Summary
The sharp increase in obesity prevalence worldwide is mainly attributable to changes in physical activity and eating behavior but the metabolic and clinical impacts of these obesogenic conditions vary between sexes and genetic backgrounds. This warrants personalized treatments of obesity and its complications, which require a thorough understanding of the diversity of metabolic responses to high-fat diet intake. By analyzing nine genetically diverse mouse strains, we show that much like humans, mice exhibit a huge variety of physiological and biochemical responses to high-fat diet. The strains exhibit various degrees of alterations in their phenotypic makeup. At the transcriptome level, we observe dysregulations of immunity, translation machinery, and mitochondrial genes. At the biochemical level, the enzymatic activity of mitochondrial complexes is affected. The diversity across mouse strains, diets, and sexes parallels that found in humans and supports the use of diverse mouse populations in future mechanistic or preclinical studies on metabolic dysfunctions.
Subject areas: Obesity medicine, Biological sciences, Endocrinology
Graphical abstract
Highlights
-
•
Strain- and sex-specific profile of metabolic dysfunction in mice
-
•
Liver mitochondrial complex activity in vivo associates with metabolic traits
-
•
Open data source for evaluating different mouse strains for metabolic disease
-
•
Interactive data exploration through an online application
Obesity medicine; Biological sciences; Endocrinology
Introduction
Today, 13% of the adult worldwide population is obese (WHO, n.d.), and the prevalence of the disease is increasing steadily, driven by changes in lifestyle (diet, physical activity, and stress) (Blüher, 2019). Obesity is often observed in conjunction with the metabolic syndrome, type-2 diabetes mellitus (Ezquerra et al., 2008), and nonalcoholic fatty liver disease (NAFLD) (Mitra et al., 2020). The hallmarks of the metabolic syndrome are insulin resistance, hypertension, and dyslipidemia, which constitute risk factors for cardiovascular diseases, cancer, and diabetes, making these three related diseases the leading cause of mortality and premature disability (Blüher, 2019). Although obesity, diabetes, and fatty liver disease often overlap, patients may present any combination of them. The wide variety of clinical presentations co-occurring with obesity is determined by environmental and genetic factors. Genome-wide association studies in humans have uncovered variants determining body fat accumulation, fat distribution, and BMI, such as the FTO gene (Goodarzi, 2018), whereas mouse studies led to identify crucial role of leptin signaling in body weight homeostasis (Pan and Myers, 2018). Despite these advances, some of the genetic modifiers of common forms of obesity remain elusive (Müller et al., 2018); this is partly because of the difficulty in estimating environmental confounders such as food intake and exercise and because external measurements of body weight or BMI are not sufficient to distinguish between different types of obesity (Müller et al., 2018).
Another approach for understanding the diverse etiology of metabolic syndrome is the use of animals, especially mouse models, in which one can easily control genetic and environmental factors (Lutz and Woods, 2012). The simplest of these models are animals fed with a fat-supplemented diet (high-fat diet, HFD). A caveat of these studies is often the focus on a single mouse strain, such as the C57BL/6J reference strain, sometimes genetically modified, which fails to model the diversity of outcomes present in the human population. Findings originating from a single strain and sex translate poorly not only to humans but even to other mouse strains (Sittig et al., 2016). Indeed, the health benefits of precision medicine in mice are highly strain dependent (Barrington et al., 2018; Montgomery et al., 2013), similar to what is observed on drug effectiveness (Recla et al., 2018) or toxicology (Church et al., 2015; Dornbos and LaPres, 2018). A recent study highlighted how a large genetically diverse population of mice could model the diversity and interindividual variability in nonalcoholic fatty liver disease (NAFLD), upon feeding with a high-sucrose and HFD to find susceptible and resistant mouse strains (de Conti et al., 2020). More generally, we hypothesize that findings that hold true in different mouse strains and subspecies are much more likely to also be conserved in humans (Sellers, 2017).
To address this, we systematically characterized the diversity and extent of the physiological, molecular, and functional changes induced by HFD across diverse Mus musculus inbred strains, subspecies, and sexes. While we observe some expected responses to the diet, most metabolic and molecular parameters affected by HFD are dependent on sex and strain. Our observations indicate that the genetic repertoire of inbred laboratory mouse strains encodes a wide range of metabolic responses to diet-induced obesity (DIO), modelling different levels of metabolic disease severity or associated comorbidities. All metabolic traits, mitochondrial activity, and transcriptome data collected in this study can be explored with an online, interactive interface at www.systems-genetics.org/CC_founders_metabolic. This resource enables researchers to quickly examine the diverse manifestations of DIO in mice and pick an appropriate mouse model for individual obesity-associated diseases.
Results
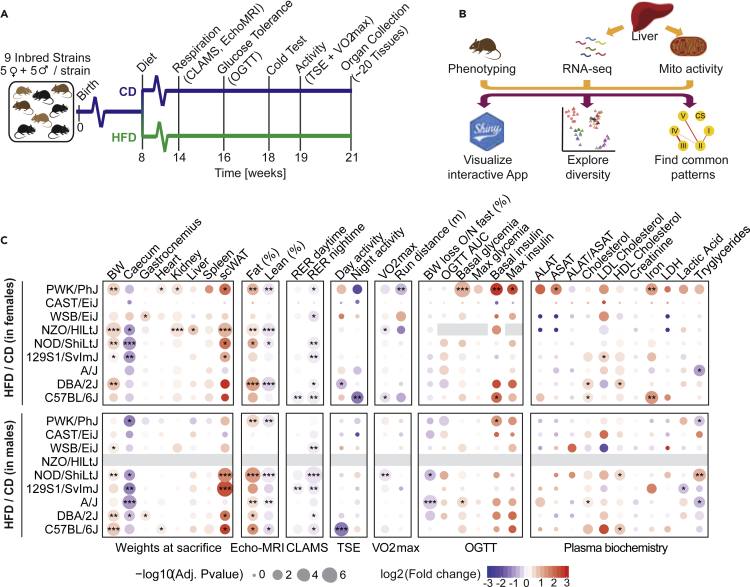
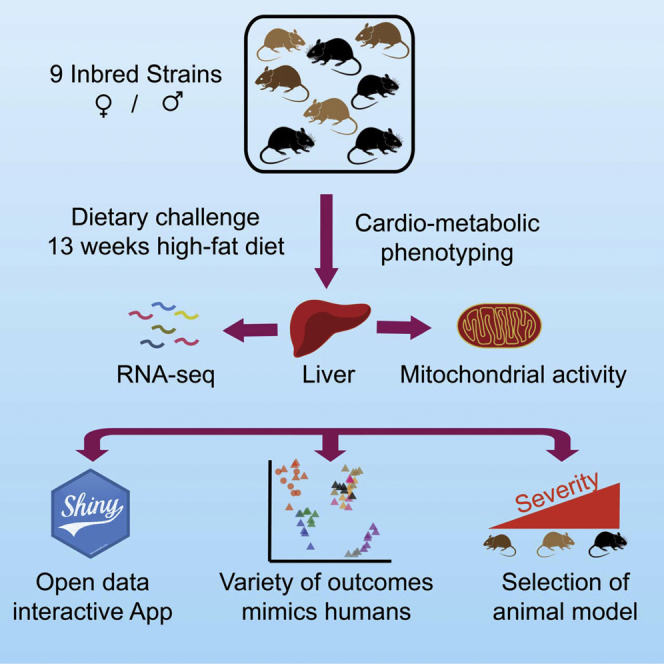
Ten female and 10 male animals from 9 inbred mouse strains were fed with either a chow diet (CD) or an HFD from the age of 8 to 21 weeks, and various metabolic and fitness tests were conducted (Figure 1A). Eight of these strains are known as the founders of the Collaborative Cross (CC) (Welsh et al., 2012) and Diversity Outbred (DO) populations (Churchill et al., 2012). The DBA/2J was added as a ninth strain to leverage our laboratory’s expertise with the BXD recombinant inbred lines (Peirce et al., 2004; Taylor et al., 1999), which are generated from crosses of the C57BL/6J and DBA/2J strains. Three distinct Mus musculus subspecies are represented in this panel: Mus musculus musculus (PWK/PhJ), Mus musculus castaneous (CAST/EiJ), and Mus musculus domesticus (all other strains including the wild-derived WSB/EiJ) (Phifer-Rixey and Nachman, 2015). In addition to the complete cardio-metabolic phenotyping of the mice, we measured the liver transcriptome and enzymatic activity of liver mitochondrial complexes (Figure 1B).
