Significance
Classic serine proteases are synthesized as inactive precursors that are proteolytically processed, resulting in irreversible activation. We report an alternative and reversible mechanism of activation that is executed by an inactive protease. This mechanism involves a protein complex between the serine protease HTRA1 and the cysteine protease calpain 2. Surprisingly, activation is restricted as it improves the proteolysis of soluble tau protein but not the dissociation and degradation of its amyloid fibrils, a task that free HTRA1 is efficiently performing. These data exemplify a challenge for protein quality control proteases in the clearing of pathogenic fibrils and suggest a potential for unexpected side effects of chemical modulators targeting PDZ or other domains located at a distance to the active site.
Keywords: HTRA1, serine protease, allostery, amyloid fibrils, tau
Abstract
Zymogen activation is a widely conserved regulatory principle across protease clans. It describes the irreversible activation by processing of the inactive zymogen precursor by an active protease. Here we report an alternative and reversible mechanism of protease activation, where the activator is an inactive protease. This mechanism involves the formation of an allosteric complex between the serine PDZ protease HTRA1 and the cysteine protease calpain 2. Surprisingly, the allosteric activation of HTRA1 is restricted to a subset of substrate conformations as it improves the proteolysis of soluble tau protein but not the dissociation and degradation of amyloid fibrils that are a prominent hallmark of Alzheimer's disease. These data exemplify an additional challenge for protein quality control factors such as HTRA1 in the clearing of pathogenic fibrils and suggest a potential for unexpected side effects of chemical modulators targeting allosteric sites.
The HtrA family of serine proteases is defined by homo-oligomeric architecture and C-terminal PDZ domains. These PDZ domains bind hydrophobic tetrapeptide motifs of exposed carboxyl termini and are implicated in the sensing of misfolded proteins, allosteric activation, substrate processing, and the dissociation of amyloid fibrils (1, 2). Deregulation of human HTRA1 is implicated in age-related eye diseases, arthritis, cancer, and familial ischemic cerebral small-vessel disease (3). In addition, HTRA1 is the only protease known to degrade key factors of Alzheimer’s disease, i.e., Aβ, tau, and ApoE4 (2, 4–6). Therefore, a critical issue for understanding the specific roles of HTRA1 in protein homeostasis concerns its regulatory features that are governed by the fundamental biochemical principles of allostery, cooperativity, and activation by oligomerization (7, 8). We have therefore explored the regulatory implications of its PDZ domain. We show that a variety of PDZ ligands serve as potent allosteric activators. In addition, our data reveal that a hetero-oligomeric complex consisting of the cysteine protease calpain 2 (CAPN2) and HTRA1 improves the degradation of soluble tau protein but not the dissociation and proteolysis of amyloid fibrils. This conformational selectivity may have wide implications for amyloid pathology and drugs targeting allosteric sites.
Results
Identification and Characterization of an HTRA1–CAPN2 Complex.
To identify candidate ligands of the PDZ domain of HTRA1, we screened a peptide library comprising 6,223 C termini of human proteins (9) (SI Appendix, Fig. S1A and Table S1). The top 200 hits of this screen included proteins that were previously reported as PDZ ligands, substrates, or inhibitors of HTRA1 such as the collagens COL1A1, COL2A1, and COL3A1 (spot numbers 681, 699, and 683, respectively) (10); tuberin (spot number 5630) (11); and macrophage migration inhibitory factor (12) (spot number 3438) (SI Appendix, Fig. S1). One interesting class of hits was the three calpain proteases (CAPN1, CAPN2, and CAPN3). CAPN2 was chosen for further analyses as this protease requires mM concentrations of Ca2+ ions for activation (13) that are rarely reached within human cells. We therefore expected that investigating the interaction between CAPN2 and HTRA1 might reveal a functional role for catalytically inactive CAPN2.
