Abstract
Long-term activation of inositol 1,4,5-trisphosphate receptors (IP3Rs) leads to their degradation by the ubiquitin–proteasome pathway. The first and rate-limiting step in this process is thought to be the association of conformationally active IP3Rs with the erlin1/2 complex, an endoplasmic reticulum–located oligomer of erlin1 and erlin2 that recruits the E3 ubiquitin ligase RNF170, but the molecular determinants of this interaction remain unknown. Here, through mutation of IP3R1, we show that the erlin1/2 complex interacts with the IP3R1 intralumenal loop 3 (IL3), the loop between transmembrane (TM) helices 5 and 6, and in particular, with a region close to TM5, since mutation of amino acids D-2471 and R-2472 can specifically block erlin1/2 complex association. Surprisingly, we found that additional mutations in IL3 immediately adjacent to TM5 (e.g., D2465N) almost completely abolish IP3R1 Ca2+ channel activity, indicating that the integrity of this region is critical to IP3R1 function. Finally, we demonstrate that inhibition of the ubiquitin-activating enzyme UBE1 by the small-molecule inhibitor TAK-243 completely blocked IP3R1 ubiquitination and degradation without altering erlin1/2 complex association, confirming that association of the erlin1/2 complex is the primary event that initiates IP3R1 processing and that IP3R1 ubiquitination mediates IP3R1 degradation. Overall, these data localize the erlin1/2 complex–binding site on IP3R1 to IL3 and show that the region immediately adjacent to TM5 is key to the events that facilitate channel opening.
Keywords: calcium channel, erlin1/2 complex, endoplasmic reticulum–associated degradation, inositol trisphosphate receptor, protein–protein interaction, ubiquitination
Abbreviations: aa, amino acid; cDNA, complementary DNA; ER, endoplasmic reticulum; ERAD, endoplasmic reticulum-associated degradation; GnRH, gonadotropin-releasing hormone; HA, hemagglutinin; IL3, intralumenal loop 3; IP, immunoprecipitation; IP3, inositol 1,4,5-trisphosphate; IP3R, inositol 1,4,5-trisphosphate receptors; SPFH, stomatin/prohibitin/flotillin/HflK/C; TM, transmembrane; UPP, ubiquitin–proteasome pathway
Inositol 1,4,5-trisphosphate (IP3) receptors (IP3Rs) form tetrameric calcium ion (Ca2+) channels in the endoplasmic reticulum (ER) membrane of mammalian cells (1). IP3R activation is initiated by the secondary messenger IP3, which can be generated via activation of cell surface receptors, such as G-protein–coupled receptors or receptor tyrosine kinases (1). In response to activation, IP3Rs undergo a conformational change and release Ca2+ from ER stores into the cytosol (1, 2, 3). IP3R-mediated calcium signaling is critical to a plethora of cellular processes, such as fertilization, secretion, and smooth muscle contraction (4, 5). The mammalian genome encodes three IP3R types (IP3R1, IP3R2, and IP3R3), which have different tissue distributions and can form homotetramers or heterotetramers, with IP3R1 being the most widely expressed and best characterized of the three types (1). IP3R1, like IP3R2 and IP3R3, is ∼2700 amino acids in length and contains six transmembrane (TM) domains near the C terminus (1). The channel pore is created by an assemblage of TM domains 5 and 6 and the intervening intralumenal loop 3 (IL3) (2, 3, 6, 7).
The ER-associated degradation (ERAD) pathway is a facet of the ubiquitin–proteasome pathway (UPP) and not only targets misfolded ER proteins for degradation but also regulates native ER proteins, such as IP3Rs, to maintain cellular homeostasis (8, 9, 10, 11). ERAD usually includes three steps: substrate recognition, extraction from the ER, and ubiquitin-dependent proteasomal degradation (8, 10, 11). It appears that the conformational change in IP3Rs that occurs upon Ca2+ channel activation (2, 6) targets them for the ERAD pathway (9). This reduces cellular IP3R levels as well as the sensitivity of ER Ca2+ stores to IP3 (12). In addition, the ERAD pathway appears to be responsible for basal IP3R turnover under resting conditions (13, 14).
We have shown previously that the erlin1/2 complex, an ∼2 MDa ER membrane complex composed of ∼40 erlin1 and erlin2 molecules, binds strongly and rapidly to activated IP3Rs (15). The erlin1/2 complex recruits RNF170, the critical ubiquitin ligase in IP3R ERAD (16), and seems to be a recognition factor specific for IP3R processing by the ERAD pathway (9). Deletion of the erlin1/2 complex inhibits RNF170 association with activated IP3Rs and ablates IP3R ubiquitination and IP3R ERAD (14). However, the molecular mechanism of how the erlin1/2 complex recognizes activated IP3Rs remains to be resolved.
We hypothesized previously (15) that the erlin1/2 complex interacts with IP3R IL3, the largest of the three intralumenal loops, because the vast majority (>90%) of the erlin1 and erlin2 polypeptides are located within the ER lumen (15), and IL3 undergoes a conformational change upon channel opening (2). Here, we have tested this idea by subjecting IL3 to mutagenesis and find that a small region of IL3 close to TM5 (amino acids 2471–2472) is critical for the erlin1/2 complex to recognize activated IP3R1 and for IP3R1 to undergo ERAD. Surprisingly, we also discovered that mutations immediately adjacent to TM5 (e.g., of D-2465) block Ca2+ channel activity, showing that this region is critical for IP3R function.
Results
Processing of endogenous IP3R1 is blocked by TAK-243
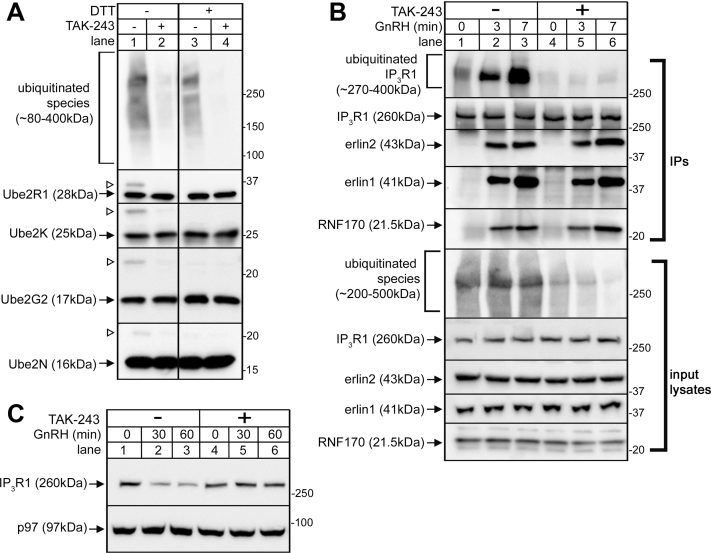
To better define the sequence of events that lead to IP3R1 ERAD, we examined the effects of TAK-243, a first-in-class inhibitor of UBE1, the mammalian E1 enzyme that charges the vast majority of E2 ubiquitin-conjugation enzymes with ubiquitin (17). We utilized αT3 mouse pituitary cells, since these cells represent an excellent model system to study IP3R1 ERAD in response to gonadotropin-releasing hormone (GnRH), which robustly stimulates IP3-dependent signaling and initiates IP3R processing (15, 18). E2 enzymes receive activated ubiquitin from UBE1 and form E2-ubiquitin intermediates via DTT-sensitive thiolester bonds (19, 20). E2-ubiquitin intermediates were detected in αT3 cell lysates prepared without DTT but were dramatically diminished by pretreatment of cells with TAK-243 for 10 min (Fig. 1A, lanes 1 and 2), and as expected, only unmodified E2 enzymes were seen in presence of DTT (lane 3 and 4). The abundance of high molecular weight ubiquitinated species in cell lysates was also greatly diminished by TAK-243 (lanes 1 and 2, top panel) but was not markedly altered by DTT (lane 3), since the isopeptide bonds via which polyubiquitinated species are formed are not DTT sensitive (20). Overall, these data show that brief TAK-243 pretreatment is sufficient to greatly inhibit the loading of multiple E2 enzymes with ubiquitin as well as the formation of ubiquitin conjugates in αT3 cells, indicating that UBE1 is efficiently inhibited. This pretreatment also completely blocked GnRH-induced IP3R1 polyubiquitination, without substantially altering association of the erlin1/2 complex and RNF170 with activated IP3R1 (Fig. 1B), indicating that erlin1/2 complex and RNF170 association occurs prior to and independently of IP3R1 polyubiquitination. Further, TAK-243 completely blocked GnRH-induced IP3R1 downregulation (Fig. 1C), indicating that it is ubiquitination rather than erlin1/2 complex and RNF170 association that directly mediates IP3R1 degradation. Overall, these data confirm our previous assumptions (9) about the sequence of events that lead to the ERAD of activated IP3Rs: first, the erlin1/2 complex associates, followed by IP3R ubiquitination, which in turn triggers IP3R degradation by the proteasome.
Figure 1.
