, and some with dopamine receptors. Further refinement of patterning methods will hopefully model other midbrain regions, such as the ventral tegmental area.
The hindbrain comprises the medulla, pons, and cerebellum. Cerebellar organoids have been generated using a TGFβ inhibitor, FGF2 and insulin for early cerebellar neuroepithelium induction [48] (Figure 2). Sequential addition of FGF19 and SDF1 produces a continuous polarized neural-tube-like structure that can be further developed into laminated cerebellar cytoarchitectures with rhombic lip-like zones. Cerebellar precursors for Purkinje cells, Golgi cells, granule cells, and DCN projection neurons were validated by immunostaining. However, it remains challenging to establish long-term cultures to generate well-organized neural networks with lobular morphogenesis. Some other hindbrain organoids were recently generated by activation of SHH and RA without pre-SMAD inhibition [49]. Immunostaining and qPCR identified several hindbrain markers (GBX2, 5-HT, CHAT and HB9) in a tubular structure at day 30. Functionality was demonstrated by increased 5-HT synthesis and TPH2 expression in response to specific metabolites of gut microbiota at day 85. However, maintenance of the structural organization and reproducibility of organoids for long-term cultures remain challenging, since variability of the 5-HT neuron proportion increased, and the tubular structure was missing at day 60. These organoid methods recapitulate some features of early-stage hindbrain and can be used for modeling diseases, such as Dandy-Walker syndrome. Further efforts are needed to generate well-organized tissues during long-term culturing as well as to model some other hindbrain regions, such as medulla and pons.
Spinal cord organoids
The spinal cord bridges the brain and body and is essential for sensory input and motor output. Its dorsal and ventral neural development is guided by high BMP/WNT and SHH signaling, respectively, while the rostro-caudal fate is patterned by high to low RA gradients. The Takahashi group [50] used TGFβ inhibition combined with bFGF and WNT activation, followed by continuous RA and BMP4 treatment at the late stage to generate dorsalized spinal cord-like organoids with enriched dorsal spinal progenitors and four types of dorsal spinal cord interneurons (Figure 2). They further generated intermediate and ventral spinal organoids by removing BMP4 and adding low or high concentrations of an SHH agonist, respectively. Another study further demonstrated that the dosage, timing and duration of BMP4 treatment modulate cell types and organization in dorsal spinal cord organoids [51]. However, these methods did not produce morphologically or functionally mature neurons. A recent study generated spinal cord organoids by guiding hiPSCs to a caudal fate with dual-SAMD inhibitors and a WNT activator in 2D, making EBs with bFGF, and then replacing bFGF with RA for neural differentiation and maturation [52]. These organoids recapitulated many aspects of spinal cord development, and produced neurons exhibiting mature markers, dendritic spines, and spontaneous and evoked neural activity with short-term plasticity.
Together, tremendous progress has been made in generating various brain region-specific organoids and we expect many more to come in the near future. Most approaches focus on generating organoids with desired cell types and diversity, which need to be better benchmarked with endogenous human cell types not only at the transcriptome level, but also epitranscriptomic [53] and epigenetic levels [54]. More attention also needs to be devoted to proper cytoarchitectural organization and long-term maintenance.
Patterning of organoids modeling subregions of the brain
Many brain subregions play critical roles in specific functions and disorders and organoids with subregion specificity can lead to a better understanding of their development, functions and pathologies. The Ming group showed the first example of organoids (ARCO) modelling the human arcuate nucleus (hARC) of the hypothalamus [55], which is essential for transmitting signals of hunger. The protocol uses dual-SMAD inhibition followed by extended WNT inhibition and triple SHH activation (SHH, SAG, and purmorphamine) (Figure 2). Immunostaining showed hypothalamic progenitor markers at day 15 and hARC markers, including OTP, DLX, TBX3, and POMC, at day 40. scRNA-seq analysis aided by machine learning showed highly similar signatures between the native neonatal hARC and hiPSC-derived ARCOs. Furthermore, Prader-Willi syndrome patient-derived hARC organoids exhibited molecular, cellular and functional deficiencies.
