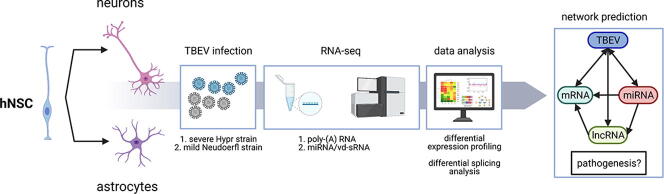
Graphical abstract
Abbreviations: A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; ACACA, Acetyl-CoA Carboxylase Alpha; AKR1C2, Aldo-Keto Reductase Family 1 Member C2; ANKS1A, Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1A; ANOS1, Anosmin 1; AOX1, Aldehyde Oxidase 1; APOBEC3G, Apolipoprotein B MRNA Editing Enzyme Catalytic Subunit 3G; APOL1/6, Apolipoprotein L1/6; ARID2, AT-Rich Interaction Domain 2; AUTS2, Activator Of Transcription And Developmental Regulator AUTS2; BCL11B, BAF Chromatin Remodeling Complex Subunit BCL11B; BCL9L, BCL9 Transcription Coactivator-like; BDKRB2, Bradykinin Receptor B2; BDNF, Brain Derived Neurotrophic Factor; BEND3, BEN Domain Containing 3; BSA, bovine serum albumin; BST2, Bone Marrow Stromal Cell Antigen 2; CALB1, Calbindin 1; CAMK2A, Calcium/Calmodulin Dependent Protein Kinase II Alpha; CD, complement determinant; CDKN1C, Cyclin Dependent Kinase Inhibitor 1C; CFAP61, Cilia And Flagella Associated Protein 61; CHRNA3, Cholinergic Receptor Nicotinic Alpha 3 Subunit; CHRNB4, Cholinergic Receptor Nicotinic Beta 4 Subunit; CLIC5, Chloride Intracellular Channel 5; CMPK2, Cytidine/Uridine Monophosphate Kinase 2; CNS, central nervous system; CNTN2, Contactin 2; CREG2, Cellular Repressor Of E1A Stimulated Genes 2; CXADR, Coxsackievirus B-Adenovirus Receptor; CYYR1, Cysteine And Tyrosine Rich 1; DACH1, Dachshund Family Transcription Factor 1; DAPI, diamidino-2-phenylindole; DCC, Netrin 1 Receptor; DCX, Doublecortin; DDX60, DExD/H-Box Helicase 60; DDX60L, DExD/H-Box 60 Like; DE, differentially expressed; DENV, Dengue virus; DIRAS2, DIRAS Family GTPase 2; DLX1/5/6, Distal-Less Homeobox 1/5/6; DNMT3B, DNA Methyltransferase 3 Beta; DPYSL2, Dihydropyrimidinase Like 2; EBF1, EBF Transcription Factor 1; EGF, Epidermal Growth Factor; ELAVL2/4, ELAV Like RNA Binding Protein 2/4; EPHB1, EPH Receptor B1; EPSTI1, Epithelial Stromal Interaction 1; ERBB4, Erb-B2 Receptor Tyrosine Kinase 4; ES, exon skipping; ESRRG, Estrogen Related Receptor Gamma; FGFb, Fibroblast Growth Factor 2; FPKM, Fragments Per Kilobase of transcript per Million mapped reads; FUT9, Fucosyltransferase 9; G2E3, G2/M−Phase Specific E3 Ubiquitin Protein Ligase; GABRG2, Gamma-Aminobutyric Acid Type A Receptor Subunit Gamma 2; GAPDH, Glyceraldehyde-3-Phosphate Dehydrogenase; GAS2L3, Growth Arrest Specific 2 Like 3; GAS7, Growth Arrest Specific 7; GATAD2B, GATA Zinc Finger Domain Containing 2B; GFAP, Glial Fibrillary Acidic Protein; GIPC2, GIPC PDZ Domain Containing Family Member 2; GLRA2, Glycine Receptor Alpha 2; GNG2, G Protein Subunit Gamma 2; GO, gene ontology; GOLGA4, Golgin A4; GRIN2A, Glutamate Ionotropic Receptor NMDA Type Subunit 2A; gRNA, genomic TBEV RNA; GSEA, gene set enrichment analysis; HERC5/6, HECT And RLD Domain Containing E3 Ubiquitin Protein Ligase 5/6; HEYL, Hes Related Family BHLH Transcription Factor With YRPW Motif Like; hNSC, human neural stem cells; HPRT1, Hypoxanthine Phosphoribosyltransferase 1; HS, hot-spot; HSPA6, Heat Shock Protein Family A (Hsp70) Member 6; HUDD (ELAV4), Hu-Antigen D/ELAV Like Neuron-Specific RNA Binding Protein 4; IFI6, Interferon Alpha Inducible Protein 6; IFIH1 (MDA5), Interferon Induced With Helicase C Domain 1/Melanoma Differentiation-Associated Protein 5; IFIT1-3, Interferon Induced Protein With Tetratricopeptide Repeats 1–3; IFITM1/2, Interferon Induced Transmembrane Protein 1/2; IFN, interferon; IGB, Integrated Genome Browser; IL6, Interleukin 6; IR, intron retention; ISG20, Interferon Stimulated Exonuclease Gene 20; ISGF3, Interferon-Stimulated Gene Factor 3 Gamma; ISGs, interferon-stimulated genes; JEV, Japanese encephalitis virus; KCND2, Potassium Voltage-Gated Channel Subfamily D Member 2; KCNK10, Potassium Two Pore Domain Channel Subfamily K Member 10; KCNS2, Potassium Voltage-Gated Channel Modifier Subfamily S Member 2; KIT, KIT Proto-Oncogene, Receptor Tyrosine Kinase; KLHDC8A, Kelch Domain Containing 8A; KLHL13, Kelch Like Family Member 13; KRR1, KRR1 Small Subunit Processome Component Homolog; LCOR, Ligand Dependent Nuclear Receptor Corepressor; LEKR1, Leucine, Glutamate And Lysine Rich 1; LGI1, Leucine Rich Glioma Inactivated 1; lncRNA, long non-coding RNA; LRRTM3, Leucine Rich Repeat Transmembrane Neuronal 3; LSV, local splicing variation; LUZP2, Leucine Zipper Protein 2; MAN1A1, Mannosidase Alpha Class 1A Member 1; MAP2, Microtubule Associated Protein 2; MBNL2, Muscleblind Like Splicing Regulator 2; MCTP1, Multiple C2 And Transmembrane Domain Containing 1; miRNA, micro RNA; MMP13, Matrix Metallopeptidase 13; MN1, MN1 Proto-Oncogene, Transcriptional Regulator; MOI, multiplicity of infection; mRNA, messenger RNA; MTUS2, Microtubule Associated Scaffold Protein 2; MX2, MX Dynamin Like GTPase 2; MYCN, MYCN Proto-Oncogene, BHLH Transcription Factor; NAV1, Neuron Navigator 1; NCAM1, Neural Cell Adhesion Molecule 1; ncRNA, non-coding RNA; NDRG4, N-Myc Downstream-Regulated Gene 4 Protein; NEK7, NIMA Related Kinase 7; NFASC, Neurofascin; NKAIN1, Sodium/Potassium Transporting ATPase Interacting 1; NMI, N-Myc And STAT Interactor 2; NRAP, Nebulin Related Anchoring Protein; NRARP, NOTCH Regulated Ankyrin Repeat Protein; NREP, Neuronal Regeneration Related Protein; NRN1, Neuritin 1; NS3, flaviviral non-structural protein 3; NXPH2, Neurexophilin 2; NYNRIN, NYN Domain And Retroviral Integrase Containing; OAS, 2′-5′-Oligoadenylate Synthetase; OASL, 2′-5′-Oligoadenylate Synthetase Like; ONECUT2, ONECUT-2 Homeodomain Transcription Factor; OPCML, Opioid Binding Protein/Cell Adhesion Molecule Like; OTX2, Orthodenticle Homeobox 2; PBS, phosphate buffer saline; PBX1, Pre-B-Cell Leukemia Transcription Factor 1; PCDH18/20, Protocadherin 18/20; pc-mRNA, protein-coding mRNA; PFKFB3, 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3; PIK3C2B, Phosphatidylinositol-4-Phosphate 3-Kinase Catalytic Subunit Type 2 Beta; PIP4P2, Phosphatidylinositol-4,5-Bisphosphate 4-Phosphatase 2; PLCH1, Phospholipase C Eta 1; POU3F4, Brain-Specific Homeobox/POU Domain Protein 4; PPM1L, Protein Phosphatase, Mg2+/Mn2+ Dependent 1L; PPP1R17, Protein Phosphatase 1 Regulatory Subunit 17; PRDM12, PR Domain Zinc Finger Protein 12; PSI, percent selective index; PSRC1, Proline And Serine Rich Coiled-Coil 1; PTPN5, Protein Tyrosine Phosphatase Non-Receptor Type 5; PTPRH, Protein Tyrosine Phosphatase Receptor Type H; qRT-PCR, quantitative reverse transcription real-time PCR; RAPGEF5, Rap Guanine Nucleotide Exchange Factor 5; RBFOX1, RNA Binding Fox-1 Homolog 1; RIG-I (DDX58), Retinoic Acid-Inducible Gene 1 Protein; RNF212, Ring Finger Protein 212; RNVU1, RNA, Variant U1 Small Nuclear; RSAD2, Radical S-Adenosyl Methionine Domain Containing 2; RTL8B, Retrotransposon Gag Like 8B; SAMD9, Sterile Alpha Motif Domain Containing 9; SEMA3E, Semaphorin 3E; SH3TC2, SH3 Domain And Tetratricopeptide Repeats 2; SHF, Src Homology 2 Domain Containing F; SHISAL1, Shisa Like 1; SIAH3, Siah E3 Ubiquitin Protein Ligase Family Member 3; SIRPA, Signal Regulatory Protein Alpha; SLITRK5, SLIT And NTRK Like Family Member 5; SNP, single-nucleotide polymorphism; snRNP, small nuclear ribonucleoproteins; SOGA1, Suppressor Of Glucose, Autophagy Associated 1; SPSB4, SplA/Ryanodine Receptor Domain And SOCS Box Containing 4; ST6GAL1, ST6 Beta-Galactoside Alpha-2,6-Sialyltransferase 1; TBC1D30, TBC1 Domain Family Member 30; TBEV, Tick-borne encephalitis virus; TFAP2A, Transcription Factor AP-2 Alpha; TFAP2B, Transcription Factor AP-2 Beta; THSD7A, Thrombospondin Type 1 Domain Containing 7A; THUMPD2, THUMP Domain-Containing Protein 2/SAM-Dependent Methyltransferase; TIPARP, TCDD Inducible Poly(ADP-Ribose) Polymerase; TM4SF18, Transmembrane 4 L Six Family Member 18; TMC8, Transmembrane Channel Like 6; TMEM229B, Transmembrane Protein 229B; TMTC1, Transmembrane O-Mannosyltransferase Targeting Cadherins 1; TNFSF10, TNF Superfamily Member 10; TRHDE, Thyrotropin Releasing Hormone Degrading Enzyme; TRIM38, Tripartite Motif Containing 38; TSHZ1, Teashirt Zinc Finger Homeobox 1; USP18, Ubiquitin Specific Peptidase 18/ISG15-Specific-Processing Protease; UTR, untranslated region; UTS2R, Urotensin 2 Receptor; vd-sRNA, virus-derived small RNA; WNV, West Nile virus; XAF1, XIAP Associated Factor 1; XRN1, 5′-3′ Exoribonuclease 1; ZIKV, Zika virus; ZMAT3, Zinc Finger Matrin-Type 3; ZMYM5, Zinc Finger MYM-Type Containing 5; ZNF124, Zinc Finger Protein 124; ZNF730, Zinc Finger Protein 730
Keywords: Alternative splicing, Astrocytes, miRNA, Neurons, Response to infection, Neuropathogenesis, Tick-borne encephalitis virus, Transcriptomics
Abstract
Tick-borne encephalitis virus (TBEV), the most medically relevant tick-transmitted flavivirus in Eurasia, targets the host central nervous system and frequently causes severe encephalitis. The severity of TBEV-induced neuropathogenesis is highly cell-type specific and the exact mechanism responsible for such differences has not been fully described yet. Thus, we performed a comprehensive analysis of alterations in host poly-(A)/miRNA/lncRNA expression upon TBEV infection in vitro in human primary neurons (high cytopathic effect) and astrocytes (low cytopathic effect). Infection with severe but not mild TBEV strain resulted in a high neuronal death rate. In comparison, infection with either of TBEV strains in human astrocytes did not. Differential expression and splicing analyses with an in silico prediction of miRNA/mRNA/lncRNA/vd-sRNA networks found significant changes in inflammatory and immune response pathways, nervous system development and regulation of mitosis in TBEV Hypr-infected neurons. Candidate mechanisms responsible for the aforementioned phenomena include specific regulation of host mRNA levels via differentially expressed miRNAs/lncRNAs or vd-sRNAs mimicking endogenous miRNAs and virus-driven modulation of host pre-mRNA splicing. We suggest that these factors are responsible for the observed differences in the virulence manifestation of both TBEV strains in different cell lines. This work brings the first complex overview of alterations in the transcriptome of human astrocytes and neurons during the infection by two TBEV strains of different virulence. The resulting data could serve as a starting point for further studies dealing with the mechanism of TBEV-host interactions and the related processes of TBEV pathogenesis.
1. Introduction
Tick-borne encephalitis virus (TBEV; genus Flavivirus, family Flaviviridae) is the most medically important tick-transmitted virus in Eurasia, affecting the lives of 10 000–12 000 diagnosed patients annually [1]. The virus is also gaining attention because of its recent emergence in new localities such as the Netherlands [2]. TBEV, like other flaviviruses, forms spherical virions (50 nm in diameter), and its monopartite genome comprises a ∼ 11 kilobases-long single-stranded RNA of positive polarity. The genomic RNA contains one ORF coding for 10 proteins, which is flanked by 5′ and 3′ untranslated regions (UTRs) [3].
Infection dissemination into the central nervous system (CNS) is a final stage in the process of TBEV pathogenesis, where neurons from different brain areas are the predominantly infected cell type and show a high level of death rate [4], [5]. TBEV also successfully replicates in astrocytes [6], [7], the neuroglial cells that provide all the necessary support for the proper neuronal function. Such as they maintain homeostasis, perform energy metabolism, regulate blood flow, support the synaptic function, and protect neurons from the infection [8]. Unlike in neurons, viability of TBEV-infected astrocytes is not negatively affected, even though similarly high viral titres in both cell types have been described [6], [7]. An identical pattern of distinctive pathogenicity in neurons and astrocytes was observed in the case of West Nile virus (WNV) [9], [10], but not in Zika virus (ZIKV) [11].
The exact factors responsible for the contrasting outcome of TBEV infection in neurons and astrocytes have not been identified yet; however, it is believed that cell type-specific response on the level of gene expression is one of the key factors involved. Indeed, a recent study by Fares et al. have described the neuron/astrocyte-specific response via a distinctive expression profile of specific innate immune response genes [12]. The phenomenon of neuron/astrocyte-specific immune response to TBEV was confirmed also on the level of cytokine and chemokine production [13]. Both studies point towards the TBEV-induced dysregulation of host gene expression with an outcome strongly dependent on the cell type-specific background.
