Abstract
Cellular metabolites play a crucial role in promoting and regulating cellular activities, but it has been difficult to monitor these cellular metabolites in living cells and in real time. Over the past decades, iterative development and improvements of fluorescent probes have been made, resulting in the effective monitoring of metabolites. In this review, we highlight recent progress in the use of fluorescent probes for tracking some key metabolites, such as adenosine triphosphate, cyclic adenosine monophosphate, cyclic guanosine 5′-monophosphate, Nicotinamide adenine dinucleotide (NADH), reactive oxygen species, sugar, carbon monoxide, and nitric oxide for both whole cell and subcellular imaging.
Keywords: fluorescent probes, metabolites, imaging
1. Introduction
Metabolites are essential components in biological systems, and as such, play a significant role in all biological processes [1, 2]. For example, numerous cellular metabolites, such as adenosine triphosphate (ATP), NAD+/NADH, amino acids, and sugars have been found to be essential in cell physiology and signalling pathways [3]. Meanwhile, the abnormal fluctuation of cell metabolites, such as redox state and nitrogen could result in inflammation and diseases in living organisms [4, 5]. Therefore, analysis of these metabolites would provide deeper insight into both physiological and pathophysiological processes. Metabolomics studies metabolites, the substrates and products of metabolism that drive important cellular functions, such as energy generation and storage, signalling, and apoptosis [5–7]. It has been applied for assessing changes of comparing cell line mutants, drug discovery, toxicology, natural product discovery, studying global effect of genetic manipulation, cancer, and nutrition [8–10]. Despite the importance of metabolomics, it is much less developed than genomics or proteomics because it is much more difficult to detect and quantify metabolites, which vary widely in speciation and concentration, but have only subtle structural differences. Lack of effective metabolomic methods, particularly for in situ and real-time detection in vivo is a major barrier to our full understanding of physiological and pathophysiological processes.
A common analytical method for most cellular metabolites is mass spectrometry, which is often combined with liquid chromatography [11, 12]. While the mass spectrometry is extremely powerful in detecting numerous metabolites simultaneously, it cannot distinguish between isomers and enantiomers, making it difficult to identify common metabolites such as L-amino acids versus D-amino acids and anomers of sugars [13]. It also can be difficult for living cell analysis of metabolites because the location and concentration of metabolites in living cells can change quickly in response to different signals [14, 15]. Therefore, there is a need to develop a complementary method for rapid, sensitive, and selective detection and quantification of metabolites in living cells and in vivo.
Fluorometric assays are such a complementary approach for analysing cell metabolites, which is based on the presence of fluorescence tags or probes [1, 16]. In general, the major advantages of fluorescence analysis of cell metabolites include high sensitivity, capabilities for performing time-based studies of concentration, experiments that are non-destructive to the cell, and high-throughput detection [17]. While some metabolites can be directly analysed in individual cells by autofluorescence via their intrinsic fluorescent compounds, only a very limited number of the metabolites display autofluorescence [18]. To overcome this limitation, fluorescent probes which combine a molecule that can bind these small-molecule metabolites and a fluorophore can be introduced in cells [1, 19–22].
After introducing the fluorescent probe into cells, a fluorescence microscope can be used to visual-ise cellular metabolites within their sub-cellular location using different fluorescence imaging analysis methods to achieve deeper understanding of biological processes [23]. Over the past decades, numerous fluorescent probes have been developed and they can be categorized into two types: whole cell and sub-cellular imaging. In this review, we highlight recent advances in the past 10 years in fluorescent probes for imaging metabolites in either whole cells or sub-cellular locations, focusing mainly on representative examples of small molecule fluorophore probes, nanomaterial based fluorescent probes, and light-up (aptamer/dye) fluorescent probes. In particular, we discuss the strengths and limitations as well as some new trends in the development with illustrative examples.
2. Whole cell imaging
Cellular metabolite fluctuations are a common feature of many diseases and therefore a promising target for diagnostics and therapeutical interventions. Therefore, a large number of fluorescent probes have been developed to image the cellular metabolites and their applications for the whole cell imaging has been the most extensively studied and widely used in biological studies. (Examples are listed in table 1, figure 1 and table 2.)
Table 1.
Examples of fluorescent probes (FP) for ATP visualization.
| Type | Name | Detection mechanism | Reference |
|---|---|---|---|
| Small molecule FP | Magnesium green | The detection of hydrolysis of MgATP; | [24] |
| Quinacrine | Fluorescent dye that binds peptide-bound ATP found in intracellular granules; | [25–27] | |
| Pincer-like benzene-bridged sensor | Form pyrene-adenine-pyrene sandwich via π-π stacking when it binds with ATP; | [28, 29] | |
| Nanomaterial based FP | P1/PDANS | Fluorophore labelled ATP aptamer release from PDANS when binds with ATP; | [30] |
| Aptamer Nano-Flares | Gold nanoparticle core functionalized with a dense monolayer of ATP aptamers; | [31] | |
| DNA TP nanoprobes | ATP binds to split ATP aptamer on the DNA triangular prism (TP) to induce the fluorescent change; | [32] | |
| Apt-Act/UCNPs | Upconversion nanoparticle conjugated with PC linker modified ATP aptamer sensor; | [33] | |
| Genetically encoded FP | ATeam | ATP binding of F1F0ATP synthase causes an increase in FRET between a CFP and a YFP; | [34] |
| QUEEN | ATP binding causes a change in the excitation spectrum of a cpEGFP; | [35] | |
| MaLion series | ATP binding of F1F0ATP synthase causes an increase fluorescence of fluorescent protein; | [36, 37] | |
| PercevalHR | ATP binding causes a change in the excitation spectrum of circularly permuted yellow fluorescent protein (cpVenus), and measuring cellular ATP/ADP ratio; | [20, 38, 39] |
Figure 1.
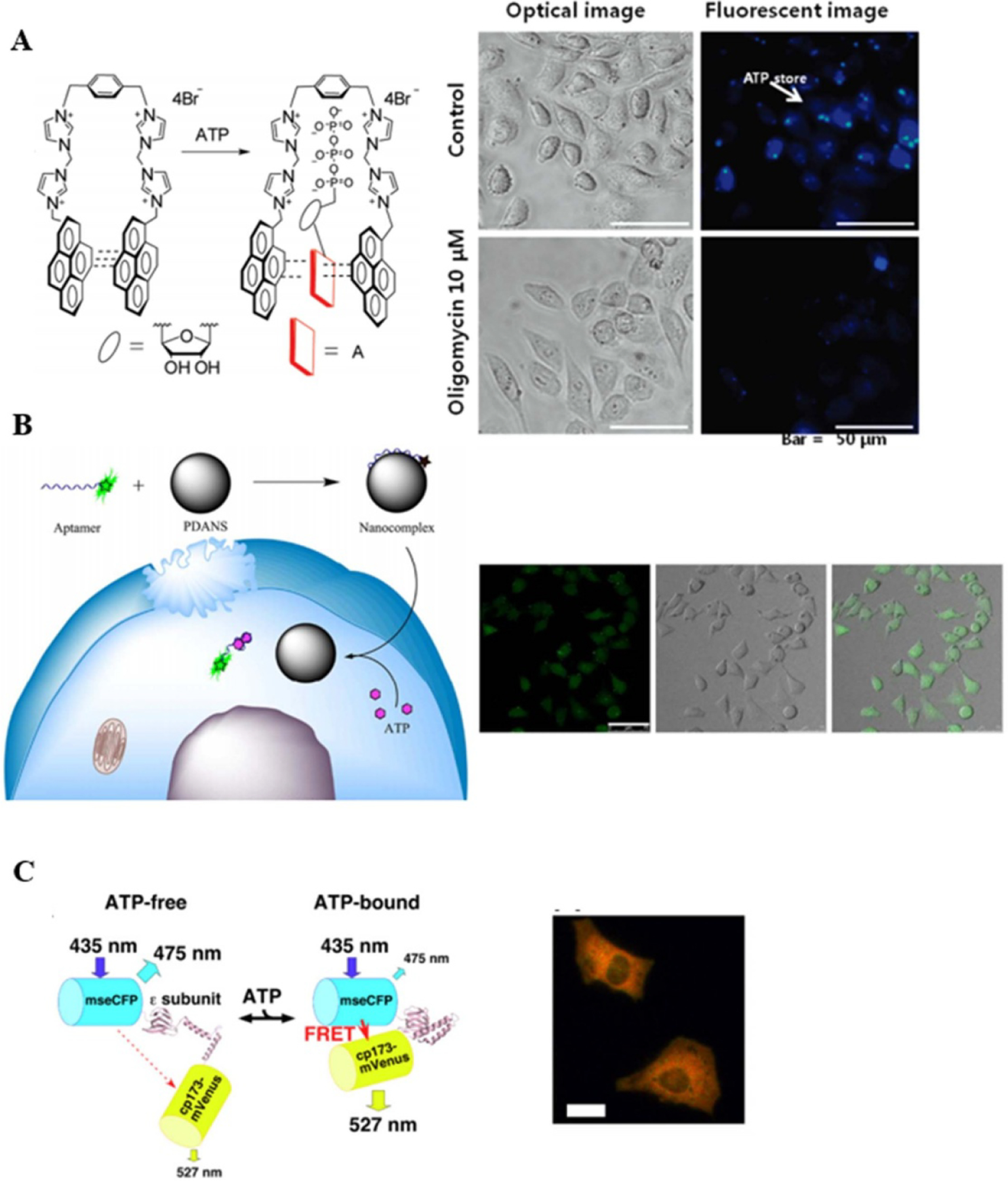
Examples of FP for ATP visualization. (A) Small molecule FP; (B) nanomaterial based FP; (C) genetically encoded FP. Reprinted with permission from [29, 30, 34]. Reprinted with permission from [29]. Copyright (2009) American Chemical Society. Reprinted with permission from [30]. Copyright (2015) American Chemical Society. Reproduced from [34]. CC BY 4.0.
Table 2.
Examples of fluorescent probes (FP) for other metabolites visualization.
| Type | Name | Target | Detection mechanism | Reference |
|---|---|---|---|---|
| Small molecule FP | EOS | Glutamate | Glutamate binds of AMPA causes a fluorescence increase of small molecule dye; | [40] |
| PTMs | H2O2 | Based on the oxidation of boronate esters; | [16,41] | |
| PFBS | Based on C–S bond cleavage of perfluoro-benzyl sulfonates; | [42] | ||
| NBzF | Based on oxidation-induced C–C bond cleavage of benzils; | [43] | ||
| SO3 H-APL | Based on C–N bond cleavage of anilines; | [44] | ||
| DPPEA-HC | Based on oxidation of phosphorous | [45] | ||
| ANRP | CO | Anchored to cell membrane and sense the CO by a metal palladium-catalysed reaction; | [46] | |
| DAF | NO | Based on the reaction with NO to furnish fluorescent triazole derivatives; | [47] | |
| Nanomaterial based FP | C3N4 Nanoribbons | Citrate | Quenched by Cu2+ and then recovered by the addition of citrate; | [48] |
| Genetically encoded FP | FICRhR | cAMP | Based on the dissociation of purified regulatory and catalytic subunits of PKA; | [49–57] |
| ICUE1 | Form Epac1-cyan-YFP sandwich when it binds with cAMP; | [58] | ||
| cADDis | cAMP binding of the regulatory region causes a large conformational change in EPAC 2; | [59, 60] | ||
| cdiA | cdiA riboswitches fused with Spinach2 aptamer; | [61] | ||
| cGES | cGMP | cGMP binding domain from PDEs; | [62, 63] | |
| cGi | cGMP binding domains from PKG and from PDEs; | [64] | ||
| CGY | cGMP binding of PKGs induced fluorescent change; | [65] | ||
| c-di-GMP-I | c-di-GMP | c-di-GMP riboswitches fused with Spinach aptamer | [66–68] | |
| Bc RNA | c-di-GMP riboswitches fused with two FP genes; | [69] | ||
| DNB sensor | c-di-GMP binding aptamer fused into DNB, Broccoli as the; | [70] | ||
| Frex | NADH | NADH binds to Rex subunit causes the conformational change of cpFP; | [71] | |
| Peredox | [NAD+]/[NADH] | The competition between NADH and NAD+ for binding to Rex subunit causes the conformational change of two FP; | [72, 73] | |
| Rex YFP | Different binding affinity of NAD+ and NADH to nucleotide-binding domains of each Rex subunit; | [74] | ||
| SoNar | Binding of NAD+ or NADH to Rex subunit both induces changes in protein conformation and fluorescence; | [75, 76] | ||
| GluSnFR/FLIPE | Glutamate | Glutamate binds to GltI causes the conformational change of two FP; | [77–82] | |
| FLIPQ-TV | Glutamine | Glutamate binds to GlnH causes the conformational change of two FP; | [82] | |
| FLIP family | Sugar | Sugar binds to PBPs causes an increase in FRET between two FP; | [83–89] | |
| roGFP/roGFP2 | ROS | ROS binds to the surface-exposed cysteine; | [90] | |
| CY-RL5 | Redox state | Binds to redox linker 5 (RL5) causes an increase in FRET between two FP; | [91] | |
| COSer | CO | CO binds to CO sensitive heme protein causes the conformational change of cpYFP; | [92] | |
| sGC | NO | Combining endogenously expressed guanylate cyclase with a FRET-based cGMP indicator; | [93] | |
| OGsor | 2OG | Binds to 2OG-binding domain GAF causes an increase in FRET between two FP; | [94] | |
| CIT | Citrate | Binds to citrate-binding domain CitA causes an increase in FRET between two FP; | [95] | |
| Lactate sensor | Lactate | Binds to lactate-binding domain of bacterial transcription factor LldR causes an increase in FRET between two FP; | [88] | |
| Lapronic | Lactate/pyruvate | Different binding affinity of lactate and pyruvate to the binding domains of transcriptional factor LutR subunit; | [96] |
2.1. Fluorescent probe for cellular ATP detection
ATP is one of the most important cellular metabolites because it is the primary energy currency in living organisms and plays critical roles in many biological processes. Many efforts to develop fluorescent probes have been made over the last several decades to visualize ATP in living cells. These probes have been developed using both direct and indirect detection mechanisms from a variety of physical formats, such as small organic indicators, nanomaterials, and fluorogenic probes
Magnesium Green is one of the best small organic indicators, developed by Leyssens et al in 1996, for indirectly detecting ATP hydrolysis [97]. Most of the intracellular ATP are complexed with Mg2+, while ADP has lower affinity for magnesium ions than ATP. Therefore, hydrolysis of MgATP can lead to an increase in free Mg2+ concentration and subsequent increase in Magnesium Green fluorescence. Shin et al then applied Magnesium Green to indirectly visualize ATP in hair cells [24]. The fluorescence of Magnesium Green could be excited with illumination in visible range, reducing the phototoxicity. However, Magnesium Green is not a ratiometric probe, showing a simple increase in fluorescence increase upon binding Mg2+, which makes it a challenge to use in quantitative studies. To directly image cellular ATP, quinacrine, another small molecule probe, stains peptide-bound ATP found in high concentrations in intracellular granules [98]. Researchers have been using quinacrine to image ATP release in endothelial and epithelial cells [25–27]. In addition, Pak et al developed an imidazolium-based, ratiometric fluorescent probe for ATP with a pyrene excimer clamp [28, 29]. This fluorescent probe will form a pyrene-adenine-pyrene sandwich via π–π stacking when it binds with ATP. Thus, the probes were applied to monitor the decrease of ATP levels in HeLa cells upon addition of an ATP synthase inhibitor (oligomycin).
