Abstract
The 16S rRNA and pmoA genes from natural populations of methane-oxidizing bacteria (methanotrophs) were PCR amplified from total community DNA extracted from Lake Washington sediments obtained from the area where peak methane oxidation occurred. Clone libraries were constructed for each of the genes, and approximately 200 clones from each library were analyzed by using restriction fragment length polymorphism (RFLP) and the tetrameric restriction enzymes MspI, HaeIII, and HhaI. The PCR products were grouped based on their RFLP patterns, and representatives of each group were sequenced and analyzed. Studies of the 16S rRNA data obtained indicated that the existing primers did not reveal the total methanotrophic diversity present when these data were compared with pure-culture data obtained from the same environment. New primers specific for methanotrophs belonging to the genera Methylomonas, Methylosinus, and Methylocystis were developed and used to construct more complete clone libraries. Furthermore, a new primer was designed for one of the genes of the particulate methane monooxygenase in methanotrophs, pmoA. Phylogenetic analyses of both the 16S rRNA and pmoA gene sequences indicated that the new primers should detect these genes over the known diversity in methanotrophs. In addition to these findings, 16S rRNA data obtained in this study were combined with previously described phylogenetic data in order to identify operational taxonomic units that can be used to identify methanotrophs at the genus level.
Methanotrophs are a group of gram-negative bacteria that can grow on methane as the sole source of carbon and energy. They are widespread in nature and have gotten increased attention in the past two decades due to their potential role in the global methane cycle (11) and their ability to cometabolize a number of environmental contaminants (15). The methanotrophs consist of eight recognized genera (3, 5–7) that fall into two major phylogenetic groups, the α subgroup of the class Proteobacteria (α-Proteobacteria) (which includes the type II methanotrophs) and the γ-Proteobacteria (which includes the type I methanotrophs). In addition, a new thermophilic genus, Methylothermus, that forms a distinct, deeply branching group within the γ-Proteobacteria has recently been described (4).
Traditionally, studies performed with natural populations of methanotrophs have focused on culture-based techniques (15) that may or may not reveal the true diversity in nature (1). More recently, however, researchers have recognized the need for culture-independent analyses of natural methanotrophic populations, and these types of analyses have been facilitated by recent advances in the molecular biology and molecular phylogeny of methanotrophs (16, 24, 28). To aid in these studies, PCR primers targeted to the 16S rRNA genes in methanotrophs have been developed (8, 17). In addition, preliminary work has been carried out to identify primers that detect pmoA, one of the genes for the diagnostic enzyme for methanotrophs, the particulate methane monooxygenase (pMMO) (16). These primers also detect amoA, which encodes the analogous subunit of the ammonia monooxygenase in nitrifying bacteria (26).
To date, most studies involving non-culture-based analyses of natural populations of methanotrophs have focused on marine and peat bog environments (17, 23, 25). In these studies, nucleic acid-based techniques have been used to obtain information on methanotrophic 16S rRNA and pmoA genes. The results of these studies have expanded the known sequence diversity for these genes and have suggested that these environments contain limited methanotroph diversity at the genus level. The environmental sequences obtained from peat environments all cluster with the type II methanotrophs (23, 25), while the two strains from marine and estuarine environments are both type I strains (17, 33).
Workers in our laboratories are interested in investigating natural populations of methanotrophs in freshwater sediments. However, it is not yet clear whether the molecular tools that are currently available detect the full range of in situ methanotroph genera in these environments. Methanotrophs in freshwater sediments are important to the global methane cycle as these environments are predicted to produce an amount of methane equivalent to approximately 40 to 50% of the annual global atmospheric methane flux (11, 18, 31). However, most of this methane never reaches the atmosphere as it is consumed by methanotrophs (18). Some data suggest that freshwater environments may contain greater methanotroph diversity than peat and marine environments since both pure-culture isolation methods and phospholipid fatty acid analyses indicate that a mixture of type I and type II strains is present (2, 9).
Currently, no data concerning the in situ populations of methanotrophs in freshwater environments as determined by using primers specific for methanotroph 16S rRNA or pmoA genes are available. In addition, it is not known whether the methanotroph primers that have been described can effectively assess the in situ methanotroph diversity in these habitats. Therefore, the objective of this study was twofold: to develop a database of methanotroph 16S rRNA and pmoA sequences for a freshwater sediment and to use this information to develop robust molecular tools for studying in situ methanotrophs in freshwater habitats. The study site chosen was Lake Washington, which we have previously analyzed to determine methanotrophic activities in carbon and oxygen cycling (19, 20).
MATERIALS AND METHODS
Collection of samples.
Sediment was collected from a 62-m-deep station in Lake Washington in Seattle, Wash., by using a box core sampler that allowed us to collect relatively undisturbed sediment. Subsections of the box cores were sectioned into 0.5-cm slices to a depth of 5 cm. Samples were kept on ice for approximately 1 to 2 h and were then used or stored at −20°C.
DNA extraction and purification.
DNA was extracted from sediment obtained in the area where peak methane oxidation occurred (1a) by using a protocol described by Gray and Herwig (14). The amount of sediment used per extraction procedure was 600 mg. The modifications of the protocol included replacing the Spin-Bind columns with Sephadex G-200 spin columns. The Sephadex G-200 spin columns were constructed by filling a 1-ml syringe with glass wool and approximately 1 to 2 cm of TE-saturated Sephadex G-200. After passage through the column, the DNA was further purified by removing residual humic acids by electrophoresis on a 1% agarose gel and purification with a Qiagen gel extraction kit (Qiagen, Inc.). DNA obtained after this treatment was used in PCR mixtures.
PCR amplification of 16S rRNA and pmoA genes.
The 16S rRNA genes were PCR amplified from total DNA extracted from sediment by using methanotroph phylogenetic group-specific primers Mb1007, Mc1005, Mm1007, and Ms1020 (17) in conjunction with bacterium-specific primer f27. Furthermore, 16S rRNA primers Mm835 (5′ GCTCCACYACTAAGTTC 3′) and Type2b (5′ CATACCGGRCATGTCAAAAGC 3′) were designed by using new and previously described sequences to specifically amplify genes from members of the genus Methylomonas and members of the genera Methylosinus and Methylocystis, respectively (Table 1). These primers were also used in subsequent PCRs with primer f27 to amplify genes from members of the genera Methylomonas, Methylosinus, and Methylocystis. All reactions were carried out in 30-μl (total volume) mixtures containing approximately 100 ng of sediment DNA, 10 pmol of each primer, 1.5 mM Mg2+, Gibco buffer, and 2.5 U of Gibco Taq polymerase. The reactions were performed in a Perkin-Elmer model 9600 GeneAmp PCR System thermal cycler by using 25 cycles consisting of 92°C for 1 min, 55°C for 1.5 min (50°C for primer Mm835), and 72°C for 1 min and a final extension step consisting of 72°C for 5 min. In addition, amplification reactions were also performed with primers specific for pmoA. To design pmoA-specific primers, pmoA and amoA sequences available from the GenBank database were aligned, and primer mb661 (5′ CCGGMGCAACGTCYTTACC 3′) was designed (Table 1). Primer mb661 was used in conjunction with primer A189gc (16). Together, primers A189gc and mb661 amplified an approximately 470-bp internal section of pmoA and produced strong signals with all of the methanotrophs tested. The methanotrophs tested included pure cultures of Methylomicrobium album BG8, Methylomonas rubra, Methylomonas methanica S1, Methylococcus capsulatus Bath, Methylosinus trichosporium OB3b, Methylocystis parvus OBBP, and the isolates obtained from Lake Washington in this study (see below). The pmoA primer pair, primers A189gc and mb661, produced no product with Nitrosomonas europaea DNA, as determined in PCRs. In addition, primer mb661 was tested in silico with additional nitrifier amoA gene sequences obtained from the GenBank database and exhibited low levels of identity (9- to 12-bp differences) with these sequences. One exception was the amoA gene of Nitrosococcus oceanus, which exhibited only a 2-bp difference. However, the amoA gene of this organism is more closely related to the pmoA genes of methanotrophs than to the amoA genes of nitrifiers so the high level of identity is not surprising (16).
