Significance
Many gram-positive organisms have evolved an elegant solution to sense and resist antimicrobial peptides that inhibit cell-wall synthesis. These organisms express an unusual “Bce-type” adenosine triphosphate–binding cassette (ABC) transporter that recognizes complexes formed between antimicrobial peptides and lipids involved in cell-wall biosynthesis. In this work, we provide the first structural snapshots of a Bce-type ABC transporter trapped in different conformational states. Our structures and associated biochemical data provide key insights into the novel target protection mechanism that these unusual ABC transporters use to sense and resist antimicrobial peptides. The studies described herein set the stage to begin developing a comprehensive molecular understanding of the diverse interactions between antimicrobial peptides and conserved resistance machinery found across most gram-positive organisms.
Keywords: bacitracin, antimicrobial, cryo-EM
Abstract
Antimicrobial peptides are diverse molecules that include powerful medications such as bacitracin and vancomycin, as well as potent bacterial signaling molecules. Several antimicrobial peptides elicit cell death in gram-positive species by binding to lipid II cycle intermediates and inhibiting the synthesis of peptidoglycan. Many gram-positive organisms have evolved an elegant mechanism to sense and resist such antimicrobial peptides. In these organisms, a “Bce-type” adenosine triphosphate–binding cassette (ABC) transporter forms a protein complex with a two-component system, and together these components sense and provide resistance to antimicrobial peptides present at the cell surface. Conformational switching of Bce-type transporters is proposed to be the stimulus that activates the associated two-component system through a novel flux-sensing mechanism. In this work, we determined the detergent-solubilized structure of the Bce-type ABC transporter BceAB from Bacillus subtilis in two distinct conformational states using cryo–electron microscopy. Together with mass spectrometry and enzymatic data, our structures reveal the overall architecture of the Bce-type transporter family, uncover a specialized lipid-binding pocket for lipid II cycle intermediates, and reveal the conformational changes that are proposed to initiate signaling through the associated two-component system.
The cell wall of gram-positive bacteria is a formidable barrier against environmental stress and physical extremes. The outer layer of peptidoglycan surrounding the gram-positive bacterial cell is essential for survival and confers structural rigidity, mechanical strength, and protection against harsh and constantly changing environments (1). Not surprisingly, the pathway used for peptidoglycan biosynthesis (lipid II cycle) is an important target of antimicrobial compounds (2, 3). Several antimicrobial peptides such as bacitracin or vancomycin bind to intermediates of the lipid II cycle, leading to inhibition of peptidoglycan synthesis and eventual cell death. Gram-positive organisms have evolved a variety of strategies to resist attack by these antimicrobial peptides, including modification of surface charge, biofilm formation, proteolytic degradation, and expression of efflux pumps (4). In Firmicute bacteria, a conserved antimicrobial peptide sensing and detoxification machinery has evolved that consists of an adenosine triphosphate (ATP)-binding cassette (ABC) transporter interfacing with a two-component system. These complexes have come to be known as “Bce modules” based on the initial discovery of the BceAB-RS system in Bacillus subtilis (5, 6). In this system, BceA forms the ATPase domains and BceB is the membrane-spanning permease domain of the ABC transporter complex. BceS is a sensor histidine kinase that phosphorylates the response regulator BceR.
Elegant studies in B. subtilis suggest that the BceAB transporter mediates resistance to bacitracin through a target protection mechanism that involves freeing the lipid II cycle intermediate undecaprenyl-pyrophosphate (UPP) from the inhibitory grasp of bacitracin (7). However, the mechanism by which BceAB recognizes UPP–bacitracin complexes in the lipid membrane and subsequently dissociates the antimicrobial peptide from UPP remains unknown. BceAB also plays another critical role by providing the stimulus to initiate signaling through the BceRS two-component system (6). Gebhard and colleagues have demonstrated that the BceAB transporter forms a complex with the BceS histidine kinase (Fig. 1 A and B) (8). Protein coevolution analysis has revealed similar interactions between Bce-type ABC transporters and histidine kinases across Firmicute bacteria (9). The BceS sensor kinase belongs to the intramembrane sensing class of histidine kinases and lacks an extracellular ligand-binding domain. Rather than binding bacitracin directly, the BceS sensor kinase is proposed to sense the conformational cycling of the BceAB transporter through a “flux-sensing” mechanism (6, 10). Activation of BceS and subsequent phosphorylation of BceR leads to increased transcription of the genes encoding the BceAB transporter, resulting in a positive feedback loop to fine-tune the levels of BceAB transporter required to detoxify bacitracin–UPP complexes at the cell membrane (Fig. 1 A and B) (11). This regulatory mechanism allows the cell to efficiently mitigate cell wall stress while minimizing the energetic cost of producing resistance machinery.
Fig. 1.
