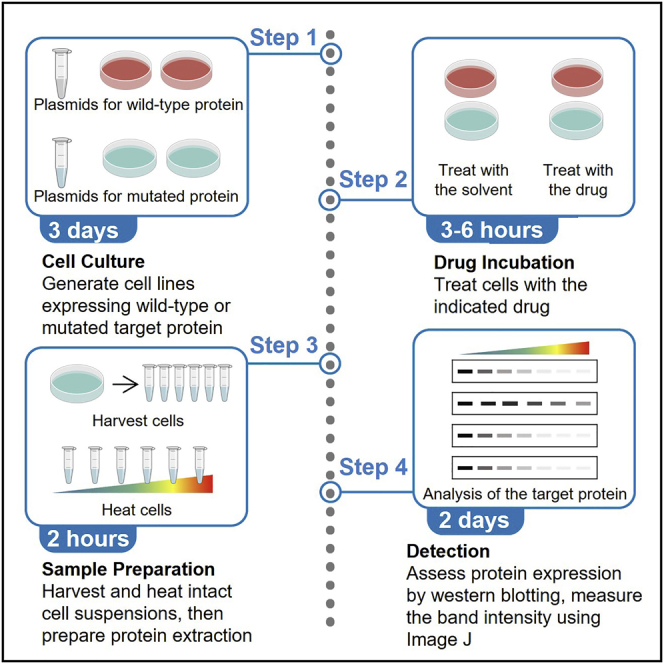
Summary
Evaluating how drugs bind to specific targets at the cellular level is essential to their development and efficacy. Here, we present a protocol to detect the amino acid sites involved in drug-target engagement using a cellular thermal shift assay, which is based on ligand-induced protein thermal stabilization. We also describe generation of cell lines expressing wild-type or mutated target protein and drug treatment. This protocol enables a convenient way to assess amino acid sites involved in drug-target interaction in situ.
For complete details on the use and execution of this protocol, please refer to Peng et al. (2021).
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Protein Biochemistry, Protein expression and purification
Graphical abstract
Highlights
-
•
Protocol for detecting drug target engagement using a cellular thermal shift assay
-
•
Generating cell lines expressing wild-type or mutated target protein for drug treatment
-
•
Assessment of amino acid sites involved in drug-target interactions
-
•
Evaluating drug-target interactions in a cellular context
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Evaluating how drugs bind to specific targets at the cellular level is essential to their development and efficacy. Here, we present a protocol to detect the amino acid sites involved in drug-target engagement using a cellular thermal shift assay, which is based on ligand-induced protein thermal stabilization. We also describe generation of cell lines expressing wild-type or mutated target protein and drug treatment. This protocol enables a convenient way to assess amino acid sites involved in drug-target interaction in situ.
Before you begin
The protocol below describes the steps for detection of the specific amino acids involved in drug-target interaction in HEK 293T cells. We have used this protocol to determine whether Cys115 and Cys161 of Cyclophilin A (CypA) are related to the engagement of CypA and its target inhibitor cyclosporine A (CsA) in a colorectal cancer cell line (RKO) (Peng et al., 2021).
Most clinically available drugs achieve their therapeutic effects through directly binding to specific target proteins, usually at the functional site of the protein (Martinez Molina et al., 2013). The extent of drug-target binding determines both the efficacy and adverse effects of a drug (Martinez Molina et al., 2013). Thus, it is essential to detect drug-target engagement in a cellular context. The cellular thermal shift assay is based on the biophysical principle that ligand binding will induce thermal stabilization of target proteins (Martinez Molina et al., 2013). This method allows the evaluation of drug-target engagement in a cellular context and was originally developed by Pär Nordlund and colleagues (Hao, 2021; Jafari et al., 2014; Martinez Molina et al., 2013).
Culture and seed HEK 293T cells
Timing: 1 day
-
1.
Culture HEK 293T cells in a 100 mm culture dish in DMEM medium in a humidified incubator at 37°C under 5% CO2.
CRITICAL: Supplement DMEM medium with 10% fetal bovine serum (BI), 100 U/mL penicillin, and 100 μg/mL streptomycin (HyClone).