The regulatory network of non-coding RNAs (ncRNAs) represents one of the most important players in the cell type-specific changes of gene expression upon infection. Among these, expression profiles of specific microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) were shown to be affected by flaviviral infection. miRNAs are a class of short (21–25 nucleotides) ncRNAs regulating gene expression at the post-transcriptional level by sequence-specific binding to mRNA [14]. Several flaviviruses, such as Japanese encephalitis virus (JEV), WNV, and ZIKV, were shown to dysregulate the expression profile of host miRNAs [15], [16], [17], [18], [19], of which some were identified to act in a proviral [20] or antiviral manner [16], [21], [22]. No miRNA-related data are currently available for TBEV [23]. lncRNAs represent another group of ncRNAs that are longer than 200 nucleotides and also contribute to gene expression regulation. In comparison to miRNAs, lncRNAs modulate the gene expression at numerous levels, including chromatin remodelling, cis-/trans-regulation of gene transcription, mRNA splicing, and translation [24]. Similarly to miRNAs, the expression pattern of lncRNAs alters upon flaviviral infection [25], [26], [27], [28], [29] and particular lncRNAs were identified as proviral [30], [31], [32] or antiviral [33] factors. Except for the host-derived ncRNAs, flavivirus-derived small RNAs (vd-sRNAs; 13–36 nt), were identified in mosquito/tick and mammalian cells [34], [35]. In arthropod vectors, the generation of vd-sRNAs results from the RNA interference process (RNAi), where vd-sRNAs are used by the host cell to target and cleave viral genomic RNA [36]. However, in the mammalian host, vd-sRNAs may play a different role by mimicking endogenous miRNA and target specific host mRNAs, thus substantially contribute to viral replication and pathogenesis as in the case of influenza A virus and human immunodeficiency virus [37], [38].
Despite numerous studies describing TBEV-induced changes in the expression profile of selected genes, no study so far has described a comprehensive transcriptomic analysis of both, ncRNAs and mRNAs in the most affected tissues of CNS. Data from such analysis would extensively broaden our understanding of TBEV-induced pathogenesis mechanism and may identify new target pathways for antiviral drug design. To fill that gap, we performed an integrative analysis describing the changes of host transcriptome on mRNA, miRNA, and lncRNA levels, including the pre-mRNA splicing evaluation, in combination with TBEV-derived vd-sRNA profiling. An in vitro model of human primary neurons and astrocytes infected by two TBEV strains of different virulence was utilised to elucidate the key players involved in TBEV-induced neuronal pathogenesis.
2. Methods
2.1. Primary cells cultivation and differentiation
Neural progenitor cells of human origin – Human Neural Stem Cells (hNSCs) purchased from Alstem (#hNSC11, Richmond, USA) were maintained in KnockOut DMEM/F-12 culture medium (#1260012, Gibco), supplemented with FGFb 20 ng/ml (#PHG0021, Thermo Fisher Scientific), EGF 20 ng/ml (#PHG0314, Thermo Fisher Scientific), 2 % StemPro Neural Supplement (#A10508-1, Thermo Fisher Scientific) and 1 % GlutaMAX-1 (2 mM) (#35050061, Gibco, Thermo Fisher Scientific) and grown in a 6-well plate at the basement membrane of Geltrex™ solution (1 % Geltrex™; #A1413302, Thermo Fisher Scientific; in KnockOut DMEM/F-12 culture medium). hNSCs were split after PBS wash by treatment with StemPro Acutase (#A11105-1, Gibco, Thermo Fisher Scientific) when reaching the 80–90 % confluency in a 1:4–1:6 ratio. Cells were cultivated in 5 % CO2 humidified atmosphere at 37 °C.
hNSCs were seeded at desired density according to the parameters of the specific experiments. Details are summarized in Table S1. After 24 h, differentiation was initiated by a transition to either Astrocyte Medium (#1801; ScienCell Research Labs, Carlsbad, CA, USA) or Neurobasal Plus Medium (#A35829-01, Gibco, Thermo Fisher Scientific), supplemented with 2 % B-27TM supplement (#17504044, Gibco, Thermo Fisher Scientific) and 2 mM GlutaMAX-1 (#35050061, Gibco, Thermo Fisher Scientific) to derive astrocytes or neurons respectively. During the differentiation process, the media were changed twice a week and the differentiation of astrocytes took 21–22 days and neurons 14–18 days before the target experiment was undertaken. For seeding of differentiated or partially differentiated cells, astrocytes or neurons were washed with PBS and detached by CTS™ TrypLE™ Select Enzyme (#A1285901, Thermo Fisher Scientific).
2.2. Viruses and infection
We used two strains of the European TBEV subtype with differing pathogenicity for cell infections. The prototype TBEV strain Neudoerfl originating from infected tick (4th passage in suckling mice brains; GenBank accession no. U27495), was provided by Prof. F.X. Heinz (Medical University of Vienna, Austria) [39]. The highly virulent TBEV strain Hypr (4th passage in suckling mice brains; GenBank accession no. U39292), isolated from a 5-year old child with a multi-tick bite history and suspect tick-borne encephalitis [40], is available at the Institute of Parasitology, Biology Centre of Czech Academy of Sciences, České Budějovice, Czech Republic. Mock-inoculated brain suspensions of suckling mice brains were used as a control in all experiments. Differentiated astrocytes and neurons were infected with both TBEV strains at the multiplicity of infection of 5 (MOI) for two hours in half volume of cultivation media to favour virus adsorption. Then, the inoculum was removed, and fresh cultivation media was replenished to the normal volume. To determine the appropriate MOI for infection, parallelly differentiated cells were counted prior to the infection in order to determine the cell numbers after differentiation.
2.3. Immunofluorescence assay
The presence of characteristic markers denoting the state of differentiation and development of infection in astrocytes and neurons was analysed by immunofluorescence assay. Cells were seeded on coated chamber slides at concentrations detailed in Table S1 and, when applicable, infected or mock-infected. At 24/72 h post-infection (p.i.), the presence of differentiation and infection markers was assayed. During sample processing, cells were fixed with 4 % paraformaldehyde for 15 min. Cells were washed in PBS and permeabilized by 0.1 % Triton X-100 in PBS, and formaldehyde auto-fluorescence was quenched by 50 mM NH4Cl in 1 % bovine serum albumin (BSA) in PBS for 10 min twice. After PBS washing, blocking in 3 % BSA in PBS was undertaken for 1 h at room temperature. Target antigens were labelled for 1 h at room temperature or overnight at 4 °C by the following primary and secondary antibodies: dsRNA mAb (#10010200, SCICONS J2 from Scicons, Biocompare), GFAP (GA5) Mouse mAb (#3670S, Cell Signaling Technology), HuC/HuD Monoclonal Antibody (16A11) (#A-21271 Invitrogen, Thermo), MAP2 (#4542S, Cell Signaling Technology), S100B Polyclonal antibody (#bs-2015R Bioss Antibodies), TBEV NS3 Langat Chicken IgY (NS3 antibodies to closely related Langat virus NS3 were kindly provided by Dr M. Bloom, 94 % homology with TBEV), Goat Chicken IgY H&L (DyLight® 488) (#ab96947, Abcam), Goat Guinea pig IgG H&L (Alexa Fluor® 594) (#ab150188, Abcam), Goat Mouse IgG H&L (DyLight® 594) preadsorbed (#ab96881 Abcam), Goat Rabbit IgG H&L (Alexa Fluor® 488) (#ab181448 Abcam). Concurrent nuclei staining and sample mounting were done with VECTASHIELD® Antifade Mounting Medium with DAPI (#H-1200-10, Vector Laboratories). Images were taken with the Olympus Fluoview FV10i confocal microscope equipped with FV10-ASW software (v.1.7).
2.4. Growth curve
To assess the replication rate of TBEV in astrocytes and neurons, cells were either mock-treated or infected with the TBEV strains Hypr and Neudoerfl at the MOI of 5 (see details of differentiation in Table S1). At 2, 12, 24, 48, 72 for both cell lines and at 120 h p.i. (astrocytes only) culture supernatant was sampled and clarified by centrifugation for TBEV titre assessment. TBEV titres were plaque assayed on the human lung adenocarcinoma monolayers (A549; kindly gifted by R. Randall, University of St. Andrews, UK) according to the modified protocol of de Madrid et al. [41]. A549 were maintained in low glucose DMEM cultivation medium (#L0064-500, Biowest, VWR) supplemented with 10 % foetal bovine serum (FBS; #S1810-500, Biowest, VWR), 1 % antibiotics (penicillin G 100 units/ml, streptomycin 100 μg/ml; #L0022-020, Biowest), and 1 % L-alanyl-L-glutamine (#X0551-100, VWR) in 5 % CO2 humidified atmosphere at 37 °C. Briefly, ten-fold dilutions of virus samples were mixed with the A549 cell suspension (1.5×105 cells/well of 24-well plate), cells were let to adhere, and virus to adsorb. After 4 h, an overlay mixture (1:1 v/v of carboxymethyl cellulose and 2× concentrated DMEM cultivation medium) was applied dropwise to the cells. After 5 days, cell monolayers were washed with physiologic solution (0.9 % NaCl), and plaques were fixed and stained with naphthalene black solution (0.1 % naphthalene black in 6 % acetic acid solution) for 45 min.
2.5. Viability assay
To detect the viability rates, neurons and astrocytes were differentiated as specified in Table S1 and infected with 5 MOI of Hypr and Neudoerfl TBEV strain. 2–3 h prior to the sampling interval (12, 24, 48, 72, 120, 168 h p.i.) incubation with AlamarBlue reagent in the cultivation media (1:10 (v/v); #DAL1025, Thermo Fisher Scientific) was performed in a dark. Viability for each cell type was assessed by fluorescence measurement (λEx = 550 nm, λEm = 590 nm) using Tecan infinite 2000Pro, (Tecan i-control, 1.11.1.0) from four biological and two technical replicates and the viability value of the mock-treated cells was set at 1 (100%).
2.6. qRT-PCR
For the analysis of TBEV replication in infected cells, viral RNA was quantified by an assay designed by Achazi et al. [42]. Total RNA (80 ng/reaction) was used for TBEV gRNA quantification with the KAPA PROBE FAST Universal One-Step qRT-PCR Master Mix (2X) (#KK4752, Sigma-Aldrich, MERCK) and relative fold induction of TBEV RNA amount was determined using the delta-delta ct (ΔΔ-ct) with the comparison to mock. For the verification of poly-(A) transcriptomic data, samples were treated with dsDNase (#EN0771, Thermo Fisher Scientific) and gene expression was analysed with the KAPA SYBR FAST Universal One-Step qRT-PCR Kit (#KK4652, Sigma-Aldrich) according to the manufacturer’s protocol. Relative expressions of HSPA-6, OASL, RNVU1, and RSAD2 (viperin) genes were processed via the ΔΔ-ct method with HPRT1 as a reference gene. All samples were analysed in biological and technical triplicates. A list of primers and probes used can be found in Table S1.
Verification of small RNA transcriptomic data was performed using miRCURY LNA miRNA PCR Assay Kit (#339306, Qiagen) according to the manufacturer’s instructions. 200 ng of total RNA was used as an input with subsequent quantification of hsa-miR-1248 (#YP00204253) and hsa-miR-145-5p (#YP00204483) in all samples. Data obtained were processed via relative quantification using the delta-delta ct (ΔΔ-ct) method with hsa-miR-103a as a reference miRNA and Sp6 as an internal spike-in control. All samples were analysed in biological and technical triplicates.
2.7. Transcriptomic analysis
2.7.1. Sample preparation
Each sample was prepared and analysed in biological triplicates. Total RNA from TBEV-infected human neurons and astrocytes was isolated at 24 and 72 h p.i. using RNA Blue reagent (Top-Bio, Vestec, Czech Republic) according to the manufacturer’s instructions. RNA concentration was measured using NanoPhotometer Pearl (Implen, München, Germany) and the quality of RNA was determined using 2100 Bioanalyzer with RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, USA).
2.7.2. Poly-(A)-enriched RNAs
Sequencing library construction, sequencing, raw read quality check and adapter trimming were done by Novogene Co., Ltd (Beijing, China). The 150 PE library was sequenced in HiSeq 4000 sequencer (Illumina, San Diego, CA, USA). Quality filtering was performed using Cutadapt v1.15 [43] and the subsequent clean read quality check was done using FastQC v0.11.5. [44]. The mapping (GRCh38.p13 reference genome), assembly and differential expression analysis were performed using the Tuxedo suite [45].
2.7.3. Small RNAs
Sequencing library construction, sequencing, raw read quality check and adapter trimming were done by Novogene Co., Ltd (Beijing, China). The 50 SE library was sequenced in HiSeq 2500 sequencer (Illumina, San Diego, CA, USA). The clean read quality check was done using FastQC v0.11.5 [44]. Adapter contamination, short and low quality reads were removed using Cutadapt v1.15 [43]. The clean reads were mapped to the reference genome (GRCh38.p13) using miRDeep 2.0.1.2 [46]. The -q option for the mapper.pl script was used to allow 1 mismatch for mapping. The miRDeep2.pl module was run using the human miRBase database (miRBase v.21) [47]. Identification of differentially expressed (DE) miRNAs was based on Benjamini-Hochberg P-value < 0.05 (unpaired Student’s t-test) and log2 fold change >1.5 or <−1.5 using the normalized fragments per kilobase of transcript per million mapped reads (FPKMs; cut-off 10 FPKM per miRNA species) ratios between the respective TBEV-infected sample and mock control.
2.7.4. Virus-derived small RNAs
The raw reads from small RNA library were also mapped to the genome of both TBEV strains, Hypr (GenBank accession number U39292) and Neudoerfl (GenBank accession number U27495), using Bowtie [48]. The read depth counts of each sample were retrieved from the corresponding bedgraphs produced using Bedtools v.2.27.1 [49]. Read distribution along the TBEV genome was evaluated and visualised using Geneious Prime v2020.0.5. (Biomatters, Ltd, Auckland, New Zealand). The sense and antisense reads were additionally discriminated by extracting individual read types from their respective bam files using Samtools v1.10 [50] and their counts were compared for each sample.
2.8. In silico miRNA/vd-sRNA target prediction
miRNA trans target prediction was performed using miRWalk [51] and LncBase v.2 [52] toolkits. Briefly, DE miRNAs were divided into two groups: (1) up-regulated and (2) down-regulated; these served as an input for the respective prediction toolkit. Generated lists of predicted miRNA targets were subsequently used to identify these targets in DE protein-coding mRNAs (pc-mRNAs)/lncRNA datasets. For up-regulated miRNAs, the datasets of down-regulated pc-mRNAs/lncRNAs were used, and vice versa, datasets of up-regulated pc-mRNAs/lncRNAs were used in the case of down-regulated miRNAs. Default settings were used for miRWalk (P-value < 0.05, miRNAs targeting 3′UTR) in combination with the miRDB prediction tool. In the case of LncBase, the prediction module with default settings was used (cut-off score > 0.90). Identification of human miRNAs targeting TBEV Hypr/Neudoerfl genomic RNA or human mRNAs being targeted by 21–23 nt long vd-sRNAs was performed using the miRDB tool with a cut-off score > 0.70 [53].
2.9. Differential splicing analysis
The identification and quantification of differential splicing were computed using MAJIQ v2.2 [54], which employs Local splicing variations (LSV) derived from the provided transcriptome annotation file and RNA-seq data. The differential splicing was considered significant for LSVs with MAJIQ default cut-off values for deltaPSI (|ΔΨ|≥0.2) supported by P(|ΔΨ]|≥0.2) with a 0.05 cut-off (Bonferroni correction).
The raw reads of each sample represented by biological triplicates were mapped to the reference genome (GRCh38.p13) with STAR 2.7.7a [55]. The resulting bam files and an annotation file of the respective human genome were supplied to a MAJIQ builder for splice graph construction using default parameters. The relative abundance of LSVs was calculated using MAJIQ Quantifier with default parameters. The quantifier module computes the abundance of each LSV using marginal percent spliced index (PSI, denoted Ψ). The PSI is calculated for each splice junction (SJ) and expresses the probability of splicing compared to other SJs in a given splicing event. The differential splicing is inferred from relative changes of LSVs among different conditions using delta PSI (dPSI, denoted ΔΨ). The summary and visualization of differential splicing were done for each sample with the MAJIQ Voila package.
2.10. Immunoblotting
Isolation of proteins was performed from the samples used for RNA isolation using RNA Blue according to the manufacturer’s instructions. The resulting protein isolates were separated on 12 % polyacrylamide gels and subsequent immunoblotting detection was performed as described previously [56]. The following antibodies were used: guinea-pig polyclonal serum against TBEV capsid protein (C) (produced in-house), anti-GAPDH antibody [EPR16891] (Abcam; #ab181602), HRP goat anti-guinea pig (Novex; #A18769), HRP goat anti-rabbit IgG antibody (Vector Laboratories; #PI-1000). Sample inputs were standardised by equal protein amounts loaded into each well (10 µg).