TABLE 1.
Methanotroph-specific primers used in this study
| Primer | Sequence (5′-3′) | Target genus or gene | Reference |
|---|---|---|---|
| Mb1007r | CACTCTACGATCTCTCACAG | Methylobacter | 17 |
| Methylomicrobium | |||
| Mc1005r | CCGCATCTCTGCAGGAT | Methylococcus | 17 |
| Mm1007r | CACTCCGCTATCTCTAACAG | Methylomonas | 17 |
| Ms1020r | CCCTTGCGGAAGGAAGTC | Methylosinus | 17 |
| Mm835 | GCTCCACYACTAAGTTC | Methylomonas | This study |
| Type2b | CATACCGGRCATGTCAAAAGC | Methylosinus-Methylocystis | This study |
| A189gc | GGNGACTGGGACTTCTGG | pmoA | 16 |
| mb661 | CCGGMGCAACGTCYTTACC | pmoA | This study |
Construction of clone banks and restriction fragment length polymorphism (RFLP) analyses.
The size and purity of each PCR product were checked on 1% agarose gels (32). The PCR products were purified with a Qiagen PCR purification kit (Qiagen, Inc.) and were ligated into the pCR2.1 vector supplied with a TA cloning kit (Invitrogen) by following the manufacturer’s instructions. Individual colonies containing inserts were suspended in 50 μl of water and boiled for 5 min, the cell debris was spun down, and 1-μl portions of the supernatant were used in PCR mixtures to reamplify the insert from the vector with the appropriate primers. The reamplified product was used in restriction digests along with tetrameric restriction enzymes. The 16S rRNA genes were digested with the enzymes MspI, HhaI, and HaeIII. The pmoA genes were digested with HhaI and a combination of MspI and HaeIII. Digests were resolved on 3% NuSieve GTG agarose (FMC) gels and were grouped manually based on the restriction patterns.
16S rRNA and pmoA genes from pure cultures.
Pure cultures requiring methane for growth were obtained from enrichment cultures by using Lake Washington sediments (1b). Chromosomal DNA was isolated from each strain by using cells grown on agarose plates. Cells were washed from the agarose surface with 500 μl of TEN (50 M Tris EDTA, 150 mM NaCl), and the liquid was collected in 1.5-ml tubes. The tubes were centrifuged for 5 min at 14,000 rpm, and the supernatant was poured off. Each pellet was resuspended by adding 500 μl of TEN supplemented with 4 mg of lysozyme per ml and was incubated at 37°C for 1 h. Next, 50 μl of 20% sodium dodecyl sulfate was added to each tube, and the tubes were incubated in a 45 to 50°C water bath for approximately 30 min. DNA was extracted with phenol and was precipitated by using ethanol and standard procedures (32). DNA from each of the isolates was used in PCR mixtures as described above. The 16S rRNA genes were amplified by using bacterium-specific primers f27 and 1492r (13). The pmoA genes from each of the isolates were amplified by using primers A189gc and mb661 as described above.
Data analyses.
Analyses and translation of DNA and DNA-derived polypeptide sequences were carried out by using Genetics Computer Group programs (Genetics Computer Group, Madison, Wis.).
Phylogenetic analysis.
16S rRNA gene sequences were compared with sequences in the small-subunit rRNA database of the Ribosomal Database Project (RDP) by using the Similarity_Rank program (22). 16S rRNA sequences were aligned manually with representative sequences of the nearest phylogenetic neighbors, as defined by the RDP, by using the SeqApp program. Dendrograms were constructed by using the programs DNADIST, DNAPARS, DNAML, NEIGHBOR, and SEQBOOT from the PHYLIP version 3.5c package (12). Tree files generated by PHYLIP were analyzed by using the program TreeView (29). The RDP program Check_Chimera was used to examine 16S rRNA gene sequences for chimeras. pmoA sequences were aligned manually with pmoA and amoA sequences obtained from the GenBank database. Dendrograms were constructed by using the programs PROTDIST, PROTPARS, NEIGHBOR, and SEQBOOT from PHYLIP, version 3.5c (12), and tree files were analyzed by using TreeView (29).
DNA sequencing.
DNA sequencing of the 16S rRNA and pmoA genes was carried out with both strands by using an ABI Prism BigDye terminator sequencing kit (Applied Biosystems). The sequences were analyzed by workers at the University of Washington Center for AIDS Research DNA Sequencing Facility and the Department of Biochemistry Sequencing Facility, who used an Applied Biosystems automated sequencer.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences determined in this study are AF150757 to AF150807.
RESULTS
RFLP analysis of known methanotrophs.
Tetrameric restriction enzymes have been shown to be useful tools for screening environmental clone libraries by RFLP analysis (10, 21, 27, 30, 34, 36). Common restriction fragments obtained from such analyses that distinguish between taxonomic groups are known as operational taxonomic units (OTUs) (27). Identification of OTUs for methanotrophs would facilitate rapid screening of both isolates and environmental clones. Therefore, a number of representative methanotrophic 16S rRNA genes available from the GenBank database were examined by performing computer-aided digestion with the tetrameric restriction enzymes MspI, HhaI, and HaeIII to determine whether OTUs could be identified. We predicted that these enzymes would produce useful patterns for regions used previously for PCR analysis (17), and a comparative computer analysis revealed that each genus could be identified by a distinct set of patterns (Table 2). To test our predictions experimentally, the same PCR products were generated by using DNA from representative strains and these PCR products were digested by the three restriction enzymes. Most of the RFLP patterns obtained for the strains tested corresponded to the patterns predicted on the basis of the previously described sequences; exceptions were the Methylomonas methanica S1, Methylomonas rubra, Methylocystis parvus OBBP, and Methylosinus trichosporium OB3b patterns. The discrepancies observed suggested that there may have been errors in the sequences deposited previously. The 16S rRNA genes from these cultures were resequenced, and significant apparent errors were identified in the original sequences. The new sequences which we obtained were 87 to 99% identical to the previously described sequences and matched the RFLP patterns obtained for the digests with chromosomal DNA, suggesting that the new sequences are correct. The RFLP patterns of the new 16S rRNA gene sequences also clearly fit into the OTUs defined for the respective genera (Table 2). The corrected sequences were especially significant for the type II Methylosinus and Methylocystis strains as only 10 16S rRNA gene sequences have been described for type II methanotrophs. It should be noted that many of the remaining eight Methylosinus and Methylocystis 16S rRNA gene sequences in the database do not produce the correct OTUs when they are analyzed in silico and may contain sequence errors in addition to ambiguous bases. All of the reference sequences used in our analyses contained genus-specific OTUs, and we were careful to choose the most accurate and complete sequence when possible.