-
2.
Aspirate the culture media when HEK 293T cells reach 80%–90% confluence, wash the cells twice with PBS.
-
3.
Add 2 mL of trypsin solution to the dish, and then incubate for 1 min at room temperature (22°C–25°C).
CRITICAL: Make sure the trypsin solution evenly covers the whole surface of the plate.
-
4.
Add 5 mL of complete DMEM medium to inactivate the trypsin, gently detach the cells and collect the medium containing cells in a 15 mL tube, spin the cells down at 350 × g for 3 min.
-
5.
Resuspend the pellet in 5 mL of complete DMEM medium, count and seed the cells in four 60 mm culture dishes (about 1.5–2 × 106 cells in 5 mL DMEM medium per dish) for transfection.
CRITICAL: Gently shake the dish to ensure the cells are uniformly seeded and then incubate at 37°C under 5% CO2.
Generate cell lines expressing wild-type or mutated target protein
Timing: 2 days
According to our study, Cys115 and Cys161 are putative amino acid sites involved in drug-target interactions between CsA and CypA (Peng et al., 2021). To validate this hypothesis, we generated HEK 293T cell lines expressing flag-tagged wild-type CypA (Flag CypA-WT) or flag-tagged mutated CypA (Flag CypA-C2A) for use in the cellular thermal shift assay.
-
6.
One day following cell seeding, transfect HEK 293T cells with wild-type or mutated CypA plasmids using the following steps.
CRITICAL: The cells should be 70% confluent prior to transfection. If the cell confluence is not appropriate, the cells for transfection should be reseeded. Do not wait until the next day prior to the transfection, or the transfection efficiency will be significantly affected.
-
7.
For each cell dish, prepare Solution 1 and Solution 2 following the steps below:
(Here we need two dishes of HEK 293T cells transfected with Flag-CypA-WT and two dishes transfected with Flag-CypA-C2A, respectively).-
a.Solution 1: Sequentially add 250 μL Opti-MEM medium and 5 μL Lipofectamine 3000 reagent to a 1.5 mL tube, gently mix well and incubate the mixture at room temperature for 5 min.
 CRITICAL: The incubation step is optional.
CRITICAL: The incubation step is optional. -
b.Solution 2: Sequentially add 250 μL Opti-MEM medium, 2 μg plasmids (Flag-CypA WT or Flag-CypA C2A, respectively) and 5 μL P3000 reagent to a 1.5 mL tube, gently mix well and incubate the mixture at room temperature for 5 min.
 CRITICAL: The concentration of plasmids should be 0.5–5 μg/μL. Excessive DNA amounts will lead to proteotoxicity. Endotoxin-free plasmids prepared with an EndoFree plasmid purification kit are recommended.
CRITICAL: The concentration of plasmids should be 0.5–5 μg/μL. Excessive DNA amounts will lead to proteotoxicity. Endotoxin-free plasmids prepared with an EndoFree plasmid purification kit are recommended.
-
a.
-
8.
Add Solution 2 to Solution 1, gently mix them well and incubate the mixture at room temperature for 10–15 min.
-
9.
Add 500 μL of the mixture from the last step to HEK 293T cells in each 60 mm culture dish.
-
10.
Gently swirl the dishes to ensure even distribution of the mixture, and then incubate the dishes at 37°C under 5% CO2.
CRITICAL: The stably transfected cell line would also be a good alternative.
CRITICAL: For efficient transduction, HEK 293T cells at 1–4 passages (2–8 days) after thaw are recommended.
Treat cells with the indicated drug
Timing: 6 h
-
11.
At 48 h following the transfection of wild-type or mutated CypA, replace the medium with 5 mL fresh DMEM containing 5 μL DMSO or 5 μL CsA and incubate for 6 h at 37°C under 5% CO2.
CRITICAL: CsA (5 mM in DMSO) must be stored at −20°C. The final concentration of DMSO is 0.1% (v/v).
CRITICAL: Keep the final concentration of DMSO constant in different groups.
