Abstract
Loss of seed shattering was a key step during cereal domestication, and it greatly facilitated seed harvest of the staple cereal foxtail millet (Setaria italica) because the cereal has very small seeds. However, the genetic basis for this loss has been largely unknown. Here, we combined comparative and association mapping to identify an 855-bp Harbinger transposable element insertion in the second exon of the foxtail millet gene shattering1 (sh1) that was responsible for the loss of seed shattering. The sh1 gene encodes zinc finger and YABBY domains. The insert prevents transcription of the second exon, causing partial loss of the zinc finger domain and then loss of natural seed shattering. Specifically, sh1 functions as a transcription repressor and represses the transcription of genes associated with lignin synthesis in the abscission zone, including CAD2. The diversity of sh1 is highly reduced in foxtail millet, consistent with either a severe domestication bottleneck or a selective sweep. Phylogenetic analysis of sh1 further revealed a single origin of foxtail millet in China. Our results support the theories that transposons were the most active factors in genome evolution driving loss of natural seed shattering during foxtail millet domestication and that sh1 underwent parallel selection during domestication across different cereal species.
Keywords: seed shattering, comparative genomics, domestication, transposable element, parallel selection
Introduction
Cereals, including rice, maize, wheat, sorghum, and foxtail millet, were domesticated thousands of years ago from wild grass progenitors in different geographic regions (Doebley et al. 2006). A number of phenotypic and physiological traits have been reshaped to give rise to cereals distinct from their wild progenitors. Wild progenitors are characterized by self-propagation, loose plant architecture, and low seed production with generally poor edibility. In contrast, crop cereals are characterized by propagation that is completely dependent on humans, compact plant statures, and high production of grain with a pleasant taste. Among the transitions during cereal domestication, known collectively as the domestication syndrome (Harlan 1992), a crucial and common step is the change from natural seed shattering—the natural and timely shedding of ripe seeds to ensure their dispersal—in wild progenitors to a lack of shattering in domesticated cereals (Lin et al. 2012). Seed shattering allows self-planting and protects the seeds from small animals, especially birds, guaranteeing the self-propagation of wild progenitors. However, seed shattering in agricultural crops causes large production losses and greatly hinders harvest. Therefore, natural seed shattering was eliminated in cereals during domestication.
Foxtail millet is a model drought-tolerant C4 cereal that provides food and animal feed in semi-arid regions (Zhang et al. 2012). Compared with the progenitors of most other cereals, which have bigger seeds, the progenitor of foxtail millet, green millet, has small seeds that easily shatter and are hard to collect. This extremely small size and ease of shattering would have been major obstacles to the domestication of green millet; thus, the elimination of natural seed shattering must have been a key early step during foxtail millet domestication. However, the genetic basis for this change in foxtail millet remains largely unknown.
Seed shattering in cereals arises from the development of an abscission layer between the seed and pedicel (Hodge and Kellogg 2016). The development of the abscission layer is synchronized with the development of the panicle; the cell wall of the abscission layer then breaks down at maturity, and the seed is then shed from the mother plant (Li et al. 2006; Lin et al. 2007). Several genes that played key roles in the elimination of seed shattering during cereal domestication have been identified and cloned. These genes control the development of the abscission layer and the breakdown of the cell wall of the abscission layer during seed shattering. Sha1 (sh4), which encodes an MYB domain, was first cloned in rice (Li et al. 2006; Lin et al. 2007). A single nucleotide change in the MYB domain of Sha1 leads to failure in the breakdown of the abscission layer cell wall. A single nucleotide variant in the promoter region located 12 kilobase (kb) away from the coding region of qSh1 with a BELL domain results in the loss of the abscission layer between the glume and pedicel in rice (Konishi et al. 2006). Variations in the wheat Q gene with an AP2 domain upregulate transcription to eliminate rachis fragility and give rise to free grain threshing (Simons et al. 2006). Sorghum sh1, which includes a YABBY domain, harbors multiple causal variants that decrease transcription and knock down gene function, thereby hindering abscission layer development (Lin et al. 2012). The homologs of sh1 also underwent parallel domestication to decrease seed shattering in rice and maize (Lin et al. 2012). Whether sh1 regulates seed shattering in other cereal species beyond these three species remains largely unknown. Additionally, the gene regulatory network of sh1 for seed shattering remains little understood.
Here, we applied comparative and association mapping to identify a transposable element insertion in sh1 that was responsible for the loss of natural seed shattering during foxtail millet domestication. The sh1 gene acts as a transcription repressor and represses the expressions of several genes in the lignin synthesis pathway of the abscission layer. During domestication, the transposable element in sh1 was fixed, potentially due to a selective sweep. Further phylogenetic analysis showed that foxtail millet might have a single origin in China. Our results suggest that transposons played an important role in foxtail millet domestication and that sh1 was under parallel selection in several cereal species during domestication.
Results
Development of the Abscission Zone Was Repressed in Foxtail Millet during Domestication
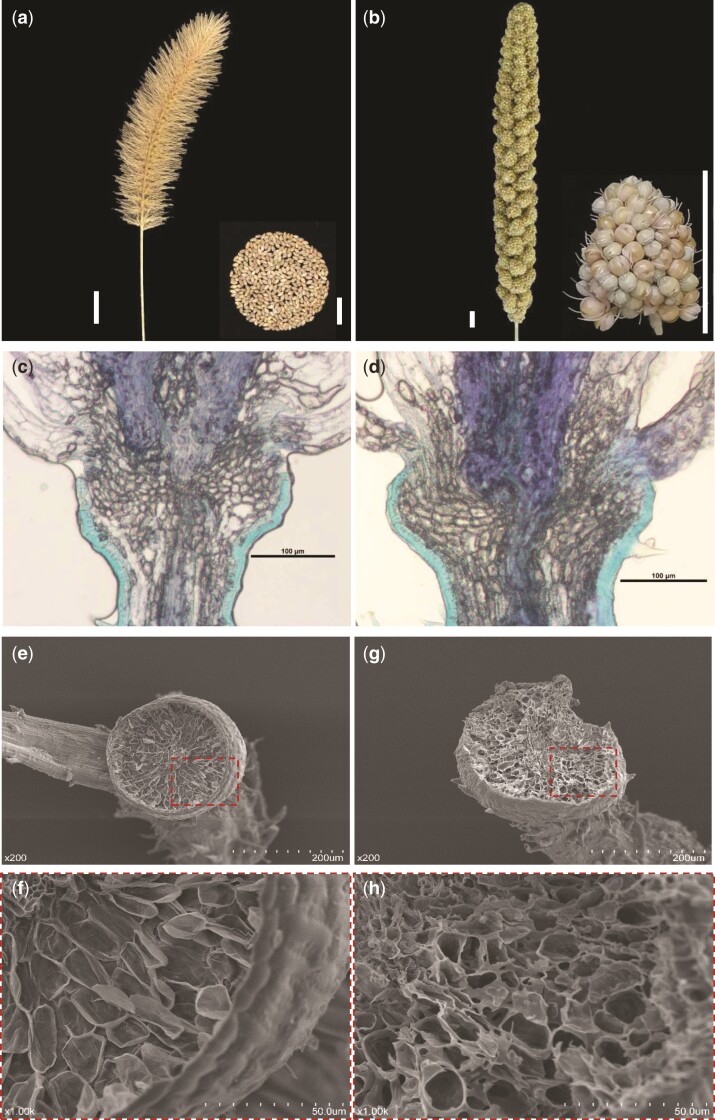
A most conspicuous and key step in domestication syndrome in cereal is the loss of seed shattering. In the wild foxtail millet progenitor, green millet (Setaria viridis), seeds are naturally shed at maturity (fig. 1a), whereas seeds from foxtail millet are not shed naturally but stay firmly attached to the head of the plant (fig. 1b). Seed shattering is correlated with the development of the abscission zone, which is present between the seed and the pedicle (Doust, Mauro-Herrera, et al. 2014). To determine how the development of the abscission zone was changed during foxtail millet domestication, we conducted a multiplex histology analysis (fig. 1 and supplementary fig. S1, Supplementary Material online). We prepared sections from spikelets of both green millet and foxtail millet obtained before heading and stained them with toluidine blue and acridine orange. Cells with high lignification are stained green by toluidine blue and false-green by acridine orange. In rice and sorghum, the abscission zone is characterized by a band of cells with low lignification between the pedicle and glume. The abscission layer cells are distinct from the adjacent cells of the glume and pedicle, which have high lignification in these two species (Konishi et al. 2006; Lin et al. 2012). However, we found that the cells in the abscission zone were not distinct from the adjacent cells of the glume and pedicle in either the wild or the domesticated foxtail millets; all three types of cells had low lignification (fig. 1c, d and supplementary fig. S1, Supplementary Material online). In the seeds naturally shed from green millet, an abscission bowl was present and the abscission layer surface of the pedicle appeared smooth in scanning electron microscopy (SEM) images (fig. 1e and f). In contrast, in the seeds forcibly detached from the pedicle in domesticated foxtail millet, no abscission bowl was present and the surface of the pedicle cell was rough (fig. 1g and h). These findings indicated that the development of the abscission zone became inhibited in foxtail millet during domestication.
