Abstract
The diversity and ecology of natural communities of the uncultivated bacterium Achromatium oxaliferum were studied by use of culture-independent approaches. 16S rRNA gene sequences were PCR amplified from DNA extracted from highly purified preparations of cells that were morphologically identified as A. oxaliferum present in freshwater sediments from three locations in northern England (Rydal Water, Jenny Dam, Hell Kettles). Cloning and sequence analysis of the PCR-amplified 16S rRNA genes revealed that multiple related but divergent sequences were routinely obtained from the A. oxaliferum communities present in all the sediments examined. Whole-cell in situ hybridization with combinations of fluorescence-labelled oligonucleotide probes revealed that the divergent sequences recovered from purified A. oxaliferum cells corresponded to genetically distinct Achromatium subpopulations. Analysis of the cell size distribution of the genetically distinct subpopulations demonstrated that each was also morphologically distinct. Furthermore, there was a high degree of endemism in the Achromatium sequences recovered from different sediments; identical sequences were never recovered from different sampling locations. In addition to ecological differences that were apparent between Achromatium communities from different freshwater sediments, the distribution of different subpopulations of Achromatium in relation to sediment redox profiles indicated that the genetically and morphologically distinct organisms that coexisted in a single sediment were also ecologically distinct and were adapted to different redox conditions. This result suggests that Achromatium populations have undergone adaptive radiation and that the divergent Achromatium species occupy different niches in the sediments which they inhabit.
Achromatium oxaliferum is a large sediment-dwelling bacterium found principally in freshwater and brackish environments (21). Notable for its large size, the bacterium also precipitates intracellular calcium carbonate, a property which makes it unique among bacteria.
Since it was first described in 1893 (32), the bacterium has remained uncultivated; however, aspects of its ecology have been inferred from its phylogenetic position, morphological characteristics, in situ activity measurements, and distribution in relation to geochemical gradients in sediments (3, 4, 14, 17, 21, 37). For instance, the presence of sulfur inclusions within the bacterium has suggested its involvement in the sulfur cycle of sediments (21). Furthermore, by artificial manipulation of the magnitude of A. oxaliferum populations in situ, it has been determined that the bacterium is capable of mediating the oxidation of reduced sulfur species to sulfate (14). This finding is consistent with phylogenetic studies that placed A. oxaliferum in the γ-subdivision of the class Proteobacteria, most closely related to a number of sulfur-oxidizing bacteria (13, 17).
Initial phylogenetic analysis of A. oxaliferum cells purified from a freshwater sediment identified several distinct but related 16S rRNA sequences from a population of cells previously thought to be homogeneous (17). On this basis, it was postulated that this A. oxaliferum population was genetically heterogeneous, comprising several related A. oxaliferum-like organisms. More recently, Achromatium sequences have been obtained from cells collected from two freshwater lakes in Germany (13). In this report, only single sequences belonging to the Achromatium clade were recovered. However, subsequent whole-cell in situ hybridization analysis with multiple fluorescence-labelled oligonucleotide probes revealed that one of the communities contained at least three genetically distinct subgroups of A. oxaliferum-like bacteria, while the second was genetically homogeneous (13).
In this study, we have explored the genetic diversity of three Achromatium communities from freshwater sediments in northern England by cloning and sequencing of 16S rRNA genes PCR amplified from purified cell preparations. Whole-cell in situ hybridization with fluorescence-labelled oligonucleotide probes targeting sequences recovered from 16S rRNA gene clone libraries was used to determine the structure and distribution of the Achromatium subpopulations in relation to different sediment redox zones.
MATERIALS AND METHODS
Sample sites and purification of cells from sediment.
Three freshwater environments in northern England that contained populations of Achromatium were studied: Jenny Dam, a shallow upland tarn located near Windermere in the English Lake District, Cumbria, United Kingdom (54°21′N, 2°51′W); a wetland area on the margins of Rydal Water, Cumbria, United Kingdom (54°27′N, 3°00′W); and Hell Kettles (38), located to the south of Darlington, County Durham, United Kingdom (54°29′N, 1°33′W). Hell Kettles, formed by the dissolution and collapse of underlying limestone strata in the year 1179, comprise two ponds (Croft Kettle and Double Kettle) situated in agricultural fenland (38). Samples were taken from the margins of the more southerly Croft Kettle.
Sediment samples containing Achromatium cells were obtained from all three sites by removing the top 3 to 5 cm of the sediment surface with a vacuum sampling device (29) or with sediment coring devices (17). Sediment samples were sequentially filtered through 100- and 64-μm-mesh-size nylon meshes to remove larger sediment particles. Crude cell suspensions obtained in this way were purified as described previously (8, 17). Briefly, the screened sediment was placed in a sterile flask. The flask was tilted until the sediment settled to its base and a white line of Achromatium cells could be observed just below the meniscus. Cells were aseptically removed with a micropipette and transferred to a sterile microcentrifuge tube (1.5-ml capacity). Cells used for DNA extraction were washed exhaustively in filter-sterilized distilled water (usually four or five times) until preparations free of contaminating bacteria, detectable by acridine orange direct counting, were obtained (17).
DNA extraction, PCR amplification, cloning, and sequencing of 16S rRNA genes.
The methods used for DNA extraction and PCR amplification have been described previously (17). PCR products were purified with either a SpinBind system (Flowgen, Lichfield, United Kingdom) or a Qiagen PCR clean-up kit (Qiagen, Crawley, United Kingdom). Cloning was carried out by two methods. TA cloning was done with the pGEM-T vector and Escherichia coli JM109 (Promega, Southampton, United Kingdom) as described previously (17); however, for some samples, forced cloning (30) was also used. PCR products were amplified with PCR primers pA and pH′ (9) modified to contain PstI (pAR; 5′-GTGCTGCAGAGAGTTTGATCCTGGCTCAG-3′) and BamHI (pHR; 5′-CACGGATCCAAGGAGGTGATCCAGCCGCA-3′) restriction sites (in bold and italic typeface) at their 5′ ends. These PCR products were cloned with pUC18 (Boehringer Mannheim, Lewes, United Kingdom). PCR products and pUC18 DNA were digested with PstI and BamHI and, following their purification with a Qiagen PCR clean-up kit, were ligated with T4 DNA ligase (Life Technologies/Gibco BRL, Paisley, United Kingdom). Ligated DNA was used to transform Epicurian Coli SURE supercompetent E. coli cells (Stratagene, Cambridge, United Kingdom). All procedures were carried out in accordance with supplier instructions.
White colonies containing inserts (22 from the Hell Kettles clone library and 20 from the Jenny Dam clone library) were selected at random, and sequence data were obtained for each with primer pE′ (9). The methods used for the PCR amplification and sequencing of cloned 16S rRNA gene inserts were described previously (17). Nearly complete 16S rRNA sequences were obtained from selected clones with the primers of Edwards et al. (9). All sequencing was conducted with the DyeDeoxy chain termination method and an ABI Prism automated DNA sequencer (PE Applied Biosystems, Warrington, United Kingdom). Sequences were aligned manually by use of the genetic data environment sequence editor (33) with reference to the E. coli secondary-structure model.
