Abstract
Culturing and molecular techniques were used to monitor changes in the bacterial flora of the avian gastrointestinal (GI) tract following introduction of genetically modified (GM) and unmodified probiotics. Community hybridization of amplified 16S ribosomal DNA demonstrated that the bacterial flora of the GI tract changed significantly in response to the probiotic treatments. The changes were not detected by culturing. Although both GM and non-GM strains of Enterococcus faecium NCIMB 11508 changed the bacterial flora of the chicken GI tract, they did so differently. Probing the community DNA with an Enterococcus faecalis-specific probe showed that the relative amount of E. faecalis in the total eubacterial population increased in the presence of the non-GM strain and decreased in the presence of the GM probiotic compared with the results obtained with an untreated control group.
Although probiotics play an important role in animal nutrition, their modes of action have not been determined yet. Microbial probiotics have been reported to have many beneficial effects when they are used in animal feeds; these effects include competitive exclusion of pathogens (6, 12) and improved digestion and absorption of nutrients (6, 19, 21, 23). It has been proposed that the efficiency of probiotics could be enhanced by genetic modifications that increase the enzymatic capacity of the gut through improvements in plant cell wall hydrolase production. However, there are concerns about using recombinant organisms in animals, particularly with respect to the transfer of antibiotic resistance genes and the possibility that a recombinant gene may move up through the food chain (9, 16, 20). To be of benefit to an animal, a probiotic organism must have an impact on the microbiology of the gut. To evaluate the usefulness of probiotics and to assess the risks, if any, associated with their use, it is important to understand the nature of the interactions. Culture techniques have not been entirely successful in assessing these interactions (26). Molecular methods, such as nucleic acid probe analysis, enzyme-linked immunoassays, and PCR, have also been used to detect probiotic strains and their effects, but the studies have been limited to detection of specific bacteria (2, 4, 10). Recent advances in the development of methods to study microbial ecology (1, 5, 7, 18, 26) have extended the range of techniques available for studying the impact of probiotics on microbial community structure in the gastrointestinal (GI) tract. In this study we used a variation of the community DNA hybridization procedures introduced by Lee and Fuhrman (7) to investigate changes in bacterial community complexity following introduction of genetically modified (GM) and non-GM strains of Enterococcus faecium into the diet of 1-day-old chicks. The aims of this study were to evaluate the impact of a GM probiotic on the bacterial flora of the chicken GI tract and to establish whether the observed changes were distinct from the changes observed when the parental, nonengineered strain was used.
MATERIALS AND METHODS
Probiotic organisms.
The GM probiotic (probiotic A) was constructed by transforming erythromycin-resistant (Eryr) plasmid pVACMC1 containing the Ruminococcus flavefaciens β-1,4-glucanase gene (27) into E. faecium NCIMB 11508. The parent strain, E. faecium NCIMB 11508, was used as the non-GM probiotic (probiotic B).
Impact of probiotics on the chicken GI tract.
Three groups of chickens were grown from 1 day old to 4 weeks old by using the following three treatments: (i) commercial diet containing probiotic A (rifampin-resistant [Rifr] E. faecium containing pVACMC1) at a concentration of 105 CFU/g; (ii) commercial diet containing probiotic B (Rifr E. faecium) at a concentration of 105 CFU/g; and (iii) commercial diet containing no probiotic (control).
At the beginning of the trial the chickens were randomly allocated to the three treatments, and each chicken was gavaged with 1 ml of either water (control) or water containing a probiotic organism at a concentration of 106 CFU/ml. To minimize cross-contamination, the three treatment groups were housed in separate rooms. After 28 days, the chickens were removed from their rings. The rings were then cleaned, and the shavings were replaced. The birds were then returned to their rings and fed the control diet (containing no probiotic) for an additional 7 days. At zero time and after 14, 28, 30, 33, and 35 days, six sample birds were removed from each treatment group, and the contents of the crops, duodena, and ceca were combined and used to monitor the effects of the probiotics on the microflora of the GI tract.
Bacterial culture.