Fig. 1.
Phenotype. (a and b) Seeds are shed freely at maturity from the wild progenitor green millet (Setaria viridis) (a) but remain firmly attached to the head of the foxtail millet (Setaria italica) plant (b). Scale bar, 1 cm. (c and d) Histology analysis revealed that no clear differences are present in longitudinal sections from pedicels of green millet and foxtail millet stained with toluidine blue. (e–h), Scanning electron microscopy of the surfaces of pedicels from green millet (e and g) and foxtail millet after forcibly detached from seed (f and h). (g and h) Close-up views of the pedicel surfaces, showing that the pedicel surface in green millet is smooth and contains an apparent abscission bowl (g), whereas the foxtail millet pedicel lacks the abscission bowl and has a rough surface (h). Red dashed lines represent the regions of close-up view.
Loss of Natural Seed Shattering in Foxtail Millet Was Convergently Controlled by sh1
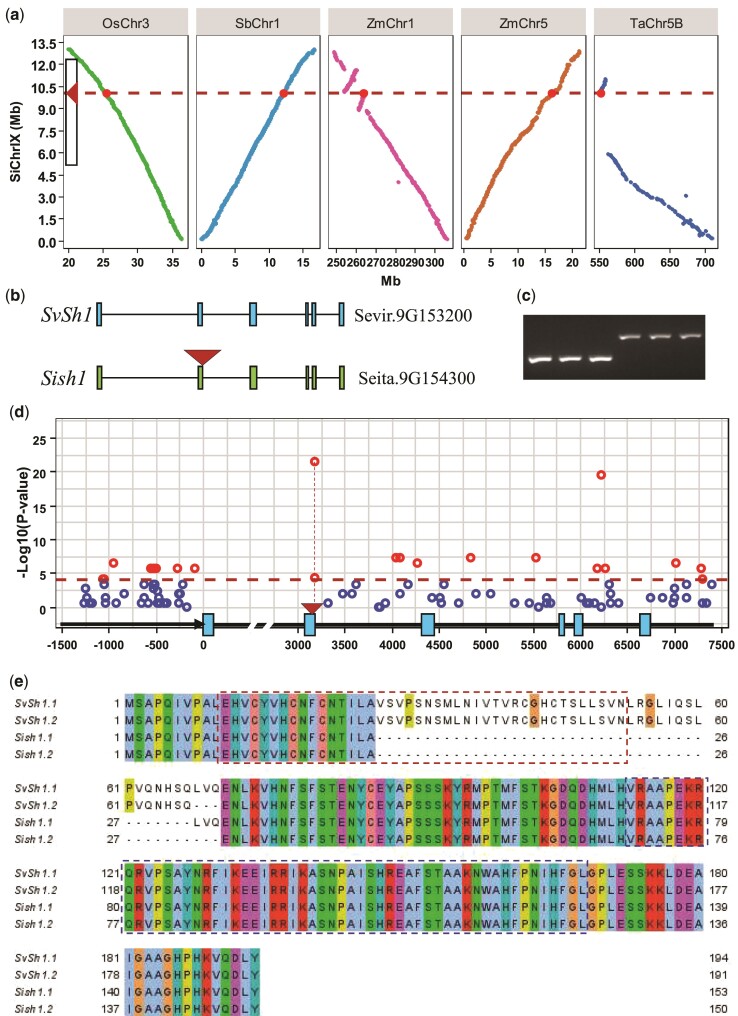
A major quantitative trait locus (QTL) for loss of seed shattering was repeatedly mapped to chromosome IX in foxtail millet (fig. 2a) (Doust, Lukens, et al. 2014; Odonkor et al. 2018). This major QTL for the loss of seed shattering was situated close to the previously identified sh1 gene (Lin et al. 2012). To determine whether the sh1 gene for the loss of seed shattering in domestication syndrome regulates seed shattering in other cereals beyond maize, rice, and sorghum, we performed a comparative genomic analysis across foxtail millet, sorghum, maize, rice, and wheat for this QTL (fig. 2a). Comparative genomic mapping identified a highly conserved syntenic chromosomal block, which contained several fragments on rice chromosome 3, sorghum chromosome 1, maize chromosomes 1 and 5, and wheat chromosome 5B. In this syntenic block, most genes and gene orders remained conserved despite two and one reversions on maize chromosome 1 and wheat chromosome 5b, respectively, and a 10-Mb deletion on wheat chromosome 5B (fig. 2a). This syntenic block was partially overlapped with the previously identified collinear genomic region based on molecular markers (Devos and Gale 2000) (fig. 2a).
Fig. 2.
The sh1 gene is responsible for the loss of seed shattering in foxtail millet. (a) Comparative mapping showed high collinearity of genome sequences at the sh1 loci among foxtail millet (Setaria italica), sorghum (Sorghum bicolor), maize (Zea mays), rice (Oryza sativa), and wheat (Triticum aestivum) (OsChr3, SbChr2, ZmChr1, ZmChr5, and Ta Chr5B, respectively). The position of the QTL for seed shattering in foxtail millet is marked on the y-axis. Blank box, QTL interval; triangle, QTL peak. (b) Sequence comparison of the sh1 genes in green millet and foxtail millet. Green and blue boxes, exons; black bars, introns; red triangle, the transposable element insertion. (c) The transposable element insertion in sh1 was absent in three random green millets and present in three random foxtail millets. (d) Association mapping showed that the transposable element insertion in the sh1 gene caused the loss of seed shattering in foxtail millet. The red dashed line represents the threshold of significance at the P = 0.01 level with multiple testing correction for association mapping. The gene structure of sh1 is shown on the x-axis, with the start codon set as position “0.” Arrow, promoter; blue boxes, exons; black bars, intron and 3′ UTR. (e) The sh1 gene encodes a protein with a zinc finger and a YABBY domain. The zinc finger and YABBY domain are highlighted in red and blue dashed line boxes, respectively.
To determine whether sh1 also controlled seed shattering in foxtail millet during domestication, we then compared sequences of sh1 orthologs in green millet A10 (Mamidi et al. 2020) and the domesticated foxtail millet Yugu1 (Bennetzen et al. 2012) with whole-genome sequences (https://phytozome.jgi.doe.gov/pz/portal.html). The two sh1 orthologs correspond to Sevir.9G153200 in A10 and Seita.9G154300 in Yugu1. Sequence comparison revealed 24 single nucleotide polymorphisms (SNPs) and seven insertions/deletions (indels) present between Sevir.9G153200 and Seita.9G154300 (supplementary dataset 1, Supplementary Material online). All of these SNPs and six of the indels were present in the introns (supplementary dataset 1, Supplementary Material online). The only indel located in the second exon was an insertion of the 855-base pair (bp) Harbinger transposable element (fig. 2b and c, supplementary fig. S2, Supplementary Material online) in the domesticated foxtail millet Yugu1. Sevir.9G153200 contains six exons and five introns and consists of 6,694 bp (fig. 2b). We then sequenced a 5,801-bp fragment of this gene from a global foxtail millet panel including 23 wild and 73 domesticated accessions (supplementary table S1, Supplementary Material online). The 5,801-bp fragment harbored a 1,269-bp promoter, an 824-bp 3′ untranslated region (3′ UTR), and a 3,708-bp gene region excluding the first intron with 2,986 bp, which was difficult to amplify due to a high AT content (61%). The large sequencing revealed 96 variants in the 5,801-bp fragment from these foxtail millet accessions (supplementary table S2, Supplementary Material online). We then performed association tests between the variants and phenotypes of seed shattering in the foxtail millet panel, finding that the gene was strongly (P < 1.0 × 10−4) associated with seed shattering in foxtail millet (fig. 2d and supplementary fig. S3a, Supplementary Material online). The strongest association signal (P = 3.75 × 10−22) was present on the transposable element inserted in the second exon (fig. 2d). Another strong signal (P = 2.81 × 10−20) occurred on the SNP at position 6,221, which was in high linkage disequilibrium (r2 = 0.89) with the transposable element (fig. 2d, supplementary fig. S3b and table S2, Supplementary Material online). All the domesticated foxtail millet accessions examined harbored this transposable element in the second exon and exhibited nonshattering, and all the green millet without this insertion showed shattering (supplementary table S2, Supplementary Material online). These results indicated that sh1 (Sevir.9G153200) controlled seed shattering and that the insertion of the 855-bp transposable element in the second exon resulted in the loss of natural seed shattering during foxtail millet domestication.
