Abstract
We used both cultivation and direct recovery of bacterial 16S rRNA gene (rDNA) sequences to investigate the structure of the bacterial community in anoxic rice paddy soil. Isolation and phenotypic characterization of 19 saccharolytic and cellulolytic strains are described in the accompanying paper (K.-J. Chin, D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen, Appl. Environ. Microbiol. 65:5042–5049, 1999). Here we describe the phylogenetic positions of these strains in relation to 57 environmental 16S rDNA clone sequences. Close matches between the two data sets were obtained for isolates from the culturable populations determined by the most-probable-number counting method to be large (3 × 107 to 2.5 × 108 cells per g [dry weight] of soil). This included matches with 16S rDNA similarity values greater than 98% within distinct lineages of the division Verrucomicrobia (strain PB90-1) and the Cytophaga-Flavobacterium-Bacteroides group (strains XB45 and PB90-2), as well as matches with similarity values greater than 95% within distinct lines of descent of clostridial cluster XIVa (strain XB90) and the family Bacillaceae (strain SB45). In addition, close matches with similarity values greater than 95% were obtained for cloned 16S rDNA sequences and bacteria (strains DR1/8 and RPec1) isolated from the same type of rice paddy soil during previous investigations. The correspondence between culture methods and direct recovery of environmental 16S rDNA suggests that the isolates obtained are representative geno- and phenotypes of predominant bacterial groups which account for 5 to 52% of the total cells in the anoxic rice paddy soil. Furthermore, our findings clearly indicate that a dual approach results in a more objective view of the structural and functional composition of a soil bacterial community than either cultivation or direct recovery of 16S rDNA sequences alone.
Rice paddy soils are one of the major sources of atmospheric methane (15). These flooded soils contain largely uncharacterized microbial populations which degrade polymeric organic matter to low-molecular-weight compounds, such as acetate, H2, and CO2. These compounds are finally converted to methane by methanogenic archaea (16). Several cultivation studies to characterize autochthonous bacterial inhabitants of rice paddy soils have been performed (18, 19, 34, 51). It is not clear how successful such studies have been in characterizing bacterial communities. It has been estimated, for example, that the portion of microbial diversity which has been obtained in pure culture by conventional cultivation techniques amounts to only 0.1 to 1% of the total diversity (1, 12). Similarly, attempts to cultivate the bacteria in soils typically yield culturable cell numbers that are less than 5% (usually less than 1%) of the total microscopically countable cell numbers (3, 29). These observations indicate the inadequacy of culture methods to characterize complex microbial communities. One reason for this inadequacy is that the growth conditions used for direct enrichment cultures (i.e., cultures grown with undiluted inoculum) often do not reflect the natural conditions in the environment examined and thus select for fast-growing bacteria which are best adapted to the growth medium (16, 29, 49, 50). Consequently, the microorganisms obtained in pure culture may not have numerical or ecological significance for the biogeochemical processes in the environment, and abundant or even dominant populations may not be detected. Underestimation of microbial diversity by culture methods has favored the use of cultivation-independent, molecular approaches to investigate the microbial diversity in natural environments. The studies which have been performed have focused mainly on retrieval and comparative analysis of 16S rRNA-encoding gene (rDNA) sequences. The results not only have dramatically changed our view of the evolution and phylogenetic diversity of prokaryotes but also have confirmed the immense underrepresentation of naturally occurring bacteria and archaea obtained so far in pure culture (4, 11, 17, 23, 24, 29, 32).
It has, however, become clear that the cultivation-independent PCR-based 16S rDNA approach is also characterized by limitations and pitfalls (29). The steps used can lead to biases in the proportions of the 16S rDNA sequence types recovered. Consequently, this approach is more qualitative than quantitative. In addition, inferring an ecological role or even a defined phenotype on the basis of the phylogenetic position of an environmental 16S rDNA sequence is often not possible.
In an attempt to address the problems mentioned above, we applied to the same sample of rice paddy soil a culture method in which we used serial liquid dilutions of inoculum and a technique to recover environmental 16S rDNA. Cultivation was carried out by using the anaerobic most-probable-number (MPN) counting format. Using this dual approach, a previous study was able to gain insight into the structural and functional composition of the methanogenic community in anoxic rice paddy soil (16). In the present study we focused on the bacterial community, which was probably more diverse. Isolation of 19 saccharolytic and cellulolytic bacteria obtained from terminal and subterminal positive dilution steps of the MPN counting procedure and phenotypic characterization of these bacteria are described in the accompanying paper (8). Below, these bacteria are referred to as MPN isolates. Here we describe the results of a comparative phylogenetic analysis of two different 16S rDNA data sets generated during this investigation; i.e., sequence data for the 19 MPN isolates were compared to 57 environmental 16S rDNA clone sequences. Close matches between the two data sets revealed that the isolates were representatives of predominant bacterial groups, and thus our findings resulted in a more objective view of the structural and functional composition of the bacterial community inhabiting the anoxic rice paddy soil which we studied.
MATERIALS AND METHODS
Source of rice paddy soil.
Rice (Oryza sativa var. Roma, type japonica) was grown and samples of anoxic paddy soil were obtained as described in the accompanying paper (8). The bulk soil which was obtained was suspended at a weight ratio of 1:1 by using deionized water which had been degassed with a vacuum pump, and the preparation was mixed well. The suspension was subsequently divided into two parts. One part was used to enumerate bacteria by the MPN count method and then to isolate 19 strains in pure culture by using the terminal and subterminal positive dilution steps (as described by Chin et al. [8]), and the other part was used as the starting material for direct recovery of environmental 16S rDNA sequences.
DNA extraction.