Aptamers are short, single stranded DNA or RNA oligonucleotides capable of specific, high-affinity molecular binding. Aptamers are widely used in studying small-molecule metabolites, which can be engineered to detect metabolite such as ATP in the nanomolar to millimolar ranges [99, 100]. However, cell permeability and oligonucleotide degradation by nucleases hinder their use in cell imaging [101, 102]. To solve this problem, nanoparticles have been used to deliver and protect aptamers from degradation by nuclease in cells. For example, Qiang et al employed a carboxyfluorescein (FAM)-labelled DNA aptamer, which binds to ATP, and polydopamine nanosphere to create a biosensor for protecting the aptamer and quenching its fluorescence [30]. The aptamer released when adding ATP to system. Zheng et al constructed an aptamer nano-flare, that can directly quantify ATP in living cells [31, 103]. The aptamer nano-flares were composed of a gold nanoparticle core, which is functionalized with a dense monolayer of aptamers. However, these fluorescence probes employed an ‘always on’ design, which lacks target-activatable nature, will inevitably result in a high-background and low signal-to-noise ratio [32, 33, 104]. To overcome the problem, Zheng et al designed a fluorescence resonance energy transfer (FRET)-based DNA nanoprism with a split aptamer design for ATP sensing in living cells [32]. The nanoprism showed high cellular permeability and successfully realised ‘FRET-off’ to ‘FRET-on’ sensing of ATP in living cells [32]. Moreover, Zhao et al developed an upconversion nanoparticle conjugated with a photocleavable linker (PC linker) modified ATP aptamer sensors, which can detect ATP in living cells in a conversion luminescence-activatable manner [33]. One disadvantage of this aptamer-based fluorescence probe is that the aptamers are selective for ATP over other nucleotides (GTP, CTP, and UTP), but cannot distinguish between adenine derivatives (ATP, ADP, and AMP) [105].
Although both small organic indicators and aptamer-based fluorescent probes have been widely used in imaging studies, they still face a challenge during sample preparation because of the need to introduce exogenous reagent by cell penetration or cell loading. On the contrary, the genetically encoded indicators such as fluorescent protein-based are in part or wholly encoded imaging reagents by a specific gene sequence. The most recently developed fluorescent protein-based probes are capable of undergoing FRET. These probes typically composed of a donor fluorescent protein and an acceptor fluorescent protein that are separated by an analyte binding domain. When binding with the analytes, this domain undergoes a conformational change that changes the distance between two fluorescent proteins, resulting in a change of FRET efficiency. Tsuboi et al reported ATP-sensitive K+ (KATP) channels fused to a cyan and yellow fluorescent protein FRET pair (ECFP–EYFP) for imaging ATP concentration changes in HEK-293 cells [106]. Imamura et al generated a series of FRET-based probes for ATP named ‘ATeam’, in which the ε subunit of the Bacillus subtilis F1F0ATP synthase acts as the ATP sensing domain [34]. Yaginuma et al reported a ratiometric single fluorescent protein probe called ‘QUEEN’ (quantitative evaluator of cellular energy) to quantify absolute ATP concentrations [35]. Recently, intensiometric single fluorescent protein probes developed by Aria et al and Lobas et al enabling the simultaneous visualization of cellular ATP dynamics [36, 37].
Since the absolute ATP, ADP, and AMP can fluctuate, the ratio of ATP/ADP ratio can be a more reliable indicator of cellular energy status. The ratiometric single fluorescent protein probe ‘Perceval’ developed by Berg et al and improved version ‘Perceval HR’ has been used for measuring cellular ATP/ADP ratio [20, 38]. Zala et al also used the Perceval to measure ATP/ADP ratio in neurons and found out that mitochondrial trafficking is dependent on mitochondrial ATP but not glycolysis [39].
2.2. Fluorescent probes for intracellular second messengers
Cyclic adenosine monophosphate (cAMP) is a second messenger of many G protein-coupled receptors (GPCRs) and regulates cAMP-dependent kinase (PKA) and the exchange protein activated by cAMP (Epac) to participate in cellular metabolism. The first cAMP fluorescent probe (FICRhR) for cellular imaging was reported by Adams et al, which was a FRET-based probe utilizing dissociation of purified regulatory and catalytic subunits of PKA, sensing the cellular cAMP by microinjection [49]. Later, the FICRhR probe was applied to investigate the link between cAMP and diverse biological activities and to monitor cAMP levels in the processes of stimulated Aplysia neurons [50, 51]. Moreover, the FICRhR probe was also introduced into single cells within brain slice preparations by perfusable patch pipettes [52]. However, the requirement for invasive loading of PKA holoenzyme in this method limits its applications. To solve this limitation, researchers developed genetically encoded versions of FICRhR, which could be introduced into cell by a routine transfection. Zaccolo et al developed FICRhR-like genetically encoded probe, which was composed of enhanced blue fluorescent protein-labelled type 2 regulatory subunits of PKA and GFP-labelled catalytic subunits [53]. Subsequently, many labs have also carried out optimization studies on this probe [54–57]. DiPilato et al constructed a fluorescent probe to monitor cellular cAMP dynamics and Epac activation by sandwiching the full-length Epac1 between cyan and yellow mutants of GFP [58]. Over the years, multiple labs developed and improved FRET-based fluorescent probe for cAMP with different characteristics regarding sensitivity, kinetics, and dynamic ranges.
As an alternative to the FRET-based methods, single-wavelength methods were developed, which the cAMP binding domain was fused to only one fluorophore. Tewson et al first developed a single-wavelength intensiometric cAMP probes cADDis [59], then Moore et al optimized the probe that cADDis fused with a 5HT6 receptor and mCherry to target cilia and measure the cAMP ratiometrically [60]. Recently, Kellenberger et al first developed a RNA-based fluorescent probe for cyclic di-AMP (cdiA, is also a second messenger in Gram-positive bacteria, some Gram-negative bacteria, and Archaea) by fusing of Spinach2 aptamer to ligand-binding domains of cdiA riboswitches, visualizing intracellular cdiA levels in live Listeria monocytogenes strains [61].
The other important second messenger is cyclic guanosine 5′-monophosphate (cGMP), which participates in many physiological processes in mammals. cGMP can be used to regulate effectors such as cGMP-specific phosphodiesterases (PDEs), cGMP-dependent protein kinases (PKGs) and cyclic nucleotide-activation ion channels. Therefore, the visualization of intracellular cGMP is critical for understanding of cGMP signalling pathway. Several different cGMP binding domains have been used as sensing units in genetically fluorescent probes. For example, the binding domain of Cygnus in cGMP energy transfer sensors (cGES) was from PDEs [62, 63] and cGMP indicators (cGi) [64] were from PKGs and PDEs. These binding domains were used to separate donor fluorescent protein and acceptor fluorescent protein. Moreover, Sato et al, reported a fluorescent probe named CGY-del1 for cGMP that contained PKG fused to single fluorescent protein [65]. Honda et al optimized the selectivity for cGMP and eliminated the constitutive kinase activity of the binding domain to reduce the disturb from the probe [107, 108].
Breaker’s group first reported the Cyclic di-GMP riboswitch (named c-di-GMP-I) [66] in eubacteria and then discovered another c-di-GMP riboswitch termed c-di-GMP-II in the Clostridium difficile [67]. Kellenberger et al designed two different probes for live cell imaging of c-di-GMP and cyclic AMP-GMP by fusing the Spinach aptamer to variants of a natural GEMM-I riboswitch (c-di-GMP-I), demonstrating the ability to change specificity of the RNA-based probes by taking advantage of rational mutations to the ligand binding domain instead of by inserting distinct aptamers [68]. Zhou et al discovered three new c-di-GMP riboswitches (Bc3, Bc4, and Bc5 RNA), which were fused between the two fluorescent protein genes amcyan and turbofp [69]. Recently, Wu et al designed a ratiometric RNA probe that comprised of dinitroaniline-binding aptamer (DNB)-based sensing domain and Broccoli domain to quantify the intracellular c-di-GMP concentration by DNB-to-Broccoli fluorescence ratio [70].
2.3. Fluorescent probes for intracellular NAD pools
Nicotinamide adenine dinucleotide (NAD) is a central cofactor involved in many enzymatic reactions, especially as a major electron carrier in redox reactions [109–111]. NAD exists in two forms, the oxidized form NAD+ and the reduced form NADH [111]. NAD+ can be reduced to NADH in the process of glycolysis and in the tricarboxylic acid (TCA) cycle [112]. NADH can be re-oxidized back to NAD+ in the electron transfer chain [111]. Meanwhile, NAD can also be phosphorylated to NADP via NAD kinases [111]. The NAD+/NADH redox couple is served as a regulator of cellular energy metabolism of glycolysis and mitochondria oxidative phosphorylation [113, 114]. While NADP+, together with its reduced form NADPH, maintain redox balance and support the biosynthesis of fatty acids and nucleic acids [114]. Therefore, similar to ATP, the NAD pool plays an important role in cellular energy balance, which is determined by the ratio of NAD+/NADH and NADP+/NADPH. Nowadays, the well-developed fluorescent probes for cellular NAD pool imaging are genetically encoded fluorescent probes. Zhao et al first inserted the circularly permuted fluorescent proteins (cpFPs) into NADH sensing domain (Rex) subunit to sense the NADH [71]. To date, there are several genetically encoded fluorescent probes that can detect NAD+/NADH ratios: Peredox [72, 73], Rex YFP [74], and SoNar [75, 76]. Peredox and Frex family probes were based on inserting a cpYPs into the Rex dimer between its subunits, detecting the NAD+/NADH ratios via Rex intersubunit interactions. While Rex YFP and SoNar were based on integrating a circularly permuted yellow fluorescent proteins (cpYFPs) into the loop between nucleotide-binding domains of each Rex subunit.
2.4. Fluorescent probes for intracellular amino acids
Glutamate plays a critical role in amino acid metabolism, participating not only signal transduction, but also regulating nitrogen circulation together with glutamine and 2-oxoglutarate [115–117]. The glutamate-sensing fluorescent reporter (GluSnFR) [77–79] and fluorescent indicator protein (FLIP) for glutamate [80–82] were the primary genetically encoded fluorescent probes, which fused the glutamate periplasmic binding protein (PBP) GltI (also known as ybeJ) to enhanced cyan fluorescent protein and a yellow fluorescent protein Citrine [118] or Venus [119]. Marvin et al also reported an intensity-based GluSnFR (iGluSnFR), fusing the binding protein GltI to cpFPs cGFP [120, 121]. Namiki et al developed a small molecule fluorescent probe, which consists of the mutated glutamate receptor GluR2 subunit of an alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor and a small molecule fluorescent dye, termed EOS [40]. Gruenwald et al studied another pivotal amino acid, glutamine, measuring the glutamine concentration in living cells by an array of FLIPQ-TV sensors with different affinities [82]. Okada et al applied bacterial PBPs to construct FRET-based probes, taking advantage of PBPs’ structure to expand the dynamic range of the probes [122].
2.5. Fluorescent probes for intracellular sugar
Sugar metabolism is involved in many types of metabolic reactions in living organisms. The PBPs have been used as the substrate-binding element of protein-based fluorescent probes by linking to fluorescent protein. These fluorescent probes are named as FLIP family probes (which are also FRET-based probes) due to the hinge-bend movement of probes leading to FRET response. Frommer et al used PBPs to design a series FLIP probes to monitor the intracellular distribution of many sugars including glucose, galactose, maltose, ribose, arabinose, sucrose, lactate, and trehalose [83–89]. Since the intracellular sugar concentrations and distribution were directly related to carbohydrate metabolism, these FLIP probes have been broadly applied in many areas, such as the food, pharmaceutical, and biofuel industries. Ballerstadt et al reported a fluorescent affinity hollow fibre probe for transdermal glucose monitoring, which consisted of the dyed beads (Safranin O and Pararosanilin) and Alexa488-Con A (Alexa fluor 488 labelled Concanavaline A) inside a hollow fiber dialysis membrane [123]. Then Heo et al developed a probe based on fluorescent hydrogel fibers for long-term monitoring of glucose in vivo [124]. Although the use of protein-based fluorescent probes in metabolite research and bioprocess visualization has progressed, there is still a need to develop new probes for other critical sugar metabolites.
2.6. Fluorescent probes for intracellular redox state
Reactive oxygen species (ROS) refers to a series of key oxygen (O2) metabolites, including hydrogen peroxide (H2O2), hydroxyl radical (•OH), hypochlorous acid (HClO), superoxide anion (O2•−), singlet oxygen (1O2), ozone (O3), and organic peroxides [125–132]. The changes of intracellular ROS will influence the redox equilibrium, resulting in macromolecular damage and is implicated in various diseases such as atherosclerosis, diabetes, neurodegeneration, cancer, and aging [126]. Numerous fluorescent probes have been developed to image the intracellular ROS or redox states. The genetically encoded fluorescent probes for sensing ROS or redox states were taking advantage of existing a sensing domain from naturally occurring protein structures by linking to fluorescent proteins. For example, redox-sensitive GFP with two surface-exposed cysteines close to the chromophore was used to monitor the molecule’s own redox states [90, 133, 134]; redox-sensitive polypeptide-flanked CFP/YFP leads to FRET signal changes response to different redox states [91]; the environmentally sensitive fluorescent protein (Venus) was fused with responsive domain of the transcriptional regulatory protein OhrR to visualize organic hydroperoxides, which constantly generate cellular stress [135]. The other types of fluorescent probe were based on small molecules. Taking H2O2 as an example, the majority of H2O2 probes were based on the oxidation of boronate esters [16, 41]. Other H2O2 sensing reactions cover C–S bond cleavage of perfluoro-benzyl sulfonates [42], oxidation-induced C–C bond cleavage of benzils [43], C–N bond cleavage of anilines [44], and direct oxidation of phosphorous [45], selenium [136, 137] and tellurium [138]. Most of the small molecule-based fluorescent probes for redox states have been extensively reviewed [16, 139–142]. Readers wishing to further advance this field would be advised to read recent discussion from Tampieri and Lu in order to address which ROS are present in solution [143, 144].
