Abstract
HIV-2, a human pathogen that causes acquired immunodeficiency syndrome, is distinct from the more prevalent HIV-1 in several features including its evolutionary history and certain aspects of viral replication. Like other retroviruses, HIV-2 packages two copies of full-length viral RNA during virus assembly and efficient genome encapsidation is mediated by the viral protein Gag. We sought to define cis-acting elements in the HIV-2 genome that are important for the encapsidation of full-length RNA into viral particles. Based on previous studies of murine leukemia virus and HIV-1, we hypothesized that unpaired guanosines in the 5’ untranslated region (UTR) play an important role in Gag:RNA interactions leading to genome packaging. To test our hypothesis, we targeted 18 guanosines located in 9 sites within the HIV-2 5’ UTR and performed substitution analyses. We found that mutating as few as three guanosines significantly reduce RNA packaging efficiency. However, not all guanosines examined have the same effect; instead, a hierarchical order exists wherein a primary site, a secondary site, and three tertiary sites are identified. Additionally, there are functional overlaps in these sites and mutations of more than one site can act synergistically to cause genome packaging defects. These studies demonstrate the importance of specific guanosines in HIV-2 5’UTR in mediating genome packaging. Our results also demonstrate an interchangeable and hierarchical nature of guanosine-containing sites, which was not previously established, thereby revealing key insights into the replication mechanisms of HIV-2.
Keywords: Retroviruses, encapsidation, RNA, Gag, guanosine, untranslated region
Graphical abstract
INTRODUCTION
Human immunodeficiency virus type 1 and type 2 (HIV-1 and HIV-2, respectively) are pathogens that were introduced into the human population through different zoonotic transmission events (1–3). HIV-1 was derived from simian immunodeficiency viruses (SIVs) that infect chimpanzees and gorillas (SIVcpz and SIVgor, respectively) (4,5), whereas HIV-2 was from SIVs that infect sooty mangabeys (SIVsm) (6). Although both viruses can cause acquired immunodeficiency syndrome (AIDS), HIV-2 is less pathogenic compared with HIV-1: only a subset of HIV-2 infected individuals develop AIDS and with a delayed onset (7,8). These two related and yet distinct viruses share ∼55% identity in nucleotide sequences (9) and have some similar features in their replication mechanisms but are different in several aspects. For example, although HIV-1 and HIV-2 have similar general genome structures, they differ in some of the encoded accessory genes; HIV-1 encodes Vpu whereas HIV-2 encodes Vpx (10).
HIV-2 packages two copies of full-length RNA as a dimeric genome into their particles (11); these RNAs carry the genetic information required for viral replication. Analyses of viral RNA content in individual particles demonstrates that most virions (>90%) contain the viral RNA genome (11). Therefore, HIV-2 RNA packaging is very efficient. This efficient genome packaging is mediated by interactions between viral protein Gag and the full-length viral RNA. In most retroviruses, sequences important for RNA packaging are located within the 5’ untranslated region (UTR) of the full-length RNA and often extend into the gag gene (12). In addition to containing the sequences important for RNA genome packaging, the 5’ UTR of HIV2 also contains multiple elements essential for other aspects of viral replication. These cis-acting elements include the trans-activation response (TAR) element, the polyadenylation (polyA) signal, and the major splice donor site (SD), all of which regulate HIV-2 RNA and gene expression, whereas the primer binding site (PBS) is required for reverse transcription.
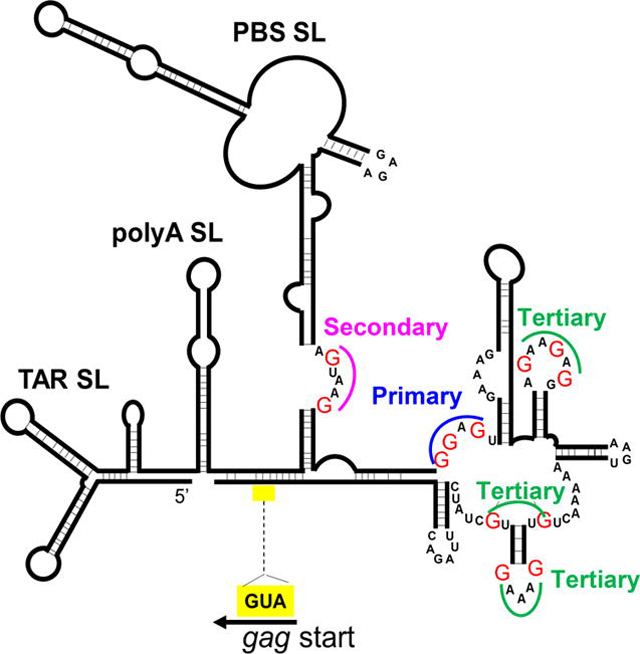
Results from chemical probing experiments have been used to develop models for the HIV-2 5’ leader RNA structure (13,14). A generally accepted model suggests that the 5’UTR forms a complex structure with multiple stem-loops (14); many of the aforementioned functional elements reside in stem-loop structures, often named after these elements. In this model, the 5’ UTR of the full-length RNA begins with a complex TAR stem loop, followed by a polyA stem loop, a short C-rich region (C-box), a PBS stem loop, stem loop 1 (SL1), after which there are four short stem loops termed psi 1, SD, psi 2, and psi 3; the 5’ UTR ends with a G-rich region (G-box) that extends into the gag reading frame. The dimerization initiation signal of HIV-2 is a 6-nucleotide palindrome located at the top of SL1, and sequence identity of this signal affects copackaging of RNA from two different proviruses, indicating RNA was dimerized prior to packaging (11). The loose dimer RNA structure, the configuration of RNA that is thought to be packaged, was also probed using in vitro transcribed RNA and Selective 2′ Hydroxyl Acylation analyzed by Primer Extension (SHAPE) analysis (15). The loose dimer structure is in general agreement with the previously proposed structure determined by chemical probing (14) except for the long-range interaction between the C-box and G-box. Long-range interactions in the HIV-2 5’ UTR have been previously suggested (16,17); specifically, interactions of the C-box and G-box were shown to affect HIV-2 RNA dimerization in vitro (17).
To assess the elements in HIV-2 5’ UTR essential for RNA packaging, multiple studies deleted a portion of the HIV-2 5’ UTR and examined genome packaging (18–20). These studies yielded varied results and conclusions probably because different deletion mutants were used. For example, one report showed that deletion of sequences between the SD and gag gene severely affected RNA packaging and concluded that the major packaging signal was located downstream of SD (18). In contrast, other reports showed that deletion mutants in this region only had a minor packaging defect (19,20) and concluded that the major packaging signal is located upstream of the SD (20). Additionally, other reports suggested that sequences important for packaging are located both upstream and downstream of SD (17,21). These conflicting results are reflected in the naming of the elements found in the 5’ UTR. In some studies, “psi”, a term commonly used to describe retroviral packaging signals, is used to describe a short sequence between PBS and SL1 (22), whereas in many other studies psi 1, psi 2, and psi 3 are used to describe a series of short stem loops flanking the SD stem loop (14,15). To avoid confusion, we have adopted the nomenclature used in the structural studies (14,15).