Two groups recently reported that upregulated RA signaling is related to mid-fetal stage human prefrontal cortex (PFC) development [56,57]. Aided by this knowledge, the Nowakowski group generated cortical organoids based on a previous protocol [34] and then added vitamin A starting from day 35. scRNA-sequencing identified a higher proportion of PFC-like excitatory neurons in RA-treated organoids, which was confirmed by histology of SATB2, CTIP2 and AUTS2 co-expression [56]. However, whether RA treatment could also produce cells of other brain regions, such as striatal neurons as reported in another study [37], has not been examined.
Generation of brain subregion-specific organoids is the next frontier for brain organoid technology and overcoming limitations of insufficient knowledge on developmental mechanisms for promoting subregion identities in humans will be essential for future progress [56,57].
Engineering multi-region CNS organoids
Another frontier of brain organoid technology is to generate organoids modeling multiple CNS regions together. Current methods include unguided cerebral organoids [26] and assembloids from fusion of individually patterned organoids of different CNS regions [30] (Figure 3a). However, the unguided nature of cerebral organoid generation leads to significant variability, whereas the lack of smooth continuums and structural intermediaries in assembloids can potentially yield inconsistent or spurious neural connections between different regions. To address these shortcomings, engineering solutions leveraging recent work in the fields of biomaterials, microfluidics and synthetic biology are increasingly being utilized [58–60].
Figure 3. Engineering of multi-region organoids.
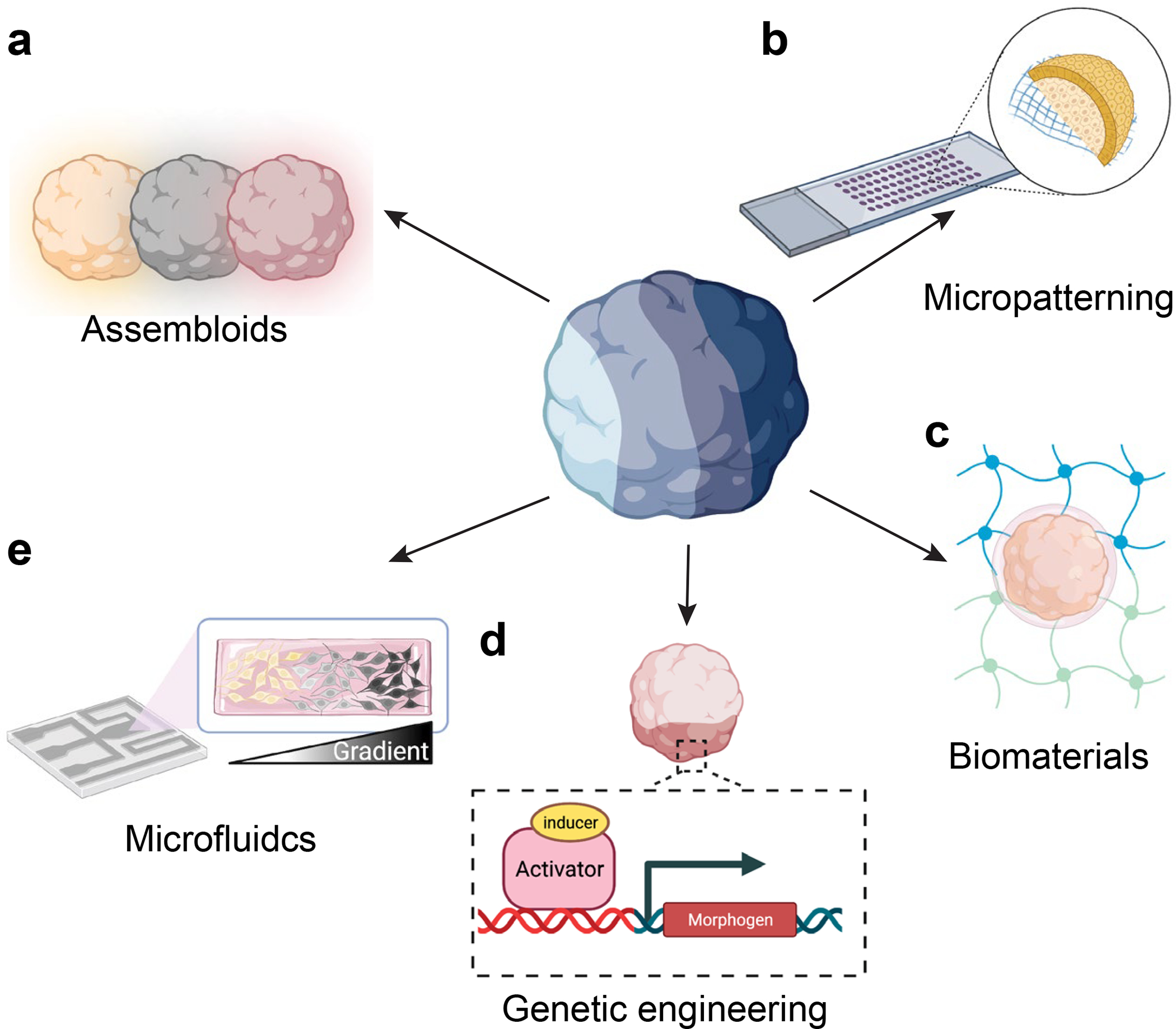
(a) Region-specific organoids can be combined to form assembloids, which can exhibit interneuron migration as well as functional long-range neural connections. (b) Micropatterning on glass slides results in geometric restriction of hiPSC colony growth, which can result in the formation of complex 3D tissues when combined with media containing the Matrigel or suspension culture. (c) By controlling the mechanical properties of the cellular environment, biomaterials can regulate the structural development of organoids. (d) Genetic engineering of cells offers a means of creating synthetic organizers within organoids that direct cell differentiation through the release of different morphogens. (e) Microfluidic devices can deliver spatial gradients of cellular morphogens or small molecules, thereby regionally patterning cells toward different fates.
Bioengineering approaches are uniquely suited for mimicking the complex array of biophysical cues [61,62] and morphogens [63,64] involved in regulating the developing CNS. A recent study used a combination of glass micropatterning of hiPSC cultures to spatially confine their growth with a 4% Matrigel environment to enable the formation of 3D cellular structures (Figure 3b). By adding BMP4 to the media, Karzbrun et al. [36] were able to induce folding of the basal tissue into a 3D structure reminiscent of the neural tube, containing PAX6+ neural tissue organized