2.11. Statistical analyses
If not mentioned otherwise default settings for built-in statistics were used in employed computational packages/tools. Statistical data analysis in the case of TBEV infection dynamics (Fig. 1) was performed in the GraphPad Prism software (version 8.3.0, GraphPad Software, San Diego, CA, USA): cell viability was analysed by Welch́s t-test with multiple testing correction by FDR (Benjamini, Krieger, and Yekutieli, q < 0.05) and TBEV titre differences were analysed by unpaired Student’s t-test (α = 0.05). Conformity of sequencing and qRT-PCR data was analysed with the Spearman correlation analysis (α = 0.05).
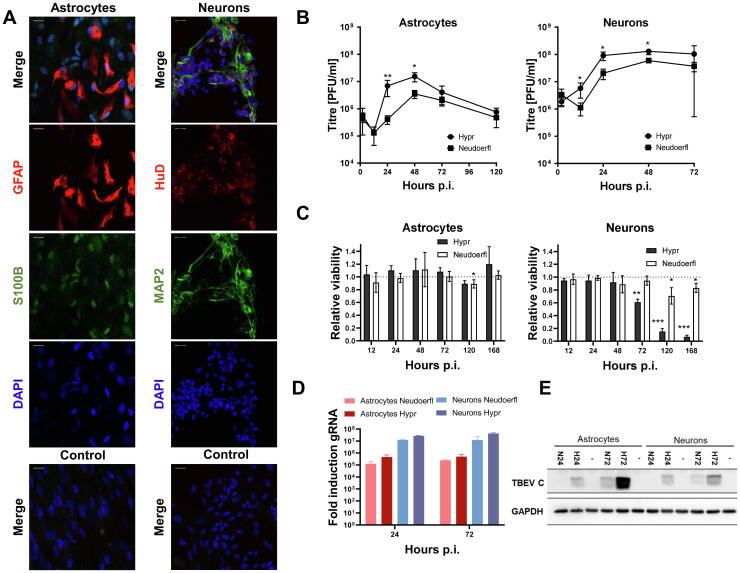
Fig. 1.
TBEV infection dynamics assessment in in vitro differentiated human neurons and astrocytes reveals higher neuronal susceptibility. Astrocytes were differentiated for 22 days, neurons for 14–17 days prior to infection with TBEV strains Hypr and Neudoerfl at MOI of 5. (A) Immunofluorescent labelling of selected differentiation markers in astrocytes (GFAP – red and S100B – green) – left and neurons (HuD – red and MAP2 -green) – right. Nuclei were stained with DAPI. Controls are labelled with secondary antibodies only. The scale bar represents 20 µm. Representative images from two independent experiments are shown. (B) TBEV growth curve of strains Hypr and Neudoerfl, virus shedding into cultivation media was quantified by plaque assay using A549 cells. Data summarise three independent experiments and values in graphs are expressed as mean ± SD. Significant difference from control was calculated by unpaired Student’s t-test (p < 0.05 (*), p < 0.01(**)). (C) Relative viability of neurons and astrocytes upon TBEV infection measured by metabolic conversion of alamarBlue and related to the viability of mock-treated cells (1.0 = 100 %, dotted line). Data summarise four biological and two technical replicates experiments, and values in graphs are expressed as mean ± SD. Welch́s t-test showed a significant difference from control with the multiple testing correction by FDR (Benjamini, Krieger, and Yekutieli, q ≤ 0.05), p ≤ 0.05 (*), p ≤ 0.01 (**), p ≤ 0.001 (***). (D) TBEV replication in astrocytes and neurons was assessed by the amount of genomic RNA quantified by qRT-PCR and related to mock with the ΔΔ-ct method. Results are represented as means ± SD of three biological and technical replicates. (E) TBEV C (capsid protein) and GAPDH (loading control) levels in infected astrocytes and neurons at 24 and 72 h p.i. were determined by immunoblotting. Representative data from three independent experiments are shown. N – Neudoerfl strain, H – Hypr strain. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Results
3.1. Infection of in vitro differentiated human neurons and astrocytes by TBEV Hypr or Neudoerfl resulted in a different pathogenic pattern
Initially, we focused on deriving a suitable experimental model to study TBEV infection in the target cells, human neurons and astrocytes. Primary human neurons and astrocytes were derived from neural progenitor cells of human origin – Human Neural Stem Cells (hNSCs). Following the differentiation, the characteristic appearance of astrocyte and neuron morphology was observed and further, the expression of cell-type-specific markers was verified using an immunofluorescence assay. Neuron identity was verified by the presence of markers HUDD and MAP2, and astrocytes identity was assayed by the expression of S100B and GFAP (Fig. 1A). In each cell type, at least 90 % of cells were labelled positively for at least one of the differentiation markers (data not shown).
Further, we intended to characterize the development of infection in neurons and astrocytes. Therefore, both cell types were infected with two European subtype strains of TBEV, the highly pathogenic Hypr and mild Neudoerfl, at the multiplicity of infection (MOI) of 5. The production of the virus in both cell lines was analysed with plaque assay on A549 cells (Fig. 1B). In addition to TBEV titres, the relative levels of TBEV genomic RNA (gRNA) were determined in the course of infection as well (Fig. 1D).
Infection of both cell types with TBEV resulted in the production of relatively high viral titres. In neurons, peak production of the virus was reached at 48 h p.i. (12.96×107 ± 3.21×107 and 5.93×107 ± 1.16×107 for Hypr and Neudoerfl, respectively) and remained steady further on. In astrocytes, the peak production of the virus was reached at 48 h p.i. (1.53×107 ± 0.46×107 and 0.35×107 ± 0.09×107 for Hypr and Neudoerfl, respectively), however, a decline in the viral titres was observed hereafter (Fig. 1B).
The viability of infected astrocytes was not significantly affected by TBEV in most of the time periods tested (12–168 h p.i.) regardless of the strain of TBEV used. The only significant reduction in viability by 10 % was seen in astrocytes infected with Neudoerfl after 5 days p.i. Dissimilarly, TBEV Hypr reduced the viability of infected neurons by 40 % at 72 h p.i. and <20 % of cells survived in the later time points. Less virulent strain Neudoerfl affected cell viability the most at 120 h p.i. by killing approximately 30 % of the neurons (Fig. 1C).
To illustrate the infection process entirely, the amount of viral genomic RNA was measured by qRT-PCR (Fig. 1D), and the amount of viral capsid protein reflecting the proteosynthesis was determined by immunoblotting at 24 and 72 h p.i. (Fig. 1E). Alike the progeny production, the amount of viral RNA in neurons exceeded the amount in astrocytes by >10-fold in both intervals examined. The more virulent TBEV strain Hypr exhibited a slightly higher replication rate than the Neudoerfl strain (Fig. 1D). A similar trend of more pronounced viral protein production in the case of TBEV Hypr infection was apparent also in the viral protein production (Fig. 1E).
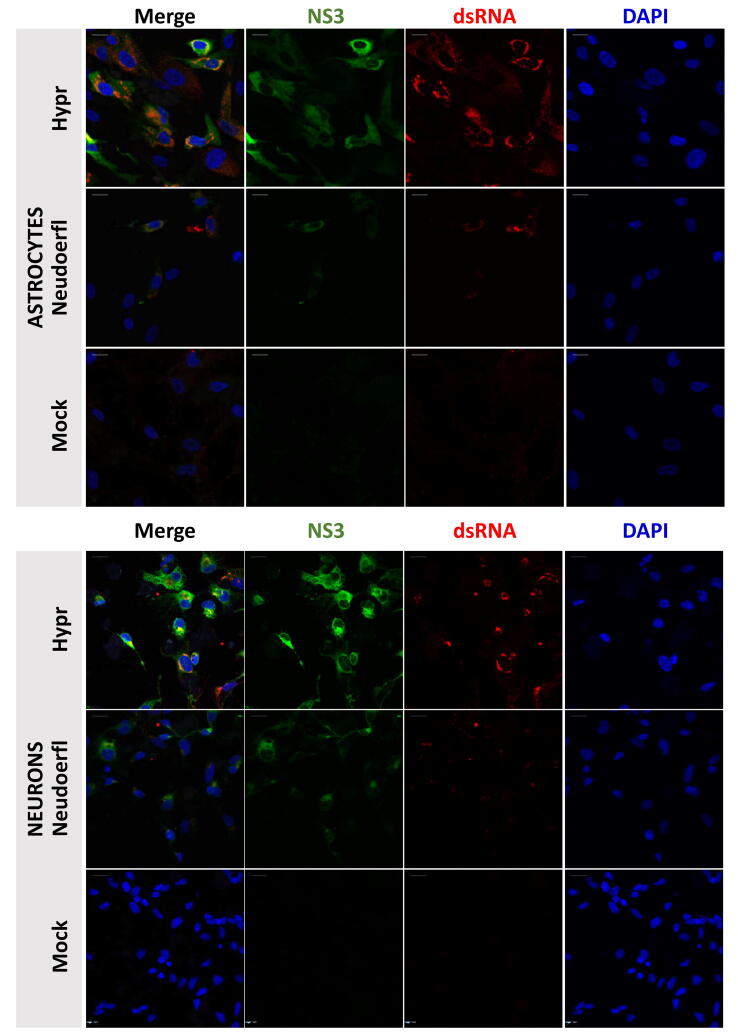
In situ labelling of viral proteins and dsRNA showed that TBEV replication was concentrated in discrete regions adjacent to nuclei, whereas TBEV non-structural protein NS3 exhibited a more diffuse, whole cytoplasmic pattern of distribution (Fig. S1).
To sum up, both cell types were highly susceptible to TBEV infection, supporting virus replication and progeny production. However, neural cell types differed in resilience, with astrocytes able to withstand the infection. We wanted to examine further the factors that underlay the difference in response between neurons and astrocytes.
3.2. Differential expression analysis of TBEV-infected human neurons and astrocytes reveals significant differences linked to the cell origin and virus virulence
Higher susceptibility of neurons to TBEV infection accompanied by higher pathogenicity than in astrocytes is a previously described phenomenon [5], [7], [12]. However, no comprehensive study combines the description of the host response on the mRNA, miRNA, and lncRNA levels. Thus, we performed an integrated differential expression analysis in both human neurons and astrocytes challenged by either TBEV Hypr or Neudoerfl infection. Fig. 2 summarizes the experimental outline for RNA-seq sample preparation (Fig. 2A) and subsequent in silico analyses (Fig. 2B). Differentiated cells were infected at MOI of 5 and total RNA was isolated at 24 and 72 h p.i. to describe the transcription dynamics of the host response. In total, 36 samples were prepared; this number includes three biological replicates for each variant (Table 1).
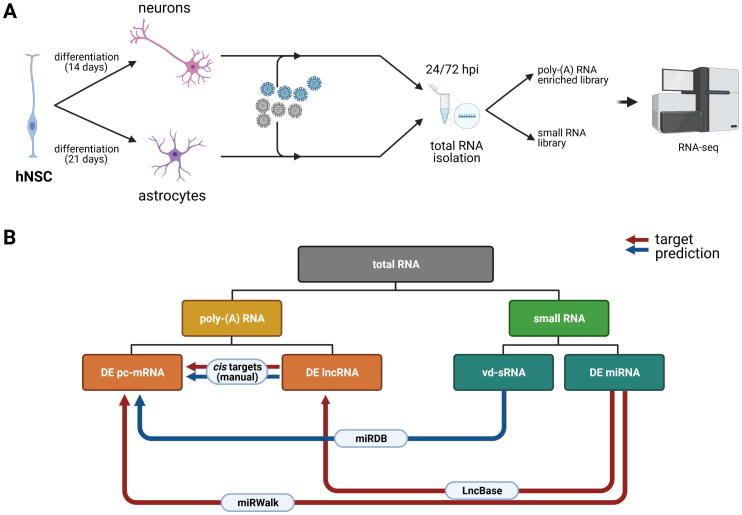
Fig. 2.
Schematic representation of an experimental design performed in this study. (A) hNSCs were differentiated into astrocytes and neurons, infected with TBEV strains Hypr and Neudoerfl at the MOI of 5 and total RNA was isolated at 24 and 72 h p.i. with RNA Blue reagent. The quality of RNA was verified using Bioanalyzer 2100 and (B) Diagram describing the generation of miRNA/mRNA/lncRNA/vd-sRNA networks. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
An overview of samples subjected to RNA-seq analysis.
| Cell type | TBEV strain | Hours p.i. | Number of samples | |
|---|---|---|---|---|
| A24_H | astrocytes | Hypr | 24 | 3 |
| A24_N | astrocytes | Neudoerfl | 24 | 3 |
| A24_M | astrocytes | 24 | 3 | |
| A72_H | astrocytes | Neudoerfl | 72 | 3 |
| A72_N | astrocytes | Hypr | 72 | 3 |
| A72_M | astrocytes | 72 | 3 | |
| N24_H | neurons | Neudoerfl | 24 | 3 |
| N24_N | neurons | Hypr | 24 | 3 |
| N24_M | neurons | 24 | 3 | |
| N72_H | neurons | Neudoerfl | 72 | 3 |
| N72_N | neurons | Hypr | 72 | 3 |
| N72_M | neurons | 72 | 3 |
3.2.1. Differential expression analysis of poly-(A) RNAs
Sequencing of poly-(A) selected library of all samples yielded on average in 80 M 150 PE reads. Quality filtering using Cutadapt dropped on average 1.8 % low-quality and/or short reads. The remaining reads were mapped to the human genome assembly (GRCh38.p13) using TopHat with 78.1 % average mapping efficiency [57]. The details of sequencing output, quality filtering, and mapping statistics per sample are given in Table S2.