Effects of TAK-243 on the processing of endogenous IP3R1. αT3 cells were preincubated without (-) or with (+) 3 μM TAK-243 for 10 min and were then treated with 0.1 μM GnRH for the times as indicated before cell lysates were prepared. A, cell lysates were subjected to SDS-PAGE without (-) or with (+) DTT and then probed in immunoblots for ubiquitin and different E2s (Ube2s) as indicated. Open arrowheads indicate E2-ubiquitin intermediates and solid arrows indicate unmodified E2s. B, anti-IP3R1 IPs and input lysates were probed for ubiquitin, IP3R1, erlin1, erlin2, and RNF170. C, cell lysates were probed for IP3R1, with p97 serving as a loading control. GnRH, gonadotropin-releasing hormone; IP, immunoprecipitation; IP3R, inositol 1,4,5-trisphosphate receptors.
Exogenous IP3R1HAWT is processed identically to endogenous IP3R1
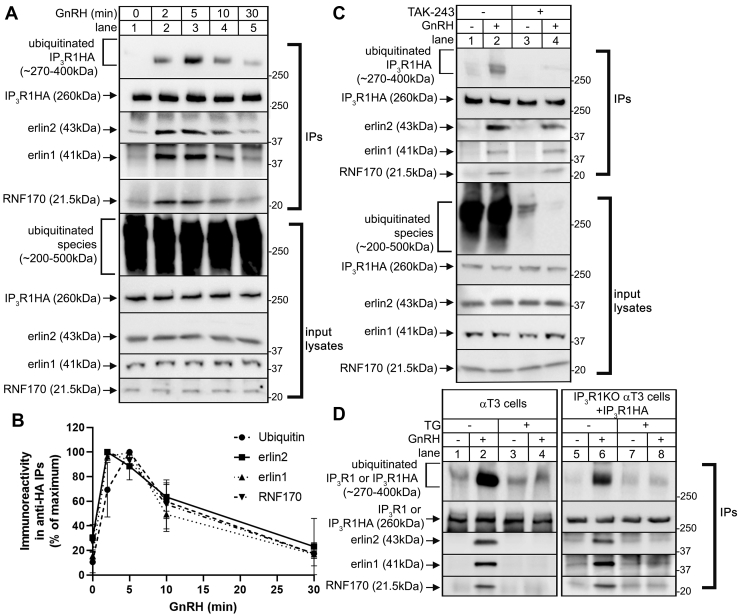
Since we intended to use IP3R1 KO αT3 cell lines (21) reconstituted with exogenous IP3R1 constructs to define the molecular determinants of the erlin1/2 complex–IP3R1 interaction, we first sought to validate this approach. WT IP3R1-hemagglutinin (HA) (IP3R1HAWT) was introduced into IP3R1KO αT3 cells and was shown by immunoprecipitation (IP) to bind to endogenous erlin1, erlin2, and RNF170, and also be polyubiquitinated following GnRH treatment (Fig. 2A) with kinetics (peaks at 2–5 min, Fig. 2B) very similar to that seen for endogenous IP3R1 (15, 16). GnRH-induced IP3R1HAWT ubiquitination was also blocked by TAK-243 pretreatment without altering erlin1, erlin2, and RNF170 association (Fig. 2C), indicating that exogenous IP3R1HAWT is processed by the UPP essentially identically to endogenous IP3R1. Likewise, inhibition of endogenous IP3R1 ERAD by acute pretreatment with the SERCA pump inhibitor, thapsigargin, which discharges ER Ca2+ stores (22) (Fig. 2D, lanes 1–4), was also seen for IP3R1HAWT expressed in IP3R1KO αT3 cells (lanes 5–8).
Figure 2.
Processing of exogenous IP3R1HAWTby the UPP in αT3 IP3R1KO cells.A and B, αT3 IP3R1KO cells were transiently transfected to express IP3R1HAWT and were incubated with 0.1 μM GnRH for 0, 2, 5, 10, and 30 min as indicated. Anti-HA IPs and input lysates were probed in immunoblots for ubiquitin, IP3R1HA, erlin1, erlin2, and RNF170. Quantitated immunoreactivity for ubiquitin, erlin1, erlin2, and RNF170 in anti-HA IPs was normalized to maximal values and graphed (mean ± SEM, n = 3). C, αT3 IP3R1KO cells were transiently transfected to express IP3R1HAWT, preincubated without (-) or with (+) 3 μM TAK-243 for 10 min, treated without (-) or with (+) 0.1 μM GnRH for 5 min, and anti-HA IPs and input lysates were probed in immunoblots as indicated. D, αT3 cells (lanes1–4) or IP3R1KO αT3 cells transiently transfected to express IP3R1HAWT (lanes 5–8) were preincubated for 10 min without (-) or with 1 μM thapsigargin (TG), followed by exposure to 0.1 μM GnRH for 5 min. Anti-IP3R1 or anti-HA IPs were then probed in immunoblots as indicated. GnRH, gonadotropin-releasing hormone; HA, hemagglutinin; IP, immunoprecipitation; IP3R, inositol 1,4,5-trisphosphate receptor.
IL3 mediates the association between IP3R1 and the erlin1/2 complex
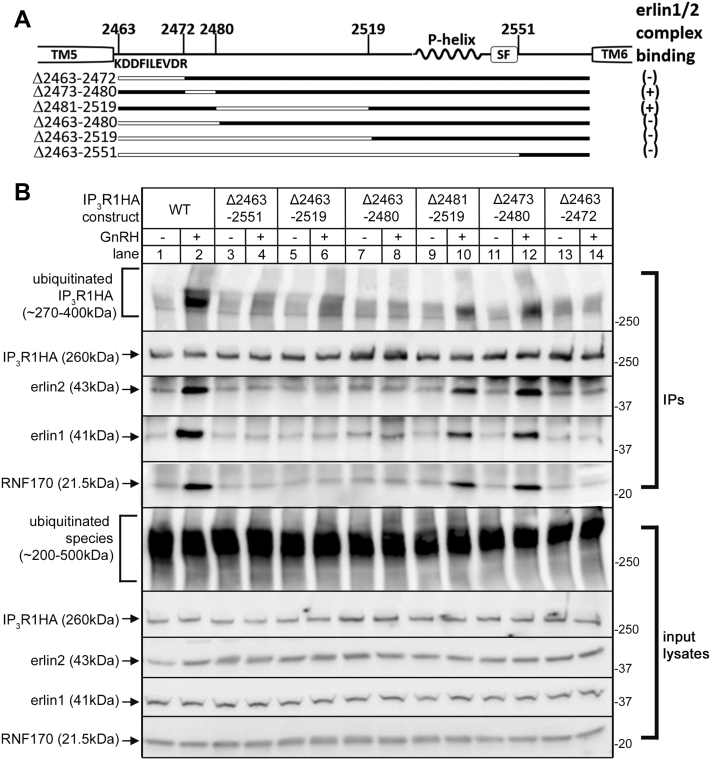
To test the role of IL3, initially large deletions to this region were made (Fig. 3A). IP3R1KO αT3 cells reconstituted with IP3R1HA mutants were stimulated with GnRH for 5 min, and erlin1/2 complex association was assessed via co-IP (Fig. 3B). The constructs with largest deletions, IP3R1HAΔ2463-2551 and IP3R1HAΔ2463-2519, both failed to associate with erlin1/2 complex after stimulation with GnRH (Fig. 3B, lanes 3–6), indicating that IL3 is indeed crucial for erlin1/2 complex binding. Interestingly, for constructs with smaller deletions, IP3R1HAΔ2481-2519 bound to the erlin1/2 complex, while IP3R1HAΔ2463-2480 did not (Fig. 3B, lanes 7–10), suggesting that the erlin1/2 complex–binding region is close to TM5. Likewise, IP3R1HAΔ2473-2480 bound to the erlin1/2 complex, while IP3R1HAΔ2463-2472 did not (Fig. 3B, lanes 11–14). Since some substantial deletions to IL3 (e.g., IP3R1HAΔ2473-2480 and IP3R1HAΔ2481-2519) do not impair erlin1/2 complex association, simply shortening length of IL3 does not explain the inability of IP3R1HAΔ2463-2472 to bind. Rather, these results (summarized in Fig. 3A, right) narrow down the putative erlin1/2 complex–binding site to the 10 amino acid (aa) region next to TM5. Additionally, RNF170 association and polyubiquitination paralleled erlin1/2 complex association for each IP3R1HA mutant, indicating that erlin1/2 complex association governs the subsequent processing of IP3R1HA mutants by the UPP.
Figure 3.
Effects of large deletions to IL3 on erlin1/2 complex binding, RNF170 binding, and IP3R ubiquitination.A, diagram of IP3R1HA mutants (deleted regions are shown as white lines), with (+) indicating strong binding of the erlin1/2 complex and (-) indicating no binding. Also shown are TM5 and TM6, the sequence of the aa 2463 to 2472 region and the positions of the pore-helix (P-helix) and selectivity filter (SF). Amino acid numbering corresponds to mouse IP3R1 (UniProtKB accession number: P11881). B, αT3 IP3R1KO cells were transiently transfected to express IP3R1HA constructs and were incubated without (-) or with (+) 0.1 μM GnRH for 5 min. Anti-HA IPs and input lysates were probed in immunoblots for ubiquitin, IP3R1HA, erlin1, erlin2, and RNF170. GnRH, gonadotropin-releasing hormone; HA, hemagglutinin; IL3, intralumenal loop 3; IP, immunoprecipitation; IP3R, inositol 1,4,5-trisphosphate receptor; TM, transmembrane.
