Abstract
Aptamer-based sensors have emerged as a major platform for detecting small-molecular targets, because aptamers can be selected to bind these small molecules with higher affinity and selectivity than other receptors such as antibodies. However, portable, accurate, sensitive, and affordable detection of these targets remains a challenge. In this work, we developed an aptasensing platform incorporating magnetic beads and a DNAzyme for signal amplification, resulting in high sensitivity. The biosensing platform was constructed by conjugating a biotin-labeled aptamer probe of small-molecular targets such as toxins and a biotin-labeled substrate strand on magnetic beads, and the DNAzyme strand hybridized with the aptamer probe to block the substrate cleavage activity. The specific binding of the small-molecular target by the aptamer probe can replace the DNAzyme strand and then induce the hybridization between the DNAzyme strand and substrate strand, and the iterative signal amplification reaction of hydrolysis and cleavage of the substrate chain occurs in the presence of a metal ion cofactor. Using invertase to label the substrate strand, the detection of small molecules of the toxin is successfully transformed into the measurement of glucose, and the sensitive analysis of small molecules such as toxins can be realized by using the household portable glucose meter as a readout. This platform is shown to detect ochratoxin, a common toxin in food, with a linear detection range of 5 orders of magnitude, a low detection limit of 0.88 pg/mL, and good selectivity. The platform is easy to operate and can be used as a potential choice for quantitative analysis of small molecules, at home or under point-of-care settings. Moreover, by changing and designing the aptamer probe and the arm of DNAzyme strand, it can be used for the analysis of other analytes.
Keywords: ochratoxin, personal glucose meter, DNAzyme amplification, aptasensor, magnetic bead
Graphical Abstract
INTRODUCTION
Aptamers are a class of single-chain nucleic acid molecules obtained by systematic evolution of ligands by exponential enrichment (SELEX) from a large DNA library of up to 1015 sequences to bind to a wide range of targets from metal ions to small molecules, proteins, and cells.1–6 Aptamers are particularly advantageous for detecting small-molecular targets, which are often difficult to detect using other types of sensing molecules such as antibodies. A primary example is mycotoxins, which are secondary metabolites produced by fungi during the production, harvesting, storage, and processing of cereals or fruits and vegetables.7 Mycotoxin contamination is a worldwide public safety issue, and the World Health Organization (WHO) and the Food and Agriculture Organization of the United Nations (FAO) have ranked mycotoxins among the top three causes of foodborne illness. To date, approximately 400 mycotoxins and their derivatives have been identified worldwide. Among these compounds, ochratoxins are of great concern because of their severe toxic effects in animals and humans.8 Ochratoxins are secondary metabolites produced by filamentous fungi such as Aspergillus and Penicillium and are found in a wide range of foods and beverages such as wheat, maize, dried fruits, coffee, wine, and beer.9 There are seven main ochratoxins that are all structurally similar, including ochratoxin A (OTA), ochratoxin B (OTB), and so on. Among them, OTA is widely distributed in agricultural products and plant-derived foods, such as cereals and cereal products, coffee beans, dried fruits, spices, legumes, wine, beer, and grape juice. OTA has a long half-life, high chemical and thermal stability, strong nephrotoxicity, hepatotoxicity, neurotoxicity, immunotoxicity, carcinogenicity, teratogenicity, and mutagenicity and can be enriched in the human body through the food chain and seriously endanger human health.10–13
Different countries have set maximum limits for OTA and other mycotoxins. For example, in the European Union, there are OTA maximum limits of 5.0 μg/kg for cereal raw materials, 3.0 μg/kg for cereal-based foods, 5.0 μg/kg for coffee, and 2.0 μg/kg for grape juice and wine.14 The traditional methods for determination of OTA in foods include thin-layer chromatography,15 high-performance liquid chromatography,16,17 liquid chromatography–mass spectrometry,18 enzyme-linked immunosorbent assays,17,19 and colloidal gold immunochromatography.14,20 Traditional thin-layer chromatography is simple and inexpensive but has poor sensitivity and reproducibility and is time-consuming and difficult to automate; thus, it cannot meet the demands of modern detection methods in terms of speed, sensitivity, reliability, and convenience. With its high sensitivity and accuracy, high-performance liquid chromatography is the most common detection method in laboratories. However, this method uses bulky and expressive instruments that need to be operated by professional technicians, so it is not suitable for rapid on-site detection. Enzyme-linked immunosorbent assays based on immunological principles are rapid, sensitive, and economical but suffer from difficult antibody preparation methods, substrate interference, and high false-positive rates. Colloidal gold immunochromatography is suitable for rapid field testing, but it uses antibodies and is qualitative rather than quantitative.
In recent years, various biosensors for OTA detection using aptamers have been developed.13,21–27 While aptamer-based biosensors have improved the specificity and stability of OTA detection, most of them use large instruments that are not suitable for rapid, on-site detection. The development of affordable point-of-care (POC) methods and devices for toxin monitoring at home is of great importance for early and rapid detection to prevent the spread of diseases and toxins, especially in remote areas with limited access to medical care.
Most POC methods are qualitative colorimetric methods (e.g., colloidal gold immunochromatography). However, accurate quantitative data are more useful for disease diagnosis and toxin analysis.28 Despite recent efforts, there are few POC devices that are widely available to the public. Among the available devices, the personal glucose meter (PGM) is one of the few success stories. Because of its small size, reliable dosing, easy operation, and low cost, the PGM has become the most widely used POC device in the world, and its combination with mobile phones will lead to even wider application.29 However, traditional PGMs can only be used to measure blood glucose levels. To broaden the application range of glucose meters, we and others have coupled an aptamer, DNAzyme, or antibodies to invertase or amylases, thus converting different nonglucose targets into glucose.29–37 Based on this principle, various sensing strategies using PGM as the readout were reported.38–45 However, because the PGMs are used to detect blood glucose that is normally in the millimolar range, the sensitivities of most PGM-based sensors are still low, especially for small molecules.38,44,46–50 To address this issue, we report herein a general aptasensing platform based on a PGM and DNAzyme signal amplification for the detection of small molecules such as mycotoxins, resulting in highly sensitive detection of small molecules by a simple PGM.
MATERIALS AND METHODS
Chemicals and Materials.
Streptavidin-coated magnetic beads (Dynabeads with average diameter of 1 μm) were obtained from Life Technologies AS (Oslo, Norway). Amicon-3K/30K centrifugal filters were bought from Millipore Inc. (Billerica, MA). Grade X Invertase from Candida utilis, Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), sulfoccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC), sucrose, and other chemicals for solvents and buffers were purchased from Sigma-Aldrich, Inc. (St. Louis, MO). The commercial personal glucose meter named Bayer Bleeze 2 was used to evaluate the performance in this work. All DNA oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA) with sequences as shown in Table S1.
Buffers used in this work are as follows:
Buffer A: 1 M sodium phosphate, pH 5.5
Buffer B: 0.1 M sodium phosphate, 150 mM NaCl, pH 7.3
Buffer C: 50 mM Tris, 150 mM NaCl, pH 7.3
Buffer D: 50 mM Tris, 150 mM NaCl, 0.01% Tween-20, pH 7.3
Synthesis of DNA–Invertase Conjugate.
A heterobifunctional linker sulfo-SMCC is used to synthesize the substrate strand–invertase conjugate (S–inv) based on the maleimide–thiol reaction. The whole procedure contained three steps: activation and purification of thiol–DNA, activation and purification of invertase, and preparation and purification of DNA–invertase conjugate. For activation and purification of thiol–DNA, 30 μL of 1 mM thiol–DNA, 2 μL of Buffer A (pH 5.5), and 2 μL of 30 mM TCEP are mixed. After incubating at room temperature for 1 h, the thiol–DNA is purified by Amicon-3K with a centrifugal speed of 13 000 rpm using Buffer B 8 times. For activation and purification of invertase, 1 mg of sulfo-SMCC is dissolved in 200 μL of ultrapure water and then mixed with 400 μL of 20 mg/mL invertase in Buffer B. After vortexing for 5 min, the solution is placed on a shaker for 1 h at room temperature. The mixture is then purified by Amicon-30K with a centrifugal speed of 13 000 rpm using Buffer B 8 times. For preparation and purification of DNA–invertase conjugate, the purified solution of sulfo-SMCC-activated invertase is mixed with the above solution of TCEP-activated thiol–DNA. The resulting solution is kept at room temperature for 48 h. To remove unreacted thiol–DNA, the solution is purified by Amicon-30K with a centrifugal speed of 13 000 rpm using Buffer C 8 times. The DNA–invertase conjugate is finally dispersed in Buffer C at a concentration of 20 mg/mL.
Preparation of Biotin–DNA-Functionalized Magnetic Beads.