around a single lumen and surrounded by an outer layer of ectodermal tissue. This microengineered system could generate a variety of tissue types, including 3D structures containing a dorsal-ventral (D-V) axis. In another approach, D-V patterning can emerge in a subset of both murine [65] and human [66] neural-tube organoids by precisely tuning the stiffness of hydrogel matrices for encapsulation (Figure 3c). Furthermore, Cederquist et al. [67] demonstrated a genetic engineering approach for D-V patterning by using small aggregates of stem cells with inducible SHH expression positioned at a single pole of their forebrain organoids to create a spatial gradient of SHH signaling, allowing ventral forebrain fates to emerge close to the induced signaling center and more dorsal forebrain regions able to develop toward to the opposite pole [67] (Figure 3d). Finally, microfluidic devices can be used for D-V patterning by delivering opposing spatial gradients of SHH and BMP4/7 for mouse ESCs embedded in Matrigel/Geltrex [68], akin to what occurs in vivo (Figure 3e).
Microfluidic devices have also been developed to recapitulate patterning of the neural tube and spinal cord along the anterior-posterior (A-P) axis (Figure 3e). Rifes et al. [69] developed a device that exposes a 2D layer of hESCs to a linear gradient of glycogen synthase kinase 3 inhibitor (GSK3i) to pattern an A-P axis [69]. This cellular monolayer expands to become around 100 μm in thickness over two weeks as cells differentiate into continuously distributed forebrain, midbrain or hindbrain progenitors as a function of their position along the GSK3i gradient [69]. Critically, the tissue contains an isthmic organizer between the midbrain and hindbrain boundaries, a group of cells that produce soluble factors that further instruct regional brain development in the neural tube [69–72]. The lack of isthmic organizers in single-cell transcriptomic data from cerebral organoids [69] highlights the strength of their microfluidic system to deterministically and spatially pattern cells along a smooth continuum. An analogous microfluidic gradient approach also successfully patterned hiPSCs along the A-P axis of the spinal cord to generate diverse types of motor neurons [73].
Moving forward, one challenge for these engineering approaches is to extend the time over which they can be applied, as brain organoid development often occurs over the course of many months. Seo et al. [74] combined initial micropatterning of stem cell cultures with subsequent suspension culture in order to form elongated spinal cord organoids with D-V-like features. In addition, methods for dynamically altering the composition or release of soluble factors from biomaterials [59] as well as synthetic biology strategies for creating improved signaling centers via gene circuits that autonomously regulate cellular aggregation [75] could prove to be invaluable for long-term organoid culture. These and future methods could potentially yield forebrain organoids containing both prefrontal and motor cortex regions, whole-brain organoids with forebrain, midbrain and hindbrain, and hypothalamic organoids with various nuclei.