2.7. Fluorescent probes for intracellular other significant metabolites
Carbon monoxide (CO) is generally regarded as a toxicant or pollutant. Nevertheless, more and more studies suggest that CO, like NO, functions as an essential second messenger [145]. Wang et al reported a genetically encoded fluorescent probe COSer for monitoring intracellular CO by fusing a dimeric CO-sensitive heme protein to cpYFP [92]. Xu et al reported a novel cell membrane-anchored fluorescent probe ANRP, which complexed ANR (a cell membrane-anchored fluorophore designed by grafting a positive charged ammonium group onto a long and linear hydrophobic Nile Red molecule.) with palladium, monitoring the release of CO from living cells [46]. Sato et al have reported a novel cell-based fluorescent probe to visualize picomolar levels of NO release from living cells, made by combining endogenously expressed guanylate cyclase with a FRET-based cGi [93]. In addition, the small molecule-based fluorescent probes for NO were well-developed. The most common method involves the use of o-Diamino aromatics under aerobic conditions, which firstly reported by Nagano’s group [47]. In the presence of O2, o-Diamino aromatics could react with NO to furnish fluorescent triazole derivatives [146–148]. Other fluorescent probes are summarized in several reviews [149, 150]. 2-Oxogluatarate (2OG) is another metabolite that plays an important role in metabolism and also serves as a signalling molecule in various organisms. Zhang et al reported FRET-based genetically encoded fluorescent probe for detecting 2OG in real-time; results showed the probe’s dynamic range appeared identical to the physiological range observed in Escherichia coli [94]. Citrate is also a critical metabolite in various biological systems, such as mitochondrial energy generation, inflammatory response, blood coagulation, and cytosolic biomacromolecular synthesis [48, 151]. Ewald et al developed FRET-based genetically encoded fluorescent probe for citrate; they optimized peptide linkers to achieve an optimal change ratio and modified the citrate-binding pocket to obtain a probe with the proper affinity for the application [95]. Hu et al also reported another fluorescent probe, which was based on carbon nitride nanoribbons for visualizing intracellular citrate anion [48]. Lactate also plays metabolic and signalling roles in healthy tissues. As the fluctuation of lactate level is associated with inflammation, hypoxia/ischemia, neurodegeneration, and cancer, visualizing intracellular lactate levels has diagnostic and therapeutic applications [88]. San Matín et al reported a genetically encoded FRET lactate probe that discriminates lactate flux in different cells; results showed T98G glioma cells have 3–5-fold higher rate of lactate production than normal cells [88]. Recently, Galaz et al developed a genetically encoded FRET probe Lapronic for imaging lactate/pyruvate ratio in living cells’ cytosolic and mitochondria matrix, allowing the assessment of glycolytic/oxidative metabolism with a straightforward fluorescent readout [96].
3. Subcellular imaging
Sub-cellular organelles are specialized subunit within cells, which are usually enclosed by their own lipid bilayer. The main eukaryotic organelles include nucleus, mitochondria, lysosome, endoplasmic reticulum (ER), and Golgi apparatus. All of these organelles play a critical and indispensable role in cellular processes [152, 153]. The dynamic fluctuations of intracellular metabolites in subcellular microenvironments determine cellular metabolism, homeostasis, signal conduction, and immunity; abnormal levels of the sub-cellular metabolites can cause disorders, which are associated with various major diseases [154–156]. Monitoring intracellular metabolites in subcellular structures is therefore important for bioanalysis and related drug discovery. Organelle fluorescent probes mainly contain three domains: localizing group, fluorophores, and recognition domain [154, 157, 158]. Fluorescent probes with diverse localizing groups can be localized in specific organelles by utilizing different physiochemical properties of diverse organelles [159]. After reaching the subcellular location of interest, they can subsequently target or react with diverse metabolites by recognition domain and further make detectable signal changes via different response mechanisms. Herein, we summarized the fluorescent probes for monitoring the fluctuation of metabolites with sub-cellular accuracy.
3.1. Fluorescent probes for imaging mitochondrial metabolites
Mitochondria, the double-membrane constructed organelles and the primary compartments for intracellular respiration in most eukaryotes, regulate energy generation, calcium circulation, protein synthesis, cell proliferation, division, and death pathways [160]. Abnormal metabolite levels in mitochondria may lead to mitochondrial dysfunction, which is related to neurodegenerative disease, malignant cancers, and cardiac diseases. Thus, fluorescent probes that specifically accumulate in mitochondria play critical roles in monitoring the mitochondrial functions and investigating various mitochondrial disease [161–166]. During mitochondrial respiration, proton pumps in the mitochondrial inner membrane transport protons into the mitochondrial membrane space, resulting in a highly negative mitochondrial transmembrane potential (MMP, approx. −180 mV) [167, 168]. Therefore, most fluorescent probes for mitochondria attract the negative potential of the mitochondrial membrane by cations. The fluorescent probes with intrinsic or post-functionalized cationic aromatic structures can be applied to image mitochondria. Delocalized lipophilic cations (DLCs) have been shown to possess the ability to localize mitochondria [169–171]. Typical DLCs ligands include triphenylphosphonium (TPP), quinoline derivatives, and positive charged pyridine; rhodamine and cyanine are the common fluorophores for the design of probes for mitochondria imaging (figure 2) [156, 172–178]. Except for MMP based fluorescent probes, mitochondria transport proteins have also been developed for targeting. The localizing ligands, such as peptides and pyruvate, possess the affinities for specific mitochondrial protein, which have been used for designing the probes for imaging mitochondria [160, 179–181].
Figure 2.
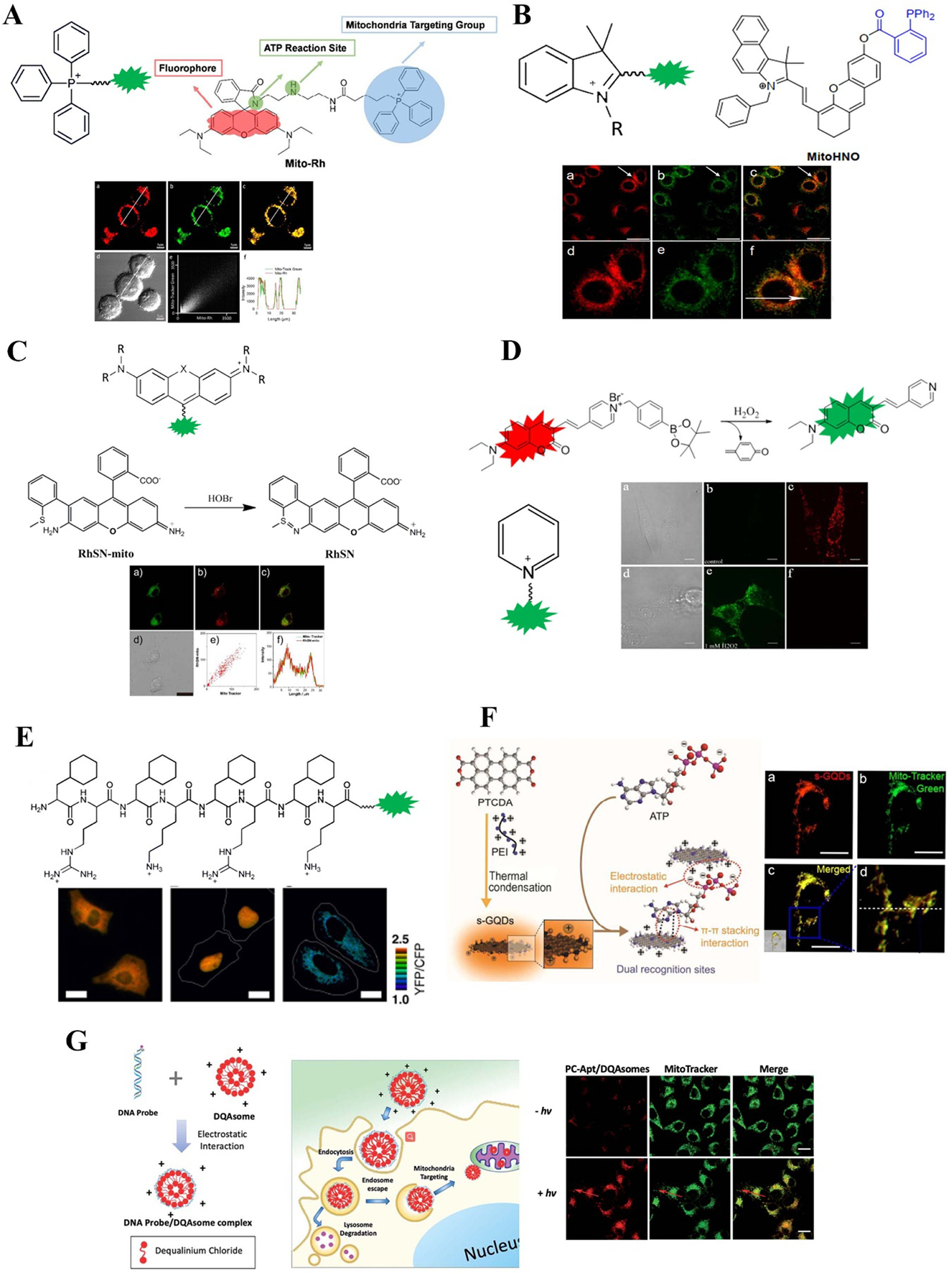
Integration of common groups and their application examples for mitochondrial metabolite probes design: (A) TPP cation. Reprinted with permission from [182]; (B) indolium cation. Reprinted with permission from [178]; (C) rhodamine cation. Reprinted with permission from [175]; (D) pyridinium cation. Reprinted with permission from [176]; (E) mitochondrial localization sequence. Reprinted with permission from [34] and selection nanomaterials for visualizing mitochondrial metabolites: (F) s-GQDs. Reprinted from [183]; (G) DQAsome. Reprinted with permission from [184]. Reprinted with permission from [182]. Copyright (2017) American Chemical Society. Reprinted from [178], Copyright (2016), with permission from Elsevier. Reprinted with permission from [175]. Copyright (2017) American Chemical Society. Reprinted from [176], Copyright (2018), with permission from Elsevier. Reproduced from [34]. CC BY 4.0. Reproduced from [183] with permission of The Royal Society of Chemistry. Reproduced from [184] with permission of The Royal Society of Chemistry.
ATP is primarily produced in mitochondria and the fluctuation of ATP will lead to, or is caused by, mitochondria dysfunction. Thus, it is essential to monitor the ATP in and around the mitochondria. Until now, several methods have been developed. The genetically encoded fluorescent probes for imaging mitochondrial ATP are based on fusing a mitochondrial localizing sequence to a fluorescent protein gene. Imamura et al fused a duplex of the mitochondrial localizing signal of cytochrome c oxidase subunit VIII to the N terminus of ATeams indicator, which made the indicator localized to the mitochondria properly [34]. Then, Depaoli et al applied this mitochondria-localizing ATeams indicator to investigate the dynamic of mitochondrial ATP pools in response to acute glucose removal, glucose substitution, as well as mitochondrial toxins [185]. Over the past decades, only few small molecule-based fluorescent probes developed for localizing mitochondrial ATP and based on MMP. Srivastava et al first developed a photoinduced electron transfer based molecular scaffolds/fluorescent probes that can monitor mitochondrial ATP [186]. Wang et al then developed a multisite-binding, switchable fluorescent probes ATP-Red 1 to monitor mitochondrial ATP levels [187]. Tan et al reported a fluorescent probe named Mito-Rh to real time monitor mitochondrial ATP, which was constructed by an ATP-sensitive fluorophore rhodamine, ATP reaction site diethylenetriamine and mitochondria-localizing site TPP [182]. Recently, Ren et al reported a novel ratiometric fluorescent probe Rh6G–ACFPN for quantitatively detecting the mitochondrial ATP levels [188]. Several nanoparticles have been developed to delivery fluorescent probes to mitochondria. Deng et al developed zeolitic imidazole frameworks to encapsulate the ATP sensitive fluorophore Rhodamine B, monitoring mitochondrial ATP fluctuation during cellular glycolysis and apoptosis [189]. Liu et al developed yellow emissive single-layered graphene quantum dots with dual recognition sites including π-conjugated single sheet to sense ATP and positively charged site to localize in mitochondria [183]. Recently, our group applied positively charged nanoparticles called DQA-somes to deliver a PC ATP aptamer sensor to mitochondria for spatiotemporally controlled monitoring of mitochondrial ATP fluctuation [184]. This approach kept the fluorescent probe inactive before reaching the mitochondria and can be activated by light to detect ATP.
To make sure cellular energy supply, diverse electron-transport chain (ETC) reactions for ATP synthesis are performed in mitochondria. The electrons leaked from ETC can react with oxygen molecules to generate O2•− and transformed into H2O2, ONOO−, •OH, 1O2 et al under the catalysis of diverse enzyme [173–175, 190, 191]. These metabolites can not only maintain the mitochondria redox homeostasis, but also regulate the cellular function. Various of fluorescent probes for mitochondria imaging have been developed for visualizing ROS levels during the cell processes. For genetically encoded fluorescent probes, the mitochondria-localizing sequences are fused to ROS sensitive fluorescent protein sequences so that the expressed protein probes can respond to mitochondrial ROS. Hyper is a classic family of genetically encoded probes for H2O2, which consists of circularly permuted YFP (cpYFP) inserted into the regulatory domain of the prokaryotic H2O2-sensing protein OxyR. Researchers developed hyper probes by fusing the mitochondria-localizing sequences so that the probes can localize to the mitochondria matrix [192, 193] and mitochondria intermembrane space [194] of HEK and other cells. Wang et al employed adenovirus-mediated gene transferred to express ROS sensitive cpYFP in the mitochondrial matrix of cultured adult cardiac myocytes using the cytochrome C oxidase subunit IV (COX IV) localizing sequence (mtcpYFP) [195]. TPP modified small molecule fluorescent probes have been widely applied in mitochondria specific metabolites imaging. For example, MFDBZH [190] and HKSOX-1 probes [196] for sensing O2•−; PMN-TPP [197], RMClO-2 [198] and RSTPP [199] probes for sensing HOCl; MNAH probe [200] for sensing 1O2. Other cationic groups are widely used in the design of probes for imaging mitochondria, such as cyan [201–203] and rhodamine [175, 198, 199, 204–206], which are also served as the fluorophore for the probe. In addition, peptides have also been developed for the design of fluorescent probes for imaging mitochondria [207]. TPP group has also been used in fluorescent nanoparticle probes. For example, Gong et al reported a mitochondrial oxidative stress amplifier to image GSH, MitoCAT-g, which consists of carbon-dot-supported atomically dispersed gold (CAT) with further surface modifications of TPP and cinnamaldehyde [208].
NADH is the important hydrogen carrier during TCA cycle (TCA cycle), and can release more energy to cells. Mitochondria-localizing sequences have also been used in genetically encoded fluorescent probes for mitochondrial NADH imaging. The genetically encoded NADH sensors, such as Frex, Peredox, RexYFP, and SoNar, are already genetically introduce to mitochondria by fusing the mitochondria-localizing sequences [74, 209–211].
Owing to the crucial biological function of mitochondria, fluorescent probes for imaging mitochondrial metabolites have been widely developed and employed. Nevertheless, most of the localizing ligands are lipophilic cationic structures, which may reduce the MMP. Furthermore, the fluorescent probes can easily leak out or become untargeted due to the MMP lost during various stimulations and cannot work precisely in these situations. Although genetically encoded fluorescent probes are able to anchor to mitochondria covalently with high selectivity and avoid the fluorescent probe leakage or poor targeting, we cannot guarantee the gene transfection efficiency, and the repeated washing steps may cause the change of mitochondrial microenvironment. In addition, the process of gene transfection makes the genetically encoded fluorescent probes difficult to apply in in vivo imaging. As we know, there are thousands of biomolecules in mitochondria, such as mitochondrial DNA, RNA, enzymes, ions, lipids, and amino acids. So, in situ imaging and measurement of specific mitochondrial metabolites at ultralow concentrations using fluorescent probes remains a challenge. A lot of reported fluorescent probes only work when cells are stimulated by various chemical agents or abundant of exogenous analytes are added. There are still relatively few probes can detect basal concentrations, which limits the further explorations of physiological and biological functions of these metabolites. Ratiometric fluorescent probes are promising for eliminating these interferences, but only limited ratiometric fluorescent probes for mitochondria imaging have been developed until now.