A 100 μL portion of the original 10 mg/mL solution of streptavidin-coated magnetic beads (MB) in a microtube is placed close to a magnetic rack for 1 min. The clear solution is discarded, and the brown magnetic beads are rinsed to replace the dissolution buffer by 1 mL of Buffer D. The buffer exchange process is repeated three times, and the magnetic beads are dispersed in 1 mL of Buffer D with a concentration of 1 mg/mL. Then, the mixture of biotin–aptamer probe–DNAzyme conjugate (AP–E) and biotin–substrate strand–invertase (S–inv) with a certain mole ratio (1:1, 1:5, 1:10, and 1:20) in Buffer D is added to the 1 mL of MB solution and mixed on a roller at room temperature (see detailed preparation procedure, including concentrations and volumes of the reagents, in Section A3.3 of the Supporting Information). The streptavidin–biotin reaction is allowed to maintain for 30 min; then, the resulting magnetic beads are washed three times with Buffer D to remove excess biotin–DNA and are ready for ochratoxin A (OTA) detection.
Detection Procedure of OTA Using Commercially Available PGM.
The OTA stock solution is diluted with Buffer D using a syringe in the biosafety cabinet to obtain a working solution of a concentration from 0 to 1 μg/mL. For each sample, 30 μL of the resulting MB solution is placed close to a magnetic rack for 1 min. The clear solution is discarded and replaced by 30 μL of Buffer D with a sufficient concentration of Zn2+ and different concentration of OTA. The reaction is allowed to maintain for 80 min; then, the solution is separated using a magnetic rack, and 10 μL of the supernatant is transferred into 3.3 μL of 2 M sucrose in Tris buffer. After incubating at room temperature for 60 min, 3 μL of the solution is measured using a commercially available PGM. The same procedures are conducted for the selectivity experiment by displacing OTA with other mycotoxins, such as ochratoxin B, aflatoxin B1, aflatoxin B2, deoxynivalenol, fumonisin B1, fumonisin B2, patulin, sterigmatocystin, and zearalenone.
RESULTS AND DISCUSSION
Sensor Design Principles and Signal Amplification Mechanism.
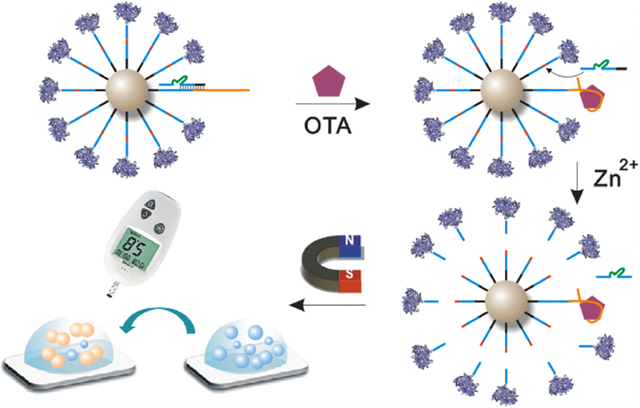
The sensor design and signal amplification mechanism are shown in Figure 1. The system contains three nucleic acid strands: the DNAzyme strand (E), the substrate strand (S), and the aptamer probe (AP). The substrate strand is the 8–17 original substrate strand with additional 12 T bases at the 5′ and 3′ ends as spacers. The key aptamer probe (Figure S1) contains the following four components: Fragment a is the original nucleic acid aptamer sequence of OTA (yellow part in Figure S1) and contains a sticky b fragment that can hybridize with the extension of the DNAzyme strand. Fragment b, which contains 8 bases, is the 3′ arm of the original substrate strand and can hybridize complementarily with the 5′ arm of the DNAzyme strand (Arm 1 in Figure 2A). Fragment c contains 11 bases and is identical to the original substrate strand except for the 3′ arm. Fragment d is a spacer sequence of 12 bases. The DNAzyme strand also consists of four components (Figure 2A). The 5′ Arm 1 (dark blue part in Figure 2A) and the 3′ Arm 2 (light blue part in Figure 2A) can hybridize to the corresponding arms of the substrate strand. The green part in Figure 2A is the catalytic core of the 8–17 DNAzyme, and the sequence of this segment cannot be changed. By contrast, the sequences of the two arms, Arm 1 and Arm 2, can be altered if needed. The two arms and the catalytic core form the original 8–17 DNAzyme strand. The black part within the red dotted box (extension) is an extended part with a different number of bases at the 5′ end compared with the original 8–17 DNAzyme strand. Together, Arm 1 and the extension form the reservoir, which can be fully complementarily hybridized with the aptamer probe to block the DNAzyme strand and prevent it from reacting with the substrate strand in the absence of the target. Both the 5′ end of the substrate strand and the 5′ end of the AP are modified with biotin, enabling them to form a spherical nucleic acid centered on the magnetic beads that is immobilized on their surface through streptavidin–biotin binding (Figure 1B). Connected to the biotin is a spacer of 12 T bases, which extends the reaction radius of this spherical nucleic acid and reduces steric hindrance. Meanwhile, because invertase is a macromolecular protein, a 12 T spacer is also included at the 3′ end of the substrate strand to further reduce steric hindrance and facilitate hybridization between the DNAzyme strand and the substrate strand.
Figure 1.
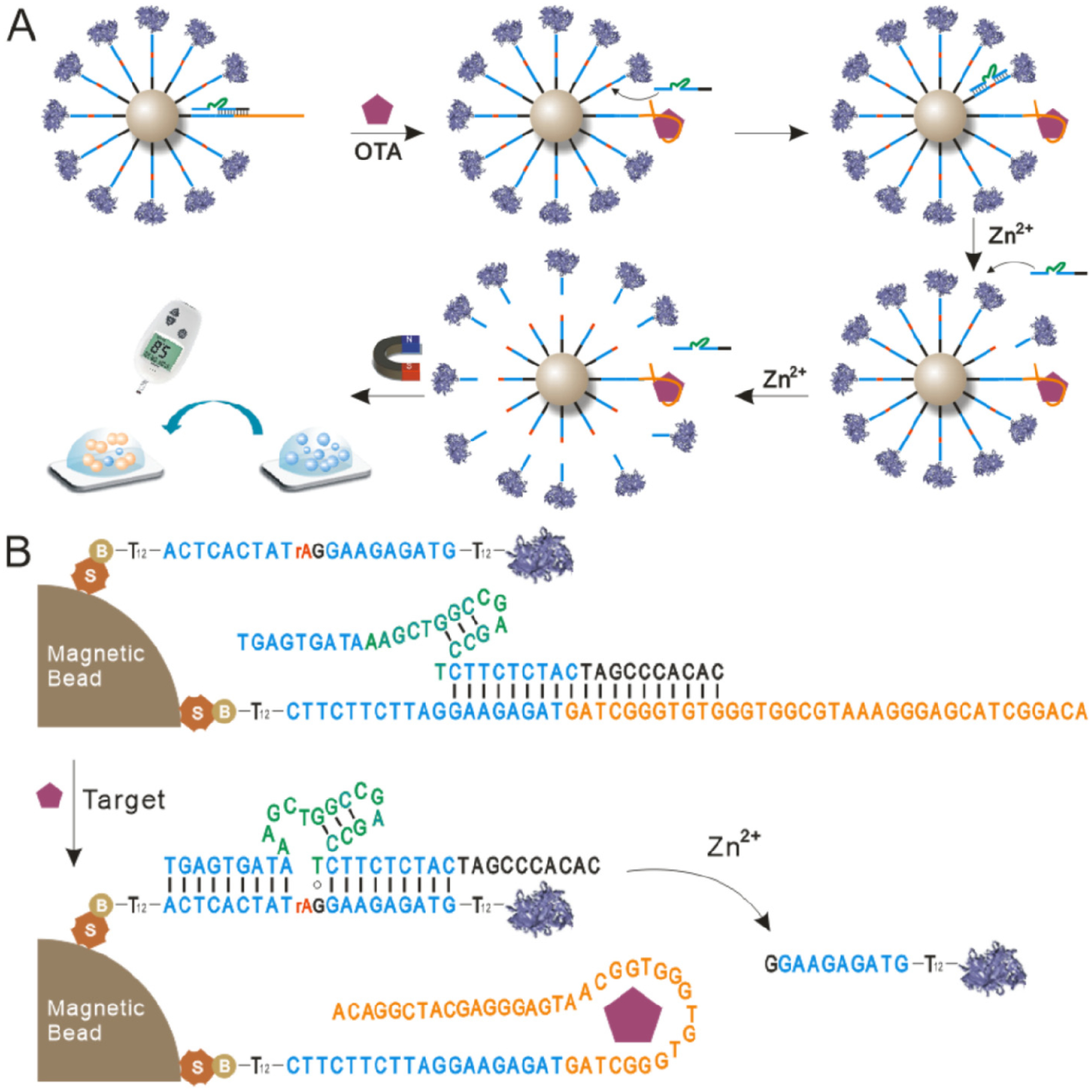
Scheme of aptasensing platform for OTA detection based on DNAzyme amplification. (A) Stepwise illustration of target recognition, signal amplification, and result readout. (B) Schematic diagram of OTA induced displacement of DNAzyme, substrate–DNAzyme hybridization, and release of the invertase-labeled substrate fragment.