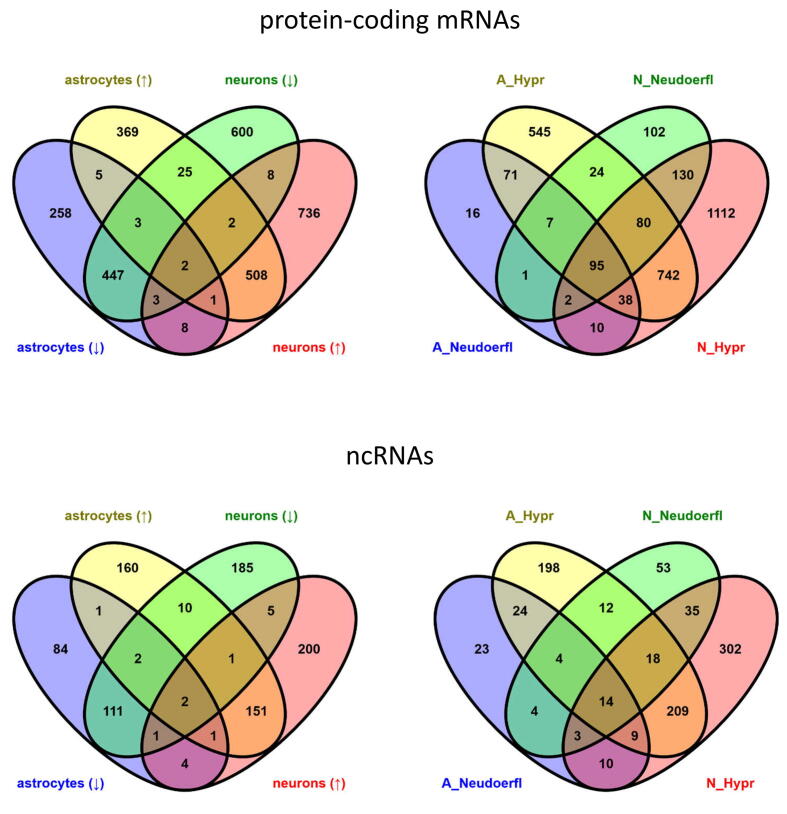
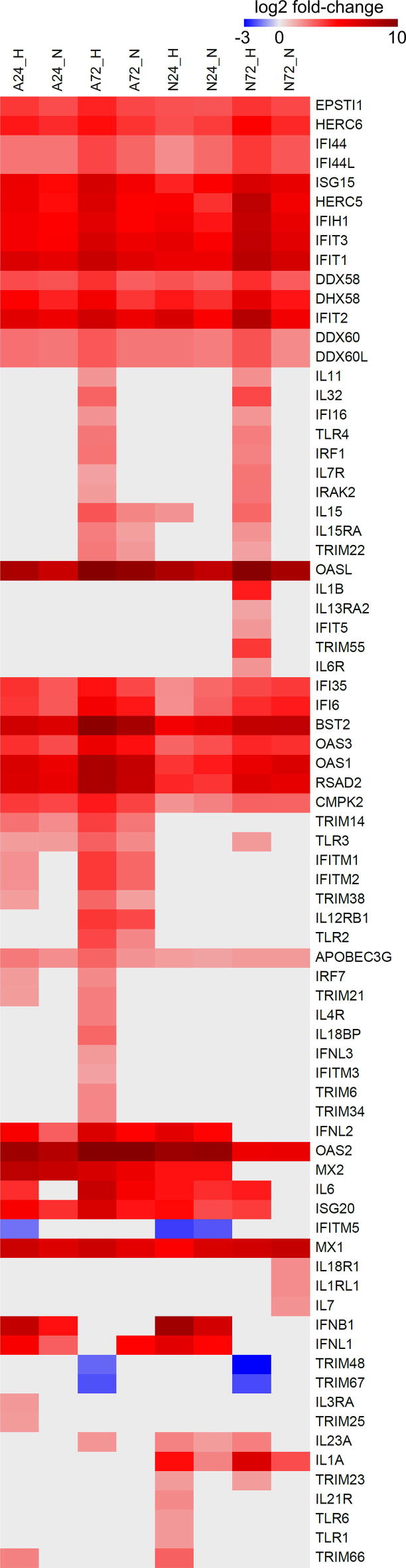
For poly-(A)-enriched datasets, a total number of 4288 differentially expressed genes (DE genes, Benjamini-Hochberg P-value < 0.05 and log2 fold change >1.5 or <−1.5) was identified in TBEV-infected cells when compared to mock control (Table S3). Of these, 2975 genes (69.4 %) represented protein-coding mRNAs (pc-mRNAs) and 923 (21.5 %) ncRNAs (Fig. 3A); pseudogenes and unclassified genes (9.1 %) were excluded from further analyses. Infection with TBEV Hypr resulted in a considerably higher number of DE genes in both, astrocytes and neurons, in comparison to TBEV Neudoerfl (Fig. 3B). Moreover, based on the expression profiles of pc-mRNAs and ncRNAs (Fig. 3C and D), a strong cell- and strain-specific response was observed, when only 1287 (33.1 %) and 591 (15.2 %) DE genes were identified in both cell types or both TBEV strains, respectively (Fig. S2). From those, a conserved group of 32 DE pc-mRNAs up-regulated in TBEV-infected datasets was identified (Table 2), which included genes coding for effectors of the innate immune response, such as receptors sensing viral infection RIG-I and MDA5, or proteins with direct antiviral effect (e.g. BST2, IFIT1-3, RSAD2, MX1, OAS1-3, OASL).
Fig. 3.
Differential expression analysis of poly-(A) enriched RNAs in TBEV-infected neurons and astrocytes reveals strain-specific and cell-specific responses. (A) Graphic summary of poly-(A) classes identified as differentially expressed upon TBEV infection. (B) Graphic summary of DE genes (protein-coding mRNAs and non-coding RNAs) in the respective dataset. (C) Heatmap visualizing the overall expression pattern of protein-coding mRNAs in analysed samples. (D) Heatmap visualizing the overall expression pattern of non-coding RNAs in analysed samples. (E) Verification of RNA-seq data by qRT-PCR. Relative expressions of HSPA-6, OASL, RNVU1, and RSAD2 (viperin) genes were processed via the ΔΔ-ct method with HPRT1 as a reference gene. All samples were analysed in biological and technical triplicates. Spearman correlation was evaluated with GraphPad Prism software. (F) Gene set enrichment analysis (DAVID tool; GO Biological processes; p < 0.001) of significantly up-regulated genes in analysed samples performed by DAVID tool. (G) Gene set enrichment analysis (DAVID tool; GO Biological processes; p < 0.001) of significantly down-regulated genes in analysed samples performed by DAVID tool.
Table 2.
Identification of a common set of genes with TBEV-induced expression in human neurons and astrocytes.
| gene_id | gene_name | A24_H | A24_N | A72_H | A72_N | N24_H | N24_N | N72_H | N72_N |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000239713 | APOBEC3G | 2.39 | 1.97 | 2.87 | 1.85 | 1.62 | 1.54 | 1.70 | 1.72 |
| ENSG00000100342 | APOL1 | 2.90 | 2.12 | 2.98 | 1.90 | 3.83 | 3.32 | 2.17 | 2.14 |
| ENSG00000221963 | APOL6 | 3.29 | 3.03 | 3.02 | 1.99 | 3.02 | 2.76 | 2.77 | 2.42 |
| ENSG00000130303 | BST2 | 7.06 | 6.40 | 9.80 | 8.65 | 5.46 | 6.16 | 7.50 | 7.60 |
| ENSG00000134326 | CMPK2 | 3.75 | 3.51 | 4.46 | 3.58 | 1.86 | 2.23 | 2.94 | 2.89 |
| ENSG00000107201 | DDX58 | 3.41 | 3.23 | 3.94 | 2.99 | 3.23 | 2.93 | 4.05 | 3.05 |
| ENSG00000137628 | DDX60 | 2.60 | 2.45 | 3.12 | 2.45 | 2.46 | 2.31 | 3.23 | 2.03 |
| ENSG00000181381 | DDX60L | 2.60 | 2.45 | 3.12 | 2.45 | 2.46 | 2.31 | 3.23 | 2.03 |
| ENSG00000108771 | DHX58 | 5.10 | 4.27 | 5.57 | 3.83 | 4.49 | 4.00 | 6.17 | 4.58 |
| ENSG00000133106 | EPSTI1 | 3.80 | 3.36 | 4.25 | 3.48 | 3.27 | 3.18 | 3.89 | 3.46 |
| ENSG00000138646 | HERC5 | 5.70 | 4.76 | 6.51 | 5.05 | 5.24 | 3.95 | 7.76 | 5.55 |
| ENSG00000138642 | HERC6 | 4.50 | 4.08 | 4.72 | 3.90 | 3.34 | 3.71 | 5.00 | 4.14 |
| ENSG00000068079 | IFI35 | 3.95 | 3.13 | 4.64 | 3.47 | 1.98 | 2.76 | 3.44 | 3.77 |
| ENSG00000137965 | IFI44 | 2.51 | 2.52 | 3.53 | 2.83 | 2.01 | 2.72 | 3.79 | 3.15 |
| ENSG00000137959 | IFI44L | 2.51 | 2.52 | 3.53 | 2.83 | 2.01 | 2.72 | 3.79 | 3.15 |
| ENSG00000126709 | IFI6 | 3.84 | 3.11 | 5.45 | 4.50 | 1.95 | 2.93 | 4.06 | 4.44 |
| ENSG00000115267 | IFIH1 | 5.47 | 4.97 | 6.24 | 5.02 | 5.58 | 4.56 | 7.48 | 5.99 |
| ENSG00000185745 | IFIT1 | 6.47 | 6.04 | 7.29 | 6.30 | 5.84 | 5.72 | 7.84 | 6.79 |
| ENSG00000119922 | IFIT2 | 6.28 | 5.72 | 6.98 | 5.72 | 6.74 | 5.23 | 8.12 | 5.57 |
| ENSG00000119917 | IFIT3 | 5.51 | 5.14 | 6.71 | 5.65 | 6.09 | 5.24 | 7.61 | 6.21 |
| ENSG00000187608 | ISG15 | 5.81 | 4.84 | 6.83 | 5.46 | 4.25 | 5.14 | 6.64 | 5.99 |
| ENSG00000157601 | MX1 | 7.16 | 6.37 | 7.12 | 6.11 | 5.11 | 6.60 | 6.93 | 7.45 |
| ENSG00000123609 | NMI | 2.85 | 2.54 | 3.87 | 2.95 | 2.39 | 1.81 | 3.14 | 2.90 |
| ENSG00000089127 | OAS1 | 6.63 | 5.84 | 8.54 | 7.47 | 3.87 | 4.44 | 5.88 | 6.44 |
| ENSG00000111335 | OAS2 | 9.00 | 8.05 | 12.07 | 11.03 | 9.10 | 9.32 | 5.68 | 5.91 |
| ENSG00000111331 | OAS3 | 3.99 | 3.37 | 5.85 | 4.68 | 2.83 | 3.31 | 4.18 | 4.01 |
| ENSG00000135114 | OASL | 8.49 | 7.18 | 11.16 | 9.47 | 8.55 | 7.59 | 9.89 | 8.58 |
| ENSG00000173193 | PARP14 | 2.37 | 2.51 | 2.85 | 2.27 | 1.73 | 1.99 | 2.26 | 2.01 |
| ENSG00000134321 | RSAD2 | 6.46 | 6.01 | 8.50 | 7.41 | 4.19 | 3.87 | 6.37 | 6.09 |
| ENSG00000205413 | SAMD9 | 4.06 | 3.99 | 5.06 | 4.10 | 3.67 | 3.84 | 5.47 | 4.94 |
| ENSG00000184979 | USP18 | 4.11 | 3.65 | 4.87 | 3.77 | 1.79 | 2.55 | 3.26 | 3.19 |
| ENSG00000132530 | XAF1 | 2.27 | 2.20 | 2.81 | 2.32 | 2.95 | 2.69 | 3.07 | 2.98 |
The differential expression analysis results were verified using real-time PCR quantification of mRNA levels for selected DE genes (RSAD2, RNVU1, HSPA6, OASL). The correlation between qPCR and NGS data is statistically significant (Spearman correlation r = 0.81, p < 0.001), therefore, the RNA-seq data were successfully validated (Fig. 3E).
To describe the cellular processes affected by the dysregulation of identified DE pc-mRNAs, we performed a gene set enrichment analysis (GSEA) using the DAVID toolkit [58]. A vast majority of significantly enriched GO classes (Benjamini-Hochberg P-value < 0.05) were related to the antiviral immune response (Fig. 3G). In addition, the same analyses of down-regulated gene sets uncovered an interesting phenomenon, where the proper neural development and assembly of the extracellular matrix are negatively affected by TBEV infection (Fig. 3G).
3.2.2. Differential expression analysis of small RNAs
Sequencing the small RNA selected library of all samples yielded an average of 41 M 50 SE reads. Quality filtering using Cutadapt dropped on average 1.8 % low-quality and/or short reads. The remaining reads were mapped to the human genome assembly (GRCh38.p13) using miRDeep2 with 58.2 % average mapping efficiency. The details of sequencing output, quality filtering and mapping statistics per sample are given in Table S4.
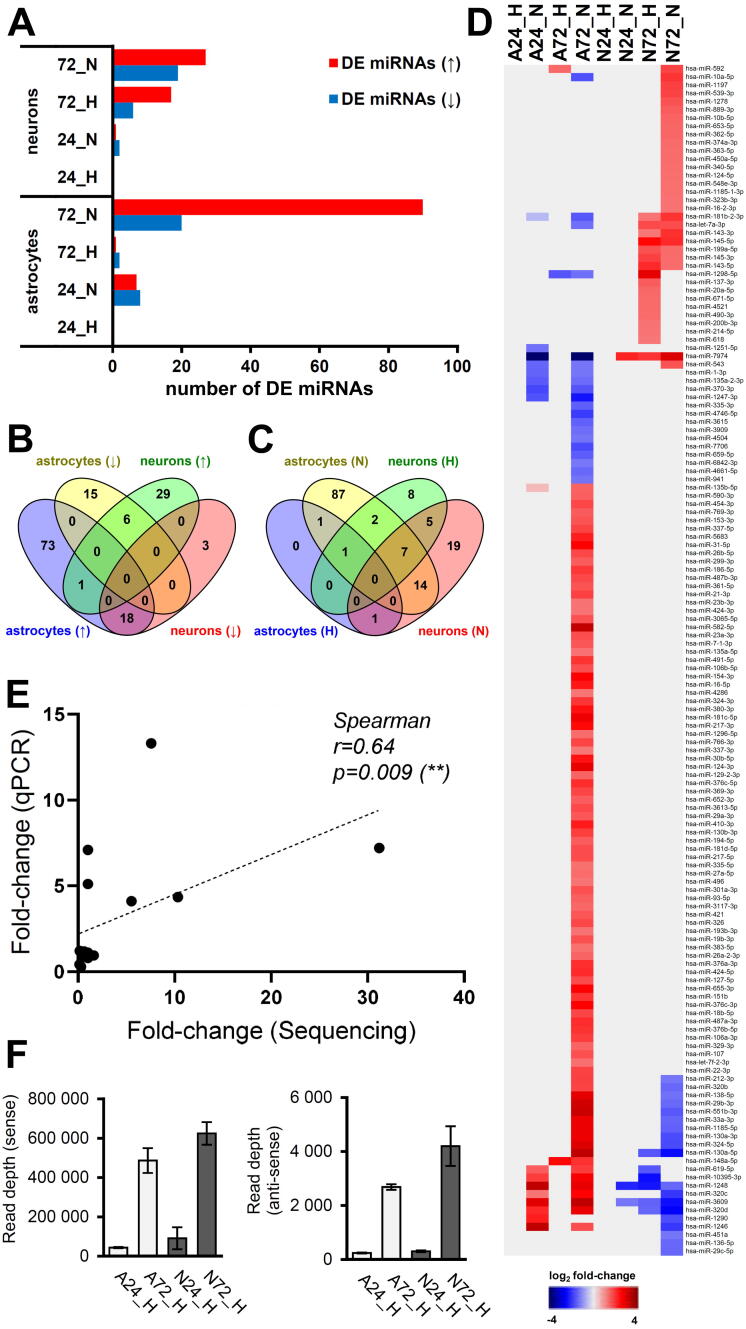
In small RNA datasets, a total of 145 miRNA species were described as significantly dysregulated upon TBEV infection (Benjamini-Hochberg P-value < 0.05 and log2 fold change >1.5 or <−1.5) (Fig. 4; Table S5). Interestingly, while infection with severe Hypr strain resulted in higher numbers of dysregulated pc-mRNAs and ncRNAs when compared to mild Neudoerfl strain, the DE miRNA pattern showed an opposite trend (Fig. 4A), that is distinct reduction of significantly affected miRNAs by more virulent TBEV strain. Furthermore, the expression profiles document a surprisingly high cell-specific response as only one miRNA species (hsa-miR-592) was found to be up-regulated in both cell types, astrocytes (A72_H) and neurons (N72_N) (Fig. 4B and D). The phenomenon of high specificity was also observed between TBEV strains, where only 14 (9.7 %) miRNA species followed the same pattern of dysregulation in the case of both, Hypr and Neudoerfl (Fig. 4C and D). The verification of miRNA-seq data was performed using qPCR quantification of hsa-miR-1248 and hsa-miR-145-5p; the positive correlation between both methods was confirmed to be statistically significant (Fig. 4E; Spearman correlation r = 0.64, p = 0.009).
Fig. 4.