3.2. Fluorescent probes for imaging nuclear metabolites
The nucleus is the crucial organelle that serves as the container of the majority of DNA in cells, maintaining the integrity of genes and regulating the gene expression to control the cell activity [212, 213]. The nucleus is enveloped by a double-layered membrane containing hundreds of nuclear pores and ribosomes. Some small molecules and ions can permeate via the nuclear membrane freely. Meanwhile, the large biomacromolecules, such as RNA and ribosomes, can transit through the nuclear pores by energy related pathway. Owing to the large amount of DNA in nucleus, small cationic fluorescent probes with two or more cationic centres and hydrophobic planar aromatic structure can be used to target to the minor grooves in negatively charged DNA double-strands to accomplish the nuclear localizing, such as commercialized Hoechst dye or DAPI [212, 214]. Dickinson et al reported another nucleus-localizing ligand modified probe, arylboronate based fluorescent probe, for imaging the nuclear H2O2 [215]. Moreover, modifying the fluorescent probes with nuclear localization signal (NLS) peptides enables the probes to bind to importins and further delivering into nucleus through nuclear pores (figure 3(A)) [213]. For example, Wen et al developed a peptide conjugated small molecule probes based on 1,8-naphthalimide and boric acid ester for imaging nuclear H2O2 [216]. Meanwhile, protein tagging is also a promising strategy for the development of fluorescent probes for imaging nucleus. The protein tagging ligands used for imaging nucleus include SNAP-tag [217, 218], HaloTag [219], coumarin [220] and HaloRT ligand. Imamura et al reported FRET-based genetically encoded fluorescent probe fused with the SV40 large T-antigen sequence to imaging the nuclear ATP [34]. However, nanomaterials are rarely used in the imaging of nuclear metabolites due to size limitation for nanomaterials to localize in nucleus [221].
Figure 3.
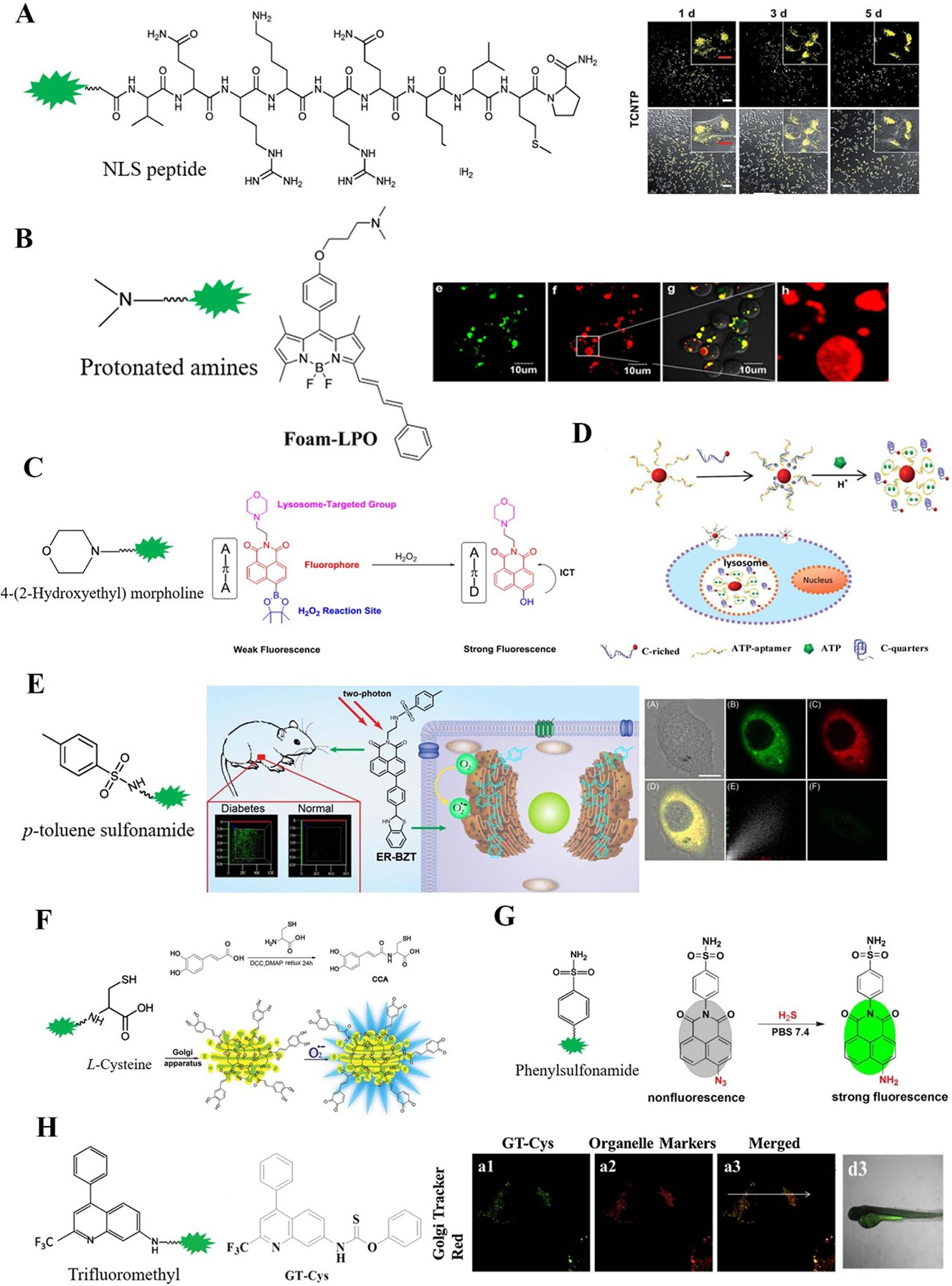
Integration of typical groups and their application examples for (A) nucleus; (B)–(D) lysosome; (E) ER; (F)–(H) Golgi apparatus localized fluorescent probes. Reprinted with permission from [213, 222–228]. Reproduced from [213] with permission of The Royal Society of Chemistry. Reprinted with permission from [222]. Copyright (2015) American Chemical Society. Reprinted from [223], Copyright (2016), with permission from Elsevier. Reproduced from [224] with permission of The Royal Society of Chemistry. Reprinted from [225], Copyright (2017), with permission from Elsevier. Reproduced from [226] with permission of The Royal Society of Chemistry. Reproduced from [227] with permission of The Royal Society of Chemistry. Reprinted from [228], Copyright (2020), with permission from Elsevier.
Several reports have shown that the modification of NLS peptides to molecules or nanomaterials can be sufficient to drive to the fluorescent probe to the nucleus specifically, and using this method, nucleus-localizing cancer therapy has been broadly studied. However, many fluorescent probes need long incubation time for positioning to nucleus due to the low targeting efficiency. Meanwhile, it is difficult to directly conjugate the probes with nucleus-localizing molecular scaffolds or NLS peptides, probably due to the minor structural alterations which can reduce the affinity of localizing ligands [154, 158]. Protein tagging appears to be an ideal way for positioning the probes to almost any organelles including the nucleus. But the synthesis process is difficult and complex, and the loss of solubility of probes after binding to the substances may hamper the application of protein tagging probes or genetically encoded fluorescent probes. In addition, non-specific or un-tagged fluorescent probes need to be washed out repeatedly before imaging to lower the background fluorescence, which can change the nucleus microenvironment of cells. Therefore, the development of novel ultrasensitive fluorescent probes for imaging nuclear metabolites should be given more attention.
3.3. Fluorescent probes for imaging lysosomal metabolites
The lysosome is the main digestive compartment in cells, where many macromolecules are delivered for degradation [159]. The main characteristics of lysosomes are acidic microenvironment (pH ~ 5.0) and an abundance of hydrolases [158]. Most importantly, lysosomes are responsible for foreign substance scavenging, digestion, and autophagy [229–231]. As the importance of lysosomes have been understood, more and more fluorescent probes for imaging lysosomal metabolites have been developed.
Similarly, the genetically encoded fluorescent probes localized in lysosome by fusing the lysosome localizing sequences. For example, McCue et al reported a genetically encoded fluorescent probes fused with lysosomal resident membrane protein LAMP1 to image lysosomal Ca2+ [232]. Other types of fluorescent probes are usually modified with lipophilic amines for driving probes into lysosomes. The protonated amines in lysosomes are membrane permeable so that the lysosomes can entrap the probes selectively (figure 3(B)) [222, 233]. The lysosome-localizing ligands morpholine has been broadly applied in the development of lysosome-localizing small molecule fluorescent probe for imaging the lysosomal ATP, H2O2, HOCl, NO, and HNO (figure 3(C)) [223, 234–240]. The other popular localizing ligand N,N-dimethylethylenediamine has been used for imaging lysosomal NO and H2S [241, 242]. Recently, Jun et al reported a ratiometric two-photon fluorescent probe Lyso-ATP for imaging lysosomal ATP by changing the core into rhodamine 6G and introducing a BODIPY at the end of the tetraamine chain, which showed the lysosome fusion process [243]. Nanomaterials have also been developed for imaging the lysosomal metabolites. For example, Jin et al constructed a nanoflare composed of AuNP, i-motif and ATP aptamer to image lysosomal ATP (figure 3(D)) [224]. The fluorescent carbon dot developed by Geng et al was also used in imaging the lysosomal ATP [244].
Although, in recent years, the development of fluorescent probes for imaging lysosomal metabolites has been made significant progress. There are still remaining certain limitations. For instance, the localizing principle of most reported fluorescent probes are based on trapping lipophilic amines and cannot be used to differentiate between endosomes, auto-phagosomes, autolysosomes and other acidic organelles. Furthermore, these localizing ligands are toxic to living cells because they can cause the alkalization of lysosome microenvironment, making them unsuitable for long-term tracing. While the genetically encoded fluorescent probes need to be washed out to lower the background fluorescent before imaging, which may also influence the lysosome microenvironment. Meanwhile, the fluorescent of some probes can be quenched owing to the acidic lysosomal microenvironment. The emission wavelengths of most of the reported probes are located in the visible region, which hinder the application in deep tissue imaging due to the poor penetration capability. These shortcomings greatly limit the development and application of lysosome-localizing fluorescent probes. Therefore, it is still a challenge to develop the fluorescent probes for imaging lysosomal metabolites with strong anti-interference capability, outstanding selectivity, and high sensitivity.
3.4. Fluorescent probes for imaging the metabolites in ER
The ER can be divided into rough ER and smooth ER based on whether they contain ribosomes. The rough ER is responsible for protein synthesis, while the smooth ER is mainly in charge of lipid and carbohydrate metabolism and calcium signalling. During these biological processes, the metabolites like ATP, ADP, ROS, NO, HNO, and H2S are essential for ER functions. Once the homeostasis of metabolites in ER is out of control, it can cause several types of disorder in ER. Therefore, the development of ER-localizing fluorescent probes and tracing of the fluctuation of metabolites become a new strategy to study the metabolism process in ER.
The ER-localizing sequences have been fused to genetically encoded fluorescent probes. Vishnu et al reported ER-localizing ATeam ERAT4.01 to record the ER ATP changes in real-time, revealing that the ATP levels within the ER were significantly lower than in the mitochondria and that Ca2+ release from the ER induced ATP increase within ER lumen [245]. Then this ER-localizing probe had also been applied in imaging ATP depletion of ATP/ADP exchanger in ER membrane [246]. The most popular ER-localizing ligand for small molecule fluorescent probes is p-toluene sulphonamide, such as ERBZT for O2•− (figure 3(E)) [225], ER-ClO for HOCl [247], ER-Nap-NO for NO [248], and Na-H2S-ER for H2S [249].
Over the past decades, a series of fluorescent probes have been developed for imaging specific metabolites selectively in ER. However, the targeting principle of these fluorescent probes still remains unknown, which hamper the development of ER-localizing fluorescent tools and the understanding of physiological and biological functions of different metabolites. Although the conjugation of ER-localizing dyes seems feasible, a slight change of molecular charge and bulk may influence the targeting capability. Meanwhile, the protein tagging methods can also be used to drive the fluorescent probes to ER, but the synthetic difficulty and complexity make this probe need to be further improvement. In addition, the emission wavelength of most of the probes are also localized in visible region, which hinder the in vivo application. Until now, probes for imaging and quantification of in situ metabolites in ER are still rare, so the development of this kind of probe could speed up the understanding of biological functions of ER.
3.5. Fluorescent probes for imaging the metabolites in Golgi apparatus
The Golgi apparatus is another crucial intracellular organelle for the modification, storage and transportation of carbohydrates, lipid and proteins. Modified and labelled cargoes in Golgi apparatus will be transferred to the final destinations, such as lysosomes and cytoplasmic membrane, and further exert the biological functions [158, 159, 226]. Notably, the stress-signalling overload in Golgi apparatus can result a series of disorders and further lead diverse diseases. However, the probes for imaging the metabolites in Golgi apparatus have not been well developed.
The developed Golgi apparatus-localizing genetically encoded fluorescent probes, by fusing amino acids 1–60 of the human galactosyl transferase truncated at position 60 in its luminal domain to N terminus of sensing domain, have been used to image Zn2+ in Golgi apparatus [250]. However, it has not been applied to the imaging of other metabolites in Golgi apparatus. Owing to a large quantity of cysteine residue recognition receptors in Golgi apparatus, the localizing ligand of some small molecule fluorescent probes are containing L-cysteine, such as CCA for O2•− (figure 3(F)) [226] and SF-1 for HOCl [251]. Recently, Zhu’s group reported two small molecule fluorescent probes for sensing H2S in Golgi apparatus, one is containing a phenylsulfonamide moiety as a localizing group and a 1,8-Naphthalimide moiety as a sensing group (figure 3(G)) [227], the other one has a trifluoromethyl moiety as a localizing group and quinoline as a sensing group (figure 3(H)) [252]. They also applied the trifluoromethyl moiety and thiobenzoate moiety to image the cysteine in Golgi apparatus [228].
Due to the lack of effective localizing ligand, fluorescent probes for imaging the Golgi apparatus have not received much attention. Although L-Cysteine has been reported to be a promising localizing ligand, we still need to do the further investigation of universality. With the development of the enzyme-activated probes, they will have the potential to apply in the Golgi apparatus-localizing fluorescent probes.
4. Summary and prospective
Metabolites serve critical roles in biology and any imbalance or fluctuation of these metabolites may result in diseases such as cancer, diabetes, obesity and neurodegeneration [2, 253]. The detecting and imaging cellular metabolites can help us to understand their roles in cellular metabolisms under both physiological and pathological conditions, thereby providing a powerful basis for diagnosis and treatment of these diseases. The development of fluorescent probes has made a remarkable contribution to biology, making it possible to observe the biochemical process directly inside living cells and sub-cellular organelles [254, 255]. Therefore, fluorescent probes for imaging subcellular metabolites have attracted much attention. Based on various localizing ligands, recognition groups, and fluorophores, many sub-cellular localized probes have been developed and applied in different areas, such as monitoring inflammation, diabetes, depression, and cancers. The development of these fluorescent probes has also greatly promoted our understanding of molecular mechanisms of different biological processes. In this review, we summarize the fluorescent probes for imaging metabolites in whole cells and subcellular systems. Localization, detection, and response principles are also discussed. Even though many fluorescent probes have been developed and applied, there are still several challenges in their application.