Figure 2.
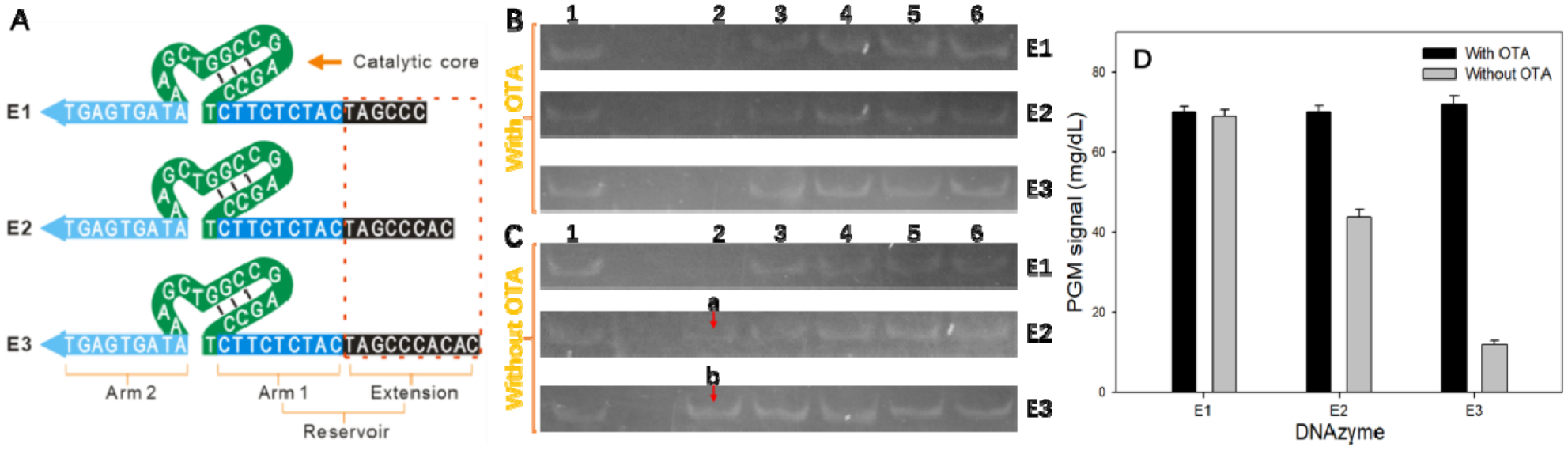
Design and optimization of the DNAzyme strand. (A) Sequence design and functional region of the DNAzyme strand. Arm 1 and Arm 2 have the same sequences with the original 8–17 DNAzyme. The 5′ end of 8–17 DNAzyme is extended by different lengths (6, 8, and 10 bases) of fragments (extension). Arm 1 combined with the extension to form the reservoir part, which is capable of hybridizing with the aptamer probe. Band change of the substrate strand in the electrophoresis experiment by 15% native PAGE using extended DNAzyme in the presence (B) and absence (C) of OTA. Lane 1: 200 nM substrate strand (S). Lanes 2–6: 200 nM AP + 200 nM E1, E2, or E3 + 200 nM S + different concentrations of Zn2+ (100 μM, 10 μM, 1 μM, 100 nM, and 10 nM, respectively) mixed with (B) or without (C) 10 ng/mL OTA. (D) DNAzyme strand optimization by PGM measurements using 1:5 magnetic beads and 10 ng/mL OTA.
In addition, the aptamer sequence of OTA is arranged to the 3′ end instead of the 5′ end because the aptamer located outside the spherical nucleic acid is more convenient for targeting of OTA molecules to the aptamer. In the presence of OTA molecules, the AP undergoes a conformational switch to specifically recognize and bind the OTA molecule. As a result, the DNAzyme strand originally blocked by hybridization with the AP is released and hybridizes to the neighboring substrate strand, undergoing a catalytic reaction in the presence of zinc ions. After the substrate strand is catalyzed, the end modified with invertase is released, while the biotinylated end remains anchored to the magnetic bead. Meanwhile, after catalysis of the substrate strand, hybridization between both arms and the DNAzyme strand becomes unstable, and the DNAzyme strand is released and hybridizes with an adjacent substrate strand, resulting in a new round of catalysis. This cycle can theoretically cut all the substrate strands on the magnetic beads and thus amplify the signal. After magnetic separation, a certain amount of the released substrate strands modified with invertase is taken and added to sucrose to catalyze the conversion of sucrose to glucose. Finally, the glucose content is analyzed by a PGM to achieve the objective of highly sensitive and portable detection of toxin molecules.
This small-molecule toxin sensing system has three main advantages: (1) Cyclic cutting of substrate strands by DNAzyme is used to achieve signal amplification and highly sensitive analysis of the target molecule. Theoretically, specific binding of one molecule of the target to an AP can induce cutting of all substrate strands on the magnetic beads. (2) The signal output is measured using a glucose meter for home use, which is easy to operate, suitable for rapid analysis in the field, and has obvious advantages for toxin analysis in food safety. (3) The sensing mechanism can be easily applied to detect other target molecules by simply replacing the aptamer sequence of OTA with a target-specific aptamer sequence. In addition, compared with other reported signal amplification methods using a personal glucose meter as the readout such as hydrogel collapses,38,51 glucose encapsulating liposome,47,52 nanoparticle-based amplification,46,48 rolling circle amplification,41,53 strand-displacement polymerization,54 exonuclease III-based amplification,50,55 hybridization chain reaction,45 and catalyzed hairpin assembly,56 our proposed DNAzyme-based aptasensing platform is capable of realizing signal amplification without the need of extra signal amplification materials (e.g., hydrogel, liposome, nanoparticles, or a tool enzyme such as polymerase and exonuclease) and complex nucleic acid strand design, endowing the sensing strategy with the merits of easy design and operation.
Characterization of Substrate Strand–Invertase Conjugates for Modification and Purification.
The production of a signal with this sensing system relies on invertase to catalyze the hydrolysis of sucrose to glucose, and this reaction allows for measurements to be made with a glucose meter for home use. The amount of invertase directly affects the measurement results, so purification of the invertase-modified substrate strand is required to ensure that the signal measured by the blood glucose meter is derived from cutting of the substrate strand–invertase conjugate. We characterized purification of the substrate strand–invertase conjugate by UV spectroscopy (Figure S2). UV absorption peaks were observed at 262 nm for substrate strand S and 278 nm for invertase and were nearly identical to those reported for pure nucleic acids (260 nm) and proteins (280 nm). The prepared substrate strand–invertase conjugate (S–inv) showed a UV absorption peak at 267 nm (stable UV absorption peak after several rounds of purification), which was red-shifted relative to the absorption peak of the substrate strand, indicating successful conjugation of the nucleic acid and invertase. The intensity of the absorption peak of the conjugate was reduced relative to that of the substrate strand, indicating that the unmodified substrate strands were separated and removed through purification.
Optimization of the DNAzyme Sequence.