Genomic DNA was extracted from pure cultures as described previously (16). Total community DNA was extracted from the bulk soil by using a protocol which included steps used in two previously described direct lysis procedures (35, 42). Briefly, 1 g of fresh material was mixed with 750 μl of sodium phosphate buffer (0.12 M, pH 8.0). An equal volume of glass beads (diameter, 0.1 mm) was added to the soil-buffer suspension, and the mixture was shaken three times for 80 s at the maximum speed with a bead beater (Mini-Bead-Beater; Biospec Products, Bartlesville, Okla.), followed each time by cooling on ice for 15 s. After three cycles of freezing and thawing (freezing at −78°C for 2 min, heating at 60°C for 2 min), 5 mg of lysozyme was added to the sample, and the suspension was incubated with intermittent mixing at 37°C for 1 h. Subsequently, sodium dodecyl sulfate (10%, wt/vol) was added to a final concentration of 2%, and this was followed by incubation for 10 min at 60°C. After 10 min of centrifugation at 13,000 × g, the supernatant was transferred to a new 2-ml tube. The pellet was suspended in 250 μl of sodium phosphate buffer and centrifuged for 10 min at 13,000 × g. The two supernatants were combined and extracted once with cold phenol-chloroform (1:1, vol/vol) and twice with chloroform-isoamyl alcohol (24:1, vol/vol). The DNA was precipitated from the aqueous phase with 0.5 volume of 7 M ammonium acetate and 1.5 volume of isopropanol. The resulting pellet was washed with 70% (vol/vol) ethanol, and after the DNA was dried, it was resuspended in 100 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0). For further purification of the DNA, 0.1 g of cesium chloride was added, and the preparation was mixed well and incubated for 3 h at room temperature. After centrifugation at 13,000 × g for 20 min, the supernatant was transferred to a new tube and purified further by two rounds of precipitation with equal volumes of isopropanol. Finally, the DNA was resuspended in 100 μl of TE buffer. The amount of DNA extracted was estimated by electrophoresing 5-μl aliquots on a 0.8% agarose gel and comparing the results to the results obtained with a HindIII digest of λ DNA. The gel was stained with ethidium bromide.
PCR amplification of bacterial 16S rDNA.
Oligonucleotide primers 27f and 1492r (27) were used to amplify 16S rDNA from both pure cultures and total community DNA. This primer system targets the 16S rDNA of a wide range of members of the domain Bacteria at positions 9 through 27 (primer 27f) and positions 1492 through 1512 (primer 1492r) (Escherichia coli 16S rRNA numbering [6]). For pure cultures, PCR amplification of 16S rDNA followed by purification of the resulting products for nonradioactive sequencing was performed as described previously (13).
16S rDNA was amplified from total community DNA in a reaction cocktail containing 1 μl (approximately 0.3 ng) of template DNA, 10 μl of PCR buffer (0.1 M Tris-HCl [pH 8.3], 0.5 M KCl, 1.5 mM MgCl2, 1 mg of bovine serum albumin per ml, 0.5% Tween 20), 20 nmol of each deoxynucleotide triphosphate (USB, Cleveland, Ohio), 20 pmol of each primer, and 2.5 U of Taq DNA polymerase (AmpliTaq DNA polymerase; Perkin-Elmer, Foster City, Calif.). The thermal profile used for amplification of the 16S rDNA sequences was as follows: 5 min at 94°C, after which the DNA polymerase was added; 29 cycles consisting of primer annealing at 48°C for 90 s, DNA elongation at 72°C for 120 s, and denaturation at 94°C for 60 s; and a final cycle consisting of 48°C for 90 s and 72°C for 6 min. Amplification was performed by using 100 μl (total volume) in 0.2-ml reaction tubes and a DNA thermal cycler (model 9600; PE Applied Biosystems, Foster City, Calif.). Aliquots (10 μl) of the 16S rDNA amplicons were visualized by electrophoresis on a 1% agarose gel after staining with ethidium bromide.
Cloning and sequencing.
The 16S rDNA PCR products obtained from the anoxic rice paddy soil were cloned by using a TA cloning kit (Invitrogen, San Diego, Calif.) as recommended by the manufacturer. To prepare randomly selected clones for sequence analysis (i.e., extraction of phagemid DNA, PCR-mediated amplification of cloned inserts, and purification of the PCR products), we basically used a previously described procedure (39). Nonradioactive sequencing of both environmental 16S rDNA clones and PCR products obtained from pure cultures was carried out by using sequencing primers described previously (27) and a PRISM ready reaction dye terminator cycle sequencing kit according to the instructions of the manufacturer (PE Applied Biosystems). The reaction mixtures were analyzed with an automatic DNA sequencer (model 373A; PE Applied Biosystems).
Phylogenetic analysis.
The 16S rDNA sequences obtained from 19 MPN isolates (8) and 57 environmental clones were added to a database consisting of about 6,000 publicly available bacterial 16S rRNA sequences (33, 37). This database is part of the ARB program package (45). The 16S rDNA sequences were integrated into the database with the automatic alignment tool of the ARB program package. The resulting alignments were manually checked and corrected if necessary. Highly variable regions of the 16S rDNA and sequence positions with possible alignment errors were excluded from the phylogenetic analysis by using only alignment positions for tree reconstruction which contained identical nucleotides in at least 50% of the 16S rDNA sequences compared (13, 14). Trees were generated by performing distance matrix analyses (evolutionary distances were calculated by using the Jukes-Cantor equation [26], and trees were constructed by using a neighbor-joining algorithm [40]). The statistical significance of interior nodes was tested by performing bootstrap analyses by the neighbor-joining method (ARB; 1,000 data resamplings). To exclude obvious chimeric 16S rDNA primary structures prior to the phylogenetic analysis (30), the “check_chimera” program of the Ribosomal Database Project (33) was used, and a separate treeing analysis of the terminal 400 nucleotide sequence positions at the 5′ and 3′ ends of the environmental 16S rDNA clones was carried out. Overall levels of similarity between 16S rDNA sequences were determined by using the appropriate tool of the ARB program package.
Nucleotide sequence accession numbers.
The 16S rDNA sequences of 57 environmental clones (clones BSV03 to BSV90) and of 19 pure cultures isolated and phenotypically characterized in a parallel study (8) have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AJ229177 through AJ229233 and AJ229234 through AJ229252.
RESULTS
DNA extraction.
Cells were lysed directly in the soil matrix by using a combined approach which included bead beating, freeze-thawing, and incubation with lysozyme-sodium dodecyl sulfate. Purification of total community DNA by extraction once with cold phenol and twice with chloroform-isoamyl alcohol followed by a cesium chloride precipitation step resulted in a brown DNA preparation which could not be used for PCR-mediated amplification of bacterial 16S rDNA, probably due to the presence of inhibitory substances like humic acids. However, DNA could be amplified without further purification after more dilute aliquots (0.3 ng) of the total community DNA were added to individual PCR mixtures. The amount of total community DNA extracted was estimated to be approximately 3 μg per g (dry weight) of rice paddy soil.
Phylogenetic placement of environmental sequences in relation to bacterial isolates.