Limitations and future directions
The last decade has witnessed the tremendous progress of brain organoid technologies. Starting from hESCs or hiPSCs, researchers could now model the development or disorders of diverse brain regions and subregions. Many key features of the developmental human brain, including the molecular signatures, cellular composition and functions, structural organization, and cross-region interaction have been recapitulated by organoids. Despite significant advances, it should be emphasized that organoid technology is still an emerging discipline and has various limitations. For example, lack of arealization and limited tissue organization hampered the study of cross-region interactions and functions in organoids. High variability complicates preclinical studies and interpretation of phenotypes. Besides, organoids only model a limited time window of early development stages and produce limited cell types and atypical physiology [76,77]. All these features have compromised certain applications of brain organoids.
With a clear understanding of these limitations, improvements are needed to fully realize the potential of brain organoid technologies. On a micro scale, orchestrating types, timing and gradients of morphogens can help generate more specific subregion organoids. On a macro scale, brain organoids with appropriately organized multiple regions and cell populations can be created by controlling spatial gradients of patterning factors to model cross-region interactions and network genesis. Most organoid protocols were unable to maintain well-organized 3D architectures over the long-term. This could also be improved via maintaining proper and dynamic gradients of morphogens and proper extracellular matrix during long-term cultivation. The reproducibility of organoids in regards to the size, structures, and cellular composition remains challenging. Standardizing controllable culture conditions, like starting cell number, organoid density, and media change frequency, will help to reduce batch-to-batch variabilities. Cultivation with defined hydrogel or ECM-free approaches could also reduce the variability caused by batch effects of the Matrigel. Robust protocols require validation with multiple cell lines and batches across laboratories. Current brain organoids mostly lack several cell types of the developing brain, such as microglia and endothelial cells, which can be reconstructed by co-culture. Lastly, current brain organoids mainly recapitulate early embryonic stages and future developments are needed to extend this model into later developmental stages with formation of functional neuronal circuits and columns as seen in the human brain. Within the next few years, we will have an increased array of brain organoids for the field to investigate basic biology of human brain development, to model various developmental brain disorders, and to test therapeutic treatments. Along with such rapid advances, ethical issues need to be considered and frequently updated [78].
Highlights.
Common steps for generation of brain organoids
Approaches for patterning of brain region-specific organoids
Generation of subregion-specific brain organoids
Engineering organoids containing multiple brain regions
Acknowledgements
We apologize for not being able to cite many important publications owing to space limitations. We thank members of Song and Ming laboratories for comments and suggestions, and Sean Park for help with preparing illustrations in Figure 2. The research in the authors’ laboratories were supported by grants from the National Institutes of Health (R35NS116843 to H.S, and R35NS097370, RF1MH123979, R01MH125528 and U19AI131130 to G-l.M.) and the Adelson Medical Research Foundation (to G-l.M.)
Footnotes
Conflict of interest statement
The author declares no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Qian X, Song H, Ming GL: Brain organoids: advances, applications and challenges. Development 2019, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lancaster MA, Knoblich JA: Organogenesis in a dish: modeling development and disease using organoid technologies. Science 2014, 345:1247125. [DOI] [PubMed] [Google Scholar]
- 3.Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park J-Y, O’Rourke NA, Nguyen KD, et al. : Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature Methods 2015, 12:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian X, Nguyen Ha N, Song Mingxi M, Hadiono C, Ogden Sarah C, Hammack C, Yao B, Hamersky Gregory R, Jacob F, Zhong C, et al. : Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165:1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, et al. : Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell 2019, 25:558–569.e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, et al. : Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benito-Kwiecinski S, Giandomenico SL, Sutcliffe M, Riis ES, Freire-Pritchett P, Kelava I, Wunderlich S, Martin U, Wray GA, McDole K, et al. : An early cell shape transition drives evolutionary expansion of the human forebrain. Cell 2021, 184:2084–2102.e2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, et al. : Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 2014, 15:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iefremova V, Manikakis G, Krefft O, Jabali A, Weynans K, Wilkens R, Marsoner F, Brändl B, Müller F-J, Koch P, et al. : An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Reports 2017, 19:50–59. [DOI] [PubMed] [Google Scholar]
- 10.Pașca AM, Park J-Y, Shin H-W, Qi Q, Revah O, Krasnoff R, O’Hara R, Willsey AJ, Palmer TD, Pașca SP: Human 3D cellular model of hypoxic brain injury of prematurity. Nature Medicine 2019, 25:784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacob F, Pather SR, Huang W-K, Zhang F, Wong SZH, Zhou H, Cubitt B, Fan W, Chen CZ, Xu M, et al. : Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27:937–950.e939. • This study used human cortical, hippocampus, hypothalamus, midbrain, and choroid plexus organoids to investigate the neurotropisum of SARS-CoV-2 infection. The study revealed a selective SARS-CoV-2 tropism for choroid plexus epithelial cells and showed that SARS-CoV-2 infection leads to increased cell death and transcriptional dysregulation. Together with #14, this highlights the application of human brain organoids in responding to a global pandemic.