Viable counts were obtained by using serial 10-fold dilutions of the gut content samples in phosphate-buffered saline that were plated in triplicate onto the appropriate selective media. The counts were determined by using the dilution that produced between 30 and 300 CFU per plate. All of the selective media were supplied by Oxoid Ltd., Basingstoke, United Kingdom, and were prepared according to the manufacturer’s instructions. All plates were incubated for 48 h as recommended by the manufacturer of the selective media, and the plates were used to determine viable counts as follows: Lactobacillus spp. were counted on MRS agar (De Man, Rogosa, Sharpe); Enterococcus spp. were counted on Slanetz-Bartley medium; coliforms were counted on cystine-lactose electrolyte-deficient medium; Salmonella spp. were counted directly on desoxycholate lactose sucrose (DCLS) agar and by using enrichment selective Rappaport-Vassiliadis broth cultures that were subcultured on brilliant green agar; Campylobacter spp. were counted on campylobacter selective media; and M17 medium containing antibiotics (50 μg of rifampin per ml and 200 μg of erythromycin per ml) was used to select for probiotic A (rifampin and erythromycin resistant) and probiotic B (rifampin resistant).
Community DNA analysis. (i) DNA extraction.
DNA was extracted from bacterial cultures by using the method of Netherwood et al. (13). DNA was extracted from gut contents by using an additional bead-beating step. Sterile glass beads (25 μg; diameter, 0.17 to 0.18 mm) were added to a guanidine thiocyanate-gut content mixture, and the preparation was shaken for 2 min at 1,600 rpm with a Mikrodismembrator U instrument (B. Braun Biotech International) before the heating step was performed.
(ii) PCR amplification of 16S ribosomal DNA (rDNA).
DNA extract (1 μl) was added to a PCR mixture containing 10 mM Tris-HCl (pH 8.8) at 25°C, 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, each deoxynucleoside triphosphate at a concentration of 500 μM, 20 pmol of primer Eub338 GC clamp (5′-CGC CCG CCG CGC CCC CGC CCC GGC CCG CCG CCC CCG CCC GCT GCC TCC CGT AGG AGT-3′), 20 pmol of primer Univ1390 (5′-ACG GGC GGT GTG TRC-3′), and 1 U of DyNAzyme II DNA polymerase (Flowgen). Amplification was carried out by using one thermal cycle consisting of 5 min at 95°C, followed by 30 cycles consisting of 1 min at 94°C, 1 min at 57°C, and 2 min at 72°C. The final cycle consisted of 72°C for 20 min. Negative controls containing no DNA and eukaryotic DNA were included in addition to a positive control containing Escherichia coli 16S ribosomal DNA.
(iii) Oligonucleotide probe hybridization.
The oligonucleotide DNA probes were radiolabelled with a synthetic oligonucleotide 5′ end labelling kit obtained from MBI Fermentas according to the manufacturer’s instructions. Hybridization was carried out by using the instructions for a Hybond-N membrane supplied by Amersham Life Sciences, Little Chalfont, United Kingdom, at 65°C for 18 h. Negative and positive controls for the probes, which consisted of DNA extracts of Enterococcus faecalis, E. faecium, Campylobacter sp., Salmonella sp., and Escherichia coli cultures, salmon sperm DNA, and no DNA, were applied to each membrane.
Group-specific probes for the following sequences were manufactured by the University of Newcastle-upon-Tyne oligonucleotide service: for E. faecalis, 5′-GAC CGC GAG GTC ATG CA-3′ (11); for bacteria, 5′-GCT GCC TCC CGT AGG AGT-3′ (1); for E. faecium, 5′-AGT CGC GAG GCT AAG CT-3′ (11); for Salmonella spp., 5′-TGC GGT TAT TAA CCA CAA CA-3′ (8); for Campylobacter spp., 5′-CGA AAA GTG TCA TCC TCC ACG CGG-3′ (3); and for E. coli, 5′-GAC CTC GGT TTA GTT CAC AGA-3′ (25).
Community hybridization.