TABLE 2.
Sizes of restriction fragments obtained from PCR-amplified products of methanotroph 16S rRNA grouped by genus
| Genusa | Sizes of restriction fragments (bp)b
|
||
|---|---|---|---|
| HhaI | MspI | HaeIII | |
| Methylomicrobium spp. | |||
| Methylomicrobium album (X72777)c | 75, 126, 160, 170, 478 | 11, 33, 67, 109, 347, 442 | 34, 53, 59, 66, 66, 85, 99, 129, 418 |
| Methylomicrobium agile (X72767)c | 76, 124, 126, 160, 170, 352 | 11, 33, 67, 109, 347, 441 | 34, 52, 66, 67, 85, 99, 128, 153, 324 |
| Environmental clone pAMC421 | 76, 126, 126, 160, 171, 350 | 67, 110, 348, 484 | 34, 53, 66, 67, 99, 129, 561 |
| Environmental clone pAMC466 | 76, 126, 126, 160, 171, 352 | 67, 110, 348, 486 | 34, 53, 66, 67, 85, 99, 129, 478 |
| Methylobacter spp. | |||
| Methylobacter whittenburyi (X72773)c | 75, 126, 126, 160, 170, 353 | 11, 33, 110, 414, 442 | 34, 53, 66, 66, 85, 99, 129, 478 |
| Methylobacter luteus (X72772)c | 75, 126, 160, 170, 479 | 11, 33, 110, 414, 442 | 34, 53, 66, 66, 85, 99, 129, 478 |
| Isolate LW1 | 75, 126, 160, 170, 478 | 11, 32, 110, 414, 442 | 34, 53, 59, 66, 85, 98, 195, 419 |
| Environmental clone pAMC405 | 76, 126, 160, 171, 477 | 32, 110, 415, 453 | 34, 59, 67, 99, 119, 129, 129, 374 |
| Environmental clone pAMC415 | 76, 126, 160, 171, 478 | 11, 32, 110, 415, 443 | 34, 53, 59, 66, 67, 85, 99, 129, 419 |
| Environmental clone pAMC417 | 76, 126, 160, 172, 476 | 11, 32, 110, 415, 442 | 34, 53, 59, 66, 67, 100, 129, 502 |
| Environmental clone pAMC419 | 76, 126, 160, 171, 478 | 32, 110, 415, 454 | 34, 66, 67, 85, 99, 182, 478 |
| Methylomonas spp. | |||
| Methylomonas methanica S1 (AF150806)d | 76, 126, 644 | 11, 32, 360, 443 | 53, 66, 67, 85, 129, 446 |
| Methylomonas rubra (AF150807)d | 26, 76, 126, 618 | 11, 32, 110, 250, 443 | 53, 66, 67, 85, 129, 446 |
| Isolate LW13 | 76, 126, 644 | 32, 360, 454 | 53, 66, 67, 85, 129, 446 |
| Isolate LW15 | 76, 125, 644 | 11, 32, 360, 442 | 66, 67, 85, 181, 446 |
| Isolate LW16 | 76, 126, 644 | 11, 32, 360, 443 | 53, 66, 67, 85, 129, 446 |
| Isolates LW19 and LW21 | 76, 126, 644 | 11, 32, 110, 250, 443 | 66, 67, 85, 182, 446 |
| Environmental clone pAMC434 | 76, 126, 644 | 11, 32, 360, 443 | 66, 67, 85, 182, 446 |
| Environmental clone pAMC435 | 76, 126, 316, 329 | 11, 32, 361, 443 | 66, 67, 85, 182, 446 |
| Environmental clone pAMC462 | 76, 126, 644 | 11, 32, 110, 250, 443 | 67, 182, 151, 446 |
| Methylosinus spp. | |||
| Methylosinus trichosporium OB3b (AF150804)d | 37, 62, 115, 172, 278, 280 | 8, 86, 151, 155, 255, 289 | 80, 85, 100, 186, 193, 300 |
| Methylosinus sp. strain LAC (M95664)c | 10, 37, 62, 115, 171, 267, 280 | 86, 151, 155, 263, 287 | 80, 85, 100, 186, 192, 299 |
| Isolate PW1 | 37, 115, 172, 280, 340 | 8, 86, 151, 155, 255, 289 | 34, 37, 66, 80, 85, 156, 186, 300 |
| Isolate LW2 | 37, 115, 170, 280, 338 | 8, 86, 149, 155, 255, 287 | 37, 80, 84, 100, 154, 186, 299 |
| Isolates LW3, LW4, and LW8 | 37, 115, 171, 280, 338 | 8, 86, 149, 155, 255, 288 | 34, 37, 66, 80, 85, 154, 186, 299 |
| Environmental clone pAMC447 | 37, 115, 172, 280, 340 | 8, 86, 151, 155, 255, 289 | 34, 37, 66, 80, 85, 156, 186, 300 |
| Environmental clone pAMC451 | 37, 62, 115, 172, 278, 280 | 8, 86, 151, 155, 255, 289 | 80, 85, 100, 186, 193, 300 |
| Environmental clone pAMC459 | 37, 62, 115, 172, 278, 280 | 8, 151, 155, 289, 341 | 80, 85, 100, 186, 193, 300 |
| Methylocystis spp. | |||
| Methylocystis sp. strain M (U81595)c | 37, 112, 114, 172, 226, 279 | 8, 149, 289, 494 | 80, 85, 100, 184, 191, 300 |
| Methylocystis parvus OBBP (AF150805)d | 37, 112, 115, 172, 226, 289 | 8, 86, 149, 155, 255, 289 | 37, 80, 85, 100, 154, 186, 300 |
| Isolate LW5 | 37, 112, 115, 172, 228, 280 | 8, 86, 151, 155, 255, 289 | 37, 80, 85, 100, 156, 186, 300 |
| Methylococcus spp. | |||
| Methylococcus sp. strain Texas (X72770)c | 2, 75, 126, 160, 165, 197, 278 | 177, 340, 486 | 34, 35, 59, 66, 151, 182, 476 |
| Methylococcus sp. strain Bath (X72771)c | 2, 76, 126, 160, 165, 197, 279 | 177, 341, 487 | 34, 35, 59, 67, 151, 182, 477 |
| Methylocaldum spp. | |||
| Methylocaldum tepidum (U89297)ce | 2, 160, 163, 197, 208, 280 | 8, 70, 177, 340, 415 | 34, 68, 92, 151, 187, 478 |
| Methylocaldum szegediense (U89300)ce | 2, 160, 164, 197, 208, 279 | 8, 66, 70, 176, 341, 349 | 34, 53, 92, 151, 202, 478 |
As determined by a phylogenetic analysis of sequences.