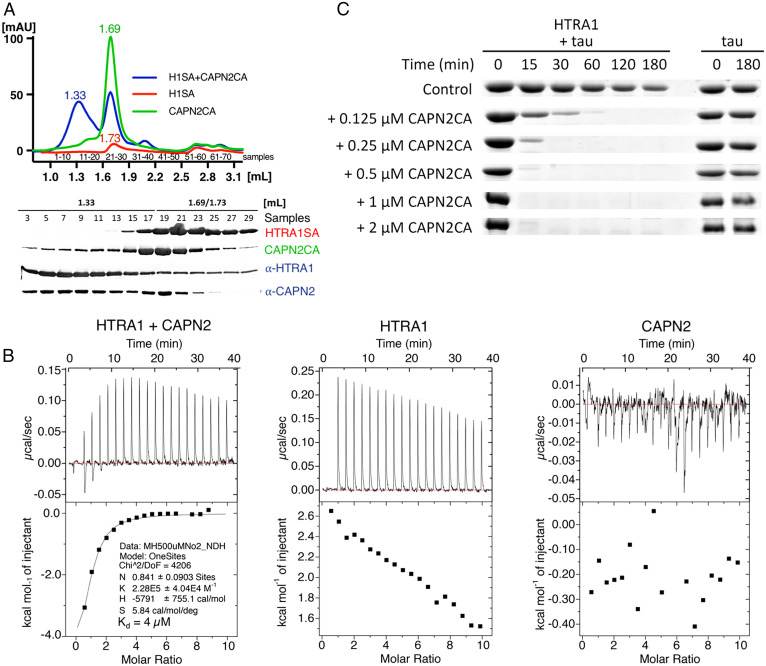
To initially explore this model, the HTRA1-CAPN2 interaction was characterized in vitro. The CAPN2 heterodimer consists of a cysteine protease and a regulatory subunit (13), while HTRA1 is a homotrimer (14). Incubation of purified proteins caused hetero-oligomeric complex formation as shown by size exclusion chromatography (Fig. 1A), and isothermal titration calorimetry (ITC) revealed a Kd of 4 µM (Fig. 1B). To investigate whether complex formation modulates HTRA1 activity, proteolytic digests were performed using a proteolytically inactive CAPN2 Cys105Ala mutant and the native HTRA1 substrate tau (2, 5). Titration experiments revealed dose-dependent activation of HTRA1 (Fig. 1C).
Fig. 1.
Characterization of the HTRA1–CAPN2 complex. (A) Top, size exclusion chromatography. Superdex200 5/150 GL column was used to determine elution profiles of HTRA1SA (H1SA) (red), CAPN2CA (green), and HTRA1SA plus CAPN2CA (blue) (HTRA1SA and CAPN2CA indicate the replacement of the active site Ser or Cys by Ala, respectively). Note that the absorption of HTRA1SA is low because it has no Trp residues. Bottom, sodium dodecylsulfate polyacrylamide gelelectrophoresis of relevant fractions followed by Western blotting. (B) ITC of HTRA1 and CAPN2. ITC measurements were performed to determine the Kd of the binding of CAPN2 to HTRA1 (Left). The individual proteins HTRA1 (Middle) and CAPN2 (Right) were used as controls. 500 µM HTRA1SA was titrated to 10 µM CAPN2CA in 50 mM bicine and 150 mM NaCl pH 9.0 at 37 °C. (C) Dose-dependent activation of HTRA1 by CAPN2CA. 0.5 µM HTRA1 was incubated with 5 µM tau in 50 mM Tris pH 8.0 at 37 °C for the time points indicated in the presence of the CAPN2 concentrations indicated.
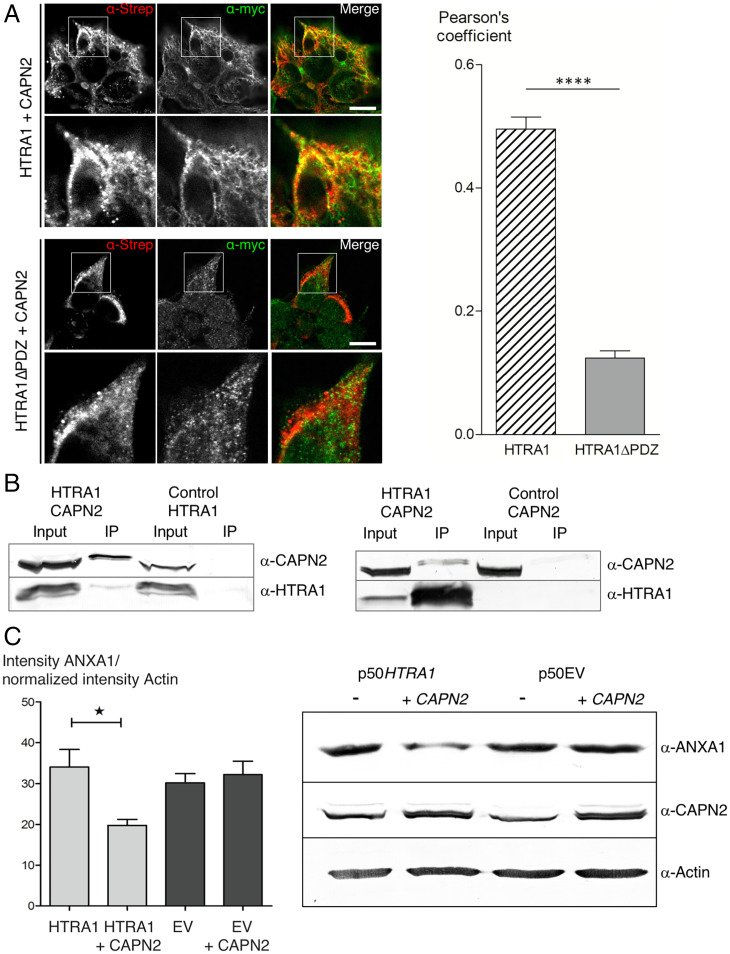
Additional experimental evidence for allosteric complex formation was obtained in cell-based assays (Fig. 2). Confocal microscopy of stably transfected cells coexpressing CAPN2 and HTRA1 or without its PDZ domain (HTRA1ΔPDZ), respectively, revealed a significant increase of colocalization in the presence of the PDZ domain (P < 10−4) (Fig. 2A and SI Appendix, Fig. S2). In addition, complex formation was supported