Abstract
Background and Objectives:
Honey has excellent antibacterial properties against various microorganisms of several different species. To date, there is no comparative evaluation of the antibacterial activity of Jarrah honey (JH), Kelulut Madu honey (KMH), Gelam honey (GH), and Acacia honey (AH) with that of Manuka honey (MH). The purpose of this study was to conduct such study and to compare the antibacterial activity of JH, KMH, GH, and AH with that of MH against Pseudomonas aeruginosa and Streptococcus pyogenes.
Materials and Methods:
Activity was assessed using broth microdilution, time kill viability, microtiter plate, scanning electron microscope (SEM) and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR).
Results:
The susceptibility tests revealed promising antibacterial activities of all honeys against both bacteria. The MICs of JH, KMH, GH, and AH ranged from 20% to 25% compared to MH (12.5%) against both bacteria. The MBCs of JH, KMH, GH, and AH ranged from 20% to 50% compared to MH (20%) against both bacteria. Treatment of both bacteria with 2× MIC (Minimum inhibitory concentration) of MH, JH, KMH, GH, and AH for 9 hours resulted in reduction in colony-forming unit (CFU/ml). SEM images showed that the morphological changes, cell destruction, cell lysis and biofilm disruption in both bacteria after exposure to all honeys. RT-qPCR analysis revealed that the expression of all genes in both bacteria were downregulated following treatment with all honeys. Among the all-tested honeys, MH showed the highest total antibacterial and antivirulence activities.
Conclusion:
Our results indicate that all honeys activity included inhibition of both bacteria due to a decrease in expression of essential genes associated with both bacteria, suggesting that all honeys could potentially be used as an alternative therapeutic agent against certain microorganisms particularly against P. aeruginosa and S. pyogenes.
Keywords: Honey, Pseudomonas aeruginosa, Streptococcus pyogenes, Gene expression profiling, Real-time polymerase chain reaction
INTRODUCTION
The biofilm trait of high antimicrobial resistance to antibiotics and disinfectants is a multifactorial and is attributed to slow antibiotic penetration, reduced microbial growth rates, persisters and unique physiology (1). Bacterial biofilms are normally pathogenic and can cause nosocomial infections. The National Institutes of Health (NIH) reported that among all microbial and chronic infections, 65% and 80%, respectively, are associated with biofilm development (2). Bacterial infections are becoming more difficult to treat due to higher numbers of patients with multiple underlying conditions and the rise in pathogens, which are resistant to modern antimicrobial treatments (3). This is difficult with a rarity of new antibiotics in development and has urged renewed interest in several novel antimicrobial therapeutics. Part of the challenge in treating bacterial infections is biofilm formation. When bacteria exist as a biofilm, they are significantly less sensitive to antibiotics; this is a result of metabolic changes to cells within the biofilm and structural features influencing drug permeability (4). The development and range of antibiotic resistance are an alarming threat to the effective treatment and inhibition of bacterial infections in humans and animals (4). Solving this problem requires searching for natural antimicrobial alternatives (5). Presently, more researchers are turning their attention to conventional medicines as a possible source of antimicrobial agents (6). Honey is one of the oldest traditional remedies that has been extremely reputed and extensively utilised for the treatment of various human infections for over 2000 years ago (7). Nowadays, different kinds of honey have been used in several nations as an alternative to pharmaceutical products for treating infected, burn wounds and contaminated. The antimicrobial properties of honey may be attributed to many factors, including high osmolarity, acidity, in addition to the presence of hydrogen peroxide (H2O2 ) and non-peroxide components, such as methylglyoxal (8). Honey’s composition is reliant on the environmental and geographical areas from which the original nectar was collected (8). This is attributed to natural variations in floral sources and climatic conditions at different locations (8). Therefore, several researchers have investigated the therapeutic effects of kinds of honey obtained from different geographical areas worldwide (9, 10). In addition, some honey varieties have been implicated in the differential expression of a number of genes essential for bacterial survival and virulence, including those involved in stress tolerance, virulence factor production, as well as multicellular behaviors, such as biofilm formation, and quorum sensing (11, 12). The present study aimed to investigate the effects of five kinds of honey on P. aeruginosa and S. pyogenes with a view to better understanding its potential to impact virulence and to compare the antibacterial activity of Jarrah honey (JH), Kelulut Madu honey (KMH), Gelam honey (GH), and Acacia honey (AH) with that of Manuka honey (UMF +10 (MH).
MATERIALS AND METHODS
Honey samples.
Manuka honey (UMF +10 (MH), Jarrah honey (JH), Kelulut Madu honey (KMH), Gelam honey (GH), and Acacia honey (AH) were purchased from commercial supplier. The samples were packed and sealed in amber glass bottles and stored at 4°C in the dark until processed (13).
Microorganisms and culture conditions.
A reference strains of P. aeruginosa ATCC 15692 and S. pyogenes ATCC 49399 were obtained from the American Type Culture Collection (ATCC). P. aeruginosa and S. pyogenes were stored at −80°C in nutrient broth (NB) medium (Oxoid, UK) with 20% (v/v) glycerol. Prior to each assay, P. aeruginosa and S. pyogenes strains were sub-cultured from the frozen stock preparations onto nutrient agar (NA) plates (Oxoid, UK). The plates were incubated at 37°C for 24 hours. Pure liquid cultures (pre-inocula) of P. aeruginosa and S. pyogenes were maintained in NB (13, 14).
Agar well diffusion assay.
The inoculum density of P. aeruginosa and S. pyogenes was adjusted to be 0.5 McFarland. A sterile cotton swab was dipped into the bacterial suspension and was rotated onto the tube with firm pressure to remove excess fluid. The swab was streaked over the entire surface of Muller Hinton agar plate (Oxoid, UK) for three times and each time the plate was rotated approximately 90°C to ensure even distribution. A sterile 9 mm cork borer was used to create six wells of agar plate. The wells of agar plate were added with 150 μL of 100%, 75%, 50%, and 25% (w/v) concentrations of MH, JH, KMH, GH and AH. Distilled water was used as a negative control. The plates were incubated at 37°C for 24 hours. Digital venire calliper was used to measure the zones of inhibition (13, 14).
Minimum inhibitory concentration (MIC).