Boldface type indicates OTUs for the genera determined by using only the portion of the 16S rRNA that would be PCR amplified with the primers used in this study.
The data in parentheses are GenBank nucleotide sequence accession numbers. Patterns were predicted by using sequences deposited in the GenBank database.
Data obtained in this study.
Restriction fragments for the first 1,010 bp of 16S rRNA.
In most cases, the RFLP patterns observed with MspI digests were sufficient to differentiate between methanotroph genera. The genus Methylomonas was the only genus whose members exhibited a clearly distinct OTU in HaeIII-digested sequences. In addition, the enzyme HhaI produced patterns that were useful for differentiating between the type II methanotrophic genera, Methylosinus and Methylocystis. Within each genus, the patterns obtained for MspI- and HhaI-digested sequences were often very similar. In these cases the patterns observed with HaeIII digests were used to differentiate between different clones and pure cultures. The sequences in Table 2 were analyzed by using only those bases that would be amplified with the genus-specific primers used in this study. The nonmethanotrophic representatives of the α- and γ-Proteobacteria tested did not exhibit any methanotrophic OTUs when they were digested in silico (data not shown).
pmoA PCR products were also analyzed both in silico and experimentally with MspI, HaeIII, and HhaI. Although these enzymes were useful for distinguishing between pmoA genes from different strains, no genus-specific OTUs could be identified.
Characterization of 16S rRNA and pmoA genes in new Lake Washington methanotrophic isolates.
Twelve pure cultures that required methane for growth were obtained from enrichment cultures established with Lake Washington sediment (33a). Sequencing of the 16S rRNA genes of these isolates revealed one Methylobacter strain, five Methylomonas strains, one Methylocystis strain, and five Methylosinus strains. The OTUs predicted for the 12 Lake Washington strains (LW and PW strains) corresponded to the expected genera (Table 2). The pmoA genes of these isolates were also sequenced and screened by performing RFLP analyses. The results of an analysis of the pmoA sequences in the database in addition to our new pmoA sequences were used to design a primer specific for pmoA that should not amplify amoA (see above). The new pmoA primer, mb661 (Table 1), was tested with more than 10 amoA sequences available in the GenBank database and exhibited low levels of identity (9 to 12 mismatches) with these sequences. No product was obtained in PCRs in which Nitrosomonas europaea DNA was used.
Characterization of 16S rRNA and pmoA genes in natural methanotroph populations. (i) 16S rRNA gene sequences.
16S rRNA PCR products obtained by using target DNA extracted from Lake Washington sediment samples were used to construct gene libraries. The primers used to construct these libraries were the methanotroph phylogenetic group-specific primers described above and shown in Table 1 (17). A total of 200 randomly selected clones containing inserts were subjected to RFLP analyses and placed into groups based on their representative RFLP patterns. The 200 clones fell into 38 groups, only 15 of which contained more than one clone. All 38 groups were examined to determine whether any of the defined methanotrophic OTUs were present (Table 2). Based on this parameter, six groups were found to be groups that contained methanotrophic sequences. Clones representing each of these six groups were used for sequencing, and the data suggested that they were methanotroph 16S rRNA genes based on a comparison with other 16S rRNA genes. Ten clones that did not contain the defined methanotrophic OTUs were also used for partial sequencing. None of the additional 10 sequences were methanotrophic 16S rRNA gene sequences based on a comparison with other sequences in the RDP, which supported the validity of the OTU analysis.
The 16S rRNA gene sequences of the six methanotroph clones included four Methylobacter sequences (pAMC405, pAMC415, pAMC417, and pAMC419) and two Methylomicrobium sequences (pAMC421 and pAMC466) (Table 3). No sequences were obtained for the remaining six genera. However, representatives of the genera Methylomonas, Methylosinus, and Methylocystis were obtained as pure cultures that were isolated from the same sediment. Based on our sequence data for these isolates, we designed new primers to specifically amplify Methylomonas sequences and Methylosinus and Methylocystis sequences (Mm835 and Type2b, respectively) (Table 1). Additional gene libraries were constructed by using these primers. For each library, 50 clones were used in RFLP and OTU analyses. For the Methylosinus-Methylocystis library, six groups were obtained, and three of these had Methylosinus- type OTUs (pAMC447, pAMC451, and pAMC459) (Table 3). The 50 clones in the Methylomonas gene library fell into five groups, and three of these had the correct OTUs (pAMC434, pAMC435, and pAMC462) (Table 3). The six clones in the Methylosinus and Methylomonas gene libraries were sequenced. For each of these libraries, the clones that did not contain the appropriate OTUs were partially sequenced. None of the clones without the appropriate OTUs contained methanotrophic 16S rRNA genes. Our analysis of the environmental clones is summarized in Table 3. An environmental clone (pAMC434) identical to a Lake Washington isolate was obtained for one Methylomonas strain, and a clone (pAMC415) that differed by only 2 nucleotides from a Lake Washington isolate was obtained for a Methylobacter strain. No other clones exhibited such close identity with any of the Lake Washington isolates.
TABLE 3.
Grouping of 16S rDNA environmental clones from Lake Washington sediment
| 16S rRNA environmental clone | RDP similarity rank
|
Pure-culture representative | |
|---|---|---|---|
| Organism | Value | ||
| pAMC405 | Methylobacter luteus | 0.861 | None |
| pAMC415 | Methylobacter luteus | 0.875 | LW1a |
| pAMC417 | Methylobacter luteus | 0.869 | None |
| pAMC419 | Methylobacter whittenburyi | 0.714 | None |
| pAMC421 | Methylomicrobium agile | 0.788 | None |
| pAMC466 | Methylomicrobium agile | 0.782 | None |
| pAMC434 | Methylomonas methanica | 0.925 | LW15 |
| pAMC435 | Methylomonas methanica | 0.925 | None |
| pAMC462 | Methylomonas methanica | 0.830 | None |
| pAMC447 | Methylosinus sp. strain B-3060 | 0.825 | None |
| pAMC451 | Methylosinus sp. strain B-3060 | 0.841 | None |
| pAMC459 | Methylosinus sp. strain B-3060 | 0.847 | None |
Sequences differ at two nucleotides.
(ii) pmoA sequences.