CRITICAL: The incubation time and drug concentration may require optimization for different drugs and proteins. The incubation time is usually 3–6 h. The drug target engagement usually gets stronger with increased incubation time and drug concentration. To determine a suitable drug concentration, an isothermal dose-response procedure should be performed initially. In detail, treat cells with different concentrations of the drug, while keep the incubation time (usually 3–6 h), heating time and temperature (50°C–60°C is the most commonly used temperature) constant. This process then generates a characteristic fingerprint of the target engagement along the drug concentration axis (Martinez Molina et al., 2013). Choose the minimum concentration at which the protein is significantly expressed for the next steps, for example, 100 μM in Figure 1.
Figure 1.
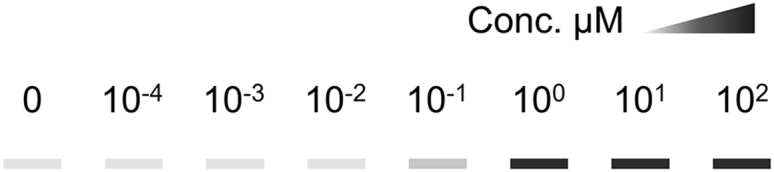
Schematic illustration showing how to determine a suitable drug concentration
Conc. means the drug concentration.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Flag tag (at 1:2000 dilution) | Cell Signaling Technology | Cat# 14793; RRID: AB_2572291 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP (at 1:5000 dilution) | Thermo Fisher Scientific | Cat# 31460; RRID: AB_228341 |
| Chemicals, peptides, and recombinant proteins | ||
| DMSO | Sigma | Cat# D2650 |
| Cyclosporine A | MedChemExpress | Cat# HY-B0579 |
| SDS | Sigma | Cat# L3771 |
| Bromophenol blue | Bio-Rad | Cat# 1610404 |
| Dithiothreitol (DTT) | Bio-Rad | Cat# 1610611 |
| Glycine | Kelong | N/A |
| Tween 20 | Kelong | N/A |
| Blotting Grade Blocker Non-Fat Dry Milk | Bio-Rad | Cat# 1706404XTU |
| Primary Antibody Dilution Buffer | Beyotime | Cat# P0023A |
| Immobilon Western Chemiluminescent HRP Substrate | Millipore | Cat# WBKLS |
| Critical commercial assays | ||
| DMEM medium (1×) | Gibco | Cat# C11995500BT |
| Trypsin-EDTA (0.25%) | Thermo Fisher Scientific | Cat# 25200056 |
| Penicillin-Streptomycin Solution | HyClone | Cat# SV30010 |
| Opti-MEM | Thermo Fisher Scientific | Cat# 31985070 |
| Lipofectamine 3000 | Thermo Fisher Scientific | Cat# L3000015 |
| Experimental models: Cell lines | ||
| HEK 293T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Recombinant DNA | ||
| pCDNA3.1-Flag-CypA WT | This paper | Addgene# 185616 |
| pCDNA3.1-Flag-CypA C2A | This paper | Addgene# 185617 |
| Software and algorithms | ||
| GraphPad Prism (v.7) | GraphPad | https://www.graphpad.com:443/ |
| ImageJ (v.2.1.0) | (Schneider et al., 2012; Yamamoto et al., 2008) | https://imagej.nih.gov/ij/ |
| Other | ||
| PVDF membrane | Millipore | Cat# IPVH00010 |
| 60 mm culture dish | WHB | Cat# WHB-60 |
| 100 mm culture dish | WHB | Cat# WHB-100 |
| Fetal bovine serum | Biological Industries | Cat# 04-001-1ACS |
Materials and equipment
Below are recipes to prepare solutions needed for this protocol:
PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 137 mM | 8 g |
| KCl | 2.7 mM | 0.2 g |
| Na2HPO4·12H2O | 10 mM | 3.62 g |
| KH2PO4 | 1.76 mM | 0.24 g |
| ddH2O | n/a | n/a |
| Total | n/a | 1 L |
Sterilize through a 0.2 μm filter and store at 20°C–25°C for 2–3 months.