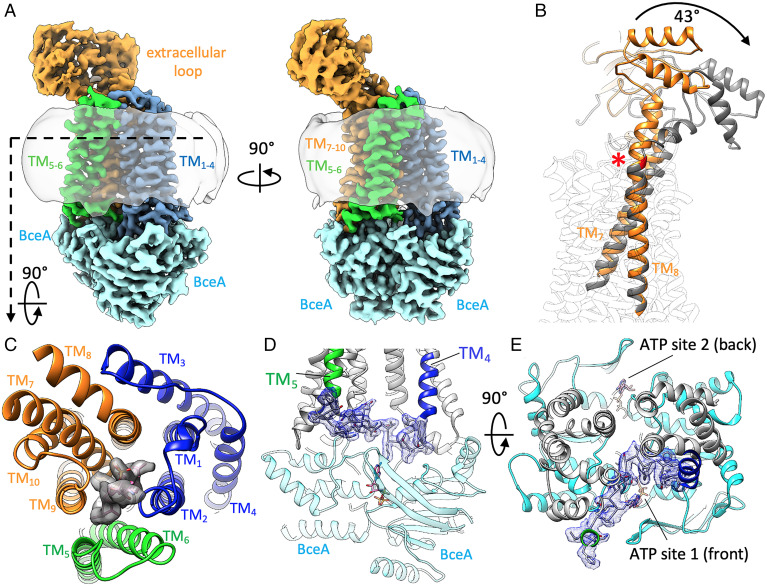
Overall structure of the BceAB complex. (A) Diagram depicting the organization and predicted membrane topology of a BceAB-RS bacitracin sensing and resistance module. BceA and BceB interact to form a complete ABC transporter complex. BceA subunits in the cytoplasm are colored cyan, and the TM helices and extracellular loop of BceB are colored blue, green, and orange. The BceAB transporter forms a complex with the BceS sensor kinase (dark gray). Conformational cycling of BceAB in response to recognition of bacitracin–UPP complexes initiates autophosphorylation of the BceS sensor kinase, which then phosphorylates (purple star with white P) the BceR response regulator (RR, light gray). Phosphorylated BceR binds to the Pbce promoter and up-regulates expression of the genes encoding the BceAB transporter. (B) Organization of BceRSAB genes in B. subtilis. BceR and BceS are constitutively expressed, whereas BceA and Bce B are controlled by the Pbce promoter that is up-regulated in response to phosphorylated BceR binding. (C) Cryo-EM structure of BceAB in a nucleotide-free conformation. BceA and BceB subunits are colored the same as depicted in (A). The detergent micelle is shown as a transparent gray outline.
Although extensive biochemical studies have established the role of Bce modules in sensing and resisting diverse antimicrobial peptides in different organisms (12–15), relatively little is known about their structure and the conformational changes that underlie their function. Revealing how the components of Bce modules work together to sense and resist antimicrobial peptides is a critical step toward developing a comprehensive understanding of how gram-positive organisms adapt to envelope stress. In this work, we determined the cryo–electron microscopy (cryo-EM) structures of the BceAB transporter from B. subtilis in two different conformational states. Our studies demonstrate that BceAB adopts a distinct architecture among the ABC transporter superfamily, reveal a critical binding pocket in the transporter for undecaprenyl lipids of the lipid II cycle, and provide insight into conformational changes that mediate target protection and activate BceS for defense against antimicrobial peptides.
Results
Overall Structure of the BceAB Transporter.
In order to express and purify the BceAB transporter for structural studies, we cloned the bceA and bceB genes from B. subtilis behind separate promoters in a pETDuet-1 vector with the addition of a 6X-His tag on the N terminus of BceA. In our initial expression and purification optimization, we found that driving expression of bceA and bceB from separate promoters, rather than as a fused operon as natively exists in B. subtilis, consistently produced more intact purified BceAB complex. The transporter complex was expressed in Escherichia coli C41(DE3), solubilized from membranes in lauryl maltose neopentyl glycol (LMNG), and purified using sequential Co3+-affinity and size-exclusion chromatography. This procedure generated highly pure and monodisperse samples amenable for structural studies (SI Appendix, Fig. S1 A and B). The structure of BceAB from B. subtilis in a nucleotide-free conformation was determined at an overall resolution of 3.8 Å using single-particle cryo-EM (Fig. 1C and SI Appendix, Figs. S2 and S3). Our cryo-EM structure confirms a previously predicted stoichiometry of the complex with two soluble BceA nucleotide-binding domains in complex with one BceB transmembrane (TM) subunit (8). Strikingly, the BceA subunits in the cytoplasmic region are arranged in a nonsymmetric fashion with one subunit significantly tilted toward the membrane plane (Figs. 1C and 2B). The asymmetric arrangement of BceA subunits is directly apparent in several side-view two-dimensional (2D) class averages of the BceAB complex (SI Appendix, Fig. S2B). The overall structure of individual BceA subunits is similar to that observed for adenosine triphosphatase (ATPase) domains in other ABC transporters such as MacB (16) and LolCDE (17, 18) with the exception of an additional C-terminal helix that faces the interface between BceA subunits (SI Appendix, Fig. S4A). In addition to other structural features described below, tilting of the BceA subunits in the cytoplasmic region makes the overall transporter complex asymmetrical in the nucleotide-free conformation.
Fig. 2.