by coimmunoprecipitations (Fig. 2B). Moreover, activation of HTRA1 was detected when determining protein levels of a native substrate, annexin A1 (ANXA1) (15). Here, cells stably overexpressing or not overexpressing HTRA1 were transfected or not transfected with a plasmid expressing CAPN2 followed by the detection of ANXA1 levels via Western blotting. These data indicated an almost twofold reduction of ANXA1 levels in the presence of overexpressed CAPN2 (P = 0.02) (Fig. 2C).
Fig. 2.
Cell based assays. (A) PDZ domain–dependent colocalization of HTRA1 with CAPN2 in HEK293 cells. Doxycycline (DOX)-inducible HEK293 cells expressing HTRA1 or HTRA1ΔPDZ, respectively, were treated with DOX and transiently transfected with CAPN2. Fixed cells were immunostained against the Strep tag of CAPN2 (red) and the Myc tag of HTRA1 (green) (Scale bar: 10 µm). Colocalization was quantified using the Pearson correlation coefficient. Data represent SEM of 15 images taken from 3 independent experiments per condition. ****P < 0.0001 (Mann–Whitney U test). (B) Coimmunoprecipitation of HTRA1 with CAPN2. Lysates (Input) of cells described in (A) were used for immunoprecipitation (IP) with Strep- or Myc-conjugated beads and immunoblotted using anti-HTRA1 and anti-CAPN2 antibodies. Representative data are shown (n = 3). (C) SW480 cells stably overexpressing HTRA1 (HTRA1) or not (EV) were transiently transfected with a CAPN2 plasmid for 24 h. Cell lysates were immunoblotted using anti-ANXA1, anti-CAPN2, and anti-actin antibodies. Protein levels were quantified by ImageJ, and the ANXA1 levels were normalized by actin. Data represent mean ± SEM of 6 experiments (*P = 0.01).
Diverse Effects of Allosteric Activation Related to Substrate Conformation.
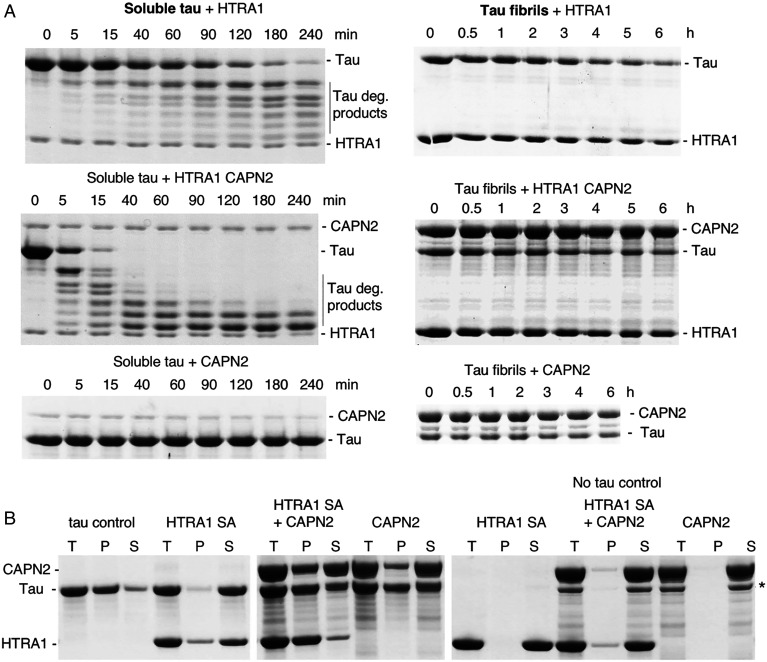
To obtain further insights into the functional implications of HTRA1 activation, we compared the proteolytic degradation of soluble and fibrillar tau conformations by HTRA1–CAPN2 complexes. While the degradation of soluble tau was improved, a slight decrease of HTRA1 activity was detected toward amyloid fibrils (Fig. 3A). HTRA1 dissociates tau fibrils for improved degradation (2). To investigate fibril dissociation