The concentrations of MH, JH, KMH, GH and AH; 50%, 25%, 12.5%, 6.25%, 3.125% and 1.562% (w/v) were freshly prepared with NB broth. The minimum inhibitory concentration (MIC) value was determined using broth microdilution method. Briefly, the cell density for both bacteria was adjusted to be 1 × 108 CFU/mL. A 100 μL was transferred into microtiter plate with 100 μL of each concentration of MH, JH, KMH, GH and AH. Broth medium only was used as negative controls and inoculum without honey was served as positive controls. The plates were incubated overnight at 37°C. Absorbance was measured by using the microtiter plate reader (Tecan Infinite 200 PRO, Austria) at 540 nm. The MIC50 and MIC90 were determined by using the following formula as mentioned below (13, 14).
Minimum bactericidal concentration (MBC).
MBC test was performed after MIC assay via streak plate method. A 20 μL from each well of the microdilution method was taken and plated onto NA plates. Subsequently, the plates were incubated for 24 hours at 37°C. MBC was considered as the lowest antimicrobial concentration that produced no colony growth (13, 14).
Time-kill studies.
The inhibitory concentration (2×MIC) of MH, JH, KMH, GH and AH that was chosen for subsequent experiments was 2×MIC, because it was two times the MIC. The effect of MH, JH, KMH, GH and AH on the viability of the cells was determined by time-kill curve studies. By inoculating 100 μL of 1×106 CFU/mL of both bacteria into 10 mL NB with and without 2×MIC of each honey. Then, the samples were incubated at 37°C in a shaking water bath (100 rpm) for 9 hours. After incubation time, the samples were collected every 3 hours up to 9 hours. Then, the mean of Log10 CFU/ml over time were plotted for each sample. Subsequently, the log reduction (LR) was calculated for each sample by subtracting the Log10 CFU at zero time and the Log10 CFU at 9 hours of incubation to determine the TVCs (13, 14).
Biofilm assessment.
Different concentrations of MH, JH, KMH, GH and AH; 15%, 30%, 45%, and 60% (w/v) in NB were freshly prepared from a stock solution of 100% (w/v). Both bacteria were adjusted to be 0.5McFarland within 0.05 to 0.10 at 600 nm wavelength using spectrophotometer. Then, 200 μl of the culture was dispensed into wells of microtiter plate and incubated at 37°C for 48 hours. After biofilms were formed, 100 μL of planktonic cells were removed and replaced with 100 μL of each honey concentrations. Then, the plates were incubated for overnight (18 hours). Biofilm without honey treatment was served as a positive control, broth only was employed as a sterility control, and honey with broth was served as a corresponding negative control. Finally, after incubation time was done, the media were then removed by invertip the plate and tapping the plate. The plate was washed three times with PBS to remove free-floating planktonic bacteria and was then drained inverted for drying. The plates were stained with 200 μl of 0.1% crystal violet for 5 minutes. Then, the plates were rinsed under running tap water to remove excess stain and were dried at room temperature before solubilizing the biofilm with 95% of ethanol. Microplate reader (Tecan Infinite 200 PRO, Austria) was used to measure the optical density at 595 nm wavelength. Percentage of biofilm degradation was calculated by following formulas as described below (13, 15).
Scanning electron microscope (SEM) of single-species biofilm.
P. aeruginosa and S. pyogenes were cultivated in NB for 24 hours at 37°C and adjusted to be equal 0.5McFarland. Centrifugation at 3,500g for 10 min at room temperature was used to collect cells and suspended in NB with MICs of MH, JH, KMH, GH and AH for 24 hours. Inoculums without adding honey were used as a positive control. Pellets were collected, fixed overnight with 2.5% (v/v) glutaraldehyde in 0.01 M phosphate buffer solution (PBS). 0.1 M sodium cacodylate buffer were used to rinse the samples. After that, 1% osmium tetroxide in 0.1 M sodium cacodylate buffer were used to rinse the samples. 0.1 M sodium cacodylate were used again to rinse the samples. Subsequently, 0.01 PBS was used to wash the samples and underwent serial dehydration with ascending concentrations of ethanol and subjected to critical point drying. The samples were coated with platinum, placed onto the copper stage holder and examined by scanning electron microscope (SEM) (JEOL 6360LA, Japan) (13).
Scanning electron microscope (SEM) of mixed-species biofilm.
The effects of MH, JH, KMH, GH and AH on mixed-species biofilm was determined using SEM. Briefly, P. aeruginosa and S. pyogenes cell suspensions were adjusted to be equal to 0.5 McFarland, 1:1 mixed-species were prepared in sterile NB and 200 μL of this standard, cell suspension was added into microtiter plate and then incubated for 24 hours at 37°C. After incubation time was done, the liquid phase was replaced by 200 μL of MIC of each honey. Biofilm mixed-species without honey treatment was used as a positive control. Then, the plates were incubated for 24 hours at 37°C. Subsequently, all samples were then centrifuged at 3500 rpm for 5 minutes. SEM procedure was followed as described earlier. The samples were then viewed by SEM (13).
RNA extraction for RT-qPCR.
A0.5 McFarland of P. aeruginosa and S. pyogenes cells were treated with MIC of MH, JH, KMH, GH and AH. Meanwhile, positive control was included inoculum without honey. Then, the incubation time was performed at 37°C for 8 hours in a shaking (100 rpm). Subsequently, one ml of treated and untreated cells was separated and centrifuged at 13,000 rpm for 2 minutes. The supernatant was discarded and the pellet was washed with PBS. Total RNA extracted using kit SV Total RNA Isolation System (Promega, UK) according to the manufacturer’s instructions. Total RNA concentrations were examined by ImplenNanoPhotometer® NP80. Total RNA samples were converted to cDNA according to the manufacturer’s instructions (Promega, UK). Samples were diluted to 100ng/μl using ultra-pure water. For each reaction, qPCR master-mix was prepared by following the manufacturer’s instructions (Promega, UK) and PCR primers were used as shown in Tables 1 and 2. The following PCR protocol was used: denaturation at 95°C for 2 minutes in one cycle, amplification at 95°C for 15 seconds in 40 cycles and a final elongation annealing at 60°C for 1 min in 40 cycles. Densitometry was performed using the Applied Biosystems StepOne Software v2.3. To determine and calculate the level of gene expression, a modified 2−ΔΔ Ct method was used (13, 16–18).
Table 1.