Although researchers have made extensive efforts in the development of fluorescent probes for imaging metabolites with subcellular accuracy during past decades, the development of fluorescent probes for whole cell imaging are significantly more than that of fluorescent probes for subcellular imaging. The main reasons for this limitation are the lack of efficient sub-cellular localizing ligands and potential toxic effects. For example, the most widely used mitochondria-localized ligand TPP can decrease MMP due to the cationic feature, and the TPP-modified fluorescent probes can leak out when the MMP is lost; the lysosome-localized ligand morpholine can cause the alkalization effect. Otherwise, the strategies that can efficiently localize sensors in other organelles, such as ER, Golgi apparatus, or the nucleus, are still rare, which severely limits the understanding of biological processes in these sub-cellular organelles. In addition, this limitation also affects the development of genetically encoded fluorescent probes for imaging the sub-cellular metabolites. Therefore, it is very important to develop the reliable localizing strategies and novel non-toxic localizing ligands for imaging the metabolites in subcellular organelles.
Furthermore, many of the cellular metabolites discussed here are present in ultralow concentration, sensitive to the environment, and have a short lifetime. Most reported probes failed in tracing the metabolites due to the poor sensitivity and/or specificity. Therefore, novel fluorescent probes for imaging the subcellular metabolites with lower detection limit and higher specificity are required. The promising solutions for these limitations are to explore higher-throughput probe screening systems, novel detection mechanisms, and accurate theoretical calculation methods. In addition, the fluorescent probes for imaging the subcellular metabolites with signal amplification may also contribute to improving detection sensitivity in future.
The signal-noise ratio is an important parameter in fluorescent imaging, because there are autofluorescent biomolecules in living cells, and they are emissive when exposure to the laser irradiation, that will interfere with the imaging resolution. Moreover, the emission wavelength of most reported fluorescent probes is in the visible region, hindering the application in background free in vivo imaging. Therefore, it is critical to develop fluorescent probes with high quantum yields, long lifetime, outstanding photostability and deep-tissue penetration capability. The fluorescent probes that can promote two or more photon excitations in near infrared emissions are promising for designing the novel probes.
Many different diseases are closely related to abnormal concentration or fluctuation of metabolites. In addition, the communication between metabolites from different sub-cellular organelles is also a promising area for investigators to explore the understanding of metabolism.
Over the past decades, considerable progress has been made to the development of fluorescent probes to image metabolites in whole cell and subcellular. The progress is mainly in the area of the improving the imaging resolution, specificity, sensitivity and better explanation for molecular mechanisms in diverse biological processes. However, several challenges remain as described in the above paragraphs. Given the tremendous progress that has been made so far, we are confident that researchers in the field will be able to meet these challenges to develop more fluorescent probes with higher performance to enhance our understanding of metabolisms that play significant roles on all biological processes.
Acknowledgments
The Lu group work described in this review was funded by the US National Institutes of Health (GM124316 and MH110975).
Data availability statement
No new data were created or analysed in this study.
References
- [1].Amantonico A, Urban PL and Zenobi R 2010. Analytical techniques for single-cell metabolomics: state of the art and trends Anal. Bioanal. Chem 398 2493–504 [DOI] [PubMed] [Google Scholar]
- [2].DeBerardinis RJ and Thompson CB 2012. Cellular metabolism and disease: what do metabolic outliers teach us? Cell 148 1132–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu JY and Wellen KE 2020. Advances into understanding metabolites as signaling molecules in cancer progression Curr. Opin. Cell Biol 63 144–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ghosh-Choudhary S, Liu J and Finkel T 2020. Metabolic regulation of cell fate and function Trends Cell Biol. 30 201–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Finkel T and Holbrook NJ 2000. Oxidants, oxidative stress and the biology of ageing Nature 408 239–47 [DOI] [PubMed] [Google Scholar]
- [6].Johnson CH, Ivanisevic J and Siuzdak G 2016. Metabolomics: beyond biomarkers and towards mechanisms Nat. Rev. Mol. Cell Biol 17 451–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Husted AS, Trauelsen M, Rudenko O, Hjorth SA and Schwartz TW 2017. GPCR-mediated signaling of metabolites Cell Metab. 25 777–96 [DOI] [PubMed] [Google Scholar]
- [8].Zhang A, Sun H, Wang P, Han Y and Wang X 2012. Modern analytical techniques in metabolomics analysis Analyst 137 293–300 [DOI] [PubMed] [Google Scholar]
- [9].Jang C, Chen L and Rabinowitz JD 2018. Metabolomics and isotope tracing Cell 173 822–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Castillo-Peinado LS and Luque de Castro MD 2016. Present and foreseeable future of metabolomics in forensic analysis Anal. Chim. Acta 925 1–15 [DOI] [PubMed] [Google Scholar]
- [11].Luan H, Wang X and Cai Z 2019. Mass spectrometry-based metabolomics: targeting the crosstalk between gut microbiota and brain in neurodegenerative disorders Mass Spectrom. Rev 38 22–33 [DOI] [PubMed] [Google Scholar]
- [12].Emwas AH 2015. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research Methods Mol. Biol 1277 161–93 [DOI] [PubMed] [Google Scholar]
- [13].Lee DY, Bowen BP and Northen TR 2010. Mass spectrometry-based metabolomics, analysis of metabolite-protein interactions, and imaging Biotechniques 49 557–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang YP and Lei QY 2018. Metabolite sensing and signaling in cell metabolism Signal Transduction Target Ther. 3 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pannkuk EL, Laiakis EC, Authier S, Wong K and Fornace AJ Jr. 2015. Global metabolomic identification of long-term dose-dependent urinary biomarkers in nonhuman primates exposed to ionizing radiation Radiat. Res 184 121–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Terai T and Nagano T 2013. Small-molecule fluorophores and fluorescent probes for bioimaging Pflug. Arch. Eur. J. Phy 465 347–59 [DOI] [PubMed] [Google Scholar]
- [17].Terai T and Nagano T 2008. Fluorescent probes for bioimaging applications Curr. Opin. Chem. Biol 12 515–21 [DOI] [PubMed] [Google Scholar]
- [18].An GH, Suh OS, Kwon HC, Kim K and Johnson EA 2000. Quantification of carotenoids in cells of Phaffia rhodozyma by autofluorescence Biotechnol. Lett 22 1031–4 [Google Scholar]
- [19].Cohen D, Dickerson JA, Whitmore CD, Turner EH, Palcic MM, Hindsgaul O and Dovichi NJ 2008. Chemical cytometry: fluorescence-based single-cell analysis Annu. Rev. Anal. Chem 1 165–90 [DOI] [PubMed] [Google Scholar]
- [20].Berg J, Hung YP and Yellen G 2009. A genetically encoded fluorescent reporter of ATP:ADP ratio Nat. Methods 6 161–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lapainis T, Scanlan C, Rubakhin SS and Sweedler JV 2007. A multichannel native fluorescence detection system for capillary electrophoretic analysis of neurotransmitters in single neurons Anal. Bioanal. Chem 387 97–105 [DOI] [PubMed] [Google Scholar]
- [22].Specht EA, Braselmann E and Palmer AE 2017. A critical and comparative review of fluorescent tools for live-cell imaging Annu. Rev. Physiol 79 93–117 [DOI] [PubMed] [Google Scholar]
- [23].Hare DJ, New EJ, de Jonge MD and McColl G 2015. Imaging metals in biology: balancing sensitivity, selectivity and spatial resolution Chem. Soc. Rev 44 5941–58 [DOI] [PubMed] [Google Scholar]
- [24].Shin JB, Streijger F, Beynon A, Peters T, Gadzala L, McMillen D, Bystrom C, Van der Zee CE, Wallimann T and Gillespie PG 2007. Hair bundles are specialized for ATP delivery via creatine kinase Neuron 53 371–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bodin P and Burnstock G 2001. Evidence that release of adenosine triphosphate from endothelial cells during increased shear stress is vesicular J. Cardiovasc. Pharmacol 38 900–8 [DOI] [PubMed] [Google Scholar]
- [26].Feranchak AP, Lewis MA, Kresge C, Sathe M, Bugde A, Luby-Phelps K, Antich PP and Fitz JG 2010. Initiation of purinergic signaling by exocytosis of ATP-containing vesicles in liver epithelium J. Biol. Chem 285 8138–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Akopova I, Tatur S, Grygorczyk M, Luchowski R, Gryczynski I, Gryczynski Z, Borejdo J and Grygorczyk R 2012. Imaging exocytosis of ATP-containing vesicles with TIRF microscopy in lung epithelial A549 cells Purinergic Signal. 8 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pak YL, Swamy KM and Yoon J 2015. Recent progress in fluorescent imaging probes Sensors 15 24374–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu Z, Singh NJ, Lim J, Pan J, Kim HN, Park S, Kim KS and Yoon J 2009. Unique sandwich stacking of pyrene-adenine-pyrene for selective and ratiometric fluorescent sensing of ATP at physiological pH J. Am. Chem. Soc 131 15528–33 [DOI] [PubMed] [Google Scholar]
- [30].Qiang W, Hu H, Sun L, Li H and Xu D 2015. Aptamer/polydopamine nanospheres nanocomplex for in situ molecular sensing in living cells Anal. Chem 87 12190–6 [DOI] [PubMed] [Google Scholar]
- [31].Zheng D, Seferos DS, Giljohann DA, Patel PC and Mirkin CA 2009. Aptamer nano-flares for molecular detection in living cells Nano Lett. 9 3258–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zheng X, Peng R, Jiang X, Wang Y, Xu S, Ke G, Fu T, Liu Q, Huan S and Zhang X 2017. Fluorescence resonance energy transfer-based DNA nanoprism with a split aptamer for adenosine triphosphate sensing in living cells Anal. Chem 89 10941–7 [DOI] [PubMed] [Google Scholar]
- [33].Zhao J, Gao J, Xue W, Di Z, Xing H, Lu Y and Li L 2018. Upconversion luminescence-activated DNA nanodevice for ATP sensing in living cells J. Am. Chem. Soc 140 578–81 [DOI] [PubMed] [Google Scholar]
- [34].Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T and Noji H 2009. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators Proc. Natl Acad. Sci. USA 106 15651–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yaginuma H, Kawai S, Tabata KV, Tomiyama K, Kakizuka A, Komatsuzaki T, Noji H and Imamura H 2014. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging Sci. Rep 4 6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Arai S et al. 2018. RGB-color intensiometric indicators to visualize spatiotemporal dynamics of ATP in single cells Angew. Chem., Int. Ed 57 10873–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lobas MA, Tao R, Nagai J, Kronschlager MT, Borden PM, Marvin JS, Looger LL and Khakh BS 2019. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP Nat. Commun 10 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tantama M, Martinez-Francois JR, Mongeon R and Yellen G 2013. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio Nat. Commun 4 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zala D, Hinckelmann MV, Yu H, Lyra Da Cunha MM, Liot G, Cordelieres FP, Marco S and Saudou F 2013. Vesicular glycolysis provides on-board energy for fast axonal transport Cell 152 479–91 [DOI] [PubMed] [Google Scholar]
- [40].Namiki S, Sakamoto H, Iinuma S, Iino M and Hirose K 2007. Optical glutamate sensor for spatiotemporal analysis of synaptic transmission Eur. J. Neurosci 25 2249–59 [DOI] [PubMed] [Google Scholar]
- [41].Miller EW and Chang CJ 2007. Fluorescent probes for nitric oxide and hydrogen peroxide in cell signaling Curr. Opin. Chem. Biol 11 620–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Maeda H, Fukuyasu Y, Yoshida S, Fukuda M, Saeki K, Matsuno H, Yamauchi Y, Yoshida K, Hirata K and Miyamoto K 2004. Fluorescent probes for hydrogen peroxide based on a non-oxidative mechanism Angew. Chem., Int. Ed 43 2389–91 [DOI] [PubMed] [Google Scholar]
- [43].Abo M, Urano Y, Hanaoka K, Terai T, Komatsu T and Nagano T 2011. Development of a highly sensitive fluorescence probe for hydrogen peroxide J. Am. Chem. Soc 133 10629–37 [DOI] [PubMed] [Google Scholar]
- [44].Kojima R, Takakura H, Kamiya M, Kobayashi E, Komatsu T, Ueno T, Terai T, Hanaoka K, Nagano T and Urano Y 2015. Development of a sensitive bioluminogenic probe for imaging highly reactive oxygen species in living rats Angew. Chem., Int. Ed 54 14768–71 [DOI] [PubMed] [Google Scholar]
- [45].Soh N, Sakawaki O, Makihara K, Odo Y, Fukaminato T, Kawai T, Irie M and Imato T 2005. Design and development of a fluorescent probe for monitoring hydrogen peroxide using photoinduced electron transfer Bioorg. Med. Chem 13 1131–9 [DOI] [PubMed] [Google Scholar]
- [46].Xu S, Liu HW, Yin X, Yuan L, Huan SY and Zhang XB 2019. A cell membrane-anchored fluorescent probe for monitoring carbon monoxide release from living cells Chem. Sci 10 320–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y and Nagano T 1998. Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins Anal. Chem 70 2446–53 [DOI] [PubMed] [Google Scholar]
- [48].Hu Y, Yang D, Yang C, Feng N, Shao Z, Zhang L, Wang X, Weng L, Luo Z and Wang L 2018. A novel ‘off-on’ fluorescent probe based on carbon nitride nanoribbons for the detection of citrate anion and live cell imaging Sensors 18 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tsien RY, Adams SR, Harootunian AT, Ji Y and Taylor SS 1991. Fluorescence ratio imaging of cyclic-AMP in single cells FASEB J. 5 A1065–A [DOI] [PubMed] [Google Scholar]
- [50].Sammak PJ, Adams SR, Harootunian AT, Schliwa M and Tsien RY 1992. Intracellular cyclic AMP not calcium, determines the direction of vesicle movement in melanophores: direct measurement by fluorescence ratio imaging J. Cell Biol 117 57–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bacskai BJ, Hochner B, Mahautsmith M, Adams SR, Kaang BK, Kandel ER and Tsien RY 1993. Spatially resolved dynamics of camp and protein kinase-a subunits in aplysia sensory neurons Science 260 222–6 [DOI] [PubMed] [Google Scholar]