5× loading buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Glycerol | 50% | 25 mL |
| 1 M Tris-HCl pH 6.8 | n/a | 15.625 mL |
| SDS | 10% | 5 g |
| Bromophenol blue | 0.05% | 25 mg |
| Dithiothreitol (DTT) | 250 mM | 1.93 g |
| ddH2O | n/a | n/a |
| Total | n/a | 50 mL |
Store at −20°C for about a year.
Running buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 25 mM | 3.03 g |
| Glycine | 190 mM | 14.26 g |
| SDS | 0.1% | 1 g |
| ddH2O | n/a | 1 L |
| Total | n/a | 1 L |
Store at 4°C–25°C for about a year.
Transfer buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 25 mM | 3.03 g |
| Glycine | 200 mM | 15 g |
| Methanol | 20% | 200 mL |
| ddH2O | n/a | 800 mL |
| Total | n/a | 1 L |
Store at 4°C–25°C for about a year. Add methanol just before use.
20×Tris-buffered saline (TBS)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris, pH 7.6 | 400 mM | 48.45 g |
| NaCl | 3 M | 175.32 g |
| ddH2O | n/a | n/a |
| Total | n/a | 1 L |
Store at 4°C–25°C for 6 months.
Tris-buffered saline with Tween 20 (TBS-T) buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 20 × TBS | n/a | 50 mL |
| Tween 20 | 0.1% | 1 mL |
| ddH2O | n/a | 950 mL |
| Total | n/a | 1 L |
Store at 4°C–25°C for 6 months.
Blocking solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Non-fat dry milk | 5% | 5 g |
| TBST buffer | n/a | 100 mL |
| Total | n/a | 100 mL |
Store at 4°C–25°C for 24 h. Prepare Blocking solution just before use.
Step-by-step method details
Harvest and heat intact cell suspensions
Timing: 1 h
In this step, cells treated with indicated drugs are harvested, then the intact cell suspension is heated in a PCR amplifier at the appropriate temperature.
-
1.
Wash cell culture plates twice with 2 mL ice-cold PBS, add 700 μL ice-cold PBS to each plate.
-
2.
Scrape adherent cells off the dishes using a cell scraper.
-
3.
Divide cell suspensions from different groups into PCR tubes (6 PCR tubes per group) with 100 μL suspension per tube.
CRITICAL: Divide the cell suspensions equally to make sure there are equal amounts of protein in the loading sample in the next step.
-
4.
Heat the cell suspensions individually at 45°C, 48°C, 51°C, 58°C, 65°C, and 69°C for 5 min in a PCR amplifier.
CRITICAL: The heating temperature and heating time vary between different proteins or drugs. Determine the appropriate heating temperature and heating time through pre-experiments. For the determination of suitable heating temperatures, set a temperature gradient from 40°C to 70°C, with intervals of 2°C–3°C. Keep incubation time (usually 3–6 h), incubate concentration and heating time constant. Then complete the full steps of the thermal shift assay to obtain a melting curve. For later experiments, choose the temperature where the protein is starting to degrade as the initial temperature point, while the temperature at which the protein is completely degraded as the ending temperature point. The temperature interval is usually 5°C–7°C. The optimization steps for the heating time are similar to that of drug concentration. The heating time is usually 3–5 min.
CRITICAL: This protocol may not be applicable to proteins that are rather unstable.
Prepare protein extraction from heated cells
Timing: 2 h
This step accomplishes protein extraction from the above-mentioned heated cells and preparation of samples for western blot analysis.
-
5.
Lyse the heated cell suspensions with three cycles of freeze-thawing with liquid nitrogen and a 37°C water bath.
-
6.
Centrifuge the cell lysate mixture at 20,000 × g for 10 min at 4°C.
-
7.
Isolate the soluble fractions from the cell debris, transfer them to microcentrifuge tubes (about 80 μL per sample).
-
8.
Determine the protein concentration of each sample using a Bradford assay.
-
9.
Add 20 μL of 5 × loading buffer to the protein extraction, mix completely and boil at 95°C–100°C for 10 min.
Pause point: The samples can be stored at −20°C for about one week. However, it is better to run as fresh as possible.