- 12.Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, et al. : Zika-Virus-Encoded NS2A Disrupts Mammalian Cortical Neurogenesis by Degrading Adherens Junction Proteins. Cell Stem Cell 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun G, Chiuppesi F, Chen X, Wang C, Tian E, Nguyen J, Kha M, Trinh D, Zhang H, Marchetto MC, et al. : Modeling Human Cytomegalovirus-Induced Microcephaly in Human iPSC-Derived Brain Organoids. Cell Rep Med 2020, 1:100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pellegrini L, Albecka A, Mallery DL, Kellner MJ, Paul D, Carter AP, James LC, Lancaster MA: SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids. Cell Stem Cell 2020, 27:951–961.e955. • Similar as #11, this study used human cerebral organoids and choroid plexus organoids to test SARS-CoV-2 neurotropism. The study showed that SARS-CoV-2 receptor ACE2 is enriched in mature choroid plexus cells rather than in neurons. SARS-CoV-2 productively infects choroid plexus cells and damages the choroid plexus epithelium.
- 15.Bian S, Repic M, Guo Z, Kavirayani A, Burkard T, Bagley JA, Krauditsch C, Knoblich JA: Genetically engineered cerebral organoids model brain tumor formation. Nature Methods 2018, 15:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, et al. : High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 2017, 21:274–283.e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. : Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 2016, 22:1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Depla JA, Sogorb-Gonzalez M, Mulder LA, Heine VM, Konstantinova P, van Deventer SJ, Wolthers KC, Pajkrt D, Sridhar A, Evers MM: Cerebral Organoids: A Human Model for AAV Capsid Selection and Therapeutic Transgene Efficacy in the Brain. Molecular Therapy - Methods & Clinical Development 2020, 18:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daviaud N, Friedel RH, Zou H: Vascularization and Engraftment of Transplanted Human Cerebral Organoids in Mouse Cortex. eneuro 2018, 5:ENEURO.0219–0218.2018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong X, Xu S-B, Chen X, Tao M, Tang X-Y, Fang K-H, Xu M, Pan Y, Chen Y, He S, et al. : Human cerebral organoids establish subcortical projections in the mouse brain after transplantation. Molecular Psychiatry 2021, 26:2964–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH: An in vivo model of functional and vascularized human brain organoids. Nature Biotechnology 2018, 36:432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoon S-J, Elahi LS, Pașca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, et al. : Reliability of human cortical organoid generation. Nature Methods 2019, 16:75–78. • This study generated human cortical organoids from a large number of iPSC cell lines across multiple experiments, and validated human cortical organoids generated with this protocol to be highly consistent.
- 23.Chen C, Rengarajan V, Kjar A, Huang Y: A matrigel-free method to generate matured human cerebral organoids using 3D-Printed microwell arrays. Bioactive Materials 2021, 6:1130–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA: Guided self-organization and cortical plate formation in human brain organoids. Nature Biotechnology 2017, 35:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Wang L, Guo Y, Zhu Y, Qin J: Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Advances 2018, 8:1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA: Cerebral organoids model human brain development and microcephaly. Nature 2013, 501:373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, Su K, Li S, Lu L, Jacob F, et al. : Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 2020, 26:766–781.e769. • This study applied a regular slicing method to prevent interior cell death of large organoids due to diffusion limits. The protocol maintained healthy growth of organoids for over 150 days and recapitulated late-stage human cortical layer developmental features identified by well-organized six-layer structures and the presence of human specific astrocytes in organoids.