- [52].Liu CY, Jamaleddin AJ, Zhang H and Christofi FL 1999. FlCRhR/cyclic AMP signaling in myenteric ganglia and calbindin-D28 intrinsic primary afferent neurons involves adenylyl cyclases I, III and IV Brain Res. 826 253–69 [DOI] [PubMed] [Google Scholar]
- [53].Zaccolo M, De Giorgi F, Cho CY, Feng LX, Knapp T, Negulescu PA, Taylor SS, Tsien RY and Pozzan T 2000. A genetically encoded, fluorescent indicator for cyclic AMP in living cells Nat. Cell Biol 2 25–9 [DOI] [PubMed] [Google Scholar]
- [54].Zaccolo M and Pozzan T 2002. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes Science 295 1711–5 [DOI] [PubMed] [Google Scholar]
- [55].Gerbino A, Ruder WC, Curci S, Pozzan T, Zaccolo M and Hofer AM 2005. Termination of cAMP signals by Ca2+ and G alpha(i) via extracellular Ca2+ sensors: a link to intracellular Ca2+ oscillations J. Cell Biol 171 303–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cardone RA, Bagorda A, Bellizzi A, Busco G, Guerra L, Paradiso A, Casavola V, Zaccolo M and Reshkin SJ 2005. Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines Mol. Biol. Cell 16 3117–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dal Molin F, Tonello F, Ladant D, Zornetta I, Zamparo I, Di Benedetto G, Zaccolo M and Montecucco C 2006. Cell entry and cAMP imaging of anthrax edema toxin Embo J. 25 5405–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].DiPilato LM, Cheng XD and Zhang J 2004. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments Proc. Natl Acad. Sci. USA 101 16513–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tewson PH, Martinka S, Shaner NC, Hughes TE and Quinn AM 2016. New DAG and cAMP sensors optimized for live-cell assays in automated laboratories J. Biomol. Screen 21 298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, Quinn AM, Hughes TE and Mirshahi T 2016. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics Proc. Natl Acad. Sci. USA 113 13069–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kellenberger CA, Chen C, Whiteley AT, Portnoy DA and Hammond MC 2015. RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP J. Am. Chem. Soc 137 6432–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Niino Y, Hotta K and Oka K 2010. Blue fluorescent cGMP sensor for multiparameter fluorescence imaging PLoS One 5 e9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nikolaev VO, Gambaryan S and Lohse MJ 2006. Fluorescent sensors for rapid monitoring of intracellular cGMP Nat. Methods 3 23–5 [DOI] [PubMed] [Google Scholar]
- [64].Russwurm M, Mullershausen F, Friebe A, Jager R, Russwurm C and Koesling D 2007. Design of fluorescence resonance energy transfer (FRET)-based cGMP indicators: a systematic approach Biochem. J 407 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sato M, Hida N, Ozawa T and Umezawa Y 2000. Fluorescent indicators for cyclic GMP based on cyclic GMP-dependent protein kinase Ialpha and green fluorescent proteins Anal. Chem 72 5918–24 [DOI] [PubMed] [Google Scholar]
- [66].Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH and Breaker RR 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP Science 321 411–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee ER, Baker JL, Weinberg Z, Sudarsan N and Breaker RR 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger Science 329 845–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kellenberger CA, Wilson SC, Sales-Lee J and Hammond MC 2013. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP J. Am. Chem. Soc 135 4906–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou H, Zheng C, Su J, Chen B, Fu Y, Xie Y, Tang Q, Chou SH and He J 2016. Characterization of a natural triple-tandem c-di-GMP riboswitch and application of the riboswitch-based dual-fluorescence reporter Sci. Rep 6 20871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wu R, Karunanayake Mudiyanselage A, Shafiei F, Zhao B, Bagheri Y, Yu Q, McAuliffe K, Ren K and You M 2019. Genetically encoded ratiometric RNA-based sensors for quantitative imaging of small molecules in living cells Angew. Chem., Int. Ed 58 18271–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hung YP, Albeck JG, Tantama M and Yellen G 2011. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor Cell Metab. 14 545–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Steinbeck J et al. 2020. In vivo NADH/NAD(+) biosensing reveals the dynamics of cytosolic redox metabolism in plants Plant Cell 32 3324–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hung YP and Yellen G 2014. Live-cell imaging of cytosolic NADH-NAD+ redox state using a genetically encoded fluorescent biosensor Methods Mol. Biol 1071 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bilan DS, Matlashov ME, Gorokhovatsky AY, Schultz C, Enikolopov G and Belousov VV 2014. Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments Biochim. Biophys. Acta 1840 951–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhao YZ et al. 2015. SoNar, a highly responsive NAD(+)/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents Cell Metab. 21 777–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhao YZ, Wang AX, Zou YJ, Su N, Loscalzo J and Yang Y 2016. In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD(+)/NADH redox state Nat. Protoc 11 1345–59 [DOI] [PubMed] [Google Scholar]
- [77].Tsien RY 2005. Building and breeding molecules to spy on cells and tumors Febs Lett. 579 927–32 [DOI] [PubMed] [Google Scholar]
- [78].Hires SA, Zhu Y and Tsien RY 2008. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters Proc. Natl Acad. Sci. USA 105 4411–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Armbruster M, Dulla CG and Diamond JS 2020. Effects of fluorescent glutamate indicators on neurotransmitter diffusion and uptake eLife 9 e54441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ and Frommer WB 2005. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors Proc. Natl Acad. Sci. USA 102 8740–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Besnard J and Okumoto S 2014. Glutamine flux imaging using genetically encoded sensors J. Vis. Exp 89 e51657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gruenwald K, Holland JT, Stromberg V, Ahmad A, Watcharakichkorn D and Okumoto S 2012. Visualization of glutamine transporter activities in living cells using genetically encoded glutamine sensors PLoS One 7 e38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Deuschle K, Okumoto S, Fehr M, Looger LL, Kozhukh L and Frommer WB 2005. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering Protein Sci 14 2304–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Fehr M, Lalonde S, Lager I, Wolff MW and Frommer WB 2003. In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors J. Biol. Chem 278 19127–33 [DOI] [PubMed] [Google Scholar]
- [85].Fehr M, Frommer WB and Lalonde S 2002. Visualization of maltose uptake in living yeast cells by fluorescent nanosensors Proc. Natl Acad. Sci. USA 99 9846–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lager I, Fehr M, Frommer WB and Lalonde S 2003. Development of a fluorescent nanosensor for ribose FEBS Lett. 553 85–9 [DOI] [PubMed] [Google Scholar]
- [87].Lager I, Looger LL, Hilpert M, Lalonde S and Frommer WB 2006. Conversion of a putative Agrobacterium sugar-binding protein into a FRET sensor with high selectivity for sucrose J. Biol. Chem 281 30875–83 [DOI] [PubMed] [Google Scholar]
- [88].San Martin A, Ceballo S, Ruminot I, Lerchundi R, Frommer WB and Barros LF 2013. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells PLoS One 8 e57712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kikuta S, Hou BH, Sato R, Frommer WB and Kikawada T 2016. FRET sensor-based quantification of intracellular trehalose in mammalian cells Biosci. Biotechnol. Biochem 80 162–5 [DOI] [PubMed] [Google Scholar]
- [90].Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY and Remington SJ 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators J. Biol. Chem 279 13044–53 [DOI] [PubMed] [Google Scholar]
- [91].Kolossov VL, Spring BQ, Sokolowski A, Conour JE, Clegg RM, Kenis PJ and Gaskins HR 2008. Engineering redox-sensitive linkers for genetically encoded FRET-based biosensors Exp. Biol. Med 233 238–48 [DOI] [PubMed] [Google Scholar]
- [92].Wang J, Karpus J, Zhao BS, Luo Z, Chen PR and He C 2012. A selective fluorescent probe for carbon monoxide imaging in living cells Angew. Chem., Int. Ed 51 9652–6 [DOI] [PubMed] [Google Scholar]
- [93].Sato M, Nakajima T, Goto M and Umezawa Y 2006. Cell-based indicator to visualize picomolar dynamics of nitric oxide release from living cells Anal. Chem 78 8175–82 [DOI] [PubMed] [Google Scholar]
- [94].Zhang C, Wei ZH and Ye BC 2013. Quantitative monitoring of 2-oxoglutarate in Escherichia coli cells by a fluorescence resonance energy transfer-based biosensor Appl. Microbiol. Biotechnol 97 8307–16 [DOI] [PubMed] [Google Scholar]
- [95].Ewald JC, Reich S, Baumann S, Frommer WB and Zamboni N 2011. Engineering genetically encoded nanosensors for real-time in vivo measurements of citrate concentrations PLoS One 6 e28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Galaz A, Cortes-Molina F, Arce-Molina R, Romero-Gomez I, Mardones GA, Barros LF and San Martin A 2020. Imaging of the lactate/pyruvate ratio using a genetically encoded FRET indicator Anal. Chem 92 10643–50 [DOI] [PubMed] [Google Scholar]
- [97].Leyssens A, Nowicky AV, Patterson L, Crompton M and Duchen MR 1996. The relationship between mitochondrial state, ATP hydrolysis, [Mg2+]i and [Ca2+]i studied in isolated rat cardiomyocytes J. Physiol 496 111–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Irvin JL and Irvin EM 1954. The interaction of quinacrine with adenine nucleotides J. Biol. Chem 210 45–56 [PubMed] [Google Scholar]
- [99].Paige JS, Nguyen-Duc T, Song W and Jaffrey SR 2012. Fluorescence imaging of cellular metabolites with RNA Science 335 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Feng C, Dai S and Wang L 2014. Optical aptasensors for quantitative detection of small biomolecules: a review Biosens. Bioelectron 59 64–74 [DOI] [PubMed] [Google Scholar]
- [101].Jo H and Ban C 2016. Aptamer-nanoparticle complexes as powerful diagnostic and therapeutic tools Exp. Mol. Med 48 e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang Y, Tang L, Li Z, Lin Y and Li J 2014. In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet-based sensing platform in living cells Nat. Protoc 9 1944–55 [DOI] [PubMed] [Google Scholar]
- [103].Torabi SF and Lu Y 2014. Functional DNA nanomaterials for sensing and imaging in living cells Curr. Opin. Biotechnol 28 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang H, Ma Y, Xie Y, An Y, Huang Y, Zhu Z and Yang CJ 2015. A controllable aptamer-based self-assembled DNA dendrimer for high affinity targeting, bioimaging and drug delivery Sci. Rep 5 10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Ozalp VC, Nielsen LJ and Olsen LF 2010. An aptamer-based nanobiosensor for real-time measurements of ATP dynamics Chembiochem 11 2538–41 [DOI] [PubMed] [Google Scholar]
- [106].Tsuboi T, Lippiat JD, Ashcroft FM and Rutter GA 2004. ATP-dependent interaction of the cytosolic domains of the inwardly rectifying K+ channel Kir6.2 revealed by fluorescence resonance energy transfer Proc. Natl Acad. Sci. USA 101 76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Honda A, Sawyer CL, Cawley SM and Dostmann WR 2005. Cygnets: in vivo characterization of novel cGMP indicators and in vivo imaging of intracellular cGMP Methods Mol. Biol 307 27–43 [DOI] [PubMed] [Google Scholar]
- [108].Honda A, Adams SR, Sawyer CL, Lev-Ram V, Tsien RY and Dostmann WR 2001. Spatiotemporal dynamics of guanosine 3′,5′-cyclic monophosphate revealed by a genetically encoded, fluorescent indicator Proc. Natl Acad. Sci. USA 98 2437–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Harris JJ, Jolivet R and Attwell D 2012. Synaptic energy use and supply Neuron 75 762–77 [DOI] [PubMed] [Google Scholar]
- [110].Ma YX, Chen HY, He X, Nie H, Hong YY, Sheng CB, Wang Q, Xia WL and Ying WH 2012. NAD(+) metabolism and NAD(+)-dependent enzymes: promising therapeutic targets for neurological diseases Curr. Drug Targets 13 222–9 [DOI] [PubMed] [Google Scholar]
- [111].Xiao WS, Wang RS, Handy DE and Loscalzo J 2018. NAD(H) and NADP(H) redox couples and cellular energy metabolism Antioxid. Redox Signal 28 251–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Canto C, Menzies KJ and Auwerx J 2015. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus Cell Metab. 22 31–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yang Y and Sauve AA 2016. NAD(+) metabolism: bioenergetics, signaling and manipulation for therapy Biosci. Biotechnol. Biochem 1864 1787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ying WH 2008. NAD(+)/NADH and NADP(+)/NADPH in cellular functions and cell death: regulation and biological consequences Antioxid. Redox Signal. 10 179–206 [DOI] [PubMed] [Google Scholar]
- [115].Paczek V, Dubois F, Sangwan R, Morot-Gaudry JF, Roubelakis-Angelakis KA and Hirel B 2002. Cellular and subcellular localisation of glutamine synthetase and glutamate dehydrogenase in grapes gives new insights on the regulation of carbon and nitrogen metabolism Planta 216 245–54 [DOI] [PubMed] [Google Scholar]
- [116].Plaitakis A and Zaganas I 2001. Regulation of human glutamate dehydrogenases: implications for glutamate, ammonia and energy metabolism in brain J. Neurosci. Res 66 899–908 [DOI] [PubMed] [Google Scholar]
- [117].Peng S, Zhang Y, Zhang J, Wang H and Ren B 2011. Glutamate receptors and signal transduction in learning and memory Mol. Biol. Rep 38 453–60 [DOI] [PubMed] [Google Scholar]
- [118].Griesbeck O, Baird GS, Campbell RE, Zacharias DA and Tsien RY 2001. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications J. Biol. Chem 276 29188–94 [DOI] [PubMed] [Google Scholar]
- [119].Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K and Miyawaki A 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications Nat. Biotechnol 20 87–90 [DOI] [PubMed] [Google Scholar]
- [120].Marvin JS et al. 2013. An optimized fluorescent probe for visualizing glutamate neurotransmission Nat. Methods 10 162–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Marvin JS et al. 2018. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR Nat. Methods 15 936–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Okada S, Ota K and Ito T 2009. Circular permutation of ligand-binding module improves dynamic range of genetically encoded FRET-based nanosensor Protein Sci. 18 2518–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ballerstadt R and Schultz JS 2000. A fluorescence affinity hollow fiber sensor for continuous transdermal glucose monitoring Anal. Chem 72 4185–92 [DOI] [PubMed] [Google Scholar]