Transcription analysis revealed that the green millet SvSh1 gene had two transcripts: SvSh1.1 and SvSh1.2 (fig. 2e). These transcripts differed by the presence or absence of a 9-bp sequence originating from alternative splicing of the 3′ end of the second intron (supplementary fig. S4 and dataset 1, Supplementary Material online). SvSh1.1 and SvSh1.2 encoded proteins with 194 and 191 amino acid residues (aa), respectively, with a zinc finger motif at the N terminus and a YABBY domain at the C terminus (fig. 2e). In contrast to green millet (A10) SvSh1, Sish1 from the domesticated foxtail millet Yugu1 also had two transcripts with the 9-bp sequence difference (supplementary fig. S4, Supplementary Material online), but the transposable element insertion resulted in the complete loss of the second exon of 123 bp in both transcripts so that more than half of the zinc finger domain was lost in the corresponding Sish1 proteins (fig. 2e). This result suggested that the transposable element insertion caused partial loss of the zinc finger domain of sh1 and thus led to the loss of natural seed shattering during foxtail millet domestication.
Transgenic Analysis Revealed that sh1 Controls Abscission Zone Development in Foxtail Millet
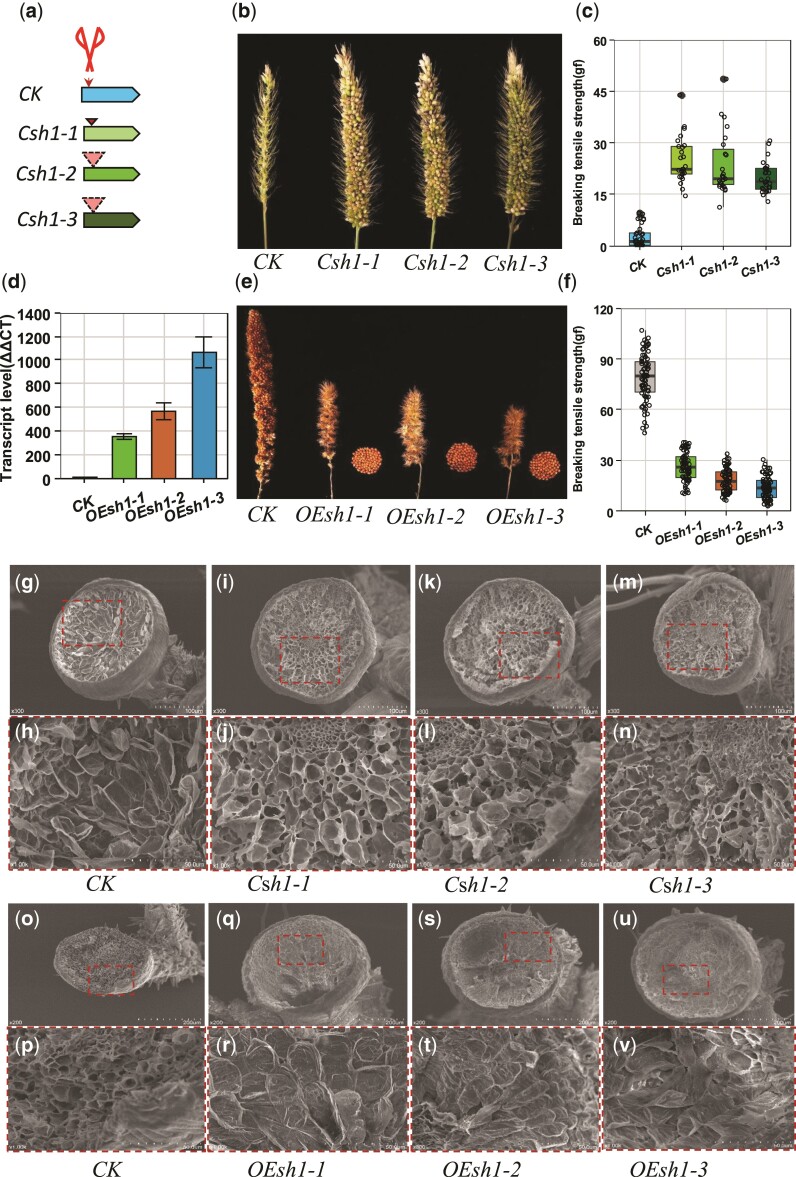
To validate whether sh1 controls seed shattering in foxtail millet, we then conducted transformation through CRISPR/Cas9 and overexpression (fig. 3). Three homozygous editing events (T1) with a 1-bp insertion and 16-bp and 19-bp deletions in the sh1 coding sequence (CDS) were obtained from an easily shattering wild foxtail millet accession, ME034v, with a highly efficient transformation (Finley et al. 2021) (fig. 3a–c and supplementary fig. S5, Supplementary Material online). All three mutations resulted in a gene-frame shift and an early stop in the translation of the sh1 gene (supplementary fig. S5b, Supplementary Material online). In comparison to wild-type ME034v, these three mutants exhibited nonshattering and the breaking tension strengths of seed detachment from the pedicle at maturity were greatly enhanced (fig. 3b and c). In parallel, we overexpressed the corresponding gene, Sh1, from a wild foxtail millet in a domesticated foxtail millet line, Ci846 (fig. 3d–f). The three overexpression transgenic lines showed extremely high expression of Sh1, more than 300-fold greater than in the nontransgenic Ci846 control plant (fig. 3d). These overexpression transgenic plants had more tillers, smaller panicles, and especially easy shattering at maturity (fig. 3e, f and supplementary fig. S6, Supplementary Material online). The breaking tension strengths of seed detachment from the pedicle were dramatically decreased in these three overexpression lines in comparison to the nontransgenic control plant (fig. 3f). The abscission bowl was generally absent from the pedicle after the detachment of the seed, and the abscission cell surface was often rougher in the three edited nonfunctional wild foxtail millet lines compared to the wild-type plants (fig. 3g–n). In contrast, the abscission bowl was restored and the abscission cell surface was smoother when Sh1 was overexpressed in the domesticated foxtail millet (fig. 3o–v). These results suggested that sh1 regulates seed shattering in foxtail millet during domestication via the control of abscission zone development.
Fig. 3.