Figure 1.
Study design and phenotypic diversity across strain, diet, and sex
(A) Scheme of the phenotyping pipeline of the study. Nine mouse inbred strains were tested (C57BL/6J, DBA/2J, A/J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HILtJ, WSB/EiJ, CAST/EiJ, and PWK/PhJ). Five females or five males per diet (CD or HFD) were measured per strain. Five mice per condition were fed an HFD from 8 to 21 weeks of age with CD matched animals. Respiration, tolerance to glucose and cold, exercise capacity, and spontaneous activity were tested. Mice were sacrificed at 21 weeks of age after an overnight fast, and organ collection was performed.
(B) Schematic of the liver molecular phenotyping and data analysis. Livers were used for RNA-Seq and measurements of mitochondrial activity.
(C) Heatmap of the effects of diet on a selection of measured metabolic traits, in males or females. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, t test (HFD versus CD), corrected for multiple testing with the Benjamin-Hochberg false discovery rate (BH-FDR) procedure.
A cohort of diet-sensitive and diet-resistant strains
We examined the effects of HFD on cardio-metabolic traits in males and females of each strain (Figures 1C, 2 and S1). Out of the 9 strains considered, 5 significantly gained weight in females and 4 in males (Figures 1C and 2A). These changes did not seem to be caused by increased food intake, although quantification of food intake under HFD was hampered by mice dispersing the food throughout the cage (Figure S1). The A/J and CAST/EiJ strains gained no additional weight upon HFD in either sex. The WSB/EiJ and CAST/EiJ are wild-derived strains known for their resistance to obesity and high percentage of lean mass (Karunakaran and Clee, 2018), but our data show that the A/J strain may exhibit a similar resistance. For all strains but the reference strain C57/BL6/J, the weight gain was greater in females than in males (Figures 1C and 2A). Subcutaneous white adipose tissue (scWAT) weight, as well as the fat percentage measured by Echo-MRI increased in most strains, generally following the same trend as body weight, although some exceptions were present. The males of the A/J strain increased fat mass and decreased lean mass despite minimal weight gain. The males of the 129S1/SvlmJ strain accumulated a large amount of scWAT despite very limited changes in body weight or fat content, perhaps indicating a genetic peculiarity in the body distribution of fat (Figures 1C and S1).
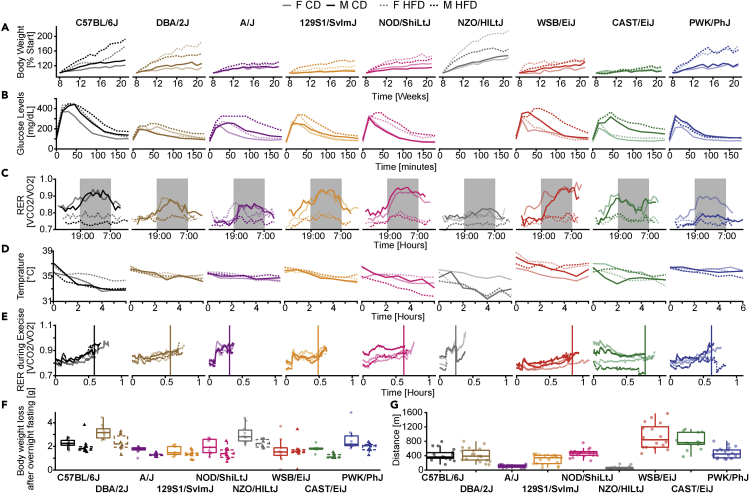
Figure 2.
Time-resolved variation of metabolic traits collected in this study
(A) Body weight (expressed as percentage from the individual starting body weight before HFD).
(B) Glucose levels during the course of an oral glucose gavage test (OGTT).
(C) Circadian respiratory exchange ratio (RER) over a day using a metabolic chamber.
(D) Rectal temperature taken during a cold test.
(E) Uphill exercise respiratory exchange ratio (RER) during a VO2max experiment. Vertical line represents the mean time run per strain.
(F) Body weight loss after an overnight fasting. Male and female mice showed similar trends and are pictured together. Left boxplots: CD; right boxplots: HFD.
(G) Distance run by the HFD-fed mice during the VO2max experiment. Male and female mice showed similar trends and are pictured together. Boxplot lower and upper hinges correspond to the first and third quartiles, and center line is the median. The whiskers extend from the hinge to the largest value no further than 1.5 ∗ inter-quartile range.
Strains present diverse metabolic defects that rarely combine into a full metabolic syndrome
The NZO/HILtJ strain, known for their glucose intolerance and susceptibility to obesity, exhibited the most severe phenotypes under HFD: males of the strain had to be sacrificed 1–2 weeks early due to 3 of the animals dying spontaneously, whereas females were excluded from the oral glucose tolerance test (OGTT) due to ethical concerns that stressful conditions (fasting, blood draws) may put them in a critical condition. To assess glucose intolerance across the other strains, we performed an oral glucose tolerance test (OGTT, Figures 1C and 2B). The PWK/PhJ strain had elevated basal glucose, yet the area under the curve (OGTT AUC) did not reveal significant glucose intolerance in any of the strains (Figure 1C). The respiratory exchange ratio (RER), defined by the ratio of CO2 to O2 consumption, is a key metabolic indicator of body substrate utilization. RER ranges from 0.7 to 1, with 0.7 indicating preferential fat consumption and higher value (up to 1) indicating carbohydrate burning. We measured the day- and night-time RER through a continuous monitoring system (Figures 1C and 2C). Most strains greatly decreased their night-time RER upon high-fat feeding, indicating an increase in fat consumption and loss of metabolic flexibility, as seen in obese humans (Hirsch et al., 2016). However, the A/J strain conserved some metabolic flexibility (relatively minor drop in RER on HFD, Figure 2C), as well as its lean mass ratio (Figure 1C), indicating remarkable resistance to DIO. Conversely, the WSB/EiJ strain lost metabolic flexibility despite not gaining weight or accumulating fat mass, showing that its metabolism was affected by the diet, despite the lack of weight gain (Figures 1C and 2C). We also measured the RER during a treadmill running exercise with the VO2max test (Figure 2D). The A/J and NZO/HILtJ mice had much lower maximum running distance during exercise (Figures 2D and 2G), perhaps because their RER peaked immediately, showing quick saturation with CO2, whereas other strain’s RER increased gradually (Figure 2D). These strains could be models of the phenomenon known as “hitting the wall,” when marathon runners slow down significantly when depleting their body’s glycogen store (Smyth, 2021). When we assessed spontaneous physical activity through the TSE system (Figure 1C, see Day and Night activity), the C57/Bl6/J strain was the only one that showed reduced activity upon HFD, while other strains did not reduce their activity despite large BW gains. Low body temperature during cold tolerance tests was associated with glucose intolerance in most strains (Figure 2E), as is also the case in humans (Atsumi et al., 2013). The lowest temperatures were observed in C57BL/6J, NOD/ShiLtJ, and NZO/HILtJ, which also reached the highest absolute glycaemia during the oral glucose tolerance test (Figure 2B). Body weight loss upon overnight fasting, an indicator of metabolic flexibility (Smith et al., 2018), was consistently lower in HFD-fed mice across strains and sexes (Figure 2F).
The females of the PWK/PhJ strain showed the strongest alterations in the blood biochemistry (Figures 1C and S1), with slightly elevated alanine and aspartate transaminases (ALAT, ASAT), as well as increased plasma iron on HFD. The ALAT/ASAT ratio of the PWK/PhJ strain did not increase, indicating a potential tissue damage, which is not necessarily liver-specific biochemistry (Figures 1C and S1). Overall, female PWK/PhJ mice had the most comprehensive response to HFD (e.g. glucose and insulin levels, RER during the night) (Figure 1C).