The amplified 16S rDNA was radiolabelled by using the nick translation method with a HexaLabel DNA labelling kit as recommended by the manufacturer (MBI Fermentas, Graiciuno, Lithuania). Reciprocal hybridizations were carried out by using the method of Lee and Fuhrman (7). Hybridization results were quantified by using a radioactivity imaging system (Instant Imager; Packard).
RESULTS
Culture analysis of the avian GI tract following addition of GM and non-GM probiotics.
Both the GM and non-GM probiotics became established in the gut at an initial concentration of 105 CFU/g of gut contents. After 4 weeks of feeding with the GM probiotic, the level increased to 107 CFU/g of gut contents (14). Within 5 days after the probiotics were eliminated from the diet, neither probiotic could be recovered from gut content samples.
Analysis of the data (Table 1) revealed no statistically significant differences among the three trial groups in terms of viable counts of Lactobacillus spp., Enterococcus spp., coliforms, and Salmonella spp. when organisms were enumerated by selective culturing. Campylobacter spp. were not isolated from the members of any of the experimental groups.
TABLE 1.
Log10 mean viable counts of microorganisms in the GI tracts of chickens fed with or without the GM strain or unmodified probiotica
| No. of weeks | Log10 viable counts
|
No. of birds containing Salmonella spp.b
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Lactobacillus spp.
|
Enterococcus spp.
|
Coliforms
|
||||||||||
| Control chickens | Probiotic B-fed chickens | GMO-fed chickensc | Control chickens | Probiotic B-fed chickens | GMO-fed chickens | Control chickens | Probiotic B-fed chickens | GMO-fed chickens | Control chickens | Probiotic B-fed chickens | GMO-fed chickens | |
| 0 | 8.0 (1.5)d | 8.0 (1.5) | 8.0 (1.5) | 8.0 (1.7) | 8.0 (1.7) | 8.0 (1.7) | 7.1 (2.3) | 7.1 (2.3) | 7.1 (2.3) | 0 | 0 | 0 |
| 2 | 6.7 (2.0) | 6.0 (0.7) | 6.3 (0.5) | 7.5 (3.2) | 7.0 (2.6) | 7.5 (1.8) | 8.5 (3.7) | 9.1 (3.6) | 8.3 (3.6) | 2.0 | 0 | 0 |
| 4 | 7.7 (1.8) | 7.3 (1.7) | 8.0 (1.5) | 8.0 (2.7) | 8.0 (1.7) | 8.3 (1.6) | 9.2 (3.5) | 9.7 (3.7) | 9.0 (3.7) | 2.0 | 3.0 | 1.0 |
| 5 | 7.7 (1.5) | 7.0 (1.6) | 7.7 (1.4) | 7.7 (1.8) | 7.3 (2.1) | 7.3 (1.8) | 7.9 (3.2) | 9.0 (3.4) | 9.0 (3.6) | 1.0 | 3.0 | 0 |
Probiotics were removed from the diets after 28 days.
Number of birds containing salmonellae isolated from an enrichment culture.
GMO, GM organism, probiotic A.
The values in parentheses are standard errors.
Community DNA analysis of the avian GI tract following addition of GM and non-GM probiotics. (i) Reciprocal hybridization.
The three replicate PCR mixtures derived from the same target DNA and obtained at each specific sampling time were combined prior to the reciprocal hybridization analysis. This reduced the effect of PCR drift, the inherent difference between replicate PCR caused by slightly different reaction conditions that affect the annealing rates of probes to different DNA species.