Lipid-binding pocket and TM arrangement in BceB. (A) Zoomed-in view of the lipid-binding pocket outlined with a dotted box in (B). Individual TM helices are labeled, along with specific residue side chains lining the lipid-binding pocket. The cryo-EM map encompassing AUP is shown as a transparent gray surface. The hydrophilic headgroup of the lipid is positioned directly beneath the extracellular domain. (B) Overall atomic model of BceAB in a nucleotide-free conformation, showing an asymmetric arrangement of BceA subunits and the protrusion of the large extracellular domain above TM5–10. (C) Sliced view from the extracellular space of the TM helix arrangement observed in BceB. Binding of AUP (magenta sticks inside gray mesh) is clearly observed between TM5,6 and TM7,9.
One of the hallmarks of Bce-type transporters is a large ∼200- to 250-residue loop region between TM helices 7 and 8 (designated TM7 and TM8). The sequence divergence of this loop region has been proposed to underlie specificity for different antimicrobial peptides between different Bce-type transporters (9, 12). In our cryo-EM structure of BceAB, the extracellular loop is positioned asymmetrically over one-half of the TM region (Figs. 1C and 2B). Three-dimensional (3D) variability analysis in CryoSPARC (19) revealed intrinsic flexibility of the extracellular loop region with respect to the TM domain (Movie S1), which resulted in a lower overall resolution in this extracellular region compared to the TM and cytoplasmic regions (SI Appendix, Fig. S3B). Despite low sequence similarity to other ABC transporters of known structure, the extracellular loop in BceB adopts a fold with Sabre and Porter subdomains (SI Appendix, Fig. S4B) similar to those seen in gram-negative mechanotransmission ABC transporters MacB and LolCDE (SI Appendix, Fig. S4C).
The TM region of BceB is composed of 10 TM helices that are arranged in a unique overall configuration (Fig. 2 B and C). TM helices 1 to 4 and 7 to 10 form individual bundles that are related by two-fold pseudosymmetry, and each represents an FtsX-domain fold like that observed in type VII mechanotransmission ABC transporters (SI Appendix, Fig. S4D) (20). TM5 and TM6 interact primarily with one another and are positioned closer to the TM7–10 bundle than the TM1–4 bundle, such that the overall TM arrangement of BceB is asymmetrical (Fig. 2C). TM7 and TM8 form long extended stalk helices that support the large extracellular domain at its base (Fig. 2B). When viewed from the extracellular space, the overall TM helix arrangement of BceB is curved, with TM2, TM4, TM6, and TM7 creating a large lipid-exposed cavity that faces the hydrophobic membrane environment (Figs. 1C and 2C). Several phospholipid-like densities filling this hydrophobic curved pocket were directly observed in our cryo-EM maps (SI Appendix, Fig. S3G). Although certain features of the BceAB complex, such as FtsX-like TM folds and an extracellular domain with Sabre and Porter subdomains, are similar to those found in other transporter systems, the TM helix arrangement and highly asymmetrical arrangement of the BceAB complex are distinct among ABC transporters of known structure.
Enzymatic Analysis of BceAB.
Previous attempts to express and purify the BceAB transporter have resulted in enzyme preparations devoid of ATPase activity (8). We suspect that this lack of activity may stem from the use of dodecyl-maltopyranoside as the detergent used for solubilization and purification of the complex, as we found BceAB to be highly unstable in this detergent. However, when solubilized and purified in LMNG detergent, BceAB remained stable and retained a high level of specific ATPase activity (Fig. 3A). Interestingly, BceAB displayed an ATPase profile consistent with substrate and/or product inhibition, where increasing concentrations of ATP resulted in slightly reduced ATPase activity (Fig. 3A). This effect was consistently seen even when an ATP-regenerating assay system was used (SI Appendix, Fig. S1C), suggesting that the slight inhibition at high ATP concentrations is not due to feedback inhibition by adenosine diphosphate (ADP) produced during the hydrolysis reaction. Ruling out the possibility of feedback ADP inhibition, the kinetics of ATP hydrolysis by detergent-solubilized BceAB suggest that ATP may bind to a lower affinity inhibitory site at high concentrations.
Fig. 3.
Biochemical characterization of BceAB. (A) ATPase activity of detergent-solubilized wild-type (WT) BceAB (open blue circles) compared to the E169Q variant (red squares). Data were fit with a model of substrate inhibition (solid lines). The E169Q substitution completely abolished ATPase activity. All data points were repeated in triplicate with the same batch of protein. Error bars represent SD from triplicate measurements. (B) ATPase activity of detergent-solubilized wild-type (WT) BceAB in the presence of increasing concentrations of Zn2+-bacitracin. Concentrations of Zn2+-bacitracin added in each reaction are indicated above each respective set of data. Data at each bacitracin concentration were fit with a model of substrate inhibition (solid lines). All data points were repeated in triplicate with the same batch of protein. Error bars represent SEM from triplicate measurements. (C) Inhibition of BceAB ATPase activity in the presence of Zn2+-bacitracin, bacitracin, or Zn2+-acetate. All data points were repeated in triplicate, and error bars represent SD from triplicate measurements. Inhibition by Zn2+-bacitracin or Zn2+-acetate was fit with a model of log[inhibitor] versus normalized response in GraphPad Prism. (D) Negative mode electrospray ionization (ESI(-)) MS/MS spectrum with m/z selection of 976 in the quadrupole and a 20- to 80-V ramp in the collision cell. m/z 976.7156 is consistent with a predicted formula of C60H99NO7P (expected m/z 976.7159, −0.7 ppm mass error). Fragment ions consistent with C55H90O4P− (undecaprenyl phosphate m/z 845.6584) and C5H11NO7P− (phospho-4-amino-4-deoxy-arabinose m/z 228.0253) are clearly visible. The green dashed box represents the zoomed in view in panel E. (E) Zoomed-in view of the 340 to 710 m/z region of the spectrum shown in D demonstrating successive neutral losses of C5H8 (68.06 Da) prenyl units.