Relative contributions of diet, strain, and sex to metabolic phenotypes
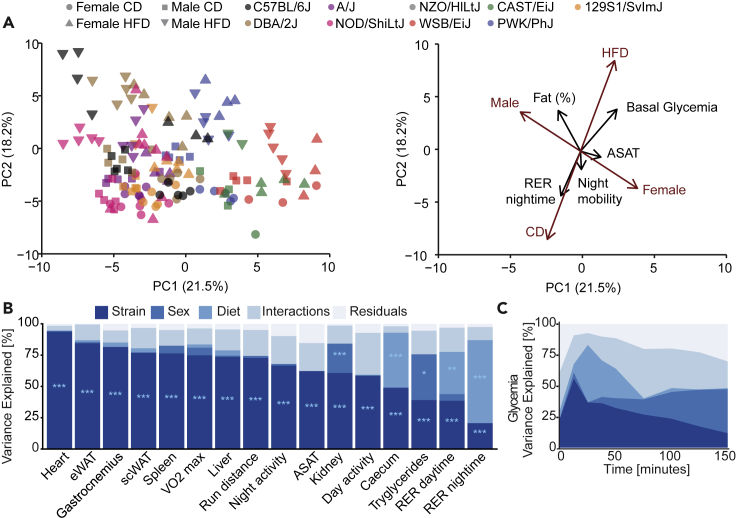
We used principal component analysis (PCA) to examine the variation of phenotypic traits across strains, sexes, and diets (Figure 3A). Principal component 1 (PC1) is associated with a combination of sex and strain, whereas principal component 2 (PC2) segregated mice primarily based on diet. Some phenotypes, such as the fat percentage and respiratory exchange ratio (RER), closely correlated with diet, whereas others, like basal glycemia and activity, were more associated with strain and sex. The covariance components of diet and sex partially overlap because body weight and organ weights are higher in both males and HFD-fed animals.
Figure 3.
Variation in metabolic traits is driven by distinct contributions of genetic background, sex, and diet
(A) Principal component analysis plot of the metabolic and clinical traits measured in the study. Individual mice are shown (left). Loadings are shown for selected phenotypes as well as gender and diet (right). Principal component 1 is associated with a combination of strain and sex, whereas PC2 is associated with the diet. Black arrows: phenotypic traits. Brown arrows: covariates.
(B) Proportion of variance of metabolic traits explained by a linear combination of strain, sex, and diet effects, their interactions, and the residuals of the linear model (Phenotype ∼ Strain ∗ Sex ∗ Diet), expressed as the mean type II sums of squares divided by the total mean type II sum of squares. Mean sum of squares is equal to the sum of squares divided by the corresponding degrees of freedom. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ANOVA test corrected for multiple testing.
(C) Variance of glycemia explained by strain, sex, and diet effects, their interactions during oral glucose gavage test (OGTT) over time.
To quantify the relative contributions of the different biological factors to metabolic outcomes, we computed the phenotypic variance explained by each factor of the study using ANOVA with type II sum of squares (Figure 3B). The effect of the diet on the RER was larger during the night than during the day (Figure 3B), reflecting the main feeding and activity period for mice. Although dynamic parameters (e.g. RER) were diet dependent, morphological measurements, such as organ weights, were mainly strain dependent, with the exception of caecum weight, which is known to be highly affected by the diet (Dalby et al., 2017) (Figure 3B). Unexpectedly, strain effects explained most of the variation of adipose tissue mass (eWAT and scWAT), highlighting the influence of genetics on fat storage. The effects on glycemia during OGTT were particularly striking (Figure 3C). Although basal glycemia (glucose 0 min) was influenced to a large extent by strain, sex, and diet interactions, the maximal glycemia (reached at 15 minutes) was predominantly a function of strain, reflecting genetic differences in glucose absorption. Finally, the values at 30, 45, and 60 minutes were much more influenced by diet, as the speed of glucose return to basal levels (clearance) is more influenced by diabetic-like phenotypes that arise in some strains under HFD. Altogether, these data highlight the complex interactions between strain, sex, and diet for various metabolic traits and show that the spectrum of variation of metabolic traits in mice partially reflects the spectrum of healthy and pathologic conditions found in humans with an obesogenic diet.
The liver transcriptome shows a context-dependent diet signature
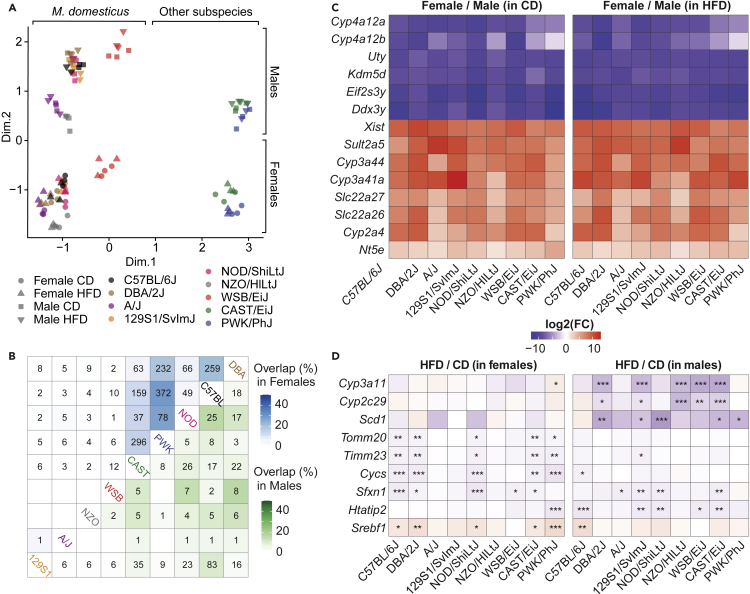
To link variation of metabolic traits with molecular data, we collected and analyzed liver RNA-Seq gene expression in 3 randomly selected mice per condition, sex, and strain. Like phenotype-based multivariate analyses, liver gene expression samples clustered mainly according to strain and sex (Figure 4A). The effects of the diet could be seen on a strain-by-strain basis but were far weaker than the sex effects (Figure S2A). This contrasted with the PCA of the phenotypes, where the effect of diet was much more pronounced and strains and sexes overlapped (Figure 3A). Although the effect of sex was conserved in both diets (Figure S2B, top), the diet effects were different between sexes (Figure S2B, bottom). To quantify differences/similarities between the transcriptomic profiles of the mice, we performed Pearson’s correlations for each pairwise combination between samples, and these comparisons were categorized according to the strain, sex, and diet similarities (Figure S2C). Pairwise transcriptome correlations consistently followed genetic relatedness, with correlations decreasing from same strain > same subspecies > different subspecies. Having the same diet only increased correlations when the strain and sex were also the same (Figure S2C, comparison on the right), showing that diet effects on the transcriptome are only reproducible within a given strain and sex. The number of overlapping differentially expressed genes (DEGs) of HFD versus CD-fed animals across genetic backgrounds contrasted across sexes (Figure 4B). In females, C57BL/6J, DBA/2J, CAST/EiJ, and PWK/PhJ mice showed the highest number of overlapping DEGs, whereas in males this was apparent mostly in 129S1/SvImJ and C57BL/6J. For example, PWK and C57BL/6J had 372 DEG in common in females and only 8 in males, whereas C57BL/6J and 129S1/SvImJ had only 2 DEG in common in females and 83 in males (Figure 4B). Therefore, differences and similarities between strains and between diets are not symmetrical between females and males, which means that the use of male or female mice needs to be considered with caution while choosing a particular strain to model a metabolic process.
Figure 4.
The transcriptome signature of HFD differ between strains and sexes
(A) Multidimensional scaling (MDS) plot of liver RNA-Seq separates animals by subspecies and sex. Euclidean distances between each animal were calculated pairwise based on the leading log2-fold changes for the 500 most differentially expressed genes. Each dot or triangle represents a single female or male mouse, respectively. Each color is assigned to a specific strain.
(B) Number of overlapping differentially expressed genes (diet effect; HFD/CD) across strains of the same sex (female: top left, male: bottom right). Color code representing the percentage of overlapping genes is based on the Jaccard index.
(C) Heatmaps of log2 fold change of the 14 most conserved sexually dimorphic genes across strains and diets. All the fold changes are significant with BH-FDR adjusted p value<0.05.
(D) Heatmaps of log2 fold change of the 9 genes affected by diet in the highest number of strains at the same time. The same color scale was used for panel C & D. Moderated t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; adjusted p value corrected for multiple testing with the BH-FDR procedure.
Conversely, sex differences are much better conserved across strains and diets (Figures 4C and S2D), with a few sexually dimorphic genes with fold changes reaching 1000-fold (log2FC∼10, Figure 4C). These genes include three Y-chromosome genes, Uty, Ddx3y, and Eif2s3y; (Walport et al., 2014), the female-specific X-inactivation gene, Xist; and several cytochrome (Cyp) genes known for their sex-dimorphism (Lu et al., 2013). In contrast, diet-influenced genes were never conserved across all strains and conditions. The most consensus diet-repressed genes (Figure 4D) consisted of ER enzymes involved in lipid metabolism (Cyp2c29, Cyp3a11, Scd1) predominantly in males. In females, the diet modulated mitochondrial protein transport (Tomm20, Timm23), mitochondrial electron chain (Cycs), and transporter (Sfxn1), as well as the cholesterol biosynthesis master regulator (Srebf1) (Figure 4D).