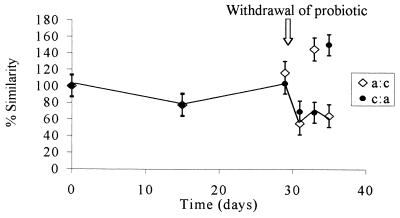
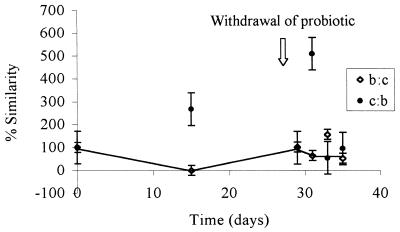
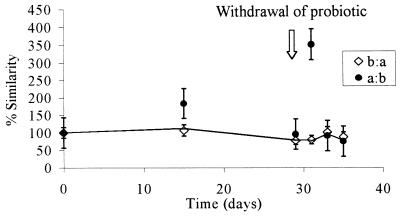
The combined PCR products obtained from each treatment group were examined by performing total community hybridization in order to estimate the fraction of common DNA in two samples obtained from the same site but subjected to different treatments or sampled at different times. DNA probes (PCR products of the GI tract DNA extracts) were radiolabelled and reciprocally hybridized to filter-bound DNA samples obtained at different times throughout the feeding trial. Levels of similarity were calculated by standardizing the values with self-hybridization values. Thus, if the probe DNA and target DNA compositions were similar, then the hybridization values were high and any changes in community structure were reflected by changes in the percentage of hybridization. rDNA from similar bacteria hybridized more strongly than rDNA from dissimilar bacteria, and thus the amount of radioactivity measured reflected the level of similarity of the bacteria in the community. Each reciprocal hybridization, in which each member of each pair of samples being compared served in turn as both a target and a probe, resulted in two values designated the observed similarity values. If the two observed similarity values were symmetrical (i.e., the level of hybridization was the same irrespective of whether the rDNA was used as the target or the probe), then the bacterial structures represented by the 16S rDNA amplicons were similar, and no change in community composition was inferred. When the values were asymmetrical, one of the samples (i.e., the probe or the target) contained a more diverse community, and thus there was a change in the community structure (7). In this case the lower of the two values was considered the true level of similarity of the communities (where the more complex community acted as the target DNA), as proposed by Lee and Fuhrman (7). Figure 1 shows the levels of similarity of the microbial communities in the control group (no probiotic) and the GM probiotic-fed chickens over the trial period. At time zero, there were no differences in the communities, and as expected, the levels of similarity were 100%. After the probiotic was removed from the diet on day 28, the reciprocal hybridization results became asymmetrical, demonstrating that a change in the diversity of the probiotic treatment group occurred. At this point the true diversity was represented by the lower of the two values; therefore, the line representing the true changes in similarity links the lower of the two points on the graph in Fig. 1. This line indicates that there was a change in the diversity of the GM-treated group compared with the diversity of the untreated control group. Similar results are shown in Fig. 2, in which changes in diversity are evident during treatment, as well as following removal of the non-GM probiotic. A comparison of the probiotic-treated groups (Fig. 3) suggested that the impact of the GM probiotic and the impact of the non-GM probiotic on the GI tract microflora were similar when they were assessed by using community DNA hybridization.
FIG. 1.
Levels of similarity of the bacterial communities in the group treated with probiotic A (GM strain) and the control (untreated) group. The two values at each sampling time are means based on three replicates of two reciprocal hybridizations. a:c, the probiotic A group was the target and the untreated group was the probe; c:a, the untreated group was the target and the probiotic A group was the probe. The true level of similarity is indicated by the line connecting the lower similarity values. The probiotic was removed from the diet after 28 days. The values are means ± standard errors of the means of results from three replicate experiments.
FIG. 2.
Levels of similarity of the bacterial communities in the group treated with probiotic B (unmodified strain) and the control (untreated) group. The two values at each sampling time are means based on three replicates of two reciprocal hybridizations. b:c, the probiotic B group was the target and the untreated group was the probe; c:b, the untreated group was the target and the probiotic B group was the probe. The true level of similarity is indicated by the line connecting the lower similarity values. The probiotic was removed from the diet after 28 days. The values are means ± standard errors of the means of results from three replicate experiments.
FIG. 3.
Levels of similarity of the bacterial communities in the group treated with probiotic A (GM strain) and the group treated with probiotic B (unmodified strain). The two values at each sampling time are means based on three replicates of two reciprocal hybridizations. a:b, the probiotic A group was the target and the probiotic B group was the probe; b:a, the probiotic B group was the target and the probiotic A group was the probe. The true level of similarity is indicated by the line connecting the lower similarity values. Probiotics were removed from the diets after 28 days. The values are means ± standard errors of the means of results from three replicate experiments.