The ATPase activity of purified BceAB was potently inhibited by common ATPase inhibitors such as the substrate analog ATP? S or sodium ortho-vanadate (SI Appendix, Fig. S1D). Furthermore, substitution of E169 to glutamine in the BceA nucleotide-binding domain completely abolished ATPase activity (Fig. 3A). Interestingly, the addition of Zn2+-bacitracin also had a pronounced inhibitory effect on BceAB ATPase activity by decreasing the maximum velocity (Vmax) and increasing the Michaelis constant (Km) of the ATP hydrolysis reaction (Fig. 3B). In order to further probe the observed inhibition, we incubated BceAB with either Zn2+-bacitracin, bacitracin alone, or Zn2+-acetate. When bacitracin was supplied without the added Zn2+ ion, no inhibition of ATPase activity was observed, whereas Zn2+-acetate produced an inhibition profile similar to that observed with Zn2+-bacitracin (Fig. 3C). This result suggests that the inhibition observed upon addition of Zn2+-bacitracin results from free Zn2+ interfering with the ATPase reaction (likely by mimicking Mg2+ in the ATP-binding site) rather than a direct effect of bacitracin on BceAB. Bacitracin is commonly sold as a Zn2+ salt, and divalent cations such as zinc mediate the interaction between bacitracin and the pyrophosphate moiety of UPP (SI Appendix, Fig. S1 E and F) (21). However, bacitracin can also utilize Mg2+ as a divalent cation to support interaction with UPP (22), and we were surprised to see that Zn2+-free bacitracin had no effect on BceAB ATPase activity even when supplied in a reaction buffer that contained excess Mg2+ (Fig. 3C). The lack of an inhibitory or stimulatory effect of bacitracin on BceAB ATPase activity was highly unanticipated and provoked a closer inspection of the lipid-binding properties of purified BceAB.
Specialized Lipid-Binding Pocket.
BceAB is proposed to recognize complexes between bacitracin and the lipid II cycle intermediate UPP. To recognize such peptide–lipid complexes, it is expected that the transporter should contain a binding site that allows for concomitant recognition of UPP in the membrane plane and bacitracin in the extracellular space. In the TM region of BceB, TM5 and TM6 are located adjacent to TM7 and TM9, where together these four helices form a V-shaped pocket facing the extracellular space (Fig. 2 B and C). The extracellular domain of BceB is positioned directly above this pocket, where it is supported at the base by stalks from TM7 and TM8 (Fig. 2B). Contained within the pocket is a long lipid-like density displaying features consistent with repeating prenyl units (Fig. 2 A and B and SI Appendix, Fig. S3H). The headgroup of this lipid extends above the BceB TM helices into the aqueous region directly beneath the extracellular domain (Fig. 2B). The location and properties of this lipid-binding pocket match closely with that expected for a bona-fide UPP-binding pocket. However, the limited resolution of the cryo-EM map in the headgroup region of the bound lipid combined with the unanticipated results of Zn2+-bacitracin inhibition in ATPase assays prompted a more thorough investigation of the lipid species bound within this pocket.
In order to help ascertain the potential identity of the lipid seen in the V-shaped pocket between TM5,6 and TM7,9, we performed liquid chromatography–tandem mass spectrometry (MS/MS) analysis on the lipids that copurified with BceAB. In addition to bacterial membrane phospholipids containing phosphoethanolamine and phosphoglycerol headgroups (SI Appendix, Figs. S5A and S6), we identified a later eluting peak with m/z 978.7301 in positive ion mode and m/z 976.7136 in negative ion mode (SI Appendix, Fig. S5A). Based on the accurate mass, MS/MS analysis (Fig. 3 D and E and SI Appendix, Fig. S5 B–E), and similarity to spectra obtained for a related lipid from Francisella novicida (23), we annotated this lipid species as 4-amino-4-deoxy-? -L-arabinopyranosyl undecaprenyl phosphate (AUP). AUP is a modified undecaprenyl-phosphate lipid common in bacteria such as E. coli, where modification of lipid A with aminoarabinose is used to resist attack by polymyxin antimicrobials (24, 25).