Phylogenetic distance analyses were conducted with the Jukes-Cantor (20) correction for multiple substitutions at a single site and the neighbor-joining method (31) as implemented in the TREECON package (35). Bootstrap resampling was conducted with 100 replicates. Parsimony analysis was conducted with the DNAPARS program, and bootstrapped data sets were generated with SEQBOOT. Consensus trees were constructed with the CONSENS program. All computer programs used in these analyses were from the PHYLIP software package (11). Maximum-likelihood analysis was conducted with fastDNAml (28). The final alignment used comprised 32 sequences and covered positions 100 to 202 and 217 to 1480 (E. coli numbering).
Whole-cell hybridization with fluorescence-labelled 16S rRNA-targeted oligonucleotide probes.
Oligonucleotide probes specific for Achromatium-derived sequence clusters recovered from different locations (Table 1) were designed from aligned 16S rRNA sequences obtained from Achromatium-derived 16S rRNA gene clone libraries. In some cases, Achromatium sequences obtained from different sampling sites contained the same target sequence (Table 1). High-pressure liquid chromatography-purified oligonucleotide probes 5′ end labelled with tetramethylrhodamine or fluorescein were purchased commercially (Genosys, Cambridge, United Kingdom). Three Achromatium 16S rRNA sequence clusters were identified at each sampling location. The structure of the populations was analyzed with (i) a cluster-specific, rhodamine-labelled probe, (ii) a cluster-specific, fluorescein-labelled probe, and (iii) an unlabelled competitor probe targeting the final cluster to ensure probe specificity (22). Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′) and nonEub (5′-ACTCCTACGGGAGGCAGC-3′) (2) were used in positive and negative control probe hybridization reactions, respectively.
TABLE 1.
Oligonucleotide probes specific for Achromatium subpopulations present in three freshwater sediments
| Probea | Probe sequence 5′→3′ | Clone(s) for which probes were specific at the following site:
|
||
|---|---|---|---|---|
| Rydal Water | Hell Kettles | Jenny Dam | ||
| ARY655a | ACCCCCCTCTCTCGTACT | RY 5 group | HK 2 group | |
| ARY655b | ––––––––––––A––––– | RY 8 group | JD 2 group | |
| ARY655c | ––––––––––A––––––– | RY 1 | ||
| AHK655a | –––––––––––C–––––– | HK 9 | ||
| AHK655b | ––––––––––TC–––––– | HK 13 | ||
| AJD655a | ––––––––––TG–––––– | JD 1 | ||
| AJD655b | –––––––––––G–––––– | JD 8 group | ||
All probes targeted positions 655 to 673 (E. coli numbering) of the 16S rRNA molecule.
Achromatium cells were fixed prior to hybridization as described previously (17). Fixed cells were pelleted (13,000 rpm, 1 min; MSE Microcentaur; Sanyo, Loughborough, United Kingdom), washed once in phosphate-buffered saline (130 mM NaCl, 10 mM sodium phosphate [pH 7.4]), and either resuspended in phosphate-buffered saline for temporary storage at 4°C (no more than 2 days) or washed in sterile distilled water and used immediately. Suspensions of fixed cells were added to sterile 1.5-ml microcentrifuge tubes and pelleted (13,000 rpm, 1 min; MSE Microcentaur), and the supernatant was removed. The cells were resuspended in 50 μl of hybridization buffer (1) containing 40% formamide and the combinations of oligonucleotide probes shown in Table 2. Hybridization mixtures contained 100 pmol of each oligonucleotide and were incubated at 42°C for 2 to 3 h. Hybridized cells were pelleted, washed three times in 50 μl of hybridization buffer for 15 min each time at 42°C, and washed once in filter-sterilized deionized water (Milli-Q50; Millipore, Watford, United Kingdom). Finally, cells were resuspended in 10 μl of an antifading agent (Citifluor, Canterbury, United Kingdom) and mounted on microscope slides. Hybridized cells were viewed with an Olympus BH-2-RFCA microscope fitted with a high-pressure mercury vapor lamp and blue and green filter sets (BP545 and BP409). Micrographs were taken with Kodak Ektachrome Elite 400 film. Automatic exposure was used for phase-contrast micrographs, and exposure times of 10 s (rhodamine fluorescence) and 30 s (fluorescein fluorescence) were used to obtain double-exposure epifluorescence micrographs.
TABLE 2.
Combinations of probes used to determine Achromatium community compositions in three freshwater sediments
| Sample site | Probe combinationa | Probe
|
||
|---|---|---|---|---|
| Rhodamine labelled | Fluorescein labelled | Unlabelled (competitor) | ||
| Rydal Water | 1 | ARY655a | ARY655c | ARY655b |
| 2 | ARY655b | ARY655c | ARY655a | |
| Hell Kettles | 1 | ARY655a | AHK655b | AHK655a |
| 2 | AHK655a | AHK655b | ARY655a | |
| Jenny Dam | 1 | ARY655b | AJD655a | AJD655b |
| 2 | ARY655b | AJD655b | AJD655a | |
Two combinations of probes were used to analyze each sample.
Determination of the composition of Achromatium populations by whole-cell in situ hybridization.
Sediment samples were obtained with a 50-ml syringe coring device (17). The top 5 cm of the core was removed, and Achromatium cells were purified from this sediment sample. Two methods of determining community composition were used. (i) Purified cells were split into two aliquots and fixed. The samples were hybridized with different combinations of oligonucleotide probes to provide independent measurements of the community composition (Table 2). For each aliquot, 200 to 400 cells from randomly selected fields of view were counted and assigned to different Achromatium-like sequences identified in clone libraries, based on their hybridization with the different fluorescence-labelled probes. (ii) Purified cells were split into three aliquots and fixed. The samples were hybridized with different combinations of Eub338 and a single Achromatium subpopulation-specific probe. For each aliquot, 200 to 400 cells from randomly selected fields of view were counted and assigned to different Achromatium-like sequences identified in clone libraries, based on their hybridization with the different fluorescence-labelled probes. A comparison of the two methods with replicate samples produced similar results (unpublished data).
Determination of the size distribution of genetically distinct Achromatium subpopulations by whole-cell hybridization.
Crude cell preparations from bulk sediment samples were hybridized with different combinations of oligonucleotide probes (Table 2). Cells corresponding to the different Achromatium lineages identified in comparative 16S rRNA analyses were photographed (Kodak Ektachrome Elite 400 film), and their dimensions were determined on projected photographic transparencies. The size distributions for genetically distinct Achromatium subpopulations obtained from this analysis were compared statistically. Analysis of variance was conducted on log10-transformed cell diameter data, followed by a multiple pairwise comparison of group means (least significant difference).
Determination of the depth distribution of genetically distinct Achromatium populations by whole-cell hybridization.