The key of this sensor system is that the presence of the target molecule induces a structural transformation of the AP, which in turn triggers release of the DNAzyme strand and subsequent cyclic cutting and amplification. In the absence of the target molecule, the DNAzyme strand should be blocked to avoid hybridization with the substrate strand and production of a nonspecific signal. In other words, when the target molecule is not present, the DNAzyme strand should be in a closed state formed by hybridization with the aptamer probe. The DNAzyme strand will only be released after addition of the target and become available for subsequent reactions. To achieve this, it is necessary to optimize the sequence of the DNAzyme strand. We added extension sequences of different lengths to the 5′ end of the original 8–17 DNAzyme strand (Figure 2A), which together with the 5′ arm (Arm 1) of the original 8–17 DNAzyme strand formed a reservoir for hybridization with the aptamer probe. Arm 1 and Arm 2 of the DNAzyme strand can hybridize with the two arms of the substrate strand. When the extension at the 5′ end of the DNAzyme strand was too short, the hybridization between the reservoir and the aptamer probe may not be stable enough, so that part of the DNAzyme strand cannot be fully blocked. Consequently, part of the DNAzyme strand hybridized with the substrate strand on the magnetic beads, resulting in a nonspecific cutting reaction. When the 5′ end extension reached a certain length, the reservoir hybridized with the aptamer probe and was blocked completely. This meant that the substrate strands could not hybridize with the DNAzyme strands, and the cutting reaction did not occur. We investigated optimization of the DNAzyme strand in detail using gel electrophoresis and glucose meter measurements. We first used denaturing gel electrophoresis to examine whether extension of the DNAzyme strand affected hybridization with the substrate strand and the cutting reaction activity. The longest extension, E3, was used as a control. Different concentrations of Zn2+ solutions were added after hybridization of the DNAzyme strand with the substrate strand. We found that the proportion of cleaved substrate strand increased with the Zn2+ concentration (Figure S3A). Most of the substrate strand was cut when the Zn2+ concentration reached 100 μM, and complete cutting with this concentration could be achieved by extending the reaction time. The extended DNAzyme strand E3 had a slower electrophoretic mobility than the original DNAzyme strand, but the cutting activities of the two strands were the same (Figure S3B). No cutting activity was observed in the presence of mutant bases in the catalytic core of the mutant DNAzyme strand (ME), even at a Zn2+ concentration of 100 μM. These results demonstrate that cutting of the substrate strand depends on the catalytic core of the DNAzyme, and that extending the arm sequence of the DNAzyme strand has no effect on the cutting activity. We then optimized the DNAzyme strand sequence by native PAGE. The AP was first hybridized with DNAzyme strands of different lengths and then mixed with substrate strands. Next, different concentrations of Zn2+ solutions were added followed by a certain concentration of OTA. The resulting products were then subjected to gel electrophoresis, and band changes of the substrate strands were analyzed. The substrate strand bands became weaker as the Zn2+ concentration increased. When the Zn2+ concentration reached 100 μM, all substrate bands disappeared in the presence of OTA (Figure 2B, complete electrophoresis images shown in Figure S4). This result indicated that the AP could strongly and specifically bind to OTA and that DNAzyme strands of different lengths could be released from the AP–DNAzyme strand conjugate and then cut the substrate strands by cyclic hybridization. When no OTA was added to the sample, part of the DNAzyme strand E1 could react with the substrate strands because of their low hybridization stability with the AP caused by the short reservoir sequence (15 bases). This resulted in a large degree of substrate strand cutting, and no obvious substrate strand band was observed for the DNAzyme strand E1 (Lane 2 in Figure 2C). When the reservoir sequence of DNAzyme strand E2 was extended to 17 bases, DNAzyme strand E2 showed more stable hybridization with the AP, and a weaker substrate strand band (Band a in Figure 2C) in Lane 2 was found, which indicated that the substrate strand was only partially cut.
In contrast, when DNAzyme strand E3 was used, the intensity of the substrate strand band (Band b in Figure 2C) was almost the same as that of the pure substrate strand, indicating that the substrate strand was barely cut even with a Zn2+ concentration of 100 μM. This is because the reservoir sequence of the DNAzyme strand E3 has up to 19 bases hybridized with the AP, and the hybridization is stable with almost no free DNAzyme strands reacting with the substrate strands. The three DNAzyme strands were used to prepare magnetic beads separately, and control experiments were performed using 10 ng/mL OTA and 1:5 magnetic beads in the presence of 500 μM Zn2+ (Figure 2D). In the presence of OTA, the signals obtained with the three types of magnetic beads were similar, indicating that when the three DNAzyme strands hybridized with the AP, the OTA molecules were able to release the DNAzyme strands and initiate subsequent cyclic hybridization and cutting reactions because of the specific binding of the OTA target to the AP. Without OTA, the reaction system prepared with DNAzyme strand E1 and E2 had a relatively high background signal, while a very low signal was detected in the absence of OTA with the magnetic beads prepared using DNAzyme strand E3. This confirmed that the reaction system developed with DNAzyme strand E3 had a very low background signal and the highest signal-to-noise ratio. Therefore, E3 was chosen to prepare magnetic beads for OTA detection in subsequent experiments.
Optimization of the Cofactor Zn2+ Concentration.
The cleavage activity of DNAzyme usually requires the existence of specific metal ions as cofactors. For example, 8–17 DNAzyme is catalytically active in the presence of Pb2+, Zn2+, and Mg2+, with its activity in the order of Pb2+ > Zn2+ > Mg2+.57 Considering that contamination with the heavy metal Pb2+ is not beneficial, we selected the metal ion with the next highest catalytic activity, Zn2+, for use in the present study. Electrophoretic characterization showed that increasing the Zn2+ concentration improved the catalytic activity of DNAzyme and resulted in more complete substrate cleavage. However, it is necessary to optimize the Zn2+ concentration because the magnetic bead-based reaction system adopts an interfacial reaction, which is different from the homogeneous reaction in the electrophoresis experiments. In the solution of substrate-modified magnetic beads, cleavage of the substrate strand by DNAzyme strand E3 occurred with different concentrations of Zn2+. As shown in Figure S5A, when the Zn2+ concentration was 10 μM, only a low signal was detected by the PGM, indicating that there was insufficient DNAzyme cleavage activity in the magnetic bead system at this Zn2+ concentration. As the concentration of Zn2+ increased, the signal detected by the PGM also increased gradually and reached its maximum when the Zn2+ concentration reached 500 μM. However, when the concentration of Zn2+ increased further, the signal detected by the PGM decreased a little. This could be attributed to the decreased effective concentration of Zn2+ caused by the hydrolysis effect that occurred with a high Zn2+ concentration. To optimize the catalytic effect of DNAzyme, a Zn2+ concentration of 500 μM was chosen for subsequent experiments with the magnetic bead reaction system.
Ratio Optimization of the Biotin-Labeled Nucleic Acids.
In this study, specific binding of the target molecule to the aptamer probe was used to release the DNAzyme, which then hybridized with the substrate strand to catalyze its hydrolysis for signal amplification. The magnetic beads were modified with both biotin-labeled aptamer probe–DNAzyme conjugate (AP–E) and biotin-labeled substrate strand–invertase (S–inv), and the AP–E/S–inv ratio directly influenced the signal amplification. We measured the signal responses with different AP–E/S–inv ratios (1:1, 1:5, 1:10, and 1:20) by fixing the amount of aptamer probe and varying the amount of substrate strand (Figure S5B). Incubation of a certain concentration of OTA with magnetic beads modified at different AP–E/S–inv ratios showed that the signal detected by the PGM became stronger as the AP–E/S–inv ratio increased. This occurred because, in the presence of sufficient reaction sites on the surface of the magnetic beads, a higher AP–E/S–inv ratio (i.e., more biotin-labeled substrate strands) resulted in cleavage of more substrate strands and production of more invertase, which catalyzed the generation of more glucose and produced a higher detection signal. The highest signal was obtained when the AP–E/S–inv ratio was 1:10. However, when the AP–E/S–inv ratio increased further to 1:20, the signal detected by the PGM decreased. This could be attributed to the fact that the number of reaction sites on the surface of the magnetic beads is limited, and when the AP–E/S–inv ratio reaches 1:20, there will be insufficient reaction sites and an excess of substrate strands. Therefore, mixing of the AP–E/S–inv mixture with magnetic beads may result in a decrease in the amount of biotin-labeled aptamer probe–DNAzyme conjugate bound to the surface of the magnetic beads. When reacting with the same concentration of OTA, the amount of DNAzyme released is reduced, resulting in a reduction of the detection signal. Remarkably, when using a set AP–E/S–inv ratio, the same results were obtained with different concentrations of OTA (10 and 0.1 ng/mL). The maximum detection signal was obtained at an AP–E/S–inv ratio of 1:10. This confirmed the stability of the magnetic bead reaction system and the validity of our reasoning for the cause of the signal changes with AP–E/S–inv ratio changes.
Investigation of Reaction Time.
The reaction time between invertase and sucrose also had a direct effect on the signal detected by the PGM. A certain amount of invertase catalyzes the continuous conversion of sucrose to glucose in the presence of an excess amount of sucrose, resulting in detection of a stronger signal by the PGM (Figure S5C). The signal detected by the PGM is linearly correlated with the reaction time when a sufficient amount of sucrose is present. Although the signal strength increases with the reaction time, a high signal that is sufficient for the sensor analysis can be obtained when the reaction time reaches 60 min. To minimize the reaction time, the invertase and sucrose reaction time in subsequent reactions was set at 60 min. In addition, the reaction time between OTA and the aptamer probe also directly affects the signal response of the sensor. A more complete reaction between OTA and the aptamer probe can release more DNAzyme, resulting in more cleaved substrate strands and detection of a higher signal by the PGM (Figure S5D). With increases in the reaction time above 80 min, the detected signal remained unchanged, which indicated that OTA had fully reacted with the aptamer probe. Therefore, 80 min was chosen as the optimal reaction time between OTA and the AP for subsequent experiments.
OTA Detection Performance.
The signal response of the sensor with different concentrations of OTA was examined under the optimized experimental conditions (Figure 3). As the OTA concentration increased, more DNAzyme strands were released, catalyzing the degradation of more substrate strands and generating a higher signal. The signal detected by the PGM showed a good linear relationship with the logarithm of the OTA concentration between 1 pg/mL and 300 ng/mL, with a linear equation of S = 19.35 log C + 75.30 and a correlation coefficient of 0.9926. The limit of detection calculated using 3 times the signal-to-noise ratio was 0.88 pg/mL. The detection limit and quantitative range of the designed sensor meet the general requirements for OTA detection, and the sensor could be used for quantitative analysis.
Figure 3.