Viral protein Gag orchestrates the assembly process; during or soon after virus assembly, viral protease cleaves Gag to yield multiple mature proteins including nucleocapsid (NC), a nucleic acid binding protein. Gag selects the full-length viral RNA from a large pool of cellular mRNAs. The mechanism by which Gag recognizes and selects viral RNA has been an important question in HIV biology. The specific sites in the 5’ UTR to which HIV-1 Gag or NC bind have been examined using several methods (23–29). Multiple recent studies have mapped the HIV-1 Gag or NC binding sites in distinct locations of the HIV-1 5’ UTR; furthermore, several studies suggest that Gag preferentially binds to unpaired guanosines in the RNA structure (24–27,29,30). Intriguingly, structural and mutational studies on murine leukemia virus (MLV) genome packaging also demonstrated that unpaired guanosines in the 5’ UTR play critical roles in RNA genome packaging (31). These studies suggest that interactions between Gag and unpaired guanosines may be a conserved feature in retroviral genome packaging. Although the binding of HIV-2 NC to in vitro transcribed short RNA has been probed (15), the mechanisms by which HIV-2 Gag selects its RNA genome has not been explored. Additionally, the packaging specificity of HIV-2 Gag is distinct from that of HIV-1 Gag because HIV-2 Gag is not capable of cross-packaging HIV-1 RNA (32,33). Therefore, it is unclear whether HIV-2 Gag: RNA molecular interactions can be predicted from those of HIV-1.
In the studies described in this report, we sought to define the cis-acting elements important for HIV-2 RNA packaging. Based on the HIV-1 and MLV studies, we hypothesized that unpaired guanosines in the HIV-2 5’ UTR play a critical role in mediating specific packaging of the HIV-2 full-length RNA. To test our hypothesis, we selected 9 sites in the 5’ UTR that are predicted to contain unpaired guanosines based on current RNA structure models and generated a mutant with 18-nt substitution mutations. As these guanosines are located in unpaired regions, these substitutions are less likely to cause large changes in the RNA structure compared with large deletions. We found that substituting 18-nt of a 9-kb genome generated a severe defect in RNA packaging without affecting virus production. Further mutational analyses identified five sites that are important for efficient RNA packaging; additionally, there is redundancy and synergism of these sites and they display a hierarchical order of importance with respect to genome packaging.
RESULTS
System employed to examine HIV-2 RNA packaging.
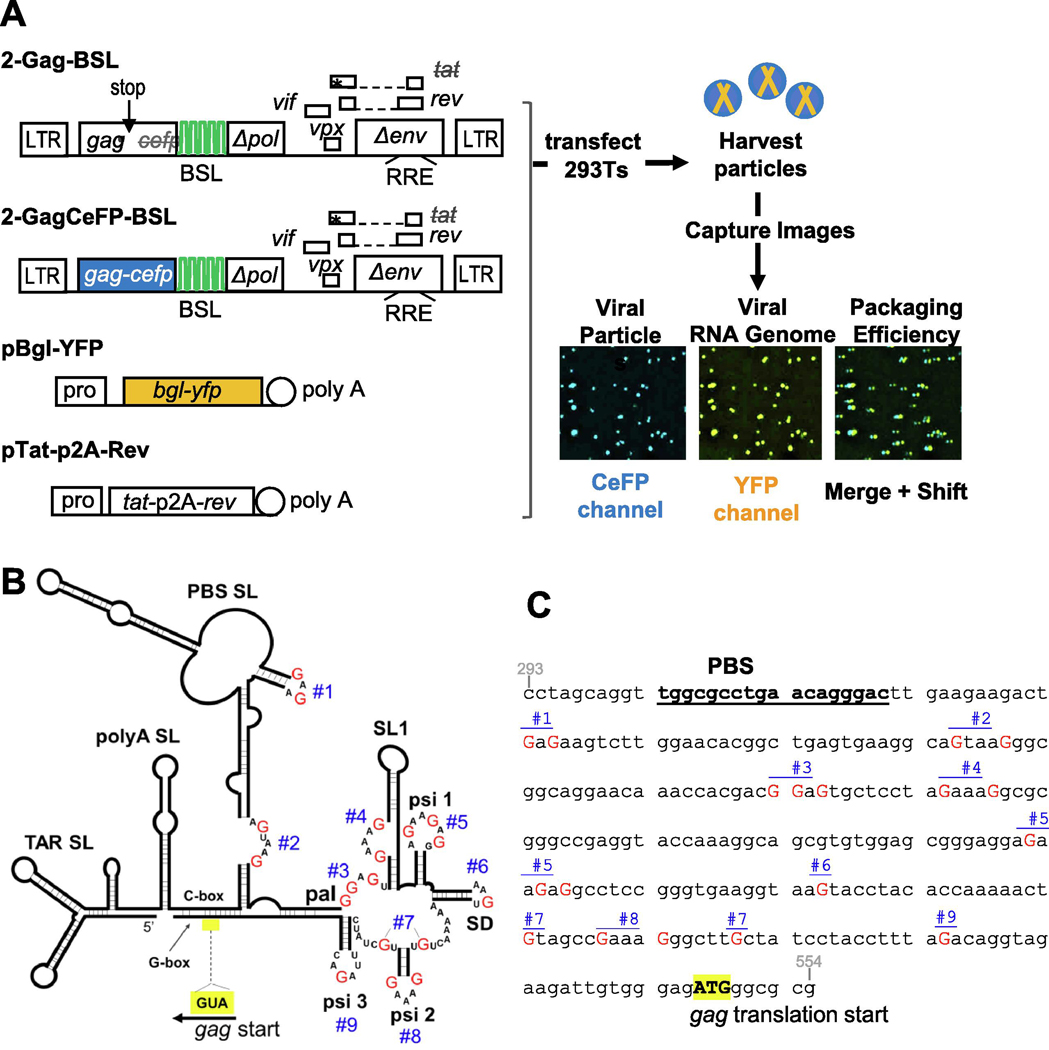
To determine the efficiency of viral RNA genome packaging, we used a previously described system termed single-virion analysis (34). This assay visualizes viral RNA in individual particles and has single-RNA detection sensitivity (11,34). Briefly, two near full-length HIV-2 constructs are used (Fig. 1A): 2-Gag-BSL expresses untagged Gag and 2-GagCeFP-BSL expresses Gag fused to cerulean fluorescent protein (CeFP) (11). Coexpression of untagged Gag and Gag tagged with fluorescent protein at a 1:1 ratio allows for the formation of morphologically normal immature particles (35). These HIV-2 constructs contain all the cis-acting elements necessary for viral replication; additionally, within the truncated pol gene, they harbor stem-loop sequences referred to as BSL that are specifically recognized by a bacterial RNA-binding protein BglG (34,36). Although these constructs contain a functional rev gene, the tat gene contains an inactivating +1 frameshift mutation.
Fig. 1.