(ii) Oligonucleotide probe analysis of 16S rDNA.
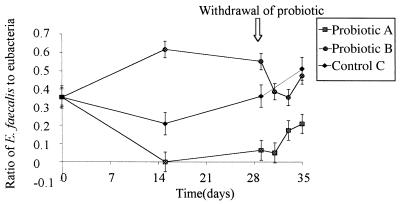
Specific probing of the bound rDNA with a probe specific for E. faecalis (which did not cross-react with E. faecium) suggested that there were significant differences between the effects of the GM and non-GM strains on the numbers of E. faecalis cells in the GI tract (Fig. 4). GM probiotic A resulted in a decrease in the relative amount of E. faecalis compared with the untreated control group, while with probiotic B the numbers (expressed relative to the binding of a bacterial probe) increased relative to the control group. No significant difference with time was observed for the hybridization results obtained with the probes for E. faecium, E. coli, Salmonella spp., and Campylobacter spp. either in individual treatment groups or between groups.
FIG. 4.
Change in the ratio of E. faecalis to bacteria in the three treatment groups. The ratio of E. faecalis to total bacteria was estimated by measuring the levels of probe hybridization to PCR-amplified regions of 16S DNA for samples treated with probiotic A (GM strain) and probiotic B (unmodified strain) and control (untreated) samples. The values are means ± standard errors of the means based on three replicates. Probiotics were removed from the diets after 28 days.
DISCUSSION
Viable counting revealed no significant differences between the groups treated with the GM and non-GM probiotics and the control group. Viable counting on selective media has been shown to recover less than 20% of bacteria (26); the remainder are viable but not culturable under the isolation conditions used (1). However, the use of molecular techniques enables the unculturable component to be studied, and here we used a modification of the community hybridization technique introduced by Lee and Fuhrman (7) to investigate changes in bacterial communities as reflected in their 16S rDNA amplicons. This is a useful modification of community analysis methods since unlike the original procedure, in which total community DNA was hybridized (7), it allows workers to analyze bacterial DNA separately from archaeal or eucaryal DNA. Furthermore, DNA extraction can be carried out more rapidly on a small scale compared with whole-community DNA hybridization analysis, and a strong hybridization signal can be produced reliably. However, there are problems with using PCR for microbial community analysis, such as errors due to variations in gene copy number, PCR drift, and primer binding bias (17, 22, 24). A recent analysis of these problems by Polz and Cavanaugh (15) indicated that gene copy number is unlikely to be a major cause of observed bias. Biases due to equipment variations, pipetting errors, and template composition can be reduced considerably by using high template concentrations, by using fewer cycles, and by mixing replicate reaction preparations. Thus, PCR error can be minimized. PCR variation may still occur, but it is likely to be reproducible, which means that comparative analyses, such as those described here, are less influenced by amplification bias since the same errors are likely to occur in both of the communities studied.
Reciprocal hybridization revealed major changes in the bacterial community structure following removal of the probiotic after 28 days. At that time, when the numbers of probiotic organisms in the gut were decreasing (14), one might have expected other bacteria to be competing for the niches previously occupied by the probiotic organisms (Fig. 1 and 2). Since major differences were not apparent when probiotic A was compared with probiotic B, the data suggest that the overall community responses to the two probiotics were similar. To resolve better the differences induced by probiotic treatment, we also used denaturing gradient gel electrophoresis to investigate the changes in 16S rDNA amplicon diversity observed during rDNA hybridization. However, with this technique, which separates DNA on a denaturing gel on the basis of sequence dissimilarities, we were not able to resolve all of the bands obtained with the consensus primers (data not shown), and the technique was not investigated further.