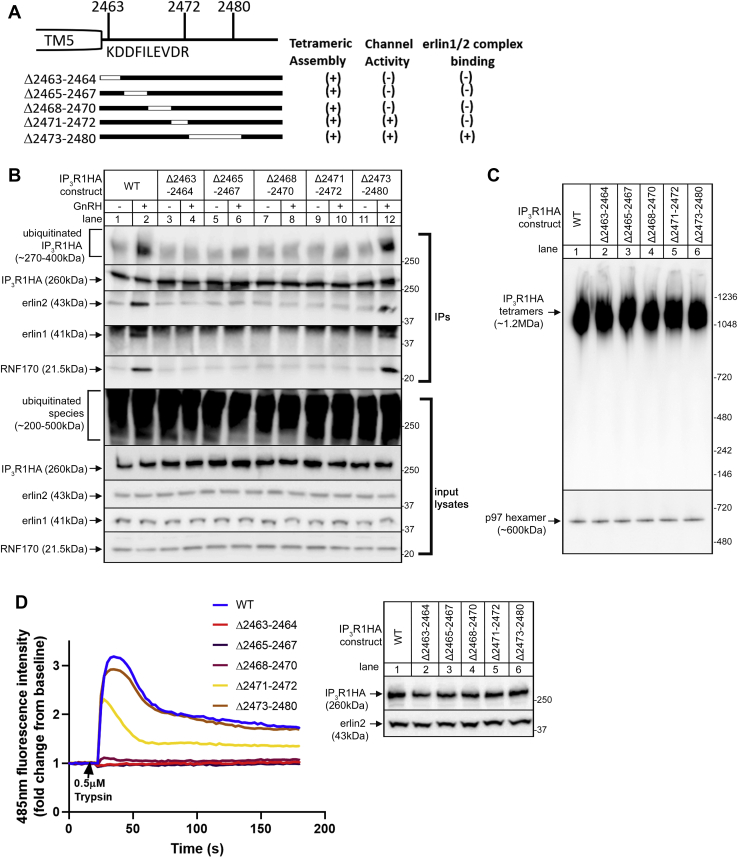
To extend this preliminary characterization, focused deletions within the 10 aa region were created and analyzed (Fig. 4A, IP3R1HAΔ2463-2464, IP3R1HAΔ2465-2467, IP3R1HAΔ2468-2470 and IP3R1HAΔ2471-2472). However, as compared to IP3R1HAWT (Fig. 4B, lanes 1 and 2) and IP3R1HAΔ2473-2480 (lanes 11 and 12), none of these mutants associated with the erlin1/2 complex after stimulation with GnRH (lanes 3–10). While this suggests that the entire aa 2463 to 2472 region may be required for erlin1/2 complex binding, failure to bind could also be due to unanticipated effects on the structural integrity of IP3R1 or the IP3-induced conformational changes to IP3R1 that normally trigger erlin1/2 complex binding. To assess these parameters, the mutants were introduced into IP3R1-3KO HEK cells (23), which exhibit high transfection efficiency, and tetramerization and Ca2+ channel activity were assessed using native PAGE and trypsin-induced Ca2+ mobilization, respectively. IP3R1HAWT and all of the mutants migrated identically at ∼1.2 MDa during native PAGE (Fig. 4C), indicating that tetramerization and overall structural integrity is normal. Interestingly, however, of the four mutants, only IP3R1HAΔ2471-2472 functioned as an IP3-activatable Ca2+ channel, exhibiting ∼73% of normal channel activity (Fig. 4D), indicating that only this mutant undergoes normal conformational changes and that failure of this mutant to bind to the erlin1/2 complex can be attributed to a specific disruption of the binding interface. In contrast, IP3R1HAΔ2463-2464, IP3R1HAΔ2465-2467, and IP3R1HAΔ2468-2470 lacked channel activity (Fig. 4D) and may be incapable of undergoing normal conformational changes, which could account for the failure of erlin1/2 complex binding. Overall, these data (summarized in Fig. 4A) indicate that the aa 2471 to 2472 region is a component of the IP3R1–erlin1/2 complex–binding site.
Figure 4.
Effects of focused deletions to IL3 on erlin1/2 complex binding, RNF170 binding, and IP3R ubiquitination.A, diagram of IP3R1 deletion mutants (deleted regions are shown as white lines), with (+) and (-) indicating success or failure in tetrameric assembly, channel activity, and erlin1/2 complex association. B, αT3 IP3R1KO cells were transiently transfected to express IP3R1HAs and were incubated without (-) or with (+) 0.1 μM GnRH for 5 min. Anti-HA IPs and input lysates were probed in immunoblots as indicated. C, native PAGE of IP3R1HAs expressed in IP3R1-3KO HEK cells, probed in immunoblots as indicated. D, Ca2+ mobilizing (channel) activity of IP3R1HAs expressed in IP3R1-3KO HEK cells in response to 0.5 μM trypsin. The immunoblot shows the expression levels of IP3R1HAs in IP3R1-3KO HEK cells, with erlin2 serving as a loading control. The peak calcium response of IP3R1HAΔ2471-2472 was 73 ± 3% (n = 5) of that seen with IP3R1HAWT. GnRH, gonadotropin-releasing hormone; HA, hemagglutinin; IL3, intralumenal loop 3; IP, immunoprecipitation; IP3R, inositol 1,4,5-trisphosphate receptor.
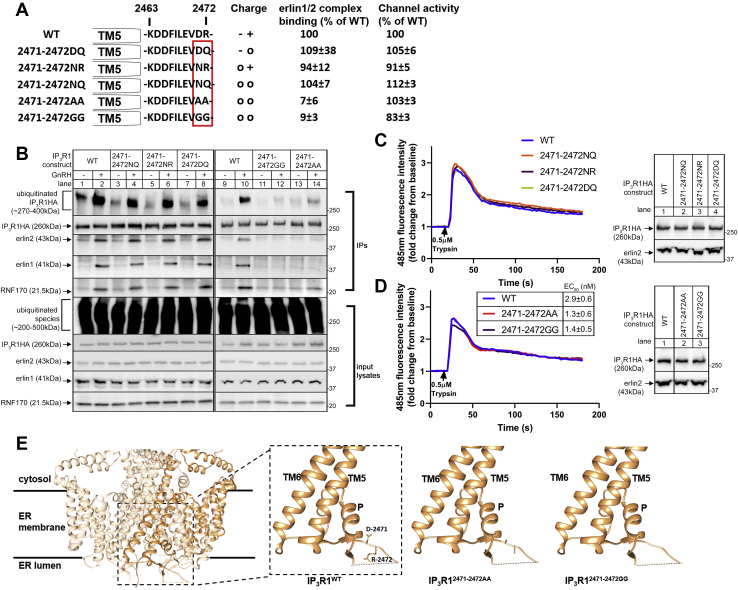
Analysis of the aa 2471 to 2472 region
To better define the requirements for erlin1/2 complex binding, IP3R1 mutants with substitutions in the aa 2471 to 2472 region were created and analyzed (Fig. 5A). Since D-2471 and R-2472 have negative and positive charges, respectively, these were replaced by neutral amino acids of similar size to examine the role of charge (Fig. 5A). Interestingly, each of the mutants created (IP3R1HA2471-2472DQ, IP3R1HA2471-2472NR, and IP3R1HA2471-2472NQ) bound to the erlin1/2 complex (Fig. 5B, lanes 3–8) and exhibited Ca2+ channel activity very similar to IP3R1HAWT (Fig. 5C), indicating that the charges of D-2471 and R-2472 are not required for erlin1/2 complex binding. To examine the role of size, D-2471 and R-2472 were also replaced by the much smaller amino acids, alanine and glycine (Fig. 5A). Remarkably, IP3R1HA2471-2472AA and IP3R1HA2471-2472GG both failed to bind to the erlin1/2 complex (Fig. 5B lanes 11–14) but exhibited channel activity very similar to IP3R1HAWT, in terms of both peak response and EC50 (Fig. 5D), indicating that these substitutions do not impair the ability of IP3R1 to undergo IP3-induced conformational changes that lead to channel opening but greatly impact the erlin1/2 complex–binding site. To gain additional insight, the highest resolution cryo-EM structure of IP3R1 was used to visualize the juxta-TM5 region (Fig. 5E) (7). D-2471 and R-2472, being part of IL3, protrude into the ER lumen and appear to be at the outer surface of the IP3R1 tetramer, which should allow for access to the erlin1/2 complex. Further, since this region of IL3 appears to lack secondary structure, the only clear difference between IP3R1WT and the nonbinding mutants is side chain length at positions 2471 and 2472. Overall, these data suggest that the size of residues in the aa 2471 to 2472 region is a crucial factor for the IP3R1–erlin1/2 complex interaction.