A Rydal Water sediment core (6-cm internal diameter; Perspex core tubes) was obtained with a Jenkin surface mud sampler (27). The core was sectioned at 5-mm intervals under oxygen-free nitrogen, and 1.5-cm3 subsamples were removed for the determination of Achromatium community composition (see above). An additional subsample (100 μl) was removed from each core section and fixed with formaldehyde (2% [wt/vol] final concentration) for the determination of total Achromatium cell counts (14). To determine sediment redox conditions, the remainder of the sediment slurry was sealed under nitrogen and stored at −20°C prior to analysis of Fe2+ and sulfate by previously described methods (14). The Achromatium community compositions in the oxidized (the top 3 cm of the sediment core) and reduced (below a 3-cm depth) zones in the sediment were compared with a Pearson chi-square analysis. The oxidized and reduced zones were operationally defined on the basis of Fe2+ concentrations.
Nucleotide sequence accession numbers.
The 12 nearly full-length 16S rRNA sequences determined in this study (4 from Jenny Dam and 8 from Hell Kettles) have been deposited in the GenBank database under accession numbers AF129548 to AF129559. The accession numbers for Achromatium sequences from other studies are L42543, L79966, L79967, L79968, AJ010593, and AJ010596 (13, 17, 18).
RESULTS
Phylogenetic diversity in Achromatium communities.
A total of 42 partial 16S rRNA sequences (ca. 480 bp) were obtained in this study (20 from Jenny Dam and 22 from Hell Kettles). Nearly complete 16S rRNA sequences were obtained for 13 selected 16S rRNA gene clones (5 from Jenny Dam and 8 from Hell Kettles), and these were compared with 6 nearly full-length sequences from previous studies (13, 17, 18). Phylogenetic analyses with different regions of the 16S rRNA sequence were conducted, and one of the sequences from Jenny Dam (JD clone 5) appeared to be chimeric. This sequence was omitted from subsequent analyses.
Phylogenetic analyses, including analyses of four Achromatium-derived sequences previously obtained from Rydal Water sediments (17, 18) and two sequences recovered from Achromatium communities inhabiting sediments in German freshwater lakes (Lake Stechlin and Lake Fuchskuhle) (13), confirmed that Achromatium-like sequences formed a strongly supported monophyletic group within the γ-subdivision of the class Proteobacteria (13, 17). Only Achromatium-like sequences were recovered from the highly purified cell preparations obtained in this study.
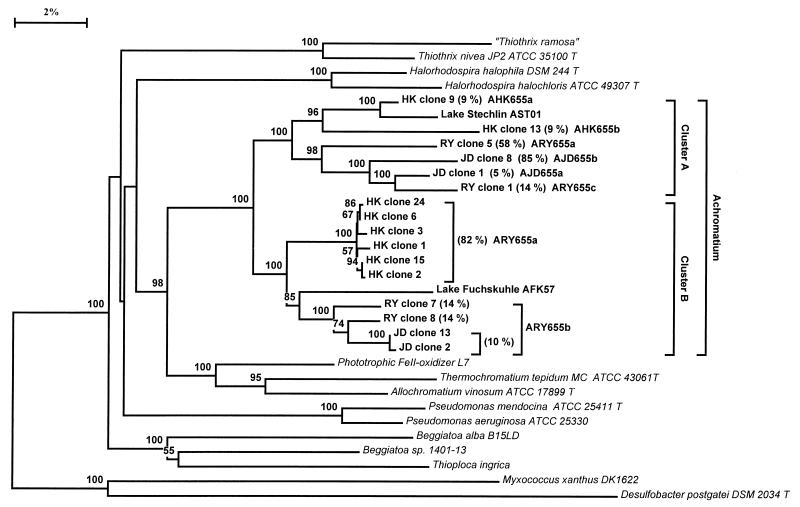
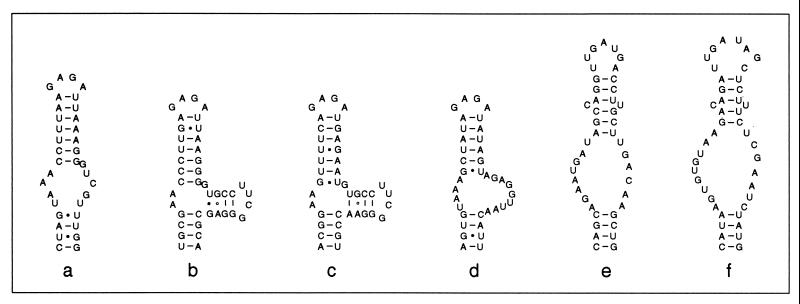
16S rRNA sequences recovered from Achromatium cells from several geographical locations demonstrated that within the cluster defined by the Achromatium-derived sequences, two strongly supported clades were present (Fig. 1). This finding supported the proposal that Achromatium-derived 16S rRNA sequences can be accommodated within two phylogenetic clusters that have been termed cluster A and cluster B (13). This bifurcation was strongly supported by distance analysis (100% bootstrap support for clusters A and B) and maximum-likelihood analysis (P < 0.01). Support for cluster A was also high in parsimony analysis (100% bootstrap support), while support for cluster B was less pronounced (76% bootstrap support). The two clusters could also be distinguished on the basis of secondary-structure elements in the V6 region of the 16S rRNA molecule (Fig. 2). This unusual structural motif was first noted in an Achromatium 16S rRNA sequence from Rydal Water (17) and was confirmed in subsequent studies (13). The V6 region typically contains three helical motifs (helices 36, 37, and 38, based on E. coli secondary structure). Sequences from Achromatium cluster A all have a characteristic deletion and lack helix 38 (positions 1024 to 1036, E. coli numbering); many Archaea and Eucarya sequences also lack this helix (Fig. 2) (16). Sequences in cluster B, in contrast, have a V6 region that resembles the E. coli 16S rRNA secondary structure and are more typical of the majority of bacteria (Fig. 2) (16).
FIG. 1.
Phylogenetic distance tree based on the comparative analysis of nearly full-length 16S rRNA gene sequences recovered from Achromatium cells purified from Rydal Water (RY), Jenny Dam (JD), and Hell Kettles (HK) sediments. Two additional Achromatium-derived sequences, from Lake Stechlin and Lake Fuchskuhle (13), were included in the analysis. The numbers in parentheses indicate the percentages of clones from the three clone libraries that were closely related to the annotated clusters or sequences (RY, n = 7; JD, n = 19; HK, n = 22). The sequences targeted by the oligonucleotide probes used in this study are also labelled. The scale bar denotes 2% sequence divergence, and the values at the nodes indicate the percentages of bootstrap trees that contained the cluster to the right of the node. Bootstrap values lower than 50 are not shown.
FIG. 2.
Secondary structure of the V6 region of small-subunit rRNA from a range of organisms (16). (a to d) Bacteria. (e) Archaea. (f) Eucarya. (a) Achromatium cluster A, RY clone 5. (b) Achromatium cluster B, JD clone 2. (c) E. coli. (d) Mycoplasma capricolum. (e) Methanobacterium formicicum. (f) Drosophila melanogaster.