HFD affects immunity and protein translation genes in a sex- and strain-dependent manner
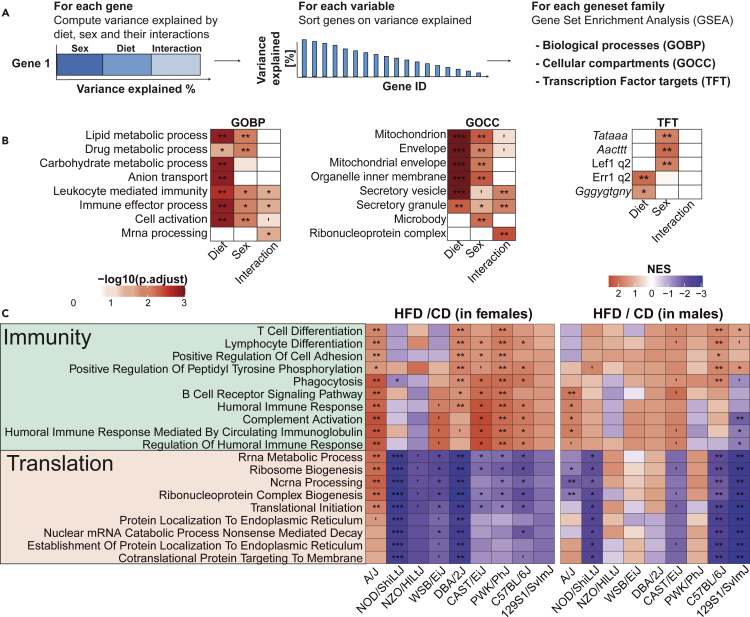
Although individual diet-dependent genes differ across strains and sexes, we investigated whether those different genes might still participate in some of the same biological pathways. Rather than analyze every combination of strain, sex, or diet separately, we used the overall variance explained by each parameter and performed gene set enrichment analysis (GSEA) to understand which pathways are driven by each covariate (Figure 5A). As expected, HFD had consistent effects on lipid and carbohydrate metabolism genes (Figure 5B). Interestingly, sex, diet, and their interactions all affected genes involved in immune processes. At the cellular component level (GOCC), mitochondrial content and secretory vesicle genes were strongly affected by sex and diet, whereas ribosomal protein (ribonucleoprotein complex) gene expression was affected by sex:diet interactions. At the transcription factor level, the target genes of lymphoid enhancer binding factor 1 (Lef1) were particularly affected by sex, whereas those of the estrogen-related receptor 1 (Err1) are affected by diet (Tripathi et al., 2020).
Figure 5.
HFD affects immunity, lipid metabolism, and liver translation in a sex- and strain-dependent manner
(A) Scheme of gene set enrichment analysis based on variance explained by diet, sex, and their interaction. (1) For each transcript, we computed the variance explained based on a linear model of strain, sex, diet, and their interactions (Transcript ∼ Strain + Diet + Sex + Diet:Sex). Then the percentage of variance explained of each transcript was assigned to specific parameters (diet, sex, and their interactions); (2) for ranking of the genes; and then (3) to perform gene set enrichment analysis for each parameter.
(B) Results of gene set enrichment analysis based on variance explained by diet, sex, and their interaction. From left to right, each enrichment was performed on gene ontology of biological processes (GOBP), cellular component (GOCC), and transcription factors. Percentage of variance explained was measured using variantPartition package.
(C) Enrichment analysis of the diet effect on liver gene expression separated by sex and strain. Gene set enrichment analysis was performed on the sign (log2 (FC)) ∗ –log10 (adjusted p value) per strain per sex. The selected gene sets add the highest score for the global response to the diet in a specific sex. GSEA permutation test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; adjusted p value corrected for multiple testing with the BH-FDR procedure.
To highlight the differences between strains, we performed another GSEA of the effect of diet on each strain/sex combination and displayed the most conserved ones (Figure 5C). Enriched gene sets fell into two main categories: immunity and translation or mRNA processing. Overall, adaptive immune responses were enriched upon HFD to a higher extent in females compared with males (Figure 5C); this reflects sex-specific manifestations of inflammation and immune response in the context of fatty liver (Lonardo et al., 2019). Gene sets associated with translation, endoplasmic reticulum (ER), and mRNA processing were reduced upon HFD in most strains, perhaps reflecting an activation of the integrated stress response (Yilmaz, 2017). In humans, the integrated stress response is activated in obese and/or patients with fatty liver (Lebeaupin et al., 2018). Although these pathways were the most conserved in the population, there were still important differences between strains. Indeed, A/J females showed a significant positive enrichment of translation-associated gene sets, opposite to the other strains (Figure 5C). Apart from A/J, the strongest immune activation was found in the PWK/PhJ strain females, which have elevated ASAT, ALAT, as well as plasma iron, all markers of potential tissue damage, which may have resulted from such inflammation in the liver or other organs.
Liver mitochondrial activity in vivo correlates with metabolic traits
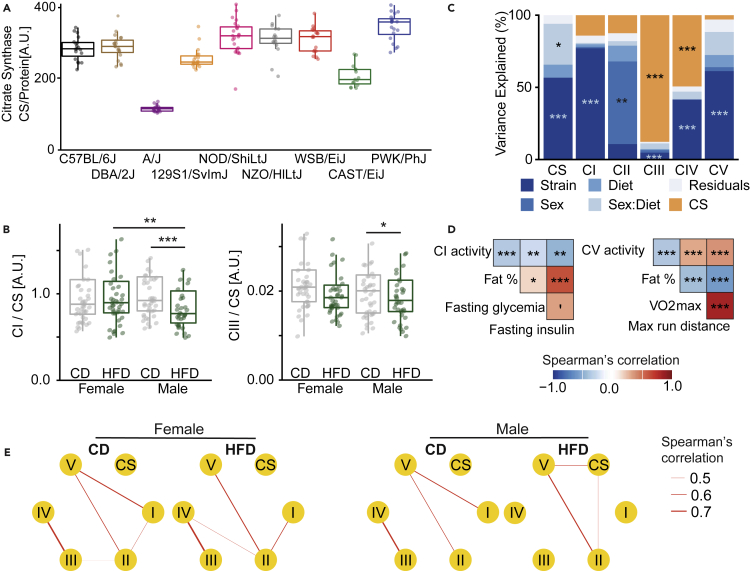
Due to the documented importance of mitochondria as a sensor and effector of metabolism and the strong overall effect of diet on mitochondrial components (Figure 5B), we measured the activity of the five main complexes of the electron transport chain (ETC; complex I–V), as well as citrate synthase (CS), the main pace-making enzyme of the tricarboxylic acid cycle (TCA). CS activity was used as a readout of entry of fuel into the TCA cycle and as a proxy of mitochondrial content (Larsen et al., 2012). Citrate synthase itself was strongly affected by strain, with the lowest activity present in A/J mice (Figure 6A), which has a known high-impact genetic variant in the coding sequence of the gene encoding citrate synthase itself (Alhindi et al., 2019; Gabriel et al., 2017; Johnson et al., 2012; Ratkevicius et al., 2010). CAST/EiJ and A/J mice are two out of three diet-resistant strains in this study, and they also had the lowest CS activity, implying a link between mitochondrial activity and resistance to DIO. We also observed a consistent reduction of complex I and complex III activity upon HFD in males only (Figure 6B). In addition, we computed the variation explained by strain, sex, diet, and their interactions on CS activity and each complex’ activity (Figure 6C). The mitochondrial complexes of the ETC are strikingly differently affected by biological variables. Indeed, although complex I activity is highly strain dependent, variation of complex II activity is predominantly explained by sex differences. Most of the variation of complex III activity can be attributed to the variation of citrate synthase activity (Figure 6C).
Figure 6.
Liver mitochondrial activity correlates with metabolic phenotypes, and correlations between complexes are disrupted by HFD in males
(A) Citrate synthase activity (A.U.). All sexes and diets are pictured together as they differed only minorly, showing that CS is principally affected by strain. Boxplot lower and upper hinges correspond to the first and third quartiles, and center line is the median. The whiskers extend from the hinge to the largest value no further than 1.5 ∗ inter-quartile range.