Differential expression analysis of miRNAs in TBEV-infected neurons and astrocytes revealed strain-specific and cell-specific responses. (A) Graphic summary of DE miRNAs in analysed samples. (↑) up-regulated miRNAs, (↓) down-regulated miRNAs. (B) Venn diagram describing the cell specificity of miRNA expression upon TBEV infection. (↑) up-regulated miRNAs, (↓) down-regulated miRNAs. (C) Venn diagram describing the strain specificity of miRNA expression upon TBEV infection. (H) – TBEV Hypr strain, (N) – TBEV Neudoerfl strain. (D) Heatmap visualizing the overall expression pattern of mi RNAs in analysed samples. (E) Verification of RNA-seq data by qRT-PCR. Relative expression of hsa-miR-1248 and hsa-miR-145-5p was calculated using the ΔΔ-ct method with hsa-miR-103a as a reference gene and Sp6 as an internal control. All samples were analysed in biological and technical triplicates. Spearman correlation was evaluated with GraphPad Prism software. (F) Frequency distribution of 21–23 nt vd-sRNAs mapped to TBEV Hypr genome in N72_H (black) and A72_H (red) samples. HS – hot-spot. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Several studies dealing with small RNA profiling in flavivirus-infected cells have reported the presence of vd-sRNAs [34], [35]. We, therefore, assessed whether TBEV infection also resulted in the generation of vd-sRNAs by mapping the small RNA datasets from RNA-seq to TBEV Hypr genomic sequence (GenBank accession no. U39292). The production of vd-sRNAs was confirmed in all TBEV Hypr-infected samples with small RNA species ranging from 16 to 50 nucleotides in size. As expected, the amounts of mapped reads increased with the infection progress in all specimens. Furthermore, vd-sRNAs were predominantly derived from the sense strand of the TBEV genome (Fig. S3) and reached higher, but not significantly different (unpaired Student’s t-test; p < 0.05), levels in neurons (6.24×105 ± 5.74×104) than in astrocytes (4.87×105 ± 6.30×104) (Fig. 4E). These findings positively correlate with higher viral titres and gRNA observed in neurons (Fig. 1). The pattern of reads distribution throughout the viral genome appears rather mosaic with several specific hot-spots. Besides, the hot-spots identified in all of the analysed samples are strikingly conserved and the main difference was observed on the level of read counts (Fig. S4). The most universal/abundant peak was identified at the end of the 3′ UTR.
3.3. Candidate gene networks responsible for a different outcome of TBEV Hypr infection in neurons vs. Astrocytes
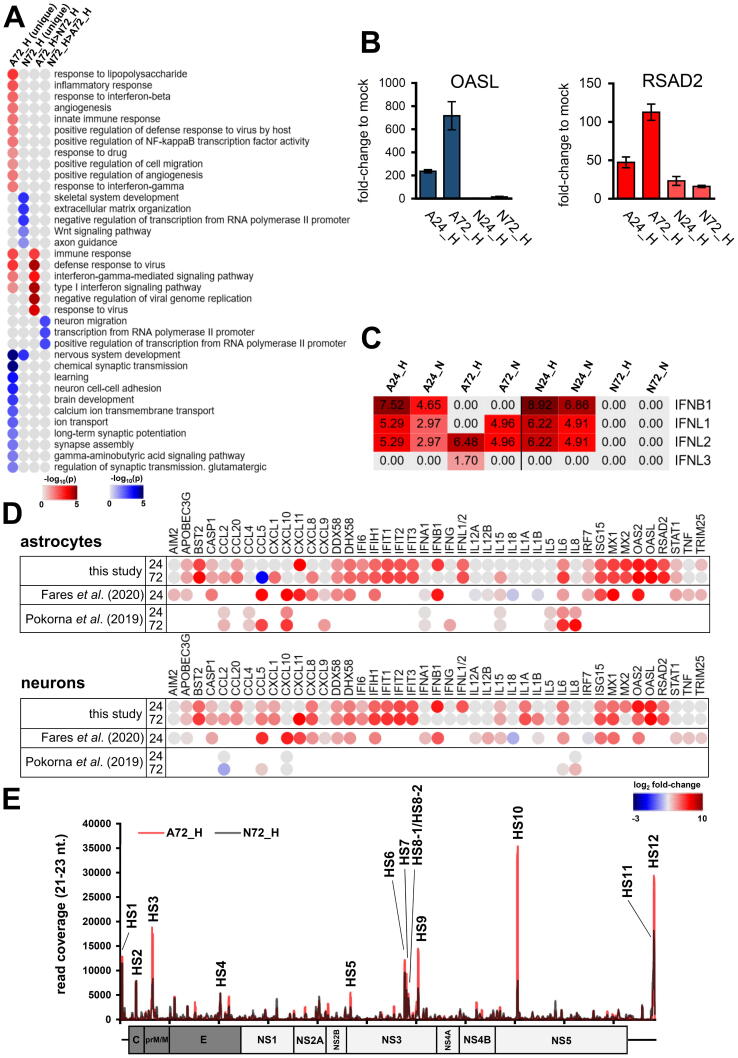
As TBEV Hypr dramatically decreased the number of living neurons but not astrocytes (Fig. 1C), we sought to characterize the differences between N72_H and A72_H datasets to identify potential key factors involved in higher neuronal susceptibility to TBEV Hypr infection. First, we selected DE pc-mRNAs, which were either uniquely up-/down-regulated in N72_H/A72_H datasets or the difference in up-/down-regulation between N72_H/A72_H datasets was higher than 1.5 log2. A total of 1849 DE pc-mRNAs were identified and further divided into four groups according to their expression profile (unique expression in neurons/astrocytes or higher expression in neurons/astrocytes). Subsequently, the GSEA analysis of all groups revealed that the main difference is a more robust immune response elicited in infected astrocytes (Fig. 5A). In more detail, either a triggering of astrocyte-specific antiviral response genes (e.g., TNFSF10, MX2, MMP13, IFITM1, or IFITM2) or higher up-regulation of immune response genes (e.g., OAS1-3, RSAD2, BST2, IL6, or ISG20) was observed (Table 2; Fig. S5). These findings correlate with qPCR data, where RSAD2 (viperin) and OASL were significantly more up-regulated in astrocytes than in neurons (Fig. 5B). A specific expression pattern was observed for IFN-β and IFN-λ genes, when IFN-β seems to be responsible for an early immune response to TBEV followed by the expression of IFN-λ (Fig. 5C). Interestingly, the late expression of IFN-λ (72 h p.i.) appeared in astrocytes only, suggesting its role as a factor associated with the neuronal susceptibility to TBEV infection. Additionally, to put our data in context with previous research, we compared our comprehensive list of DE immune response-related genes with altered protein levels of selected cytokines [13] and mRNAs [12] retrieved for TBEV by others (Fig. 5D). Our data showed a high level of coherence with these previously published expression patterns and considerably extended the previous ones.
Fig. 5.
TBEV Hypr infection in astrocytes results in a stronger innate immune response than in neurons. (A) Heatmap summarizing results from gene set enrichment analysis (DAVID tool; GO Biological processes; p < 0.05) of DE genes 1) uniquely expressed in either A72_H or N72_H, and 2) genes expressed with >1.5 log2 difference in one of the samples. (B) Relative expression of OASL and RSAD2 (viperin) genes in TBEV infected cells. Calculations were performed via the ΔΔ-ct method with HPRT1 as a reference gene. All samples were analysed in biologcal and technical triplicates. (C) Expression pattern of IFN-β and IFN-λ in TBEV-infected cells based on RNA-seq data. (D) Overview and comparison of the expression pattern of selected immune response-related genes from this study and works of Fares et al. [12] and Pokorna et al. [13] (E) Number of mapped reads for vd-sRNAs (16–50 nt) in the respective sample. Data summarise three independent experiments, and values in graphs are expressed as mean ± SD.
The DE miRNAs and lncRNAs were selected based on the same unique or more pronounced response criteria used to evaluate pc-mRNAs. This way, the miRNA trans target prediction was performed for the selected datasets of pc-mRNAs (miRWalk; [51]) and lncRNAs (LncBase v.2; [52]). Prediction results summarized in Table S6 show that 173 genes were identified as possible trans targets (133 pc-mRNAs and 40 lncRNAs), including genes involved in immune response (e.g., OAS1, TRIM38, SPSB4), proper neuronal development and function (e.g., NRARP, KCNK10, SIRPA, CLIC5, TMC8, KIT, CNTN2, DCC) as well as transcription factors (e.g., TFAP2B, HEYL, GATAD2B, BEND3). In order to get a complete overview of the possible interactions, potential cis targets (100 kb upstream and downstream) were predicted for DE lncRNAs with log2-fold change >4 or <−4 as well as for lncRNAs identified as potential targets of DE miRNAs (Table S7). In total, 45 cis targets were predicted, including immune response-related genes (loci RSAD2/CMPK2 and IFNL1-4; IFI6), receptors (e.g., DCC, UTS2R), and transcription factors (e.g., DLX5, OTX2).
The differences in the vd-sRNA mapping profile between the two cell types may substantially influence the enhanced neuronal sensitivity to Hypr infection. One of the possible scenarios suggests that vd-sRNAs could mimic host miRNAs, leading to a lowered expression of a target gene [37]. Therefore, we extracted the 21–23 nt long vd-sRNAs and selected hotspots HS1-HS12 which were present uniquely or in higher/lower numbers in N72_H and A72_H samples (Table 3; Fig. 5E). Surprisingly, HS3, HS7, HS9, HS10, and HS12 had higher read counts in Hypr-infected astrocytes, and only HS11 had a higher read count in Hypr-infected neurons. Subsequently, the prediction of vd-sRNA targets was performed using the miRDB tool in combination with DE pc-mRNA lists. In total, 51 host DE pc-mRNAs were identified (Table 4). Interestingly, vd-sRNAs derived from HS10 and HS12, the two hot-spots with the highest read count in astrocytes, were both predicted to target RNA splicing factors MBNL2 and RBFOX1, whose mRNA levels were significantly lower in A72_H in comparison to N72_H (Table S3). For HS11-derived vd-sRNA, nine mRNA targets with the corresponding expression pattern (N72_H < A72_H) were predicted (LEKR1, DLX1, AOX1, MYCN, PCDH18, DPYSL2, TSHZ1, PFKFB3, BCL9L). Notably, four out of nine identified target genes were characterized as transcription factors (DLX1, MYCN, TSHZ1, BCL9L), with DLX1 and DPYSL2 further considered as factors contributing to proper neural development [59], [60].
Table 3.
Identification of hot-spots for 21–23 nucleotides long vd-sRNAs in TBEV Neudoerfl and Hypr genome. SNPs characteristic for either Hypr or Neudoerfl strains are marked in bold.
| vd-sRNA | sequence (5′-3′) | start | end |
|---|---|---|---|
| HS1 | TGCTTCGGACAGCATTAGCAGC | 26 | 47 |
| HS2 | AAGGCGTTCTGGAACTCAGTCCC | 310 | 332 |
| HS3 | AGGAGAAGAGCCTGTTGACGTG | 645 | 666 |
| HS4 | ATCTCCAGATGTGAACGTGGCC | 2016 | 2037 |
| HS5 | TAAGGACGGTGTCTACAGGATT | 4659 | 4680 |
| HS6 | AAAGTGTGATCTGTTTGAACAG (Hypr) | 5756 | 5777 |
| AAAGTGTGATTTGTTTGAACAG (Neu) | |||
| HS7 | TGAAAAGGACTACTCAAGAGT (Hypr) | 5787 | 5807 |
| TGAAAAGGACTACTCCAGAGT (Neu) | |||
| HS8-1 | TGTGGTGACGACTGATATCTC | 5829 | 5849 |
| HS8-2 | TGACGACTGATATCTCGGAGATG | 5834 | 5856 |
| HS9 | TGGACAGTGTGATGATGATGAC | 6030 | 6051 |
| HS10 | ATTTAGATCAGGAATGGACGTG (Hypr) | 8040 | 8061 |
| ATTTCGATCAGGAATGGACGTG (Neu) | |||
| HS11 | GACACAGATAGTCTGACAAGGA (Hypr) | 10,784 (Hypr), 11,090 (Neu) | 10,805 (Hypr), 11,111 (Neu) |
| GACACAGGTAGTCTGACAAGGA (Neu) | |||
| HS12 | TGATGTGTGACTCGGAAAAAC | 10,808 (Hypr), 11,114 (Neu) | 10,828 (Hypr), 11,134 (Neu) |
Table 4.
List of miRDB tool-predicted host targets of vd-sRNAs. A/N ratio – mapped read count ratio between astrocytes and neurons.
| vd-sRNA | Σ read count |
A/N ratio | predicted gene targets (miRDB) | |
|---|---|---|---|---|
| A72_H | N72_H | |||
| HS1 | 279 497 | 243 019 | 1.15 | N/A |
| HS2 | 171 548 | 169 885 | 1.01 | N/A |
| HS3 | 373 158 | 159 523 | 2.34 | OPCML, THUMPD2, LPXN, GAS7, CDH7, ERBB4, ZMAT3, ANKS1A, GIPC2 |
| HS4 | 92 596 | 96 652 | 0.96 | N/A |
| HS5 | 108 436 | 50 154 | 2.16 | no targets |
| HS6 | 246 963 | 193 317 | 1.28 | N/A |
| HS7 | 193 436 | 120 863 | 1.60 | TBC1D30, PLCH1, G2E3, PBX1, ESRRG, TIPARP, MAN1A1 |
| HS8-1 | 91 952 | 72 746 | 1.26 | N/A |
| HS8-2 | 92 748 | 81 201 | 1.14 | N/A |
| HS9 | 309 580 | 125 640 | 2.46 | BDNF, KCNS2, SHISAL1, GOLGA4, LGI1 |
| HS10 | 751 154 | 150 617 | 4.99 | PCHD20, TM4SF18, TMTC1, MCTP1, RBFOX1, TRHDE, ZMYM5, SEMA3E, MBNL2, LCOR, CXADR, CALB1, ARID2, GABRG2, DIRAS2, NEK7, FUT9 |
| HS11 | 124 140 | 202 003 | 0.61 | LEKR1, DLX1, AOX1, MYCN, PCDH18, DPYSL2, TSHZ1, PFKFB3, BCL9L |
| HS12 | 597 869 | 338 876 | 1.76 | RBFOX1, RNF212, MBNL2, GRIN2A, CREG2, KRR1 |
| vd-sRNA | Σ read count | N/H ratio | predicted gene targets (miRDB) | |
| N72_N | N72_H | |||
| HS1 | 5 594 | 243 019 | 0.02 | no targets |
| HS2 | 26 642 | 169 885 | 0.16 | no targets |
| HS3 | 39 241 | 159 523 | 0.25 | SIAH3, NKAIN1, EBF1, KLHDC8A, ST6GAL1, DNMT3B, SOGA1, SHF, KLHL13 |
| HS4 | 25 886 | 96 652 | 0.27 | PIK3C2B, EPHB1, NYNRIN, ELAVL2, SH3TC2, ZNF124, SLITRK5, SIRPA, ONECUT2, BDKRB2, ACACA, RTL8B, ZNF730 |
| HS5 | 11 486 | 50 154 | 0.23 | no targets |
| HS6 | 27 970 | 193 317 | 0.14 | AKR1C2, NCAM1 |
| HS7 | 22 292 | 120 863 | 0.18 | TFAP2A, ELAVL4, BCL11B, POU3F4, AUTS2, MN1, SH3TC2, DACH1, ONECUT2, PIP4P2, TMEM229B |
| HS8-1 | 20 918 | 72 746 | 0.29 | NFASC, NCAM1 |
| HS8-2 | 22 262 | 81 201 | 0.27 | no targets |
| HS9 | 34 893 | 125 640 | 0.28 | NAV1, GLRA2, GAS2L3 |
| HS10 | 52 035 | 150 617 | 0.35 | PRDM12, NRN1, ELAVL4, CDKN1C, ANOS1, NXPH2, PSRC1, PPM1L, CYYR1, LUZP2, LRRTM3 |
| HS11 | 19 224 | 202 003 | 0.10 | PTPN5, DCX, MYCN, DLX1, NREP, THSD7A, NXPH2, PIK3C2B, CAMK2A, EPHB1, RAPGEF5, PCDH18, DPYSL2, KCND2, GNG2, TSHZ1, KLHL13, ONECUT2, ISGF3, PFKFB3, BCL9L, PPP1R17 |
| HS12 | 46 500 | 338 876 | 0.14 | DACH1, NDRG4 |
3.4. Candidate gene networks responsible for distinct TBEV infection outcomes in neurons during infection with strains of varying severity
The comparison of infection dynamics and cell death rate between severe Hypr and mild Neudoerfl strains in infected neurons confirmed a higher pathogenic effect for Hypr (Fig. 1). Therefore, observed differences between Hypr and Neudoerfl infection manifestation in human neuronal cells drove us to search for differences between N72_H and N72_N datasets on the pc-mRNA/miRNA/ncRNA level. Using an in silico approach, we employed an analogous pipeline to A72_H and N72_H datasets.