Gene specific primers of P. aeruginosa used for RT-qPCR analysis
| Gene name | Amplicon Size (bp) | Annealing temp (C°) | Direction | Primer sequence (5’ → 3’) | References |
|---|---|---|---|---|---|
| fliA | 132 | 55 | Forward | CTCCAATTGAGCCTCGAAGA | (13, 19 ) |
| Reverse | TTCGTTGTGACTGAGGCTGG | ||||
| fliC | 121 | 55 | Forward | GCTTCGACAACACCATCAAC | (13, 19 ) |
| Reverse | AGCACCTGGTTCTTGGTCAG | ||||
| flhF | 127 | 54 | Forward | CGAGCCTGAACGTGAAGAAT | (13, 19 ) |
| Reverse | GCCTCGTCCAGCTTAGTCA | ||||
| fleN | 137 | 56 | Forward | GAGCCGTATACGAGGCATTC | (13, 19 ) |
| Reverse | GTGTTGGACCAGTCGTTCG | ||||
| fleQ | 134 | 54 | Forward | AAGGACTACCTGGCCAACCT | (13, 19 ) |
| Reverse | CCGTACTTGCGCATCTTCTC | ||||
| fleR | 109 | 55 | Forward | ACAGCCGCAAGATGAACCT | (13, 19 ) |
| Reverse | TGGATGGCGTTGTCGAGTT | ||||
| rpoD* | 146 | 53 | Forward | GCGACGGTATTCGAACTTGT | (13, 19 ) |
| Reverse | CGAAGAAGGAAATGGTCGAG |
Reference gene
Table 2.
Gene specific primers of S. pyogenes used for RT-qPCR analysis
| Gene name | Amplicon Size (bp) | Annealing temp (C°) | Direction | Primer sequence (5’ → 3’) | References |
|---|---|---|---|---|---|
| Sof | 873 | 57 | Forward | ACTTAGAAAGTTATCTGTAGGG | (13, 19 ) |
| Reverse | TCTCTCGAGCTTTATGGATAG | ||||
| sfbI | 960 | 55 | Forward | AACTGCTTTAGGAACAGCTTC | (13, 19 ) |
| Reverse | CCACCATAGCCACAATGCT | ||||
| scpA | 622 | 55 | Forward | GCTCGGTTACCTCACTTGTCC | (13, 19 ) |
| Reverse | CAATAGCAGCAAACAAGTCACC | ||||
| ftsY | 97 | 54 | Forward | TCGAAAATTCTTTGGCCTGT AT- | (13, 19 ) |
| Reverse | CAAACGTGTTGTGCCAGA | ||||
| glr * | 797 | 54 | Forward | ATGGATACAAGACCAATTGG | (13, 19 ) |
| Reverse | TCATAAGGTGACATGCTCCAC |
Reference gene
Statistical analysis.
For all assays, all experiments were carried out in triplicate. All data were expressed as mean ± standard deviation. Independent student t-test from (SPSS version 20) was used to compare between treated and untreated groups. The statistical analyses performed were considered significant when P< 0.05.
RESULTS
Agar well diffusion assay.
Inhibition zone for MH, JH, KMH, GH and AH against P. aeruginosa and S. pyogenes is mentioned in Tables 3 and 4. All tested honeys were observed to have antibacterial activity against both bacteria. In general, all tested honeys showed a measurable antibacterial activity on both bacteria with different values. MH, JH, KMH, GH and AH showed a significant inhibition zone against both bacteria at 100%, 75%, 50% and 25% concentrations.
Table 3.
Antibacterial activity (Inhibition Zone (mm) ± SD) of all tested honeys at different concentrations against P. aeruginosa
| Honey samples | 100% | 75% | 50% | 25% |
|---|---|---|---|---|
| MH | 25.3 ± 0.6 | 21.4 ± 0.1 | 18.6 ± 0.2 | 14.6 ± 0.3 |
| JH | 19.2 ± 0.4 | 17.2 ± 0.4 | 14.1 ± 0.6 | 13.0 ± 1.0 |
| KMH | 25.1 ± 0.6 | 21.3 ± 0.5 | 17.1 ± 0.4 | 11.6 ± 0.3 |
| GH | 20.2 ± 0.4 | 16.0 ± 0.6 | 12.1 ± 0.5 | 11.5 ± 0.2 |
| AH | 19.7 ± 0.5 | 18.0 ± 0.4 | 12.1 ± 0.6 | 11.4 ± 0.1 |
Table 4.
Antibacterial activity (Inhibition Zone (mm) ± SD) of all tested honeys at different concentrations against S. pyogenes
| Honey samples | 100% | 75% | 50% | 25% |
|---|---|---|---|---|
| MH | 25.1 ± 0.5 | 21.2 ± 0.1 | 18.4 ± 0.3 | 14.1 ± 0.4 |
| JH | 18.4 ± 0.5 | 16.1 ± 0.2 | 13.1 ± 0.5 | 12.0 ± 0.8 |
| KMH | 24.1 ± 0.2 | 20.1 ± 0.3 | 16.7 ± 0.3 | 10.8 ± 0.7 |
| GH | 19.4 ± 0.2 | 15.4 ± 0.1 | 11.7 ± 0.4 | 10.6 ± 0.5 |
| AH | 18.6 ± 0.7 | 17.2 ± 0.2 | 11.6 ± 0.3 | 10.3 ± 0.3 |
Determination of MICs, MICs90, MICs50 and MBCs.
As shown in Table 5, the MIC value for MH, JH, KMH, GH and AH against planktonically grown P. aeruginosa was 12.5%, 25%, 20%, 20% and 20% (w/v) respectively. The MBC value for MH, JH, KMH, GH and AH against planktonically grown P. aeruginosa was 20%, 50%, 25%, 25% and 50% (w/v) respectively. In addition, the MIC90 for MH, JH, KMH, GH and AH against planktonically grown P. aeruginosa was 20%, 50%, 25–50%, 25% and 50% (w/v) respectively. The MIC50 for MTH against planktonically grown P. aeruginosa was 12.5%, 20%, 20–25%, 20% and 25% (w/v) respectively.
Table 5.