(B) Complex II and complex III activity (normalized by citrate synthase activity) is affected by diet in sex-specific manner.
(C) Proportion of variance of liver mitochondrial activity explained by strain, sex, diet, sex-by-diet interactions (sex:diet), and citrate synthase (CS) activity. Each bar corresponds to the activity of the different complexes of the electron transport chain as well as the citrate synthase activity or a model that takes all the complexes together into account (ALL). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ANOVA test.
(D) Spearman’s correlations between the activity of liver mitochondrial complexes and essential cardio-metabolic traits. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, correlation test, corrected for multiple testing with the BH-FDR procedure.
(E) Correlation network of liver mitochondrial complex activities in each condition. The width of the edges corresponds to the magnitude of positive correlation between complexes. Only significant edges are shown (Benjamini-Hochberg adjusted-p <0.05).
We assessed whether the activity of individual complexes could associate with some of the metabolic traits measured previously. Complex I and complex V activity both negatively correlated with fat percentage, suggesting that activation of specific mitochondrial complexes may be protective against weight gain or be negatively impacted by HFD. Liver complex I activity further correlated negatively with fasting glycemia and insulin, whereas complex V activity positively correlated with VO2 max and running distance, indicators of endurance and stamina (Figure 6D). Thus, the activity of mitochondrial complexes I and V, the main entry and exit points of mitochondrial electron transport, may reflect the type of response to obesogenic diet. This result also indicates that the activity of different mitochondrial complexes may be relevant in distinct processes in vivo and that the activity of the ETC is not fully characterized when using only citrate synthase activity as a readout.
We then computed the correlation between ETC complex activities for each experimental group (Figure 6E). Complex II and complex V activity are consistently correlated across conditions, whereas correlation between complex I and complex V activity was lost upon HFD. In addition, correlations of complex I and complex II activity were sex specific, present in females and not in males. Finally, complex III and complex IV activity associations are not present in males fed HFD, which potentially highlights the differences of the response to HFD between males and females (Figure 6E). In the future, the correlation map (Figure 6E) of the activity of mitochondrial complexes could be used as a type of metabolic “fingerprint,” predicting metabolic responses, although this warrants further investigation.
Toward a strain-specific profile of metabolic dysfunction
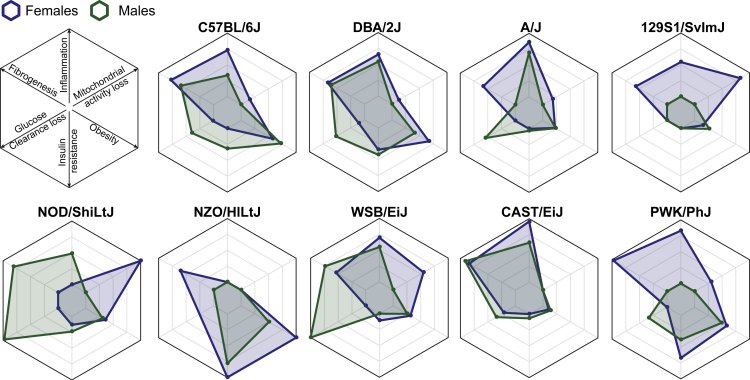
To summarize the different parameters measured in this study, we show key phenotypic and molecular parameters associated with the response to HFD in each mouse strain (Figure 7). Each parameter was assessed relative to the other strains. Much like humans, each mouse strain responds differently to an obesogenic diet. The NZO/HILtJ strain had the more severe obesity and insulin resistance phenotype, yet transcript results of GSEA related to liver fibrogenesis and inflammation were comparatively lower. Conversely, the CAST/EiJ strain gained almost no weight and displayed no signs of glucose intolerance and insulin resistance, and yet it had some of the more pronounced transcript expressions of fibrogenesis and inflammation markers in the liver. Although females in general displayed most severe symptoms, this was not the case in every strain, as female NOD/ShiLtJ mice were generally less affected by HFD than males. Our overview of these strain-specific profiles (based on their phenotypic, transcriptomic, and mitochondrial signatures) may guide researchers in choosing an appropriate model for correctly representing the different measurable outcomes in metabolic syndromes.
Figure 7.
Strain-specific response signatures to obesogenic diet
Radar plots of some hallmarks reflecting predisposition to metabolic disease in each strain. Each parameter is expressed as a percentage of the highest strain. Fibrogenesis and inflammation scores are based on the normalized enrichment score of the GSEA for “collagen-containing extracellular matrix” and “humoral immune response,” respectively. Insulin resistance is based on Homa-IR value under HFD. Obesity is based on body weight gain relative to the starting weight before HFD. Finally, glucose clearance loss is the change in the slope of glucose clearance in the OGTT test upon HFD (missing data in the NZO strain, due to a too severe phenotype).
Discussion
A study that gave identical diets to a large human cohort found highly variable glycemic responses to identical meals (Zeevi et al., 2015), showing that even with tightly controlled dietary intake, humans could exhibit huge variations in their metabolic responses. Our study aimed to refine this observation in mice and understand how much genetics, sex, and diet can influence metabolic outcomes in a highly controlled environment. One of the most striking findings of this study is the different importance of diet, sex, and strain across different biological scales or layers. At the morphological scale (body and tissue weights), we found that the genetic effect of the strain predominated over any other. Even the fat weight and percentage, which we would expect to be primarily driven by environmental factors, were mostly a function of the strain, highlighting the importance of genetics in determining morphological traits, including fat tissues. Conversely, functional phenotypes representing metabolism or physical performance were much more affected by the diet and sex.
When we zoom in toward individual gene expression in the liver transcriptome, sex becomes by far the most impactful and consistent parameter, overtaking diet and strain, with the second most impactful parameter being the mouse sub-species, with the Mus musculus musculus (PWK/PhJ) and Mus musculus castaneous (CAST/EiJ) strains strongly diverging from the other Mus musculus domesticus strains at the gene expression level, but not at the phenotypic level. When we zoom out, away from individual genes and toward pathways, the effect of sex, driven by a small number of sexually dimorphic genes, disappears, and the effects of the diet re-emerge, pointing to immune activation and a decrease in translation, which may be a prelude to more severe liver disease.
To complete the study, we attempted to connect the different layers of physiological and biochemical traits through the central hub of metabolism, the mitochondria. The activity of mitochondrial complexes, particularly I and V, was positively correlated with indicators of physical fitness, such as maximal aerobic capacity (VO2 Max) and maximum run distance, indicating that optimal liver metabolic activity may be an important determinant of organismal fitness. These results suggest that specific ETC complexes measured in vivo or mitochondrial complex assembly may play a role in the liver response to dietary conditions, adding to the growing body of evidence for the central role of mitochondria in obesity and its complications (Bournat and Brown, 2010).
The strains chosen in this study are the founders of several genetic reference populations (GRP), such as the collaborative cross (CC), diversity outbred (DO), and BXD populations (Swanzey et al., 2021). Establishing a baseline for the response to dietary challenge in the founder strains of these populations can be useful when choosing strains, conditions, time points, and phenotyping tests in future studies. In addition, our data reveal that some of these strains exhibit resistance or sensitivity to metabolic disease. Although the NZO/HILtJ strain is a known model of autoimmune diabetes, we find that the PWK/PhJ strain may develop more severe metabolic defects upon HFD than the reference C57BL/6J. On the other end of the spectrum, the CAST/EiJ strain does not gain weight or exhibit phenotypic signs of metabolic abnormalities on the systemic level, despite a prototypical activation of inflammatory and fibrogenesis genes in the liver. The genetic basis for these unique characteristics can be studied directly or through their genetic offspring in the DO or CC mouse populations and may provide unique insights into the resistance or susceptibility to metabolic disease.
Our work highlights the importance of considering genetic background and sex before drawing conclusions from HFD feeding experiments. By looking at various strains of mice, some of which come from distinct subspecies or from the wild, we can approach the phenotypic variety present in human populations and find conserved features, strengthening our conclusions; this is particularly important considering the number of discoveries that are affected by hidden biological and experimental parameters.
The data in this paper are openly accessible and easily explorable with an online interface, available at www.systems-genetics.org/CC_founders_metabolic. This resource will be valuable to the community, to help researchers choose one or preferably more than one appropriate mouse model for a given study. Indeed, the combined use of several strains and sexes in tandem is more likely to achieve generalizable results that will be more translatable across species.