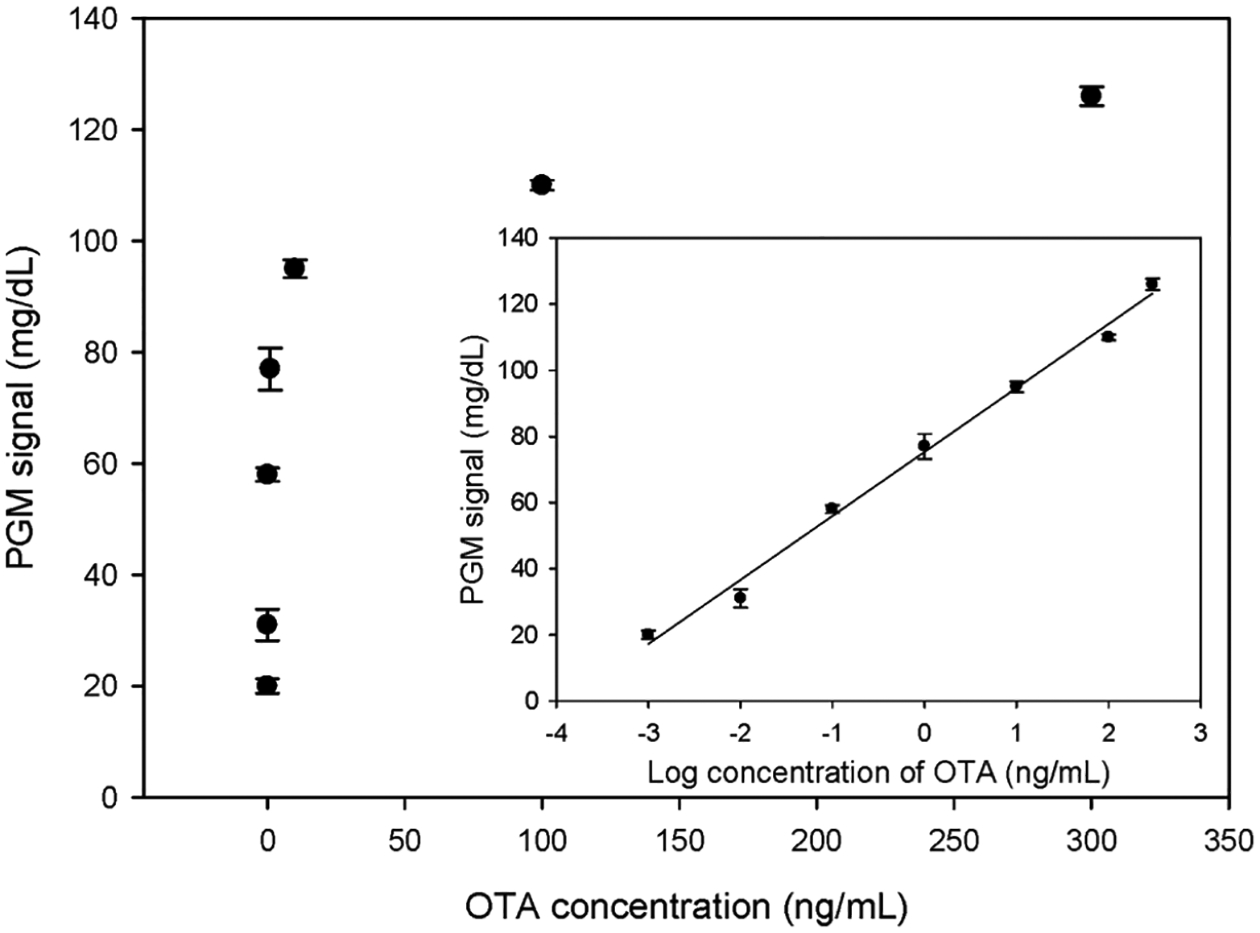
Sensor response to different concentrations of OTA. Inset: linear relationship between the PGM signal and log concentration of OTA. The error bars denote the standard derivation of three repeated measurements.
Compared with conventional OTA detection methods, such as instrumental methods including high-performance liquid chromatography (HPLC) and liquid chromatography–mass spectrometry (LC–MS), immunoassay methods such as enzyme-linked immunosorbent assays and gold immunochromatographic assays, and other newly developed detection techniques using nucleic acid aptamers, such as electrochemical,22,58–60 chemiluminescent,61 colorimetric,62,63 fluorescent,13,64,65 and surface-enhanced Raman scattering,12,66,67 the present sensor system has a lower detection limit and a linear detection range of more than 5 orders of magnitude, showing better analytical performance for OTA detection. In addition, compared with the dissociation constant Kd of 200 nM for the original OTA aptamer,68 an apparent dissociation constant Kd of 0.169 nM was obtained when fitting Figure 3 to a binding curve (shown in Figure S6), which is much smaller because of the DNAzyme-based amplification effect. The selectivity of our sensing strategy was also examined. OTA at 300 and 1.0 ng/mL produced much higher signals than the blank sample and higher concentrations (500 ng/mL) of other mycotoxins, such as OTB, aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), deoxynivalenol (DON), fumonisin B1 (FB1), fumonisin B2 (FB2), patulin (PAT), sterigmatocystin (ST), and zearalenone (ZEA) (Figure 4).
Figure 4.
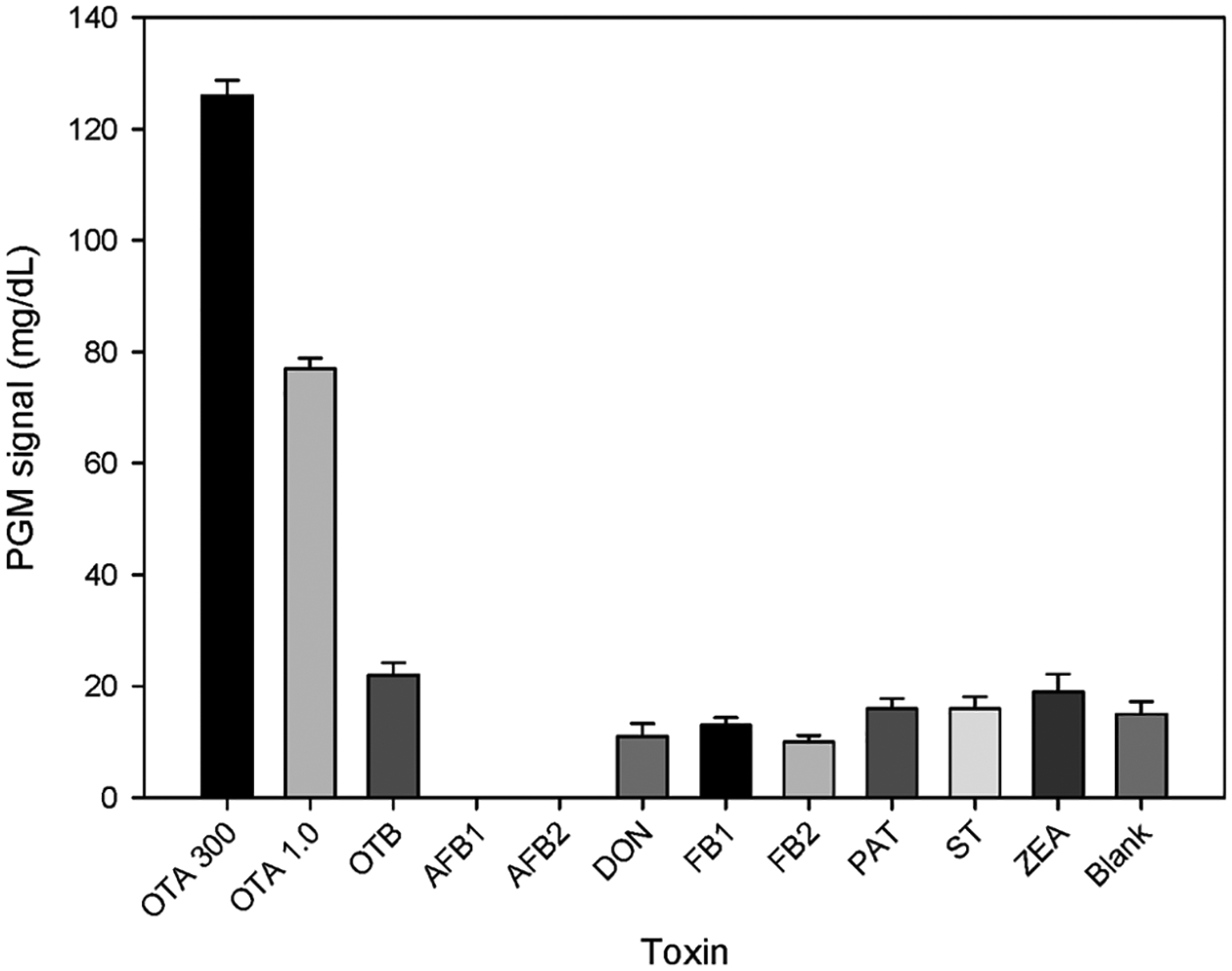
Selectivity investigation of the DNAzyme amplified aptasensing platform using different kinds of toxins. The concentrations of other mycotoxins are 500 ng/mL, and the error bars represent the standard deviation of three repeated measurements.