Assess protein expression by western blotting
Timing: 2 days
This step accomplishes the separation of proteins by SDS-PAGE, the transfer of proteins onto the PVDF membrane, imaging, and data analysis. Detection of no protein expression change in the mutant group after drug treatment means that the amino acid sites are involved in the drug-target engagement.
Run SDS-PAGE
Timing: 1.5–2 h
-
10.
Prepare pre-made (12% polyacrylamide) gels.
CRITICAL: Determine the appropriate polyacrylamide percentage to best resolve the specific protein of interest based on molecular weight.
Alternatives: Purchased gels are excellent alternatives here.
-
11.
Load the gels into the electrophoresis apparatus, fill it with 1 L Running buffer.
CRITICAL: The volume of Running buffer varies depending on the gel systems used.
-
12.
Load samples containing equal amounts of protein (10–50 μg/lane) into the lanes of the SDS-PAGE gels.
CRITICAL: Include a molecular weight marker in one of the lanes to indicate the molecular weight. Set up gels for Coomassie blue, Ponceau S or Revert Total Protein Stan (LICOR) staining as a loading control.
-
13.
Run the gels at 120 V for 1–2 h, until the bromophenol blue reaches the bottom of the gels.
CRITICAL: Time and voltage may require optimization for different proteins and polyacrylamide gel percentage.
Transfer protein from the gel into PVDF membrane
Timing: 1–2 h
-
14.
Take out the gel from the electrophoresis apparatus, put it in a tray with Transfer buffer.
-
15.
Prepare PVDF membrane: wet PVDF membrane in methanol for 30 s and then soak it in the tray with Transfer buffer.
CRITICAL: Handle the membrane carefully, avoid scratching or puncturing the surface.
-
16.Assemble the transfer sandwich:
-
a.Sequentially assemble the transfer sandwich in the following order:
-
i.Sponge layer.
-
ii.Filter paper layer.
-
iii.Gel layer.
-
iv.PVDF membrane layer.
-
v.Filter paper layer.
-
vi.Sponge layer.
-
i.
-
b.Gently remove any air bubbles with a scraper or roller, make sure no air bubbles are trapped in any layer.
-
a.
CRITICAL: Make sure to remove any air bubbles, or the transfer of proteins to the membrane will be inhibited.
-
17.Perform wet transfer:
-
a.Fill the transfer apparatus with Transfer buffer.
-
b.Place the transfer sandwich into it.
-
c.Run at 300 mA for 1 h.
-
a.
CRITICAL: Transfer time and voltage may require optimization according to the molecular weight of proteins.
CRITICAL: Keep the PVDF membrane wet at all time during the western blotting process.
Block the membrane and incubate with primary/secondary antibody
Timing: 16–20 h
-
18.
Rinse the PVDF membrane in TBST.
-
19.
Incubate the membrane in Blocking solution for 2 h at room temperature (20°C–25°C) with gentle shaking.
-
20.
Rinse the membrane briefly in TBST to remove residual milk.
-
21.
Incubate the membrane with the primary antibody against Flag tag overnight (for 10–12 h) at 4°C with gentle rocking.
CRITICAL: Dilute the primary antibody with Primary antibody dilution buffer at the appropriate concentration. Here we used anti-Flag antibody for Flag-CypA WT or Flag-CypA C2A protein at 1:2000 dilution in Primary antibody dilution buffer.
-
22.
Aspirate the primary antibody solution and wash the membrane 3 times with TBST (10 min each with gentle rocking).
-
23.
Incubate the membrane with HRP conjugated secondary antibody solution for 2 h at room temperature (20°C–25°C) with gentle rocking.
CRITICAL: Dilute the secondary antibody in TBST at the appropriate concentration. May include 5% milk. Here we used Goat anti-Rabbit IgG (H+L) at 1:5000 dilution in TBST.
CRITICAL: The secondary antibody must recognize the host species of the primary antibody.
-
24.
Wash the membrane 6 times with TBST (10 min each with gentle rocking).
Acquire the image and analyze the data
Timing: 1–2 h
-
25.