Limitations of the study
Our study uses inbred mouse strains, including several laboratory strains that represent only a fraction of the genetic diversity found in wild mice, and may present specific adaptations to a sedentary and protected laboratory environment (Yang et al., 2011). We addressed this by including three “wild-derived” strains: the CAST/EiJ, WSB/EiJ, and PWK/PhJ strains. We found that they presented a wider variability of phenotypes and gene expression patterns, and the strain-driven diversity of phenotypes observed in this study suggests that multiple genetic variants with strong effects on cardio-metabolic traits can be found in this panel. Another limitation of this study was the use of a chow diet, which is not perfectly matched in composition to the HFD. Therefore, components of the diet other than fat composition may have influenced the observed phenotypes.
Data and materials availability
The authors declare that all the data supporting the findings of this study are available within the paper and its supplemental information.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol | Invitrogen | Cat#15596026 |
| Direct-zol-96 RNA kits | Zymo | Cat# R2056 |
| Critical commercial assays | ||
| Metabolic Modular Treadmill | Columbus Instruments | N/A |
| Metabolic cages | TSE systems | N/A |
| Oxymax/CLAMS | Columbus Instruments | N/A |
| Echo-MRI 3-in-1 | Echo Medical Systems | N/A |
| Mitochondrial function assay | IntegraCell | N/A |
| Deposited data | ||
| RNAseq | Gene Expression Omnibus | GEO: GSE182668 |
| Metabolic traits | Mouse Phenome Database | MPD: Auwerx2 identifier |
| Experimental models: Organisms/strains | ||
| C57BL6/J | Jackson Laboratory | RRID:IMSR_JAX:000664 |
| DBA2/J | Jackson Laboratory | RRID:IMSR_JAX:000671 |
| A/J | Jackson Laboratory | RRID:IMSR_JAX:000646 |
| 129S1/SvlmJ | Jackson Laboratory | RRID:IMSR_JAX:002448 |
| NOD/ShiLtJ | Jackson Laboratory | RRID:IMSR_JAX:001976 |
| NZO/HILtJ | Jackson Laboratory | RRID:IMSR_JAX:002105 |
| WSB/EiJ | Jackson Laboratory | RRID:IMSR_JAX:001145 |
| CAST/EiJ | Jackson Laboratory | RRID:IMSR_JAX:000928 |
| PWK/PhJ | Jackson Laboratory | RRID:IMSR_JAX:003715 |
| Software and algorithms | ||
| R software version 3.5.2 | R project | https://www.r-project.org/ |
| Rstudio Pro version 1.4.1717-3 | Rstudio | https://www.rstudio.com/ |
| variantPartition version: 1.12.3 | Bioconductor | http://bioconductor.org/packages/release/bioc/html/variancePartition.html |
| limma version 3.38.3 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/limma.html |
| data.table version 1.13.0 | CRAN | https://cran.r-project.org/web/packages/data.table/ |
| clusterprofiler version 3.10.1 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| ggplot2 version 2 2.0.0 | CRAN | https://cran.r-project.org/web/packages/ggplot2/index.html |
| cowplot version 1.1.1 | Github | https://github.com/wilkelab/cowplot |
| Other | ||
| Interactive application | Systems-genetics.org | www.systems-genetics.org/CC_founders_metabolic |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Prof. Johan Auwerx (admin.auwerx@epfl.ch).
Materials availability
All mouse tissue samples generated in this study are available under a collaboration agreement with the lead contact.
Experimental model and subject details
Mouse strains were imported from Charles River and bred at the École Polytechnique Fédérale de Lausanne (EPFL) animal facility for more than two generations before incorporation into the study. We examined 15 cohorts of 9 mouse inbred strains fed a CD or HFD, separated randomly into two groups of five mice of each sex for each diet. Strains were entered into the phenotyping program randomly and had staggered entry, typically by 1 or 2 weeks. HFD feeding started at 8 weeks of age. CD is Teklad Global 18% Protein Rodent Diet (18% kCal of fat, 24% kCal of protein, and 58% kCal of carbohydrates), and HFD is Teklad TD.06414 (60% kCal of fat, 20% kCal of protein, and 20% kCal of carbohydrates). All mice were housed under 12 hours of light alternated with 12 hours of dark, with ad libitum access to food and water at all times. Body weight was measured weekly from 8 weeks of age until killing. All research was approved by the Swiss cantonal veterinary authorities of Vaud under license 2257.2. In accordance with the ethical license’s protocol, some animals were sacrificed early due to displaying signs of severe illness. Most notably, all but one of the NZO/HILtJ mice under HFD were sacrificed due to severe diabetes. In addition, the oral glucose tolerance test (OGTT) was omitted in the NZO/HILtJ mice, due to concerns about the stress of repeated blood collection. A visual summary of the phenotyping program is also included in (Figure 1A).
Method details
Echo-MRI
Body composition was recorded by Echo-MRI at week 14. To do so, each mouse was placed briefly in an Echo-MRI (magnetic resonance imaging) machine (the 3-in-1, Echo Medical Systems), where lean and fat mass are recorded, along with total body weight, taking ∼1 min per individual. Lean mass is used as a corrective factor for respiratory calculations from the Comprehensive Lab Animal Monitoring System (CLAMS). All other tests are normalized to total body weight in our analyses.
Metabolic cages
At 14 weeks of age, after the MRI, the cohorts underwent their first phenotyping test: 48 hours of respiration measurements in individual metabolic cages (Oxymax/CLAMS, Columbus Instruments). The first 24 hours were considered adaptation, and the second 24 hours were used for data analysis, including analysis of movement, the volume of oxygen inhaled, the volume of carbon dioxide exhaled, and derived parameters of these two, such as the respiratory exchange ratio (RER).
Oral glucose tolerance test
At 16 weeks of age, all cohorts underwent an oral glucose tolerance test. Mice were fasted overnight before the test, and fasted glucose was tested with a glucometer at the tail vein. All individuals were then weighed and given an oral gavage of 20% glucose solution at 10 mL per kg of weight. Glucometer strips were used at 15, 30, 45, 60, 90, 120, 150, and 180 min after the gavage to examine glucose response over time. Blood was also collected at 0 (pregavage), 15, and 30 min to examine insulin levels.
Cold test
At 18 weeks of age, we performed a cold response test. The basal body temperatures of mice were examined rectally, after which mice were placed individually in prechilled cages in a room at 4°C. The cages were the standard housing cages but with only simple wood chip bedding. Body temperature was checked every hour for 6 hours, after which the mice were returned to their normal housing cage.
Basal activity recording
At 19 weeks of age, the mice were placed individually in regular housing cages for basal activity recording. The housing cages were then placed in laser detection grids developed by TSE Systems (Bad Homburg, Germany). Within the cages, woodchip bedding was retained. Food and water were as normal throughout the standard housing, both of which require rearing to reach. The detection grid has two layers: one for detecting horizontal (X-Y) movement (“ambulation”) the other for vertical (Z) movement (“rearing”). Mice were housed individually for the 48-hour experiment starting at about 10 a.m., with the night cycles (7 p.m. to 7 a.m., with 30 min of both dawn and dusk) used for movement calculations.
VO2max
Immediately after the 2-day basal activity recording, all mice performed a VO2max treadmill experiment using the Metabolic Modular Treadmill (Columbus Instruments). During the first 15 min in the machine for each individual, the treadmill was off while basal respiratory parameters were calculated. The last 2 minutes of data before the treadmill turned on are considered basal levels (most mice spend the first few minutes exploring the device). The treadmill then started at a pace of 4.8 m per minute (m/min), followed by a gradual increase over 60 s to 9 m/min, then 4 min at that pace before increasing to 12 m/min over 60 s, then four min at that pace before increasing to 15 m/min over 60 s, then 4 min at that pace, then the speed increased continuously by 0.015 m per second2 (or +0.9 m/min2) thereafter until the end of the experiment at 63.5 min, 1354.5 m, or when the mouse is exhausted. CD cohorts ran against a 10° incline, whereas HFD cohorts were set at 0°. For this test, all mice were taken out when exhausted or when physical constraints were too high. The distance, maximum VO2, and maximum RER were recorded. Maximums must be consistent across multiple measurements, and not single-measurement spikes, which were removed.