Although copurification of BceAB with an undecaprenyl-phosphate lipid containing an alternate headgroup is likely the result of expressing a gram-positive transporter in a gram-negative host, several lines of evidence suggest that the lipid-binding pocket observed in our cryo-EM structure is the native binding location for UPP. First, several residues lining the pocket, such as G215, S219, and A301, have previously been shown through random chemical mutagenesis studies to be involved in bacitracin sensing and/or resistance by BceAB (26). Moreover, the orientation of the lipid observed in our structure would orient the pyrophosphate moiety of UPP directly beneath the extracellular domain. As bacitracin is known to bind the pyrophosphate moiety of UPP (21), this would position the UPP-bound bacitracin complex in direct vicinity for interaction with the extracellular domain. Thus, our cryo-EM maps and mass spectrometry analysis in conjunction with previous biochemical experiments suggest that the V-shaped pocket formed between TM5–7 and TM9 is the preferred binding location for UPP and other undecaprenyl-phosphate–containing lipids of the lipid II cycle.
Structure of BceAB in an ATP-Bound Conformation.
The conformational changes of BceAB elicited by ATP binding and hydrolysis are required to mediate resistance to bacitracin in B. subtilis, as well as to initiate signaling through the BceS sensor kinase (6). To understand the nature of these conformational changes, we utilized the ATPase-deficient E169Q variant of BceA to determine the cryo-EM structure of BceAB in an ATP-bound conformation at an overall resolution of ∼3.7 Å (Fig. 4A and SI Appendix, Figs. S7 and S8). Binding of ATP induces the BceA subunits to transition into a symmetrical dimer arrangement (Fig. 4A), and clear density for ATP is observed between the signature and Walker A motifs in both potential ATP-binding sites between BceA monomers (SI Appendix, Fig. S8G).
Fig. 4.
ATP binding induced conformational changes in BceAB. (A) Cryo-EM structure of BceAB (formed using E169Q BceA variant) in an ATP-bound conformation. Subunits are colored the same as depicted in previous figures. The detergent micelle is shown as a transparent gray outline. (B) Structures of BceAB in the nucleotide-free (orange cartoon) and ATP-bound (gray cartoon) conformation were aligned at the intracellular region of TM7,8. ATP binding induces helical bending at G525 (red cartoon and asterisk) and causes the extracellular domain to rotate ∼40° away from the central z axis of the transporter. (C) View from the extracellular space of the TM helices of BceAB in an ATP-bound conformation. AUP is shown in magenta sticks and gray surface density, where it is captured on all sides by the collapse of the TM helices inward in the ATP-bound conformation. (D) View of the cytosolic linker between TM4 and TM5 that becomes ordered in the ATP-bound conformation. The cryo-EM map for the linker is shown in blue mesh just above the bound ATP (magenta sticks). (E) View from the membrane of the two ATP-binding sites between the BceA monomers. The front ATP-binding site 1 is covered by the ordered TM4-TM5 linker, which substantially reduces solvent accessibility to the bound ATP compared to the back ATP-binding site 2.
In the ATP-bound conformation, the cytosolic linker between TM4 and TM5 encompassing residues 178 to 196 of BceB becomes ordered and visible in the cryo-EM map (Fig. 4 D and E). The linker spans across and interacts with the top of both BceA subunits and is in close proximity to one of the two ATP-binding sites. The linker position makes it such that the ATP-binding site in the front of the transporter is more buried and shielded from solvent than the opposite site in the rear (Fig. 4E). Thus, although the two ATP-binding sites in a BceA dimer are composed of identical binding interfaces, interaction of the TM4–5 linker with the BceA subunits in the front of the transporter complex generates asymmetry between the two ATP-binding sites in the overall complex. Although from our cryo-EM structure alone we cannot distinguish if the two observed ATPase sites are equally functional, the different solvent accessibility and asymmetry induced by the cytosolic linker between TM4 and TM5 may underlie the substrate inhibition profiles observed in our ATPase assays (Fig. 3A and SI Appendix, Fig. S1C). At high concentrations of ATP when both potential ATP-binding sites are saturated, interaction of the TM4–5 linker with the top of the BceA monomers could potentially slow the dissociation of the BceA subunits after the ATP hydrolysis reaction is complete. This in turn could slow the dissociation of ADP and/or phosphate from the more obstructed site, resulting in the substrate inhibition profile we observed. However, further work is needed to dissect the potential role of each ATP-binding site in the overall function of the BceAB complex, particularly in an in vivo context.
Dimerization of BceA subunits in response to ATP binding also induced large-scale conformational shifts in the TM and extracellular regions of BceB. Most strikingly, in the ATP-bound conformation, the extracellular domain was rotated ∼43° away from its position in the nucleotide-free structure. This rotation was primarily a result of bending in the middle of the TM8 at residue G525 (Fig. 4B). G525 was previously identified in a chemical mutagenesis screen as a residue involved in resistance to bacitracin and initiation of signaling through the BceS sensor kinase (26). Our cryo-EM structure confirmed that the flexibility and hinge-like properties of G525 allowed the extracellular domain to undergo large-scale conformational shifts in response to ATP binding. Although the G525 hinge allowed the extracellular domain to pivot atop the TM region, the internal structure of the extracellular domain itself remained stable, and the domain moved primarily as a rigid body between nucleotide-free and ATP-bound conformations.