MIC, MIC90, MIC50 and MBC of all tested honeys against P. aeruginosa
| Honey samples | MIC % (w/v) | MIC 90 % (w/v) | MIC 50 % (w/v) | MBC % (w/v) |
|---|---|---|---|---|
| MH | 12.5% | 20% | 12.5% | 20% |
| JH | 25% | 50% | 20% | 50% |
| KMH | 20% | 25–50% | 20–25% | 25% |
| GH | 20% | 25% | 20% | 25% |
| AH | 20% | 50% | 25% | 50% |
From Table 6, the MIC value for MH, JH, KMH, GH and AH against planktonically grown S. pyogenes was 12.5%, 25%, 20%, 20% and 20% (w/v) respectively. The MBC value for MH, JH, KMH, GH and AH against planktonically grown S. pyogenes was 20%, 50%, 25%, 50% and 50% (w/v) respectively. In addition, the MIC90 for MH, JH, KMH, GH and AH against planktonically grown S. pyogenes was 25%, 50%, 25–50%, 25–50% and 50% (w/v) respectively. The MIC50 for MTH against planktonically grown S. pyogenes was 20%, 20%, 20–25%, 20% and 25% (w/v) respectively.
Table 6.
MIC, MIC90, MIC50 and MBC of all tested honeys against S. pyogenes
| Honey samples | MIC % (w/v) | MIC 90 % (w/v) | MIC 50 % (w/v) | MBC % (w/v) |
|---|---|---|---|---|
| MH | 12.5% | 25% | 20% | 20% |
| JH | 25% | 50% | 20% | 50% |
| KMH | 20% | 25–50% | 20–25% | 25% |
| GH | 20% | 25–50% | 20% | 50% |
| AH | 20% | 50% | 25% | 50% |
Time-kill studies.
The total number of P. aeruginosa cells significantly decreased when exposed to 2×MIC MH, JH, KMH, GH, and AH. However, P. aeruginosa incubated with 2×MIC MH, JH, KMH, GH, and AH demonstrated rapid loss of viability. Therefore, after exposure to 2×MIC of MH, JH, KMH, GH and AH, P. aeruginosa resulted in 1.7-log10, 1.3-log10, 1.5-log10, 1.4-log10 and 1.2-log10 reduction in CFU/ml compared to untreated cells at 6 hours incubation (P<0.05) respectively. P. aeruginosa incubated with 2×MIC MH, JH, KMH, GH, and AH demonstrated that the greatest bactericidal activity at 9 h incubation with >3-log10 killing unit for MH and JH and with >2.5-log10 for KMH, GH and AH. The change in cell count in P. aeruginosa treated and untreated cells was statistically significant (Fig. 1).
Fig. 1.
Time-kill studies of P. aeruginosa and S. pyogenes after exposed to all honeys. Asterisks; *P<0.05 indicate statistically significant difference between treated and control samples.
The number of S. pyogenes cells decreased after following treatment with 2×MIC MH, JH, KMH, GH, and AH with 1.7-log10, 1.3-log10, 1.3-log10, 1.5-log10, and 1.3-log10, reduction in CFU/ml (≈99% killing) respectively at 6 h. The mean difference between treated and untreated S. pyogenes cells was statistically significant (P <0.05). However, 2×MIC MH, JH, KMH, GH, and AH achieved a 2.5-log10, 2.3-log10, 2.2-log10, 2.4-log10, and 2.7-log10 reduction (≈99% killing) in S. pyogenes population at 9 h (Fig. 1).
Sub-inhibitory concentrations of all tested honeys decreased the biofilm biomass.
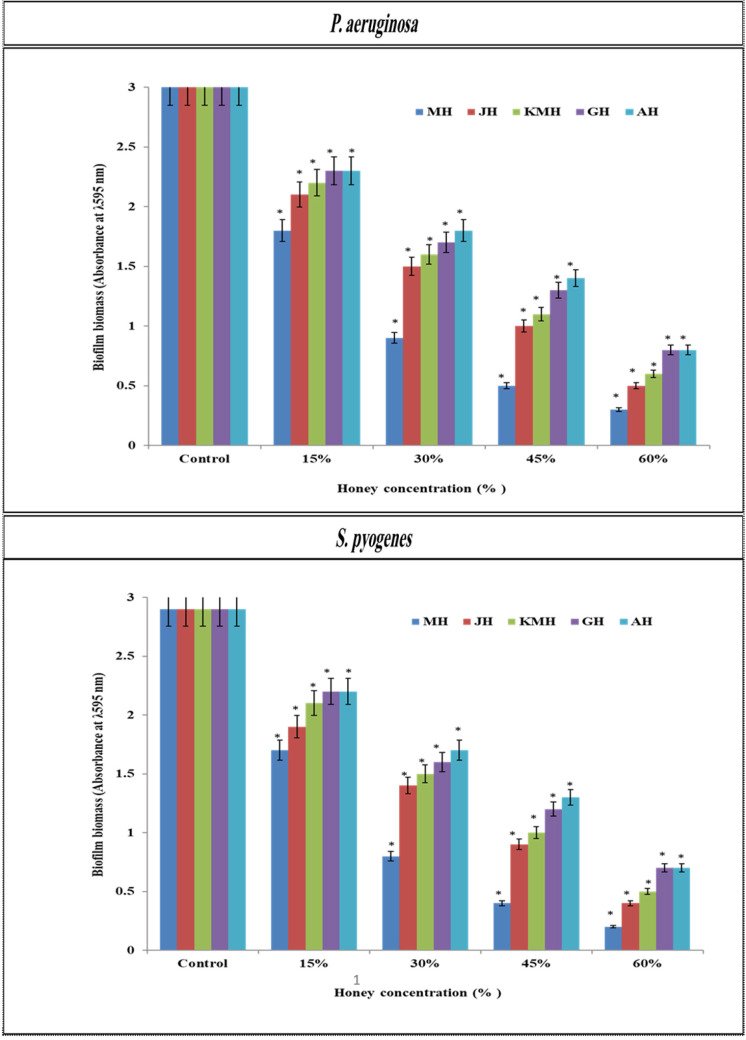
The average of optical density (OD) for control sample and tested sample of biofilm mass was calculated. The effect of MH, JH, KMH, GH and AH on biofilm biomass varied depending on the MH, JH, KMH, GH and AH concentrations. A statistically significant (P<0.05). When MH, JH, KMH, GH and AH were used at the 15% (w/v), the optical density of P. aeruginosa biofilm biomass was reduced to 1.8, 2.1, 2.2, 2.3, and 2.3 respectively compared to untreated biofilm. However, at 30% (w/v) MH, JH, KMH, GH and AH, the optical density of P. aeruginosa biofilm biomass was reduced to 0.9, 1.5, 1.6, 1.7, and 1.8 respectively compared to untreated biofilm. Meanwhile, the optical density of P. aeruginosa biofilm biomass was reduced to 0.5, 1.0, 1.1, 1.3, and 1.4 respectively compared to untreated biofilm at 45% (w/v). At 60% (w/v) MH, JH, KMH, GH and AH was more effective at reducing the optical density of P. aeruginosa biofilm biomass by 0.3, 0.5, 0.6, 0.8 and 0.8 respectively compared to untreated biofilm (Fig. 2).