Approach used to examine the roles of unpaired guanosines in the HIV-2 5’UTR on RNA packaging. A. General structures of constructs and experimental protocol used to study RNA packaging. BSL, stem-loop sequences recognized by bacterial protein BglG; asterisk denotes inactivating mutation; stop with arrow indicates introduced stop codon. Poly A, polyadenylation signal; pro, promoter; LTR, long terminal repeat; RRE, Rev response element; p2A, p2A self-cleaving peptide. Asterisks indicate stop codons. Representative images of viral particles captured using fluorescence microscopy are shown. Merge and Shift, CeFP and YFP signals were overlaid and YFP signals were shifted to the right by 3 pixels to visualize colocalization. B. Schematic of HIV-2 leader RNA structure as described by Purzycka et al. 2011 (15). For simplicity, only the nucleotides within the single-stranded sites targeted for mutations are shown; red uppercase Gs indicate specific guanosines mutated; blue numbers (#1 to #9), denote sites of mutations; yellow box denotes position of Gag translation start. Site 3 is embedded within a 10-nucleotide palindromic sequence (pal). C. Sites of mutations in the context of the HIV-2 leader DNA sequence used in this study. Red uppercase G, guanosines targeted for mutation; blue numbers, individual sites. Indicated guanosines were mutated to adenosines, except in the case of site #2 in which the AGTAAG sequence was mutated to ACTAAA to avoid introducing a major polyadenylation signal and site #3 in which the GGAGTG sequence was mutated to AAATTG to avoid introducing an ATG upstream of the gag open reading frame. For reference, the PBS sequence is shown in bold and underlined, and the Gag ATG translation start codon is shown in bold uppercase with yellow highlight.
To analyze viral RNA packaging, Gag- and GagCeFP-expressing HIV-2 constructs were cotransfected into human 293T cells along with a plasmid expressing BglG protein fused with yellow fluorescent protein (YFP) and a plasmid expressing HIV-1 Tat and Rev (Fig. 1A). Supernatants were harvested from transfected cells, clarified by filtration, plated on slides, and imaged using fluorescent microscopy. Viral particles were detected by the CeFP signals from the GagCeFP fusion protein, whereas viral RNA genomes were detected by the YFP signals mediated by specific binding of BglG-YFP to the BSL in the viral RNA (Fig. 1A). Because BSL is located in the pol gene, only full-length viral RNAs were detected. RNA packaging efficiency was determined by quantifying the proportion of Gag particles (CeFP signal) that contained full-length viral RNA (YFP signal). Particle production was quantified as the average number of CeFP particles per field captured in the imaging experiments normalized by the volume of supernatant plated on the slide. To determine RNA packaging efficiency, more than 1,000 viral particles were analyzed in each sample of an experiment and a minimum of three independent experiments were performed.
Substituting predicted unpaired guanosines in the HIV-2 5’ UTR results in severe packaging defects.
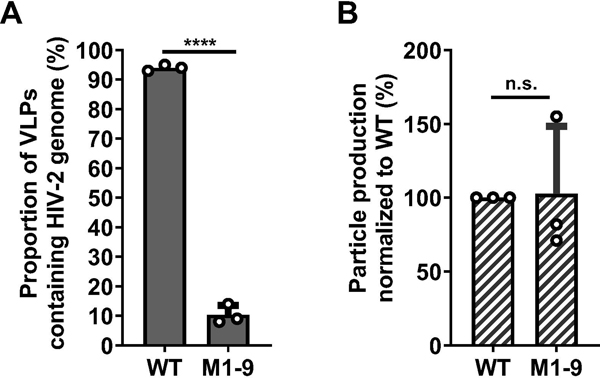
A consensus of the secondary structure of the HIV-2 5’UTR has been established, confirmed by recent SHAPE study, and predicts multiple unpaired guanosines (14,15) (Fig. 1B). We hypothesized that the unpaired guanosines play an important role in Gag:RNA interactions that lead to RNA packaging. To test our hypothesis, we replaced unpaired guanosines in 9 sites predicted by the SHAPE study (15); these sites are located between the PBS and the translational start codon of gag. There are 18 unpaired guanosines in these 9 sites; the red Gs in Fig. 1B and 1C denote guanosines that were mutated, and the blue numbers indicate sites in which these guanosines reside. We modified 2-Gag-BSL and 2GagCeFP-BSL to generate a pair of mutants, M1-9, in which all 18 of the unpaired guanosines were replaced with adenosines except in sites 2 and 3. In site 2, the sequence AGUAAG was changed to ACUAAA to avoid the generation of a polyadenylation signal. Site 3 has the sequence GGAGT, which is flanked by a guanosine at the 3’ end; this GGAGT(G) sequence was changed to AAATT(G) to avoid introducing an ATG start codon upstream of gag translation start codon (Fig 1C). We have performed RNA structure modeling and found that the 5’UTRs from wild-type HIV-2 and from mutant M1-9 fold into similar general structures with comparable delta-G free energies (Supplementary Figure 1). These results suggest that the substitutions introduced in the 18 guanosines do not cause major changes in the overall RNA folding of the 5’UTR. Although mutations were made in both Gag- and GagCeFP-expression constructs, for simplicity we refer to them as one mutant. We cotransfected the Gag- and Gag- CeFP-expressing plasmids with Bgl-YFP and Tat/Rev-expression constructs, harvested particles, and performed single-virion analyses. Most of the viral particles (94%) generated from the wild-type control with unaltered 5’ UTR contained the viral RNA, indicating that viral RNA was packaged efficiently (Fig. 2A). In contrast, only 10% of the particles generated from mutant M1-9 contained viral RNA (Fig. 2A). Importantly, the M1-9 mutant generated similar amounts of viral particles as the wild-type controls (Fig. 2B), indicating that the substitutions did not affect gene expression or viral particle production. In this system, the same RNA species is used for Gag translation and genome packaging; therefore, these results suggest that substituting the 18 guanosines significantly affected RNA packaging efficiency without affecting other aspects of HIV-2 RNA.
Fig. 2.
Effects of mutating putative unpaired guanosines in the HIV-2 5’UTR on RNA packaging and viral particle production. A. Proportions of viral particles containing HIV-2 RNA genome. Particles derived from HIV-2 constructs containing unaltered 5’UTR (WT) or mutated 5’ UTR in which guanosines in 9 target sites were mutated (M1-9). B. Particle production of WT and M1-9 constructs. WT particle production is set to be 100%; equal amounts of WT or M1-9 DNA were used in each transfection. Results shown are averages of three independent experiments; error bars indicate standard deviations and open circles indicate values obtained from each independent experiment. P-values were calculated using the unpaired t-test; n.s., not significant; ****, p-value < 0.0001.
Site 3 contains guanosines important for HIV-2 RNA packaging.