To determine whether the probiotics changed the composition of the gut microbial community, probes specific for E. faecium, E. faecalis, Salmonella spp., Campylobacter spp., and E. coli and a general probe for all bacteria were hybridized with the bound 16S rDNA. The results of these studies suggested that there were significant changes in the relative amounts of E. faecalis following addition of the GM probiotic and the non-GM probiotic to the diet. In the presence of the non-GM probiotic, the relative amount of E. faecalis (as assessed relative to the binding of a bacterial probe) increased compared with the control, while with the GM probiotic the relative amount of E. faecalis decreased (Fig. 4). This suggests that despite the similar responses observed at the community level, the structural changes actually involved different components of the bacterial population. Although not conclusive, the data suggest that E. faecalis and E. faecium may occupy similar niches or even have a synergistic relationship. Although this possibility is perhaps predictable due to similarities in the physiology and growth of the two organisms, it awaits confirmation by in situ hybridization. When the probiotics were removed from the diet, the gut flora shifted again. As the probiotic gradually left the GI tract, the relative amount of E. faecalis changed such that it returned to the level observed in the untreated control group. This indicates that the effects observed were reversible and dependent on the continued presence of the viable probiotic in the gut. Importantly, the data show that the probiotic does not become established in the GI tract and that it would have to be supplied continuously in the diet in order to provide any beneficial effect.
The probe for E. faecium did not reveal a significant difference in the signals obtained. This indicates either that the relative amount of E. faecium in the enterococcal component of the gut was too low to produce statistically significant differences, that the microbial composition of the gut was not changed by the probiotic treatments, or that the relatively low-level signals observed were due to a lack of access of the probe to the target site (1). The probes for Salmonella spp., Campylobacter spp., and E. coli produced very low-intensity results, which indicates that the proportions of these organisms in the microbial community of the avian GI tract are relatively low. This is reflected in the very low viable counts or the lack of viable counts for Salmonella spp. and Campylobacter spp., and although the coliform viable count was as high as 109 CFU/ml of gut contents, it appears that E. coli accounts for a minor proportion of the coliform population. Thus, the community hybridization approach followed by specific probe analysis proposed here provides a useful and cost-effective means for screening PCR products of community DNA and for investigating changes in community structure during succession.
Finally, there was no evidence of any detrimental or beneficial effect on the health of the chickens used in these studies due to the changes in the microbial flora of the gut. Pathogens were not detected in greater numbers in the chickens fed diets supplemented with GM probiotics, and we observed no significant differences in the health or growth of the three trial groups. However, the chickens were kept under optimal conditions and were not under the sorts of stresses imposed by commercial production. Thus, any extrapolation to the safety of GM probiotics in the commercial rearing of chickens awaits further investigation.
ACKNOWLEDGMENTS
This work was supported by a grant from the United Kingdom Ministry of Agriculture, Fisheries and Food as part of the Novel Foods Programme.
We thank Harry Flint, Rowett Research Institute, Aberdeen, United Kingdom, for donation of the pVACMC1 plasmid.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charteris W P, Kelly P M, Morelli L, Collins J K. Selective detection, enumeration and identification of potentially probiotic Lactobacillus and Bifidobacterium species in mixed bacterial populations. Int J Food Microbiol. 1997;35:1–27. doi: 10.1016/s0168-1605(96)01222-6. [DOI] [PubMed] [Google Scholar]
- 3.Dewhirst F E, Seymour C, Fraser G J, Paster B J, Fox J G. Phylogeny of Helicobacter isolates from bird and swine feces and description of Helicobacter pametensis sp. nov. Int J Syst Bacteriol. 1994;44:553–560. doi: 10.1099/00207713-44-3-553. [DOI] [PubMed] [Google Scholar]