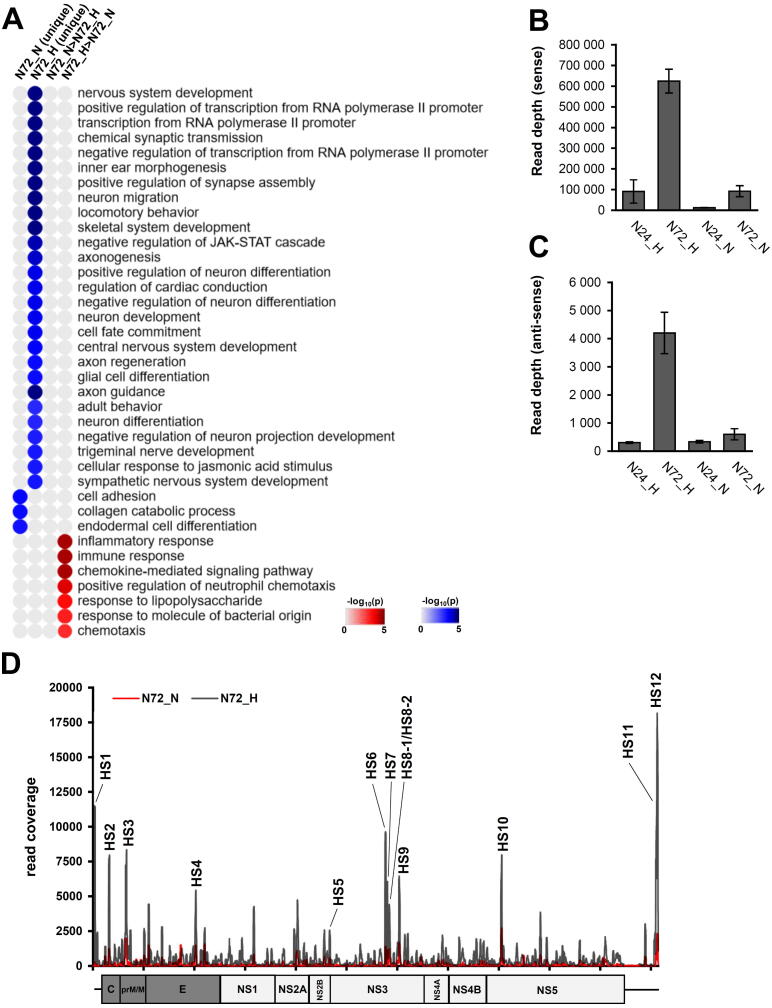
We identified 1935 DE pc-mRNAs either uniquely up-/down-regulated in N72_H/N72_N datasets or differentially expressed (both up and down-regulated) between N72_H/N72_N datasets with a fold change higher than 1.5 log2. The GSEA analysis found significant differences in infection between the two TBEV strains in (i) the extent of the negative impact of Hypr infection on neuronal development, (ii) the level of Hypr-induced dysregulated activity of RNA polymerase II promoters, and (iii) higher activation of host immune response to Hypr infection (Fig. 6A).
Fig. 6.
Comparison of TBEV Hypr/Neudoerfl infection in neurons reveals disruption of proper neural development. (A) Heatmap summarizing results from gene set enrichment analysis (DAVID tool; GO Biological processes; p < 0.05) of DE genes 1) uniquely expressed in either N72_H or N72_N, and 2) genes expressed with >1.5 log2 difference in one of the samples. (B) Number of mapped reads for vd-sRNAs (16–50 nt) in the respective sample. Data summarise three independent experiments and values in graphs are expressed as mean ± SD. (C) Frequency distribution of 21–23 nt vd-sRNAs mapped to TBEV Hypr/Neudoerfl genome in N72_H (black) and N72_N (red) samples. HS – hot-spot. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We identified 1935 DE pc-mRNAs either uniquely up-/down-regulated in N72_H/N72_N datasets or differentially expressed (both up and down-regulated) between N72_H/N72_N datasets with a fold change higher than 1.5 log2. The GSEA analysis found significant differences in infection between the two TBEV strains in (i) the extent of the negative impact of Hypr infection on neuronal development, (ii) the level of Hypr-induced dysregulated activity of RNA polymerase II promoters, and (iii) higher activation of host immune response to Hypr infection (Fig. 6A).
Identifying the complex regulatory network of small regulatory RNAs also requires the identification of their target sites. Thus, we selected DE expressed miRNAs and lncRNAs under the same conditions as the pc-mRNAs selection. Using the miRWalk and lncBase v.2 databases, the miRNA targets were identified for pc-mRNAs and lncRNAs, respectively. The predicted 268 trans targets for both small RNA classes (207 pc-mRNAs and 61 lncRNAs) are listed in Table S8. The complete overview of the possible interactions was predicted by the identification of potential cis targets (100 kb upstream and downstream) for DE lncRNAs with log2-fold change >4 or <−4 and lncRNAs identified as potential targets of DE miRNAs (Table S9). In total, 47 cis targets were predicted, including transcription factors (e.g. DLX5, DLX6, TFAP2A) and various receptors (e.g. CHRNA3, CHRNB4, DCC, PTPRH).
The assessment of vd-sRNA presence in TBEV Neudoerfl-infected cells revealed a similar pattern to Hypr vd-sRNA data – increasing number of vd-sRNAs through the infection and higher numbers of sense reads (Fig. S4). Comparison between N72_N and N72_H datasets further documented higher numbers of mapped reads during Hypr infection compared to Neudoerfl (Fig. 6B and C), which is in agreement with higher titres and viral gRNA levels (Fig. 1). The distribution pattern of 21–23 nt long vd-sRNAs was identical for 12 hot-spots with higher abundance in N72_H samples (Table 3; Fig. 6D). Prediction of host mRNA targets was performed using the miRDB tool in combination with DE pc-mRNA datasets with 66 identified host mRNA targets (Table 4).
We further focused on HS11, the hot-spot with the highest number of mapped 21–23 nt reads in N72_H samples. The vd-sRNA derived from HS11 was predicted to target 22 mRNAs with the corresponding expression pattern (N72_H < N72_N), out of which seven pc-mRNAs (DLX1, MYCN, PCDH18, DPYSL2, TSHZ1, PFKFB3, BCL9L) were also identified as less expressed in A72_H samples compared to N72_H. We suggest that these genes represent a candidate set of markers of increased pathogenicity that Hypr strain confers towards neurons.
3.5. TBEV infection results in altered pre-mRNA splicing
Both, neuron/astrocyte-specific response to infection and pathogenicity-related response to infection, led us to examine the potential impairment of host pre-mRNA splicing events since several lncRNAs and vd-sRNAs were predicted to interact with partners involved in this pathway. The idea was also supported by previous studies describing DENV- and ZIKV-induced alterations in host pre-mRNA splicing [27], [61], [62]. Thus, RNA-seq data of astrocytes and neurons infected with TBEV were submitted to a differential splicing analysis using MAJIQ v2.2 [54]. The corresponding mock controls were used as a reference for differential splicing calculation. Identified LSVs were further classified by MAJIQ to binary and complex events. Binary LSVs include exon skipping (ES), intron retention (IR), alternative 3′/5′ splice site (A3SS/A5SS) and involve only two exons or two splice sites in the same exon. Complex LSVs combine several binary events originating from or targeting the same site. In total, differential splicing analysis revealed 4009 LSVs in all datasets (Table S10) and the vast majority of detected LSVs (3639; 90.7 %) were documented for N72_H (2390; 59.6 %) and A72_H (1249; 31.2 %) datasets; interestingly, infection with Neudoerfl strain resulted in a total of only 94 (2.3 %) identified LSVs. The detailed characterization of identified LSVs revealed that exon skipping was the most frequent LSV type (Fig. 7B). The biological pathways affected by TBEV-induced differential splicing were determined using a GSEA analysis for A72_H, N72_H, and N72_N samples using the list of differentially spliced genes. Significantly enriched GO terms (biological process; Benjamini-Hochberg P-value < 0.05) were identified only in the case of A72_H and N72_H samples (Fig. 7C). Only a single GO term “mRNA splicing process” was significantly enriched in both samples. The remaining biological processes were significantly enriched only in the case of N72_H. The most highly ranking terms were found among DNA repair, G2/M transition of mitotic cell cycle, protein transport, and membrane fusion biological processes. MTUS2 and CFAP61 were the top-ranked genes recognized in the list of TBEV-induced alterations in host pre-mRNA splicing in A72_H and N72_H samples, respectively. Therefore, we prepared sashimi plots of LSVs identified as significantly spliced in these two genes using the MAJIQ Voila tool (Fig. 7D). As a control, the visualization of bam files used for LSV identification in MTUS2 and CFAP61 genes was done using Integrated Genome Browser (IGB) v9.1.8 [71] to support the MAJIQ LSV detection and quantification capacity. Both IGB graphs also support the presence of newly included exons in LSV detection by MAJIQ in these two genes in Hypr-infected cells (Fig. 7E).
Fig. 7.
TBEV infection induces changes in the splicing of host pre-mRNAs. (A) Graphical summary of identified LSVs (local splicing variants) in RNA-seq data using MAJIQ tool. Binary LSVs include exon skipping (ES), intron retention (IR), alternative 3′/5′ splice site (A3SS/A5SS) and involve only two exons or two splice sites in the same exon. Complex LSVs combine several binary events originating from or targeting the same site. (B) Classification of binary and complex LSVs identified in TBEV-infected astrocytes and neurons (combined). (C) Heatmap summarizing results from gene set enrichment analysis (DAVID tool; GO Biological processes; p < 0.05) of genes in which > 1 LSV was identified (for samples A72_H, N72_H, and N72_N). (D) MAJIQ-generated Sashimi plots of top-ranked genes in N72_H (CFAP61) and A72_H (MTUS2) samples. Numbers above splicing event arcs represent the number of mapped junction reads. For the sake of readability, cropped areas with identified LSVs are shown. (E) Analysis of read depth distribution across CFAP61 and MTUS2 genes (areas with identified LSVs) in N72_H/M and A72_H/M samples, respectively, using the integrated genome browser.
4. Discussion
TBEV, a neurotropic flavivirus, targets the host CNS and frequently causes severe encephalitis in infected patients. The exact mechanism responsible for TBEV-induced neuropathogenesis has not been fully understood to date, although several studies suggest the combination of virus-induced neuronal cell death and the immunopathogenic effect of activated host immune response [[10], [12]]. The complexity of the whole process is further supported by the different outcomes of TBEV infection in neurons and astrocytes [5], [6], [7], [12]. One of the suggested mechanisms underlying the distinct susceptibility of both cell types to TBEV infection is the neuron-/astrocyte-specific expression pattern of immune response-related genes [12], [13]. However, both studies assayed only a limited number of pre-selected genes in a microarray-based analysis. Here, we present a thorough transcriptomic study describing differential expression of both small RNAs as well as poly-(A) RNAs in human neurons and astrocytes infected by mild and severe strains of TBEV in an early and late stage infection. Thus, the combination of all factors (infection time, cell type, TBEV strain) gives a complex overview of host-cell response at the RNA level and provides new significant data for a better understanding of TBEV neuropathogenesis.
4.1. Cell-type specific response in neurons and astrocytes challenged by TBEV
In silico pc-mRNA-miRNA-lncRNA interactome analysis identified major differences in response between TBEV challenged astrocytes and neurons. The key feature was the expression pattern of immune response-related genes (Fig. 5; Table S3; Fig. S3), which is in concordance with studies of Fares et al. and Pokorna et al. [12], [13]. The phenomenon of cell type-specific immune response to flaviviral infection in CNS was also described for WNV [63], which further highlights its relevance for the flaviviral pathogenic effect. Besides, Hypr infection negatively affects extracellular matrix organization and RNAPII promoter activity pathways in infected neurons, which may also play a substantial role in the impairment of the neuronal tissue.
The altered expression pattern of pc-mRNAs, which belong to the aforementioned pathways, could result from the miRNA/lncRNA-driven regulation process. Thus, we further focused on the linkage of the DE miRNAs/lncRNAs to their possible targets in the DE pc-mRNA datasets. The in silico prediction revealed several networks possibly involved in the differential expression pattern documented in astrocytes and neurons.
Interferon signalling is considered a key regulatory cascade of the innate immune system triggering the expression of a wide panel of ISGs with a direct or indirect antiviral effect [64]. Our data revealed that in addition to IFN-β, IFN-λ1/2 expression is induced in both cell types in an early TBEV infection (24 h p.i.). However, only astrocytes maintained the IFN-λ1/2 expression later during the infection (72 h p.i.). Interestingly, the IFN-λ cluster was predicted to be a cis target regulated by AC011445.2 lncRNA (ENSG00000269246), which was highly up-regulated in A72_H, but not in N72_H samples (Fig. 5, Table S3). Our previous study demonstrates that IFN-λ1 is the predominantly expressed interferon in response to TBEV infection in cells of neuronal origin [28], and its significance was also demonstrated in the case of WNV infection [65]. Thus, the deficiency of IFN-λ1/2 production in neurons is a potential contributor to the decreased expression of specific ISGs, which are necessary for a successful antiviral response to TBEV in astrocytes. Similarly, astrocyte-specific up-regulation of AL445490.1 lncRNA (ENSG00000225886) may contribute to the elevated expression of IFI6, a predicted cis-target of this lncRNA (Table S7).
When we focused on the neural cell-type-specific response in the case of host miRNAs, the hsa-miR-1298 is the only miRNA species which was significantly up-regulated in Hypr-infected neurons, while significantly down-regulated in Hypr-infected astrocytes (Fig. 4D). Among the predicted hsa-miR-1298 mRNA targets with the corresponding DE profile (LUZP2, SPSB4, VCAN, TFAP2B, MAST3, RSAD2, CMPK2, and DLX5), the RSAD2 and CMPK2 we found the most important since both were documented to interfere with flavivirus infection [66]. Additionally, hsa-miR-7974, the second most up-regulated miRNA in Hypr-infected neurons, was predicted to target OAS1, whose DE profile also showed a decreasing induction rate in Hypr-infected neurons and which has already been proven to possess strong antiviral activity against flaviviruses [67]. Interestingly, a post mortem analysis of brain tissue from children with congenital ZIKV syndrome revealed elevated levels of hsa-miR-145 and hsa-miR-148a [68]. Similarly, here we documented that hsa-miR-145-5p was up-regulated exclusively in Hypr- and Neudoerfl-infected neurons, whereas hsa-miR-148a-5p was up-regulated exclusively in Hypr- and Neudoerfl-infected astrocytes (Fig. 4).
Besides dysregulated host miRNAs, small RNAs derived from the viral genome may also interfere with host mRNA levels [37], [38]. Our data document an accumulation of TBEV vd-sRNAs in infected neurons and astrocytes; the distribution pattern of vd-sRNAs across the TBEV genome was remarkably similar among the samples, with coverage rate as the only variable factor. Interestingly, in the case of TBEV Hypr-infected astrocytes, vd-sRNAs (21–23 nt long) derived from the two hot-spots with the highest coverage (HS10, HS12) were both predicted to target MBNL2 and RBFOX1. These genes are involved in pre-mRNA splicing regulation necessary for proper neural development [69], [70], [71], [72], and their depletion may thus contribute to the observed dysregulation of the splicing process in astrocytes. This hypothesis is supported by a finding that one of the RBFOX1 targets, SNAP25 [69], was identified among differently spliced mRNAs in TBEV Hypr-infected astrocytes. Analogously, vd-sRNA derived from HS11 (hot-spot with the highest coverage in TBEV Hypr-infected neurons) was predicted to target 22 genes in total. However, only seven genes (DLX1, MYCN, PCDH18, DPYSL2, TSHZ1, PFKFB3, BCL9L) met the criterion of being expressed at a lower rate in N72_H in comparison to A72_H and N72_N samples. The dysregulation of mRNA levels through HS11-derived vd-sRNA for these genes could be thus responsible for the severe pathogenic effect in Hypr-infected neurons when compared to Hypr-infected astrocytes and Neudoerfl-infected neurons.