Figure 5.
Effects of substitutions to the aa 2471 to 2472 region on erlin1/2 complex binding, RNF170 binding, and IP3R ubiquitination.A, diagram of IP3R1 mutants, with the red box designating the aa 2471 to 2472 region, and showing the amino acid charges in that region (-, negative; +, positive; or o, neutral), together with quantitated erlin1/2 complex binding and channel activity for each construct. B–D, protein association and channel activity characteristics of WT and mutant IP3R1s were examined in IP3R1KO αT3 cells and IP3R1-3KO HEK cells, respectively, as in Figure 4. Erlin1/2 complex binding in the presence of GnRH was quantitated by measuring erlin2 immunoreactivity in anti-HA IPs, relative to that for IP3R1HAWT (mean ± SEM, n ≥ 5); this is appropriate, as erlin2 constitutes the majority of the erlin1/2 complex and is essential for its function (14). Channel activity was quantitated by measuring the peak calcium response of mutant IP3R1HAs, relative to that of IP3R1HAWT (mean ± SEM, n ≥ 3). D, trypsin EC50 values for peak responses are shown and differences between IP3R1HAWT and the mutants were not significant (mean ± SEM, n ≥ 3, p> 0.1 by one-way ANOVA followed by Tukey’s post hoc test). E, structure of the TM region of an IP3R1 tetramer with one subunit highlighted in gold (derived from PDB ID: 7LHE) (7). The zoomed-in region shows the positions of TM5, TM6, the pore helix (P), and the juxta-TM5 region that forms the first part IL3 and contains the aa 2471 to 2472 region. Structures of IP3R1 mutants were created to illustrate the differences in side chain size, using UCSF chimera. Note: dotted lines indicate unresolved parts of IL3 and the zoomed-in region was tilted for clarity. GnRH, gonadotropin-releasing hormone; HA, hemagglutinin; IL3, intralumenal loop 3; IP, immunoprecipitation; IP3R, inositol 1,4,5-trisphosphate receptor; PDB, Protein Data Bank; TM, transmembrane.
To confirm the role of the aa 2471 to 2472 region, IP3R1HAWT and IP3R1HA2471-2472AA were stably expressed in IP3R1KO αT3 cells. The highest expressing cell lines contained ∼10% of the IP3R1 levels seen in unmodified αT3 cells (Fig. 6A). Ca2+-mobilizing activity was almost fully restored in these reconstituted IP3R1KO cell lines, with the peak responses very similar to that seen in unmodified αT3 cells (Fig. 6B), indicating that both IP3R1HAWT and IP3R1HA2471-2472AA form functional channels and undergo normal conformational changes upon stimulation. Note that the low level of Ca2+ mobilization in IP3R1KO cells is likely due to the presence of residual IP3R2 and IP3R3 (less than 1% of total IP3R levels) (21). As expected, stably expressed IP3R1HAWT bound to the erlin1/2 complex upon GnRH stimulation but IP3R1HA2471-2472AA did not (Fig. 6C), and RNF170 association, polyubiquitination, and downregulation of IP3R1HAs followed in parallel (Fig. 6 C and D). These data confirm the importance of the aa 2471 to 2472 region for erlin1/2 complex association and for IP3R1 processing by the ERAD pathway.
Figure 6.
Analysis of IP3R1HAs stably expressed in IP3R1KO αT3 cells.A, lysates from unmodified and IP3R1KO αT3 cells and IP3R1KO cell lines stably expressing IP3R1HAWT or IP3R1HA2471-2472AA were probed in immunoblots for IP3R1, with erlin2 serving as a loading control. B, Ca2+ mobilizing activity (top) and peak Ca2+ response (bottom) in the different αT3 cell lines after stimulation with 0.1 μM GnRH. One-way ANOVA followed by Tukey’s post hoc test was used for statistical analysis, with ∗∗ indicating p < 0.01, ∗∗∗ indicating p < 0.001, and N.S. indicating not significant (p > 0.05). C, protein association characteristics of IP3R1HAs stably expressed in αT3 IP3R1KO cells. Cells were incubated without (-) or with (+) 0.1 μM GnRH for 5 min and anti-HA IPs together with input lysates were probed in immunoblots as indicated. D, αT3 IP3R1KO cells stably expressing IP3R1HAs were treated with 0.1 μM GnRH for the times as indicated. Cell lysates were probed in immunoblots for IP3R1HA, with erlin2 serving as a loading control. GnRH, gonadotropin-releasing hormone; HA, hemagglutinin; IP, immunoprecipitation; IP3R, inositol 1,4,5-trisphosphate receptor.
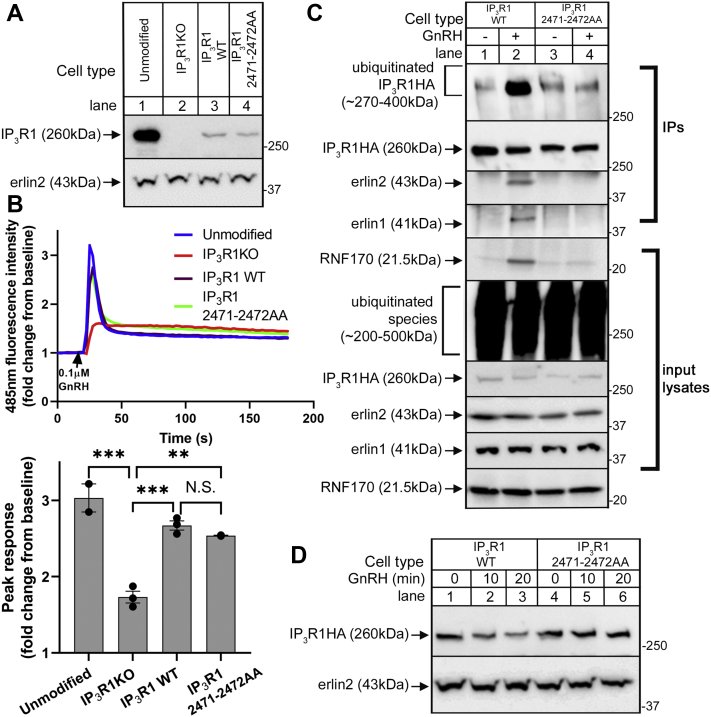
Role of the juxta-TM5 region in channel activity
Sequence alignment of the N-terminal end of IL3 shows that the juxta-TM5 region is highly conserved (Fig. 7A), indicating that it might be indispensable to IP3R function. To gain more insight into why the deletion mutants in this region (IP3R1HAΔ2463-2464, IP3R1HAΔ2465-2467, and IP3R1HAΔ2468-2470) lack channel activity (Fig. 4A), substitution mutants were constructed and characterized (Fig. 7, B–D). IP3R1HA2463-2464AA, with substitutions immediately adjacent to TM5, mobilized Ca2+ well (Fig. 7C), showing that the replaced K-2463 and D-2464 are not critical for channel activity and that the structural integrity of this mutant is still maintained, despite the loss of charged residues, which could potentially affect the proper positioning of TM5 in the ER membrane (24). In contrast, IP3R1HA2465-2467AAA completely lacked channel activity (Fig. 7C), and IP3R1HA2464-2465NN and IP3R1HAD2465N, with charge neutralizing D to N substitutions, lost either all or the vast majority of Ca2+ channel activity (Fig. 7C), indicating charges in this region, especially that of D-2465, are essential to IP3R1 channel activity. A nonspecific effect of these mutations on the positioning of TM5 is unlikely in view of the ability of IP3R1HA2463-2464AA to function normally. Finally, among the mutants, only IP3R1HA2463-2464AA was able to bind to the erlin1/2 complex (Fig. 7D), which correlates with its near normal channel activity.
Figure 7.
Role of the juxta-TM5 region in channel activity.A, sequence alignment of IP3R1-3 from Mus musculus (m) and IP3R1 from Rattus norvegicus (r), and Homo sapiens (h) (UniProtKB accession numbers: P11881, Q9Z329, P70227, P29994, and Q14643, respectively), with the red box designating the aa 2463 to 2472 region and black indicating identity between IP3Rs. B, diagram of IP3R1 mutants with the red boxes designating the substitution sites, with quantitated channel activity and erlin1/2 complex binding for each construct. C and D, channel activity and erlin2 association of WT and mutant IP3R1HAs were examined in IP3R1-3KO HEK cells and IP3R1KO αT3 cells, respectively, as in Figure 4. Channel activity (% of WT) and erlin1/2-binding ability (% of WT) were quantitated as in Figure 5 (mean ± SEM, n ≥ 3). E, structure of IP3R1 channel pore (derived from PDB: 7LHE) (7), with a side view (two subunits shown for clarity) and a sectional view (indicated by the dashed line box), showing TM5 and TM6 (light blue), D-2465 (red), the selectivity filter (yellow), and the pore helix (green). HA, hemagglutinin; IP3R, inositol 1,4,5-trisphosphate receptor; PDB, Protein Data Bank; TM, transmembrane.