Representative sequences from both clusters were identified at all three sites investigated in the current study. Within clusters A and B, further structure was evident. Sequences recovered from Jenny Dam and Rydal Water formed monophyletic groups that were distinct from the clades harboring Hell Kettles sequences (Fig. 1). These relationships were also evident in parsimony and maximum-likelihood analyses, and the monophyletic nature of the Rydal Water-Jenny Dam and Hell Kettles sequences within clusters A and B was supported by bootstrap values in the range of 94 to 100%. An Achromatium sequence from Lake Stechlin (13) was most closely related to the cluster A sequences from Hell Kettles, whereas an Achromatium sequence from Lake Fuchskuhle (13) may represent a distinct lineage affiliated with the cluster B sequences from Rydal Water and Jenny Dam (Fig. 1).
Occurrence of phylogenetically distinct subpopulations within Achromatium communities.
There are a number of possible explanations for the recovery of multiple related Achromatium-like sequences in PCR-derived clone libraries of 16S rRNA genes produced from purified preparations of Achromatium cells. The sequence diversity may correspond to genetically distinct populations of Achromatium cells present in a single sample. Alternatively, the observed sequence diversity may be due to the presence of multiple divergent 16S rRNA genes present within individual cells of a single species of Achromatium. Heterogeneous 16S rRNA operons are known to occur in the genomes of many bacterial species (7, 23, 26), and it has been suggested that heterogeneity in rRNA operons could, in some cases, contribute to the diversity observed in bacterial communities studied by rRNA-based techniques (12). A third explanation is that the sequences came from bacteria that were phylogenetically related to Achromatium but were not morphologically identifiable as Achromatium cells. Whole-cell hybridization with fluorescence-labelled oligonucleotide probes specific for the different Achromatium-derived sequences recovered from the three geographical locations studied here was used to establish which explanation was correct.
Oligonucleotide probes were designed to target the more deeply branching lineages within the Achromatium clades (i.e., those exhibiting less than 97% sequence similarity) (Table 3 and Fig. 1). On this basis, three probes were used to discriminate genetically distinct Achromatium subpopulations at each study site. For example, to analyze the Achromatium population in Rydal Water sediments, two different probes were used to identify cells containing RY clone 1 or RY clone 5 sequences (93.75% similarity), and a single probe was used to identify both the RY clone 7 and the RY clone 8 sequences (97.26% similarity).
TABLE 3.
Similarity among nearly complete 16S rRNA gene sequences from Achromatiuma
| Sequence | Clone | Cluster | % Similarity of sequence:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |||
| 1 | RY 8 | B | |||||||||||||||||
| 2 | RY 7 | B | 97.26 | ||||||||||||||||
| 3 | RY 5 | A | 91.46 | 92.15 | |||||||||||||||
| 4 | RY 1 | A | 91.93 | 93.12 | 93.75 | ||||||||||||||
| 5 | HK 1 | B | 95.58 | 94.79 | 91.97 | 91.71 | |||||||||||||
| 6 | HK 2 | B | 95.72 | 94.94 | 92.05 | 91.86 | 99.44 | ||||||||||||
| 7 | HK 3 | B | 95.58 | 94.79 | 92.04 | 91.71 | 99.37 | 99.37 | |||||||||||
| 8 | HK 6 | B | 95.79 | 95.01 | 92.05 | 91.79 | 99.44 | 99.58 | 99.65 | ||||||||||
| 9 | HK 15 | B | 95.72 | 94.94 | 92.05 | 91.86 | 99.44 | 99.86 | 99.37 | 99.58 | |||||||||
| 10 | HK 24 | B | 95.65 | 95.01 | 92.05 | 91.79 | 99.30 | 99.44 | 99.51 | 99.86 | 99.44 | ||||||||
| 11 | HK 9 | A | 92.72 | 93.48 | 93.62 | 92.86 | 92.87 | 92.87 | 93.08 | 93.23 | 92.87 | 93.08 | |||||||
| 12 | HK 13 | A | 90.31 | 91.21 | 92.48 | 91.17 | 91.24 | 91.25 | 91.46 | 91.60 | 91.25 | 91.60 | 94.72 | ||||||
| 13 | JD 2 | B | 97.26 | 97.75 | 91.33 | 92.07 | 95.23 | 95.38 | 95.23 | 95.45 | 95.38 | 95.31 | 92.94 | 90.47 | |||||
| 14 | JD 13 | B | 97.40 | 97.89 | 91.47 | 92.21 | 95.37 | 95.52 | 95.37 | 95.59 | 95.52 | 95.45 | 93.08 | 90.61 | 99.86 | ||||
| 15 | JD 8 | A | 91.59 | 91.71 | 93.98 | 95.84 | 91.74 | 91.68 | 91.88 | 91.89 | 91.68 | 91.89 | 93.59 | 91.55 | 91.74 | 91.88 | |||
| 16 | JD 1 | A | 92.49 | 93.26 | 94.11 | 97.60 | 92.15 | 92.30 | 92.29 | 92.37 | 92.30 | 92.23 | 94.35 | 92.38 | 92.79 | 92.93 | 96.34 | ||
| 17 | Stechlin | A | 92.50 | 93.55 | 93.54 | 92.78 | 92.58 | 92.59 | 92.79 | 92.94 | 92.59 | 92.94 | 98.87 | 94.21 | 92.79 | 92.93 | 93.44 | 94.06 | |
| 18 | Fuchskuhle | B | 95.01 | 95.42 | 91.60 | 91.77 | 94.38 | 94.67 | 94.45 | 94.67 | 94.74 | 94.67 | 92.65 | 90.31 | 94.66 | 94.80 | 91.45 | 92.99 | 92.85 |
Analysis was conducted by pairwise comparisons of nearly complete 16S rRNA sequences (1,479 positions). Boxed data show similarities among sequences from the same sampling location; data outside the boxes show similarities among sequences from the different sampling locations.
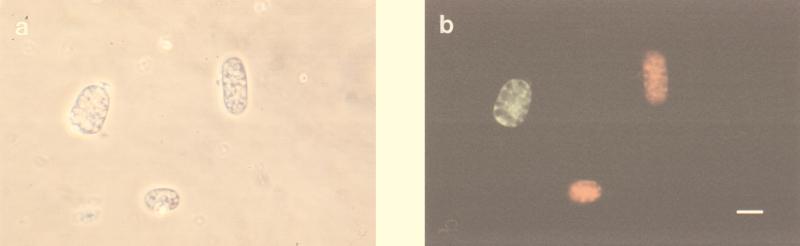
Simultaneous whole-cell hybridization with fluorescence-labelled oligonucleotide probes targeting different Achromatium-derived sequences clearly demonstrated that the divergent sequences recovered in 16S rRNA gene clone libraries represented phylogenetically distinct subgroups present within natural Achromatium populations (Fig. 3). This finding was confirmed for all of the sediments investigated in this study. In whole-cell hybridization experiments, Achromatium populations comprised fluorescein-labelled, rhodamine-labelled, and unlabelled cells. The binding of more than one probe to a single cell was never observed with any combination of probes used. Consequently, different target sequences are unlikely to represent multiple rRNA operons occurring within a single Achromatium cell. Furthermore, all of the Achromatium-specific probes bound to cells that could be identified, on the basis of their morphology, as Achromatium.
FIG. 3.