A total of 62 environmental 16S rDNA sequences, designated BSV (bulk soil Vercelli) clones, were randomly selected and used for further analysis. The primary structure was determined for at least 1,000 nucleotide sequence positions, and most sequences were nearly full-length sequences. None of the 62 BSV clones was identical to any of the bacterial 16S rDNA sequences available in public databases. A search for chimeric structures did not reveal any ambiguous clone sequences with significantly different phylogenetic assignments based on the separately analyzed terminal 400-bp stretches at the 5′ and 3′ ends.
Our phylogenetic analyses placed 57 clone sequences in the following six major groups of the domain Bacteria: (i) the division Verrucomicrobia (21) (2 clones); (ii) the Cytophaga-Flavobacterium-Bacteroides (CFB) group (3 clones); (iii) the low-G+C-content gram-positive bacteria, which included Clostridium-like sequences (32 clones) and Bacillus-like sequences (13 clones); (iv) the class Actinobacteria (44) (1 clone); (v) the family Chlorobiaceae (3 clones); and (vi) the α subdivision of the division Proteobacteria (3 clones). Detailed information concerning these environmental sequences is given in Table 1. The remaining five clone sequences grouped within the phylogenetic radiation of the family Geobacteraceae (31) in the δ subdivision of the division Proteobacteria. Detailed phylogenetic characterization of these five clones will be given when a study of the diversity and activity of dissimilatory Fe(III)-reducing bacteria in flooded rice microcosms is described (22).
TABLE 1.
Nucleotide sequence positions determined for 57 environmental 16S rDNA clones, the cultured organisms most closely related to the clones, and the EMBL, GenBank, and DDBJ accession numbers of the nucleotide sequences of the clones
| Clone | Region sequenced (total no. of bases)a | Closest phylogenetic relative(s)
|
Accession no. | |
|---|---|---|---|---|
| Taxon | % Similarityb | |||
| BSV64c | 28-1476 (1465) | MPN isolates PB90-1, PB90-3, ACB90 | 98.6 | AJ229212 |
| BSV69c | 28-1476 (1465) | MPN isolates PB90-1, PB90-3, ACB90 | 98.6 | AJ229213 |