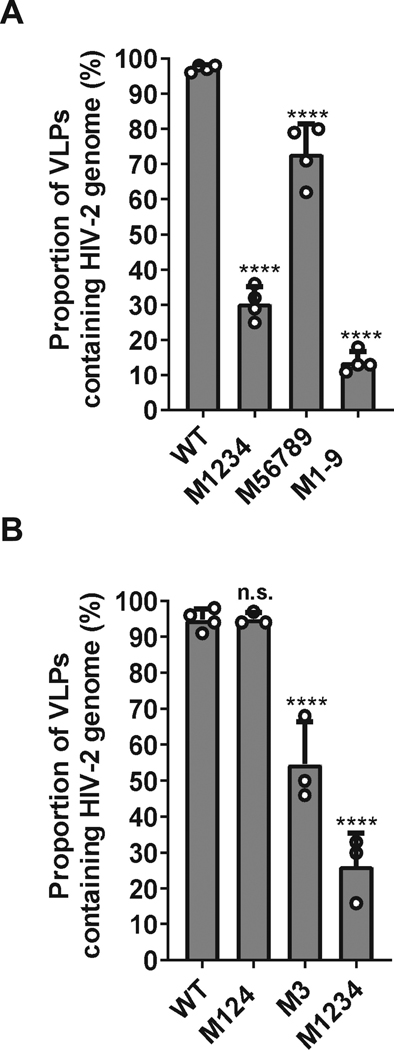
To determine the substitutions that caused the packaging defect in M1-9, we first generated two mutants, M1234 and M56789, each carrying a portion of the mutations in M1-9. For clarity, the names of the mutants correspond to the mutated sites; for example, M1234 contains mutations in sites 1 to 4, whereas M56789 contains mutations in sites 5 to 9 as shown in the Fig. 1B and 1C. We performed single-virion analyses using these mutants along with the wild-type control and M1-9 mutant. We observed that ∼30% (31% ± 5%; mean and standard deviation from three experiments) of the particles generated by M1234 contained the viral RNA genome (Fig. 3A). This finding indicates the presence of guanosine(s) within sites 1, 2, 3, and 4 that are important for HIV-2 RNA packaging. Mutant M56789 exhibits a mild packaging defect, 74% ± 8 of its particles contained viral RNA, indicating that guanosines altered in mutant M56789 contribute to RNA genome packaging. This conclusion is consistent with the observation that M1234 exhibits a significantly better RNA packaging efficiency compared with that of M1-9 (Fig. 3A; P = 0.003; ANOVA with Bonferroni correction), indicating that restoring guanosines in sites 56789 partly recover viral RNA packaging efficiency.
Fig. 3.
Identifying sites in M1-9 that affect HIV-2 genome packaging. A. Effects of mutations in the first four (M1234) or last five (M56789) sites on HIV-2 RNA packaging. B. Effects of mutating sites 1,2, and 4 (M124) or site 3 (M3) on HIV-2 RNA packaging. Results shown are averages of at least three independent experiments. Error bars, standard deviations; open circles indicate values obtained from each independent experiment. P-values were calculated by one-way ANOVA with Bonferroni correction for multiple pairwise comparisons; n.s., not significant; ****, p-value < 0.0001.
Among the four sites mutated in M1234, site 3 has been proposed to play important roles in RNA packaging and viral replication (22,37,38). Thus, to further delineate the roles of the mutated guanosines, we generated two mutants, M3 and M124, and performed single virion analyses. Our results showed that ∼50% (54% ± 10%) of the particles generated by mutant M3 contained the viral RNA genome (Fig. 3B). Hence, compared with wild-type control, substituting three guanosines in the site 3 resulted in a significant 2-fold decrease in packaging efficiency of a 9-kB genome (P = 0.0003; ANOVA with Bonferroni correction). In contrast, mutant M124 packaged the viral RNA genome at levels comparable to that of the wild-type control (94% ± 1%) (Fig. 3B). These results indicate that site 3 is the major determinant in viral genome packaging. However, the packaging defect displayed by M3 is significantly less than that from M1234 (P < 0.001; ANOVA with Bonferroni correction). Thus, although mutations in M124 do not cause detectable defects by themselves, combining them with substitutions in M3 exacerbate packaging defects.
Delineating mutations required to recapitulate the packaging defect observed in M1-9.
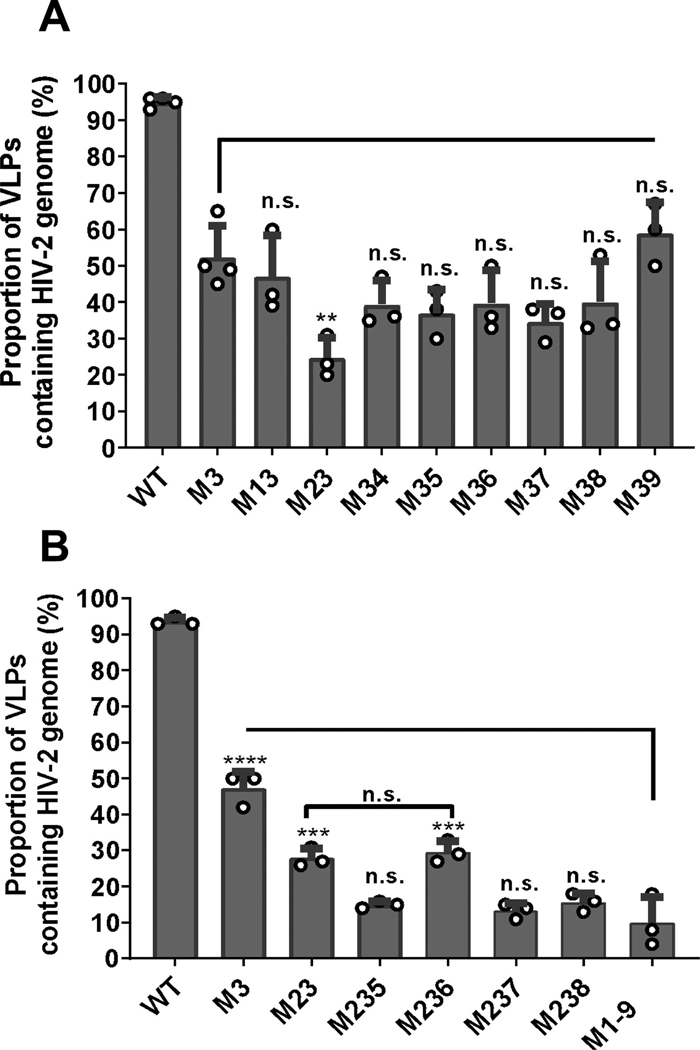
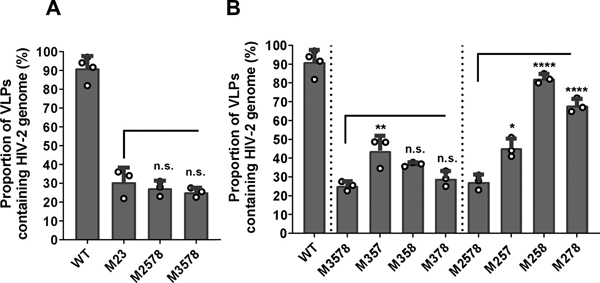
Our study identified that guanosines in site 3 are important for RNA packaging; however, the M3 mutant only exerts moderate packaging defects. We hypothesized that site 3 together with other sites coordinate Gag binding leading to RNA packaging; thus, combining site 3 mutations with other guanosine substitutions would cause more severe packaging defects. To identify the minimum substitutions required for such effects, we generated 8 mutants, each containing mutations in site 3 and another site; the names of these mutants reflect the sites mutated for example, M13 contained mutations of sites 1 and 3. We performed single virion analyses using these mutants along with the M3 mutant and wild-type control. To assess the effects of mutating site 3 plus an additional site, we compared the RNA packaging efficiencies of these mutants with that of M3 (Fig. 4A). Of these mutants, M39 did not show a reduction of genome packaging compared to M3 (59% ± 7% and 52% ± 8% respectively); whereas M13, M34, M35, M36, M37, and M38 appeared to have a slightly lower, but not significantly different, RNA packaging efficiencies compared with M3 (Fig. 4A). Compared with M3, the only mutant that had a significantly lower RNA packaging efficiency was M23 (25% ± 4%). Furthermore, the RNA packaging efficiency of M23 is similar to that of M1234 (Fig. 3A), suggesting that mutations in site 2 and 3 account for most of the packaging defects in M1234.