- [124].Heo YJ, Shibata H, Okitsu T, Kawanishi T and Takeuchi S 2011. Long-term in vivo glucose monitoring using fluorescent hydrogel fibers Proc. Natl Acad. Sci. USA 108 13399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Circu ML and Aw TY 2010. Reactive oxygen species, cellular redox systems, and apoptosis Free Radical Biol. Med 48 749–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ray PD, Huang BW and Tsuji Y 2012. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling Cell Signal. 24 981–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Finkel T 2011. Signal transduction by reactive oxygen species J. Cell Biol 194 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yudhistira T, Mulay SV, Kim Y, Halle MB and Churchill DG 2019. Imaging of hypochlorous acid by fluorescence and applications in biological systems Chem. Asian J 14 3048–84 [DOI] [PubMed] [Google Scholar]
- [129].Huang J, Li J, Lyu Y, Miao Q and Pu K 2019. Molecular optical imaging probes for early diagnosis of drug-induced acute kidney injury Nat. Mater 18 1133–43 [DOI] [PubMed] [Google Scholar]
- [130].Huang J and Pu K 2020. Activatable molecular probes for second near-infrared fluorescence, chemiluminescence, and photoacoustic imaging Angew. Chem., Int. Ed 59 11717–31 [DOI] [PubMed] [Google Scholar]
- [131].Cheng P, Miao Q, Li J, Huang J, Xie C and Pu K 2019. Unimolecular chemo-fluoro-luminescent reporter for crosstalk-free duplex imaging of hepatotoxicity J. Am. Chem. Soc 141 10581–4 [DOI] [PubMed] [Google Scholar]
- [132].Huang J, Lyu Y, Li J, Cheng P, Jiang Y and Pu K 2019. A renal-clearable duplex optical reporter for real-time imaging of contrast-induced acute kidney injury Angew. Chem., Int. Ed 58 17796–804 [DOI] [PubMed] [Google Scholar]
- [133].Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ and Tsien RY 2004. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators J. Biol. Chem 279 22284–93 [DOI] [PubMed] [Google Scholar]
- [134].Ostergaard H, Henriksen A, Hansen FG and Winther JR 2001. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein Embo J. 20 5853–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Zhao BS, Liang Y, Song Y, Zheng C, Hao Z and Chen PR 2010. A highly selective fluorescent probe for visualization of organic hydroperoxides in living cells J. Am. Chem. Soc 132 17065–7 [DOI] [PubMed] [Google Scholar]
- [136].Liao YX, Li K, Wu MY, Wu T and Yu XQ 2014. A selenium-contained aggregation-induced ‘turn-on’ fluorescent probe for hydrogen peroxide Org. Biomol. Chem 12 3004–8 [DOI] [PubMed] [Google Scholar]
- [137].Yu F, Li P, Li G, Zhao G, Chu T and Han K 2011. A near-IR reversible fluorescent probe modulated by selenium for monitoring peroxynitrite and imaging in living cells J. Am. Chem. Soc 133 11030–3 [DOI] [PubMed] [Google Scholar]
- [138].Koide Y, Kawaguchi M, Urano Y, Hanaoka K, Komatsu T, Abo M, Terai T and Nagano T 2012. A reversible near-infrared fluorescence probe for reactive oxygen species based on Te-rhodamine Chem. Commun 48 3091–3 [DOI] [PubMed] [Google Scholar]
- [139].Jiang X, Wang L, Carroll SL, Chen J, Wang MC and Wang J 2018. Challenges and opportunities for small-molecule fluorescent probes in redox biology applications Antioxid. Redox Signal 29 518–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Xiao H, Zhang W, Li P, Zhang W, Wang X and Tang B 2020. Versatile fluorescent probes for imaging the superoxide anion in living cells and in vivo Angew. Chem., Int. Ed 59 4216–30 [DOI] [PubMed] [Google Scholar]
- [141].Chan J, Dodani SC and Chang CJ 2012. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging Nat. Chem 4 973–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Guo Z, Park S, Yoon J and Shin I 2014. Recent progress in the development of near-infrared fluorescent probes for bioimaging applications Chem. Soc. Rev 43 16–29 [DOI] [PubMed] [Google Scholar]
- [143].Tampieri F, Cabrellon G, Rossa A, Barbon A, Marotta E and Paradisi C 2019. Comment on ‘water-soluble fluorescent probe with dual mitochondria/lysosome targetability for selective superoxide detection in live cells and in zebrafish embryos’ ACS Sens. 4 3080–3 [DOI] [PubMed] [Google Scholar]
- [144].Lu X, Lin G, Dong X, Zhao W and Chen Z 2019. Reply to comment on ‘water-soluble fluorescent probe with dual mitochondria/lysosome targetability superoxide detection in live cells and in zebrafish embryos’ ACS Sens. 4 3084–7 [DOI] [PubMed] [Google Scholar]
- [145].Yuan L, Lin W, Tan L, Zheng K and Huang W 2013. Lighting up carbon monoxide: fluorescent probes for monitoring CO in living cells Angew. Chem., Int. Ed 52 1628–30 [DOI] [PubMed] [Google Scholar]
- [146].Kojima H, Urano Y, Kikuchi K, Higuchi T, Hirata Y and Nagano T 1999. Fluorescent indicators for imaging nitric oxide production Angew. Chem., Int. Ed 38 3209–12 [DOI] [PubMed] [Google Scholar]
- [147].Gabe Y, Urano Y, Kikuchi K, Kojima H and Nagano T 2004. Highly sensitive fluorescence probes for nitric oxide based on boron dipyrromethene chromophore-rational design of potentially useful bioimaging fluorescence probe J. Am. Chem. Soc 126 3357–67 [DOI] [PubMed] [Google Scholar]
- [148].Sasaki E, Kojima H, Nishimatsu H, Urano Y, Kikuchi K, Hirata Y and Nagano T 2005. Highly sensitive near-infrared fluorescent probes for nitric oxide and their application to isolated organs J. Am. Chem. Soc 127 3684–5 [DOI] [PubMed] [Google Scholar]
- [149].Chen Y 2020. Recent developments of fluorescent probes for detection and bioimaging of nitric oxide Nitric Oxide 98 1–19 [DOI] [PubMed] [Google Scholar]
- [150].Wang L, Zhang J, An X and Duan H 2020. Recent progress on the organic and metal complex-based fluorescent probes for monitoring nitric oxide in living biological systems Org. Biomol. Chem 18 1522–49 [DOI] [PubMed] [Google Scholar]
- [151].Akram M 2014. Citric acid cycle and role of its intermediates in metabolism Cell Biochem. Biophys 68 475–8 [DOI] [PubMed] [Google Scholar]
- [152].Harrison VS, Carney CE, MacRenaris KW, Waters EA and Meade TJ 2015. Multimeric near IR-MR contrast agent for multimodal in vivo imaging J. Am. Chem. Soc 137 9108–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Li H, Parigi G, Luchinat C and Meade TJ 2019. Bimodal fluorescence-magnetic resonance contrast agent for apoptosis imaging J. Am. Chem. Soc 141 6224–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Xu W, Zeng Z, Jiang JH, Chang YT and Yuan L 2016. Discerning the chemistry in individual organelles with small-molecule fluorescent probes Angew. Chem., Int. Ed 55 13658–99 [DOI] [PubMed] [Google Scholar]
- [155].Jiao X, Li Y, Niu J, Xie X, Wang X and Tang B 2018. Small-molecule fluorescent probes for imaging and detection of reactive oxygen, nitrogen, and sulfur species in biological systems Anal. Chem 90 533–55 [DOI] [PubMed] [Google Scholar]
- [156].Xu Z and Xu L 2016. Fluorescent probes for the selective detection of chemical species inside mitochondria Chem. Commun 52 1094–119 [DOI] [PubMed] [Google Scholar]
- [157].Zhang R, Song B and Yuan J 2018. Bioanalytical methods for hypochlorous acid detection: recent advances and challenges Trends Analyt. Chem 99 1–33 [Google Scholar]
- [158].Gao P, Pan W, Li N and Tang B 2019. Fluorescent probes for organelle-targeted bioactive species imaging Chem. Sci 10 6035–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Zhu H, Fan J, Du J and Peng X 2016. Fluorescent probes for sensing and imaging within specific cellular organelles Acc. Chem. Res 49 2115–26 [DOI] [PubMed] [Google Scholar]
- [160].Jean SR, Ahmed M, Lei EK, Wisnovsky SP and Kelley SO 2016. Peptide-mediated delivery of chemical probes and therapeutics to mitochondria Acc. Chem. Res 49 1893–902 [DOI] [PubMed] [Google Scholar]
- [161].Yang Z et al. 2013. A self-calibrating bipartite viscosity sensor for mitochondria J. Am. Chem. Soc 135 9181–5 [DOI] [PubMed] [Google Scholar]
- [162].Henze K and Martin W 2003. Evolutionary biology: essence of mitochondria Nature 426 127–8 [DOI] [PubMed] [Google Scholar]
- [163].Lin MT and Beal MF 2006. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases Nature 443 787–95 [DOI] [PubMed] [Google Scholar]
- [164].Krumova K, Greene LE and Cosa G 2013. Fluorogenic alpha-tocopherol analogue for monitoring the antioxidant status within the inner mitochondrial membrane of live cells J. Am. Chem. Soc 135 17135–43 [DOI] [PubMed] [Google Scholar]
- [165].Lei EK and Kelley SO 2017. Delivery and release of small-molecule probes in mitochondria using traceless linkers J. Am. Chem. Soc 139 9455–8 [DOI] [PubMed] [Google Scholar]
- [166].Leung CW, Hong Y, Chen S, Zhao E, Lam JW and Tang BZ 2013. A photostable AIE luminogen for specific mitochondrial imaging and tracking J. Am. Chem. Soc 135 62–5 [DOI] [PubMed] [Google Scholar]
- [167].Dickinson BC and Chang CJ 2008. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells J. Am. Chem. Soc 130 9638–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Matsushita M, Mishima K, Esaki R, Ishiguro N, Ohno K and Kitoh H 2017. Maternal administration of meclozine for the treatment of foramen magnum stenosis in transgenic mice with achondroplasia J. Neurosurg. Pediatr 19 91–5 [DOI] [PubMed] [Google Scholar]
- [169].Johnson LV, Walsh ML and Chen LB 1980. Localization of mitochondria in living cells with rhodamine 123 Proc. Natl Acad. Sci. USA 77 990–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Horton KL, Stewart KM, Fonseca SB, Guo Q and Kelley SO 2008. Mitochondria-penetrating peptides Chem. Biol 15 375–82 [DOI] [PubMed] [Google Scholar]
- [171].Chen LB 1988. Mitochondrial membrane potential in living cells Annu. Rev. Cell Biol 4 155–81 [DOI] [PubMed] [Google Scholar]
- [172].Jiang N, Fan J, Xu F, Peng X, Mu H, Wang J and Xiong X 2015. Ratiometric fluorescence imaging of cellular polarity: decrease in mitochondrial polarity in cancer cells Angew. Chem., Int. Ed 54 2510–4 [DOI] [PubMed] [Google Scholar]
- [173].Sarkar AR, Heo CH, Lee HW, Park KH, Suh YH and Kim HM 2014. Red emissive two-photon probe for real-time imaging of mitochondria trafficking Anal. Chem 86 5638–41 [DOI] [PubMed] [Google Scholar]
- [174].Wen Y, Liu K, Yang H, Liu Y, Chen L, Liu Z, Huang C and Yi T 2015. Mitochondria-directed fluorescent probe for the detection of hydrogen peroxide near mitochondrial DNA Anal. Chem 87 10579–84 [DOI] [PubMed] [Google Scholar]
- [175].Liu X, Zheng A, Luan D, Wang X, Kong F, Tong L, Xu K and Tang B 2017. High-quantum-yield mitochondria-targeting near-infrared fluorescent probe for imaging native hypobromous acid in living cells and in vivo Anal. Chem 89 1787–92 [DOI] [PubMed] [Google Scholar]
- [176].Shen Y, Zhang X, Zhang Y, Wu Y, Zhang C, Chen Y, Jin J and Li H 2018. A mitochondria-targeted colorimetric and ratiometric fluorescent probe for hydrogen peroxide with a large emission shift and bio-imaging in living cells Sens. Actuators B 255 42–8 [Google Scholar]
- [177].Zhang G, Gruskos JJ, Afzal MS and Buccella D 2015. Visualizing changes in mitochondrial Mg(2+) during apoptosis with organelle-targeted triazole-based ratiometric fluorescent sensors Chem. Sci 6 6841–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Gong X, Yang X-F, Zhong Y, Chen Y and Li Z 2016. A mitochondria-targetable near-infrared fluorescent probe for imaging nitroxyl (HNO) in living cells Dyes Pigm. 131 24–32 [Google Scholar]
- [179].Song H, Li W, Qi R, Yan L, Jing X, Zheng M and Xiao H 2015. Delivering a photosensitive transplatin prodrug to overcome cisplatin drug resistance Chem. Commun 51 11493–5 [DOI] [PubMed] [Google Scholar]
- [180].Wang H. et al. Targeted production of reactive oxygen species in mitochondria to overcome cancer drug resistance. Nat. Commun. 2018;9:562. doi: 10.1038/s41467-018-02915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Field LD, Delehanty JB, Chen Y and Medintz IL 2015. Peptides for specifically targeting nanoparticles to cellular organelles: quo vadis? Acc. Chem. Res 48 1380–90 [DOI] [PubMed] [Google Scholar]
- [182].Tan KY, Li CY, Li YF, Fei J, Yang B, Fu YJ and Li F 2017. Real-time monitoring ATP in mitochondrion of living cells: a specific fluorescent probe for ATP by dual recognition sites Anal. Chem 89 1749–56 [DOI] [PubMed] [Google Scholar]
- [183].Liu JH, Li RS, Yuan B, Wang J, Li YF and Huang CZ 2018. Mitochondria-targeting single-layered graphene quantum dots with dual recognition sites for ATP imaging in living cells Nanoscale 10 17402–8 [DOI] [PubMed] [Google Scholar]
- [184].Hong S, Zhang X, Lake RJ, Pawel GT, Guo Z, Pei R and Lu Y 2020. A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of ATP in the mitochondria of living cells Chem. Sci 11 713–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Depaoli MR et al. 2018. Real-time imaging of mitochondrial ATP dynamics reveals the metabolic setting of single cells Cell Rep. 25 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Srivastava P, Razi SS, Ali R, Srivastav S, Patnaik S, Srikrishna S and Misra A 2015. Highly sensitive cell imaging ‘off-on’ fluorescent probe for mitochondria and ATP Biosens. Bioelectron 69 179–85 [DOI] [PubMed] [Google Scholar]
- [187].Wang L et al. 2016. A multisite-binding switchable fluorescent probe for monitoring mitochondrial ATP level fluctuation in live cells Angew. Chem., Int. Ed 55 1773–6 [DOI] [PubMed] [Google Scholar]
- [188].Ren TB, Wen SY, Wang L, Lu P, Xiong B, Yuan L and Zhang XB 2020. Engineering a reversible fluorescent probe for real-time live-cell imaging and quantification of mitochondrial ATP Anal. Chem 92 4681–8 [DOI] [PubMed] [Google Scholar]
- [189].Deng J, Wang K, Wang M, Yu P and Mao L 2017. Mitochondria targeted nanoscale zeolitic imidazole Framework-90 for ATP imaging in live cells J. Am. Chem. Soc 139 5877–82 [DOI] [PubMed] [Google Scholar]
- [190].Li P, Zhang W, Li K, Liu X, Xiao H, Zhang W and Tang B 2013. Mitochondria-targeted reaction-based two-photon fluorescent probe for imaging of superoxide anion in live cells and in vivo Anal. Chem 85 9877–81 [DOI] [PubMed] [Google Scholar]
- [191].Li H, Li X, Wu X, Shi W and Ma H 2017. Observation of the generation of ONOO(−) in mitochondria under various stimuli with a sensitive fluorescence probe Anal. Chem 89 5519–25 [DOI] [PubMed] [Google Scholar]