Transformation through overexpression and CRISPR/Cas9 editing. (a) Gene editing of Sh1 from green millet through CRISPR/Cas9 with two gRNA targets. Three editing events (T1, Csh1-1, Csh1-2, and Csh1-3) contained a 1-bp insertion, a 16-bp deletion, and a 19-bp deletion in the CDS of Sh1, respectively, which resulted in gene-frame shifts and early stoppage of translation. (b) The seeds remained firmly on the panicles of the plants from the three editing events (T1), while all seeds were shed freely from the head of the control green millet plants. (c) The breaking tensile strength of seed detachment from the pedicel remained low in the control green millet plant but was greatly enhanced (P < 0.001) in the plants descended from the three editing events. The breaking tensile strengths were recorded in gravitational units of force (gf). (d) Three overexpression transgenic events were obtained resulting in plants (T1, Osh1-1, Osh1-2, and Osh1-3) with much higher expressions in comparison to the control foxtail millet plant. (e) Plants resulting from the three overexpression events exhibited easy shattering, while the control foxtail millet was nonshattering. (f) Breaking tensile strengths of seed detachment from the pedicel were greatly decreased (P < 0.001) in the transgenic overexpression plants as compared to the nontransgenic foxtail millet plants. (g–n) Scanning electron microscopy images of the surfaces of the pedicels from the edited and control green millet plants (g, i, k, and m) and close-up views of the surfaces (h, j, l, and n). (o–v) Scanning electron microscopy images of the surfaces of the pedicels from the overexpression transgenic and control foxtail millet plants (o, q, s, and u) and close-up views of the surfaces (p, r, t, and v).
sh1 Functions as a Transcription Repressor
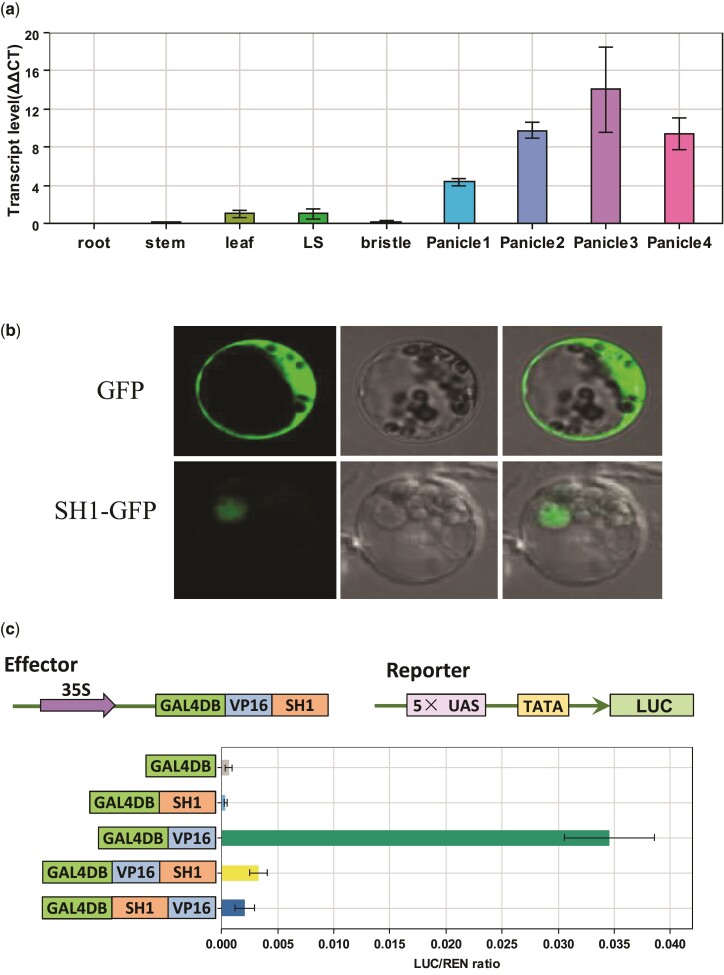
The sh1 gene had extremely low expression levels in the root, stem, and panicle bristle, low expression in the leaf and leaf sheath, and strong expression in the panicle (fig. 4a). sh1 transcripts accumulated in the panicle before anthesis, reached a maximum at the anthesis stage, and then dropped during the grain-filling stage (fig. 4a).
Fig. 4.
Gene function analysis of sh1 in foxtail millet. (a) Expression levels of sh1 in multiple organs including the root, stem, leaf, leaf sheath (LS), bristle, and panicles at different stages (Panicle1, before heading; Panicle2, 3 d after heading; Panicle3, 8 d after heading; Panicle4, grain-filling stage). (b) Subcellular localization of the SH1–GFP fusion protein in foxtail millet leaf cells. (c) Dual-luciferase transient activity assays determined that SH1 functioned as a transcriptional repressor. The GAL4DB–VP16–SH1 fusion protein dramatically (P = 6.9 × 10−5) repressed luciferase activity in comparison to the control protein GAL4DB–VP16. **, strongly significant; error bars, SD (n = 3).
Sh1 encodes a transcription factor with a zinc finger and a YABBY domain. To identify the subcellular localization of SH1, we transformed a construct with an SH1–GFP fusion protein into foxtail millet leaf protoplasts from Yugu1 (fig. 4b). We detected fluorescent signals from SH1–GFP in the nuclei of leaf protoplast cells (fig. 4b). To investigate whether SH1 acts as a transcription factor, we then conducted transcriptional activity assays using the luciferase and yeast two-hybrid systems (fig. 4c and supplementary fig. S7, Supplementary Material online). The effector construct consisted of chimeric proteins to fuse SH1 to the DNA-binding domain from the yeast GAL4 gene (GAL4DB) and the activation domain from herpes simplex virus protein 16 (VP16). The reporter constructs harbored the luciferase reporter gene controlled by a synthetic promoter with five copies of the upstream activating sequence (UAS) from GAL4 and a TATA box (fig. 4c). GAL4DB–VP16 greatly enhanced luciferase activities, whereas the fusion protein GAL4DB–VP16–SH1 strongly repressed the activities of the same reporter by over 7-fold (fig. 4c). The results from the yeast two-hybrid system were consistent with those from the luciferase system: The fusion protein of SH1 and the GAL4 DNA-binding domain did not activate the transcription of the reporter in yeast (supplementary fig. S7, Supplementary Material online). These results suggested that SH1 functions as a transcription repressor.
sh1 Represses Expression of Lignin Synthesis Genes in the Abscission Zone
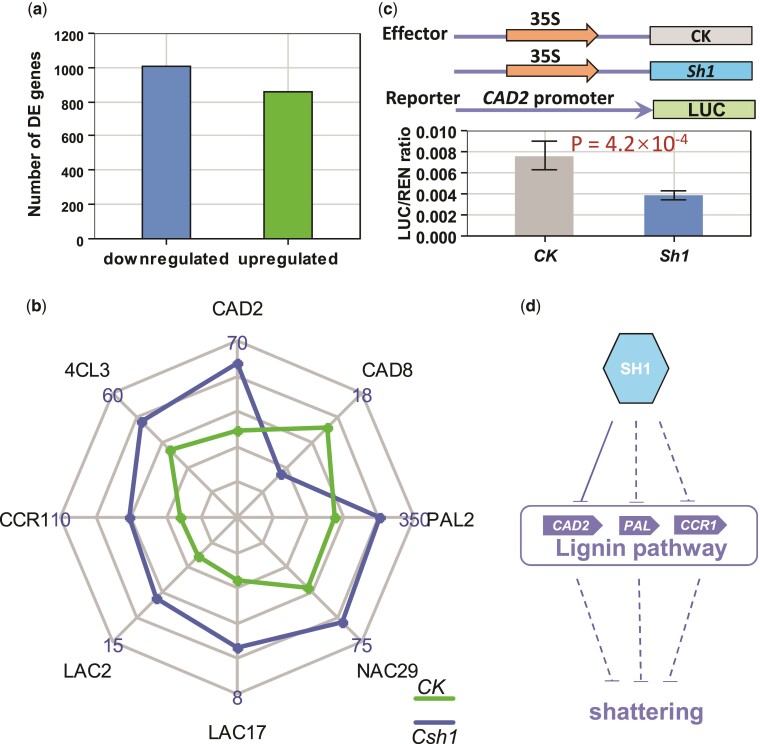
To identify how sh1 controls the downstream genes related to shattering, we then conducted RNA sequencing (RNA-seq) of samples from the panicles of a transgenic edited plant (Csh1-1) with a 1-bp-insertion mutation and the nontransgenic control plant ME034v. RNA-seq revealed that 1,002 and 860 differentially expressed genes (DEGs) were downregulated and upregulated, respectively, in the edited plant relative to the control (fig. 5a, supplementary table S3, Supplementary Material online). A Gene Ontology (GO) analysis (supplementary table S4, Supplementary Material online) through agriGO (Tian et al. 2017) then indicated that a top enriched GO term (FDR = 8.7 × 10−5) was the single-organism metabolic process. Lignin deposition in the abscission cell wall was strongly negatively correlated with shattering (Yoon et al. 2014, 2017). A careful search of the 1,862 DEGs revealed eight DEGs in the lignin synthesis pathway regulated by sh1: CAD2, 4CL3, CCR1, LAC2, LAC17, NAC29, PAL2, and CAD8 (fig. 5b and supplementary fig. S8, Supplementary Material online). Seven of these DEGs (the exception being CAD8) were upregulated in the Csh1-1 plant compared with the control. We also performed dual-luciferase transient expression assays to detect the effect of sh1 on these eight DEGs (fig. 5c). The sh1 gene showed no direct effect on the transcriptions of any of these eight DEGs except CAD2. Compared with the control construct, the effector of sh1 controlled by the 35S promoter significantly repressed luciferase activity under the control of the CAD2 promoter (fig. 5c). This result indicated that sh1 directly downregulated the expression of CAD2 via banding to its promoter. Together, the above results indicated that sh1 represses the expressions of lignin genes, resulting in seed shattering in foxtail millet (fig. 5d).