Fig. 4.
Delineating the minimal number of mutations required to recapitulate the M1-9 phenotype. A. Effects of mutating site 3 in combination with another site on genome packaging efficiency. The RNA packaging efficiencies of double mutants were compared to that of M3. B. Effects of mutating sites 2 and 3 in combination with select single sites on RNA packaging. Statistical comparisons to M1-9 are shown for the mutants. All results are averages of at least three independent experiments; error bars, standard deviations, with open circles indicating values obtained from each individual experiment. P-values for all experiments were calculated by one-way ANOVA with Bonferroni correction for multiple pairwise comparisons: n.s., not significant; **, p-value < 0.01; ***, p-value < 0.005; ****, p-value < 0.0001.
Although mutant M23 exhibited a packaging defect, it packages RNA significantly better than mutant M1-9 (Fig. 4B), suggesting that there are additional sites that contribute to genome packaging. Compared to wild-type control, mutant M56789 exhibited a moderate RNA packaging defect, suggesting that sequences altered within this mutant contribute to specific RNA packaging (Fig. 3A). To further delineate these potential sequences, we added substitution of an additional site to M23 to generate M235, M236, M237, and M238. We did not test the effects of site 9 because M39 did not exhibit a lower packaging efficiency compared with M3. Single virion analyses showed that the viral RNA packaging efficiency of M236 is comparable to that of M23 and is significantly different than M1-9 (P > 0.999 and P = 0.0001, respectively; one-way ANOVA with Bonferroni correction). These results indicate that adding the site 6 mutation to M23 does not enhance packaging defects. In contrast, mutants M235, M237, and M238 all packaged viral RNA significantly lower than mutant M23 (P = 0.0114, P = 0.0036, and P = 0.0184, respectively; one-way ANOVA with Bonferroni correction). Furthermore, these three mutants package viral RNA at levels comparable to that of M1-9 mutant (P > 0.999 for all three mutants; one-way ANOVA with Bonferroni correction), indicating that adding mutation of site 5, 7 or 8 to M23 generates a mutant with defects similar to that of M1-9 (Fig. 4B). Thus, a minimum of three mutated sites are required to recapitulate the defects of M1-9.
Characterizing the interplay between mutating various sites on RNA packaging efficiencies.
Thus far, we have identified site 3 and site 2 as important elements in RNA packaging, and adding mutations in site 5, 7, or 8 to M23 resulted in phenotypes similar to that of M1-9. These findings raised the question whether combining mutations of these three sites could recapitulate the effects of mutations of site 3 or site 2 in M23. This possibility is supported by our results on mutant M56789, which includes mutations in site 5, 7, and 8, displayed a mild packaging defect (∼75%, Fig 3A). To examine the effects of combining site 5,7, and 8, we generated two mutants, M2578 and M3578, and compared their RNA packaging efficiencies with that of M23. Our single virion analyses showed that M2578 and M3578 packaged HIV-2 RNA genomes at levels comparable to that of M23 (Fig 5A; P > 0.999 for both M2578 and M3578, one-way ANOVA with Bonferroni correction). Thus, when all three 5,7, and 8 sites were mutated, these mutations can indeed replicate the effects of mutating sites 2 or 3 independently.
Fig. 5.
Effects of sites 5, 7, and 8 mutations on genome packaging efficiency. A. Effect of combining sites 2 or 3 with sites 5,7, and 8 mutations on RNA packaging. B. Effects of restoring sites 5, 7, or 8 in M2578 and M3578 on RNA packaging. The same data sets on WT, M2578, and M3578 are shown in both A and B panels. All results are averages of at least three independent experiments; error bars, standard deviations, with open circles indicating values obtained from each individual experiment. P-values were calculated by one-way ANOVA with Bonferroni correction for multiple pairwise comparisons: n.s., not significant; *, p-value < 0.05; **, p-value < 0.01; ****, p-value < 0.0001.
To further delineate whether mutations in all the three sites are required to cause the observed phenotypes, we systematically restored one of the sites, either 5, 7, or 8, in the M2578 and M3578 mutants. We then compared the RNA packaging efficiencies of M357, M358, and M378 with that of M3578. We found that two of the mutants, M358 and M378, packaged RNA at similar efficiencies as M3578; whereas the third mutant, M357, exhibited slightly, but significantly improved RNA packaging efficiency (Fig. 5B; P = 0.009; one-way ANOVA with Bonferroni correction). We then compared the packaging efficiencies of M257, M258, and M278 with that of M2578, and found that all three mutants exhibited improved RNA packaging efficiencies (Fig 5B; P = 0.014 for M257 and P < 0.0001 for both M258 and M278, one-way ANOVA with Bonferroni correction). Thus, to recapitulate the M23 phenotype, all three 5,7, and 8 sites need to be mutated when combining with site 2 substitutions whereas mutations in two sites can be sufficient when combined with M3. These results further underscore the importance of the site 3 in mediating HIV-2 RNA packaging. Intriguingly, reverting a given site can yield very different results when combined with site 2 or site 3 mutations. For example, the site 7 mutation was reverted in M3578 to generate M358, and these two mutants package RNA at similar efficiencies. In contrast, the site 7 mutation was reverted in M2578 to generate M258, which packaged RNA at almost wild-type efficiencies (Fig. 5B). These findings further illustrate the complex interplay between Gag and various sites in the viral 5’ UTR during the process of packaging the viral genomes.
Determining the relationship between the numbers of guanosine mutated and the RNA packaging efficiencies.
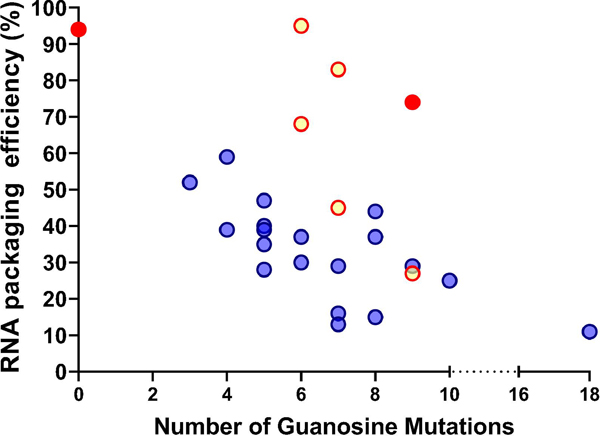
We have examined the effects of mutating 18 guanosines in the 5’ UTR and determined the RNA packaging efficiencies of the mutants. To assess whether the number of guanosine mutations is directly correlated with genome packaging defects, we plotted the wild-type control along with the 25 mutants (shown as yellow, red, or blue dots) in Fig. 6, with the number of guanosines mutated in the x-axis and the RNA packaging efficiencies in the y-axis. The graph illustrates that RNA packaging is highly efficient without mutation and severely defective when all 18 guanosines are mutated; however, the phenotypes between these two data points are more complex. There is weak or little association between the number of guanosine mutations and RNA packaging efficiencies (R2=0.22); furthermore, there are large variations among mutants with the same numbers of guanosine mutations. For example, the RNA packaging efficiencies of 7 mutants, each with 7 guanosine substitutions, varied from 75% to 15%. These results indicate that the number of guanosine mutations is not the sole determinant of the levels of genome packaging defects. Instead, these findings suggest that individual guanosines have different effects on RNA packaging efficiencies.