Binding of ATP and dimerization of the BceA subunits also induced a conformational rearrangement of the TM helices in BceB (compare Figs. 2C and 4C). In the ATP-bound conformation, the TM5–10 helical bundle rotated inward toward the opposite TM1–4 bundle, closing off the large hydrophobic groove observed at the front of the complex in the nucleotide-free state (compare Figs. 1C and 4A). Although the TM5–10 helical bundle underwent rotation largely as a rigid body, the rotation axis was centered more toward TM7–10 than TM5,6. For this reason, the rotation caused significant translation of TM5,6 toward TM1–4 in the opposite half of the transporter (SI Appendix, Fig. S8E). In the ATP-bound conformation, TM2 contacted TM6 at the apex near the extracellular space (Fig. 4C), effectively sealing off the UPP-binding pocket from the lipid environment. Interestingly, clear lipid density corresponding to AUP was observed within the center of the TM helix bundle, where it was surrounded by several TM helices and shielded from the bulk lipid membrane (Fig. 4C and SI Appendix, Fig. S8 F and H). In this configuration, the aminoarabinosyl headgroup of AUP was pointed into the extracellular space, where it was almost completely solvent exposed and located far from the tilted extracellular domain (SI Appendix, Fig. S8H). Thus, the overall conformational transition of BceAB induced by binding of ATP serves to sequester the bound AUP tightly within the TM helices and move the extracellular domain far from the headgroup of the lipid.
Discussion
Bce modules are widespread components of cell envelope stress response systems in gram-positive bacteria. Despite their importance to cell survival and resistance to a variety of medically relevant antibiotics, the precise mechanism by which Bce modules sense and respond to antimicrobial peptides has remained obscure. Our cryo-EM structures begin to shed light on the architecture and mechanism of these unusual membrane protein complexes. Compared to other ABC transporters, BceAB possesses a unique overall architecture with 10 TM helices and a single large extracellular domain. However, several structural features of the transporters are reminiscent of type VII mechanotransmission ABC transporters from gram-negative bacteria. The extracellular domain with Sabre and Porter subdomains, along with FtsX-like folds in the TM helix bundles, bear similarity to ABC transporters such as MacB and LolCDE (SI Appendix, Fig. S4D) (16–18). Much like these transporters in gram-negative bacteria, our structural analysis suggests that Bce-type ABC transporters do not transport any molecules across lipid membranes. Rather, the structure and conformational changes of BceAB are consistent with a previously proposed mechanism of target protection (7) wherein the conformational cycling of the transporter frees lipid II cycle intermediates from the inhibitory grasp of antimicrobial peptides.
Our cryo-EM structures suggest a plausible overall mechanism for bacitracin recognition and resistance (Fig. 5). The structure of BceAB in a nucleotide-free conformation revealed a critical binding pocket for undecaprenyl lipid II intermediates such as UPP, with the headgroup of the bound lipid positioned directly beneath the large extracellular domain (Fig. 5, State 1). The bound lipid was located such that if bacitracin were to bind to the pyrophosphate moiety of UPP, it would be positioned to interact with the large extracellular domain of BceB (Fig. 5, State 2). While the precise points of interaction between bacitracin and BceAB have not yet been identified, the role of the extracellular domain in antimicrobial peptide recognition is supported by experiments in which the extracellular domain of Staphylococcus aureus Bce module transporter VraG was swapped with that of the closely related transporter VraE, resulting in altered ligand specificity (15). Subsequent binding of ATP between BceA subunits induces a conformational change of the entire complex such that the bound lipid is sandwiched tightly by the TM helices and remains in the plane of the membrane, while at the same time helical bending at G525 in TM8 tilts the extracellular domain to facilitate removal of bacitracin from UPP (Fig. 5, State 3). The model presented in Fig. 5 is consistent with previous cell-based biochemical studies that suggest BceAB releases bacitracin from UPP (7).
Fig. 5.
Model of bacitracin recognition and removal. Shown is a cartoon model of how BceAB recognizes bacitracin-bound UPP and removes the bacitracin from the lipid to mediate target protection. In the resting state (State 1), UPP (magenta) binds with the lipid tail between the TM5,6 and TM8–10 helical bundles and the pyrophosphate headgroup extending into the extracellular space just beneath the large extracellular loop domain. The BceA subunits are tilted in an asymmetric fashion. In State 2, bacitracin (white open donut) binds and wraps around the pyrophosphate moiety of UPP. The bound bacitracin is gripped by the extracellular domain. Subsequent binding of ATP (red square) binding induces dimerization of the BceA subunits, which collapses the TM helices around UPP in the membrane plane (State 3). Dimerization of BceA results in TM8 helical bending (black asterisk), which tilts the extracellular domain and removes bacitracin from UPP. ATP hydrolysis causes the BceA subunits to dissociate from one another and relax the complex back to the resting state.