Fig. 5.
Regulatory network of sh1. (a) 1,002 and 860 differentially expressed genes (DEGs) were downregulated and upregulated, respectively, in the panicles from the plants carrying a CRISPR/Cas9-edited sh1 as compared to the unedited control plant based on RNA-seq. (b) Radar chart indicated that seven genes (CAD2, 4CL3, CCR1, LAC2, LAC17, PAL2, and NAC29) and one gene (CAD8) related to lignin synthesis in the abscission zone whose transcription was upregulated and downregulated, respectively, in the edited plant (Csh1) compared with the control plant (CK). The numbers on the chart represent the transcription level (fragments per kilobase of exon model per million mapped fragments (FPKM)) based on RNA-seq. (c) Dual-luciferase transient activity assay revealed that the transcription of CAD2 of lignin synthesis in the abscission zone was directly repressed (P = 4.2 × 10−4) by the sh1 gene. Error bars, SD (n = 5). (d) Gene regulatory network of Sh1. Sh1 directly and indirectly represses the transcriptions of the genes of lignin synthesis and then activates seed shattering in foxtail millet. Solid and dashed line “T” bars represent direct and indirect repression, respectively.
sh1 Shows a Severe Loss of Genetic Diversity and the Transposable Element Insertion Becomes Fixed during Foxtail Millet Domestication
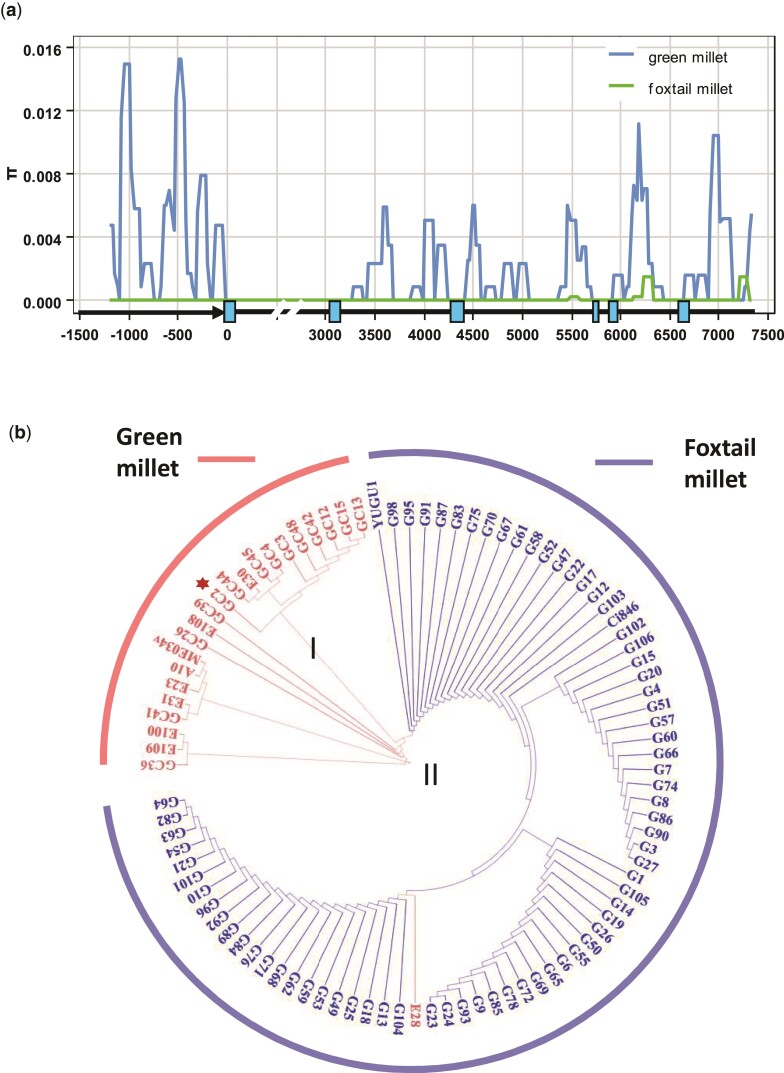
In this study, we identified an 855-bp transposable element insertion in the shattering gene sh1 in foxtail millet. To determine whether sh1 was under selection during foxtail millet domestication, we performed a DNA diversity analysis of global foxtail millet accessions including 23 wild and 73 domesticated foxtail millets. Wild foxtail millets had abundant diversity in the promoter and coding regions and the 3′ UTR in the sh1 gene (fig. 6a). However, no DNA diversity was present in any gene region except for a low diversity in the last intron and the 3′ UTR (fig. 6a). In total, only 2.3% of the DNA diversity of sh1 remained in domesticated foxtail millets compared to wild accessions. Tajima’s D test (D = –2.1, P < 0.05) suggested that selection might have shaped the sh1 gene diversity of foxtail millet. Notably, further genome-wide diversity analysis will be needed to confirm this DNA diversity pattern because a severe domestication bottleneck originating from other regions would also induce such a diversity pattern in sh1. All 73 of the investigated domesticated foxtail millets harbored this transposable element insertion in sh1. These results indicated that sh1 might be subject to human selection during the domestication of the small-seeded cereal foxtail millet.
Fig. 6.
DNA diversity analysis of sh1. (a) DNA diversity comparison in the sh1 gene between green millet and foxtail millet. Gene structure is shown on the x-axis. Arrow, promoter; blue boxes, exons; black bars, intron, and 3′ UTR. (b) Phylogenetic tree based on the sequence of sh1 revealed a single origin of foxtail millet. The foxtail millet cluster was derived from a Chinese green millet accession (GC2; red star). Green millet and foxtail millet accessions are marked in pink and blue, respectively. The cluster of green millet split into cluster I and II.
Discussion
Transposon Drove the Domestication of Foxtail Millet
Foxtail millet was domesticated approximately 9,000 years ago (Diao and Jia 2017). As this is the cereal crop with small seeds, the first obstacle during its domestication would have been seed shattering, which would have made it extremely difficult to harvest the seeds. The loss of seed shattering must have greatly reduced seed loss and enhanced harvest efficiency and thus been a key early step toward domestication. Plant genomes generally contain numerous repeat sequences, composed mainly of different transposable elements. In maize, transposable elements comprise 85% of the genome (Schnable et al. 2009) and, as the most active factor in the maize genome, they played important roles in the domestication and improvement of this outcrossing species (Clark et al. 2006; Studer et al. 2011; Zhang et al. 2020). In contrast, in foxtail millet, only approximately 50% of the genome consists of transposable elements (Zhang et al. 2012; Mamidi et al. 2020), and it has been less clear whether transposable elements played a role during the domestication of this selfing species. In this study, we identified a transposon that was inserted into the second exon of sh1 and was responsible for the loss of natural seed shattering during foxtail millet domestication. This indicates that, as in maize, transposons did play an important role during domestication in foxtail millet: a Harbinger transposon drove the loss of seed shattering and thus probably initiated the domestication of this species. When the transposon insertion occurred in the foxtail millet sh1 gene, human selection might slowly sweep the diversity of sh1 and the transposable element insertion finally became fixed in this cereal during domestication.
Even though foxtail millet is a selfing species, we detected a gene flow from domesticated foxtail millet to wild green millet in this study: one wild green millet, E28, was grouped with domesticated foxtail millets based on the sh1 sequence (fig. 6b and supplementary table S3, Supplementary Material online). This green millet harbored the transposable element insertion in sh1 and had nonshattering seeds (supplementary fig. S9, Supplementary Material online).