To better understand the importance of the juxta-TM5 region and particularly the charge of D-2465, the highest resolution cryo-EM model of IP3R1 was used to visualize the channel core (Fig. 7E) (7). Relative to the selectivity filter sequences, which help form the narrow ion conduction pathway, D-2465 is found slightly laterally and toward the ER lumen but still within ∼9 Å of the filter. It is, thus, possible that like the negative charges in the pore of other cation channels (25, 26, 27, 28), D-2465 serves to concentrate Ca2+ ions to facilitate Ca2+ ion conduction.
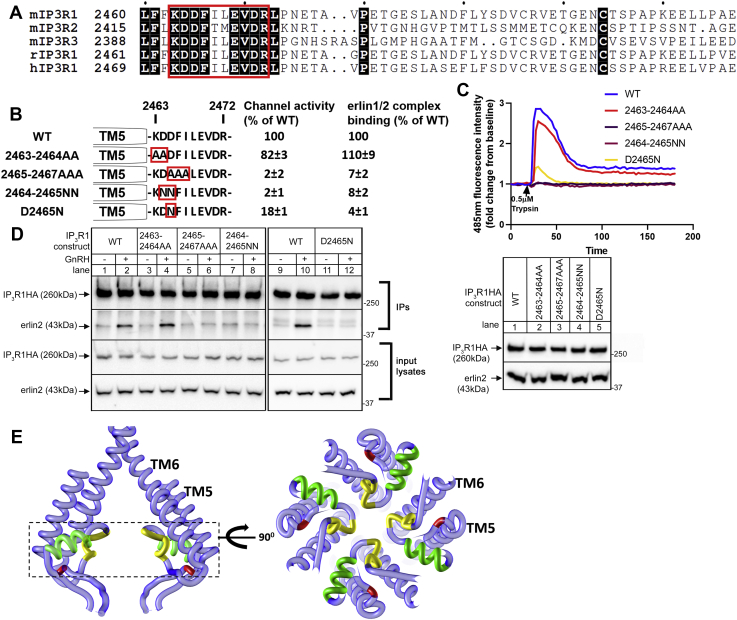
The relationship between IP3R1 channel activity and erlin1/2 complex binding
Of all the IP3R1 mutants examined in Figure 4, Figure 5, Figure 6, Figure 7, those that lack channel activity also fail to bind to the erlin1/2 complex, suggesting that the IP3-induced conformational changes that lead to channel activity are necessary for erlin1/2 complex binding. To further test this correlation, we examined three constructs with inactivating mutations remote from the N-terminal of IL3, suppressor domain deletion (ΔSD), R2596A and D2550A, all of which have been reported to lack Ca2+ channel activity (29, 30, 31, 32, 33). Surprisingly, while IP3R1HAΔSD and IP3R1HAR2596A indeed lacked channel activity, IP3R1HAD2550A was still clearly active (∼51% of WT, Fig. 8, A and B), showing that IP3R1HAD2550A actually possesses residual channel activity. A similar result was also seen upon comparison of rat IP3R1WT and IP3R1D2550A (Fig. 8C). As expected, erlin1/2 complex–binding ability correlated with channel activity, with IP3R1HAD2550A binding the erlin1/2 complex but IP3R1HAΔSD and IP3R1HAR2596A not binding. Clearly though, some IP3R1 mutants (e.g., IP3R1HA2471-2472AA and IP3R1HA2471-2472GG) have normal channel activity but do not bind to the erlin1/2 complex (Figs. 5 and 6). Therefore, Ca2+ channel activity seems to be necessary but not sufficient for erlin1/2 complex binding.
Figure 8.
The relationship between IP3R1 channel activity and erlin1/2 complex binding.A, summary of IP3R1 mutants with quantitated channel activity and erlin1/2 complex binding for each construct. B–D, channel activity of mouse IP3R1HAs (WT, ΔSD, D2550A, R2596A) and rat IP3R1s (WT and D2550A) was examined in IP3R1-3KO HEK cells, and erlin2 association with IP3R1HAs was assessed in IP3R1KO αT3 cells as in Figure 4. Channel activity (% of WT) and erlin1/2-binding ability (% of WT) for each mouse IP3R1HA construct was quantitated as in Figure 5 (mean ± SEM, n ≥ 3). HA, hemagglutinin; IP3R, inositol 1,4,5-trisphosphate receptor.
Discussion
It is clear that the erlin1/2 complex interacts with activated IP3Rs (9, 12, 15), but the molecular determinants of this interaction have yet to be resolved. Here, we show, using IP3R1 mutants, that the erlin1/2 complex binds to IL3 and, in particular, to two amino acids (D-2471 and R-2472) in a region close to TM5. Interestingly, these two amino acids and the residues immediately adjacent to TM5 that are critical for Ca2+ channel activation are highly conserved between IP3R1-3, suggesting that this region of IL3 plays an important role in the normal functioning of IP3Rs.
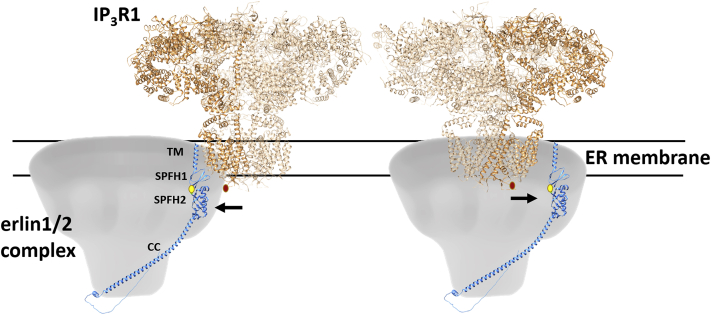
The key result that locates the erlin1/2 complex–binding site to the D-2471/R-2472 region is that replacement of these amino acids with alanine or glycine ablates stimulus-induced erlin1/2 complex binding, without altering Ca2+ mobilization (which shows that the mutant IP3Rs can undergo normal conformational changes). This indicates that the erlin1/2 complex–binding interface is specifically perturbed in IP3R1HA2471-2472AA and IP3R1HA2471-2472GG. High resolution cryo-EM images of IP3R1 tetramers (7) show that D-2471 and R-2472 are within the ER lumen close to the channel pore and are relatively exposed and thus could create a binding site for the erlin1/2 complex. It remains a possibility, however, that the binding site is in fact immediately adjacent to the D-2471/R-2472 region and that the reduction in side chain size and/or increased flexibility resulting from the introduction of alanine or glycine at positions 2471/2472 causes a local structural change that disrupts that adjacent binding site. Currently, there is no equivalent high-resolution cryo-EM structure for the erlin1/2 complex, but a recently obtained structure of the bacterial stomatin/prohibitin/flotillin/HflK/C (SPFH) domain–containing proteins HflK and HflC, which share homology with erlin1 and 2 (34, 35), provides insight into how the erlin1/2 complex might interface with IP3R1. HflK and HflC form a cup-shaped, ∼1 MDa complex linked to the bacterial inner membrane by their N-terminal TM regions, with the junction between their SPFH1 and SPFH2 domains located close to the membrane and providing the binding site for client FtsH hexamers, which the HflK/C complex encircle (34, 35). Since the erlin1/2 complex forms a similar ∼2 MDa structure linked to the ER membrane (9, 12, 15), models can be constructed for how it interacts with activated IP3Rs (Fig. 9). Interestingly, the SPFH1–SPFH2 domain junction in erlin2 may well contribute to the binding site, as mutation of the T-65 residue, which is found at the junction (Fig. 9) (34), prevents the erlin1/2 complex from interacting with activated IP3Rs (14). Clearly, the binding interface will be better understood when high-resolution structures are defined for the erlin1/2 complex and, if possible, for active IP3R1s in association with the erlin1/2 complex.
Figure 9.
Model of how an activated IP3R1 tetramer and the erlin1/2 complex interact. Diagrams of IP3R1 tetramers (7) and erlin1/2 complexes, based on the dimensions and membrane topology of HflC/K (34, 35), with one AlphaFold-derived erlin2 subunit (52) fitted into each complex. Also, shown are the aa 2471 to 2472 region of one IP3R1 subunit (red oval) and for erlin2, the positions of the TM region, the SPFH1 and SPFH2 domains, the long coil–coil (CC) domain, and T-65 (yellow oval), that is key to interactions with activated IP3R1 (15). Activated IP3R1 tetramers could bind with the erlin1/2 complex through interactions with the outer surface of the complex (left) or even be encircled by the complex (right). IP3R, inositol 1,4,5-trisphosphate receptor; TM, transmembrane.