| BSV13 | 28-1431 (1392) | MPN isolate XB45 | 88.4 | AJ229182 |
| BSV73 | 38-1491 (1445) | MPN isolate XB45 | 88.2 | AJ229217 |
| BSV85c | 28-1491 (1438) | MPN isolate XB45 | 99.9 | AJ229229 |
| BSV04c | 28-1348 (1317) | “Bacillus pseudomegaterium”d | 98.0 | AJ229178 |
| BSV05c | 28-1491 (1471) | “Bacillus pseudomegaterium”d | 97.5 | AJ229179 |
| BSV06c | 28-1340 (1321) | “Bacillus pseudomegaterium”d | 97.6 | AJ229180 |
| BSV39c | 28-1200 (1181) | “Bacillus pseudomegaterium”d | 97.7 | AJ229195 |
| BSV45c | 28-1376 (1357) | “Bacillus pseudomegaterium”d | 97.9 | AJ229200 |
| BSV46c | 28-1474 (1455) | “Bacillus pseudomegaterium”d | 96.7 | AJ229201 |
| BSV55c | 28-1363 (1344) | “Bacillus pseudomegaterium”d | 96.5 | AJ229207 |
| BSV71c | 28-1384 (1353) | “Bacillus pseudomegaterium”d | 97.3 | AJ229215 |
| BSV77c | 45-1276 (1240) | “Bacillus pseudomegaterium”d | 97.7 | AJ229221 |
| BSV84c | 28-1339 (1320) | “Bacillus pseudomegaterium”d | 97.6 | AJ229228 |
| BSV44 | 31-1491 (1466) | Paenibacillus gordonae | 93.3 | AJ229199 |
| BSV52 | 28-1393 (1369) | Paenibacillus gordonae | 93.0 | AJ229204 |
| BSV79 | 28-1457 (1443) | Paenibacillus alvei | 94.5 | AJ229223 |
| BSV51 | 28-1491 (1456) | MPN isolate XB90 | 96.1 | AJ229203 |
| BSV61c | 28-1491 (1448) | MPN isolate XB90 | 95.3 | AJ229210 |
| BSV70c | 28-1491 (1446) | MPN isolate XB90 | 96.8 | AJ229214 |
| BSV20c | 28-1473 (1407) | Strain RPec1 | 96.1 | AJ229185 |
| BSV21c | 28-1450 (1383) | Strain RPec1 | 96.6 | AJ229186 |
| BSV41c | 28-1367 (1300) | Strain RPec1 | 96.2 | AJ229197 |
| BSV74c | 28-1350 (1283) | Strain RPec1 | 96.4 | AJ229218 |
| BSV80 | 28-1361 (1295) | Clostridium pascui | 93.4 | AJ229224 |
| BSV86 | 28-1491 (1424) | Clostridium pascui | 93.7 | AJ229230 |
| BSV62 | 28-1348 (1291) | Clostridium aurantibutyricum | 98.8 | AJ229211 |
| BSV82 | 28-1411 (1354) | Clostridium aurantibutyricum | 98.8 | AJ229226 |
| BSV07 | 28-1368 (1308) | Oxobacter pfennigii | 92.2 | AJ229181 |
| BSV50 | 28-1491 (1440) | Oxobacter pfennigii | 89.4 | AJ229202 |
| BSV78 | 28-1365 (1307) | Oxobacter pfennigii | 91.2 | AJ229222 |
| BSV34 | 28-1452 (1398) | Thermobrachium celere | 90.1 | AJ229194 |
| BSV54 | 28-1450 (1390) | Thermobrachium celere | 90.4 | AJ229206 |
| BSV83 | 28-1051 (998) | Thermobrachium celere | 90.0 | AJ229227 |
| BSV87 | 28-1375 (1328) | MPN isolate FCB90-3 | 92.0 | AJ229231 |
| BSV72 | 28-1475 (1418) | Acetivibrio cellulolyticus | 92.8 | AJ229216 |
| BSV75 | 28-1321 (1269) | Acetivibrio cellulolyticus | 90.6 | AJ229219 |
| BSV89 | 31-1249 (1190) | Clostridium termitidis | 92.9 | AJ229232 |
| BSV16c | 28-1491 (1478) | Strain DR1/8 | 95.7 | AJ229183 |
| BSV90 | 28-1462 (1451) | MPN isolate SB90 | 91.6 | AJ229233 |
| BSV43 | 28-1366 (1348) | Clostridium quercicolum | 86.9 | AJ229198 |
| BSV24 | 28-1439 (1405) | Moorella thermoautotrophica | 86.8 | AJ229187 |
| BSV27 | 28-1473 (1408) | “Clostridium aminobutyricum” | 94.6 | AJ229189 |
| BSV76 | 298-1491 (1168) | Clostridium lituseburense | 98.2 | AJ229220 |
| BSV29 | 35-1453 (1401) | MPN isolate FCB45 | 88.6 | AJ229191 |
| BSV30 | 28-1475 (1431) | MPN isolate FCB45 | 88.6 | AJ229192 |
| BSV31 | 28-1456 (1412) | MPN isolate FCB45 | 88.5 | AJ229193 |
| BSV28 | 28-1474 (1420) | Caloramator indicus | 85.1 | AJ229190 |
| BSV81 | 28-1375 (1309) | Caloramator indicus | 84.2 | AJ229225 |
| BSV58 | 41-1375 (1321) | Streptomyces albus | 96.4 | AJ229209 |
| BSV03 | 28-1453 (1366) | Devosia riboflavina | 94.6 | AJ229177 |
| BSV53 | 28-1409 (1327) | Methylosinus sporium | 96.4 | AJ229205 |
| BSV56 | 28-1473 (1389) | Beijerinckia indica | 95.6 | AJ229208 |
| BSV19 | 28-1441 (1404) | Chlorobium limicola | 79.5 | AJ229184 |
| BSV26 | 28-1369 (1331) | Chlorobium limicola | 79.2 | AJ229188 |
| BSV40 | 28-1472 (1438) | Chlorobium limicola | 80.6 | AJ229196 |
E. coli 16S rRNA numbering (6).
Overall level of 16S rDNA sequence similarity, as determined by using the appropriate tool of the ARB software package (45).
Environmental BSV clone that exhibits more than 95% overall 16S rDNA sequence similarity to strains isolated either in a parallel study (MPN isolates) (8) or in previous studies (9, 38).
Clones assigned to “Bacillus pseudomegaterium” are also related to MPN isolate SB45 with levels of 16S rDNA similarity of more than 95%.
Below we describe the results of the comparative phylogenetic analysis of the two 16S rDNA sequence data sets generated in this study. The first data set included data for the environmental 16S rDNA clones which were recovered directly from the rice paddy soil, and the second data set included data for the nearly full-length sequences obtained for the 19 MPN isolates (8) plus 16S rDNA sequence data for strains isolated from the same habitat in previous studies. The latter strains were referred to as strains that were isolated from Vercelli rice paddy soil.
(i) Division Verrucomicrobia.
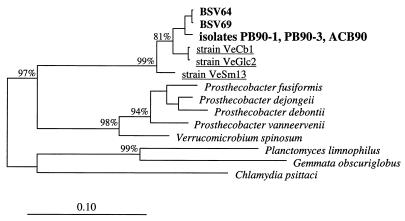
Clones BSV64 and BSV69 were closely affiliated with MPN isolates PB90-1, PB90-3, and ACB90. The latter three strains had identical 16S rDNA sequences. Strains PB90-1 and PB90-3 were isolated from the terminal (PB90-1) and subterminal (PB90-3) positive dilution steps of the same MPN counting experiment performed with pectin, while strain ACB90 was obtained from the terminal positive dilution step of an MPN counting experiment performed with amorphous cellulose as the growth substrate (8). The level of 16S rDNA similarity between the two BSV clones and the three MPN isolates was more than 98% (Table 1 and Fig. 1). This tight cluster belonged to the recently proposed subdivision 4 (24) of the division Verrucomicrobia (21). This cluster had 16S rDNA dissimilarity values ranging from 19 to 22% and was clearly distinct from the validly named members of the division Verrucomicrobia (24, 52) (i.e., Verrucomicrobium spinosum [41] and the fusiform caulobacter-like Prosthecobacter spp. [20, 21], which were assigned to subdivision 1 of the Verrucomicrobia [24]).
FIG. 1.
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones BSV64 and BSV69 to MPN isolates PB90-1, PB90-3, and ACB90 (8) and to representative members of the division Verrucomicrobia (21), the order Planctomycetales, and Chlamydia psittaci. Strains VeCb1, VeGlc2, and VeSm13 (underlined) were isolated from Vercelli rice paddy soil in a previous study (25). Based on a 50% invariance criterion for 51 sequences from members of the Verrucomicrobia, Planctomycetales, and the genus Chlamydia, 1,192 nucleotide sequence positions (E. coli 16S rRNA positions 31 to 1476) were used for tree construction. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The scale bar represents a 10% estimated difference in nucleotide sequences.
So far, subdivision 4 environmental 16S rDNA clones have been detected only in peat bog soil (36) and in rice paddy soil (25). Strains VeCb1, VeGlc2, and VeSm13, which were isolated from Vercelli rice paddy soil and were characterized as anaerobic ultramicrobacteria (25), are closely affiliated with MPN isolates PB90-1, PB90-3, and ACB90 (Fig. 1). Assignment of environmental clone PAD7 (33), which was recovered from rice paddy soil in Japan, to this distinct Verrucomicrobia lineage indicated that the members of this lineage are widely distributed in anoxic rice paddy soil (25) (clone PAD7 is not shown in Fig. 1 due to the rather short sequence consisting of 276 nucleotides which was determined).
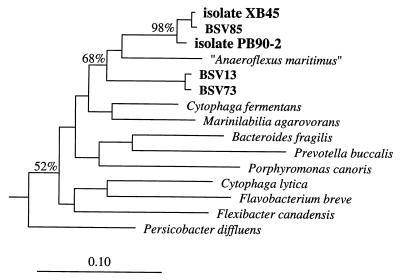
(ii) CFB group.
The closest affiliation between one of the environmental clone sequences and one of the MPN isolates was the affiliation between clone BSV85 and strain XB45; there were only four nucleotide differences between the two 16S rDNA sequences, which corresponded to positions 29 through 1491 of the E. coli 16S rRNA (Table 1 and Fig. 2). Together with MPN isolate PB90-2, strain XB45 and clone BSV85 form a distinct branch which shares a common interior node with BSV13 and BSV73, as well as the tentatively named organism “Anaeroflexus maritimus” (33) in the CFB group. Members of the genus Marinilabilia (e.g., Marinilabilia agarovorans) and Cytophaga fermentans are the cultured organisms that are most closely related to MPN isolates XB45 and PB90-2 (Fig. 2); the levels of 16S rDNA sequence similarity between the new isolates and the previously described species are 87 to 90%.