While our structural studies begin to paint a clear picture of the target protection mechanism mediated by BceAB, more in-depth structural and biochemical work will be required to understand some key aspects of the model proposed above. In particular, the precise molecular determinants that underlie interaction of bacitracin or other antimicrobial peptides with the extracellular domain of BceB remain unclear. In our ATPase assays, bacitracin failed to elicit an effect on BceAB activity, and free Zn2+ had a pronounced inhibitory effect. The fact that bacitracin lacked an effect on BceAB ATPase activity is seemingly at odds with the proposed detoxifying activity of the complex. However, this result is most readily explained by the unanticipated presence of an aminoarabinose-modified form of undecaprenyl-phosphate in the V-shaped pocket of BceB. Replacement of the pyrophosphate of UPP with the aminoarabinosyl headgroup of the copurified lipid likely prevents proper interaction between bacitracin and the lipid target and prevents BceAB from recognizing a proper ternary complex of Zn2+–bacitracin–UPP. In order to obtain structural and functional insight into the interaction between antimicrobial peptides, lipid II cycle intermediates, and the extracellular domains of Bce-type transporters, future efforts will require altered protein expression and purification procedures to isolate complexes from more native host organisms with pools of unmodified lipid species.
Future emphasis should also be placed on understanding substrate specificity of Bce modules. Previous studies have shown that Bce modules within a single species each respond to a different suite of antimicrobial peptides. For instance, BceAB can interact with bacitracin–UPP complexes and also with similar complexes such as that formed between mersacidin and lipid II (27). How Bce-type transporters distinguish between apparently similar antimicrobial peptides is unclear. Our studies presented here provide the structural basis to begin untangling this complicated network of transporter–lipid–antimicrobial peptide interactions. Revealing the overall structure, specialized lipid-binding properties, and nucleotide-binding induced conformational changes of BceAB represents an important step forward in developing a comprehensive molecular understanding of antimicrobial peptide sensing and resistance mechanisms across gram-positive bacteria.
Materials and Methods
Protein Expression and Purification.
The overlapped genes encoding BceA (28kDa; Uniprot ID: O34697) and BceB (72kDa; Uniprot ID: O34741) were initially PCR amplified from B. subtilis genomic DNA (primers 1 and 2, SI Appendix, Table S2) and cloned between the Nde1 and Kpn1 sites of a pETDUET-1 vector. This procedure added a 6x-His tag to the N terminus of BceA. After initial expression and purification trials with this plasmid (or the same construct subcloned into a pET28a vector), we sought to separate the overlapped bceA and bceB genes and place each gene under the control of an individual promoter in petDUET-1. To construct this plasmid, the bceA gene with an N-terminal 6x-His tag (primers 3 and 4, SI Appendix, Table S2) and the bceB gene (primers 5 and 6, SI Appendix, Table S2) were separately PCR amplified from the initial pETDUET-1 expression plasmid. These two PCR fragments were then combined with a gBLOCK fragment encoding the second T7 promoter region of pETDUET-1 and assembled into a single DNA fragment (6x-His bceA – T7 – bceB) using NEB HiFi Assembly according to the manufacturer’s protocol. Following assembly, the resulting DNA fragment was digested with Nco1 and Xho1 and cloned into a fresh pETDUET-1 vector that had been digested with the same enzymes. This procedure generated an expression plasmid containing DNA encoding N-terminally 6x-HIS tagged bceA with expression driven from one T7 promoter of petDUET-1 and DNA encoding bceB driven from the second T7 promoter. This plasmid with bceA and bceB behind separate T7 promoters in pETDUET-1 was used to produce BceAB for all structural and enzymatic studies.
The expression plasmid was transformed into C41(DE3) cells, and a starter culture from a single colony was grown at 37 °C in Luria Broth (LB) medium with 100 μg/mL ampicillin. The starter culture was used to inoculate 4 L of LB medium with 100 μg/mL ampicillin and Antifoam-204 in baffled Fernbach flasks. Cultures were grown at 37 °C to an optical density (OD600) of ∼0.8 before reducing the temperature to 30 °C and inducing protein expression with 0.7 mM isopropyl-β-D-thiogalactoside. After 4 h of induction, the cultures were harvested by centrifugation, and the cell pellets were flash frozen in liquid nitrogen and stored at −80 °C.
Frozen cell pellets were thawed and resuspended in lysis buffer (25 mM Tris [pH 8], 150 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol) supplemented with 1 μg/mL pepstatin A, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 0.6 mM benzamidine, and 5,000 units of Dr. Nuclease (Syd Labs). Cells were lysed by sonication, and membranes were isolated by ultracentrifugation at ∼100,000 × g for 1 h. Isolated membranes were resuspended in lysis buffer and solubilized with 1% LMNG by stirring for 1 h at 4 °C. Insoluble material was removed by ultracentrifugation at 100,000 × g for 1 h, and the resulting supernatant was applied to Co3+-Talon resin. The resin was washed with ∼10 column volumes of buffer A (25 mM Tris [pH 8], 150 mM NaCl, 20 mM imidazole, 10% glycerol, and 0.005% LMNG) before eluting bound protein with buffer A containing 250 mM imidazole. The eluted protein was concentrated in a 100-kDa MWCO spin concentrator and injected onto a Superdex 200 Increase 10/300 GL column equilibrated in 25 mM Tris (pH 8), 150 mM NaCl, and 0.005% LMNG. Peak fractions corresponding to intact BceAB complex (peak 1, SI Appendix, Fig. S1A, elution volume of ∼11 to 13 mL) were pooled, concentrated in a 100-kDa MWCO spin concentrator, and used immediately for cryo-EM studies or flash frozen in liquid nitrogen and stored at −80 °C for use in biochemical assays.