Among these mycotoxins, five of them (AFB1, AFB2, DON, FB1, and FB2) produced lower signals than the blank sample. Mycotoxins AFB1 and AFB2 even showed a “LOW” signal on the glucose meter, which might be due to the effects of these toxins on the activity of invertase at high concentrations. Although OTB has a similar structure to the target molecule OTA, the sensing system still showed good selectivity for OTA. Notably, the Bayer Breeze 2 glucose meter detects glucose in the range 0.6–33.3 mmol/L (mass concentration range 10–600 mg/dL). When the detected concentration is less than 10 mg/dL, the system displays the result “LOW” instead of a specific value.
Practical Sample Analysis and Comparison with the Standard Method.
HPLC-MS is one of the most common methods for OTA detection. HPLC-MS can be used for both qualitative and quantitative analysis of OTA and is the standard method for OTA detection in many countries. To investigate the feasibility of the method proposed in this study for the detection of OTA in practical samples, the standard addition method was used with red wine samples, and the results were compared with those obtained by HPLC-MS (Table 1).
Table 1.
Comparison of the Proposed Sensing Platform with the Standard HPLC-MS Method
| sample | OTA added (ng/mL) | OTA found (ng/mL), proposed method | recovery, proposed method | OTA found (ng/mL), HPLC-MS method | relative error |
|---|---|---|---|---|---|
| 1 | 300 | 279.2 | 93.1% | 298.3 | −6.4% |
| 2 | 10 | 10.83 | 108.3% | 10.65 | +1.7% |
| 3 | 0.1 | 0.1034 | 103.4% | 0.1032 | +0.2% |
Different concentrations of OTA in the linear detection range (0.1, 10, and 300 ng/mL) were added to red wine, and the recoveries obtained by the proposed method in this study ranged from 93.1% to 108.3%. The error of the proposed method ranged from −6.4% to 1.7% relative to the HPLC-MS standard method, indicating that there were no significant differences between the results from this method and those from HPLC-MS. Therefore, this method could be used for accurate quantitative analysis of OTA in practical samples. Moreover, compared with the HPLC-MS standard method, the present sensing strategy does not require expensive analytical instruments or professional operators. Consequently, it could be used as a useful supplement to the HPLC-MS standard method, especially for rapid analysis in the field.
CONCLUSIONS
In this study, we have reported a sensing platform that uses a nucleic acid aptamer and DNAzyme signal amplification. Specific binding of OTA to the aptamer probe releases the DNAzyme strand, which in turn triggers a series of cyclic hybridization and cutting reactions to achieve signal amplification and highly sensitive analysis of OTA. After optimization of the DNAzyme sequence, cofactor (Zn2+) concentration, ratio of the two biotin-labeled DNA strands, and incubation time, the resulting sensor displays a linear relationship between 1.0 pg/mL and 300 ng/mL, and a limit of detection for OTA was as low as 0.88 pg/mL. This method is selective and easy to perform and does not require expensive instruments or professional operators, so it is a promising choice for the detection of the small-molecule mycotoxin OTA. In addition, this method can be applied to the detection of other target molecules by modifying the aptamer probe sequences and hybridized DNAzyme sequences.
Supplementary Material
ACKNOWLEDGMENTS
This study is financially supported by the National Key Research and Development Program of China (Grant 2016YFD0400902), Research Foundation of Education Bureau of Hunan Province (Grant 19B384), Beijing Agricultural Forestry Academy Youth Foundation (Grant QNJJ201903), Open Project of State Key Laboratory of Chemo/biosensing and Chemometrics (Grant 2016010), China Postdoctoral Science Foundation (Grant 2015M582339), Changde Science and Technology Innovation Project (Grant 2019ZD25, 2020ZD29), and US National Institute of Health (GM124316 and MH110975).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.0c20417.
Experimental procedures including UV spectrum characterization of substrate strand–invertase conjugate, electrophoresis characterization procedure, and optimization of experimental conditions; oligonucleotide sequences used in this work; additional figures including schemes of the designed aptamer probe, UV spectrum of S–inv, and PAGE for investigation of DNAzyme activity; and optimization of experimental conditions (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsami.0c20417
The authors declare the following competing financial interest(s): Y. Lu is a co-founder of GlucoSentient, Inc., a startup company in Champaign, IL that has licensed the background IP of this work.
Contributor Information
Songbai Zhang, Hunan Provincial Key Laboratory of Water Treatment Functional Materials, Hunan Province Engineering Research Center of Electroplating Wastewater Reuse Technology, Hunan Province Cooperative Innovation Center for The Construction & Development of Dongting Lake Ecological Economic Zone, College of Chemistry and Chemical Engineering, Hunan University of Arts and Science, Changde 415000, P. R. China; Department of Chemistry, University of Illinois at Urbana–Champaign, Urbana, Illinois 61801, United States.
Yunxia Luan, Beijing Research Center for Agricultural Standards and Testing, Agricultural Product Quality and Safety Risk Assessment Laboratory of the Department of Agriculture, Beijing Municipal Key Laboratory of Agriculture Environment Monitoring, Beijing 100097, P. R. China; Department of Chemistry, University of Illinois at Urbana–Champaign, Urbana, Illinois 61801, United States.
Mengyi Xiong, Department of Chemistry, University of Illinois at Urbana–Champaign, Urbana, Illinois 61801, United States.
Jingjing Zhang, Department of Chemistry, University of Illinois at Urbana–Champaign, Urbana, Illinois 61801, United States.
Ryan Lake, Department of Chemistry, University of Illinois at Urbana–Champaign, Urbana, Illinois 61801, United States.
Yi Lu, Department of Chemistry, University of Illinois at Urbana–Champaign, Urbana, Illinois 61801, United States.
REFERENCES
- (1).Ellington AD; Szostak JW In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346 (6287), 818–822. [DOI] [PubMed] [Google Scholar]
- (2).Tuerk C; Gold L Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249 (4968), 505–510. [DOI] [PubMed] [Google Scholar]
- (3).Shangguan D; Li Y; Tang Z; Cao ZC; Chen HW; Mallikaratchy P; Sefah K; Yang CJ; Tan W Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc. Natl. Acad. Sci. U. S. A 2006, 103 (32), 11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fang X; Tan W Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc. Chem. Res 2010, 43 (1), 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhang S; Hu X; Yang X; Sun Q; Xu X; Liu X; Shen G; Lu J; Shen G; Yu R Background Eliminated Signal-on Electrochemical Aptasensing Platform for Highly Sensitive Detection of Protein. Biosens. Bioelectron 2015, 66, 363–369. [DOI] [PubMed] [Google Scholar]
- (6).Zhang S; Hu X; Geng H; Huang M; Qiu Y; Shen C; Wang S; Shen G; Yang M A Highly Sensitive and Signal-on Electrochemical Aptasensor Based on Restriction Endonuclease Induced Background Elimination. J. Electrochem. Soc 2016, 163 (8), B411–B416. [Google Scholar]
- (7).Stoev SD Food Security and Foodborne Mycotoxicoses, Risk Assessment, Preventive Measures, and Underestimated Hazard of Masked Mycotoxins or Joint Mycotoxin Interaction. Food Toxicol 2016, 9, 169–199. [Google Scholar]