Whole-cell in situ hybridization of Achromatium cells purified from Rydal Water sediments. Cell were simultaneously hybridized with three oligonucleotide probes: ARY655a (rhodamine-labelled, RY clone 5-specific probe), ARY655b (unlabelled [competitor] RY clone 7- or RY clone 8-specific probe), and ARY655c (fluorescein-labelled, RY clone 1-specific probe). Phase contrast (a) and epifluorescence (b) micrographs of the same microscopic field are shown. Two RY clone 5 cells (red fluorescence) and one RY clone 1 cell (green fluorescence) are shown. The scale bar represents 20 μm and applies to both micrographs.
Using different combinations of fluorescein-labelled, rhodamine-labelled, and unlabelled probes (Table 2), we obtained direct counts of specific subgroups of Achromatium for cells collected from Rydal Water, Jenny Dam, and Hell Kettle sediments. The compositions of the Achromatium communities were determined independently with different probe combinations (Table 4). For example, in a sample of cells collected from Jenny Dam (July 1998), the proportions of cells identified as containing JD clone 1-like (or JD clone 8-like) sequences were very similar whether they were identified on the basis of hybridization with a JD clone 1-specific (or JD clone 8-specific) probe or on the basis of the fact that they did not bind probes specific for the other two groups analyzed (Table 4). Furthermore, at least 98% of Achromatium cells (n = 200) identified by phase-contrast microscopy hybridized with fluorescence-labelled Achromatium-specific probes. This value is comparable to the 97.8% of cells (n = 267) which bound the general bacterial probe Eub338. Similar values were obtained with samples from all three sites investigated; thus, the diversity encompassed by the majority of Achromatium cells was characterized with PCR amplification and cloning of 16S rRNA genes.
TABLE 4.
Community compositions of different Achromatium subpopulations in Jenny Dam sediments (July 1998) determined with different combinations of labelled and unlabelled competitor probesa
| Specificity of probes (probe name) that were:
|
% of cells (clone) that were:
|
||||
|---|---|---|---|---|---|
| Rhodamine labelled | Fluorescein labelled | Unlabelled | Rhodamine labelled | Fluorescein labelled | Unlabelled |
| JD 2 (ARY655b) | JD 1 (AJD655a) | JD 8 (AJD655b) | 56 (JD 2) | 5 (JD 1) | 39 (JD 8) |
| JD 2 (ARY655b) | JD 8 (AJD655b) | JD 1 (AJD655a) | 58 (JD 2) | 37 (JD 8) | 5 (JD 1) |
The percentages of cells identified as JD clone 1 or JD clone 8 were approximately the same whether they were enumerated on the basis of hybridization with a specific labelled probe or lack of hybridization with the other two probes used. The sample size was 200.
Morphological differentiation of phylogenetically distinct subpopulations within Achromatium communities.
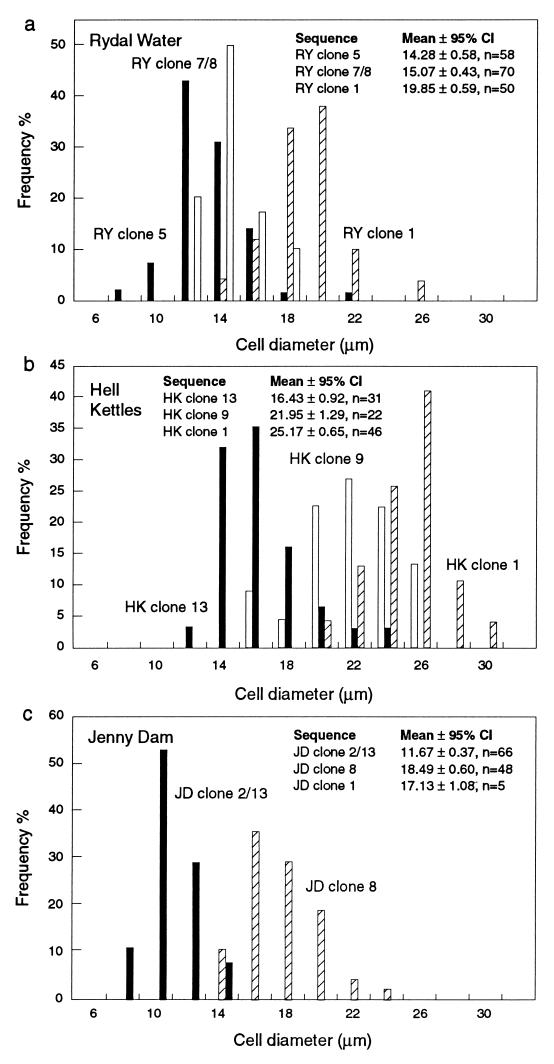
The frequency distribution of cell diameter in Achromatium communities from different geographical locations was analyzed. Cell diameter rather than cell volume was found to be a better descriptor of differences in cell size, presumably because length or volume measurements of Achromatium cells, which grow along their long axis and divide perpendicular to it, include growth-related variations.
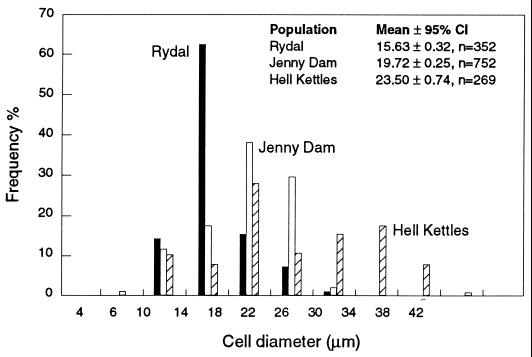
The range of cell diameters observed for Achromatium communities from Rydal Water, Jenny Dam, and Hell Kettles sediments (Fig. 4) fell within the range of cell dimensions reported for Achromatium cells from other habitats (3, 5, 10, 24, 32, 36, 37). Moreover, the mean cell diameters observed for Achromatium communities from the different study sites investigated here were found to be significantly different (P < 0.05) (Fig. 4). This finding suggested that different species or strains of Achromatium with different mean cell diameters were present in the different sediments studied. However, while this conclusion may be true, until now no detailed analysis of the size distribution of Achromatium cells has been reported, and the considerable overlap in the dimensions of Achromatium cells has confounded the use of cell size as a criterion for the differentiation of Achromatium species (10). This situation is well illustrated in the analysis of Achromatium cells from Rydal Water, Jenny Dam, and Hell Kettles sediments. Although the mean cell diameters for the communities were statistically significantly different, the considerable overlap in the size distribution of each population would make it impossible to assign an individual cell to any particular population, except perhaps the larger cells present in Hell Kettles sediments.
FIG. 4.
Frequency distribution of the diameters of Achromatium cells from sediment samples taken from Rydal Water, Jenny Dam, and Hell Kettles. The Achromatium communities present at each sampling site had a characteristic site distribution, and the mean diameters of cells from these locations were significantly different. CI, confidence interval.
Phylogenetic analysis of cells from different geographical locations (Fig. 1) confirmed not only that the Achromatium communities present in a range of freshwater sediments were genetically distinct but that each community comprised several genetically distinct subpopulations (Fig. 1 and 3). To determine if there was morphological differentiation (on the basis of cell size) of Achromatium subpopulations from a single sediment, identification of genetically distinct cells by whole-cell hybridization was combined with measurements of cell diameter.