The new pmoA-specific primers were used to amplify partial pmoA gene products from DNA extracted from Lake Washington sediment, and these PCR products were used to construct gene libraries. A total of 200 clones containing inserts were subjected to RFLP analysis with the tetrameric restriction enzymes MspI plus HaeIII and HhaI. The 200 clones fell into 34 groups, and only 8 of these groups contained more than one clone. Clones representing 24 of the groups were sequenced, and 15 of these clones were pmoA gene sequences. No amoA sequences were obtained. Pairwise comparisons of translated amino acid sequences for the pmoA PCR products obtained from environmental samples and from pure cultures indicated levels of identity ranging from 63.9 to 100% (Table 4). An examination of the nucleotide sequences from the same region revealed levels of identity ranging from 63 to 99.6% (Table 4). Analysis of this larger data set confirmed that it was not possible to identify OTUs for pmoA by using these RFLP profiles.
TABLE 4.
Levels of identity for the pmoA products of environmental clones and pure cultures of methanotrophs
| Clone or culture | Genus | % Identitya
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | ||
| 1. pAMC503 | Methylobacter | 97 | 95.9 | 93.5 | 98.2 | 78 | 79.3 | 85.8 | 88.2 | 88.2 | 87.6 | 87.6 | 89.3 | 89.3 | 66.3 | 69.2 | 68.6 | 66.9 | 66.9 | 88.8 | 88.8 | 88.2 | 87.6 | 87.1 | 78.5 | 63 | 51.6 | |
| 2. pAMC511 | Methylobacter | 93.3 | 95.3 | 96.4 | 95.3 | 78.1 | 79.9 | 83.4 | 85.8 | 85.8 | 85.2 | 85.2 | 87 | 87 | 65.1 | 68 | 67.5 | 65.7 | 65.7 | 86.4 | 86.4 | 86.4 | 85.2 | 84.7 | 79.1 | 61.7 | 52.2 | |
| 3. pAMC523 | Methylobacter | 87.8 | 87.4 | 93.5 | 95.3 | 79.3 | 79.3 | 84 | 86.4 | 86.4 | 85.8 | 85.8 | 87.6 | 87.6 | 65.7 | 68.6 | 68 | 66.3 | 66.3 | 87 | 87 | 86.4 | 85.8 | 85.3 | 80.4 | 63 | 52.9 | |
| 4. pAMC524 | Methylobacter | 87.6 | 94.3 | 86.8 | 91.7 | 78.7 | 78.1 | 84.6 | 87 | 87 | 86.4 | 86.4 | 85.8 | 85.8 | 66.9 | 69.2 | 68.6 | 67.5 | 67.5 | 85.2 | 85.2 | 85.2 | 86.4 | 85.9 | 79.1 | 63.6 | 51.6 | |
| 5. pAMC528 | Methylobacter | 94.7 | 90.6 | 88.2 | 87.8 | 76.3 | 78.1 | 85.2 | 87.6 | 87.6 | 87 | 87 | 89.3 | 89.3 | 65.1 | 68 | 67.5 | 65.7 | 65.7 | 88.8 | 88.8 | 88.2 | 87 | 86.5 | 77.3 | 61.7 | 52.9 | |
| 6. pAMC501 | Methylococcus | 70.9 | 69.9 | 70.7 | 70.1 | 69.7 | 92.3 | 74 | 74.6 | 74.6 | 75.1 | 75.1 | 75.1 | 75.1 | 67.5 | 65.1 | 64.5 | 68 | 68 | 74.6 | 74.6 | 74.6 | 75.1 | 73.6 | 92 | 63.6 | 54.1 | |
| 7. pAMC512 | Methylococcus | 68.5 | 70.7 | 68.7 | 69.5 | 68.9 | 78.2 | 73.4 | 74 | 74 | 74.6 | 74.6 | 74 | 74 | 66.3 | 65.1 | 64.5 | 66.9 | 66.9 | 73.4 | 73.4 | 73.4 | 74.6 | 73.6 | 92.6 | 61.7 | 56.1 | |
| 8. pAMC507s | Methylomicrobium | 79.5 | 79.5 | 76.8 | 78.5 | 79.1 | 72.4 | 69.7 | 97.6 | 97.6 | 98.2 | 98.2 | 84.6 | 84.6 | 65.1 | 66.9 | 66.3 | 64.5 | 65.7 | 84.6 | 85.2 | 84 | 98.2 | 97.5 | 74.2 | 61.7 | 49.7 | |
| 9. pAMC509 | Methylomicrobium | 79.7 | 80.1 | 77.4 | 79.5 | 79.3 | 72 | 69.7 | 98.8 | 100 | 99.4 | 99.4 | 85.8 | 85.8 | 66.9 | 68.6 | 68 | 67.5 | 67.5 | 85.8 | 86.4 | 85.2 | 99.4 | 98.8 | 74.8 | 63.6 | 51.6 | |
| 10. pAMC519 | Methylomicrobium | 79.5 | 79.7 | 77.2 | 79.1 | 78.9 | 72 | 69.7 | 98.4 | 99.6 | 99.4 | 99.4 | 85.8 | 85.8 | 66.9 | 68.6 | 68 | 67.5 | 67.5 | 85.8 | 86.4 | 85.2 | 99.4 | 98.8 | 74.8 | 63.6 | 51.6 | |
| 11. pAMC521 | Methylomicrobium | 79.7 | 79.7 | 77.4 | 79.1 | 79.3 | 72.4 | 70.3 | 99 | 99.4 | 99 | 100 | 85.8 | 85.8 | 66.9 | 68.6 | 68 | 67.5 | 67.5 | 85.8 | 86.4 | 85.2 | 100 | 99.4 | 75.5 | 63.6 | 51.6 | |
| 12. pAMC526 | Methylomicrobium | 79.9 | 80.3 | 77.2 | 79.3 | 79.5 | 72.4 | 69.7 | 99.2 | 99.6 | 99.2 | 99.4 | 85.8 | 85.8 | 66.9 | 68.6 | 68 | 67.5 | 67.5 | 85.8 | 86.4 | 85.2 | 100 | 99.4 | 75.5 | 63.6 | 51.6 | |
| 13. pAMC507 | Methylomonas | 82.5 | 80.3 | 79.9 | 79.1 | 81.9 | 71 | 67.9 | 77.6 | 78.1 | 78.1 | 78.1 | 78 | 100 | 63.9 | 65.1 | 64.5 | 64.5 | 64.5 | 99.4 | 98.2 | 97.6 | 85.8 | 85.3 | 74.2 | 60.5 | 51 | |
| 14. pAMC514 | Methylomonas | 82.7 | 80.5 | 80.1 | 79.3 | 82.5 | 71.4 | 67.9 | 78 | 78.1 | 78.1 | 78.1 | 78.3 | 99.4 | 63.9 | 65.1 | 64.5 | 64.5 | 64.5 | 99.4 | 98.2 | 97.6 | 85.8 | 85.3 | 74.2 | 60.5 | 51 | |