Presence of a Transposon in sh1 Supports a Single Origin of Foxtail Millet in China
Foxtail millet was domesticated thousands of years ago. The origin(s) of foxtail millet and the location(s) where foxtail millet was domesticated were debated in the past, with origins in China, Europe, and West Africa variously proposed (Jones 2004; Nasu et al. 2007; Diao and Jia 2017). Recently, many studies consistently revealed a single origin in northern China for foxtail millet (d'Ennequin et al. 2000; Jia, Huang, et al. 2013; Jia, Shi, et al. 2013). Our identification of the role of sh1 in the loss of shattering in foxtail millet provides a chance to test whether foxtail millet was singly originated in northern China. In this study, we investigated wild green millets collected from China, Europe (including Portugal and Russia), Central Asia (including Mongolia, Afghanistan, and Kazakhstan), West Asia (including Iran and Turkey), and the Americas (including the United States, Canada, and Chile), along with domesticated foxtail millets collected from more than 15 countries around the world (supplementary table S1, Supplementary Material online). A phylogenetic tree based on the sh1 gene revealed one main clade in wild green foxtail millets and a second main clade in domesticated foxtail millets. This supports a single origin for foxtail millet. The main wild green millet cluster was then further split into two subgroups, I and II. Subgroup I contained 11 green millets, all from northern China. The main clade of domesticated foxtail millet apparently descended from subgroup I of wild green millet, supporting that foxtail millet was first domesticated in northern China.
Whether Convergent Phenotypic Changes during Cereal Domestication, Diversification, and Improvement Have a Similar Genetic Basis
Staple cereals including rice, wheat, maize, sorghum, and foxtail millet belong to separate species. The genomes of all these species have been well sequenced, and comparative genomic analysis has revealed that many genomic regions are highly conserved across species (Schnable and Lyons 2011; Schnable et al. 2011; Jackson 2016), although these species vary widely in various phenotypes. However, over thousands of years of domestication, diversification, and improvement, human selections have reshaped these cereals into increasingly “similar” ideotypes. However, whether the parallel phenotypic changes during cereal domestication, diversification, and improvement share a similar genetic basis remains largely unknown. In this study, we investigated one common transition during cereal domestication, from seed shattering in wild grasses to nonshattering in domesticated cereals. In previous studies, sh1 was shown to be responsible for the loss of seed shattering during sorghum, maize, and Asian and African rice domestication (Lin et al. 2012; Lv et al. 2018). Here, our comparative mapping showed that the sh1 loci have high collinearity across foxtail millet, sorghum, maize, rice, and wheat. An 855-bp transposable element inserted in sh1 caused nonshattering during foxtail millet domestication, indicating that sh1 was under parallel selection during domestication across different cereal species. Cereals became better adapted to local environments and evolved into many landraces during diversification. These landraces are generally insensitive to the varying photoperiods in different regions around the world. The key genes ZmCCT in maize (Yang et al. 2013) (its ortholog Ghd7 in rice; Xue et al. 2008); Hd1, encoding a CCT domain, in rice, sorghum, and foxtail millet (Yano et al. 2000; Liu, Liu, et al. 2015); and Zcn8 in maize (Meng et al. 2011) (hd3a in rice; Kojima et al. 2002) were generally involved in reshaping flowering time during diversification across different species. Grain yield was the key target of human selection and has been greatly improved during domestication and improvement. Several key genes related to yield improvement, including tb1 (Studer et al. 2011; Lyu et al. 2020), tga1 (Wang et al. 2005) (gw8 in rice; Wang et al. 2012), tin1 (Zhang et al. 2019) (prog1 in rice; Tan et al. 2008), krn1 (Wang et al. 2019) (Q in wheat; Simons et al. 2006), and krn4 (Liu, Du, et al. 2015) (ipa1 in rice; Jiao et al. 2010), have undergone parallel selection across the staple cereals. All these above studies support the likelihood that the parallel phenotypic changes during cereal domestication, diversification, and improvement have a similar genetic basis.
Methods
Plant Materials
A foxtail millet panel consisting of 23 wild and 73 domesticated accessions (supplementary table S1, Supplementary Material online) from around the world was grown for phenotyping with three replicates at the China Agricultural University experimental station in Beijing in 2018. Natural shattering and nonshattering phenotypes were investigated 2 weeks after maturity. All green millet and foxtail millet accessions from different regions of the world were obtained from the Germplasm Resources Information Network (http://www.ars-grin.gov/) and the Chinese Crop Germplasm Resources Information System (http://www.cgris.net).
Comparative Mapping
The foxtail millet genomic sequence (of the line Yugu1, id28806) at the sh1 locus on chromosome IX was compared with maize (B73, id333), rice (Nipponbare, id3), wheat (Chinese Spring, id54192), and sorghum (Tx623, id331) genomes in the comparative genomics database CoGe (http://genomevolution.org/CoGe/). A syntenic map was plotted using R based on the BLAST results for these comparative genomes at the sh1 locus.
DNA Diversity Analysis
The whole sh1 gene, including a 1,269-bp promoter region, a 3,708-bp gene body excluding the first intron, and an 824-bp 3′ UTR, was amplified from 23 wild and 73 domesticated accessions. The first intron was excluded for sequencing because of its high AT content (61%), which resulted in the failure of amplification. The resulting PCR products were cleaned with a QIAquick PCR Purification Kit (Qiagen) and then sequenced with Sanger sequencing. The obtained sequences were imported into ClustalW and DnaSPV5.1 to analyze DNA diversity (π) with a sliding window of 100 bp and a step size of 25 bp without a gap, and Tajima’s D tests were conducted with DnaSPV5.1 (Librado and Rozas 2009) to determine whether the sequences were subjected to selection.
Association Mapping
To determine whether sh1 was responsible for the loss of shattering in foxtail millet, associations between the variants from 23 wild and 73 domesticated foxtail millet accessions based on sequencing and phenotypes were tested with Fisher’s exact test in R because the natural seed shattering trait exhibited a typical quality trait. The significance threshold was determined for multiple testing through Bonferroni correction based on the following equation: α′ ≈ α/n = 1.04 × 10−4, where α is the nominal significance threshold (α = 0.01) and n is the number of variants (n = 96).
Transformation
The wild-type Sh1 gene from ME034v was edited with two gRNAs (TAGCACACATGCTCCAGCGCCGG and CTGCTGTCAGTGAACTTGAGAGG), targeting the first and second exons of Sh1 through the pRLG103 vector with the CRISPR/Cas9 system (Čermák et al. 2017) in a wild foxtail millet accession (ME034v) suitable for transformation with natural seed shattering (Finley et al. 2021). The overexpression construct harboring the CDS of Sh1 driven by the promoter of maize Ubiquitin was transformed into a domesticated foxtail millet cultivar (Ci846). The resulting three editing and three overexpressed T0 transformation events were self-pollinated to generate homozygous T1 plants. The Sh1 sequences of all homozygous T1 transformant plants were determined through Sanger sequencing.
The homozygous T1 transformation plants were phenotyped 2 weeks after maturity. Each seed was pulled down by a clip linked to a force gauge. The breaking tensile strength was measured at the moment of seed detachment and recorded in gravitational units of force (gf).
Sectioning and Microscopy
Spikelets with pedicels from young panicles (3–4 cm) of wild green millet ME034v and Yugu1 before heading were fixed in 3.7% FAA and then dehydrated in an ethanol series for 1 h each, followed by a Histo-Clear series with ethanol as the solvent. Samples were further treated by replacing one-fourth of the volume of Histoclear/Paraplast mix with new molten Paraplast (Sigma, MO, USA) twice a day for 3 days in a 60 °C oven. Samples were then embedded, sectioned (8 µm thickness) with a Leica RM2265 automated microtome, spread in 37 °C water with a Leica HI1210 water bath, and mounted on microscope slides at 42 °C on a Leica HI1210 slide warmer. Sections were then deparaffinized by a Histo-Clear series, rehydrated in an ethanol series, and then stained with toluidine blue O and acridine orange. The sections were stained with 0.5% toluidine blue O for 30 min, rinsed with water, differentiated in 0.5% glacial acetic acid, rinsed with water again, dehydrated in ethanol, and cleared with Histo-Clear. Images were taken using a Leica biological microscope. In parallel, the sections were stained with 0.01% (w/v) acridine orange for 10 min in the dark, rinsed with water, and then observed using an Olympus FV1000 laser scanning microscope with 488- and 543-nm laser lines.