- (8).Ünüsan N Systematic Review of Mycotoxins in Food and Feeds in Turkey. Food Control 2019, 97, 1–14. [Google Scholar]
- (9).Pagkali V; Petrou PS; Salapatas A; Makarona E; Peters J; Haasnoot W; Jobst G; Economou A; Misiakos K; Raptis I; Kakabakos SE Detection of Ochratoxin A in Beer Samples with a Label-Free Monolithically Integrated Optoelectronic Biosensor. J. Hazard. Mater 2017, 323, 75–83. [DOI] [PubMed] [Google Scholar]
- (10).Sun AL; Zhang YF; Sun GP; Wang XN; Tang D Homogeneous Electrochemical Detection of Ochratoxin A in Foodstuff Using Aptamer–Graphene Oxide Nanosheets and DNase I-Based Target Recycling Reaction. Biosens. Bioelectron 2017, 89, 659–665. [DOI] [PubMed] [Google Scholar]
- (11).Tian J; Wei W; Wang J; Ji S; Chen G; Lu J Fluorescence Resonance Energy Transfer Aptasensor between Nanoceria and Graphene Quantum Dots for the Determination of Ochratoxin A. Anal. Chim. Acta 2018, 1000, 265–272. [DOI] [PubMed] [Google Scholar]
- (12).Jing X; Chang L; Shi L; Liu X; Zhao Y; Zhang W Au Film-Au@Ag Core-Shell Nanoparticle Structured Surface-Enhanced Raman Spectroscopy Aptasensor for Accurate Ochratoxin A Detection. ACS Appl. Bio Mater 2020, 3 (4), 2385–2391. [DOI] [PubMed] [Google Scholar]
- (13).Zhu X; Li W; Lin L; Huang X; Xu H; Yang G; Lin Z Target-Responsive Ratiometric Fluorescent Aptasensor for OTA Based on Energy Transfer between [Ru(Bpy)3]2+ and Silica Quantum Dots. Microchim. Acta 2020, 187 (5), 270. [DOI] [PubMed] [Google Scholar]
- (14).Liu L; Xu L; Suryoprabowo S; Song S; Kuang H Development of an Immunochromatographic Test Strip for the Detection of Ochratoxin A in Red Wine. Food Agric. Immunol 2018, 29 (1), 434–444. [Google Scholar]
- (15).Lin L; Zhang J; Wang P; Wang Y; Chen J Thin-Layer Chromatography of Mycotoxins and Comparison with Other Chromatographic Methods. J. Chromatogr. A 1998, 815, 3–20. [DOI] [PubMed] [Google Scholar]
- (16).Moez E; Noel D; Brice S; Benjamin G; Pascaline A Aptamer Assisted Ultrafiltration Cleanup with High Performance Liquid Chromatography-fluorescence Detector for the Determination of OTA in Green Coffee. Food Chem 2020, 310, 125851. [DOI] [PubMed] [Google Scholar]
- (17).Dohnal V; Dvořák V; Malíř F; Ostrý V; Roubal T A Comparison of ELISA and HPLC Methods for Determination of Ochratoxin A in Human Blood Serum in the Czech Republic. Food Chem. Toxicol 2013, 62, 427–431. [DOI] [PubMed] [Google Scholar]
- (18).Al-Taher F; Banaszewski K; Jackson L; Zweigenbaum J; Ryu D; Cappozzo J Rapid Method for the Determination of Multiple Mycotoxins in Wines and Beers by LC-MS/MS Using a Stable Isotope Dilution Assay. J. Agric. Food Chem 2013, 61 (10), 2378–2384. [DOI] [PubMed] [Google Scholar]
- (19).Sun Z; Wang X; Tang Z; Chen Q; Liu X Ecotoxicology and Environmental Safety Development of a Biotin-Streptavidin-Ampli Fi Ed Nanobody-Based ELISA for Ochratoxin A in Cereal. Ecotoxicol. Environ. Saf 2019, 171, 382–388. [DOI] [PubMed] [Google Scholar]
- (20).Tian M; Xie W; Zhang T; Liu Y; Lu Z; Ming C; Liu Y A Sensitive Lateral Flow Immunochromatographic Strip with Prussian Blue Nanoparticles Mediated Signal Generation and Cascade Ampli Fi Cation. Sens. Actuators, B 2020, 309, 127728. [Google Scholar]
- (21).Huang X-B; Wu SH; Hu HC; Sun JJ AuNanostar@4-MBA@Au Core-Shell Nanostructure Coupled with Exonuclease III-Assisted Cycling Amplification for Ultrasensitive SERS Detection of Ochratoxin A. ACS Sensors 2020, 5 (8), 2636–2643. [DOI] [PubMed] [Google Scholar]
- (22).Suea-Ngam A; Howes PD; Stanley CE; Demello AJ An Exonuclease I-Assisted Silver-Metallized Electrochemical Aptasensor for Ochratoxin A Detection. ACS Sensors 2019, 4 (6), 1560–1568. [DOI] [PubMed] [Google Scholar]
- (23).Qian J; Ren C; Wang C; Chen W; Lu X; Li H; Liu Q; Hao N; Li H; Wang K Magnetically Controlled Fluorescence Aptasensor for Simultaneous Determination of Ochratoxin A and Aflatoxin B1. Anal. Chim. Acta 2018, 1019, 119–127. [DOI] [PubMed] [Google Scholar]
- (24).Song C; Hong W; Zhang X; Lu Y Label-Free and Sensitive Detection of Ochratoxin A Based on DsDNA-Templated Copper Nanoparticles and Exonuclease-Catalyzed Target Recycling Amplification. Analyst 2018, 143, 1829–1834. [DOI] [PubMed] [Google Scholar]
- (25).Liu C; Guo Y; Luo F; Rao P; Fu C; Wang S Homogeneous Electrochemical Method for Ochratoxin A Determination Based on Target Triggered Aptamer Hairpin Switch and Exonuclease III-Assisted Recycling Amplification. Food Analytical Methods 2017, 10 (6), 1982–1990. [Google Scholar]
- (26).Modh H; Scheper T; Walter JG Detection of Ochratoxin A by Aptamer-Assisted Real-Time PCR-Based Assay (Apta-QPCR). Eng. Life Sci 2017, 17 (8), 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Dai S; Wu S; Duan N; Wang Z A Luminescence Resonance Energy Transfer Based Aptasensor for the Mycotoxin Ochratoxin A Using Upconversion Nanoparticles and Gold Nano-rods. Microchim. Acta 2016, 183 (6), 1909–1916. [Google Scholar]
- (28).Xiang Y; Lu Y Portable and Quantitative Detection of Protein Biomarkers and Small Molecular Toxins Using Antibodies and Ubiquitous Personal Glucose Meters. Anal. Chem 2012, 84, 4174–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zhang J; Shen Z; Xiang Y; Lu Y Integration of Solution-Based Assays onto Lateral Flow Device for One-Step Quantitative Point-of-Care Diagnostics Using Personal Glucose Meter. ACS Sensors 2016, 1 (9), 1091–1096. [Google Scholar]
- (30).Xiang Y; Lu Y Using Personal Glucose Meters and Functional DNA Sensors to Quantify a Variety of Analytical Targets. Nat. Chem 2011, 3 (9), 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Xiang Y; Lu Y Using Commercially Available Personal Glucose Meters for Portable Quantification of DNA. Anal. Chem 2012, 84 (4), 1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Xiang Y; Lu Y Portable and Quantitative Detection of Protein Biomarkers and Small Molecular Toxins Using Antibodies and Ubiquitous Personal Glucose Meters. Anal. Chem 2012, 84 (9), 4174–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Xiang Y; Lu Y An Invasive DNA Approach toward a General Method for Portable Quantification of Metal Ions Using a Personal Glucose Meter. Chem. Commun 2013, 49 (6), 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Xiang Y; Lan T; Lu Y Using the Widely Available Blood Glucose Meter to Monitor Insulin and HbA1c. J. Diabetes Sci. Technol 2014, 8 (4), 855–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhang J; Xiang Y; Novak DE; Hoganson GE; Zhu J; Lu Y Using a Personal Glucose Meter and Alkaline Phosphatase for Point-of-Care Quantification of Galactose-1-Phosphate Uridyltransferase in Clinical Galactosemia Diagnosis. Chem. - Asian J 2015, 10 (10), 2221–2227. [DOI] [PubMed] [Google Scholar]
- (36).Gu C; Lan T; Shi H; Lu Y Portable Detection of Melamine in Milk Using a Personal Glucose Meter Based on an in Vitro Selected Structure-Switching Aptamer. Anal. Chem 2015, 87 (15), 7676–7682. [DOI] [PubMed] [Google Scholar]
- (37).Zhang J; Xiang Y; Wang M; Basu A; Lu Y Dose-Dependent Response of Personal Glucose Meters to Nicotinamide Coenzymes: Applications to Point-of-Care Diagnostics of Many Non-Glucose Targets in a Single Step. Angew. Chem., Int. Ed 2016, 55 (2), 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yan L; Zhu Z; Zou Y; Huang Y; Liu D; Jia S; Xu D; Wu M; Zhou Y; Zhou S; Yang C Target-Responsive “Sweet” Hydrogel with Glucometer Readout for Portable and Quantitative Detection of Non-Glucose Targets. J. Am. Chem. Soc 2013, 135 (10), 3748–3751. [DOI] [PubMed] [Google Scholar]