independent of degradation, the proteolytically inactive Ser328Ala mutant (HTRA1SA) was used in fibril sedimentation assays. Fibrils were incubated with or without HTRA1SA alone, the HTRA1SA–CAPN2 complex, or CAPN2 alone and subjected to ultracentrifugation. Note that CAPN2 was inactive because Ca2+ ions were omitted from the buffer. The pellet fractions contain fibrils, while the supernatants contain solubilized tau (Fig. 3B). While HTRA1SA solubilized the tau fibrils, the HTRA1SA–CAPN2 complex was even less efficient compared to CAPN2 alone. Partial fibril dissociation by CAPN2 might be explained by the presence of tau binding sites because CAPN2 proteolyzes fibrils in the presence of Ca2+ (16). CAPN2 binding to fibrils is the likely reason for the observed co-sedimentation of CAPN2 and fibrils (Fig. 3B).
Fig. 3.
Proteolysis of tau protein. (A) Left, proteolysis of soluble tau. 5 µM tau was incubated with 0.5 µM HTRA1 or 0.5 µM CAPN2 or with HTRA1 CAPN2 at 37 °C in 50 mM Tris HCl, pH 8. Samples were taken at the time points indicated and analyzed by SDS-PAGE. Right, proteolysis of tau fibrils. Digests were performed as described for soluble tau except that 2.5 µM final concentration of tau fibrils and HTRA1/HTRA1-CAPN2 were used. (B) Sedimentation assay of tau fibrils. Tau fibrils were incubated with equimolar amounts of proteolytically inactive HTRA1 S328A (HTRA1SA) or CAPN2 or HTRA1 CAPN2 followed by ultracentrifugation. Subsequently, samples before centrifugation (T, total) of pellet (P) and supernatant (S) fractions were subjected to SDS-PAGE and Coomassie staining. *CAPN2 fragment. Note that CAPN2 does not digest tau because the buffers used did not contain Ca2+.
Mechanistic Insights into Allosteric Activation of HTRA1.
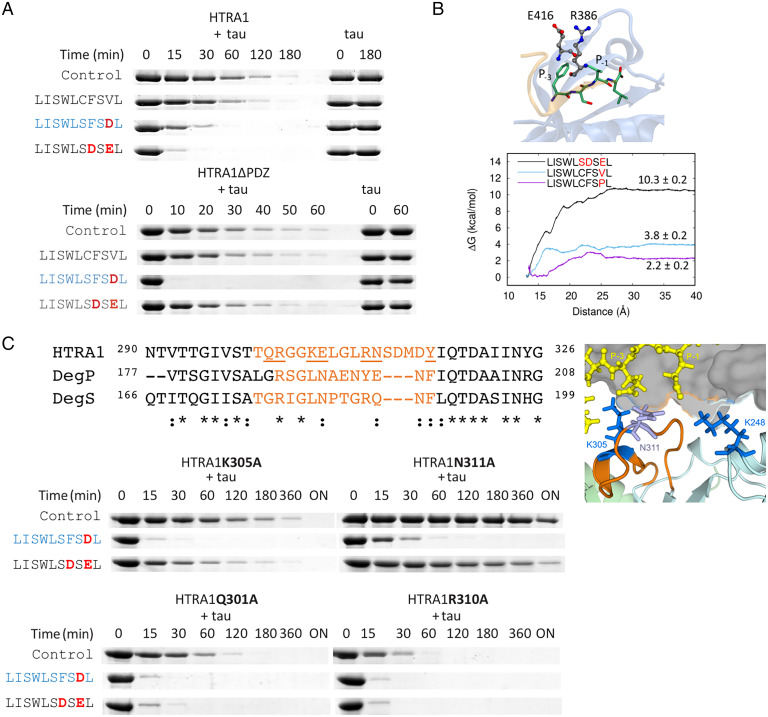
To obtain mechanistic insights, we used decameric peptides corresponding to the C terminus of CAPN2 (LISWLCFSVL) to study allosteric activation in detail (Fig. 4). As HTRA1 lacking its PDZ domain (HTRA1ΔPDZ) was not activated by the CAPN2 peptide, allostery was the most likely mechanism of activation (Fig. 4A). Correspondingly, PDZ binding sites were postulated for CAPN2, but binding partners remained elusive (17).