- 4.Durant J A, Young C R, Nisbet D J, Stanker L H, Ricke S C. Detection and quantification of poultry probiotic bacteria in mixed culture using monoclonal antibodies in an enzyme-linked immunosorbent assay. Int J Food Microbiol. 1997;38:181–189. doi: 10.1016/s0168-1605(97)00102-5. [DOI] [PubMed] [Google Scholar]
- 5.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haschke F, Wang W, Ping G Z, Varavithya W, Podhipak A, Rochat F, LinkAmster H, Pfeifer A, DialloGinstl E, Steenhout P. Clinical trials prove the safety and efficacy of the probiotic strain Bifidobacterium Bb12 in follow-up formula and growing-up milks. Monatsschr Kinderheilkd. 1998;146:S26–S30. [Google Scholar]
- 7.Lee S, Fuhrman J A. DNA hybridization to compare species compositions of natural bacterioplankton assemblages. Appl Environ Microbiol. 1990;56:739–746. doi: 10.1128/aem.56.3.739-746.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin C K, Tsen H Y. Development and evaluation of two novel oligonucleotide probes based on 16S-ribosomal RNA sequence for the identification of Salmonella in foods. J Appl Bacteriol. 1995;78:507–520. doi: 10.1111/j.1365-2672.1995.tb03093.x. [DOI] [PubMed] [Google Scholar]
- 9.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucchini F, Kmet V, Cesena C, Coppi L, Bottazzi V, Morelli L. Specific detection of a probiotic Lactobacillus strain in faecal samples by using multiplex PCR. FEMS Microbiol Lett. 1998;158:273–278. doi: 10.1111/j.1574-6968.1998.tb12832.x. [DOI] [PubMed] [Google Scholar]
- 11.Monstein H J, Quednau M, Samuelsson A, Ahrne S, Isaksson B, Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144:1171–1179. doi: 10.1099/00221287-144-5-1171. [DOI] [PubMed] [Google Scholar]
- 12.Morishita T Y, Aye P P, Harr B S, Cobb C W, Clifford J R. Evaluation of an avian-specific probiotic to reduce the colonization and shedding of Campylobacter jejuni in broilers. Avian Dis. 1997;41:850–855. [PubMed] [Google Scholar]
- 13.Netherwood T, Binns M, Townsend H, Wood J L N, Mumford J A, Chanter N. The Clostridium perfringens enterotoxin from equine isolates; its characterization, sequence and role in foal diarrhoea. Epidemiol Infect. 1998;120:193–200. doi: 10.1017/s0950268897008534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netherwood T, Bowden R, Harrison P, O’Donnell A G, Parker D S, Gilbert H J. Gene transfer in the gastrointestinal tract. Appl Environ Microbiol. 1999;65:5139–5141. doi: 10.1128/aem.65.11.5139-5141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redenbaugh K, Hiatt W, Martineau B, Emlay D. Determination of the safety of genetically engineered crops. Am Chem Soc Symp Ser. 1995;605:72–87. [Google Scholar]
- 17.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosado A S, Duarte G F, Seldin L, VanElsas J D. Molecular microbial ecology: a minireview. Rev Microbiol. 1997;28:135–147. [Google Scholar]
- 19.Scheinbach S. Probiotics: functionality and commercial status. Biotechnol Adv. 1998;16:581–608. doi: 10.1016/s0734-9750(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 20.Schlundt J, Saadbye P, Lohmann B, Jacobsen B L, Nielsen E M. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microbiol Ecol Health Dis. 1994;7:59–69. [Google Scholar]
- 21.Sissons J W. Potential of probiotic organisms to prevent diarrhoea and promote digestion in farm animals—a review. J Sci Food Agric. 1989;49:1–13. [Google Scholar]
- 22.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomke S, Elwinger K. Growth promotants in feeding pigs and poultry. III. Alternatives to antibiotic growth promotants. Ann Zootech (Paris) 1998;47:245–271. [Google Scholar]
- 24.Wagner A, Blackstone N, Cartwright P, Dick M, Misof B, Snow P, Wagner G P, Bartels J, Murtha M, Pendleton J. Surveys of gene families using polymerase chain reaction—PCR selection and PCR drift. Syst Biol. 1994;43:250–261. [Google Scholar]
- 25.Wang R F, Cao W W, Cernigilia C E. PCR detection and quantification of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward D M, Weller R, Bateson M M. 16S ribosomal RNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 27.Whitehead T R, Flint H J. Heterologous expression of an endoglucanase gene (EndA) from the ruminal anaerobe Ruminococcus flavefaciens 17 in Streptococcus bovis and Streptococcus sanguis. FEMS Microbiol Lett. 1995;126:165–169. doi: 10.1111/j.1574-6968.1995.tb07411.x. [DOI] [PubMed] [Google Scholar]