- (39).Wang Q; Wang H; Yang X; Wang K; Liu F; Zhao Q; Liu P; Liu R Multiplex Detection of Nucleic Acids Using a Low Cost Microfluidic Chip and a Personal Glucose Meter at the Point-of-Care. Chem. Commun 2014, 50 (29), 3824–3826. [DOI] [PubMed] [Google Scholar]
- (40).Lin B; Liu D; Yan J; Qiao Z; Zhong Y; Yan J; Zhu Z; Ji T; Yang CJ Enzyme-Encapsulated Liposome-Linked Immunosorbent Assay Enabling Sensitive Personal Glucose Meter Readout for Portable Detection of Disease Biomarkers. ACS Appl. Mater. Interfaces 2016, 8 (11), 6890–6897. [DOI] [PubMed] [Google Scholar]
- (41).Xue Q; Kong Y; Wang H; Jiang W Liposome-Encoded Magnetic Beads Initiated by Padlock Exponential Rolling Circle Amplification for Portable and Accurate Quantification of Micro-RNAs. Chem. Commun 2017, 53 (78), 10772–10775. [DOI] [PubMed] [Google Scholar]
- (42).Zhang Y; Xue Q; Liu J; Wang H Magnetic Bead-Liposome Hybrids Enable Sensitive and Portable Detection of DNA Methyltransferase Activity Using Personal Glucose Meter. Biosens. Bioelectron 2017, 87, 537–544. [DOI] [PubMed] [Google Scholar]
- (43).Hong L; Zhou F; Shi D; Zhang X; Wang G Portable Aptamer Biosensor of Platelet-Derived Growth Factor-BB Using a Personal Glucose Meter with Triply Amplified. Biosens. Bioelectron 2017, 95, 152–159. [DOI] [PubMed] [Google Scholar]
- (44).Ming J; Fan W; Jiang TF; Wang YH; Lv ZH Portable and Sensitive Detection of Copper(II) Ion Based on Personal Glucose Meters and a Ligation DNAzyme Releasing Strategy. Sens. Actuators, B 2017, 240, 1091–1098. [Google Scholar]
- (45).Zhu X; Kou F; Xu H; Lin L; Yang G; Lin Z A Highly Sensitive Aptamer-Immunoassay for Vascular Endothelial Growth Factor Coupled with Portable Glucose Meter and Hybridization Chain Reaction. Sens. Actuators, B 2017, 253, 660–665. [Google Scholar]
- (46).Liang X; Wang L; Wang D; Zeng L; Fang Z Portable and Quantitative Monitoring of Mercury Ions Using DNA-Gated Mesoporous Silica Nanoparticles Using a Glucometer Readout. Chem. Commun 2016, 52 (10), 2192–2194. [DOI] [PubMed] [Google Scholar]
- (47).Tang J; Huang Y; Liu H; Zhang C; Tang D Novel Glucometer-Based Immunosensing Strategy Suitable for Complex Systems with Signal Amplification Using Surfactant-Responsive Cargo Release from Glucose-Encapsulated Liposome Nanocarriers. Biosens. Bioelectron 2016, 79, 508–514. [DOI] [PubMed] [Google Scholar]
- (48).Zhang J; Tang Y; Teng L; Lu M; Tang D Low-Cost and Highly Efficient DNA Biosensor for Heavy Metal Ion Using Specific DNAzyme-Modified Microplate and Portable Glucometer-Based Detection Mode. Biosens. Bioelectron 2015, 68, 232–238. [DOI] [PubMed] [Google Scholar]
- (49).Chen S; Zhang J; Gan N; Hu F; Li T; Cao Y; Pan D An On-Site Immunosensor for Ractopamine Based on a Personal Glucose Meter and Using Magnetic β-Cyclodextrin-Coated Nanoparticles for Enrichment, and an Invertase-Labeled Nanogold Probe for Signal Amplification. Microchim. Acta 2015, 182 (3–4), 815–822. [Google Scholar]
- (50).Zeng L; Gong J; Rong P; Liu C; Chen J A Portable and Quantitative Biosensor for Cadmium Detection Using Glucometer as the Point-of-Use Device. Talanta 2019, 198, 412–416. [DOI] [PubMed] [Google Scholar]
- (51).Gao X; Li X; Sun X; Zhang J; Zhao Y; Liu X; Li F DNA Tetrahedra-Cross-Linked Hydrogel Functionalized Paper for Onsite Analysis of DNA Methyltransferase Activity Using a Personal Glucose Meter. Anal. Chem 2020, 92 (6), 4592–4599. [DOI] [PubMed] [Google Scholar]
- (52).Zhao Y; Du D; Lin Y Glucose Encapsulating Liposome for Signal Amplification for Quantitative Detection of Biomarkers with Glucometer Readout. Biosens. Bioelectron 2015, 72, 348–354. [DOI] [PubMed] [Google Scholar]
- (53).Jia Y; Sun F; Na N; Ouyang J Detection of P53 DNA Using Commercially Available Personal Glucose Meters Based on Rolling Circle Amplification Coupled with Nicking Enzyme Signal Amplification. Anal. Chim. Acta 2019, 1060, 64–70. [DOI] [PubMed] [Google Scholar]
- (54).Xue-Tao X; Kai-Yi L; Jia-Ying Z Portable and Sensitive Quantitative Detection of DNA Based on Personal Glucose Meters and Isothermal Circular Strand-Displacement Polymerization Reaction. Biosens. Bioelectron 2015, 64, 671–675. [DOI] [PubMed] [Google Scholar]
- (55).Xue-Tao X; Kai-Yi L; Jia-Ying Z Portable and Sensitive Quantitative Detection of DNA Using Personal Glucose Meters and Exonuclease III-Assisted Signal Amplification. Analyst 2014, 139 (19), 4982–4986. [DOI] [PubMed] [Google Scholar]
- (56).Liu C; An Y; Zhang Y; Li X; Xue Q; Wang H Digital Quantitative Detection of Serum Circulating MiRNAs Using Dual-Enhanced Magnetobiosensors Based on Cascaded Nucleic Acid Circuits. Chem. Commun 2019, 55 (91), 13733–13736. [DOI] [PubMed] [Google Scholar]
- (57).Li J; Lu Y A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J. Am. Chem. Soc 2000, 122 (27), 10466–10467. [Google Scholar]
- (58).Zhang J; Xu X; Qiang Y Ultrasensitive Electrochemical Aptasensor for Ochratoxin A Detection Using AgPt Bimetallic Nanoparticles Decorated Iron-Porphyrinic Metal-Organic Framework for Signal Amplification. Sens. Actuators, B 2020, 312, 127964. [Google Scholar]
- (59).Zhu C; Liu D; Li Y; Shen X; Ma S; Liu Y; You T Ratiometric Electrochemical Aptasensor for Ultrasensitive Detection of Ochratoxin A Based on a Dual Signal Amplification Strategy: Engineering the Binding of Methylene Blue to DNA. Biosens. Bioelectron 2020, 150, 111814. [DOI] [PubMed] [Google Scholar]
- (60).Taghdisi SM; Danesh NM; Ramezani M; Alibolandi M; Nameghi MA; Gerayelou G; Abnous K A Novel Electrochemical Aptasensor for Ochratoxin a Sensing in Spiked Food Using Strand-Displacement Polymerase Reaction. Talanta 2021, 223, 121705. [DOI] [PubMed] [Google Scholar]
- (61).Yan XL; Xue XX; Luo J; Jian YT; Tong L; Zheng XJ Construction of Chemiluminescence Aptasensor Platform Using Magnetic Microsphere for Ochratoxin A Detection Based on G Bases Derivative Reaction and Au NPs Catalyzing Luminol System. Sens. Actuators, B 2020, 320, 128375. [Google Scholar]
- (62).Hao N; Lu J; Zhou Z; Hua R; Wang K A PH-Resolved Colorimetric Biosensor for Simultaneous Multiple Target Detection. ACS Sensors 2018, 3, 2159–2165. [DOI] [PubMed] [Google Scholar]
- (63).Santovito E; Greco D; Ascanio VD; Marianna S Development of a DNA-Based Biosensor for the Fast and Sensitive Detection of Ochratoxin A in Urine. Anal. Chim. Acta 2020, 1133, 20–29. [DOI] [PubMed] [Google Scholar]
- (64).Chen R; Sun Y; Huo B; Zhao X; Huang H; Li S; Bai J; Liang J; Gao Z A Copper Monosulfide-Nanoparticle-Based Fluorescent Probe for the Sensitive and Specific Detection of Ochratoxin A. Talanta 2021, 222, 121678. [DOI] [PubMed] [Google Scholar]
- (65).Han B; Fang C; Sha L; Jalalah M; Al-Assiri MS Cascade Strand Displacement Reaction-Assisted Aptamer-Based Highly Sensitive Detection of Ochratoxin A. Food Chem 2021, 338, 127827. [DOI] [PubMed] [Google Scholar]
- (66).Huang X-B; Wu SH; Hu HC; Sun JJ AuNanostar@4-MBA@Au Core-Shell Nanostructure Coupled with Exonuclease III-Assisted Cycling Amplification for Ultrasensitive SERS Detection of Ochratoxin A. ACS Sensors 2020, 5 (8), 2636–2643. [DOI] [PubMed] [Google Scholar]
- (67).Zheng F; Ke W; Shi L; Liu H; Zhao Y Plasmonic Au-Ag Janus Nanoparticle Engineered Ratiometric Surface-Enhanced Raman Scattering Aptasensor for Ochratoxin A Detection. Anal. Chem 2019, 91, 11812–11820. [DOI] [PubMed] [Google Scholar]
- (68).Cruz-Aguado JA; Penner G Determination of Ochratoxin A with a DNA Aptamer. J. Agric. Food Chem 2008, 56 (22), 10456–10461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