Scanning Electron Microscopy
Pedicels were collected from wild and domesticated foxtail millet as well as edited and overexpression transformation plants 2 weeks after maturity. The samples were then sputter-coated with gold and palladium for 60 s. After these treatments, the tissues were observed at 15 kV with a SEM (Hitachi S-3400N).
Phylogenetic Analysis
The phylogenetic tree was generated using MEGA7 (Kumar et al. 2016) with the maximum likelihood method based on the DNA alignment of the foxtail millet sh1 gene.
RNA-seq Analysis
RNA was prepared from the panicles (∼5 cm) of the edited transformed plants and the control plants (ME034v) with three biological replicates 2 days after heading. The RNA was then treated with RNase-Free DNase I (D2215, Takara) to remove DNA. The DNA-free RNA samples were subjected to library construction and sequencing on an Illumina HiSeq-2500 platform and resulted in 50 Gb of raw sequencing data. The raw RNA-seq data were treated with a common RNA-seq pipeline. Briefly, the raw RNA-seq reads were trimmed with Trimmomatic (Bolger et al. 2014), cleaned with fastq_clean (Zhang et al. 2014), and then aligned to the green millet reference genome (A10) with STAR (Dobin et al. 2013). Gene expression was further identified based on fragments per kilobase of exon per million fragments mapped (FPKM) with Cufflinks and cuffdiff2 (Trapnell et al. 2014). Finally, DEGs between the two edited plants and the control plant were determined based on their corrected P-values (q-value < 0.05).
RT-qPCR
Total RNA was extracted from roots, stems, leaves, leaf sheaths, bristle, and young panicles before heading (3–4 cm), 3 days after heading, 8 days after heading, and at the grain-filling stage using an RNA extraction kit (TianGen Biotech). After cDNA was synthesized with the TransScript-Uni cDNA Synthesis SuperMix (TransGen Biotech), quantitative PCR (qPCR) in three biological and three technical replications was conducted on a Bio-Rad CFX Maestro system using the foxtail millet Actin gene (Zhao et al. 2020) as an internal control. The relative expression levels were eventually determined through the ΔΔCT relative quantification method (Livak and Schmittgen 2001).
Luciferase Transient Expression Assay
To identify how the Sh1 gene regulates the transcription of the genes related with lignin synthesis, promoters from CAD2, CCR1, 4CL3, PAL2, LAC2, LAC17, NAC29, and CAD8 (2.5–3-kb upstream fragments from the start codons of these genes from ME034v) were respectively fused with the firefly luciferase reporter gene (LUC) in the vector pGreenII 0800-LUC to generate reporter constructs, using the luciferase reporter gene from Renilla reniformis as an internal control driven by the 35S cauliflower mosaic virus (CaMV) promoter. The full-length CDS of Sh1 was cloned into the pGreenII 62-SK vector to generate the effector construct, driven by the 35S CaMV promoter. These effector and reporter constructs were cotransformed into foxtail millet Yugu1 etiolated leaf protoplasts at the two-leaf stage using the empty effector pGreenII 62-SK and respective reporter construct as a control.
Freshly isolated protoplasts were mixed with 20 μg DNA of the reporter construct in a PEG transfer solution for 18 min at room temperature. The transformed protoplasts were incubated for 16 h at 25 °C, then harvested by centrifugation, lysed in passive lysis buffer (PLB, Promega), and assayed according to the Dual-Luciferase Reporter Assay System (Promega). Three biological replicates of each construct were performed, and all assays were repeated three times.
Subcellular Localization
The CDS of Sh1 was introduced into the pCAMBIA1300-GFP vector. The construct with the SH1–GFP fusion protein under the control of the 35S promoter was then introduced into Yugu1 leaf protoplasts by PEG. The signal of SH1–GFP was detected with a 488-nm laser line using an Olympus FV1000 laser scanning microscope.
Transcriptional Activity Assay
To identify the transcriptional activity of the SH1 protein, a dual-luciferase transient expression assay and a Matchmaker GAL4 Two-Hybrid assay (Clontech) were conducted. In the dual-luciferase transient assay, the full-length CDS of Sh1 was fused with GAL4DB and VP16 to construct the effector GAL4DB–VP16–SH1, and a promoter with 5× GAL4 UAS sequence and a TATA box was cloned into pGreenII 0800-LUC to generate the reporter. Under the control of the empty effector construct, the reporter and effector constructs were cointroduced into foxtail millet (Yugu1) leaf protoplasts. In the two-hybrid assay, the full-length CDSs of Sh1 and sh1 were fused with the DNA-binding domain of GAL4 (GAL4BD) in the pGBKT7 vector using the construct containing a transcription factor ZmCCT fused with GAL4BD as a positive control and empty pGBKT7 as a negative control. These constructs were transformed into yeast strain AH109 based on the manufacturer’s instructions, and the colonies were then diluted and grown onto yeast synthetic drop-out medium without Trp or without Trp, Ade, and His.
Primers
All primers used in this study are listed in supplementary table S5, Supplementary Material online, for details.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the 2022 Research Program of Sanya Yazhou Bay Science and Technology City (SYND-2022-18 to Z.L.), the National Natural Science Foundation of China (31871632 to Z.L.), and the 2115 Talent Development Program of China Agricultural University.
Contributor Information
Hangqin Liu, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Xiaojian Fang, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Leina Zhou, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Yan Li, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Can Zhu, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Jiacheng Liu, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Yang Song, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Xing Jian, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Min Xu, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Li Dong, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China.
Zhongwei Lin, National Maize Improvement Center, Department of Crop Genetics and Breeding, China Agricultural University, Beijing, China; Sanya Institute of China Agricultural University, Sanya, Hainan, China.
Author Contributions
Z.L.: designed the study. H.L., X.F., L.Z., Y.L., C.Z., J.L., Y.S., X.J., M.X., and L.D.: performed the research. H.L., X.F., and Z.L.: analyzed the data. H.L. and Z.L.: wrote the manuscript.
Data Availability
RNA-seq data were deposited under the accession number PRJNA780032 at the National Center for Biotechnology Information (NCBI).