Fig. 2.
Biofilm formation of P. aeruginosa and S. pyogenes grown with and without all honeys. Asterisks; * P<0.05 indicate statistically significant difference between treated and control samples.
In the presence of 15%, 30%, 45% and 60% (w/v) MH, JH, KMH, GH and AH concentrations, the optical density of established S. pyogenes biofilms was significantly (P < 0.05) decreased compared to untreated biofilm. After MH, JH, KMH, GH and AH was used at the 15% (w/v), the optical density of established S. pyogenes biofilms was reduced to 1.7, 1.9, 2.1, 2.2, and 2.2 respectively compared to untreated biofilm. However, at 30% (w/v) MH, JH, KMH, GH and AH, the optical density of established S. pyogenes biofilms was reduced to 0.8, 1.4, 1.5, 1.6, and 1.7 respectively compared to untreated biofilm. Meanwhile, at 45% (w/v) MH, JH, KMH, GH and AH, the optical density of established S. pyogenes biofilms was reduced to 0.4, 0.9, 1.0, 1.2, and 1.3 respectively compared to untreated biofilm. In addition, at 60% (w/v) MH, JH, KMH, GH and AH was more effective at reducing the optical density of established S. pyogenes biofilms by 0.2, 0.4, 0.5, 0.7, and 0.7 respectively compared to untreated bio-film. It was observed that the lowest concentration of MH, JH, KMH, GH and AH prevented S. pyogenes to establish biofilm was found to be 15% (w/v). Remarkably, MH was the most effective in preventing formation of S. pyogenes and P. aeruginosa biofilm. The inhibiting effect of MH, at low concentrations (15%), on the formation of S. pyogenes and P. aeruginosa biofilm was greater than that of the other honeys (Fig. 2).
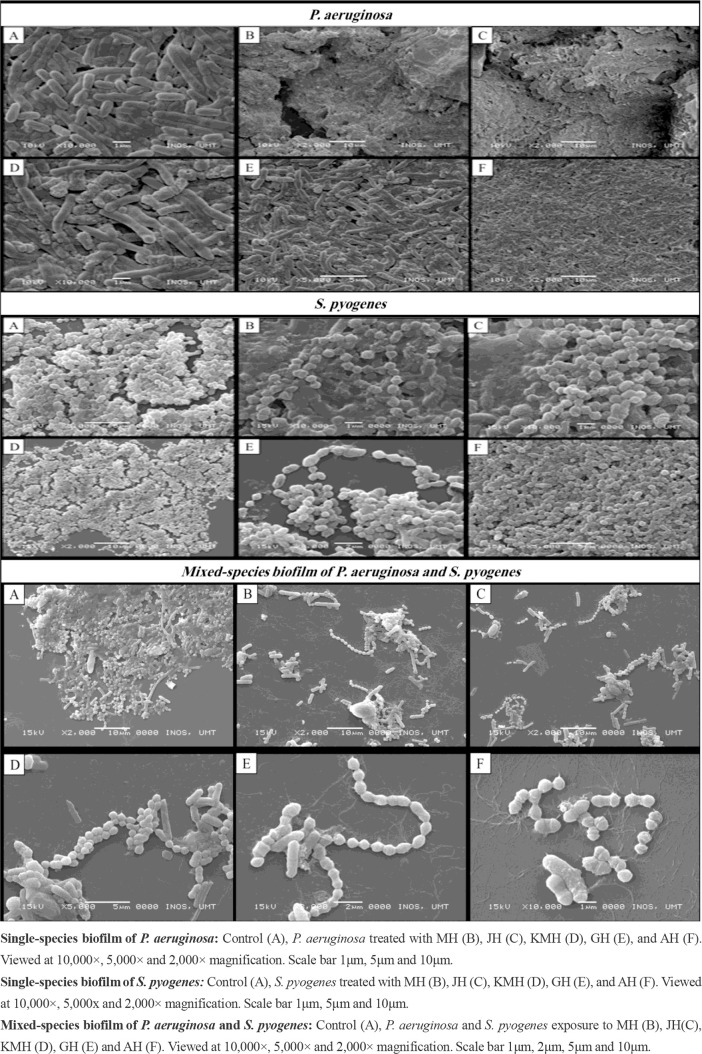
Scanning electron microscope (SEM) of single-species biofilm.
SEM micrographs of untreated P. aeruginosa cells showed that the cells appeared to be rod-shaped with regular structure and hundreds of bacterial cells are connected by a substantial amount of extracellular matrix. Extensive structural changes in biofilms were seen following treatment with all honeys and loss of viability was found, in addition to loss of biofilm structure. SEM images provided reasonable evidence of damage and disruption of the integrity of biofilm after exposure to all honeys. In addition, rough cell surfaces were observed after treated with all honeys (Fig. 3).
Fig. 3.
SEM of single and mixed-species biofilm of P. aeruginosa and S. pyogenes after exposure to all honeys
SEM micrographs of untreated S. pyogenes demonstrated the regular cocci with chain structure and S. pyogenes biofilm shows numerous cocci cells and diverse thickness connected each other. When S. pyogenes following treatment with all honeys the density of biofilm formed were reduced compared to untreated. In addition, several morphological changes, such as changes of cocci shape, abnormal cell division and ruptured cell structure were observed after exposed to all honeys (Fig. 3).
Scanning electron microscope (SEM) of mixed-species biofilm.
SEM showed that the surface structure and morphology of mixed-species biofilms formed by P. aeruginosa and S. pyogenes without honey treatment. The control group of mixed-species biofilms showed the typical multilayer growth of bacterial biofilms, while the group treated with MH, JH, KMH, GH and AH demonstrated that exhibited a reduction of mixed-species biofilm, reduction of cell density, and decrease extracellular matrix compared to control cells for both bacteria. Altogether, the findings provide evidence that MH, JH, KMH, GH and AH have a potent antibiofilm action against the mixed-species biofilm (Fig. 3).