| 15. pAMC510 | Methylosinus | 63.5 | 63.4 | 63.9 | 64.4 | 64.2 | 71.2 | 67.6 | 66.3 | 66.9 | 67.3 | 67.3 | 66.7 | 64.8 | 64.8 | 91.7 | 92.3 | 98.8 | 98.8 | 63.9 | 64.5 | 63.9 | 66.9 | 65.4 | 67.9 | 90.8 | 48.1 | |
| 16. LW2 | Methylocystis | 64.5 | 64.1 | 65.2 | 64 | 65 | 67.4 | 68.4 | 65.9 | 66.3 | 66.7 | 66.9 | 66.3 | 65.5 | 65.5 | 88.2 | 98.2 | 91.7 | 91.7 | 65.1 | 65.7 | 65.1 | 68.6 | 67.9 | 65.4 | 87.1 | 47.5 | |
| 17. LW5 | Methylocystis | 64.2 | 63.3 | 64.5 | 63.1 | 64.1 | 68.1 | 69.2 | 65 | 65.4 | 65.8 | 65.9 | 65.4 | 65.4 | 65.4 | 88 | 93.9 | 92.3 | 92.3 | 64.5 | 65.1 | 64.5 | 68 | 67.3 | 65.7 | 89 | 48.2 | |
| 18. PW1 | Methylosinus | 63.7 | 63.3 | 64.3 | 64.1 | 63.9 | 71.4 | 67 | 66.5 | 67.1 | 67.5 | 67.5 | 66.9 | 64.4 | 64.4 | 97.8 | 87.8 | 88.2 | 98.8 | 64.5 | 65.1 | 64.5 | 67.5 | 66 | 68.5 | 90.8 | 48.7 | |
| 19. LW3 | Methylosinus | 63.4 | 63 | 64 | 63.8 | 63.6 | 71.5 | 66.9 | 67.3 | 67.9 | 68.3 | 68.3 | 67.7 | 64.9 | 64.9 | 97.4 | 87.8 | 88 | 98.4 | 64.5 | 65.1 | 64.5 | 67.5 | 66 | 68.5 | 90.8 | 48.7 | |
| 20. LW21 | Methylomonas | 83.3 | 82.1 | 80.9 | 81.3 | 82.7 | 71.4 | 67.1 | 78.1 | 78.3 | 78 | 78.3 | 78.5 | 92.3 | 92.9 | 63.4 | 64.3 | 64 | 63.9 | 63.7 | 98.2 | 97.6 | 85.8 | 85.3 | 73.6 | 60.5 | 51 | |
| 21. LW13, LW16, LW19 | Methylomonas | 82.5 | 81.5 | 79.7 | 80.5 | 82.1 | 70.1 | 67.3 | 78 | 78.1 | 78.1 | 78.1 | 78.3 | 90 | 90.6 | 63.5 | 63.7 | 63.4 | 63.3 | 63.1 | 94.5 | 98.2 | 86.4 | 85.9 | 73.6 | 61.1 | 52.2 | |
| 22. LW15 | Methylomonas | 83.5 | 82.5 | 80.5 | 81.3 | 82.7 | 71 | 67.9 | 77.8 | 78.7 | 78.3 | 78.3 | 78.5 | 90.9 | 91.1 | 63.6 | 64.3 | 64.4 | 63.3 | 63.4 | 95.1 | 93.9 | 85.2 | 84.7 | 73.6 | 60.5 | 51.6 | |
| 23. LW1 | Methylomicrobium | 79.9 | 80.3 | 77.6 | 79.7 | 79.5 | 72.2 | 69.9 | 98.8 | 99.6 | 99.2 | 99.4 | 99.6 | 78.3 | 78.3 | 66.9 | 66.5 | 65.6 | 67.1 | 67.9 | 78.5 | 78.3 | 78.9 | 99.4 | 75.5 | 63.6 | 51.6 | |
| 24. BG8 | Methylomicrobium | 79.5 | 79 | 75.8 | 78 | 78.4 | 71.2 | 68.5 | 98.8 | 99.2 | 98.4 | 98.6 | 99.2 | 77.6 | 76.9 | 65.4 | 65.2 | 63.9 | 65.8 | 66.4 | 77.1 | 76.7 | 77.1 | 98.8 | 75.2 | 62.8 | 51.6 | |
| 25. Mc | Methylococcus | 69.9 | 70.7 | 71.3 | 71.9 | 69.3 | 87 | 82.3 | 72.4 | 72.5 | 72.4 | 73 | 72.4 | 70.9 | 70.9 | 71.5 | 68.6 | 68.9 | 72 | 71.3 | 71.3 | 70.3 | 71.1 | 72.6 | 71.4 | 64 | 55.4 | |
| 26. OB3b | Methylosinus | 61 | 60.8 | 61.4 | 61 | 61.6 | 70.6 | 65.5 | 64 | 64 | 64.4 | 64.6 | 64.4 | 63.4 | 63.6 | 89.2 | 84.7 | 84.7 | 88.1 | 88.4 | 61.6 | 60.9 | 60.2 | 64.2 | 63.7 | 70.3 | 44.9 | |
| 27. N. europaea | Nitrosomonas | 60.2 | 59 | 57.8 | 57.8 | 60.3 | 60 | 62.6 | 59.6 | 60.2 | 60.3 | 60.2 | 60.2 | 60.9 | 60.6 | 60.7 | 59.6 | 58.1 | 61.8 | 60.7 | 61.3 | 60.6 | 62.4 | 60.4 | 58.8 | 62.6 | 57.1 | |
The values on the upper right are levels of amino acid identity for translated pmoA products, and the values on the lower left are levels of nucleotide identity.
The 15 environmental pmoA sequences were compared to previously described pmoA sequences and were found to group with sequences from members of previously described genera (Fig. 1). These sequences included one Methylosinus sequence, two Methylococcus sequences, five Methylomicrobium sequences, two Methylomonas sequences, and five Methylobacter sequences. When these sequences were examined, we identified two clones that exhibited 100% amino acid identity with a type I methanotrophic isolate from Lake Washington (LW1). The amino acid sequences of some clones were identical, but the nucleotide sequences were different. In these cases, both clones are shown in Table 3. For all of the environmental clones and Lake Washington isolates, the pmoA gene obtained exhibited a higher level of identity with other pmoA genes than with a homologous gene, amoA from Nitrosomonas europaea (Table 4). The levels of nucleotide sequence identity with amoA ranged from 57.8 to 62.6%, while the levels of amino acid identity with the amoA product were 47.5 to 56.1%.
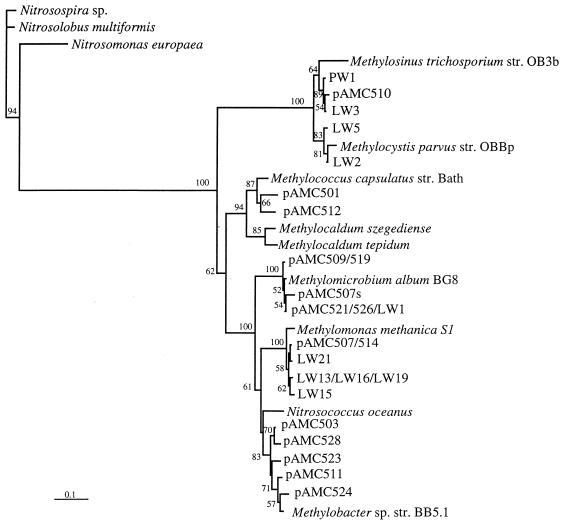
FIG. 1.