Fig. 4.
Effects of peptides derived from the C terminus of CAPN2 on HTRA1 activity. (A) Digests of soluble tau with HTRA1 and HTRA1ΔPDZ. 0.5 µM HTRA1 was incubated with 5 µM tau in 50 mM Tris pH 8.0 at 37 °C for the time points indicated in the absence (control) or presence of 50 µM of the peptides indicated. Deviations from the native CAPN2 sequence are indicated in red. The active-site ligand LISWLSFSDL is shown in blue. (B) Computational modeling and optimization of peptide sequences. Interaction between CAPN2 peptide and the PDZ domain. The mutation of residues P-1 and/or P-3 to Glu or Asp favors the formation of a salt bridge with Arg386 enhancing the binding of the peptide to the PDZ domain. The cartoon representation depicts the PDZ domain (blue) and peptide (beige), while the licorice representation shows the carbon atoms of residues of the PDZ domain (gray) and the peptide (green). Potential of mean force profiles, the reaction coordinate is the distance between the centers of mass of the PDZ domain and the peptides. (C) Loop L3 mutants implicated in the activation of HTRA1. Sequence alignment of the loop L3 sequences of HTRA1, DegP, and DegS. Activity of mutants determined by tau digests. 50 µM peptides were added when indicated. Cartoon model (Right) of LISWLSDSEL bound to the PDZ domain and its potential interactions with K305 and N311 of loop L3 and K248 of the protease domain based on high-resolution structures of the protease domain (PDB code: 3NZI) (loop L3, orange) and of the PDZ domain (gray) with the peptide DSRIWWV (yellow) (PDB code: 2JOA). Residues of loops of the protease domain pointing toward the bound peptide are shown as sticks.
To address the important question of sequence specificity, the CAPN2 peptide was characterized using docking calculations, molecular dynamics simulations, and free-energy calculations (SI Appendix, Fig. S3 and Supplementary Text). These computational studies suggested that negatively charged residues at the −1 and −3 positions of the CAPN2 peptide could favor the interactions with the Arg386 of the PDZ domain (Fig. 4B). To explore this model, we tested derivatives of the CAPN2 peptide, in which the −1 residue Val and the −3 residue Phe were exchanged by Asp or Glu. To avoid the formation of interpeptide S-S bonds, the −4 Cys residue was additionally replaced by Ser. Tau digests revealed that the ligand LISWLSDSEL was a more potent activator compared to the original CAPN2 peptide and showed that this peptide activated wild-type but not HTRA1ΔPDZ (Fig. 4A). Surprisingly, the −1 Val-to-Asp variant activated both wild-type and HTRA1ΔPDZ, suggesting that this peptide was converted from a PDZ into an active-site ligand. Note that ITC could not be performed with these CAPN2 peptides because of solubility issues.
The mechanism of allosteric activation is well understood for the bacterial HtrA proteases DegS and DegP (18, 19). A critical element is the sensor loop L3 of the protease domain, which repositions upon binding to the allosteric PDZ peptide. The interaction of the repositioned loop L3 with loop LD* (originating from an adjacent protomer) stabilizes the active conformation across protomers. However, one peculiarity of HTRA1 is that loop L3 can directly interact with substrates bound to the active site (14), while its involvement in allosteric activation cannot be addressed by X-ray crystallography because the PDZ domain cannot be visualized due to its flexibility (14, 20). To test the involvement of loop L3 in the allosteric activation of HTRA1, sequence alignments and available crystal structures were inspected to introduce point mutations (Fig. 4C). The resulting mutations, Lys305Ala and Asn311Ala, impaired allosteric activation but still allowed activation by the active-site ligand LISWLSFSDL. Therefore, this part of loop L3 is a candidate for direct interaction with allosteric peptides. One resulting mechanistic hypothesis of the improved allosteric activation by peptides harboring negatively charged residues at the −1 and −3 positions is that these charges may support the initial engagement of the peptide with the PDZ domain via their interaction with Arg386 of the PDZ domain. A subsequent tilt of the bound peptide toward loop L3 mediates allosteric activation, which is more pronounced when negatively charged residues at the −1 and −3 positions interact with residues Lys305 and Asn311 of loop L3 as well as with Lys248, located at another loop of the protease domain (Fig. 4C).