Characterization of virulence factor activity indicated that honeys were able to reduce activity of several key virulence factors. In the present study, RT-qPCR was used to assess and compare the expression of six genes in P. aeruginosa that have been previously shown to be involved in the flagella regulon proteins, biofilm formation, motility and virulence of the microorganism and four genes in S. pyogenes that have been previously shown to be involved in the fibronectin binding proteins, surface adhesins and biofilm formation after exposure to all honeys.
Effects of five tested honeys on the mRNA expression of P. aeruginosa.
Following treatment of P. aeruginosa with MIC of MH, JH, KMH, GH and AH there were significant reductions (P<0.05, P≤ 0.01 and P≤ 0.001) in the relative abundance of mRNA for fliA, fliC, flhF, fleN, fleQ and fleR when compared to untreated cells. When P. aeruginosa treated with MH, the fold change ranged from 3.2-fold to 6.3-fold (for fleR, fliA, flhF, fleQ, fliC and fleN respectively). Also, when P. aeruginosa treated with JH, the fold change ranged from 2.7-fold to 4.5-fold (for fliA, fleR, flhF, fleQ, fliC and fleN respectively). In addition, when P. aeruginosa treated with KMH, the fold change ranged from 1.9-fold to 4.1-fold (for fleR, fliA, fleQ, flhF, fleN, and fliC respectively). Furthermore, when P. aeruginosa treated with GH, the fold change ranged from 1.5-fold to 3.8-fold (for fleR, fliA, fleQ, flhF, fleN, and fliC respectively). When P. aeruginosa treated with AH, the fold change ranged from 1.3-fold to 2.8-fold (for fleR, fliA, fleQ, flhF, fleN, and fliC respectively). Comparing MH honey treated P. aeruginosa cell samples with JH, KMH, GH and AH, the expression of mRNA transcripts for each gene tested with MH was decreased more than other honeys (Fig. 4).
Fig. 4.
Changes in gene expression profiles of P. aeruginosa and S. pyogenes after treated with all tested honeys as determined by RT-qPCR. Mean values of fold changes (± SD) are shown in relation to untreated S. pyogenes cells. Asterisks; * P<0.05; **P ≤ 0.01; and ***P≤ 0.001 indicate statistically significant difference in the expression of each gene between treated and untreated samples.
Effects of five tested honeys on the mRNA expression of S. pyogenes.
The major genes encoding the surface adhesins for scpA, ftsY, sfbI, and sof in S. pyogenes were downregulated after exposure to MIC of MH, JH, KMH, GH and AH. Following treatment of S. pyogenes with MIC of MH, JH, KMH, GH and AH there were significant reductions (P<0.05, P≤ 0.01 and P≤ 0.001) in the relative abundance of mRNA for scpA, ftsY, sfbI, and sof when compared to untreated cells. When S. pyogenes treated with MH, the fold change ranged from 4.4-fold to 6.8-fold (for scpA, ftsY, sfbI, and sof respectively). Also, when S. pyogenes treated with JH, the fold change ranged from 3.6-fold to 5-fold (for scpA, ftsY, sfbI, and sof respectively). In addition, when S. pyogenes treated with KMH, the fold change ranged from 3.7-fold to 6.1-fold (for scpA, ftsY, sof and sfbI respectively). When S. pyogenes treated with GH, the fold change ranged from 3.4-fold to 4.8-fold (for scpA, ftsY, sfbI, and sof respectively). Whereas, when S. pyogenes treated with GH, the fold change ranged from 2.8-fold to 4.3-fold (for scpA, ftsY, sfbI, and sof respectively). Comparing MH honey treated S. pyogenes cell samples with JH, KMH, GH and AH, the expression of mRNA transcripts for each gene tested with MH was decreased more than other honeys (Fig. 4).
DISCUSSION
Antibacterial activity of honey has been broadly discussed among researchers worldwide. It is postulated to be closely on several factors such as, osmolarity, pH and other major constituents such as phenolic acids and flavonoids (20). Previous study showed that 25% concentration of honey exhibited lower antibacterial action (21). The similarly or divergence of results might be due to several reasons such contain different level of active compounds including phenolic acids and flavonoids (22). Limitations of some antibacterial assay such as agar well-diffusion test were reported including the insensitivity in detecting low level of antimicrobial activity, variation in the experimental conditions and permeability of non-polar components (23, 24). Agar well-diffusion test may not be the most appropriate method to evaluate the antibacterial activity of honey. Micro-broth dilution was performed to determine the MIC for antibacterial activity of honey toward all the tested bacteria (25). The lowest concentration of honey solution needed to inhibit 99% of bacterial growth is considered to be MIC. The lowest concentration of honey required to kill at least 99% of the tested bacterial strains is defined as MBC (25). In the current study, the MIC values ranging from 12.5% to 20% against both bacteria and the MBC values ranging from 20% to 50% against both bacteria. Previous studies showed that the MIC for Algerian, Manuka and Egyptian clover honeys against P. aeruginosa was at 20% and MBC was at 25% (25–28). Recently, other studies revealed that MIC for Manuka honey on S. pyogenes was at 20% and MBC was at 25% (29, 30). A previous study showed that the MIC for Manuka honey against P. aeruginosa was at 12% and MBC was at 16% (31). Time-kill studies were used to determine the bactericidal or bacteriostatic actions of antimicrobials (32). It is investigated by plotting log10 CFU/mL over time (13). The log10 CFU/ml for P. aeruginosa and S. pyogenes exposure to all honeys were seen at 9 hours which is about >3-log10 of both bacteria were killed. All tested honeys were able to decrease biofilm biomass in both bacteria. However, this