- [192].Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV and Lukyanov S 2006. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide Nat. Methods 3 281–6 [DOI] [PubMed] [Google Scholar]
- [193].Malinouski M, Zhou Y, Belousov VV, Hatfield DL and Gladyshev VN 2011. Hydrogen peroxide probes directed to different cellular compartments PLoS One 6 e14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Ermakova YG. et al. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat. Commun. 2014;5:5222. doi: 10.1038/ncomms6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Wang W et al. 2008. Superoxide flashes in single mitochondria Cell 134 279–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Hu JJ et al. 2015. Fluorescent probe HKSOX-1 for imaging and detection of endogenous superoxide in live cells and in vivo J. Am. Chem. Soc 137 6837–43 [DOI] [PubMed] [Google Scholar]
- [197].Xiao H, Li J, Zhao J, Yin G, Quan Y, Wang J and Wang R 2015. A colorimetric and ratiometric fluorescent probe for ClO(−) targeting in mitochondria and its application in vivo J. Mater. Chem. B 3 1633–8 [DOI] [PubMed] [Google Scholar]
- [198].Hou JT, Li K, Yang J, Yu KK, Liao YX, Ran YZ, Liu YH, Zhou XD and Yu XQ 2015. A ratiometric fluorescent probe for in situ quantification of basal mitochondrial hypochlorite in cancer cells Chem. Commun 51 6781–4 [DOI] [PubMed] [Google Scholar]
- [199].Zhou J, Li L, Shi W, Gao X, Li X and Ma H 2015. HOCl can appear in the mitochondria of macrophages during bacterial infection as revealed by a sensitive mitochondrial-targeting fluorescent probe Chem. Sci 6 4884–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Liu HW, Xu S, Wang P, Hu XX, Zhang J, Yuan L, Zhang XB and Tan W 2016. An efficient two-photon fluorescent probe for monitoring mitochondrial singlet oxygen in tissues during photodynamic therapy Chem. Commun 52 12330–3 [DOI] [PubMed] [Google Scholar]
- [201].Xu F, Li H, Yao Q, Fan J, Wang J and Peng X 2016. A NIR fluorescent probe: imaging endogenous hydrogen peroxide during an autophagy process induced by rapamycin J. Mater. Chem. B 4 7363–7 [DOI] [PubMed] [Google Scholar]
- [202].Xie X, Yang X, Wu T, Li Y, Li M, Tan Q, Wang X and Tang B 2016. Rational design of an alpha-ketoamide-based near-infrared fluorescent probe specific for hydrogen peroxide in living systems Anal. Chem 88 8019–25 [DOI] [PubMed] [Google Scholar]
- [203].Liu F, Du J, Song D, Xu M and Sun G 2016. A sensitive fluorescent sensor for the detection of endogenous hydroxyl radicals in living cells and bacteria and direct imaging with respect to its ecotoxicity in living zebra fish Chem. Commun 52 4636–9 [DOI] [PubMed] [Google Scholar]
- [204].Hou JT, Wu MY, Li K, Yang J, Yu KK, Xie YM and Yu XQ 2014. Mitochondria-targeted colorimetric and fluorescent probes for hypochlorite and their applications for in vivo imaging Chem. Commun 50 8640–3 [DOI] [PubMed] [Google Scholar]
- [205].Li K, Hou JT, Yang J and Yu XQ 2017. A tumor-specific and mitochondria-targeted fluorescent probe for real-time sensing of hypochlorite in living cells Chem. Commun 53 5539–41 [DOI] [PubMed] [Google Scholar]
- [206].Kim S, Tachikawa T, Fujitsuka M and Majima T 2014. Far-red fluorescence probe for monitoring singlet oxygen during photodynamic therapy J. Am. Chem. Soc 136 11707–15 [DOI] [PubMed] [Google Scholar]
- [207].Si F, Liu Y, Yan K and Zhong W 2015. A mitochondrion targeting fluorescent probe for imaging of intracellular superoxide radicals Chem. Commun 51 7931–4 [DOI] [PubMed] [Google Scholar]
- [208].Gong N et al. 2019. Carbon-dot-supported atomically dispersed gold as a mitochondrial oxidative stress amplifier for cancer treatment Nat. Nanotechnol 14 379–87 [DOI] [PubMed] [Google Scholar]
- [209].Zhao Y and Yang Y 2016. Real-time and high-throughput analysis of mitochondrial metabolic states in living cells using genetically encoded NAD(+)/NADH sensors Free Radical Biol. Med 100 43–52 [DOI] [PubMed] [Google Scholar]
- [210].Zhao Y and Yang Y 2012. Frex and FrexH: indicators of metabolic states in living cells Bioeng. Bugs 3 181–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Patterson GH, Knobel SM, Arkhammar P, Thastrup O and Piston DW 2000. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells Proc. Natl Acad. Sci. USA 97 5203–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Nakamura A, Takigawa K, Kurishita Y, Kuwata K, Ishida M, Shimoda Y, Hamachi I and Tsukiji S 2014. Hoechst tagging: a modular strategy to design synthetic fluorescent probes for live-cell nucleus imaging Chem. Commun 50 6149–52 [DOI] [PubMed] [Google Scholar]
- [213].Cheng Y, Sun C, Ou X, Liu B, Lou X and Xia F 2017. Dual-targeted peptide-conjugated multifunctional fluorescent probe with AIEgen for efficient nucleus-specific imaging and long-term tracing of cancer cells Chem. Sci 8 4571–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Lukinavicius G. et al. SiR-Hoechst is a far-red DNA stain for live-cell nanoscopy. Nat. Commun. 2015;6:8497. doi: 10.1038/ncomms9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Dickinson BC, Tang Y, Chang Z and Chang CJ 2011. A nuclear-localized fluorescent hydrogen peroxide probe for monitoring sirtuin-mediated oxidative stress responses in vivo Chem. Biol 18 943–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Wen Y, Liu K, Yang H, Li Y, Lan H, Liu Y, Zhang X and Yi T 2014. A highly sensitive ratiometric fluorescent probe for the detection of cytoplasmic and nuclear hydrogen peroxide Anal. Chem 86 9970–6 [DOI] [PubMed] [Google Scholar]
- [217].Srikun D, Albers AE, Nam CI, Iavarone AT and Chang CJ 2010. Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-Tag protein labeling J. Am. Chem. Soc 132 4455–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Wang C, Song X, Han Z, Li X, Xu Y and Xiao Y 2016. Monitoring nitric oxide in subcellular compartments by hybrid probe based on rhodamine spirolactam and SNAP-tag ACS Chem. Biol 11 2033–40 [DOI] [PubMed] [Google Scholar]
- [219].Wang J, Zhao Y, Wang C, Zhu Q, Du Z, Hu A and Yang Y 2015. Organelle-specific nitric oxide detection in living cells via halotag protein labeling PLoS One 10 e0123986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Chen J, Zhao M, Jiang X, Sizovs A, Wang MC, Provost CR, Huang J and Wang J 2016. Genetically anchored fluorescent probes for subcellular specific imaging of hydrogen sulfide Analyst 141 1209–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Ma X, Gong N, Zhong L, Sun J and Liang XJ 2016. Future of nanotherapeutics: targeting the cellular sub-organelles Biomaterials 97 10–21 [DOI] [PubMed] [Google Scholar]
- [222].Zhang X, Wang B, Wang C, Chen L and Xiao Y 2015. Monitoring lipid peroxidation within foam cells by lysosome-targetable and ratiometric probe Anal. Chem 87 8292–300 [DOI] [PubMed] [Google Scholar]
- [223].Ren M, Deng B, Wang JY, Kong X, Liu ZR, Zhou K, He L and Lin W 2016. A fast responsive two-photon fluorescent probe for imaging H(2)O(2) in lysosomes with a large turn-on fluorescence signal Biosens. Bioelectron 79 237–43 [DOI] [PubMed] [Google Scholar]
- [224].Jin F, Zheng J, Liu C, Yang S, Li Y, Li J, Lian Y and Yang R 2014. Dual-stimuli responsive i-motif/nanoflares for sensing ATP in lysosomes Analyst 139 3714–7 [DOI] [PubMed] [Google Scholar]
- [225].Xiao H, Liu X, Wu C, Wu Y, Li P, Guo X and Tang B 2017. A new endoplasmic reticulum-targeted two-photon fluorescent probe for imaging of superoxide anion in diabetic mice Biosens. Bioelectron 91 449–55 [DOI] [PubMed] [Google Scholar]
- [226].Zhang W, Zhang J, Li P, Liu J, Su D and Tang B 2019. Two-photon fluorescence imaging reveals a Golgi apparatus superoxide anion-mediated hepatic ischaemia-reperfusion signalling pathway Chem. Sci 10 879–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Zhu H, Liu C, Liang C, Tian B, Zhang H, Zhang X, Sheng W, Yu Y, Huang S and Zhu B 2020. A new phenylsulfonamide-based Golgi-targeting fluorescent probe for H2S and its bioimaging applications in living cells and zebrafish Chem. Commun 56 4086–9 [DOI] [PubMed] [Google Scholar]
- [228].Zhang X. et al. A highly specific Golgi-targetable fluorescent probe for tracking cysteine generation during the Golgi stress response. Sens. Actuators B. 2020;310:127820. [Google Scholar]
- [229].Wang C, Zhao T, Li Y, Huang G, White MA and Gao J 2017. Investigation of endosome and lysosome biology by ultra pH-sensitive nanoprobes Adv. Drug Delivery Rev 113 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Lan H, Xue F, Liu Z, Chen L, Huang C and Yi T 2016. Evolution of rhodamine B into near-infrared dye by phototriggered radical reaction and its application for lysosome-specific live-cell imaging Adv. Opt. Mater 4 1367–72 [Google Scholar]
- [231].Wu Z and Tang X 2015. Visualizing fluoride ion in mitochondria and lysosome of living cells and in living mice with positively charged ratiometric probes Anal. Chem 87 8613–7 [DOI] [PubMed] [Google Scholar]
- [232].McCue HV, Wardyn JD, Burgoyne RD and Haynes LP 2013. Generation and characterization of a lysosomally targeted, genetically encoded Ca(2+)-sensor Biochem. J 449 449–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Wan Q, Chen S, Shi W, Li L and Ma H 2014. Lysosomal pH rise during heat shock monitored by a lysosome-targeting near-infrared ratiometric fluorescent probe Angew. Chem., Int. Ed 53 10916–20 [DOI] [PubMed] [Google Scholar]
- [234].Reja SI, Gupta M, Gupta N, Bhalla V, Ohri P, Kaur G and Kumar M 2017. A lysosome targetable fluorescent probe for endogenous imaging of hydrogen peroxide in living cells Chem. Commun 53 3701–4 [DOI] [PubMed] [Google Scholar]
- [235].Yuan L, Wang L, Agrawalla BK, Park SJ, Zhu H, Sivaraman B, Peng J, Xu QH and Chang YT 2015. Development of targetable two-photon fluorescent probes to image hypochlorous Acid in mitochondria and lysosome in live cell and inflamed mouse model J. Am. Chem. Soc 137 5930–8 [DOI] [PubMed] [Google Scholar]
- [236].Zhu B, Li P, Shu W, Wang X, Liu C, Wang Y, Wang Z, Wang Y and Tang B 2016. Highly specific and ultrasensitive two-photon fluorescence imaging of native HOCl in lysosomes and tissues based on thiocarbamate derivatives Anal. Chem 88 12532–8 [DOI] [PubMed] [Google Scholar]
- [237].Huang BH, Geng ZR, Ma XY, Zhang C, Zhang ZY and Wang ZL 2016. Lysosomal ATP imaging in living cells by a water-soluble cationic polythiophene derivative Biosens. Bioelectron 83 213–20 [DOI] [PubMed] [Google Scholar]
- [238].Yu H, Xiao Y and Jin L 2012. A lysosome-targetable and two-photon fluorescent probe for monitoring endogenous and exogenous nitric oxide in living cells J. Am. Chem. Soc 134 17486–9 [DOI] [PubMed] [Google Scholar]
- [239].Jing X, Yu F and Chen L 2014. Visualization of nitroxyl (HNO) in vivo via a lysosome-targetable near-infrared fluorescent probe Chem. Commun 50 14253–6 [DOI] [PubMed] [Google Scholar]
- [240].Swamy KMK, Eom S, Liu Y, Kim G, Lee D and Yoon J 2019. Rhodamine derivatives bearing thiourea groups serve as fluorescent probes for selective detection of ATP in mitochondria and lysosomes Sens. Actuators B 281 350–8 [Google Scholar]
- [241].Dai Z, Tian L, Song B, Liu X and Yuan J 2017. Development of a novel lysosome-targetable time-gated luminescence probe for ratiometric and luminescence lifetime detection of nitric oxide in vivo Chem. Sci 8 1969–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Wu Z, Liang D and Tang X 2016. Visualizing hydrogen sulfide in mitochondria and lysosome of living cells and in tumors of living mice with positively charged fluorescent chemosensors Anal. Chem 88 9213–8 [DOI] [PubMed] [Google Scholar]
- [243].Jun YW, Wang T, Hwang S, Kim D, Ma D, Kim KH, Kim S, Jung J and Ahn KH 2018. A ratiometric two-photon fluorescent probe for tracking lysosomal ATP: direct in cellulo observation of lysosomal membrane fusion processes Angew. Chem., Int. Ed 57 10142–7 [DOI] [PubMed] [Google Scholar]
- [244].Geng X, Sun Y, Guo Y, Zhao Y, Zhang K, Xiao L, Qu L and Li Z 2020. Fluorescent carbon dots for in situ monitoring of lysosomal ATP levels Anal. Chem 92 7940–6 [DOI] [PubMed] [Google Scholar]
- [245].Vishnu N, Jadoon Khan M, Karsten F, Groschner LN, Waldeck-Weiermair M, Rost R, Hallstrom S, Imamura H, Graier WF and Malli R 2014. ATP increases within the lumen of the endoplasmic reticulum upon intracellular Ca2+ release Mol. Biol. Cell 25 368–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Klein MC. et al. AXER is an ATP/ADP exchanger in the membrane of the endoplasmic reticulum. Nat. Commun. 2018;9:3489. doi: 10.1038/s41467-018-06003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Hou JT, Kim HS, Duan C, Ji MS, Wang S, Zeng L, Ren WX and Kim JS 2019. A ratiometric fluorescent probe for detecting hypochlorite in the endoplasmic reticulum Chem. Commun 55 2533–6 [DOI] [PubMed] [Google Scholar]
- [248].Li SJ, Zhou DY, Li Y, Liu HW, Wu P, Ou-Yang J, Jiang WL and Li CY 2018. Efficient two-photon fluorescent probe for imaging of nitric oxide during endoplasmic reticulum stress ACS Sens. 3 2311–9 [DOI] [PubMed] [Google Scholar]
- [249].Tang Y, Xu A, Ma Y, Xu G, Gao S and Lin W 2017. A turn-on endoplasmic reticulum-targeted two-photon fluorescent probe for hydrogen sulfide and bio-imaging applications in living cells, tissues, and zebrafish Sci. Rep 7 12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Hessels AM and Merkx M 2015. Genetically-encoded FRET-based sensors for monitoring Zn(2+) in living cells Metallomics 7 258–66 [DOI] [PubMed] [Google Scholar]
- [251].Ali F, Aute S, Sreedharan S, Anila HA, Saeed HK, Smythe CG, Thomas JA and Das A 2018. Tracking HOCl concentrations across cellular organelles in real time using a super resolution microscopy probe Chem. Commun 54 1849–52 [DOI] [PubMed] [Google Scholar]
- [252].Zhu H et al. 2020. Rational design of a targetable fluorescent probe for visualizing H2S production under golgi stress response elicited by monensin Anal. Chem 92 1883–9 [DOI] [PubMed] [Google Scholar]
- [253].Kaelin WG Jr and McKnight SL 2013. Influence of metabolism on epigenetics and disease Cell 153 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Tsien RY 2009. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew. Chem., Int. Ed 48 5612–26 [DOI] [PubMed] [Google Scholar]
- [255].Day RN and Davidson MW 2009. The fluorescent protein palette: tools for cellular imaging Chem. Soc. Rev 38 2887–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analysed in this study.