The size distribution of genetically distinct Achromatium subpopulations identified by whole-cell in situ hybridization indicated that cells corresponding to the different Achromatium sequence types fall into distinct size classes (Fig. 5). It should be noted that the frequency distributions of cell diameters presented were obtained by measurement of cells selected from the different subpopulations until sufficient cells had been measured to obtain a meaningful distribution. Thus, the sum of the frequency distributions is not representative of a naturally occurring Achromatium community. Multiple pairwise comparisons (least significant difference) of the mean cell diameters of the genetically distinct Achromatium subpopulations indicated significant differences (P < 0.01) between almost all of the phylogenetically distinct Achromatium subpopulations identified by whole-cell in situ hybridization. Statistically significant differences in mean cell diameters were evident both in subpopulations coexisting in a single sediment and among all the subpopulations identified at different geographical locations, with the exception of cells identified as JD clone 1 from Jenny Dam. Comparisons of JD clone 1 with JD clone 8 and of JD clone 1 with HK clone 13 (Fig. 5) revealed no significant differences in cell diameters. However, only five cells corresponding to JD clone 1 were identified in this analysis (Fig. 5c); thus, a much smaller sample size was used in the statistical comparison. If a larger number of JD clone 1-like cells had been available for cell diameter measurements, a more definitive statistical analysis of this Achromatium subpopulation would have been possible.
FIG. 5.
Frequency distributions of genetically distinct Achromatium cells from Rydal Water (a), Hell Kettles (b), and Jenny Dam (c) sediments. Cells were discriminated with fluorescence-labelled oligonucleotide probes specific for different Achromatium-derived sequences identified in clone libraries from the different sediments (Tables 1 and 2). Cells corresponding to JD clone 1 were rather rare, and only a small number of cells were measured (n = 5); consequently, data from this subpopulation were not plotted. CI, confidence interval.
Temporal variations in the compositions of Achromatium communities.
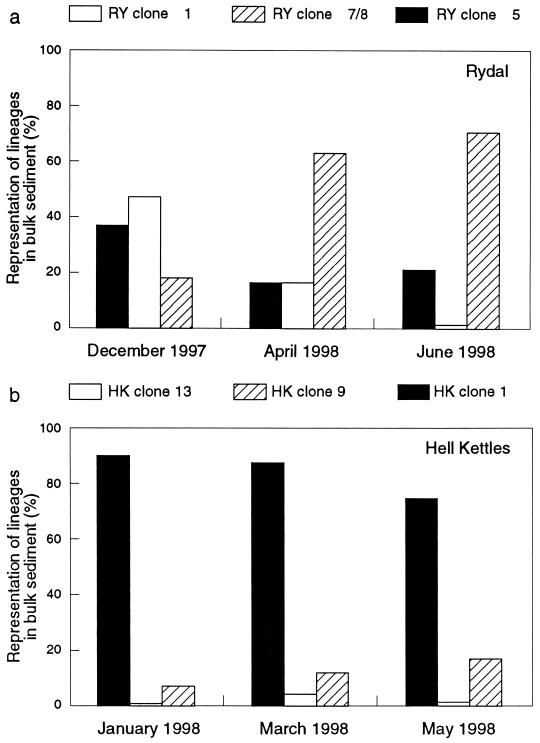
Using whole-cell hybridization with specific fluorescence-labelled oligonucleotide probes, we obtained direct counts of Achromatium subpopulations from Rydal Water and Hell Kettles sediment cores sampled at different times of the year. The structure of the Achromatium communities within sediments was dynamic, and changes in the numerical dominance of different subpopulations were observed at different sampling dates (Fig. 6). Changes in community structure were clearly demonstrated for Achromatium communities from Rydal Water sediments. Between December 1997 and June 1998, there was a relative decrease in the larger RY clone 1 cells, while there was a relative increase in the RY clone 7 or RY clone 8 cells. Less pronounced population changes were observed at Hell Kettles, where the HK clone 1 cell type remained dominant throughout the period of sampling (Fig. 6).
FIG. 6.
Relative abundance of genetically distinct Achromatium subpopulations in sediment samples taken at different times of the year from Rydal Water and Hell Kettles. Cells belonging to the different subpopulations were discriminated with fluorescence-labelled oligonucleotide probes specific for different Achromatium-derived sequences (Tables 1 and 2). The total number of cells (the sum of cells counted from each aliquot) counted in each sample was as follows: Rydal Water—December 1997, n = 913, April 1998, n = 384, and June 1998, n = 395; Hell Kettles—January 1998, n = 300, March 1998, n = 300, and May 1998, n = 300.
Depth distributions of Achromatium subpopulations.
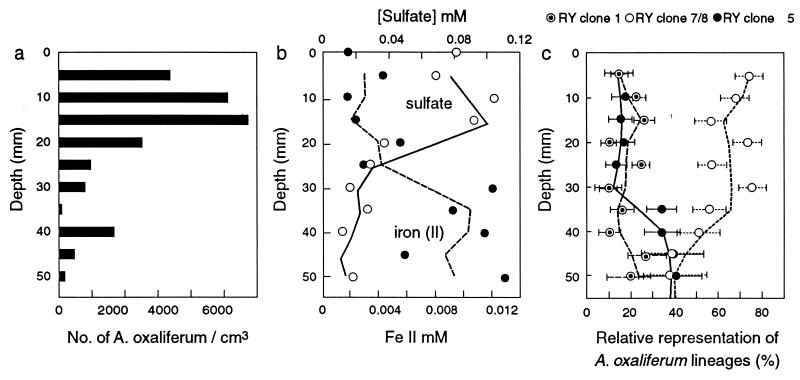
The depth distribution of Achromatium cells in a sediment core from Rydal Water indicated that Achromatium cells were present throughout the top 5 cm of the sediment (Fig. 7a). The majority of cells were located in the top 3 cm, where high sulfate and low Fe2+ concentrations indicated oxidizing conditions (Fig. 7b) (6). Below 3 cm, where Achromatium cells were less abundant, low sulfate and high Fe2+ concentrations indicated more reducing conditions. The distribution of Achromatium across both oxidized and reduced zones in sediments was consistent with previous observations (14). Analysis of the composition of the Achromatium population at each depth was conducted by whole-cell in situ hybridization. This analysis demonstrated that each subpopulation was present at all depths (Fig. 7c). However, while the proportions of the RY clone 1 cell type remained similar regardless of depth, a relative change in the proportions of RY clone 7 or RY clone 8 and RY clone 5 cell types corresponded with the transition between oxidized and reduced zones. The RY clone 7 or RY clone 8 cell type, which comprised 67.8% of cells (n = 1,137) in the oxidized zone, represented a statistically significantly smaller proportion of the population (50.1%, n = 373) in the reduced zone (Pearson chi-square value, 56.69; df, 2; P < 0.001). Conversely, the RY clone 5 cell type represented 14.8% (n = 1,089) of the Achromatium population in the oxidized zone but represented a significantly larger proportion of the population (34.6%, n = 399) in the reduced zone (Pearson chi-square value, 72.55; df, 2; P < 0.001). Thus, the subpopulations present within a single Achromatium community exhibited different distributions under different redox conditions, providing evidence of niche adaptation in the Achromatium subpopulations.