FIG. 2.
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolates PB90-2 and XB45 (8) and to representative members of the CFB group. Based on a 50% invariance criterion for 176 sequences from members of the CFB group, 1,190 nucleotide sequence positions (E. coli 16S rRNA positions 110 to 1431) were considered for tree construction. The percentages are the significance values for the interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of E. coli as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
(iii) Low-G+C-content gram-positive bacteria.
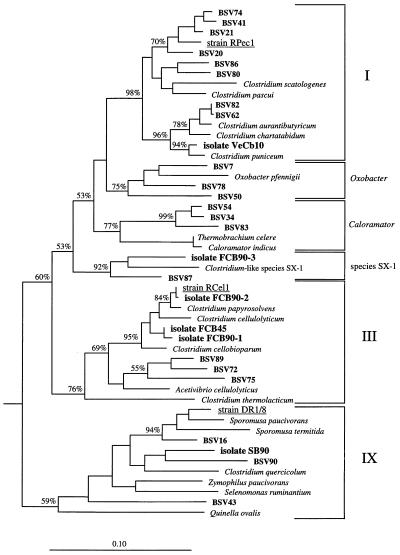
Most of the BSV clones that were randomly selected and used for our analysis grouped in the phylogenetic radiation of the low-G+C-content gram-positive bacteria. Most of these clones could be assigned either to one of the clostridial clusters (clostridial clusters I through XIVa) which have been proposed by Collins et al. (10) (Fig. 3 and 4) or to the family Bacillaceae (Fig. 5).
FIG. 3.
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolate XB90 (8) and to representative members of clostridial cluster XIVa of Collins et al. (10). Based on a 50% invariance criterion for 326 Clostridium-like sequences, 1,238 nucleotide sequence positions (E. coli 16S rRNA positions 31 to 1386) were considered for tree construction. The percentage is the significance value for an interior node, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Clostridium fallax as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 4.
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolates FCB45, FCB90-1, FCB90-2, FCB90-3, SB90, and VeCb10 (8) and to representatives of clostridial clusters I, III, and IX of Collins et al. (10), as well as related groups. Strains RPec1, RCel1, and DR1/8 (underlined) were isolated from Vercelli rice paddy soil in previous studies (9, 38). Based on a 50% invariance criterion for 357 Clostridium-like sequences, 1,040 nucleotide sequence positions (E. coli 16S rRNA positions 100 to 1249) were considered for tree construction. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Fervidobacterium islandicum as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
FIG. 5.
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clones (BSV clones) to MPN isolate SB45 (8) and to representative members of the genera Bacillus and Paenibacillus. Based on a 50% invariance criterion for 462 sequences from members of the Bacillaceae, 1,146 nucleotide sequence positions (E. coli 16S rRNA positions 101 to 1274) were considered for tree construction. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Clostridium fallax as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
(a) Clostridium-like sequences.
MPN isolate XB90 and three clones, clones BSV51, BSV61, and BSV70, formed a distinct branch within clostridial cluster XIVa (Fig. 3). The close affiliation between strain XB90 and the three BSV clones was revealed by 16S rDNA sequence similarity values greater than 95% (Table 1).
MPN isolate VeCb10 and eight environmental clone sequences were related to members of clostridial cluster I (Fig. 4). Strain VeCb10 grouped very closely with Clostridium puniceum (level of 16S rDNA similarity, 99.1%) but showed no clear affiliation with any of the BSV clones. BSV20, BSV21, BSV41, and BSV74 formed a distinct group that was not affiliated with any of the previously described cluster I clostridia. However, they were closely related to strain RPec1, an isolate obtained by direct enrichment culturing from Vercelli rice paddy soil (9) (Fig. 4). The levels of 16S rDNA similarity between strain RPec1 and these four BSV clones ranged from 96 to 97% (Table 1). The almost identical sequences of clones BSV62 and BSV82 were closely affiliated with Clostridium aurantibutyricum (level of 16S rDNA similarity, 98.8%).
BSV7, BSV50, and BSV78 formed a branch with Oxobacter pfennigii, while BSV34, BSV54, and BSV83 grouped with Thermobrachium celere and Caloramator indicus. However, the BSV clones were only distantly related to the cultured organisms (Table 1 and Fig. 4).
MPN isolate FCB90-3 and clone BSV87 exhibited a moderately distant relationship and belonged to a clostridial lineage characterized so far only by the Clostridium-like species SX-1 and SX-2 (28) (Fig. 4).
MPN isolates FCB45, FCB90-1, and FCB90-2, as well as three clones (BSV72, BSV75, and BSV89), grouped in clostridial cluster III. The 16S rDNA sequence of MPN isolate FCB90-2 was almost identical to the 16S rDNA sequence of strain RCel1, an isolate obtained by direct enrichment culturing from Vercelli rice paddy soil (9). These two strains are closely related to Clostridium papyrosolvens (level of 16S rDNA similarity, 99.1%). Closely affiliated MPN isolates FCB45 and FCB90-1 could not be assigned to any of the previously described members of cluster III but belonged to the same group as Clostridium cellobioparum and Clostridium cellulolyticum, as well as MPN isolate FCB90-2 and C. papyrosolvens. Clones BSV72, BSV75, and BSV89 formed another distinct branch in clostridial cluster III, which was clearly separated from the MPN isolates (Fig. 4). In addition, the almost identical clones BSV29, BSV30, and BSV31 were affiliated with clostridial cluster III but formed a deeply rooted branch that was clearly separated from the previously described members of this cluster (the latter group is not shown in the tree).
MPN isolate SB90 and two clones (BSV16 and BSV90) belonged to clostridial cluster IX (gram-positive bacteria with a gram-negative cell wall structure). The 16S rDNA sequence of strain SB90 was most similar to the 16S rDNA sequence of Zymophilus paucivorans (level of similarity, 91.9%), but strain SB90 grouped in the tree next to Clostridium quercicolum and clone BSV90 (Fig. 4). Clone BSV16 exhibited a closer affiliation with Sporomusa paucivorans and especially strain DR1/8 (level of 16S rDNA similarity, 95.7%). Strain DR1/8 was isolated from the terminal positive dilution step of an MPN counting experiment performed to determine the level of homoacetogenic bacteria in Vercelli rice paddy soil (38). The level of strain DR1/8-like organisms was calculated to be about 108 cells per g (dry weight) of soil. Clone BSV43 (Fig. 4) could also be assigned to clostridial cluster IX but exhibited no clear affiliation with any of the previously described clostridial species.