study found that the higher concentration of all honeys was necessary to complete elimination of established biofilm. Regarding to the results obtained for P. aeruginosa and S. pyogenes biofilms, a significant reduction was observed after 24 hour’s exposure to all honey at all concentrations were used. In the current study, SEM revealed that the morphological changes of cells, cells destruction, cells lysis and biofilm disruption in both bacteria following treatment with all honeys. In addition, SEM images showed that the treated mixed-species biofilm presented several damaged cells in both bacteria compared to untreated mixed-species biofilm. The previous study demonstrated that the structure of P. aeruginosa was influenced using Manuka honey (33). A study by Enany et al. (2015) pointed that Sidr honey disrupted the cell of S. aureus (34). As demonstrated by RT-qPCR, a number of genes fliA, fliC, flhF, fleN, fleQ and fleR have been previously shown to be involved in the process of microcolony, biofilm formation and motility in P. aeruginosa (13, 16). Also, a set of genes have been previously shown to play an important role in the adhesion and biofilm formation and quorum-sensing network of S. pyogenes, such as the sof, sfbl, emm13, scpA and ftsY genes (29). The current results revealed that all selected genes in both bacteria were downregulated following exposure to all honeys. Our results are in agreement with those of (16, 31), who reported down-regulation of multiple genes involved in microcolony, motility and biofilm formation in P. aeruginosa strain following exposure to manuka honey. Study by Maddocks et al. (2012) reported that downregulation of sof, and sfbl genes in S. pyogenes after exposure to Manuka honey (29). Previous study showed that five genes; fleN, fleQ, fleR, fliA and fliC in P. aeruginosa and five genes; sof, sfbl, scpA, ftsY and emm13 in S. pyogenes were reduced in gene expression following treatment with Tualang honey (13). Another study reported that ycfR (BhsA) and evgA genes of E. coli were upregulated in expression in the range of 2.2–4.19-fold and 1.09-fold respectively after treated with 25% concentration of Egyptian honey (6). Study by Roberts et al. (2014) showed that fliA, fliC, flhF, fleN, fleQ and fleR genes in P. aeruginosa were reduced in gene expression after treated with manuka honey (16). Previous study showed that tnaA and yjfO (bsmA) genes were downregulated in expression of E. coli in the range of 12.5-16.2-fold after treated with 25% concentration of Egyptian honey (6). It was noticed that all these studies that mentioned above are in agreement with our results. This indicates that the honey-induced alterations in the expression of this group of genes are most probably due to particular molecules contained in honey and not only due to their sugar content. Previous study suggested that the osmotic action of sugar combined with hydrogen peroxide and bee-derived antibacterial peptide defensin-1 is crucial for the antibiofilm activity of honey (35). In addition, this change in expression pattern may indicate variations in the phytochemical components and/or differences in the antimicrobial mechanisms of all honey on both bacteria (30). It is evident that honey is effective at inhibiting the growth of both bacteria, causing abnormal cell by reducing structural integrity to the point of cell lysis as mentioned in SEM results. To our knowledge, this is the first attempt to compare the impacts of Jarrah honey (JH), Kelulut Madu honey (KMH), Gelam honey (GH), and Acacia honey (AH) with that of Manuka honey (MH) on the tested organisms at both structural and molecular levels.
CONCLUSION
This is the first attempt study to compare the impacts of Jarrah honey (JH), Kelulut Madu honey (KMH), Gelam honey (GH), and Acacia honey (AH) with that of Manuka honey (MH) on the tested organisms at both structural and molecular levels. A reduction of P. aeruginosa and S. pyogenes cell growth in both planktonic and biofilm state was observed with all honey treatment. Comparing all honeys tested, for planktonic and biofilm cultures, Manuka honey (MH) had a higher effect on both bacteria. In this study, results indicate that the JH, KMH, GH, and AH may represent promising antibacterial, antibiofilm and anti-virulence agents for treatment and modulation of infections caused by P. aeruginosa and S. pyogenes compared with MH. Antibacterial and antibiofilm activities of all tested honeys against both bacteria, which were further supported by the morphological and structural investigations. However, understanding the behavior of P. aeruginosa and S. pyogenes species in polymicrobial biofilms is an important step in the clinical context and for the selection of the most efficient treatment. Because of this, the effect of all honeys was assessed on structure of mixed P. aeruginosa and S. pyogenes biofilms. The honeys were able to reduce both species in the mixed biofilm and were demonstrated to be a promising alternative for the treatment of infections caused by mixed species biofilms. The use of a natural product such as honey may be used in clinical practice, to prevent or even treat P. aeruginosa and S. pyogenes infections. This study, suggest that each honey could have a crucial derivatives compound that have the ability to effectively inhibit the biofilms of P. aeruginosa and S. pyogenes.
ACKNOWLEDGEMENTS
This work was supported by Universiti Sultan Zainal Abidin (UniSZA). We thank all the staff members of the Faculty of Health Sciences and Faculty of Pharmacy at Universiti Sultan Zainal Abidin and Al-Zaytoonah University of Jordan for their support and commitment.