Phylogenetic analysis of the derived amino acid sequences encoded by pmoA genes. Bootstrap values greater than 50% based on 100 replicates are shown near the branch points. The bar represents 10% sequence divergence as determined by measuring the lengths of the horizontal lines connecting two species.
Phylogenetic analyses.
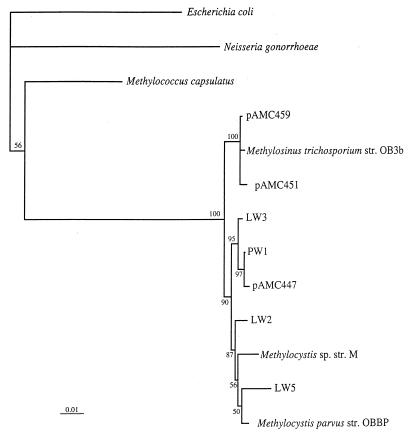
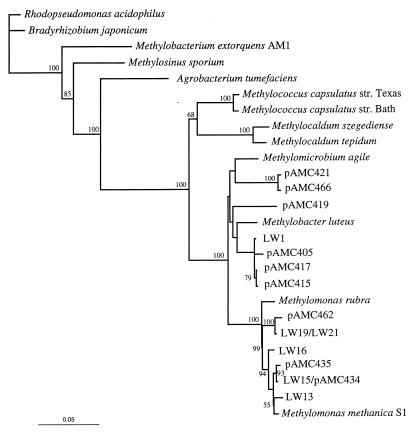
The 16S rRNA and pmoA sequences obtained from pure cultures and environmental clones were subjected to phylogenetic analyses by using PHYLIP. In general, most of the new sequences grouped within the range of the previously described sequences (Fig. 1 through 3). However, one group of 16S rRNA sequences formed a distinct new cluster in the type II methanotrophs, which was supported by bootstrap values (Fig. 3). This group comprised isolates LW3 and PW1 and clone pAMC447. The diversity of both the 16S rRNA and pmoA representatives was much greater than the diversity found previously in peat or marine environments and spanned the known diversity of methanotrophs, except that we found no 16S rRNA sequences that represented the genera Methylococcus, Methylosphaera, and Methylocaldum. However, we identified two environmental pmoA clones that grouped with the genus Methylococcus, although no Methylocaldum- or Methylosphaera-like pmoA sequences were found.
FIG. 3.
Phylogenetic analysis of 16S rRNA genes from type II methanotrophs. Bootstrap values greater than 50% based on 100 replicates are shown near the branch points. The bar represents 1% sequence divergence as determined by measuring the lengths of the horizontal lines connecting two species.
DISCUSSION
Methanotrophic bacteria are important environmentally due to their role in carbon and oxygen cycling, as well as their use in bioremediation strategies. In order to more fully apply molecular techniques associated with these important bacteria, more information regarding the diversity of in situ populations in various environments is needed. Molecular tools are especially important because many methanotrophs are difficult to isolate on agar plates, which makes growth-based assessment of natural populations problematic (15). The ability to rapidly assess and monitor natural populations of methanotrophs by using molecular techniques holds great promise for understanding the complex role of these bacteria in nature.
Although there are currently primers for studying both 16S rRNA and pmoA genes of methanotrophs, these primers have some disadvantages for studying natural populations of the organisms. The 16S rRNA primers currently available were based on a relatively small sequence database. In addition, our study showed that some of the previously described sequences on which the primers were based contain errors that make accurate primer design difficult. In our study, these primers detected only a small subset of the existing methanotroph diversity in Lake Washington samples, and there was specific underrepresentation of the type I Methylomonas strains and all of the type II strains (both Methylosinus and Methylocystis strains). The previously described type I primers, Mb1007r and Mc1005r (17), were found to be sufficient for detecting these groups of methanotrophs. The pmoA primers that are available have a disadvantage opposite that of the 16S rRNA primers in that they amplify both amoA and pmoA, which makes them too nonspecific for methanotroph-specific studies. Based on the sequences generated in this study, we designed new primers for methanotroph 16S rRNA and pmoA genes that appear to be more useful for studying methanotroph diversity in freshwater environments.
Using the newly developed primers (in addition to 16S rRNA primers Mb1007r and Mc1005r), we analyzed the 16S rRNA and pmoA genes in pure cultures isolated from Lake Washington and in environmental clone libraries obtained from the same sediment. We identified a broad diversity of both of these genes, including 13 new type I 16S rRNA genes, 7 new type II 16S rRNA genes, and 18 new pmoA genes, 5 of which grouped with pmoA sequences from type II strains. It is especially important to have additional type II gene data, as the database contains fewer type II sequences than type I sequences. However, it is equally important to have added environmental type I sequences to the database, as only two such sequences, both from marine environments, have been described. We did not detect any 16S ribosomal DNA (rDNA) sequences that grouped with the thermophilic methanotrophs belonging to the genera Methylococcus, Methylocaldum, and Methylothermus, nor did we detect any Methylosphaera-like sequences. Since Lake Washington sediment is a freshwater environment that stays at moderately low temperatures year-round (10 to 12°C), these results were not surprising.
So far, the phylogeny of the pmoA genes that have been described has mimicked the 16S rRNA phylogeny of the methanotrophs from which the pmoA genes were obtained. We observed the same correlation for the genes from new Lake Washington isolates described here. These combined results suggest that pmoA gene sequences may be useful in inferring 16S rRNA phylogeny of methanotrophs in situ (28). A comparison of the sequences from the environmental libraries of the methanotroph 16S rRNA and pmoA genes showed that the two types of sequences cover similar ranges of diversity, except that we did detect two pmoA sequences that are most similar to Methylococcus pmoA, even though no Methylococcus 16S rDNA sequences were detected.
In addition to the new methanotroph primers, we also identified genus level OTUs for methanotrophs. Since all of the strains and sequences tested in this study exhibited complete correlation with the OTUs, it seems likely that these OTUs will be useful tools for screening methanotrophic isolates and environmental clone libraries from a wide range of environments. In addition, the OTUs can also be useful for screening enrichment cultures for the presence of nonmethanotrophs as an aid in facilitating isolation and purification of methanotrophs. Even though all of the methanotroph-specific primers used in this study showed no other close matches with any of the other organisms in the database, nonmethanotrophic sequences were obtained with all of the primers when environmental DNA templates were used. In this study, many of the nonmethanotrophic 16S rRNA sequences obtained were chimeric. As yet, no reliable protocol to circumvent these problems is in use. However, in the case of the methanotrophs, our data suggest that the OTUs defined in this study can be used as initial screening tools to distinguish between methanotroph and nonmethanotroph sequences in 16S rRNA gene libraries constructed from environmental samples.