- 28. •.Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, et al. : Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nature Neuroscience 2019, 22:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szebényi K, Wenger LMD, Sun Y, Dunn AWE, Limegrover CA, Gibbons GM, Conci E, Paulsen O, Mierau SB, Balmus G, et al. : Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nature Neuroscience 2021, 24:1542–1554. • These two studies (#28, #29) cultured sliced organoids at the air–liquid interface to avoid generation of hypoxic core and enhance long-term viability of neurons in large organoids. By maintaining organoids over several months under this condition, they observed improved neural morphological and functional maturity.
- 30.Marton RM, Pasca SP: Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol 2020, 30:133–143. [DOI] [PubMed] [Google Scholar]
- 31.Lindeboom RGH, van Voorthuijsen L, Oost KC, Rodriguez-Colman MJ, Luna-Velez MV, Furlan C, Baraille F, Jansen PWTC, Ribeiro A, Burgering BMT, et al. : Integrative multi-omics analysis of intestinal organoid differentiation. Molecular Systems Biology 2018, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkbeiner C, Ortuno-Lizaran I, Sridhar A, Hooper M, Petter S, Reh TA: Single-cell ATAC-seq of fetal human retina and stem-cell-derived retinal organoids shows changing chromatin landscapes during cell fate acquisition. Cell Reports 2022, 38. [DOI] [PubMed] [Google Scholar]
- 33.Sakaguchi H, Ozaki Y, Ashida T, Matsubara T, Oishi N, Kihara S, Takahashi J: Self-Organized Synchronous Calcium Transients in a Cultured Human Neural Network Derived from Cerebral Organoids. Stem Cell Reports 2019, 13:458–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y: Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell–derived neocortex. Proceedings of the National Academy of Sciences 2013, 110:20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming G-l: Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nature Protocols 2018, 13:565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karzbrun E, Khankhel AH, Megale HC, Glasauer SMK, Wyle Y, Britton G, Warmflash A, Kosik KS, Siggia ED, Shraiman BI, et al. : Human neural tube morphogenesis in vitro by geometric constraints. Nature 2021, 599:268–272. •• This study cultured hPSCs on micropatterned glass surfaces and in the presence of Matrigel to form novel 3D stem cell structures, including an in vitro model of neural tube folding and D-V patterned spheroids. This study highlights the power of applying an engineering approach to optimize brain organoid generation.
- 37.Wang Y, Chiola S, Yang G, Russell C, Armstrong CJ, Wu Y, Spampanato J, Tarboton P, Chang AN, Harmin DA, et al. : Modeling autism-associated SHANK3 deficiency using human cortico-striatal organoids generated from single neural rosettes. bioRxiv 2021:2021.2001.2025.428022. [Google Scholar]
- 38.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA: Fused cerebral organoids model interactions between brain regions. Nature Methods 2017, 14:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang Y, Tanaka Y, Patterson B, Kang Y-J, Govindaiah G, Roselaar N, Cakir B, Kim K-Y, Lombroso AP, Hwang S-M, et al. : Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017, 21:383–398.e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miura Y, Li M-Y, Birey F, Ikeda K, Revah O, Thete MV, Park J-Y, Puno A, Lee SH, Porteus MH, et al. : Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nature Biotechnology 2020, 38:1421–1430. • This study generated human striatum organoids containing medium spiny neurons and fused them with cortical organoids. Rabies tracing and functional assays showed the unidirectional synaptic connections from the excitatory neurons in cortical organoids to GABAergic medium spiny neurons in striatum organoids. Assembloids generated from patient-derived cells showed defects in cellular calcium activity related with Phelan-McDermid syndrome. This work showed organoids can be used to study the development and functional assembly of multiple brain regions and related disorders.
- 41. Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim K-Y, Sun P, Kang Y-J, Zhong M, Liu X, Patra P, et al. : hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 2019, 24:487–497.e487. • This study generated organoids with thalamic-specific progenitors and neurons with diverse identities. Fusing these thalamic organoids with cortical organoids modeled reciprocal thalam-ocortical axonal projections similar as in vivo. This work demonstrated the application of organoids in modeling interactions between multiple brain regions.