REFERENCES
- 1.Al-Bakri AG, Mahmoud NN. Photothermal-induced antibacterial activity of gold nanorods loaded into polymeric hydrogel against Pseudomonas aeruginosa biofilm. Molecules 2019;24:2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamal M, Ahmad W, Andleeb S, Jalil F, Imran M, Nawaz MA, et al. Bacterial biofilm and associated infections. J Chin Med Assoc 2018;81:7–11. [DOI] [PubMed] [Google Scholar]
- 3.Dhingra S, Rahman NAA, Peile E, Rahman M, Sartelli M, Hassali MA, et al. Microbial resistance movements: an overview of global public health threats posed by antimicrobial resistance, and how best to counter. Front Public Health 2020;8: 535668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jolivet-Gougeon A, Bonnaure-Mallet M. Biofilms as a mechanism of bacterial resistance. Drug Discov Today Technol 2014;11:49–56. [DOI] [PubMed] [Google Scholar]
- 5.Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 2019;37:177–192. [DOI] [PubMed] [Google Scholar]
- 6.Wasfi R, Elkhatib WF, Khairalla AS. Effects of selected Egyptian honeys on the cellular ultrastructure and the gene expression profile of Escherichia coli. PLoS One 2016;11 (3):e0150984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pećanac M, Janjić Z, Komarčević A, Pajić M, Dobanovački D, Mišković SS. Burns treatment in ancient times. Med Pregl 2013;66:263–267. [PubMed] [Google Scholar]
- 8.Mavric E, Wittmann S, Barth G, Henle T. Identification and quantification of methylglyoxal as the dominant antibacterial constituent of Manuka (Leptospermum scoparium) honeys from New Zealand. Mol Nutr Food Res 2008;52:483–489. [DOI] [PubMed] [Google Scholar]
- 9.Tsuruda JM, Chakrabarti P, Sagili RR. Honey bee nutrition. Vet Clin North Am Food Anim Pract 2021;37:505–519. [DOI] [PubMed] [Google Scholar]
- 10.Anthimidou E, Mossialos D. Antibacterial activity of Greek and Cypriot honeys against Staphylococcus aureus and Pseudomonas aeruginosa in comparison to manuka honey. J Med Food 2013;16:42–47. [DOI] [PubMed] [Google Scholar]
- 11.Ng WJ, Ken KW, Kumar RV, Gunasagaran H, Chandramogan V, Lee YY. In-vitro screening of Malaysian honey from different floral sources for antibacterial activity on human pathogenic bacteria. Afr J Tradit Complement Altern Med 2014;11:315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins R, Burton N, Cooper R. Proteomic and genomic analysis of methicillin-resistant Staphylococcus aureus (MRSA) exposed to manuka honey in vitro demonstrated down-regulation of virulence markers. J Antimicrob Chemother 2014;69:603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-kafaween MA, Al-Jamal HAN, Hilmi ABM, Elsahoryi NA, Jaffar N, Zahri MK. Antibacterial properties of selected Malaysian Tualang honey against Pseudomonas aeruginosa and Streptococcus pyogenes. Iran J Microbiol 2020;12:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zainol MI, Mohd Yusoff K, Mohd Yusof MY.. Antibacterial activity of selected Malaysian honey. BMC Complement Altern Med 2013;13:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarawneh O, Abu Mahfouz H, Hamadneh L, Deeb AA, Al-Sheikh I, Alwahsh W, et al. Assessment of persistent antimicrobial and anti-biofilm activity of p-HEMA hydrogel loaded with rifampicin and cefixime. Sci Rep 2022;12:3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts AE, Maddocks SE, Cooper RA. Manuka honey reduces the motility of Pseudomonas aeruginosa by suppression of flagella-associated genes. J Antimicrob Chemother 2015;70:716–725. [DOI] [PubMed] [Google Scholar]
- 17.Jarrar YB, Jarrar Q, Abaalkhail SJ, Moh’d Kalloush H, Naser W, Zihlif M, et al. Molecular toxicological alterations in the mouse hearts induced by sub-chronic thiazolidinedione drugs administration. Fundam Clin Pharmacol 2022;36:143–149. [DOI] [PubMed] [Google Scholar]
- 18.Jarrar Y, Jarrar Q, Abu-Shalhoob M, Abed A, Sha’ban E. Relative expression of mouse Udp-glucuronosyl transferase 2b1 gene in the livers, kidneys, and hearts: the influence of nonsteroidal anti-inflammatory drug treatment. Curr Drug Metab 2019;20:918–923. [DOI] [PubMed] [Google Scholar]
- 19.A Al-Kafaween M, Mohd Hilmi AB, Al-Jamal HAN, A Elsahoryi N, Jaffar N, Zahri MK. Pseudomonas aeruginosa and Streptococcus pyogenes exposed to Malaysian Trigona honey in vitro demonstrated downregulation of virulence factor. Iran J Biotechnol 2020;18 (4):e2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornara L, Biagi M, Xiao J, Burlando B. Therapeutic properties of bioactive compounds from different honeybee products. Front Pharmacol 2017;8:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almasaudi S. The antibacterial activities of honey. Saudi J Biol Sci 2021;28:2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albaridi NA. Antibacterial potency of honey. Int J Microbiol 2019;2019:2464507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bang LM, Buntting C, Molan P. The effect of dilution on the rate of hydrogen peroxide production in honey and its implications for wound healing. J Altern Complement Med 2003;9:267–273. [DOI] [PubMed] [Google Scholar]
- 24.Molan PC. The antibacterial activity of honey: 1. The nature of the antibacterial activity. Bee world 1992;73:5–28. [Google Scholar]
- 25.Zainol MI, Mohd Yusoff K, Mohd Yusof MY. Antibacterial activity of selected Malaysian honey. BMC Complement Altern Med 2013;13:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas HA. Comparative antibacterial and antibiofilm activities of manuka honey and Egyptian clover honey. Asian J Appl Sci 2014;2:110–115. [Google Scholar]
- 27.Bouacha M, Ayed H, Grara N. Honey bee as alternative medicine to treat eleven multidrug-resistant bacteria causing urinary tract infection during pregnancy. Sci Pharm 2018;86:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenoy VP, Ballal M, Shivananda P, Bairy I. Honey as an antimicrobial agent against Pseudomonas aeruginosa isolated from infected wounds. J Glob Infect Dis 2012;4:102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddocks SE, Lopez MS, Rowlands RS, Cooper RA. Manuka honey inhibits the development of Streptococcus pyogenes biofilms and causes reduced expression of two fibronectin binding proteins. Microbiology (Reading) 2012;158:781–790. [DOI] [PubMed] [Google Scholar]
- 30.Mandal MD, Mandal S. Honey: its medicinal property and antibacterial activity. Asian Pac J Trop Biomed 2011;1:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts AEL, Maddocks SE, Cooper RA. Manuka honey is bactericidal against Pseudomonas aeruginosa and results in differential expression of oprF and algD. Microbiology (Reading) 2012;158:3005–3013. [DOI] [PubMed] [Google Scholar]
- 32.Boorn KL, Khor YY, Sweetman E, Tan F, Heard TA, Hammer KA. Antimicrobial activity of honey from the stingless bee Trigona carbonaria determined by agar diffusion, agar dilution, broth microdilution and time- kill methodology. J Appl Microbiol 2010;108:1534–43. [DOI] [PubMed] [Google Scholar]
- 33.Henriques AF, Jenkins RE, Burton NF, Cooper RA. The effect of manuka honey on the structure of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 2011;30:167–171. [DOI] [PubMed] [Google Scholar]
- 34.Zakaria AS. Mechanism of antibacterial action of honey on pathogenic wound bacterial strains: A proteomic analysis. Int Res J Pharm 2015;6:778–788. [Google Scholar]
- 35.Proaño A, Coello D, Villacrés-Granda I, Ballesteros I, Debut A, Vizuete K. The osmotic action of sugar combined with hydrogen peroxide and bee-derived antibacterial peptide Defensin-1 is crucial for the antibiofilm activity of eucalyptus honey. LWT 2021;136:110379. [Google Scholar]