The use of the new tools, new sequences, primers, and OTUs developed in this study demonstrated that the methanotrophs in Lake Washington sediment samples that could be detected by the methods which we used exhibit diversity as broad as the diversity of the known methanotrophs from all mesophilic environments. These results contrast with the results of studies of peat environments, which appear to contain only a limited group of type II strains (23, 25), and marine environments, which appear to be dominated by a limited group of type I strains (17, 33). The genes from two of the Lake Washington strains isolated from enrichment cultures were also found in the environmental clone libraries, suggesting that these two strains may be significant in the in situ populations. This is especially true for strain LW1 since both a 16S rDNA sequence and a pmoA sequence that exhibited high levels of identity to the same genes in this strain were found in the clone libraries.
The types of analyses carried out in this study cannot provide information concerning the dominant groups of methanotrophs in situ due to the known problems associated with PCR-based approaches, including differential amplification, artifactual PCR products, and inhibition of PCR amplification by contaminants (35). However, we are now in a position to develop and test hybridization probes for assessing the relative importance of methanotroph subgroups and specific strains (such as strain LW1) in detectable methanotroph populations.
FIG. 2.
Phylogenetic analysis of 16S rRNA genes from type I methanotrophs. Bootstrap values greater than 50% based on 100 replicates are shown near the branch points. The bar represents 5% sequence divergence as determined by measuring the lengths of the horizontal lines connecting two species.
ACKNOWLEDGMENTS
This work was supported by a subcontract to DOE grant DE-AC05-960R22464 with Oak Ridge National Laboratory, managed by Lockheed Martin Energy Research Corp. A.M.C. was supported in part by a National Science Foundation graduate fellowship.
We thank John Murray, University of Washington, and Michaeleen Callahan, California Institute of Technology, for their assistance during this study.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Auman, A., and M. E. Lidstrom. Unpublished data.
- 1b.Auman, A., S. Stolyar, and M. E. Lidstrom. Unpublished data.
- 2.Bender M, Conrad R. Methane oxidation activity in various soils and freshwater sediments: occurrence, characteristics, vertical profiles and distribution on grain size fractions. J Geophys Res. 1994;99:16531–16540. [Google Scholar]
- 3.Bodrossy L, Holmes E M, Holmes A J, Kovacs K L, Murrell J C. Analysis of 16S rRNA and methane monooxygenase gene sequences reveals a novel group of thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch Microbiol. 1997;168:493–503. doi: 10.1007/s002030050527. [DOI] [PubMed] [Google Scholar]
- 4.Bodrossy L, Kovacs K L, McDonald I R, Murrell J C. A novel thermophilic methane-oxidising γ-Proteobacterium. FEMS Microbiol Lett. 1999;170:335–341. [Google Scholar]
- 5.Bowman J P, McCammon S A, Skerratt J H. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology. 1997;143:1451–1459. doi: 10.1099/00221287-143-4-1451. [DOI] [PubMed] [Google Scholar]
- 6.Bowman J P, Sly L I, Nichols P D, Hayward A C. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol. 1993;43:735–753. [Google Scholar]
- 7.Bowman J P, Sly L I, Stackebrandt E. The phylogenetic position of the family Methylococcaceae. Int J Syst Bacteriol. 1995;45:182–185. doi: 10.1099/00207713-45-1-182. [DOI] [PubMed] [Google Scholar]
- 8.Brusseau G A, Bulygina E S, Hanson R S. Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol. 1994;60:626–636. doi: 10.1128/aem.60.2.626-636.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz L A, Valklump J, Collins M L P, Brantner C A, Remsen C C. Activity of methanotrophic bacteria in Green-Bay sediments. FEMS Microbiol Ecol. 1995;16:1–8. [Google Scholar]
- 10.Chandler D P, Li S-M, Spadoni C M, Drake G R, Balkwill D L, Fredrickson J K, Brockman F J. A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol Ecol. 1997;23:131–144. [Google Scholar]
- 11.Cicerone R J, Oremland R S. Biogeochemical aspects of atmospheric methane. Global Biogeochem Cycles. 1988;1:61–86. [Google Scholar]
- 12.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 13.Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons; 1991. pp. 177–203. [Google Scholar]
- 14.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson R S, Hanson T E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes A J, Costello A, Lidstrom M E, Murrell J C. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- 17.Holmes A J, Owens N J P, Murrell J C. Detection of novel marine methanotrophs using phylogenetic and functional gene probes after methane enrichment. Microbiology. 1995;141:1947–1955. doi: 10.1099/13500872-141-8-1947. [DOI] [PubMed] [Google Scholar]
- 18.King G M. Ecological aspects of methane oxidation, a key determinant of global methane dynamics. In: Marshall K C, editor. Advances in microbial ecology. New York, N.Y: Plenum Press; 1992. pp. 431–474. [Google Scholar]
- 19.Kuivila K M, Murray J W, Devol A H, Lidstrom M E, Reimers C E. Methane cycling in the sediments of Lake Washington. Limnol Oceanogr. 1988;33:571–581. [Google Scholar]
- 20.Lidstrom M E, Somers L. Seasonal study of methane oxidation in Lake Washington. Appl Environ Microbiol. 1984;47:1255–1260. doi: 10.1128/aem.47.6.1255-1260.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald I R, Hall G H, Pickup R W, Murrell J C. Methane oxidation potential and preliminary analysis of methanotrophs in blanket bog peat using molecular ecology techniques. FEMS Microbiol Ecol. 1996;21:197–211. [Google Scholar]
- 24.McDonald I R, Holmes A J, Kenna E M, Murrell J C. Molecular methods for the detection of methanotrophs. In: Sheehan D, editor. Methods in biotechnology. 2. Bioremediation protocols. Totowa, N.J: Humana Press, Inc.; 1998. pp. 111–126. [Google Scholar]
- 25.McDonald I R, Murrell J C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol Lett. 1997;156:205–210. doi: 10.1111/j.1574-6968.1997.tb12728.x. [DOI] [PubMed] [Google Scholar]
- 26.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene encoding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murrell J C, McDonald I R, Bourne D G. Molecular methods for the study of methanotroph ecology. FEMS Microbiol Ecol. 1998;27:103–114. [Google Scholar]
- 29.Page R D M. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 30.Rath J, Wu K Y, Herndl G J, Delong E F. High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol. 1998;14:261–269. [Google Scholar]
- 31.Reeburgh W S. “Soft spots” in the global methane budget. In: Lidstrom M E, Tabita F R, editors. Microbial growth on C1 compounds. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 334–342. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Smith K S, Costello A M, Lidstrom M E. Methane and trichloroethylene oxidation by an estuarine methanotroph, Methylobacter sp. strain BB5.1. Appl Environ Microbiol. 1997;63:4617–4620. doi: 10.1128/aem.63.11.4617-4620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Stolyar, S., A. Auman, and M. E. Lidstrom. Unpublished data.
- 34.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 36.Weidner S, Arnold W, Pühler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]