Sample collection
The sacrifices took place from 8:30 a.m. until 10:30 a.m., after an overnight fasting with isoflurane anesthesia followed by a complete blood draw (∼1 mL) from the vena cava, followed by perfusion with phosphate-buffered saline. Half of the blood was placed into lithium-heparin (LiHep)–coated tubes and the other half in EDTA-coated tubes; then both were shaken and stored on ice, followed immediately by collection of the liver. The LiHep blood taken for plasma analysis was also centrifuged at 4500 revolutions per minute (rpm) for 10 min at 4°C before being flash-frozen in liquid nitrogen. Whole blood taken for cellular analysis was processed immediately after the killing (i.e., after ∼1 to 2 hours on ice). All organs were weighted fresh after collection. Gallbladders were removed, and the livers were cut into small pieces before freezing in liquid nitrogen until preparation into mRNA, protein, or metabolite samples. Organs and blood serum were stored at −80°C until analysis.
Sample preparation
For mRNA, liver tissue were crushed in liquid nitrogen and then 10 mg of tissues were suspended in TRIzol (Invitrogen) and homogenized with stainless steel beads using a TissueLyser II (Qiagen) at 30 Hz for 2 min. RNA was extracted and purified using Direct-zol-96 RNA kits (Zymo Research). mRNA concentration was measured for all samples. All samples passed a quality check of purity (NanoDrop) and fragmentation (FragmentAnalyzer).
Mitochondrial activity measurements
Measurement of mitochondrial activity were performed by IntegraCell (formerly Metabiolab France). Whole tissue powder was lysed and mitochondrial activity of the different complexes was measured by a Roche/Hitachi-ModularP analyzer. Mitochondrial complex activity was normalized by citrate synthase activity when not stated otherwise.
Quantification and statistical analysis
RNA-sequencing and mapping
The STAR aligner (Dobin et al., 2013) was used for mapping the RNA-Seq data to the C57BL/6J reference genome and determining gene counts. We did not use distinct genomes for each strain due to various genome quality differences between mouse strains that could create bigger artefacts than mapping all strains on the same reference genome in terms of mapping efficiency and gene count estimation. All data analysis was performed using R version 3.5.2 and Rstudio Pro. Data formatting was perfomed using the data.table package.
Bioinformatic and statistical analysis
All the bioinformatics and statistical analysis were performed in R 3.5.2 and Rstudio Pro. Effect size was computed as follows: difference of the means divided by the global standard deviation. Correlation are based on Spearman’s correlation (except when stated otherwise). When needed, p-values were corrected for multiple testing with the Benjamin-Hochberg false discovery rate.
Differential gene expression was done using the limma R package and the voom method (Law et al., 2014). Gene set enrichment analysis (GSEA) was done using the GSEA method of the clusterprofiler R package (Yu et al., 2012). Genes were ranked according the signed –log10 (BH adjusted p-value) obtained by looking at the diet effect in female or male in each strain separately. We selected the genesets from the biological process of the gene ontology which had the highest significance levels in the overall response to the diet in each sex. Figures were generated using the ggplot2 (Villanueva and Chen, 2019, p. 2) and cowplot packages. For variance analysis, we used the type II sum of squares, which shows the variance explained by a parameter conditional on the presence of all other main effects in the model. The type II sum of squares preserve the principle of marginality and are independent of the listing order of the model parameters. variantPartition package was used to compute the variance explained by defined parameters for transcriptomic data.
Acknowledgments
We thank the members of J. Auwerx’s laboratory for their help with sacrifices and sample processing and the staff of EPFL’s Center of Phenogenomics (CPG) for help with phenotyping. We thank all members of J. Auwerx and K. Schoonjans laboratories for helpful discussions. We thank Metabiolab for their help in measuring the activity of mitochondrial complexes. Funding: this project received funding from the EPFL, the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC-AdG-787702), the Swiss National Science Foundation (SNSF 31003A_179435), and a GRL grant of the National Research Foundation of Korea (NRF 2017K1A1A2013124). LJEG was supported by the European Union’s Horizon 2020 research and innovation program through the Marie Skłodowska-Curie Individual Fellowship “AmyloAge” (grant agreement No. 896042) and GB was supported by the grant #2018-422 of the Strategic Focal Area “Personalized Health and Related Technologies (PHRT)” of the ETH Domain.
Author contributions
A.M.B., M.B.S., and J.A. conceived the project. A.M.B., S.R., G.B., T.I.L., S.L., and M.C. performed the experiments. J-D.M., A.M.B., and L.J.E.G. performed data analysis. G.E.A. worked on the online website. J.D.M., M.B.S., and J.A. supervised the study. J-D.M., A.M.B., and J.A. wrote the manuscript with comments from all authors.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104468.
Supplemental information
Data and code availability
-
•
Data: All metabolic traits, mitochondrial activity and transcriptome data collected in this study can be explored with an online, interactive interface at www.systems-genetics.org/CC_founders_metabolic. The website is separated into 5 parts (description of the experimental design, the metadata, the phenotypic traits, the transcriptomic data and the mitochondrial activity). In addition to the exploration of the raw data it is possible to see the trait-trait correlations as well as the effect of diet and sex on each strain separately. At the transcript level, individual genes and gene sets can be visualized as well as the effects of the diet and sex in any subset of strains. The raw transcriptomic data has been submitted to GEO expression database under the accession GEO:GSE182668. The individual phenotype data have been submitted to the mouse phenome database at https://phenome.jax.org/, where they will be available under the dataset identifier MPD:Auwerx2. For now, they are provided on Table S1.
-
•
Code: This study did not generate original code, and instead used standard R libraries described in the methods.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- Alhindi Y., Vaanholt L.M., Al-Tarrah M., Gray S.R., Speakman J.R., Hambly C., Alanazi B.S., Gabriel B.M., Lionikas A., Ratkevicius A. Low citrate synthase activity is associated with glucose intolerance and lipotoxicity. J. Nutr. Metab. 2019;2019:1–14. doi: 10.1155/2019/8594825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi A., Ueda K., Irie F., Sairenchi T., Iimura K., Watanabe H., Iso H., Ota H., Aonuma K. Relationship between cold temperature and cardiovascular mortality, with assessment of effect modification by individual characteristics. Circ. J. 2013;77:1854–1861. doi: 10.1253/circj.cj-12-0916. [DOI] [PubMed] [Google Scholar]
- Barrington W.T., Wulfridge P., Wells A.E., Rojas C.M., Howe S.Y.F., Perry A., Hua K., Pellizzon M.A., Hansen K.D., Voy B.H., et al. Improving metabolic health through precision dietetics in mice. Genetics. 2018;208:399–417. doi: 10.1534/genetics.117.300536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M. Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019;15:288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- Bournat J.C., Brown C.W. Mitochondrial dysfunction in obesity. Curr. Opin. Endocrinol. Diabetes Obes. 2010;17:446–452. doi: 10.1097/MED.0b013e32833c3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church R.J., Gatti D.M., Urban T.J., Long N., Yang X., Shi Q., Eaddy J.S., Mosedale M., Ballard S., Churchill G.A., et al. Sensitivity to hepatotoxicity due to epigallocatechin gallate is affected by genetic background in diversity outbred mice. Food Chem. Toxicol. 2015;76:19–26. doi: 10.1016/j.fct.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G.A., Gatti D.M., Munger S.C., Svenson K.L. The diversity outbred mouse population. Mamm. Genome. 2012;23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby M.J., Ross A.W., Walker A.W., Morgan P.J. Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Rep. 2017;21:1521–1533. doi: 10.1016/j.celrep.2017.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Conti A., Tryndyak V., Willett R.A., Borowa-Mazgaj B., Watson A., Patton R., Khare S., Muskhelishvili L., Olson G.R., Avigan M.I., et al. Characterization of the variability in the extent of nonalcoholic fatty liver induced by a high-fat diet in the genetically diverse Collaborative Cross mouse model. FASEB J. 2020;34:7773–7785. doi: 10.1096/fj.202000194R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornbos P., LaPres J.J. Incorporating population-level genetic variability within laboratory models in toxicology: from the individual to the population. Toxicology. 2018;395:1–8. doi: 10.1016/j.tox.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezquerra E.A., Vázquez J.M.C., Barrero A.A. Obesity, metabolic syndrome, and diabetes: cardiovascular implications and therapy. Rev. Esp. Cardiol. Engl. Ed. 2008;61:752–764. doi: 10.1016/S1885-5857(08)60212-1. [DOI] [PubMed] [Google Scholar]