An additional significant finding from the analysis of IP3R1 mutants is that the region of IL3 immediately adjacent to TM5 is critical for channel activation. This was revealed by deletion and substitution mutants that dramatically inhibit channel activity and implicate the negative charges of D-2464 and particularly D-2465 as being key, since IP3R1HAD2465N possess only residual Ca2+ channel activity. D-2464 and D-2465 are found close to the channel pore and selectivity filter, and it is possible that the negative charges serve to concentrate Ca2+ ions, as has been proposed for other negatively charged residues in IP3R1 (e.g., E-2469) (7) and in other cation channels (25, 26, 27, 28). Alternatively, these residues could contribute to the putative Ca2+-binding regulatory site (1, 3) that may be present between aa 2463 to 2528 of IL3 (36). To the best of our knowledge, the importance of this juxta-TM5 region in IP3R1 channel activation has not been previously examined, but clearly, our data indicate that it warrants further investigation.
It is intriguing that the erlin1/2 complex is not the only protein that interacts with IL3 (1, 3). Chromogranins A and B interact with the C-terminal end of IL3 near the selectivity filter and increase channel open probability (37, 38). Conversely, ERp44 binds to the N-terminal half of IL3 and inhibits IP3R1 activity (39), and GRP78 interacts with the same region and supports channel activity and tetramer assembly (40), although the exact binding sites have yet to be defined. All of these interactions were observed with nonactivated IP3Rs (37, 38, 39, 40). In contrast, the erlin1/2 complex interacts with IL3 only when IP3Rs are activated and targets them for ubiquitination and processing by the ERAD pathway (9, 12, 15). Our data also show that the integrity of the juxta-TM5 region of IL3 is critical for Ca2+ channel activity. Thus, IL3 is an important regulatory locus, with the regions adjacent to TM5 and TM6 being most important, a finding supported by our observations that deletions in the central part of IL3 (IP3R1HAΔ2473-2480 and IP3R1HAΔ2481-2519) do not affect channel activity or erlin1/2 complex association.
There appears to be a complicated relationship between the functionality of IP3Rs and their ability to recruit the erlin1/2 complex that can be best summarized by the statement that Ca2+ channel activity is “necessary, but not sufficient” for erlin1/2 complex recruitment. Examples are the range of mutants (e.g., IP3R1HAΔ2463-2464, IP3R1HAΔSD, IP3R1HA2464-2465NN, and IP3R1HAR2596A, etc) that both fail to act as Ca2+ channels and fail to recruit erlin1/2 complex, presumably because they cannot undergo normal conformational changes (i.e., activity is “necessary”), versus IP3R1HA2471-2472AA and IP3R1HA2471-2472GG, which act as Ca2+ channels but fail to recruit erlin1/2 complex, presumably because the binding interface is disrupted (i.e., activity is “not sufficient”). This concept would be invalidated if mutants existed that failed to act as a Ca2+ channels but could still recruit the erlin1/2 complex, but despite our best efforts, no such mutants have been found. Data from other sources are consistent with the “not sufficient” aspect; mimicking IP3R-mediated Ca2+ mobilization by releasing Ca2+ from the ER with thapsigargin does not cause erlin1/2 complex recruitment and in fact blocks it (Fig. 2D), and raising cytosolic Ca2+ concentration through plasma membrane depolarization also does not induce recruitment (41). Overall, these data support the notion that the conformational changes that accompany Ca2+ channel opening, but not Ca2+ mobilization itself, provides the trigger for erlin1/2 complex recruitment.
A surprising result from our analysis of IP3R mutants is that IP3R1HAD2550A is not “pore-dead” as has been widely reported (31, 32, 33) but rather exhibits considerable channel activity (∼50% of that seen with IP3R1HAWT). This was observed in IP3R1-3KO HEK cells, so pure tetramers of IP3R1HAD2550A can be assumed to be responsible and was seen using both mouse and rat IP3R1 constructs. Furthermore, IP3R1HAD2550A activation recruits the erlin1/2 complex, indicating that it can adopt an active conformation. These data suggest that the D2550A mutation does not obliterate channel activity and that care should be taken in interpreting data from this mutant. Many of the studies showing D2550A mutants to be inactive have been performed in DT40 3KO cells (32, 33), and it is possible that cell-specific factors or high expression levels obtained in IP3R1-3KO HEK cells allow for residual channel activity of the mutants to be observed.
By using the novel UBE1 and UPP inhibitor TAK-243 (17), we were able to gain insight into the sequence of events that mediate IP3R ERAD. In αT3 cells, this compound very rapidly inhibits loading of E2 enzymes with ubiquitin and the ubiquitination of generic substrates and blocks GnRH-induced IP3R1 ubiquitination, without affecting recruitment of the erlin1/2 complex. This shows that these two events in IP3R ERAD are independent and that erlin1/2 complex recruitment occurs first. Further, as TAK-243 also blocks IP3R1 downregulation, binding of the erlin1/2 complex to activated IP3R1s per se is insufficient to trigger IP3R1 transfer to the proteasome—it really does require that IP3R1s be ubiquitinated. The ability of TAK-243 to rapidly inhibit ubiquitination should make this compound very useful for probing mechanisms within the UPP, since its mechanism of action contrasts with existing and widely used UPP inhibitors that block steps after substrate ubiquitination, for example, p97 and proteasome inhibitors (42, 43).
Overall, we have identified the site in IP3Rs to which the erlin1/2 complex binds when IP3Rs are activated. This highly specific recognition mechanism appears to be triggered by conformation changes to IP3R1 that accompany channel activation and is not typical of how other substrates are recognized for ERAD, which tend to be more generic (8, 9, 10, 11). Erlin1/2 complex binding is the critical initiation step for IP3R ERAD and mediates IP3R downregulation during cell activation (9, 14, 15) and also the basal turnover of IP3Rs, since erlin1/2 complex deletion causes accumulation of IP3Rs (9, 14). In view of the “necessary, but not sufficient” requirement for Ca2+ channel activity, disease-causing IP3R1 mutants that are inactive channels (e.g., K2563del, G2506R, etc) (44, 45), will likely be resistant to ERAD and will accumulate in the ER membrane, leading to perturbations beyond just abnormal Ca2+ homeostasis. For example, IP3R accumulation will increase the levels of the Bcl-2 family member Bok (21) and perhaps other Bcl-2 family members that interact with IP3Rs (1, 3) and may affect apoptotic signaling (21). IP3R accumulation could also reduce free Beclin1 levels and inhibit autophagy as IP3R contains a Beclin1 docking site (46, 47) and/or alter the bioavailability of some of the many known IP3R-interacting proteins (1, 3).
Experimental procedures
Materials
αT3 cells, IP3R1KO αT3 cells (21), and IP3R1-3KO HEK cells (23) were cultured as described (21). Antibodies used were: rabbit polyclonal anti-IP3R1 (48), anti-erlin1 (15), anti-erlin2 (49), anti-RNF170 (16), anti-HA epitope (16) (used for IP), anti-Ube2K/E2-25K (#UG 5920; Affiniti Research Products Ltd) and anti-Ube2G2/Ubc7 (a generous gift from Dr Yihong Ye, National Institutes of Health); mouse monoclonal anti-HA clone HA11 (#MMS-101R; Covance, used for immunoblot), antiubiquitin (#BML-PW8810; BioMol International), anti-p97 (#10R-P104A; Fitzgerald), anti-Ube2N/Ubc13 (#37-1100; Invitrogen), and anti-Ube2R1/Cdc34 (#25820; Transduction Laboratories). Protease inhibitors, Triton X-100CHAPS, GnRH, trypsin, thapsigargin, and horseradish peroxidase–conjugated secondary antibodies were from Sigma. DTT and reagents for SDS-PAGE were from Bio-Rad. TAK-243 was from MedChemExpress. Protein A-Sepharose CL-4B was from GE Healthcare and PEI (PEI 25K) was from Polysciences Inc.
Plasmids
A pcDNA3.1 based vector encoding WT mouse IP3R1 tagged at the C terminus with a HA epitope (IP3R1HA) (50) was utilized as a template for generating IP3R1HA mutants by inverse PCR (primer sequences available upon request). All IP3R1HA mutants were confirmed by DNA sequencing (Genewiz). IP3R1HAD2550A was prepared as described (22) and rat WT and D2550A IP3R1s were generous gifts from Dr David Yule’s lab (University of Rochester).
Cell lysis, IP, electrophoresis, and immunoblotting
For cell lysis to assess protein expression level, cells were collected with HEPES buffered saline with EDTA (155 mm NaCl, 10 mm HEPES, 1 mm EDTA, pH 7.4), resuspended in ice-cold Triton lysis buffer (150 mM NaCl, 50 mM Tris–HCl, 1 mM EDTA, 1% Triton X-100, 10 μM pepstatin A, 0.2 μM soybean trypsin inhibitor, 0.2 mM PMSF, 1 mM DTT, pH 8.0), incubated on ice for 30 min, centrifuged at 16,000g for 10 min at 4 °C, and supernatants were collected for analysis. For analysis of IP3R1 ubiquitination and protein association, αT3 cells were lysed with ice-cold CHAPS lysis buffer (150 mM NaCl, 50 mM Tris–HCl, 1 mM EDTA, 1% CHAPS, 10 μM pepstatin A, 0.2 μM soybean trypsin inhibitor, 0.2 mM PMSF, pH 8.0) supplemented with 5 mM N-ethylmaleimide, incubated on ice for 30 min, followed by addition of 5 mM DTT, and centrifugation at 16,000g for 10 min at 4 °C. Supernatants were incubated with anti-IP3R1 and protein A-Sepharose CL-4B beads for ∼16 h at 4 °C, and IPs were then washed three times with CHAPS lysis buffer, resuspended in gel loading buffer (final concentration: 1% SDS, 0.05% bromphenol blue, 5% glycerol, 100 mM DTT, and 50 mM Tris–HCl pH 6.8), incubated at 37 °C for 30 min, and subjected to SDS-PAGE, transfer to nitrocellulose membranes, and immunoblotting. Immunoreactivity was detected with Supersignal chemiluminescence reagents (Thermo Fisher) and a Chemidoc imager (Bio-Rad).