FIG. 7.
Depth profiles of Achromatium cells (a), redox-sensitive chemical species (Fe2+ and SO42−) (b), and relative abundance of the three main Achromatium lineages identified in a sediment core from Rydal Water (c). The trend lines show a three-point moving average of the data, and error bars represent 95% confidence intervals for counts of the Achromatium subpopulations.
DISCUSSION
Phylogenetic diversity in Achromatium populations.
The presence of phylogenetic diversity in Achromatium communities has now been demonstrated unequivocally. Of the five Achromatium communities from different geographical locations studied to date (three in this study and two studied by Glöckner et al. [13]), four have been shown to comprise several genetically distinct subgroups, while only one, the community present in Lake Fuchskuhle sediments, appears to be homogeneous. Moreover, the genetically distinct Achromatium subpopulations present at each site were endemic to those lake sediments (Fig. 1). Only Lake Fuchskuhle and Lake Stechlin potentially share a common Achromatium subpopulation (13). A total of 28% of the cells in Lake Stechlin bound two oligonucleotide probes designed to target an Achromatium sequence recovered from Lake Fuchskuhle (AFK192 [positions 192 to 211, E. coli numbering] and AFK433 [positions 433 to 450, E. coli numbering); nonetheless, a sequence corresponding to the genotype found in Lake Fuchskuhle was not detected in 16S rRNA gene clone libraries obtained from cells purified from Lake Stechlin. However, as Glöckner et al. (13) pointed out, the phylogenetic affinity of the cells from Lake Fuchskuhle sediments and AFK192/AFK433-positive cells in Lake Stechlin is only speculative, since their identification was based on probe binding alone and the presence of two identical target sequences in different 16S rRNA molecules does not necessarily imply high similarity elsewhere in the molecules (13). This point is well illustrated when the divergent sequences obtained in this study are considered. If one were to construct oligonucleotide probes targeting the same positions in the 16S rRNA molecules as AFK192 and AFK433 but specific for Achromatium rRNA sequences obtained from Jenny Dam and Rydal Water, cells containing the JD clone 1 and RY clone 1 sequences would bind both probes, yet these sequences have only 97.6% sequence identity (Table 3). Likewise, cells belonging to JD clone 2 or JD clone 13 and to RY clone 7 or RY clone 8 (97.3 to 97.9% similarity) would also bind both probes but are clearly phylogenetically distinct (Fig. 1). It is also evident that much more divergent sequences also contain common diagnostic oligonucleotide sequences; for example, RY clone 5 and HK clone 2 sequences have only 92.1% homology in their 16S rRNAs, yet both contain the target sequence for probe ARY655a (Table 1).
The occurrence of genetically distinct subpopulations in Achromatium communities provides evidence to support the delineation of Achromatium into different taxonomic groupings. For instance, many of the 16S rRNA sequences obtained from Achromatium cells had less than 97.5% 16S rRNA sequence homology. This level of sequence divergence has been used to delineate species (34). On this basis, most of the Achromatium subpopulations identified may reasonably be defined as separate species. No members of the genus Achromatium have been isolated in culture; consequently, relationships among different Achromatium spp. can be inferred only by phylogenetic analysis of environmental samples, morphology, and possibly habitat. On these bases, the Achromatium population native to Lake Fuchskuhle sediments has been described as a new species, “Candidatus Acromatium minus” (13). The genetic characterization of Achromatium and the discovery that genetically distinct cells are differentiated morphologically on the basis of cell size in the current study indicate that several more Achromatium spp. remain to be described.
Significance of morphology (cell diameter) in the classification of Achromatium.
From a historical perspective, the different cell size distributions of phylogenetically distinct Achromatium subpopulations observed in this study clarify one of the earliest debates surrounding the taxonomy of Achromatium. Without access to cultured organisms or molecular techniques, early classification relied on morphological methods alone. These early studies (5, 10, 24, 32, 36, 37) and some later contributions (e.g., 3) reported that Achromatium cells from different locations had different dimensions. These early reports indicated that the bacterium exhibited cell dimensions ranging from 7 to 36 μm in diameter and 7 to 102 μm in length. On this basis, Nadson and Visloukh (24) suggested that a number of different forms of A. oxaliferum existed and proposed the additional epithets minus, medium, majus, elongatum, and gigas to describe them (see also reference (5)). Classification based on cell size has been applied to filamentous sulfur bacteria related to Achromatium. For example, different Thioploca spp. and Beggiatoa spp. can be distinguished on the basis of filament diameter, a characteristic which can vary by as much as 1 order of magnitude within these genera (25). However, in the genus Achromatium, there is a considerable overlap in the cell size distributions of different species. Given this continuous distribution of cell size and the natural variations that populations of bacteria are likely to exhibit when located in different environments, one author (10) considered that delineation of different Achromatium species on the basis of cell size was not valid. The data presented here provide the first direct evidence that different Achromatium species are morphologically distinct and support the original proposal of Nadson and Visloukh (24). However, although different Achromatium spp. have now been shown to have characteristic cell sizes, in isolation cell size cannot be recommended as a criterion for their identification. Recovery of 16S rRNA sequences and whole-cell in situ hybridization provide the only reliable means of identifying new Achromatium species.
Ecological significance of genetically distinct subpopulations of Achromatium.
Achromatium species from different geographical locations show some degree of site specificity; i.e., Achromatium sequences recovered from Rydal Water and Jenny Dam are more closely related to each other than they are to sequences from Hell Kettles (Fig. 1). Furthermore, this relationship is apparent within both cluster A and cluster B sequences and appears to correlate with the nutritional characteristics of the Achromatium communities found at the different sites. Recent studies on the uptake of inorganic and organic substrates by Achromatium cells in Rydal Water and Hell Kettles sediments, determined by microautoradiography (15), have indicated that some cells in both populations can assimilate simple organic compounds. However, while the majority of cells from Rydal Water were shown to assimilate inorganic carbon, none of the cells from Hell Kettles had this ability. Furthermore, homologues of the ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit gene (rbcL), involved in CO2 fixation, and adenosine-5′-phosphosulfate reductase genes (aprBA), associated with energy-conserving sulfur oxidation pathways (19), could be amplified from highly purified preparations of Achromatium cells from Rydal Water and Jenny Dam sediments but not from cells collected at Hell Kettles (18). The presence of these genes in genomic DNA from Achromatium species present at Rydal Water is consistent with the autotrophic potential exhibited by at least some of the cells in this population.