- 42.Tieng V, Stoppini L, Villy S, Fathi M, Dubois-Dauphin M, Krause K-H: Engineering of Midbrain Organoids Containing Long-Lived Dopaminergic Neurons. Stem Cells and Development 2014, 23:1535–1547. [DOI] [PubMed] [Google Scholar]
- 43.Jo J, Xiao Y, Sun Alfred X, Cukuroglu E, Tran H-D, Göke J, Tan Zi Y, Saw Tzuen Y, Tan C-P, Lokman H, et al. : Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 2016, 19:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, Jarazo J, Walter J, Brüggemann I, Boussaad I, et al. : Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports 2017, 8:1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim H, Park HJ, Choi H, Chang Y, Park H, Shin J, Kim J, Lengner CJ, Lee YK, Kim J: Modeling G2019S-LRRK2 Sporadic Parkinson’s Disease in 3D Midbrain Organoids. Stem Cell Reports 2019, 12:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smits LM, Reinhardt L, Reinhardt P, Glatza M, Monzel AS, Stanslowsky N, Rosato-Siri MD, Zanon A, Antony PM, Bellmann J, et al. : Modeling Parkinson’s disease in midbrain-like organoids. npj Parkinson’s Disease 2019, 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohamed N-V, Sirois J, Ramamurthy J, Mathur M, Lépine P, Deneault E, Maussion G, Nicouleau M, Chen CX-Q, Abdian N, et al. : Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy. Brain Communications 2021, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y: Self-Organization of Polarized Cerebellar Tissue in 3D Culture of Human Pluripotent Stem Cells. Cell Reports 2015, 10:537–550. [DOI] [PubMed] [Google Scholar]
- 49.Valiulahi P, Vidyawan V, Puspita L, Oh Y, Juwono VB, Sittipo P, Friedlander G, Yahalomi D, Sohn J-W, Lee YK, et al. : Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs. Stem Cell Reports 2021, 16:1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogura T, Sakaguchi H, Miyamoto S, Takahashi J: Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells. Development 2018, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duval N, Vaslin C, Barata TC, Frarma Y, Contremoulins V, Baudin X, Nedelec S, Ribes VC: BMP4 patterns Smad activity and generates stereotyped cell fate organization in spinal organoids. Development 2019, 146. [DOI] [PubMed] [Google Scholar]
- 52.Seo K, Cho S, Lee J-H, Kim JH, Lee B, Jang H, Kim Y, Cho HM, Lee S, Park Y, et al. : Symmetry breaking of hPSCs in micropattern generates a polarized spinal cord-like organoid (pSCO) with dorsoventral organization. bioRxiv 2021:2021.2009.2018.460734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. : Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 2017, 171:877–889 e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchís-Calleja F, Guijarro P, Sidow L, Fleck JS, Han D, et al. : Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 2019, 574:418–422. [DOI] [PubMed] [Google Scholar]
- 55. Huang W-K, Wong SZH, Pather SR, Nguyen PTT, Zhang F, Zhang DY, Zhang Z, Lu L, Fang W, Chen L, et al. : Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell 2021, 28:1657–1670.e1610. •• This study generated the first subregion brain organoids that model the arcuate nucleus of the hypothalamus. The organoids express arcuate progenitor and neural markers and showed similar molecular signatures as the neontal human arcuate nucleus cell types. Arcuate organoids generated from patient-derived cells modeled molecular, cellular and functional defects of Prader-Willi syndrome.
- 56. Ziffra RS, Kim CN, Ross JM, Wilfert A, Turner TN, Haeussler M, Casella AM, Przytycki PF,Keough KC, Shin D, et al. : Single-cell epigenomics reveals mechanisms of human cortical development. Nature 2021, 598:205–213. •• This study generated prefrontal cortex subregion organoids guided by their scATAC-seq of the human fetal tissue, which suggsted the important role of RA for patterning prefrontal cortex neurons during development. This work provides a promising direction for guiding the generation of different subregion-specific brain organoids in the future.