- Gabriel B.M., Al-Tarrah M., Alhindi Y., Kilikevicius A., Venckunas T., Gray S.R., Lionikas A., Ratkevicius A. H55N polymorphism is associated with low citrate synthase activity which regulates lipid metabolism in mouse muscle cells. PLoS One. 2017;12:e0185789. doi: 10.1371/journal.pone.0185789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi M.O. Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 2018;6:223–236. doi: 10.1016/S2213-8587(17)30200-0. [DOI] [PubMed] [Google Scholar]
- Hirsch K.R., Smith-Ryan A.E., Blue M.N.M., Mock M.G., Trexler E.T., Ondrak K.S. Metabolic characterization of overweight and obese adults. Phys. Sportsmed. 2016;44:362–372. doi: 10.1080/00913847.2016.1248222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.R., Gagnon L.H., Longo-Guess C., Kane K.L. Association of a citrate synthase missense mutation with age-related hearing loss in A/J mice. Neurobiol. Aging. 2012;33:1720–1729. doi: 10.1016/j.neurobiolaging.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran S., Clee S.M. Genetics of metabolic syndrome: potential clues from wild-derived inbred mouse strains. Physiol. Genomics. 2018;50:35–51. doi: 10.1152/physiolgenomics.00059.2017. [DOI] [PubMed] [Google Scholar]
- Larsen S., Nielsen J., Hansen C.N., Nielsen L.B., Wibrand F., Stride N., Schroder H.D., Boushel R., Helge J.W., Dela F., Hey-Mogensen M. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 2012;590:3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaupin C., Vallée D., Hazari Y., Hetz C., Chevet E., Bailly-Maitre B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2018;69:927–947. doi: 10.1016/j.jhep.2018.06.008. [DOI] [PubMed] [Google Scholar]
- Lonardo A., Nascimbeni F., Ballestri S., Fairweather D., Win S., Than T.A., Abdelmalek M.F., Suzuki A. Sex differences in nonalcoholic fatty liver disease: state of the art and identification of research gaps. Hepatol. Baltim. Md. 2019;70:1457–1469. doi: 10.1002/hep.30626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.-F., Jin T., Xu Y., Zhang D., Wu Q., Zhang Y.-K.J., Liu J. Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol. Int. 2013;30:1135–1143. doi: 10.3109/07420528.2013.805762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz T.A., Woods S.C. Overview of animal models of obesity. Board S.J.E., editor. Curr. Protoc. Pharmacol. 2012;58:5–61. doi: 10.1002/0471141755.ph0561s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., De A., Chowdhury A. Epidemiology of non-alcoholic and alcoholic fatty liver diseases. Transl. Gastroenterol. Hepatol. 2020;5:16. doi: 10.21037/tgh.2019.09.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery M.K., Hallahan N.L., Brown S.H., Liu M., Mitchell T.W., Cooney G.J., Turner N. Mouse strain-dependent variation in obesity and glucose homeostasis in response to high-fat feeding. Diabetologia. 2013;56:1129–1139. doi: 10.1007/s00125-013-2846-8. [DOI] [PubMed] [Google Scholar]
- Müller M.J., Geisler C., Blundell J., Dulloo A., Schutz Y., Krawczak M., Bosy-Westphal A., Enderle J., Heymsfield S.B. The case of GWAS of obesity: does body weight control play by the rules? Int. J. Obes. 2018;42:1395–1405. doi: 10.1038/s41366-018-0081-6. [DOI] [PubMed] [Google Scholar]
- Pan W.W., Myers M.G. Leptin and the maintenance of elevated body weight. Nat. Rev. Neurosci. 2018;19:95–105. doi: 10.1038/nrn.2017.168. [DOI] [PubMed] [Google Scholar]
- Peirce J.L., Lu L., Gu J., Silver L.M., Williams R.W. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M., Nachman M.W. Insights into mammalian biology from the wild house mouse Mus musculus. Elife. 2015;4:e05959. doi: 10.7554/eLife.05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratkevicius A., Carroll A.M., Kilikevicius A., Venckunas T., McDermott K.T., Gray S.R., Wackerhage H., Lionikas A. H55N polymorphism as a likely cause of variation in citrate synthase activity of mouse skeletal muscle. Physiol. Genomics. 2010;42A:96–102. doi: 10.1152/physiolgenomics.00066.2010. [DOI] [PubMed] [Google Scholar]
- Recla J.M., Bubier J.A., Gatti D.M., Ryan J.L., Long K.H., Robledo R.F., Glidden N., Hou G., Churchill G.A., Maser R.S., et al. Genetic mapping in Diversity Outbred mice identifies a Trpa1 variant influencing late phase formalin response. bioRxiv. 2018 doi: 10.1101/362855. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers R.S. Translating mouse models: immune variation and efficacy testing. Toxicol. Pathol. 2017;45:134–145. doi: 10.1177/0192623316675767. [DOI] [PubMed] [Google Scholar]
- Sittig L.J., Carbonetto P., Engel K.A., Krauss K.S., Barrios-Camacho C.M., Palmer A.A. Genetic background limits generalizability of genotype-phenotype relationships. Neuron. 2016;91:1253–1259. doi: 10.1016/j.neuron.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.L., Soeters M.R., Wüst R.C.I., Houtkooper R.H. Metabolic flexibility as an adaptation to energy resources and requirements in health and disease. Endocr. Rev. 2018;39:489–517. doi: 10.1210/er.2017-00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth B. How recreational marathon runners hit the wall: a large-scale data analysis of late-race pacing collapse in the marathon. PLoS One. 2021;16:e0251513. doi: 10.1371/journal.pone.0251513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanzey E., O’Connor C., Reinholdt L.G. Mouse genetic reference populations: cellular platforms for integrative systems genetics. Trends Genet. 2021;37:251–265. doi: 10.1016/j.tig.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B.A., Wnek C., Kotlus B.S., Roemer N., MacTaggart T., Phillips S.J. Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm. Genome. 1999;10:335–348. doi: 10.1007/s003359900998. [DOI] [PubMed] [Google Scholar]
- Tripathi M., Yen P.M., Singh B.K. Estrogen-related receptor alpha: an under-appreciated potential target for the treatment of metabolic diseases. Int. J. Mol. Sci. 2020;21:E1645. doi: 10.3390/ijms21051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva R.A.M., Chen Z.J. ggplot2: elegant graphics for data analysis. Meas. Interdiscip. Res. Perspect. 2019;17:160–167. doi: 10.1080/15366367.2019.1565254. (2nd ed.) [DOI] [Google Scholar]
- Walport L.J., Hopkinson R.J., Vollmar M., Madden S.K., Gileadi C., Oppermann U., Schofield C.J., Johansson C. Human UTY (KDM6C) is a male-specific nε-methyl lysyl demethylase. J. Biol. Chem. 2014;289:18302–18313. doi: 10.1074/jbc.M114.555052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh C.E., Miller D.R., Manly K.F., Wang J., McMillan L., Morahan G., Mott R., Iraqi F.A., Threadgill D.W., de Villena F.P.-M. Status and access to the collaborative cross population. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2012;23:706–712. doi: 10.1007/s00335-012-9410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, n.d. WHO fact sheet: obesity and overweight [WWW Document]. URL https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed 2.11.22)
- Yang H., Wang J.R., Didion J.P., Buus R.J., Bell T.A., Welsh C.E., Bonhomme F., Yu A.H.-T., Nachman M.W., Pialek J., et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz E. In: Obesity and Lipotoxicity, Advances in Experimental Medicine and Biology. Engin A.B., Engin A., editors. Springer International Publishing; Cham: 2017. Endoplasmic reticulum stress and obesity; pp. 261–276. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A., Ben-Yacov O., Lador D., Avnit-Sagi T., Lotan-Pompan M., et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: All metabolic traits, mitochondrial activity and transcriptome data collected in this study can be explored with an online, interactive interface at www.systems-genetics.org/CC_founders_metabolic. The website is separated into 5 parts (description of the experimental design, the metadata, the phenotypic traits, the transcriptomic data and the mitochondrial activity). In addition to the exploration of the raw data it is possible to see the trait-trait correlations as well as the effect of diet and sex on each strain separately. At the transcript level, individual genes and gene sets can be visualized as well as the effects of the diet and sex in any subset of strains. The raw transcriptomic data has been submitted to GEO expression database under the accession GEO:GSE182668. The individual phenotype data have been submitted to the mouse phenome database at https://phenome.jax.org/, where they will be available under the dataset identifier MPD:Auwerx2. For now, they are provided on Table S1.
-
•
Code: This study did not generate original code, and instead used standard R libraries described in the methods.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.