References
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, et al. 2012. Reference genome sequence of the model plant Setaria. Nat Biotechnol. 30:555–561. 10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. 2017. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217. 10.1105/tpc.16.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J. 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat Genet. 38:594–597. 10.1038/ng1784 [DOI] [PubMed] [Google Scholar]
- d'Ennequin ML, Panaud O, Toupance B, Sarr A. 2000. Assessment of genetic relationships between Setaria italica and its wild relative S. viridis using AFLP markers. Theor Appl Genet. 100:1061–1066. 10.1007/s001220051387 [DOI] [Google Scholar]
- Devos KM, Gale MD. 2000. Genome relationships: the grass model in current research. Plant Cell 12:637–646. 10.1105/tpc.12.5.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao X, Jia G. 2017. Origin and domestication of foxtail millet. In: Doust A, Diao X, editors. Genetics and genomics of Setaria. Cham: Springer International Publishing. p. 61–72. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. 2006. The molecular genetics of crop domestication. Cell 127:1309–1321. 10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Doust AN, Lukens L, Olsen KM, Mauro-Herrera M, Meyer A, Rogers K. 2014. Beyond the single gene: how epistasis and gene-by-environment effects influence crop domestication. Proc Natl Acad Sci USA. 111:6178–6183. 10.1073/pnas.1308940110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust AN, Mauro-Herrera M, Francis AD, Shand LC. 2014. Morphological diversity and genetic regulation of inflorescence abscission zones in grasses. Am J Bot. 101:1759–1769. 10.3732/ajb.1400186 [DOI] [PubMed] [Google Scholar]
- Finley T, Chappell H, Veena V. 2021. Agrobacterium-mediated transformation of Setaria viridis, a model system for cereals and bioenergy crops. Curr Protoc. 1:e127. 10.1002/cpz1.127 [DOI] [PubMed] [Google Scholar]
- Harlan JR. 1992. Crop and man. Madison, WI: American Society of Agronomy. [Google Scholar]
- Hodge JG, Kellogg EA. 2016. Abscission zone development in Setaria viridis and its domesticated relative, Setaria italica. Am J Bot. 103:998–1005. 10.3732/ajb.1500499 [DOI] [PubMed] [Google Scholar]
- Jackson SA. 2016. Rice: the first crop genome. Rice 9:14. 10.1186/s12284-016-0087-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia GQ, Huang XH, Zhi H, Zhao Y, Zhao Q, Li WJ, Chai Y, Yang LF, Liu KY, Lu HY, et al. 2013. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genet. 45:957–961. 10.1038/ng.2673 [DOI] [PubMed] [Google Scholar]
- Jia GQ, Shi SK, Wang CF, Niu ZG, Chai Y, Zhi H, Diao XM. 2013. Molecular diversity and population structure of Chinese green foxtail [Setaria viridis (L.) Beauv.] revealed by microsatellite analysis. J Exp Bot. 64:3645–3656. 10.1093/jxb/ert198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, Liu GF, Dong GJ, Zeng DL, Lu ZF, Zhu XD, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 42:541–544. 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Jones M. 2004. Between fertile crescents: minor grain crops and agricultural origins. In: Jones M, editor. Traces of ancestry: studies in honour of Colin Renfrew. Cambridge: McDonald Institute for Archaeological Research. p. 127–135. [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43:1096–1105. 10.1093/pcp/pcf156 [DOI] [PubMed] [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M. 2006. An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396. 10.1126/science.1126410 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CB, Zhou AL, Sang T. 2006. Rice domestication by reducing shattering. Science 311:1936–1939. 10.1126/science.1123604 [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lin Z, Li X, Shannon LM, Yeh CT, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, et al. 2012. Parallel domestication of the Shattering1 genes in cereals. Nat Genet. 44:720–724. 10.1038/ng.2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZW, Griffith ME, Li XR, Zhu ZF, Tan LB, Fu YC, Zhang WX, Wang XK, Xie DX, Sun CQ. 2007. Origin of seed shattering in rice (Oryza sativa L.). Planta 226:11–20. 10.1007/s00425-006-0460-4 [DOI] [PubMed] [Google Scholar]
- Liu HH, Liu HQ, Zhou LN, Zhang ZH, Zhang X, Wang ML, Li HX, Lin ZW. 2015. Parallel domestication of the Heading Date 1 gene in cereals. Mol Biol Evol. 32:2726–2737. 10.1093/molbev/msv148 [DOI] [PubMed] [Google Scholar]
- Liu L, Du YF, Shen XM, Li MF, Sun W, Huang J, Liu ZJ, Tao YS, Zheng YL, Yan JB, et al. 2015. KRN4 controls quantitative variation in maize kernel row number. PLoS Genet. 11:e1005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lv SW, Wu WG, Wang MH, Meyer RS, Ndjiondjop MN, Tan LB, Zhou HY, Zhang JW, Fu YC, Cai HW, et al. 2018. Genetic control of seed shattering during African rice domestication. Nat Plants 4:331–337. [DOI] [PubMed] [Google Scholar]
- Lyu J, Huang LY, Zhang SL, Zhang YS, He WM, Zeng P, Zeng Y, Huang GF, Zhang J, Ning M, et al. 2020. Neo-functionalization of a Teosinte branched 1 homologue mediates adaptations of upland rice. Nat Commun. 11:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi S, Healey A, Huang P, Grimwood J, Jenkins J, Barry K, Sreedasyam A, Shu S, Lovell JT, Feldman M, et al. 2020. A genome resource for green millet Setaria viridis enables discovery of agronomically valuable loci. Nat Biotechnol. 38:1203–1210. 10.1038/s41587-020-0681-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. 2011. The FT-Like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 23:942–960. 10.1105/tpc.110.081406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu H, Momohara A, Yasuda Y, He JJ. 2007. The occurrence and identification of Setaria italica (L.) P. Beauv. (foxtail millet) grains from the Chengtoushan site (ca. 5800 cal B.P.) in central China, with reference to the domestication centre in Asia. Veg History Archaeobot. 16:481–494. 10.1007/s00334-006-0068-4 [DOI] [Google Scholar]
- Odonkor S, Choi S, Chakraborty D, Martinez-Bello L, Wang XW, Bahri BA, Tenaillon MI, Panaud O, Devos KM. 2018. QTL mapping combined with comparative analyses identified candidate genes for reduced shattering in Setaria italica. Front Plant Sci. 9:918. 10.3389/fpls.2018.00918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Lyons E. 2011. Comparative genomics with maize and other grasses: from genes to genomes!. Maydica 56:183–199. [Google Scholar]
- Schnable JC, Springer NM, Freeling M. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA. 108:4069–4074. 10.1073/pnas.1101368108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei FS, Pasternak S, Liang CZ, Zhang JW, Fulton L, Graves TA, et al. 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang ZC, Tai YS, Gill BS, Faris JD. 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172:547–555. 10.1534/genetics.105.044727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 43:1160–1163. 10.1038/ng.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LB, Li XR, Liu FX, Sun XY, Li CG, Zhu ZF, Fu YC, Cai HW, Wang XK, Xie DX, et al. 2008. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 40:1360–1364. 10.1038/ng.197 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan HY, You Q, Yi X, Du Z, Xu WY, Su Z. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucl Acids Res. 45:W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2014. Erratum: Corrigendum: Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 9:2513. 10.1038/nprot1014-2513a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li BL, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF. 2005. The origin of the naked grains of maize. Nature 436:714–719. 10.1038/nature03863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lin ZL, Zhang X, Liu HQ, Zhou LN, Zhong SY, Li Y, Zhu C, Lin ZW. 2019. krn1, a major quantitative trait locus for kernel row number in maize. New Phytol. 223:1634–1646. 10.1111/nph.15890 [DOI] [PubMed] [Google Scholar]
- Wang SK, Wu K, Yuan QB, Liu XY, Liu ZB, Lin XY, Zeng RZ, Zhu HT, Dong GJ, Qian Q, et al. 2012. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 44:950:954. 10.1038/ng.2327 [DOI] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 40:761–767. 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- Yang Q, Li Z, Li WQ, Ku LX, Wang C, Ye JR, Li K, Yang N, Li YP, Zhong T, et al. 2013. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc Natl Acad Sci USA. 110:16969–16974. 10.1073/pnas.1310949110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2483. 10.1105/tpc.12.12.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Cho LH, Antt HW, Koh HJ, An G. 2017. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 174:312–325. 10.1104/pp.17.00298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Cho LH, Kim SL, Choi H, Koh HJ, An G. 2014. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 79:717–728. 10.1111/tpj.12581 [DOI] [PubMed] [Google Scholar]
- Zhang GY, Liu X, Quan ZW, Cheng SF, Xu X, Pan SK, Xie M, Zeng P, Yue Z, Wang WL, et al. 2012. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 30:549–554. 10.1038/nbt.2195 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhan F, Sun HH, Gong XJ, Fei ZJ, Gao S. 2014. Fastq_clean: an optimized pipeline to clean the Illumina sequencing data with quality control. 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2014 Nov 2–5; Belfast, UK. IEEE. 10.1109/BIBM.2014.6999309 [DOI] [Google Scholar]
- Zhang X, Lin ZL, Wang J, Liu HQ, Zhou LN, Zhong SY, Li Y, Zhu C, Liu JC, Lin ZW. 2019. The tin1 gene retains the function of promoting tillering in maize. Nat Commun. 10:5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZH, Zhang X, Lin ZL, Wang J, Liu HQ, Zhou LN, Zhong SY, Li Y, Zhu C, Lai JS, et al. 2020. A large transposon insertion in the stiff1 promoter increases stalk strength in maize. Plant Cell 32:152–165. 10.1105/tpc.19.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MC, Tang S, Zhang HS, He MM, Liu JH, Zhi H, Sui Y, Liu XT, Jia GQ, Zhao ZY, et al. 2020. DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc Natl Acad Sci USA. 117:21766–21774. 10.1073/pnas.2002278117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited under the accession number PRJNA780032 at the National Center for Biotechnology Information (NCBI).