The selectivity of the loop L3 mutants Lys305Ala and Asn311Ala was addressed by control mutations within loop L3, i.e., Arg302Ala, Gln301Ala, and Arg310Ala. The proteolytic activity of none of these three mutants was affected by allosteric peptides. As Arg302 mediates the essential interaction of loops L3 and LD*, this mutant was completely inactive (SI Appendix, Fig. S4), while Gln301Ala and Arg310Ala were more active compared to wild-type HTRA1 even in the absence of ligands (Fig. 4C). The increased activity of Gln301Ala might be best explained by the disruption of the intramolecular interaction between Gln301 and Ser270 that stabilizes the position of loop L3 in the inactive conformation (14). Similar effects are expected for Arg310Ala; however, this part of loop L3 is invisible in the known crystal structures. Together, these data suggest that the involvement of loop L3 in allosteric activation is conserved between bacterial and mammalian HtrA proteases. However, the specific feature of HTRA1 is a larger loop L3 that allows the binding of both active-site and PDZ ligands to mediate the reversible switch between the inactive and active conformations.
General Aspects of Allosteric Activation.
Given the many interaction partners of HTRA1, it is likely that the allosteric interactions described above represent a general regulatory mechanism for fine-tuning the activity of HTRA1. In the cellular context, this mechanism is expected to involve the carboxyl-termini of various proteins that bind with different affinities to the PDZ domain and interact more or less productively with loop L3, leading to variable levels of activation. To test this hypothesis, additional hits of the peptide screen were analyzed, i.e., the carboxyl-termini of complement factor D (CFD) and cyclin H (CCNH) (SI Appendix, Table S1). In ITC, the CFD (SYAAWIDSVLA) and CCNH (EWTDDDLVESL) peptides exhibited Kds of approximately 4 µM and 40 µM, respectively, for HTRA1 and the isolated PDZ domain, while binding to HTRA1ΔPDZ was not detectable. In addition, both peptides activated HTRA1 but not HTRA1ΔPDZ in tau digests (SI Appendix, Fig. S5 A and B). Notably, the CCNH peptide, despite its weaker affinity, activated HTRA1 more strongly compared to the CFD peptide, supporting the model that binding affinity is not the only determinant of the level of activation.
To further examine the sequence preferences for activation, hybrid peptides of CFD and CCNH were generated in which the C-terminal 4 residues that interact with the ligand binding site of the PDZ domain were switched (SI Appendix, Fig. S5A and B). While ITC indicated that all hybrid peptides were PDZ ligands, the transfer of the C-terminal 4 residues of CFD onto CCNH and of CCNH onto CFD reduced the Kds and the levels of activation of CCNH and CFD in most cases. To explore whether the positive effect of negatively charged residues at the −1 position in PDZ binding sequences is a general phenomenon (Fig. 4A and C), Leu at the −1 position in the CFD and Ser in the CCNH peptides were each replaced by Asp, respectively. Increased levels of activation were observed for the CAPN2 peptides, suggesting that indeed, negatively charged residues at the −1 position have positive effects on activation.
Finally, the selectivity of three strongly activating peptides (two derivatives of CAPN2 and the unmodified CCNH peptide) was assessed toward the human homologs HTRA2 and HTRA3 in tau digests (SI Appendix, Fig. S6). Of these, LISWLSFSDL activated HTRA1 by interaction with the active site, while the others addressed its PDZ domain. The PDZ domain ligands had only minor or no effects on HTRA2 and HTRA3 activity, suggesting that the PDZ domain of HTRA1 may represent a promising target for chemical modulation of its proteolytic activity. In contrast, the active-site ligand activated HTRA3 strongly and HTRA2 weakly, supporting the general notion that PDZ domains represent preferred targets for future synthetic intervention.
Discussion
The C terminus of free CAPN2 has an alpha-helical conformation and interacts with the small regulatory subunit. As such, it cannot bind to the PDZ domain of HTRA1. Binding to the PDZ domain is thought to proceed in two steps. First, the PDZ domain of HTRA1 acts as an alternative and probably higher-affinity binding site. Second, the interaction of the alpha helix with the binding pocket of the PDZ domain is expected to occur via a beta-augmentation process that typically occurs during the binding of a substrate in helical conformation to the active site of a serine protease (1, 21). In addition, our docking simulations of the HTRA1–CAPN2 complex displayed multiple interaction sites between both proteins (SI Appendix, Fig. S7 and Table S2), which could explain the improved activation of HTRA1 by CAPN2 compared to the isolated C-terminal decapeptide (Figs. 1C and 4A).