Prepare the Chemiluminescent HRP Substrate mixture: mix equal amounts of solution A and solution B provided by the manufacturer.
CRITICAL: About 2–3 mL of the mixture is required for a 12 cm × 6 cm membrane.
-
26.
Remove excess buffer from the membrane, incubate the membrane in the Chemiluminescent HRP Substrate mixture at room temperature for 10 s.
-
27.
Expose the membrane with the chemiluminescent imaging system Clinx.
CRITICAL: The incubation time may require optimization. More sensitive substrates require shorter incubation times to achieve optimal signal and avoid overexposure. LI-COR NIR secondary antibodies are better alternatives to avoid overexposure.
CRITICAL: Use multiple exposure lengths to identify the optimal exposure time.
-
28.
Measure the band intensity using image analysis software such as Image J.
-
29.
Calculate the relative band intensity and draw the cellular thermal shift assay curve using GraphPad.
CRITICAL: We use total Flag-tagged CypA without any heating as the loading controls. A housekeeping protein is a good alternative.
Expected outcomes
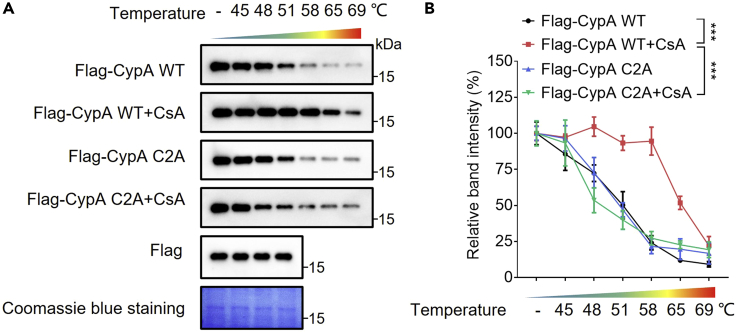
The cellular thermal shift assay determines whether a protein of interest is binding to an indicated drug. If the protein of interest interacts with an indicated drug, the protein from the drug-treated group will be more stabilized after heating compared to the control group. Here, in cells transfected with Flag-CypA WT plasmid, the protein was stabilized against the thermal increase in the CsA group (Figure 2A, the Flag-CypA WT band and the Flag-CypA WT+CsA band). This result validated that CypA could interact with CsA. However, in cells transfected with Flag-CypA C2A plasmid, no change in protein stabilization against thermal changes was observed after CsA treatment (Figure 2A, the Flag-CypA C2A band and the Flag-CypA C2A+CsA band), indicating that Cys115 and Cys161 are involved in the interaction between CypA and CsA.
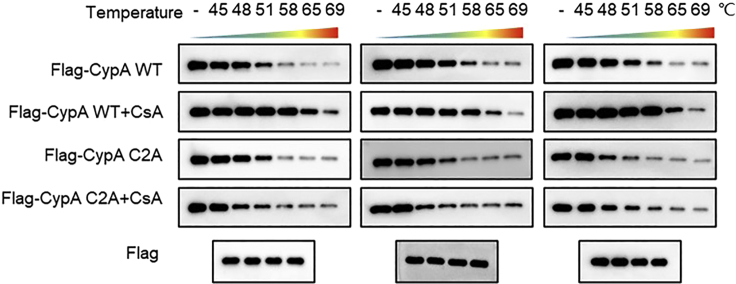
CRITICAL: Three biological repetitions exhibiting the repeatability between batches are presented (Figure 3).
Figure 2.
Analysis of amino acid sites involved in drug-target engagement between CypA and CsA
(A) Cellular thermal shift assay of WT CypA or mutated CypA in HEK 293T cells treated with or without CsA. The first lane shows the four samples without any heating. The Flag band and the Coomassie blue staining are used as loading controls.
(B) The relative immunoblotting band density is analyzed. All data are shown as the mean ± SD and are representative of three independent experiments. ∗∗∗p < 0.001.
Figure 3.
The results of three sample batches subjected to thermal shift assay
The first lane shows the four samples without any heating. The Flag band is used as a loading control.