Three other environmental clones could be assigned to one of the clostridial clusters (Table 1). Clones BSV27 and BSV76 were identified as members of clostridial cluster XI, and “Clostridium aminobutyricum” and Clostridium lituseburense, respectively, were the most closely related cultured relatives. Clone BSV24 grouped next to the thermophilic and homoacetogenic representatives of the genus Moorella but was clearly distinct from these organisms (levels of 16S rDNA similarity, 85 to 86%).
(b) Bacillus-like sequences.
Ten BSV clones were closely related to “Bacillus pseudomegaterium” (levels of 16S rDNA similarity, 96.5 to 98.0%) and also were affiliated with MPN isolate SB45 (levels of similarity, 95.8 to 96.6%). Three other clones (BSV44, BSV52, and BSV79) grouped within the phylogenetic radiation of the genus Paenibacillus (Table 1 and Fig. 5).
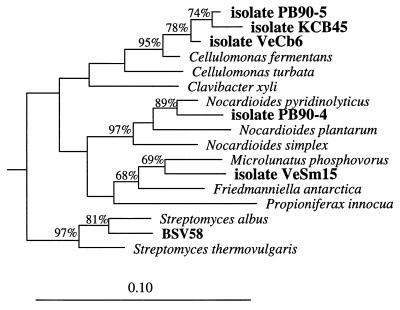
(iv) Class Actinobacteria.
Five MPN isolates were high-G+C-content gram-positive bacteria, but only one of the environmental 16S rDNA clones could be assigned to this group (Fig. 6). MPN isolates KCB45, PB90-5, and VeCb6 formed a tight cluster that was most closely related to Cellulomonas fermentans, and the levels of 16S rDNA similarity were 97.7, 97.1, and 96.6%, respectively. MPN isolate VeSm15 grouped with Microlunatus phosphovorus (level of 16S rDNA similarity, 94.6%). MPN isolate PB90-4 was most closely affiliated with Nocardioides pyridinolyticus (level of 16S rDNA similarity, 97.5%). Clone BSV58 was not affiliated with any of these MPN isolates but grouped next to Streptomyces albus.
FIG. 6.
16S rDNA-based dendrogram showing the phylogenetic relationship of environmental clone BSV58 to MPN isolates KCB45, PB90-4, PB90-5, VeCb6, and VeSm15 (8) and to representative members of the class Actinobacteria (44). Based on a 50% invariance criterion for 859 sequences from members of the class Actinobacteria, 1,296 nucleotide sequence positions (E. coli 16S rRNA positions 41 to 1375) were considered for tree construction (except for isolate KCB45). Isolate KCB45 was inserted into the tree based on a continuous sequence stretch corresponding to E. coli 16S rRNA positions 41 to 1060. The percentages are the significance values for interior nodes, as derived from a bootstrap test in which 1,000 data resamplings were used. The root was determined by using the 16S rDNA sequence of Clostridium fallax as the outgroup reference sequence. The scale bar represents a 10% estimated difference in nucleotide sequences.
(v) Family Chlorobiaceae.
Of all of the BSV clones analyzed in this study, clones BSV19, BSV26, and BSV40 exhibited the lowest levels of 16S rDNA similarity (between 79 and 81%) to the most closely related cultured relatives, which were representatives of the family Chlorobiaceae, such as Chlorobium limicola (Table 1) (data not shown). The closely related clones BSV19 and BSV26 and clone BSV40 formed two individual lines of descent which differed by 16S rDNA dissimilarity values of between 19 and 20%. No pure cultures belonging to this group were isolated in the parallel cultivation study (8).
(vi) Alpha subdivision of the division Proteobacteria.
MPN isolate KCB90 and three clones (BSV03, BSV53, and BSV56) grouped in the α subdivision of the division Proteobacteria (Table 1) (data not shown). Strain KCB90 exhibited a moderate level of affiliation with clone BSV56 and Rhodopseudomonas acidophila (levels of 16S rDNA similarity, 92.7 and 91%, respectively). However, clone BSV56 itself was most closely related to the nitrogen-fixing species Beijerinckia indica. Clones BSV03 and BSV53 were affiliated with Devosia riboflavina and Methylosinus sporium, respectively.
DISCUSSION
Cultivation-independent studies carried out during the past decade clearly revealed the vastness of microbial diversity which is not represented by pure cultures (11, 17, 23, 24) and confirmed the view that only a minor portion of the naturally occurring microbial diversity has been obtained in pure culture (1). Only a few studies have been conducted to compare the microbial diversity detectable by cultivation with the microbial diversity detectable by direct recovery of 16S rDNA in a defined environment (7, 46, 49, 50). Chandler et al. (7) found, for a subsurface sediment core, close correlations at the genus level between the culturable portion of aerobic heterotrophic bacteria and the data obtained from a 16S rDNA approach. However, these correlations were detected after aerobic treatment of various sediment samples for 1 to 21 weeks at the in situ temperature (17°C) but not with the untreated soil core. The treatments might have caused a selective shift towards enrichment of defined bacterial groups in the samples analyzed compared to the original sediment core. Studies of hot spring microbial mats that were probably less complex resulted in several close matches between 16S rDNA of organisms obtained by culture methods and directly recovered 16S rDNA, but only after serial liquid dilutions of the inoculum were used for cultivation instead of direct enrichment based on undiluted inoculum (49, 50). The major conclusions of the investigations of microbial mat communities were as follows: (i) for the most part direct enrichment techniques select for populations which are more fit under the enrichment conditions used and may not be numerically significant, and (ii) the growth of numerically relevant populations may be favored by using inoculum diluted to extinction, especially in growth medium which reflects the natural resources and conditions in the habitat being studied.
These conclusions are consistent with the results of another comparative analysis of 127 bacterial isolates and 58 environmental 16S rDNA clones which were recovered from the same seawater sample (46). The two 16S rDNA data sets generated showed little overlap, probably due to direct plating of the undiluted inoculum onto solidified medium, which may have resulted in isolation of community members that were not numerically significant (46).