The allosteric activation of HTRA1 by an inactive protease belonging to a different catalytic type expands our insights into the vast variety of regulatory processes employed by biological systems to maintain proteostasis. The particular case of the observed HTRA1–CAPN2 complex is reminiscent of the previously reported conformational switch in S-adenosylmethionine decarboxylase of Trypanosoma brucei triggered by the binding of its catalytically dead paralogous partner prozyme, resulting in allosteric activation (22). Future studies addressing the modulation of CAPN2 activity by posttranslational modifications such as phosphorylation and their effects on the activity and physiological roles of the HTRA1–CAPN2 complex would be of considerable interest (23).
There are several implications of our data. In the context of protein quality control, the HTRA1–CAPN2 complex results in the improved degradation of soluble tau proteins, potentially delaying the formation of amyloid fibrils. Our docking simulations of the HTRA1–CAPN2 complex displayed multiple interaction sites between both proteins (SI Appendix, Fig. S7 and Table S2), which could propitiate a tighter binding of HTRA1 to CAPN2 with respect to the isolated C-terminal decapeptide. However, allosteric activation is restricted to soluble substrate conformations as the dissociation and degradation of amyloid fibrils is reduced. The most likely underlying mechanism of this effect is that the PDZ binding sites of HTRA1 that are required for fibril dissociation (2) are occupied by the C termini of allosteric ligands such as CAPN2. It is therefore of interest to determine in future studies whether the improved degradation of soluble tau or the perturbing competition between allosteric ligands and amyloid fibrils for binding to the PDZ domains of HTRA1 has greater effects on the resulting levels of amyloid deposits. In any case, the clarification of these issues is likely to significantly expand existing models explaining the onset of amyloid diseases.
Allosteric drugs became popular because they are highly specific and rather modulate than abolish a target’s activity, features likely to coincide with reduced side effects (24). However, a so-far unexpected class of side effects should be considered as we show that the occupation of an allosteric site can have negative consequences on a subset of a target’s functions. Despite these restrictions, allosteric activators might be further developed into research tools that have several advantages. They would allow the artificial activation of HTRA1 without overproduction of the protease. Such activating ligands would complement the existing portfolio of synthetic inhibitors such as cyclodepsipeptides and peptidic boronic acids (14, 25). Moreover, because allosteric activators stabilize the active conformation, they could even be coupled to active site inhibitors and activity-based probes to thereby improve slow on-rates.
Materials and Methods
Protein purification, proteolytic digests of soluble tau and amyloid fibrils, ITC, immunoblots, and fluorescence microscopy were done as described previously (2). They are described in detail in the SI Appendix, which includes materials and methods and computational characterization of a peptidic allosteric activator.
Co-Immunoprecipitation.
We incubated 1 mg lysates of human embryonic kidney HEK293 cells expressing Myc-tagged HTRA1 and Strep-tagged CAPN2 at 4 °C with magnetic Strep or Myc beads for 24 h or 1 h, respectively. Subsequently, beads were washed 3 times with 50 mM Hepes pH 7.5, 300 mM NaCl, or 50 mM Tris pH 8.0, 150 mM NaCl. Bound proteins were eluted by boiling in sample buffer containing 100 µM DTT for 10 min. Samples were subjected to immunoblotting using antibodies against CAPN2 (Cell Signaling, #2539) and HTRA1 (Triple Point Biologics). In control experiments, only CAPN2 or HTRA1 were overproduced and subsequently immunoprecipitated using Strep or Myc beads, respectively, to show that these proteins do not bind to beads.
Computational Methods.
The interactions between the PDZ domain of HTRA1 and the peptides were studied using molecular dynamics (MD) simulations. The starting coordinates of the PDZ–DSRIWWV complex were taken from the Protein Data Bank (PDB) entry 2JOA. The initial coordinates of the complexes between the PDZ domain and the other peptides were obtained by docking calculations using AutoDock Vina. The MD simulations (120 ns each, 2 fs time step) were carried out with NAMD 2.9 and the CHARMM22 force field (including energy grid correction map, CMAP) as well as particle mesh Ewald (PME) method for electrostatic interactions. The transferable intermolecular potential 3-P water (TIP3P) model was used for water, and the water box had a minimum distance of 20 Å between the system and the walls of the box.
Umbrella sampling simulations were carried out to estimate the free-energy changes associated with the binding of peptides to the PDZ domain using the distance between the centers of mass of the PDZ domain and the peptides as a collective variable.