Fig. 6.
Delineating the effects of guanosine mutations on HIV-2 RNA packaging. X-axis, number of guanosines mutated; y-axis, RNA packaging efficiency. Blue dots, mutants containing site 3 mutations; yellow dots, mutants without site 3 mutation but containing site 2 mutations; red dots, wild-type and a mutant (M56789) without sites 2 or 3 mutations.
In our study, we observed that mutation of site 3 alone caused the RNA packaging efficiency to decrease to ∼50%. In contrast, the mutant containing sites 1, 2, and 4 mutations packaged RNA at a wild-type level, whereas the mutant with substitutions in all of the 5,6,7,8 and 9 sites displayed a packaging efficiency of 75%. Thus, of the 9 sites we examined, mutation of site 3 had the most severe effect on RNA packaging and was designated the “primary site”. Of the other 8 sites, when added individually to M3, only site 2 significantly further hindered genome packaging. Thus, site 2 is designated as the “secondary site”. Addition of mutations in site 5, 7, or 8 to M23 worsened packaging defects; thus, these three sites are designated as “tertiary sites”.
In the current study, our mutants are based on a molecular clone, ROD12. To investigate whether the sites described in this report are conserved in HIV-2 variants, we aligned 41 full-length HIV-2 group A and B sequences available in Los Alamos HIV database and analyzed genetic variation in the guanosine positions of sites 2, 3, 5, 7, and 8. Our analyses revealed that all guanosines in sites 3 and 2 are fully conserved in all analyzed sequences, which is consistent with their role as primary and secondary site, respectively. However, not all the guanosines in the tertiary sites are fully conserved. Site 5 contains three guanosines; of these, two guanosines are fully conserved whereas 20 of 41 sequences (49%) encode guanosine on the third nucleotide. Site 7 contains two guanosines, and guanosine is the predominant nucleotide in both positions (38 and 40 out of 41). Site 8 has two guanosines, and guanosine is encoded by 26 and 23 of 41 sequences (63% and 56% respectively). Taken together, we found that in the analyzed sequences, guanosines are fully conserved for the positions in the primary and secondary sites and highly conserved in the positions in tertiary sites.
We then examined how the primary and secondary site mutations affect the correlation between the number of guanosines mutated and RNA packaging defects. In Fig. 6, mutants containing site 3 mutations are shown in blue, and mutants containing site 2 mutations but without site 3 mutations are shown in yellow. The wild-type control and M56789, which does not have mutations in site 2 and site 3, are shown in red. Among the four mutants with 6 guanosine substitutions, two mutants without site 3 mutations exhibited less severe packaging defects. Similarly, among the mutants with 7 guanosine substitutions, two mutants without site 3 mutations also displayed less severe packaging defects. These results further support the importance of site 3 for RNA packaging. However, not all mutants with an unchanged site 3 exhibited improved packaging. Three mutants had 9 guanosine substitutions: M1234, M2578, and M56789; among these mutants, M1234 and M2578 packaged RNA at similarly reduced efficiencies, whereas M56789 (red dot) has the least packaging defects among the three mutants and encapsidated RNA at ∼75%. These results further support the importance of site 2 and site 3 in mediating RNA packaging. Together, these mutational analyses illustrate that specific guanosines in the HIV-2 5’UTR are important for RNA packaging; however, not all guanosines impact packaging equally. Instead, there is a hierarchical order of sites that have varied effects in mediating genome packaging.
DISCUSSION
During assembly of nascent particles, HIV-2 must package its full-length RNA to carry the genetic information. Thus, RNA packaging is an essential process for generating infectious virions. In this report, we sought to determine elements in the 5’ UTR of HIV-2 full-length RNA that affect genome packaging. We performed targeted mutagenesis on 18 guanosines located in 9 different sites of the 5’ UTR and found that substituting these guanosines strongly affected HIV-2 genome packaging. However, not all the guanosine mutations had the same effects; mutating specific sets of guanosines caused severe packaging defects whereas mutating other guanosines have little effects on RNA packaging. Our analysis revealed three previously unknown aspects of HIV-2 RNA packaging elements. First, there is a hierarchical order to the multiple sites that affect HIV-2 RNA packaging: site 3 and 2 are the primary and secondary sites, respectively; whereas three sites flanking the major splice donor site are the tertiary sites. Second, there are synergistic effects to mutating sites. For example, M1234 and M56789 have packaging efficiencies of 31% and 74%, respectively; however, M1-9 has a packaging efficiency of 10%, which is more defective than expected from additive effects (31% × 74% = 23%). Similarly, M3 and M124 have packaging efficiencies of 54% and 94%, respectively; and yet M1234 has an efficiency of 31%, lower than expected from combined effects (54% × 94% =51%). Our findings demonstrate that these mutations act synergistically to cause packaging defects. Finally, there is functional overlap among some guanosine sites; for example, M23, M2578, and M3578 exhibited similar packaging defects, indicating that mutating sites 5, 7, and 8 together can replace site 2 or site 3 mutation in the dual mutant. These results suggest that the HIV-2 packaging signal contains redundant elements important for RNA packaging.
Our findings indicate that HIV-2 RNA contains multiple Gag binding sites, more than minimally required to mediate packaging, thereby creating functional overlap; furthermore, these sites have varied affinities. Because multiple Gag proteins need to bind HIV-2 RNA to mediate packaging, we hypothesize that the presence of more than the minimally required sites allow multiple paths to be taken to achieve sufficient Gag:RNA interactions necessary for efficient packaging. Thus, mutating any one site, even in the case of the primary site, does not completely abrogate packaging. However, mutating multiple sites can eliminate the more efficient paths to achieve sufficient Gag:RNA interactions for packaging, thereby creating the observed synergistic effects of multiple mutations on RNA packaging. The presence of more Gag binding sites than minimally required ensures efficient genome packaging, an essential step in viral replication. Furthermore, the functional overlap allows genome packaging to occur even when one or two sites are mutated, a feature that also serves as a layer of protection against substitution mutations in the 5’ UTR.
Studies of MLV showed that two UCUG-UR-UCUG motifs in the 5’ UTR of full-length RNA are the major Gag binding sites (31). Mutation of all four guanosines in both motifs caused severe RNA packaging defects (31). In contrast, studies of HIV-1 Gag:RNA interactions suggest that there are multiple Gag/NC binding sites at the 5’ UTR of the full-length RNA (24,27,39); furthermore, these binding sites contained exposed guanosines. We and others have examined the role of the unpaired guanosines in HIV-1 RNA packaging (27,29). In our extensive mutational study, we identified multiple sites that are important for RNA packaging; of those, mutations in two specific NC binding sites caused more severe defects, reminiscent of the M3 mutant described in this report. Similar to HIV-2, combining guanosine mutations in HIV-1 also has synergistic effects on RNA packaging (29).