When the previously published data and the relatively small number of MPN isolates and 16S rDNA clones analyzed in our investigation were taken into account, the following two factors might have been decisive for obtaining close matches between the two 16S rDNA data sets generated in this study: (i) abundant community members were recovered in pure culture from the complex rice paddy soil microbial community by isolating bacteria enriched in the terminal and subterminal positive steps of dilution series (8), and (ii) both cultivation and direct recovery of 16S rDNA were used with the same environmental sample. The importance of these two factors was indicated by the results of a previous study in which Methanosaeta spp. and Methanobacterium spp. were found to be dominant methanogenic populations in the rice paddy soil system when both cultivation and direct recovery of 16S rDNA were used (16). Nonetheless, it should be kept in mind that individual dilution steps in MPN dilution series experiments still represent enrichment cultures, and only those bacteria which are able to grow under the culture conditions used are isolated, even from the tubes containing dilute inocula.
The comparative phylogenetic placement of 19 bacterial isolates (8) and 57 environmental 16S rDNA clones revealed close matches for the isolates from culturable populations determined by MPN counting to be large (3 × 107 to 2.5 × 108 cells per g [dry weight] of soil), including MPN isolates PB90-1 (as well as PB90-3 and ACB90 [Verrucomicrobia]), XB45 and PB90-2 (CFB group), XB90 (clostridial cluster XIVa), and SB45 (Bacillaceae) and strain DR1/8 (clostridial cluster IX). Strain DR1/8 was isolated previously from Vercelli rice paddy soil (38). Other than strain ACB90, the MPN isolates originating from counting experiments that indicated that lower numbers (smaller populations) were present showed only moderate or distant affiliations with the respective clones (Table 1). This observation corresponds well to the general idea that the 16S rDNA approach cannot reveal the full range of microbial diversity in a given habitat, especially the microbial populations present in lower numbers (7, 46, 50). Although MPN isolates PB90-1 and ACB90 had identical 16S rDNA sequence types, these two strains are phenotypically different since strain ACB90 is able to utilize cellulose while strain PB90-1 is not able to utilize cellulose (8). Thus, strain ACB90, which was isolated with amorphous cellulose as the substrate and had a population size of less than 107 cells per g (dry weight) of soil, may represent (at the species or subspecies level) a closely related but distinct subpopulation of strain PB90-1 (43).
Close matches, such as those found in the present study, between the 16S rDNA data sets created by two independent approaches suggest an actual abundance of those populations defined by the organismic isolates and the affiliated clones (7). Thus, these matches provide strong evidence that the MPN isolates obtained from counting experiments which indicate that the populations are large can be considered geno- and phenotypic representatives of abundant and functionally relevant bacterial groups in the rice paddy soil examined. This view is also supported by the fact that identical 16S rDNA sequence types (PB90-1 and ACB90) or at least highly related sequence types (XB45 and PB90-2) were independently isolated from the terminal positive dilution steps in the MPN counting experiments carried out with different substrates (8). More evidence of population abundance supported by the close affiliation of an MPN isolate with environmental sequences was provided by strain XB90, which exhibited levels of 16S rDNA similarity with clones BSV51, BSV61, and BSV70 of 95 to 97%. Such levels of relatedness can be exhibited by microorganisms which may or may not have major differences in their phenotypic traits. This is especially true for the various sublineages within the Proteobacteria. However, for the most part clostridial strains that exhibit levels of 16S rDNA similarity of more than 95% have similar phenotypic characteristics. The assumption that this is also the case for strain XB90 and the phenotypes represented by clones BSV51, BSV61, and BSV70 is supported by the finding that the taxa formed a phylogenetically coherent cluster whose members were detected in the same environmental sample (Fig. 3).
The conclusion that populations represented by MPN isolates PB90-1, XB45, and XB90 belong to predominant bacterial groups in the rice paddy soil which we examined is also supported by the total number of cells detected by DAPI (4′,6-diamidino-2-phenylindole) staining (4.8 × 108 cells per g [dry weight] of soil) and by fluorescent in situ hybridization performed with a universal oligonucleotide probe for members of the domain Bacteria (2.8 × 108 cells per g [dry weight] of soil) (8). These three strains were isolated in MPN counting experiments which suggested that the culturable population sizes were about 2.5 × 108 cells per g (dry weight) of soil. Additional support was also provided by an estimate of the total number of cells per gram (dry weight) of rice paddy soil based on the amount of total community DNA extracted. Amounts of genomic DNA between 1.6 and 8.4 fg per cell have been reported previously for soil bacteria recovered by using various cell extraction methods (47). Assuming that the average amount of DNA was about 5 fg of genomic DNA per cell (2), we concluded that the population size was 5.4 × 108 cells per g (dry weight) of soil based on our DNA recovery data; this value agrees rather well with the data obtained by MPN counting and staining techniques.
The close matches between MPN isolates and environmental 16S rDNA clones also suggest that the molecular diversity represented by the clone library largely reflects the composition of the dominant members of the bacterial community in the rice paddy soil examined. Apart from members of the Verrucomicrobia and the CFB group, this community includes mainly lineages of low-G+C-content gram-positive bacteria (Table 1 and Fig. 4). Only three clones (BSV19, BSV26, and BSV40) (Table 1) exhibited more than 15% overall dissimilarity to the 16S rDNA sequence of the most closely related cultured microorganism. The treeing analyses grouped these three clones, which exhibited levels of 16S rDNA similarity to Chlorobium limicola of 78 to 81%, as two deeply branching sublineages within the Chlorobiaceae (data not shown).
Identification of members of several Clostridium-like clusters and of members of the Bacillaceae as abundant organisms in the rice paddy soil (most of these organisms are capable of forming spores) appears to correlate well with the major environmental factors which determine the levels of defined microbial groups in rice paddy soil (5), including alternate periods of flooding and dryness, anoxic conditions during the flooded stage, and the fact that the main portion of the substrates is in the form of polymers, such as xylan, pectin, and cellulose.
Together, the dual approach consisting of cultivation and direct retrieval of 16S rDNA confirmed the inadequacy of culture methods for determining the composition of complex microbial communities in defined habitats (48, 50). However, this approach resulted in characterization of strains isolated from the terminal and subterminal positive steps of serial dilution series as geno- and phenotypic representatives of predominant bacterial groups which may account for a substantial portion of the rice paddy soil microbial community (i.e., 5 to 52% of the total number of cells). Thus, these representatives (e.g., strains PB90-1, XB45, and XB90) should be excellent candidates for detailed genomic and ecophysiological studies performed to determine in more detail the mechanisms which lead to an abundance or even dominance of microbial populations in a defined environment, such as rice paddy soil.