Limitations
It has been reported that larger proteins such as protein complexes are likely to have weak or no ligand-induced response in the cellular thermal shift assay (Jafari et al., 2014). Thus, this protocol may be limited to smaller proteins.
When detecting the specific amino acid sites involved in drug-target engagement, we transfected exogenous plasmids into HEK 293T cells, whose state of drug-target engagement may be different from the endogenous protein.
Troubleshooting
Problem 1
The protein concentration is too high or too low (step 8 of the step-by-step method details section).
Potential solution
Optimize the cell number seeded (step 5 of the before you begin section) or the volume of PBS added (step 1 of the step-by-step method details section).
Decrease or increase the cycles of freeze-thawing with liquid nitrogen (step 5 of the step-by-step method details section).
Problem 2
The intensity of the protein band is too strong or too weak (steps 27–28 of the step-by-step method details section).
Potential solution
Optimize the amount of protein loaded onto the gel.
Decrease or increase the time of exposure.
Dilute the Chemiluminescent HRP Substrate mixture with TBST if the protein band intensity is too strong.
Check the transfection efficacy and the activity of the primary/secondary antibody.
Problem 3
The cellular thermal shift assay curve is not regular (step 29 of the step-by-step method details section).
Potential solution
Optimize the heating temperature or heating time.
Adjust the amount of protein loaded onto the gel and the exposure time to achieve the proper band intensity of the protein.
Problem 4
The mutant expression is significantly less than WT (steps 27–28 of the step-by-step method details section).
Potential solution
Adjust the protein loading to keep the protein concentration of Flag-tagged wild-type CypA and Flag-tagged mutated CypA constant.
Problem 5
The drug molecule is not so soluble, e.g., requires higher DMSO content (step 11 of the before you begin section).
Potential solution
Change the solvent. For example, if the drug molecule is hydrophilic, PBS could be used as a solvent. If the drug molecule is lipophilic, ethanol may be a potential alternative.
Facilitate drug dissolution through appropriate ultrasound or heating.
Problem 6
Protein has a small temperature window (i.e., it degrades quickly within a small range of temperatures) (step 4 of the step-by-step method details section).
Potential solution
Increase the drug concentration or incubation time to promote protein-drug engagement.
Decrease the heating time or supplement the buffer with a protease inhibitor cocktail to attenuate degradation.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Canhua Huang (hcanhua@hotmail.com).
Materials availability
Plasmids generated in this study will be made available on request, but we may require payment and/or a complete Materials Transfer Agreement if there is potential for commercial application.
Acknowledgments
This work was supported by grants from the National Key Research and Development Project of China (2020YFA0509400), Guangdong Basic and Applied Basic Research Foundation (2019B030302012), National Natural Science Foundation of China (81821002, 82130082, 81790251, and 82003098), and China Postdoctoral Science Foundation (2020TQ0214 and 2020M673252).
Author contributions
C.H. conceived the manuscript. L.P. and J.J. drafted the manuscript. L.Z. scrutinized technical details. E.C.N. revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This study did not generate datasets and code.
References
- Hao J. Thermal shift assay for exploring interactions between fatty acid-binding protein and inhibitors. Methods Mol. Biol. 2021;2261:395–409. doi: 10.1007/978-1-0716-1186-9_24. [DOI] [PubMed] [Google Scholar]
- Jafari R., Almqvist H., Axelsson H., Ignatushchenko M., Lundback T., Nordlund P., Martinez Molina D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Peng L., Jiang J., Chen H.N., Zhou L., Huang Z., Qin S., Jin P., Luo M., Li B., Shi J., et al. Redox-sensitive cyclophilin A elicits chemoresistance through realigning cellular oxidative status in colorectal cancer. Cell Rep. 2021;37:110069. doi: 10.1016/j.celrep.2021.110069. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Takiwaki H., Arase S., Ohshima H. Derivation and clinical application of special imaging by means of digital cameras and Image J freeware for quantification of erythema and pigmentation. Skin Res. Technol. 2008;14:26–34. doi: 10.1111/j.1600-0846.2007.00256.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate datasets and code.