In addition, docking simulations for studying the interaction between HTRA1 and CAPN2 were carried out using the HADDOCK 2.4 server. The structure predicted by AlphaFold for human HTRA1 was used after removing the N-terminal domain. Detailed descriptions of the umbrella sampling simulations and the docking calculations as well as additional details and references concerning the computational methods can be found in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Peter Davies for plasmids encoding CAPN2 and the regulatory subunit. We thank Michael Meltzer for discussions and Simon Poepsel for comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.E. (EH 100/18-1) and E.S.-G. (CRC1430 and 436586093). The Research Institute of Molecular Pathology is funded by Boehringer Ingelheim. E.S.-G. acknowledges a Plus-3 grant by the Boehringer Ingelheim Foundation, the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy (EXC 2033–390677874–RESOLV), and the computational time provided by the Computing and Data Facility of the Max Planck Society. K.B.-R. and E.S.-G. acknowledge the Gauss Centre for Supercomputing e.V. (GCS; https://www.gauss-centre.eu) for funding this project by providing computing time on the GCS Supercomputer JUQUEEN at the Juelich Supercomputing Centre.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113520119/-/DCSupplemental.
Data Availability
The authors declare no primary datasets and computer codes linked to this study.
References
- 1.Clausen T., Kaiser M., Huber R., Ehrmann M., HTRA proteases: Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 12, 152–162 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Poepsel S., et al. , Determinants of amyloid fibril degradation by the PDZ protease HTRA1. Nat. Chem. Biol. 11, 862–869 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Zurawa-Janicka D., et al. , Structural insights into the activation mechanisms of human HtrA serine proteases. Arch. Biochem. Biophys. 621, 6–23 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Grau S., et al. , Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc. Natl. Acad. Sci. U.S.A. 102, 6021–6026 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tennstaedt A., et al. , Human high temperature requirement serine protease A1 (HTRA1) degrades tau protein aggregates. J. Biol. Chem. 287, 20931–20941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu Q., et al. , HtrA1 proteolysis of ApoE in vitro is allele selective. J. Am. Chem. Soc. 138, 9473–9478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merdanovic M., et al. , Activation by substoichiometric inhibition. Proc. Natl. Acad. Sci. U.S.A. 117, 1414–1418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monod J., Wyman J., Changeux J. P., On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 (1965). [DOI] [PubMed] [Google Scholar]
- 9.Cushing P. R., Fellows A., Villone D., Boisguérin P., Madden D. R., The relative binding affinities of PDZ partners for CFTR: A biochemical basis for efficient endocytic recycling. Biochemistry 47, 10084–10098 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murwantoko, et al. , Binding of proteins to the PDZ domain regulates proteolytic activity of HtrA1 serine protease. Biochem. J. 381, 895–904 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campioni M., et al. , The serine protease HtrA1 specifically interacts and degrades the tuberous sclerosis complex 2 protein. Mol. Cancer Res. 8, 1248–1260 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Fex Svenningsen Å., et al. , Macrophage migration inhibitory factor (MIF) modulates trophic signaling through interaction with serine protease HTRA1. Cell. Mol. Life Sci. 74, 4561–4572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell R. L., Davies P. L., Structure-function relationships in calpains. Biochem. J. 447, 335–351 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Truebestein L., et al. , Substrate-induced remodeling of the active site regulates human HTRA1 activity. Nat. Struct. Mol. Biol. 18, 386–388 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Schillinger J., Severin K., Kaschani F., Kaiser M., Ehrmann M., HTRA1-dependent cell cycle proteomics. J. Proteome Res. 17, 2679–2694 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Yang L. S., Ksiezak-Reding H., Calpain-induced proteolysis of normal human tau and tau associated with paired helical filaments. Eur. J. Biochem. 233, 9–17 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Baudry M., Bi X., Calpain-1 and calpain-2: The Yin and Yang of synaptic plasticity and neurodegeneration. Trends Neurosci. 39, 235–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T., Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell 117, 483–494 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Merdanovic M., et al. , Determinants of structural and functional plasticity of a widely conserved protease chaperone complex. Nat. Struct. Mol. Biol. 17, 837–843 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Eigenbrot C., et al. , Structural and functional analysis of HtrA1 and its subdomains. Structure 20, 1040–1050 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Hedstrom L., Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Volkov O. A., et al. , Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog. eLife 5, e20198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spinozzi S., Albini S., Best H., Richard I., Calpains for dummies: What you need to know about the calpain family. Biochim. Biophys. Acta. Proteins Proteomics 1869, 140616 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Nussinov R., Tsai C. J., Allostery in disease and in drug discovery. Cell 153, 293–305 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Köcher S., et al. , Tailored Ahp-cyclodepsipeptides as potent non-covalent serine protease inhibitors. Angew. Chem. Int. Ed. Engl. 56, 8555–8558 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare no primary datasets and computer codes linked to this study.