Ubiquitin conjugation to E2s
αT3 cells were harvested with HEPES buffered saline with EDTA and resuspended in ice-cold Triton lysis buffer without DTT, followed by incubation on ice for 30 min and centrifugation at 16,000g for 10 min at 4 °C. Supernatants were split and transferred into two new tubes and mixed with gel loading buffer without or with 100 mM DTT (20), incubated at 37 °C for 30 min, and subjected to SDS-PAGE and probed in immunoblots for E2s and ubiquitin.
Analysis of exogenous IP3R1HAs in IP3R1KO αT3 cells
IP3R1KO αT3 cells were transfected with IP3R1HA complementary DNAs (cDNAs) via electroporation using the Neon Transfection System (Invitrogen). Briefly, cells were trypsinized, mixed with culture medium, centrifuged at ∼400g for 5 min at room temperature, resuspended in PBS at a density of 3 × 107/ml together with 100 μg/ml cDNA, and 5 × 100 μl aliquots per condition were electroporated (1 pulse, 20 ms, 1500 V). To improve transfection efficiency and cell survival, electroporated cells were collected in open sterile microcentrifuge tubes and placed in a humidified 37 °C/5% CO2 incubator for ∼20 min before being transferred back to culture medium in 10 cm diameter dishes (51). To assess ubiquitination and the protein association characteristics of exogenous IP3R1HAs, cells were harvested 24 to 48 h later in CHAPS lysis buffer; cell lysates were incubated with anti-HA to IP IP3R1HAs, and samples were processed for immunoblotting as already described.
Analysis of exogenous IP3R1HAs in IP3R1-3KO HEK cells
IP3R1-3KO HEK cells were seeded at 5 × 105 cells/well in 6-well plates and transfected with 2 μg IP3R1HA cDNAs and 6 μl of 1 mg/ml PEI. Cells were subcultured ∼24 h later and transferred either to poly-D-lysine–treated 96-well microplates (Greiner) to measure free cytosolic Ca2+ or to new 6-well plates for analysis of IP3R1HA expression ∼24 h later. FLIPR Calcium 6 assay Kit (Molecular Devices) was used, and fluorescent signals were detected using a FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices). To determine IP3R1HA expression levels, cells from 6-well plates were lysed with Triton lysis buffer for SDS-PAGE and immunoblotting. To assess tetrameric assembly of IP3R1HAs, cells were disrupted with CHAPS lysis buffer and subjected to native PAGE (Invitrogen) and transferred to polyvinylidene fluoride membranes for immunoblotting as described (14).
Stable expression of IP3R1HAs
Stably reconstituted cell lines were obtained by transfecting IP3R1KO αT3 cells with IP3R1HAWT or IP3R1HA2471-2472AA cDNAs using the Neon Transfection System, followed by selection in 1.3 mg/ml G418 for 72 h. After recovery in G418-free culture medium for 24 h, the cells were seeded at a density of 1 cell/well in 96-well plates, expanded, and screened for expression in immunoblots with both anti-IP3R1 and anti-HA. For each DNA construct, two clones were selected for analysis and yielded essentially the same results.
Data presentation
All experiments were performed at least twice and representative images of gels with molecular markers (in kDa) on the side are presented. Immunoreactivity was quantitated using ImageJ (https://imagej.nih.gov/ij/). Calcium response traces shown are the average of fluorescence signals from 2 to 3 wells from one representative experiment. All quantitated data are expressed as mean ± SEM (n= the number of independent experiments).
Data availability
All data described and discussed are located within the article.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Drs Kamil Alzayady and David Yule (University of Rochester) for providing IP3R1-3KO HEK cells and rat IP3R1 constructs, Dr Yihong Ye (National Institutes of Health) for providing anti-Ubc7, Dr Forrest Wright for initiating experiments on IL3, Ms Katherine Keller for technical support, and Dr Stephan Wilkens, Dr Alaji Bah and Ms Fanghui Hua for help with protein structure modeling. This work was primarily supported by National Institutes of Health Grants DK107944 and GM121621.
Author contributions
X. G. and R. W. conceptualization; X. G., C. B., and R. W. methodology; X. G. and R. W. validation; X. G. formal analysis; X. G. and C. B. investigation; X. G. data curation; X. G. and R. W. writing–original draft; C. B. writing–review and editing; X. G. and C. B. visualization; R. W. supervision; R. W. project administration; R. W. funding acquisition.
Funding and additional information
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by George DeMartino
References
- 1.Prole D.L., Taylor C.W. Structure and function of IP3 receptors. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan G., Baker M.R., Wang Z., Seryshev A.B., Ludtke S.J., Baker M.L., et al. Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating. Cell Res. 2018;28:1158–1170. doi: 10.1038/s41422-018-0108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woll K.A., Van Petegem F. Calcium-release channels: structure and function of IP3 receptors and ryanodine receptors. Physiol. Rev. 2021;102:209–268. doi: 10.1152/physrev.00033.2020. [DOI] [PubMed] [Google Scholar]
- 4.Mikoshiba K. Role of IP3 receptor signaling in cell functions and diseases. Adv. Biol. Regul. 2015;57:217–227. doi: 10.1016/j.jbior.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Lin Q., Zhao G., Fang X., Peng X., Tang H., Wang H., et al. IP(3) receptors regulate vascular smooth muscle contractility and hypertension. JCI insight. 2016;1 doi: 10.1172/jci.insight.89402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan G., Baker M.L., Wang Z., Baker M.R., Sinyagovskiy P.A., Chiu W., et al. Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature. 2015;527:336–341. doi: 10.1038/nature15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker M.R., Fan G., Seryshev A.B., Agosto M.A., Baker M.L., Serysheva Cryo-EM structure of type 1 IP(3)R channel in a lipid bilayer. Commun. Biol. 2021;4:625. doi: 10.1038/s42003-021-02156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christianson J.C., Carvalho P. Order through destruction: how ER-associated protein degradation contributes to organelle homeostasis. EMBO J. 2022;41 doi: 10.15252/embj.2021109845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wojcikiewicz R.J.H. The making and breaking of inositol 1,4,5-trisphosphate receptor tetramers. Messenger (Los Angeles) 2018;6:45–49. doi: 10.1166/msr.2018.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemberg M.K., Strisovsky K. Maintenance of organellar protein homeostasis by ER-associated degradation and related mechanisms. Mol. Cell. 2021;81:2507–2519. doi: 10.1016/j.molcel.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Kumari D., Brodsky J.L. The targeting of native proteins to the endoplasmic reticulum-associated degradation (ERAD) pathway: an expanding repertoire of regulated substrates. Biomolecules. 2021;11:1185. doi: 10.3390/biom11081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright F.A., Wojcikiewicz R.J.H. Chapter 4 - inositol 1,4,5-trisphosphate receptor ubiquitination. Prog. Mol. Biol. Transl. Sci. 2016;141:141–159. doi: 10.1016/bs.pmbts.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Wright F.A., Lu J.P., Sliter D.A., Dupré N., Rouleau G.A., Wojcikiewicz R.J.H. A point mutation in the ubiquitin ligase RNF170 that causes autosomal dominant sensory ataxia destabilizes the protein and impairs inositol 1,4,5-trisphosphate receptor-mediated Ca2+ signaling. J. Biol. Chem. 2015;290:13948–13957. doi: 10.1074/jbc.M115.655043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright F.A., Bonzerato C.G., Sliter D.A., Wojcikiewicz R.J.H. The erlin2 T65I mutation inhibits erlin1/2 complex-mediated inositol 1,4,5-trisphosphate receptor ubiquitination and phosphatidylinositol 3-phosphate binding. J. Biol. Chem. 2018;293:15706–15714. doi: 10.1074/jbc.RA118.004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce M.M.P., Wormer D.B., Wilkens S., Wojcikiewicz R.J.H. An endoplasmic reticulum (ER) membrane complex composed of SPFH1 and SPFH2 mediates the ER-associated degradation of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 2009;284:10433–10445. doi: 10.1074/jbc.M809801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J.P., Wang Y., Sliter D.A., Pearce M.M.P., Wojcikiewicz R.J.H. RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J. Biol. Chem. 2011;286:24426–24433. doi: 10.1074/jbc.M111.251983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyer M.L., Milhollen M.A., Ciavarri J., Fleming P., Traore T., Sappal D., et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 2018;24:186–193. doi: 10.1038/nm.4474. [DOI] [PubMed] [Google Scholar]