4.2. Alternative splicing as an important co-factor of high TBEV Hypr virulence in neurons
Based on the previous studies describing flavivirus-induced changes in the host pre-mRNA splicing process [61], [62] in combination with observed dysregulation of various splicing factors upon TBEV infection (e.g., MBNL2, RBFOX1, or CELF3/4/5; Table S3), we hypothesized that TBEV infection alters the splicing of host mRNAs. Indeed, the differential splicing analysis revealed significant alterations in TBEV-infected cells. Since more than half of differently spliced LSVs were identified in Hypr-infected neurons (64.5 %), we contemplate that this phenomenon may also contribute substantially to the high pathogenic effect of Hypr strain in infected neurons. This assumption is further corroborated by the list of affected significantly enriched biological pathways, including DNA repair, G2/M transition during the mitotic cycle, protein transport, and membrane fusion. Malfunction of these processes, especially DNA repair and G2/M checkpoint, results in an elevated apoptotic rate and neuronal dysfunction [73], [74].
Earlier, we reported a diverse rate of pathogenesis between TBEV Hypr and Neudoerfl strains in a human cell line of neuronal origin [56]. Similar findings were documented in the human primary neurons in this work, where Hypr infection resulted in a substantially more severe pathogenic effect (Fig. 1C). Therefore, we tried to extrapolate the link between the intensity of pathogenesis, replication rate, and host transcriptional response. Even though both strains reached high titres in neurons, Hypr strain replication and progeny production were more efficient, generating almost 10-times more mature virions (Fig. 1B), gRNA (Fig. 1D), and vd-sRNAs (Fig. 6B-D). Moreover, a lower replication rate of the Neudoerfl strain correlates with low numbers of identified DE poly-(A) RNAs (Fig. 3B) and altered splicing events (Fig. 7A), which suggests a direct dependency on virus replication rate and intensity of the host response. Almost identical findings were described in the study of Överby et al., where the Hypr strain replicated with higher efficiency and induced a stronger immune response than the Neudoerfl strain [75]. On the contrary, the study of Sessions et al. described an inverse trend, where an attenuated DENV1 strain caused three-fold more changes in the host transcriptome than the wild-type DENV1 [61].
4.3. Diverse strain virulence in neurons
The lower replication and pathogenesis rates observed for the mild Neudoerfl strain can also be perceived as a consequence of a more successful host response. However, Neudoerfl-infected cells showed a lower rate of immune response activation in the case of pc-mRNAs (Table S3 and Fig. S3). Thus, the dysregulation of miRNA with proviral or antiviral properties may be the factor involved. We, therefore, compared the list of DE miRNAs in N72_H and N72_N samples with the list of experimentally verified proviral and antiviral miRNAs in the case of ZIKV [76] and DENV [77]. However, no corresponding matches were found. We, therefore, searched for novel antiviral miRNAs targeting Hypr or Neudoerfl gRNA using the miRDB tool. The hsa-miR-592 was identified as the only miRNA species targeting Hypr and Neudoerfl gRNA. At the same time, this miRNA was up-regulated in samples with lower TBEV titres (A72_H and N72_N), but not in samples with higher TBEV titres (N72_H) (Fig. 4). These correlations thus suppose that hsa-miR-592 may negatively regulate TBEV replication in human neurons and astrocytes. Except for the direct interaction with TBEV gRNA, hsa-miR-592 may indirectly affect TBEV replication as well. He et al. [78] experimentally proved that hsa-miR-592 targets SPRY2 mRNA, which is a negative regulator of interferon signalling [79], and in our study was shown to be up-regulated only in N72_H samples. We thus identified a new potential circuit of host cell countermeasure against TBEV involving hsa-miR-592 unique among flaviviruses.
5. Conclusions
Altogether, we have characterized the alterations in poly-(A) RNA and small RNA expression profile of human neurons and astrocytes upon TBEV infection using two TBEV strains of distinctive virulence (mild Neudoerfl and severe Hypr). Subsequent integrative in silico analysis of miRNA/mRNA/lncRNA/vd-sRNA networks and pre-mRNA splicing found significant changes in inflammatory and immune response pathways, nervous system development and regulation of mitosis in TBEV Hypr-infected neurons. Candidate mechanisms include specific regulation of host mRNA levels via differentially expressed miRNAs/lncRNAs or vd-sRNAs mimicking endogenous miRNAs and virus-driven modulation of host pre-mRNA splicing. Thus, our data provide a valuable source of information for any research that aims to investigate and further characterise the mechanism of TBEV-host interactions and the related process of TBEV pathogenesis. Moreover, strain-specific expression pattern of selected host genes also represents a list of potential biomarkers, which can be used for improved TBEV diagnostics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank dr. R. Randall (University of St. Andrews, UK) for providing us with A549 cell line and dr. M. Bloom (National Institute of Allergy and Infectious Diseases, USA) for NS3 antibodies.
Funding
This research was supported by the Ministry of Education, Youth, and Sports of the Czech Republic INTER-ACTION (projects LTARF 18021 and LTAUSA 18040), and the Czech Science Foundation (GA18-27204S).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.05.052.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
Supplementary figure 4.
Supplementary figure 5.
References
- 1.Gritsun TS, Lashkevich VA, Gould EA. Tick-borne encephalitis. Antiviral research. 2003;57(1-2):129-46. Epub 2003/03/05. doi: 10.1016/s0166-3542(02)00206-PubMed PMID: 12615309. [DOI] [PubMed]
- 2.de Graaf JA, Reimerink JH, Voorn GP, Bij de Vaate EA, de Vries A, Rockx B, et al. First human case of tick-borne encephalitis virus infection acquired in the Netherlands, July 2016. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2016;21(33). Epub 2016/08/27. doi: 10.2807/1560-7917.Es .2016.21.33.30318. PubMed PMID: 27562931; PubMed Central PMCID: PMCPMC4998423. [DOI] [PMC free article] [PubMed]
- 3.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nature reviews Microbiology. 2018;16(3):125-42. Epub 2018/02/1doi: 10.1038/nrmicro.2017.170. PubMed PMID: 29430005; PubMed Central PMCID: PMCPMC7097628. [DOI] [PMC free article] [PubMed]
- 4.Gelpi E, Preusser M, Garzuly F, Holzmann H, Heinz FX, Budka H. Visualization of Central European tick-borne encephalitis infection in fatal human cases. Journal of neuropathology and experimental neurology. 2005;64(6):506-12. Epub 2005/06/28. doi: 10.1093/jnen/64.6.506. PubMed PMID: 15977642. [DOI] [PubMed]
- 5.Bílý T, Palus M, Eyer L, Elsterová J, Vancová M, Růžek D. Electron Tomography Analysis of Tick-Borne Encephalitis Virus Infection in Human Neurons. Scientific reports. 2015;5:1074Epub 2015/06/16. doi: 10.1038/srep1074PubMed PMID: 26073783; PubMed Central PMCID: PMCPMC4466586. [DOI] [PMC free article] [PubMed]
- 6.Potokar M, Korva M, Jorgačevski J, Avšič-Županc T, Zorec R. Tick-borne encephalitis virus infects rat astrocytes but does not affect their viability. PloS one. 2014;9(1):e86219. Epub 2014/01/28. doi: 10.1371/journal.pone.0086219. PubMed PMID: 24465969; PubMed Central PMCID: PMCPMC3896472. [DOI] [PMC free article] [PubMed]
- 7.Palus M, Bílý T, Elsterová J, Langhansová H, Salát J, Vancová M, et al. Infection and injury of human astrocytes by tick-borne encephalitis virus. The Journal of general virology. 2014;95(Pt 11):2411-26. Epub 2014/07/09. doi: 10.1099/vir.0.068411-0. PubMed PMID: 25000960. [DOI] [PubMed]
- 8.Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta neuropathologica. 2010;119(1):7-35. Epub 2009/12/17. doi: 10.1007/s00401-009-0619-PubMed PMID: 20012068; PubMed Central PMCID: PMCPMC2799634. [DOI] [PMC free article] [PubMed]
- 9.Diniz J.A., Da Rosa A.P., Guzman H., Xu F., Xiao S.Y., Popov V.L., et al. West Nile virus infection of primary mouse neuronal and neuroglial cells: the role of astrocytes in chronic infection. The American journal of tropical medicine and hygiene. 2006;75(4):691–696. Epub 2006/10/14 PubMed PMID: 17038696. [PubMed] [Google Scholar]
- 10.van Marle G, Antony J, Ostermann H, Dunham C, Hunt T, Halliday W, et al. West Nile virus-induced neuroinflammation: glial infection and capsid protein-mediated neurovirulence. Journal of virology. 2007;81(20):10933-49. Epub 2007/08/03. doi: 10.1128/jvi.02422-06. PubMed PMID: 17670819; PubMed Central PMCID: PMCPMC2045515. [DOI] [PMC free article] [PubMed]
- 11.Jorgačevski J, Korva M, Potokar M, Lisjak M, Avšič-Županc T, Zorec R. ZIKV Strains Differentially Affect Survival of Human Fetal Astrocytes versus Neurons and Traffic of ZIKV-Laden Endocytotic Compartments. Scientific reports. 2019;9(1):8069. Epub 2019/05/31. doi: 10.1038/s41598-019-44559-8. PubMed PMID: 31147629; PubMed Central PMCID: PMCPMC6542792. [DOI] [PMC free article] [PubMed]
- 12.Fares M, Cochet-Bernoin M, Gonzalez G, Montero-Menei CN, Blanchet O, Benchoua A, et al. Pathological modeling of TBEV infection reveals differential innate immune responses in human neurons and astrocytes that correlate with their susceptibility to infection. Journal of neuroinflammation. 2020;17(1):76. Epub 2020/03/05. doi: 10.1186/s12974-020-01756-x. PubMed PMID: 32127025; PubMed Central PMCID: PMCPMC7053149. [DOI] [PMC free article] [PubMed]
- 13.Pokorna Formanova P, Palus M, Salat J, Hönig V, Stefanik M, Svoboda P, et al. Changes in cytokine and chemokine profiles in mouse serum and brain, and in human neural cells, upon tick-borne encephalitis virus infection. Journal of neuroinflammation. 2019;16(1):205. Epub 2019/11/09. doi: 10.1186/s12974-019-1596-z. PubMed PMID: 31699097; PubMed Central PMCID: PMCPMC6839073. [DOI] [PMC free article] [PubMed]
- 14.O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Frontiers in endocrinology. 2018;9:402. Epub 2018/08/21. doi: 10.3389/fendo.2018.00402. PubMed PMID: 30123182; PubMed Central PMCID: PMCPMC6085463. [DOI] [PMC free article] [PubMed]
- 15.Kumar M, Nerurkar VR. Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology. 2014;452-453:143-51. Epub 2014/03/13. doi: 10.1016/j.virol.2014.01.004. PubMed PMID: 24606691; PubMed Central PMCID: PMCPMC3959158. [DOI] [PMC free article] [PubMed]
- 16.Slonchak A, Shannon RP, Pali G, Khromykh AA. Human MicroRNA miR-532-5p Exhibits Antiviral Activity against West Nile Virus via Suppression of Host Genes SESTD1 and TAB3 Required for Virus Replication. Journal of virology. 2015;90(5):2388-402. Epub 2015/12/18. doi: 10.1128/jvi.02608-15. PubMed PMID: 26676784; PubMed Central PMCID: PMCPMC4810706. [DOI] [PMC free article] [PubMed]
- 17.Rastogi M, Srivastava N, Singh SK. Exploitation of microRNAs by Japanese Encephalitis virus in human microglial cells. Journal of medical virology. 2018;90(4):648-54. Epub 2017/11/18. doi: 10.1002/jmv.24995. PubMed PMID: 29149532. [DOI] [PubMed]
- 18.Dang JW, Tiwari SK, Qin Y, Rana TM. Genome-wide Integrative Analysis of Zika-Virus-Infected Neuronal Stem Cells Reveals Roles for MicroRNAs in Cell Cycle and Stemness. Cell reports. 2019;27(12):3618-28.e5. Epub 2019/06/20. doi: 10.1016/j.celrep.2019.05.059. PubMed PMID: 31216479; PubMed Central PMCID: PMCPMC6687627. [DOI] [PMC free article] [PubMed]
- 19.Seong RK, Lee JK, Cho GJ, Kumar M, Shin OS. mRNA and miRNA profiling of Zika virus-infected human umbilical cord mesenchymal stem cells identifies miR-142-5p as an antiviral factor. Emerging microbes & infections. 2020;9(1):2061-75. Epub 2020/09/10. doi: 10.1080/22221751.2020.1821581. PubMed PMID: 32902370; PubMed Central PMCID: PMCPMC7534337. [DOI] [PMC free article] [PubMed]
- 20.Hazra B., Kumawat K.L., Basu A. The host microRNA miR-301a blocks the IRF1-mediated neuronal innate immune response to Japanese encephalitis virus infection. Sci Signaling. 2017;10(466):eaaf5185.;Epub 2017/02/16 doi: 10.1126/scisignal.aaf5185. PubMed PMID: 28196914. [DOI] [PubMed] [Google Scholar]
- 21.Wu N, Gao N, Fan D, Wei J, Zhang J, An J. miR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926 cells. Microbes and infection. 2014;16(11):911-22. Epub 2014/09/03. doi: 10.1016/j.micinf.2014.08.011. PubMed PMID: 25181337; PubMed Central PMCID: PMCPMC7110837. [DOI] [PMC free article] [PubMed]
- 22.Diosa-Toro M, Echavarría-Consuegra L, Flipse J, Fernández GJ, Kluiver J, van den Berg A, et al. MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression. PLoS neglected tropical diseases. 2017;11(10):e0005981. Epub 2017/10/19. doi: 10.1371/journal.pntd.0005981. PubMed PMID: 29045406; PubMed Central PMCID: PMCPMC5662241. [DOI] [PMC free article] [PubMed]
- 23.Cai W, Pan Y, Cheng A, Wang M, Yin Z, Jia R. Regulatory Role of Host MicroRNAs in Flaviviruses Infection. Frontiers in microbiology. 2022;13:869441. Epub 2022/04/29. doi: 10.3389/fmicb.2022.869441. PubMed PMID: 35479613; PubMed Central PMCID: PMCPMC9036177. [DOI] [PMC free article] [PubMed]
- 24.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nature reviews Molecular cell biology. 2021;22(2):96-118. Epub 2020/12/doi: 10.1038/s41580-020-00315-9. PubMed PMID: 33353982; PubMed Central PMCID: PMCPMC7754182. [DOI] [PMC free article] [PubMed]
- 25.Bhattacharyya S, Vrati S. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Scientific reports. 2015;5:17794. Epub 2015/12/05. doi: 10.1038/srep17794. PubMed PMID: 26634309; PubMed Central PMCID: PMCPMC4669524. [DOI] [PMC free article] [PubMed]
- 26.Wang XJ, Jiang SC, Wei HX, Deng SQ, He C, Peng HJ. The Differential Expression and Possible Function of Long Noncoding RNAs in Liver Cells Infected by Dengue Virus. The American journal of tropical medicine and hygiene. 2017;97(6):1904-12. Epub 2017/10/11. doi: 10.4269/ajtmh.17-0307. PubMed PMID: 29016307; PubMed Central PMCID: PMCPMC5805055. [DOI] [PMC free article] [PubMed]
- 27.Hu B, Huo Y, Yang L, Chen G, Luo M, Yang J, et al. ZIKV infection effects changes in gene splicing, isoform composition and lncRNA expression in human neural progenitor cells. Virology journal. 2017;14(1):217. Epub 2017/11/09. doi: 10.1186/s12985-017-0882-6. PubMed PMID: 29116029; PubMed Central PMCID: PMCPMC5688814. [DOI] [PMC free article] [PubMed]