- 57.Shibata M, Pattabiraman K, Lorente-Galdos B, Andrijevic D, Kim S-K, Kaur N, Muchnik SK, Xing X, Santpere G, Sousa AMM, et al. : Regulation of prefrontal patterning and connectivity by retinoic acid. Nature 2021, 598:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garreta E, Kamm RD,Chuva de Sousa Lopes SM, Lancaster MA, Weiss R, Trepat X, Hyun I, Montserrat N: Rethinking organoid technology through bioengineering. Nat Mater 2021, 20:145–155. [DOI] [PubMed] [Google Scholar]
- 59.Roth JG, Huang MS, Li TL, Feig VR, Jiang Y, Cui B, Greely HT, Bao Z, Pasca SP, Heilshorn SC: Advancing models of neural development with biomaterials. Nat Rev Neurosci 2021, 22:593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hofer M, Lutolf MP: Engineering organoids. Nat Rev Mater 2021:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heisenberg CP, Bellaiche Y: Forces in tissue morphogenesis and patterning. Cell 2013, 153:948–962. [DOI] [PubMed] [Google Scholar]
- 62.Javier-Torrent M, Zimmer-Bensch G, Nguyen L: Mechanical Forces Orchestrate Brain Development. Trends Neurosci 2021, 44:110–121. [DOI] [PubMed] [Google Scholar]
- 63.Wilson L, Maden M: The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev Biol 2005, 282:1–13. [DOI] [PubMed] [Google Scholar]
- 64.Sansom SN, Livesey FJ: Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harb Perspect Biol 2009, 1:a002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP: Neural tube morphogenesis in synthetic 3D microenvironments. Proc Natl Acad Sci U S A 2016, 113:E6831–E6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Y, Xue X, Resto-Irizarry AM, Li Z, Shao Y, Zheng Y, Zhao G, Fu J: Dorsal-ventral patterned neural cyst from human pluripotent stem cells in a neurogenic niche. Sci Adv 2019, 5:eaax5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, Studer L: Specification of positional identity in forebrain organoids. Nat Biotechnol 2019, 37:436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Demers CJ, Soundararajan P, Chennampally P, Cox GA, Briscoe J, Collins SD, Smith RL: Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development 2016, 143:1884–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rifes P, Isaksson M, Rathore GS, Aldrin-Kirk P, Moller OK, Barzaghi G, Lee J, Egerod KL, Rausch DM, Parmar M, et al. : Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat Biotechnol 2020, 38:1265–1273. •• This study reports a novel microfluidic device capable of multi-week long culture of hPSCs under a spatial gradient of WNT activator akin to the A-P axis development in the neural tube. Cells acquired a range of fates continously from forebrain to hindbrain and formed an isthmic organizer at the midbrain-hidbrain boundary of their system. This study highlights using an engineering approach to deterministically and spatially pattern cells along a smooth continuum.
- 70.Kiecker C, Lumsden A: The role of organizers in patterning the nervous system. Annu Rev Neurosci 2012, 35:347–367. [DOI] [PubMed] [Google Scholar]
- 71.Kiecker C, Lumsden A: Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci 2005, 6:553–564. [DOI] [PubMed] [Google Scholar]
- 72.Wurst W, Bally-Cuif L: Neural plate patterning: upstream and downstream of the isthmic organizer. Nat Rev Neurosci 2001, 2:99–108. [DOI] [PubMed] [Google Scholar]
- 73.Lim GS, Hor JH, Ho NRY, Wong CY, Ng SY, Soh BS, Shao H: Microhexagon gradient array directs spatial diversification of spinal motor neurons. Theranostics 2019, 9:311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seo K, Cho S, Lee J-H, Kim JH, Lee B, Jang H, Kim Y, Cho HM, Lee S, Park Y, et al. : Symmetry breaking of hPSCs in micropattern generates a polarized spinal cord-like organoid (pSCO) with dorsoventral organization. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glykofrydis F, Cachat E, Berzanskyte I, Dzierzak E, Davies JA: Bioengineering Self-Organizing Signaling Centers to Control Embryoid Body Pattern Elaboration. ACS Synth Biol 2021, 10:1465–1480. [DOI] [PubMed] [Google Scholar]
- 76.Bhaduri A, Andrews MG, Kriegstein AR, Nowakowski TJ: Are Organoids Ready for Prime Time? Cell Stem Cell 2020, 27:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andrews MG, Kriegstein AR: Challenges of Organoid Research. Annu Rev Neurosci 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farahany NA, Greely HT, Hyman S, Koch C, Grady C, Pasca SP, Sestan N, Arlotta P, Bernat JL, Ting J, et al. : The ethics of experimenting with human brain tissue. Nature 2018, 556:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]