- 18.Wojcikiewicz R.J.H., Xu Q., Webster J.M., Alzayady K., Gao C. Ubiquitination and proteasomal degradation of endogenous and exogenous inositol 1,4,5-trisphosphate receptors in αT3-1 anterior pituitary cells. J. Biol. Chem. 2003;278:940–947. doi: 10.1074/jbc.M206607200. [DOI] [PubMed] [Google Scholar]
- 19.Liu W., Tang X., Qi X., Fu X., Ghimire S., Ma R., et al. The ubiquitin conjugating enzyme: an important ubiquitin transfer platform in ubiquitin-proteasome system. Int. J. Mol. Sci. 2020;21:2894. doi: 10.3390/ijms21082894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Q., Farah M., Webster J.M., Wojcikiewicz R.J.H. Bortezomib rapidly suppresses ubiquitin thiolesterification to ubiquitin-conjugating enzymes and inhibits ubiquitination of histones and type I inositol 1,4,5-trisphosphate receptor. Mol. Cancer Ther. 2004;3:1263–1269. [PubMed] [Google Scholar]
- 21.Schulman J.J., Wright F.A., Han X., Zluhan E.J., Szczesniak L.M., Wojcikiewicz R.J.H. The stability and expression level of Bok are governed by binding to inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 2016;291:11820–11828. doi: 10.1074/jbc.M115.711242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alzayady K.J., Wojcikiewicz R.J.H. The role of Ca2+ in triggering inositol 1,4,5-trisphosphate receptor ubiquitination. Biochem. J. 2005;392:601–606. doi: 10.1042/BJ20050949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alzayady K.J., Wang L., Chandrasekhar R., Wagner L.E., 2nd, Van Petegem F., Yule D.I. Defining the stoichiometry of inositol 1,4,5-trisphosphate binding required to initiate Ca2+ release. Sci. Signal. 2016;9:ra35. doi: 10.1126/scisignal.aad6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker J.A., Wong W.-C., Eisenhaber B., Warwicker J., Eisenhaber F. Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule. BMC Biol. 2017;15:66. doi: 10.1186/s12915-017-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corry B. Understanding ion channel selectivity and gating and their role in cellular signalling. Mol. Biosyst. 2006;2:527–535. doi: 10.1039/b610062g. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Xu L., Pasek D.A., Gillespie D., Meissner G. Probing the role of negatively charged amino acid residues in ion permeation of Skeletal muscle ryanodine receptor. Biophys. J. 2005;89:256–265. doi: 10.1529/biophysj.104.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naranjo D., Moldenhauer H., Pincuntureo M., Díaz-Franulic I. Pore size matters for potassium channel conductance. J. Gen. Physiol. 2016;148:277–291. doi: 10.1085/jgp.201611625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brelidze T.I., Niu X., Magleby K.L. A ring of eight conserved negatively charged amino acids doubles the conductance of BK channels and prevents inward rectification. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9017–9022. doi: 10.1073/pnas.1532257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchida K., Miyauchi H., Furuichi T., Michikawa T., Mikoshiba K. Critical regions for activation gating of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 2003;278:16551–16560. doi: 10.1074/jbc.M300646200. [DOI] [PubMed] [Google Scholar]
- 30.Bhanumathy C., da Fonseca P.C.A., Morris E.P., Joseph S.K. Identification of functionally critical residues in the channel domain of inositol trisphosphate receptors. J. Biol. Chem. 2012;287:43674–43684. doi: 10.1074/jbc.M112.415786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehning D., Joseph S.K. Functional properties of recombinant type I and type III inositol 1, 4,5-trisphosphate receptor isoforms expressed in COS-7 cells. J. Biol. Chem. 2000;275:21492–21499. doi: 10.1074/jbc.M001724200. [DOI] [PubMed] [Google Scholar]
- 32.Wagner L.E., 2nd, Joseph S.K., Yule D.I. Regulation of single inositol 1,4,5-trisphosphate receptor channel activity by protein kinase A phosphorylation. J. Physiol. 2008;586:3577–3596. doi: 10.1113/jphysiol.2008.152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Rossum D.B., Patterson R.L., Kiselyov K., Boehning D., Barrow R.K., Gill D.L., et al. Agonist-induced Ca2+ entry determined by inositol 1,4,5-trisphosphate recognition. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2323–2327. doi: 10.1073/pnas.0308565100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma C., Wang C., Luo D., Yan L., Yang W., Li N., et al. Structural insights into the membrane microdomain organization by SPFH family proteins. Cell Res. 2022;32:176–189. doi: 10.1038/s41422-021-00598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daumke O., Lewin G.R. SPFH protein cage — one ring to rule them all. Cell Res. 2022;32:117–118. doi: 10.1038/s41422-021-00605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sienaert I., De Smedt H., Parys J.B., Missiaen L., Vanlingen S., Sipma H., et al. Characterization of a cytosolic and a luminal Ca2+ binding site in the type I inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 1996;271:27005–27012. doi: 10.1074/jbc.271.43.27005. [DOI] [PubMed] [Google Scholar]
- 37.Thrower E.C., Choe C.U., So S.H., Jeon S.H., Ehrlich B.E., Yoo S.H. A functional interaction between chromogranin B and the inositol 1,4,5-trisphosphate receptor/Ca2+ channel. J. Biol. Chem. 2003;278:49699–49706. doi: 10.1074/jbc.M309307200. [DOI] [PubMed] [Google Scholar]
- 38.Thrower E.C., Park H.Y., So S.H., Yoo S.H., Ehrlich B.E. Activation of the inositol 1,4,5-trisphosphate receptor by the calcium storage protein chromogranin A. J. Biol. Chem. 2002;277:15801–15806. doi: 10.1074/jbc.M110139200. [DOI] [PubMed] [Google Scholar]
- 39.Higo T., Hattori M., Nakamura T., Natsume T., Michikawa T., Mikoshiba K. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell. 2005;120:85–98. doi: 10.1016/j.cell.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 40.Higo T., Hamada K., Hisatsune C., Nukina N., Hashikawa T., Hattori M., et al. Mechanism of ER stress-induced brain damage by IP(3) receptor. Neuron. 2010;68:865–878. doi: 10.1016/j.neuron.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Wojcikiewicz R.J., Nakade S., Mikoshiba K., Nahorski S.R. Inositol 1,4,5-trisphosphate receptor immunoreactivity in SH-SY5Y human neuroblastoma cells is reduced by chronic muscarinic receptor activation. J. Neurochem. 1992;59:383–386. doi: 10.1111/j.1471-4159.1992.tb08916.x. [DOI] [PubMed] [Google Scholar]
- 42.Anderson D.J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manasanch E.E., Orlowski R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017;14:417–433. doi: 10.1038/nrclinonc.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terry L.E., Alzayady K.J., Wahl A.M., Malik S., Yule D.I. Disease-associated mutations in inositol 1,4,5-trisphosphate receptor subunits impair channel function. J. Biol. Chem. 2020;295:18160–18178. doi: 10.1074/jbc.RA120.015683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerber S., Alzayady K.J., Burglen L., Brémond-Gignac D., Marchesin V., Roche O., et al. Recessive and dominant de novo ITPR1 mutations cause gillespie Syndrome. Am. J. Hum. Genet. 2016;98:971–980. doi: 10.1016/j.ajhg.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kania E., Roest G., Vervliet T., Parys J.B., Bultynck G. IP3 receptor-mediated calcium signaling and its role in autophagy in cancer. Front. Oncol. 2017;7:140. doi: 10.3389/fonc.2017.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicencio J.M., Ortiz C., Criollo A., Jones A.W., Kepp O., Galluzzi L., et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16:1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 48.Wojcikiewicz R.J.H. Type I, II, and III inositol 1,4,5-trisphosphate receptors are unequally susceptible to down-regulation and are expressed in markedly different proportions in different cell types. J. Biol. Chem. 1995;270:11678–11683. doi: 10.1074/jbc.270.19.11678. [DOI] [PubMed] [Google Scholar]
- 49.Pearce M.M.P., Wang Y., Kelley G.G., Wojcikiewicz R.J.H. SPFH2 mediates the endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors and other substrates in mammalian cells. J. Biol. Chem. 2007;282:20104–20115. doi: 10.1074/jbc.M701862200. [DOI] [PubMed] [Google Scholar]
- 50.Zhu C.C., Furuichi T., Mikoshiba K., Wojcikiewicz R.J. Inositol 1,4,5-trisphosphate receptor down-regulation is activated directly by inositol 1,4,5-trisphosphate binding. Studies with binding-defective mutant receptors. J. Biol. Chem. 1999;274:3476–3484. doi: 10.1074/jbc.274.6.3476. [DOI] [PubMed] [Google Scholar]
- 51.Lesueur L.L., Mir L.M., André F.M. Overcoming the specific toxicity of large plasmids electrotransfer in primary cells in vitro. Mol. Ther. Nucl. Acids. 2016;5 doi: 10.1038/mtna.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data described and discussed are located within the article.