Results from this report, together with previous studies on MLV and HIV-1, suggest a common theme of retroviral Gag proteins preferentially binding to unpaired guanosines within a highly structured RNA region. However, there are many differences among these three viruses. The major sites of MLV Gag binding that lead to RNA packaging are defined in two 10-nt motifs; furthermore, these motifs do not appear to have a hierarchical order. Our study demonstrates that the guanosines that influence RNA packaging in HIV-2 do not reside within one conserved motif but are scattered throughout a 190-nt region; furthermore, there is a hierarchical order and redundancy among these sites. Intriguingly, the mutational analyses of HIV-1 unpaired guanosines demonstrated features similar to HIV-2; the Gag binding sites mapped in HIV-1 reside within a longer region in the 5’UTR, have a hierarchical order, and display redundancy and synergism (29). These comparisons suggest that although retroviral Gag proteins recognize unpaired guanosines in the 5’ UTR, different retroviruses may have evolved distinct features of Gag:RNA interactions. Compared with MLV, HIV-1 and HIV-2 may share more similarities in the RNA elements and their mechanistic interactions with Gag that govern genome packaging.
Previous studies have proposed that cis-acting elements important for packaging reside either upstream, downstream, or both upstream and downstream of the major splice donor site of HIV-2 RNA leader (17,18,20,21). Our results are in agreement with proposals that elements both upstream and downstream of the major splice donor site are important for genome packaging. Additionally, we have confirmed the role of a ‘GGAG’ element in site 3 on RNA packaging as proposed by several studies (37,38). This ‘GGAG’ element is part of a larger 10-nt palindromic sequence upstream of the major splice donor site. When deleted or substituted in previous studies, ∼2-fold packaging defects were observed (38). Using a different experimental system compared to all previous studies, we also observe a ∼2-fold packaging defect, with only guanosine substitutions in the GGAG element (M3). These studies firmly establish that the ‘GGAG’ minimal subsequence of the 10-nt palindrome as a key site important for HIV-2 viral genome packaging.
The 5’ UTR of retroviral RNA is rich in regulatory elements for gene expression, genome encapsidation, and reverse transcription. To serve these functions, RNA often folds into complex structures and an element can have more than one function. Understanding the intricate roles of the 5’ UTR sequences and structures on HIV genome packaging not only furthers our understanding of viral replication but also opens the possibility of targeting these elements for potential antiviral strategies.
MATERIALS AND METHODS
Viral vectors and plasmids construction.
The HIV-2 constructs, 2-GagCeFP-BSL and 2-Gag-BSL, were modified from previously described p2-GagCeFP-BSL and p2-Gag-BSL, respectively (11). These constructs were derived from the ROD12 molecular clone and contain all the cis-acting elements necessary for viral replication, express untagged Gag or Gag tagged with CeFP, and in the truncated pol gene, contain 18 copies of stem-loop sequences (BSL) recognized by Escherichia coli BglG protein (34). Additionally, these constructs have inactivating deletions in vpr, and env, and an inactivating insertion in nef containing the internal ribosomal entry site (IRES) from the encephalomyocarditis virus (EMCV) and the mouse heat stable antigen (hsa); for simplicity, IRES-hsa is not shown in the Fig. 1A. Two modifications distinguish 2-GagCeFP-BSL and 2-Gag-BSL from previously described p2-GagCeFP-BSL and p2-Gag-BSL, respectively: first, an inactivating +1 frameshift was introduced in the first exon of tat; second, a silent one-base substitution was introduced in the matrix (MA)-encoding region of the gag gene to generate a unique AgeI site. DNA fragments containing mutations in the 5’ UTR were synthesized (IDT) and introduced into 2-GagCeFP-BSL and 2-Gag-BSL by replacing the existing SfoI to AgeI fragment. In all experiments, untagged Gag- and GagCeFP-expressing plasmids are transfected at a 1:1 molar ratio to maintain normal immature viral particle morphology (35). Plasmid Bgl-YFP has been previously described (40) and expresses a truncated version of the BglG protein fused to YFP. Plasmid Tat-p2A-Rev is a CMV-driven codon-optimized tat-rev expression construct with a p2A self-cleaving peptide from porcine teschovirus (41) between HIV-1 tat and rev genes. All newly generated constructs were verified by sequencing of the cloned region.
Cell culture, transfections and virus production.
Human embryonic kidney 293T cells were cultured in a humidified 37°C incubator with 5% CO2 and maintained in clear Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). DNA transfections were performed using TransIT-LT1 transfection reagent (Mirus) at 3:1 μl reagent: total μg DNA. In all experiments, GagCeFP- and Gag-expressing plasmids were cotransfected at a 1:1 molar ratio and a total of 100 ng Gag-expressing plasmids were used per well of 6-well plates. Virus supernatant was harvested 24 hrs post-transfection, filtered through a 0.45-μm-pore-size filter (Millipore), and either used immediately for image acquisition or briefly stored at 4°C prior to image acquisition.
Single-virion analyses.
Filtered supernatant was mixed with Polybrene (50 μg/ml), plated onto μ-Slide ibiTreat 8-well slides (Ibidi) and spun in a Beckman Coulter Allegra™ 21R centrifuge with a Beckman Coulter S2096 rotor for 90 mins, 1200 × G at 15°C. The process was repeated to increase the number of particles per observation field for microscopy.
Epifluorescence microscopy was performed with an inverted Nikon Eclipse TE 2000 microscope and a 100X 1.49 numerical aperture oil objective, using a SOLA light engine® light source (Lumencor) for illumination. Digital images were acquired using a Hammamatsu ORCA-ERA camera and NIS-Elements software (Nikon) with the excitation and emission filter sets 427/10 nm and 472/30 nm for CeFP and 504/12 nm and 542/27 nm for YFP. The diffraction-limited spots were detected, and their positions were determined in each image using Localize (42). Colocalization of the Gag (CeFP) and RNA (YFP) signals was determined using a custom MATLAB program (Mathworks) (34,43); signals were considered colocalized if their centers were within 3 pixels (∼0.39 µm).
One-way ANOVA multiple comparison tests with Bonferroni correction, and correlation analysis were performed using GraphPad Prism version 7 for Windows (GraphPad Software, La Jolla, California, USA, www.graphpad.com).
RNA structure modeling and sequence alignment.
RNA secondary structure prediction of HIV-2 wild-type (ROD12) and mutant 5′ UTR sequences (nucleotides 185 – 555) were performed with RNAStructure V6.0.1 (https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html) (44) Multiple sequence alignment of 41 full-length HIV-2 sequences (Group A & B) from the HIV Los Alamos Sequence Database (http://www.hiv.lanl.gov) was performed with the Clone Manager program (Sci-Ed Software).
Supplementary Material
HIGHLIGHTS.
Mutating specific guanosines in the HIV-2 leader RNA abrogated genome packaging
Five guanosine-containing sites were identified with varied importance to packaging
There are functional overlaps and synergistic effects among sites with guanosines
We propose that synergistic Gag:RNA interactions mediate HIV-2 RNA packaging
ACKNOWLEDGEMENTS
We thank Eric Freed and Alan Rein for helpful discussions. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research, by NIH Intramural AIDS Targeted Antiviral Program grant funding (to W.-S. H. and to V.K.P.), and by the Innovation Fund, Office of AIDS Research, NIH (to W.-S. H. and to V.K.P.).
Footnotes
DECLARATION OF INTERESTS: None.