While different geographical locations appear to harbor genetically and nutritionally distinct Achromatium populations, presumably as a result of ecosystem-imposed selection pressures, phylogenetically and morphologically distinct Achromatium subpopulations also occur together within the same ecosystem; at a single location, the Achromatium population structure can be dynamic (Fig. 6). These results suggest that conditions which favor the proliferation of one Achromatium subpopulation do not necessarily apply to all. In this study, it was not possible to determine physiological differences among coexisting Achromatium subpopulations; however, depth- and redox-related changes in community composition were detected (Fig. 7c). This finding indicated that different Achromatium subpopulations are better adapted to different redox conditions and probably occupy distinct ecological niches within the sediment environment. Thus, adaptive radiation in response to environmental heterogeneity may have resulted in the evolution of several coexisting but genetically and ecologically distinct Achromatium subpopulations that have optimal activities under different redox conditions. One source of heterogeneity in sediments is the depth-related succession of respiratory processes (6) that gives rise to a number of physiochemical and depth-defined niches in close proximity. The presence of these niches may have been the driving force behind the diversification observed for Achromatium communities.
ACKNOWLEDGMENTS
We thank Nick Davis for providing data on cell size distributions from Rydal Water. We also thank Frank Oliver Glöckner and Rudolf Amann for providing Achromatium sequence data from Lake Fuchskuhle and Lake Stechlin prior to publication and for stimulating discussion.
This work was supported by the IFE and the FBA. Financial support was provided by the Natural Environment Research Council (grant GR3/9148 to I.M.H., R.W.P., and J.G.J. and studentship GT4/95/235/F to R.H.).
REFERENCES
- 1.Amann R, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babenzien H-D. Achromatium oxaliferum and its ecological niche. Zentralbl Mikrobiol. 1991;146:41–49. [Google Scholar]
- 4.Babenzien H-D, Sass H. The sediment-water interface—habitat of the unusual bacterium Achromatium oxaliferum. Arch Hydrobiol. 1997;48:247–251. [Google Scholar]
- 5.Bavendamm W. Die Farblosen und roten Schwefelbakterien des Süß und Salzwassers. Pflanzenforschung. 1924;2:7–156. [Google Scholar]
- 6.Berner R A. A new geochemical classification of sedimentary environments. J Sediment Petrol. 1981;51:359–365. [Google Scholar]
- 7.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 8.de Boer W E, La Rivière J W M, Schmidt K. Some properties of Achromatium oxaliferum. Antonie Leeuwenhoek. 1971;37:553–563. doi: 10.1007/BF02218525. [DOI] [PubMed] [Google Scholar]
- 9.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct complete nucleotide determination of entire genes. Characterisation of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis D. Sulphur bacteria: a monograph. London, England: Longmans, Green and Co.; 1932. [Google Scholar]
- 11.Felsenstein J. PHYLIP: phylogeny inference package. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 12.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glöckner F R, Babenzien H-D, Wulf J, Amann R. Phylogeny and diversity of Achromatium oxaliferum. Syst Appl Microbiol. 1999;22:28–38. doi: 10.1016/S0723-2020(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 14.Gray N D, Pickup R W, Jones J G, Head I M. Ecophysiological evidence that Achromatium oxaliferum is responsible for the oxidation of reduced sulfur species to sulfate in a freshwater sediment. Appl Environ Microbiol. 1997;63:1905–1910. doi: 10.1128/aem.63.5.1905-1910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray N D, Howarth R, Pickup R W, Jones J G, Head I M. Substrate uptake by uncultured bacteria from the genus Achromatium determined by microautoradiography. Appl Environ Microbiol. 1999;65:5100–5106. doi: 10.1128/aem.65.11.5100-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutell R R. Collection of small-subunit (16S and 16S-like) ribosomal-RNA structures. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Head I M, Gray N D, Clarke K J, Pickup R W, Jones J G. The phylogenetic position and ultrastructure of the uncultured bacterium Achromatium oxaliferum. Microbiology. 1996;142:2341–2354. doi: 10.1099/00221287-142-9-2341. [DOI] [PubMed] [Google Scholar]
- 18.Head, I. M., N. D. Gray, R. Howarth, K. J. Clarke, R. W. Pickup, and J. G. Jones. Achromatium oxaliferum: understanding the unmistakable. Adv. Microb. Ecol., in press.
- 19.Hipp W M, Pott A S, Thum-Schmitz N, Faath I, Dahl C, Trüper H G. Towards the phylogeny of APS reductases and sirohaem sulfite reductases in sulfate-reducing and sulfur-oxidizing prokaryotes. Microbiology. 1997;143:2891–2902. doi: 10.1099/00221287-143-9-2891. [DOI] [PubMed] [Google Scholar]
- 20.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 21.La Rivière J W M, Schmidt K. Morphologically conspicuous sulfur-oxidizing eubacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes: a handbook on the biology of bacteria; ecophysiology, isolation, identification, applications. 2nd ed. Vol. 4. New York, N.Y: Springer-Verlag; 1992. pp. 3934–3947. [Google Scholar]
- 22.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of the proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 23.Mylvaganan S, Dennis P P. Sequence heterogeneity between the 2 genes encoding 16S ribosomal-RNA from the halophilic Archaebacterium Haloarcula marismortu. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadson G A, Visloukh S M. La structure et la vie de la bactérie géante Achromatium oxaliferum. Schew Bull Jard Imp Bot St-Petersbourg. 1923;22(Suppl. 1):1–37. [Google Scholar]
- 25.Nelson D C, Wirsen C O, Jannasch H W. Characterization of large, autotrophic Beggiatoa spp. abundant at hydrothermal vents of the Guaymas Basin. Appl Environ Microbiol. 1989;55:2909–2917. doi: 10.1128/aem.55.11.2909-2917.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohnstad F R, Jones J G. The Jenkin Surface-Mud Sampler user manual. Occasional publication no. 15. Freshwater Biological Association. Kendal, Cumbria, United Kingdom: Titus Wilson & Son; 1982. [Google Scholar]
- 28.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 29.Pickup R W. Sampling and detecting bacterial populations in natural environments. In: Baumberg S, Young J P W, Saunders J R, Wellington E M, editors. Population genetics of bacteria. Cambridge, England: Cambridge University Press; 1995. pp. 295–315. [Google Scholar]
- 30.Rochelle P A, Will J A K, Fry J C, Jenkins G J S, Parkes R J, Turley C M, Weightman A J. Extraction and amplification of 16S rRNA genes from deep marine sediments and seawater to assess bacterial community diversity. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment: methods and applications. Berlin, Germany: Springer-Verlag KG; 1995. pp. 219–239. [Google Scholar]
- 31.Saitou N, Nei M. The neighbour joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Schewiakoff W. Über einen neuen bakterienähnlichen Organismus des Süßwassers. Heidelberg, Germany: Habilitationsschrift; 1893. [Google Scholar]
- 33.Smith S W, Overbeek R, Woese C R, Gilbert W, Gillevet P M. The genetic data environment: an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 34.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 35.van de Peer Y, De Wachter R. TREECON for windows—a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 36.West G S, Griffiths B M. Hillhousia mirabilis, a giant sulphur bacterium. Proc R Soc London Ser B. 1909;81:389–409. [Google Scholar]
- 37.West G S, Griffiths B M. The lime sulphur bacteria of the genus Hillhousia. Ann Bot. 1913;27:83–91. [Google Scholar]
- 38.Wheeler D B, Whitton B A. Ecology of Hell Kettles. 1. Terrestrial and sub-aquatic vegetation. Vasculum. 1971;56:25–37. [Google Scholar]