ACKNOWLEDGMENTS
We thank Sonja Fleissner for excellent technical assistance.
This study was supported by grants from the Deutsche Forschungsgemeinschaft and from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (contract 0311121) awarded to W.L.
REFERENCES
- 1.Amann R, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1995. [Google Scholar]
- 3.Bakken L R, Olsen R A. The relationship between cell size and viability of soil bacteria. Microb Ecol. 1987;13:103–114. doi: 10.1007/BF02011247. [DOI] [PubMed] [Google Scholar]
- 4.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossio D A, Scow K M. Impact of carbon and flooding on the metabolic diversity of microbial communities in soils. Appl Environ Microbiol. 1995;61:4043–4050. doi: 10.1128/aem.61.11.4043-4050.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler D P, Li S-M, Spadoni C M, Drake G R, Balkwill D L, Fredrickson J K, Brockman F J. A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol Ecol. 1997;23:131–144. [Google Scholar]
- 8.Chin K-J, Hahn D, Hengstmann U, Liesack W, Janssen P H. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl Environ Microbiol. 1999;65:5042–5049. doi: 10.1128/aem.65.11.5042-5049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin K-J, Rainey F A, Janssen P H, Conrad R. Methanogenic degradation of polysaccharides and the characterization of polysaccharolytic clostridia from anoxic rice field soil. Syst Appl Microbiol. 1998;21:185–200. [Google Scholar]
- 10.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 11.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson R L, Buckley E N, Palumbo A V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984;47:49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finster K, Liesack W, Thamdrup B. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol. 1998;64:119–125. doi: 10.1128/aem.64.1.119-125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich M, Springer N, Ludwig W, Schink B. Phylogenetic positions of Desulfofustis glycolicus gen. nov., sp. nov., and Syntrophobotulus glycolicus gen. nov., sp. nov., two new strict anaerobes growing with glycolic acid. Int J Syst Bacteriol. 1996;46:1065–1069. doi: 10.1099/00207713-46-4-1065. [DOI] [PubMed] [Google Scholar]
- 15.Galchenko V F, Lein A, Ivanov M. Biological sinks of methane. In: Andreae M O, Schimel D S, editors. Exchange of trace gases between terrestrial ecosystems and the atmosphere. Chichester, United Kingdom: John Wiley & Sons; 1989. pp. 59–71. [Google Scholar]
- 16.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Großkopf R, Stubner S, Liesack W. Novel euryarchaeotal lineages detected on rice roots and in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1998;64:4983–4989. doi: 10.1128/aem.64.12.4983-4989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hattori T, Mitsui H, Haga H, Wakao N, Shikano S, Gorlach K, Kasahara Y, El-Beltagy A, Hattori R. Advances in soil microbial ecology and the biodiversity. Antonie Leeuwenhoek. 1997;72:21–28. doi: 10.1023/a:1000201017238. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi S, Furusaka C. Studies on Propionibacterium isolated from paddy soils. Antonie Leeuwenhoek. 1979;45:565–574. doi: 10.1007/BF00403656. [DOI] [PubMed] [Google Scholar]
- 20.Hedlund B P, Gosink J J, Staley J T. Phylogeny of Prosthecobacter, the fusiform caulobacters: members of a recently discovered division of the Bacteria. Int J Syst Bacteriol. 1996;46:960–966. doi: 10.1099/00207713-46-4-960. [DOI] [PubMed] [Google Scholar]
- 21.Hedlund B P, Gosink J J, Staley J T. Verrucomicrobia div. nov., a new division of the Bacteria containing three new species of Prosthecobacter. Antonie Leeuwenhoek. 1997;72:29–38. doi: 10.1023/a:1000348616863. [DOI] [PubMed] [Google Scholar]
- 22.Hengstmann, U., and W. Liesack. Unpublished data.
- 23.Hershberger K, Barns S M, Reysenbach A-L, Dawson S C, Pace N R. Wide diversity of Crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 24.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen P H, Schuhmann A, Mörschel E, Rainey F A. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol. 1997;63:1382–1388. doi: 10.1128/aem.63.4.1382-1388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press Inc.; 1969. pp. 21–132. [Google Scholar]
- 27.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 28.Li T, Bisaillon J-G, Villemur R, Létourneau L, Bernard K, Lépine F, Beaudet R. Isolation and characterization of a new bacterium carboxylating phenol to benzoic acid under anaerobic conditions. J Bacteriol. 1996;178:2551–2558. doi: 10.1128/jb.178.9.2551-2558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liesack W, Janssen P H, Rainey F A, Ward-Rainey N, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker Inc.; 1997. pp. 375–439. [Google Scholar]
- 30.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 31.Lonergan D J, Jenter H L, Coates J D, Phillips E J P, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 33.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelidov S N. Microbiology of the flooded soils of rice paddies. Eurasian Soil Sci. 1994;26(8):41–56. [Google Scholar]
- 35.Ogram A, Sayler G C, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 36.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 37.Rodriguez-Tomé P, Stoehr P J, Cameron G N, Flores T P. The European Bioinformatics Institute (EBI) databases. Nucleic Acids Res. 1996;24:6–12. doi: 10.1093/nar/24.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosencrantz D, Rainey F A, Janssen P H. Culturable populations of Sporomusa spp. and Desulfovibrio spp. in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1999;65:3526–3533. doi: 10.1128/aem.65.8.3526-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Schlesner H. Verrucomicrobium spinosum gen. nov., sp. nov.: a fimbriated prosthecate bacterium. Syst Appl Microbiol. 1987;10:54–56. [Google Scholar]
- 42.Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A, van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 43.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 44.Stackebrandt E, Rainey F A, Ward-Rainey N L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 45.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K. H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. Technische Universität München, Munich, Germany.
- 46.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trevors J T. DNA extraction from soil. Microb Releases. 1992;1:3–9. [Google Scholar]
- 48.Wagner M, Amann R, Lemmer H, Schleifer K-H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward D M, Ferris M J, Nold S C, Bateson M M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe I, Furusaka C. Microbial ecology of flooded rice soils. Adv Microb Ecol. 1980;4:125–168. [Google Scholar]
- 52.Zwart G, Huismans R, van Agterveld M P, Van de Peer Y, De Rijk P, Eenhoorn H, Muyzer G, van Hannen E J, Gons H J, Laanbroek H J. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol Ecol. 1998;25:159–169. [Google Scholar]