Conceptualization: CNU, AD, OAN, VKP, and WSH
Methodology: CNU, AD, OAN, and JC
Formal analysis: CNU and AD
Resources: CNU, AD, OAN, SAR, YL, JC, ST, and VKP
Writing-original draft: CNU, AD, and WSH
Writing-Review and editing: CNU, AD, OAN, SAR, YL, JC, ST, VKP, and WSH
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO and Hahn BH (2001) The origins of acquired immune deficiency syndrome viruses: where and when? Philosophical transactions of the Royal Society of London, 356, 867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp PM and Hahn BH (2011) Origins of HIV and the AIDS pandemic. Cold Spring Harbor perspectives in medicine, 1, a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peeters M, Jung M and Ayouba A (2013) The origin and molecular epidemiology of HIV. Expert Rev Anti Infect Ther, 11, 885–896. [DOI] [PubMed] [Google Scholar]
- 4.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature, 397, 436–441. [DOI] [PubMed] [Google Scholar]
- 5.Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y et al. (2006) Human immunodeficiency viruses: SIV infection in wild gorillas. Nature, 444, 164. [DOI] [PubMed] [Google Scholar]
- 6.Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Greene BM, Sharp PM, Shaw GM and Hahn BH (1992) Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature, 358, 495–499. [DOI] [PubMed] [Google Scholar]
- 7.Nyamweya S, Hegedus A, Jaye A, Rowland-Jones S, Flanagan KL and Macallan DC (2013) Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev Med Virol, 23, 221–240. [DOI] [PubMed] [Google Scholar]
- 8.Tchounga B, Ekouevi DK, Balestre E and Dabis F (2016) Mortality and survival patterns of people living with HIV-2. Curr Opin HIV AIDS, 11, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motomura K, Chen J and Hu WS (2008) Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. Journal of virology, 82, 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed EO and Martin MA (2013) In Knipe DM and Howley PM (eds.), Fields Virology. 6th ed. Lippincott, Williams, & Wilkins, Philadelphia, PA, USA, Vol. II, pp. 1502 – 1560. [Google Scholar]
- 11.Ni N, Nikolaitchik OA, Dilley KA, Chen J, Galli A, Fu W, Prasad VV, Ptak RG, Pathak VK and Hu WS (2011) Mechanisms of Human Immunodeficiency Virus Type 2 RNA Packaging: Efficient trans Packaging and Selection of RNA Copackaging Partners. Journal of virology, 85, 7603–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Souza V and Summers MF (2005) How retroviruses select their genomes. Nature reviews, 3, 643–655. [DOI] [PubMed] [Google Scholar]
- 13.Berkhout B and Schoneveld I (1993) Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic acids research, 21, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkhout B (1996) Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol, 54, 1–34. [DOI] [PubMed] [Google Scholar]
- 15.Purzycka KJ, Pachulska-Wieczorek K and Adamiak RW (2011) The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic acids research, 39, 7234–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirac AM, Huthoff H, Kjems J and Berkhout B (2002) Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic acids research, 30, 2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanchy JM, Rentz CA, Ivanovitch JD and Lodmell JS (2003) Elements located upstream and downstream of the major splice donor site influence the ability of HIV-2 leader RNA to dimerize in vitro. Biochemistry, 42, 2634–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poeschla E, Gilbert J, Li X, Huang S, Ho A and Wong-Staal F (1998) Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. Journal of virology, 72, 6527–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCann EM and Lever AM (1997) Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. Journal of virology, 71, 4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin SD, Allen JF and Lever AM (2001) The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. Journal of virology, 75, 12058–12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arya SK, Zamani M and Kundra P (1998) Human immunodeficiency virus type 2 lentivirus vectors for gene transfer: expression and potential for helper virus-free packaging. Human gene therapy, 9, 1371–1380. [DOI] [PubMed] [Google Scholar]
- 22.Lanchy JM, Ivanovitch JD and Lodmell JS (2003) A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA (New York, N.Y, 9, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damgaard CK, Dyhr-Mikkelsen H and Kjems J (1998) Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and −2 untranslated leader regions. Nucleic acids research, 26, 3667–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC and Weeks KM (2008) High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS biology, 6, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abd El-Wahab EW, Smyth RP, Mailler E, Bernacchi S, Vivet-Boudou V, Hijnen M, Jossinet F, Mak J, Paillart JC and Marquet R (2014) Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat Commun, 5, 4304. [DOI] [PubMed] [Google Scholar]
- 26.Bernacchi S, Abd El-Wahab EW, Dubois N, Hijnen M, Smyth RP, Mak J, Marquet R and Paillart JC (2017) HIV-1 Pr55Gag binds genomic and spliced RNAs with different affinity and stoichiometry. RNA Biol, 14, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J et al. (2015) Structure of the HIV-1 RNA packaging signal. Science (New York, N.Y, 348, 917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M and Bieniasz PD (2014) Global Changes in the RNA Binding Specificity of HIV-1 Gag Regulate Virion Genesis. Cell, 159, 1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolaitchik OA, Somoulay X, Rawson JMO, Yoo JA, Pathak VK and Hu WS (2020) Unpaired Guanosines in the 5’ Untranslated Region of HIV-1 RNA Act Synergistically to Mediate Genome Packaging. Journal of virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb JA, Jones CP, Parent LJ, Rouzina I and Musier-Forsyth K (2013) Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA (New York, N.Y, 19, 1078–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW Jr., Gorelick RJ, Rein A and Weeks KM (2010) Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proceedings of the National Academy of Sciences of the United States of America, 107, 19248–19253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye JF and Lever AM (1998) Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. Journal of virology, 72, 5877–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dilley KA, Ni N, Nikolaitchik OA, Chen J, Galli A and Hu WS (2011) Determining the Frequency and Mechanisms of HIV-1 and HIV-2 RNA Copackaging by Single Virion Analysis. Journal of virology, 85, 10499–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK and Hu WS (2009) High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proceedings of the National Academy of Sciences of the United States of America, 106, 13535–13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson DR, Johnson MC, Webb WW and Vogt VM (2005) Visualization of retrovirus budding with correlated light and electron microscopy. Proceedings of the National Academy of Sciences of the United States of America, 102, 15453–15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houman F, Diaz-Torres MR and Wright A (1990) Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell, 62, 11531163. [DOI] [PubMed] [Google Scholar]
- 37.Baig TT, Lanchy JM and Lodmell JS (2009) Randomization and in vivo selection reveal a GGRG motif essential for packaging human immunodeficiency virus type 2 RNA. Journal of virology, 83, 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.L’Hernault A, Greatorex JS, Crowther RA and Lever AM (2007) Dimerisation of HIV-2 genomic RNA is linked to efficient RNA packaging, normal particle maturation and viral infectivity. Retrovirology, 4, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keane SC, Van V, Frank HM, Sciandra CA, McCowin S, Santos J, Heng X and Summers MF (2016) NMR detection of intermolecular interaction sites in the dimeric 5’-leader of the HIV-1 genome. Proceedings of the National Academy of Sciences of the United States of America, 113, 13033–13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Grunwald D, Sardo L, Galli A, Plisov S, Nikolaitchik OA, Chen D, Lockett S, Larson DR, Pathak VK et al. (2014) Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proceedings of the National Academy of Sciences of the United States of America, 111, E5205–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA and Choi SY (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one, 6, e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zenklusen D, Larson DR and Singer RH (2008) Single-RNA counting reveals alternative modes of gene expression in yeast. Nature structural & molecular biology, 15, 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burdick RC, Delviks-Frankenberry KA, Chen J, Janaka SK, Sastri J, Hu WS and Pathak VK (2017) Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLoS pathogens, 13, e1006570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuter JS and Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.