- 28.Selinger M, Wilkie GS, Tong L, Gu Q, Schnettler E, Grubhoffer L, et al. Analysis of tick-borne encephalitis virus-induced host responses in human cells of neuronal origin and interferon-mediated protection. The Journal of general virology. 2017;98(8):2043-60. Epub 2017/08/09. doi: 10.1099/jgv.0.000853. PubMed PMID: 28786780; PubMed Central PMCID: PMCPMC5817271. [DOI] [PMC free article] [PubMed]
- 29.Zhong XL, Liao XM, Shen F, Yu HJ, Yan WS, Zhang YF, et al. Genome-wide profiling of mRNA and lncRNA expression in dengue fever and dengue hemorrhagic fever. FEBS open bio. 2019;9(3):468-77. Epub 2019/03/15. doi: 10.1002/2211-5463.12576. PubMed PMID: 30868055; PubMed Central PMCID: PMCPMC6396354. [DOI] [PMC free article] [PubMed]
- 30.Wang P, Xu J, Wang Y, Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science (New York, NY). 2017;358(6366):1051-5. Epub 2017/10/28. doi: 10.1126/science.aao0409. PubMed PMID: 29074580. [DOI] [PubMed]
- 31.Barriocanal M, Carnero E, Segura V, Fortes P. Long Non-Coding RNA BST2/BISPR is Induced by IFN and Regulates the Expression of the Antiviral Factor Tetherin. Frontiers in immunology. 2014;5:655. Epub 2015/01/27. doi: 10.3389/fimmu.2014.00655. PubMed PMID: 25620967; PubMed Central PMCID: PMCPMC4288319. [DOI] [PMC free article] [PubMed]
- 32.Kambara H, Niazi F, Kostadinova L, Moonka DK, Siegel CT, Post AB, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic acids research. 2014;42(16):10668-80. Epub 2014/08/15. doi: 10.1093/nar/gku713. PubMed PMID: 25122750; PubMed Central PMCID: PMCPMC4176326. [DOI] [PMC free article] [PubMed]
- 33.Qian X., Xu C., Zhao P., Qi Z. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology. 2016;Epub 2016/03/08(492):155–165. doi: 10.1016/j.virol.2016.02.020. PubMed PMID: 26945984. [DOI] [PubMed] [Google Scholar]
- 34.Schnettler E, Tykalová H, Watson M, Sharma M, Sterken MG, Obbard DJ, et al. Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic acids research. 2014;42(14):9436-46. Epub 2014/07/24. doi: 10.1093/nar/gku657. PubMed PMID: 25053841; PubMed Central PMCID: PMCPMC4132761. [DOI] [PMC free article] [PubMed]
- 35.Schirtzinger EE, Andrade CC, Devitt N, Ramaraj T, Jacobi JL, Schilkey F, et al. Repertoire of virus-derived small RNAs produced by mosquito and mammalian cells in response to dengue virus infection. Virology. 2015;476:54-60. Epub 2014/12/22. doi: 10.1016/j.virol.2014.11.019. PubMed PMID: 25528416; PubMed Central PMCID: PMCPMC4323773. [DOI] [PMC free article] [PubMed]
- 36.Schuster S, Miesen P, van Rij RP. Antiviral RNAi in Insects and Mammals: Parallels and Differences. Viruses. 2019;11(5). Epub 2019/05/19. doi: 10.3390/v11050448. PubMed PMID: 31100912; PubMed Central PMCID: PMCPMC6563508. [DOI] [PMC free article] [PubMed]
- 37.Li X, Fu Z, Liang H, Wang Y, Qi X, Ding M, et al. H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2. Cell research. 2018;28(2):157-71. Epub 2018/01/13. doi: 10.1038/cr.2018.3. PubMed PMID: 29327729; PubMed Central PMCID: PMCPMC5799819. [DOI] [PMC free article] [PubMed]
- 38.Ouellet DL, Vigneault-Edwards J, Létourneau K, Gobeil LA, Plante I, Burnett JC, et al. Regulation of host gene expression by HIV-1 TAR microRNAs. Retrovirology. 2013;10:86. Epub 2013/08/14. doi: 10.1186/1742-4690-10-86. PubMed PMID: 23938024; PubMed Central PMCID: PMCPMC3751525. [DOI] [PMC free article] [PubMed]
- 39.Heinz FX, Kunz C. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. The Journal of general virology. 1981;57(Pt 2):263-74. Epub 1981/12/01. doi: 10.1099/0022-1317-57-2-263. PubMed PMID: 6172553. [DOI] [PubMed]
- 40.Pospisil L., Jandasek L., Pesek J. Isolation of new strains of meningoencephalitis virus in the Brno region during the summer of 1953. Lekarske listy. 1954;9(1):3–5. Epub 1954/01/01 PubMed PMID: 13131921. [PubMed] [Google Scholar]
- 41.De Madrid AT, Porterfield JS. A simple micro-culture method for the study of group B arboviruses. Bulletin of the World Health Organization. 1969;40(1):113-21. Epub 1969/01/01. PubMed PMID: 4183812; PubMed Central PMCID: PMCPMC2554446. [PMC free article] [PubMed]
- 42.Achazi K, Nitsche A, Patel P, Radonić A, Donoso Mantke O, Niedrig M. Detection and differentiation of tick-borne encephalitis virus subtypes by a reverse transcription quantitative real-time PCR and pyrosequencing. Journal of virological methods. 2011;171(1):34-9. Epub 2010/10/12. doi: 10.1016/j.jviromet.2010.09.026. PubMed PMID: 20933016. [DOI] [PubMed]
- 43.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17(1):3. Epub 2011-08-02. doi: 10.14806/ej.17.1.200.
- 44.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010;Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 45.Ghosh S, Chan CK. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods in molecular biology (Clifton, NJ). 2016;1374:339-61. Epub 2015/11/01. doi: 10.1007/978-1-4939-3167-5_18. PubMed PMID: 26519415. [DOI] [PubMed]
- 46.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic acids research. 2012;40(1):37-52. Epub 2011/09/14. doi: 10.1093/nar/gkr688. PubMed PMID: 21911355; PubMed Central PMCID: PMCPMC3245920. [DOI] [PMC free article] [PubMed]
- 47.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34(Database issue):D140-4. Epub 2005/12/31. doi: 10.1093/nar/gkj112. PubMed PMID: 16381832; PubMed Central PMCID: PMCPMC1347474. [DOI] [PMC free article] [PubMed]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10(3):R25. Epub 2009/03/06. doi: 10.1186/gb-2009-10-3-r25. PubMed PMID: 19261174; PubMed Central PMCID: PMCPMC2690996. [DOI] [PMC free article] [PubMed]
- 49.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England). 2010;26(6):841-2. Epub 2010/01/30. doi: 10.1093/bioinformatics/btq033. PubMed PMID: 20110278; PubMed Central PMCID: PMCPMC2832824. [DOI] [PMC free article] [PubMed]
- 50.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England). 2009;25(16):2078-9. Epub 2009/06/10. doi: 10.1093/bioinformatics/btp352. PubMed PMID: 19505943; PubMed Central PMCID: PMCPMC2723002. [DOI] [PMC free article] [PubMed]
- 51.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PloS one. 2018;13(10):e0206239. Epub 2018/10/20. doi: 10.1371/journal.pone.0206239. PubMed PMID: 30335862; PubMed Central PMCID: PMCPMC6193719. [DOI] [PMC free article] [PubMed]
- 52.Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic acids research. 2016;44(D1):D231-8. Epub 2015/11/28. doi: 10.1093/nar/gkv1270. PubMed PMID: 26612864; PubMed Central PMCID: PMCPMC4702897. [DOI] [PMC free article] [PubMed]
- 53.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic acids research. 2015;43(Database issue):D146-52. Epub 2014/11/08. doi: 10.1093/nar/gku1104. PubMed PMID: 25378301; PubMed Central PMCID: PMCPMC4383922. [DOI] [PMC free article] [PubMed]
- 54.Vaquero-Garcia J, Barrera A, Gazzara MR, González-Vallinas J, Lahens NF, Hogenesch JB, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife. 2016;5:e11752. Epub 2016/02/02. doi: 10.7554/eLife.11752. PubMed PMID: 26829591; PubMed Central PMCID: PMCPMC4801060. [DOI] [PMC free article] [PubMed]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 2013;29(1):15-21. Epub 2012/10/30. doi: 10.1093/bioinformatics/bts635. PubMed PMID: 23104886; PubMed Central PMCID: PMCPMC3530905. [DOI] [PMC free article] [PubMed]
- 56.Selinger M, Tykalová H, Štěrba J, Věchtová P, Vavrušková Z, Lieskovská J, et al. Tick-borne encephalitis virus inhibits rRNA synthesis and host protein production in human cells of neural origin. PLoS neglected tropical diseases. 2019;13(9):e0007745. Epub 2019/09/29. doi: 10.1371/journal.pntd.0007745. PubMed PMID: 31560682; PubMed Central PMCID: PMCPMC6785130. [DOI] [PMC free article] [PubMed]
- 57.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562-78. Epub 2012/03/03. doi: 10.1038/nprot.2012.016. PubMed PMID: 22383036; PubMed Central PMCID: PMCPMC3334321. [DOI] [PMC free article] [PubMed]
- 58.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44-57. Epub 2009/01/10. doi: 10.1038/nprot.2008.211. PubMed PMID: 19131956. [DOI] [PubMed]
- 59.Pla R, Stanco A, Howard MA, Rubin AN, Vogt D, Mortimer N, et al. Dlx1 and Dlx2 Promote Interneuron GABA Synthesis, Synaptogenesis, and Dendritogenesis. Cerebral cortex (New York, NY : 1991). 2018;28(11):3797-815. Epub 2017/10/14. doi: 10.1093/cercor/bhx241. PubMed PMID: 29028947; PubMed Central PMCID: PMCPMC6188538. [DOI] [PMC free article] [PubMed]
- 60.Ip JP, Fu AK, Ip NY. CRMP2: functional roles in neural development and therapeutic potential in neurological diseases. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2014;20(6):589-98. Epub 2014/01/10. doi: 10.1177/1073858413514278. PubMed PMID: 24402611. [DOI] [PubMed]
- 61.Sessions OM, Tan Y, Goh KC, Liu Y, Tan P, Rozen S, et al. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS neglected tropical diseases. 2013;7(3):e2107. Epub 2013/03/22. doi: 10.1371/journal.pntd.0002107. PubMed PMID: 23516652; PubMed Central PMCID: PMCPMC3597485. [DOI] [PMC free article] [PubMed]
- 62.De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS pathogens. 2016;12(8):e1005841. Epub 2016/08/31. doi: 10.1371/journal.ppat.1005841. PubMed PMID: 27575636; PubMed Central PMCID: PMCPMC5004807. [DOI] [PMC free article] [PubMed]
- 63.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Jr., Diamond MS. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nature medicine. 2013;19(4):458-64. Epub 2013/03/05. doi: 10.1038/nm.3108. PubMed PMID: 23455712; PubMed Central PMCID: PMCPMC3618596. [DOI] [PMC free article] [PubMed]
- 64.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annual review of immunology. 2014;32:513-45. Epub 2014/02/22. doi: 10.1146/annurev-immunol-032713-120231. PubMed PMID: 24555472; PubMed Central PMCID: PMCPMC4313732. [DOI] [PMC free article] [PubMed]
- 65.Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, et al. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Science translational medicine. 2015;7(284):284ra59. Epub 2015/04/24. doi: 10.1126/scitranslmed.aaa4304. PubMed PMID: 25904743; PubMed Central PMCID: PMCPMC4435724. [DOI] [PMC free article] [PubMed]
- 66.Rivera-Serrano EE, Gizzi AS, Arnold JJ, Grove TL, Almo SC, Cameron CE. Viperin Reveals Its True Function. Annual review of virology. 2020;7(1):421-46. Epub 2020/07/01. doi: 10.1146/annurev-virology-011720-095930. PubMed PMID: 32603630; PubMed Central PMCID: PMCPMC8191541. [DOI] [PMC free article] [PubMed]
- 67.Soveg FW, Schwerk J, Gokhale NS, Cerosaletti K, Smith JR, Pairo-Castineira E, et al. Endomembrane targeting of human OAS1 p46 augments antiviral activity. eLife. 2021;10. Epub 2021/08/04. doi: 10.7554/eLife.71047. PubMed PMID: 34342578; PubMed Central PMCID: PMCPMC8357416. [DOI] [PMC free article] [PubMed]
- 68.Castro FL, Geddes VEV, Monteiro FLL, Gonçalves R, Campanati L, Pezzuto P, et al. MicroRNAs 145 and 148a Are Upregulated During Congenital Zika Virus Infection. ASN neuro. 2019;11:1759091419850983. Epub 2019/06/20. doi: 10.1177/1759091419850983. PubMed PMID: 31213064; PubMed Central PMCID: PMCPMC6585135. [DOI] [PMC free article] [PubMed]
- 69.Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43(7):706-11. Epub 2011/05/31. doi: 10.1038/ng.841. PubMed PMID: 21623373; PubMed Central PMCID: PMCPMC3125461. [DOI] [PMC free article] [PubMed]
- 70.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20(12):1434-42. Epub 2013/11/12. doi: 10.1038/nsmb.2699. PubMed PMID: 24213538; PubMed Central PMCID: PMCPMC3918504. [DOI] [PMC free article] [PubMed]
- 71.Wang ET, Ward AJ, Cherone JM, Giudice J, Wang TT, Treacy DJ, et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome research. 2015;25(6):858-Epub 2015/04/18. doi: 10.1101/gr.184390.114. PubMed PMID: 25883322; PubMed Central PMCID: PMCPMC4448682. [DOI] [PMC free article] [PubMed]
- 72.Su CH, D D, Tarn WY. Alternative Splicing in Neurogenesis and Brain Development. Frontiers in molecular biosciences. 2018;5:12. Epub 2018/02/28. doi: 10.3389/fmolb.2018.00012. PubMed PMID: 29484299; PubMed Central PMCID: PMCPMC5816070. [DOI] [PMC free article] [PubMed]
- 73.Wang Y, Ji P, Liu J, Broaddus RR, Xue F, Zhang W. Centrosome-associated regulators of the G(2)/M checkpoint as targets for cancer therapy. Molecular cancer. 2009;8:8. Epub 2009/02/17. doi: 10.1186/1476-4598-8-8. PubMed PMID: 19216791; PubMed Central PMCID: PMCPMC2657106. [DOI] [PMC free article] [PubMed]
- 74.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40(2):179-204. Epub 2010/10/23. doi: 10.1016/j.molcel.2010.09.019. PubMed PMID: 20965415; PubMed Central PMCID: PMCPMC2988877. [DOI] [PMC free article] [PubMed]
- 75.Overby AK, Popov VL, Niedrig M, Weber F. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. Journal of virology. 2010;84(17):8470-83. Epub 2010/06/18. doi: 10.1128/jvi.00176-10. PubMed PMID: 20554782; PubMed Central PMCID: PMCPMC2919015. [DOI] [PMC free article] [PubMed]
- 76.Polonio CM, Peron JPS. ZIKV Infection and miRNA Network in Pathogenesis and Immune Response. Viruses. 2021;13(10). Epub 2021/10/27. doi: 10.3390/v13101992. PubMed PMID: 34696422; PubMed Central PMCID: PMCPMC8541119. [DOI] [PMC free article] [PubMed]
- 77.Wong RR, Abd-Aziz N, Affendi S, Poh CL. Role of microRNAs in antiviral responses to dengue infection. Journal of biomedical science. 2020;27(1):4. Epub 2020/01/04. doi: 10.1186/s12929-019-0614-x. PubMed PMID: 31898495; PubMed Central PMCID: PMCPMC6941309. [DOI] [PMC free article] [PubMed]
- 78.He Y, Ge Y, Jiang M, Zhou J, Luo D, Fan H, et al. MiR-592 Promotes Gastric Cancer Proliferation, Migration, and Invasion Through the PI3K/AKT and MAPK/ERK Signaling Pathways by Targeting Spry2. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2018;47(4):1465-81. Epub 2018/06/28. doi: 10.1159/000490839. PubMed PMID: 29949784. [DOI] [PubMed]
- 79.Sharma B, Joshi S, Sassano A, Majchrzak B, Kaur S, Aggarwal P, et al. Sprouty proteins are negative regulators of interferon (IFN) signaling and IFN-inducible biological responses. The Journal of biological chemistry. 2012;287(50):42352-60. Epub 2012/10/18. doi: 10.1074/jbc.M112.400721. PubMed PMID: 23074222; PubMed Central PMCID: PMCPMC3516778. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.