BceAB containing the E169Q variant of BceA was generated by site-directed mutagenesis using the Q5 Mutagenesis Kit (New England Biolabs) according to the manufacturer’s protocol. The protein complex containing the BceA-E169Q variant was expressed and purified as described above. Before preparing grids for cryo-EM analysis, the protein complex was incubated with 5 mM ATP-Mg2+ for ∼30 min at room temperature.
ATPase Assay.
Colorimetric ATPase assays were modified from a previously described protocol (28) and performed in 96-well plates. To each well 2 μg of purified BceAB was added, followed by buffer containing the indicated concentration of ATP, 4 mM MgCl2 and, when appropriate, the indicated concentration of Zn2+-bacitracin, Zn2+-acetate, free bacitracin, ATPγS, or sodium-orthovanadate to a total reaction volume of 50 μL. Samples were incubated at 37 °C for 30 min before stopping the reaction by addition of 50 μL of a 12% (wt/vol) sodium dodecyl-sulfate solution. Then 100 μL of a 1:1 mixture of 12% ascorbic acid (wt/vol) and 2% ammonium molybdate (wt/vol) was added, followed by 150 μL of a solution containing 2% sodium citrate (wt/vol), 2% sodium (meta)arsenite (wt/vol), and 2% acetic acid (vol/vol). Absorbance at 850 nm was measured in a SpectraMax M5 plate reader and converted to total ATP consumed using a standard curve generated with K2HPO4 solutions. The rate of ATP consumption (μmol/min/mg) was plotted in GraphPad Prism, and curves were fit with a model of substrate inhibition {Velocity = Vmax*[ATP]/(Km + [ATP]*(1+[ATP]/Ki))}.
MS.
To extract copurified lipids from BceAB, 200 μL of gel filtration–purified BceAB (0.7 mg/mL) was mixed with 600 μL of a 50:50 (vol/vol) mixture of isopropanol and methyl tertiary-butyl ether containing 0.01% butylated hydroxytoluene. A buffer control was run in parallel by replacing the BceAB protein solution with gel filtration buffer. Samples were vortexed and centrifuged to pellet the precipitated protein, and the supernatant was transferred to LC autosampler vials. Then, 5 μL of each sample was injected onto a Waters Acquity BEH-C18 column (2.1 × 100 mm) operated with a column temperature of 55 °C and a flow rate of 0.25 mL/min using a Waters Acquity UPLC. Compounds were separated using the following gradient: initial conditions were 50% mobile phase A (10 mM ammonium formate in water) and 50% mobile phase B (isopropanol/methanol, 90:10 vol/vol containing 5 mM ammonium formate), held at 50% B until an elapsed period of 0.5 min, ramped to 100% B at 7 min and held at 100% B until 8 min, returned to 50% B at 8.01 min, and held at 50% B until 10 min. Mass spectra were obtained on a Waters Xevo G2-XS quadrupole Time-of-Flight (TOF) mass spectrometer operated in both positive and negative electrospray ionization modes using a data-independent MSe (Waters) method having alternating scans without collision energy or with a 20- to 80-V collision energy ramp. For follow-up MS/MS analyses, the m/z 978 (positive ion mode) or m/z 976 (negative ion mode) species was selected in the quadrupole, followed by fragmentation in the collision cell with a 20- to 80-V energy ramp and subsequent scanning of daughter ions in the TOF analyzer. Data were processed using Waters Masslynx software.
Cryo-EM Imaging and Data Processing.
Grids for cryo-EM imaging were prepared on a Vitrobot Mark IV by applying 2.5 μL of purified BceAB at ∼7 mg/mL to Quantifoil R2/2 200-mesh grids that had been glow-discharged for 45 seconds at 15 mA in a Pelco EasyGlow. Grids were blotted for 5 s at 4 °C, 100% humidity, and a blot-force of 1 before being plunge frozen in liquid ethane cooled by liquid nitrogen. Frozen grids were screened for ice quality and particle distribution on a Talos Arctica equipped with a Falcon-3 detector. Final data collection was performed using Leginon (29) on a Titan Krios with a K3 direct electron detector and Gatan Quantum GIF energy filter set to a 20-eV slit width. Movies were collected in counting mode with a pixel size of 0.872 Å and a total dose of 60.5 electrons/Å2.
Movies were corrected for beam-induced motion by performing patch motion correction in cryoSPARC (19), and contrast transfer function (CTF) parameters were determined by patch CTF estimation in cryoSPARC. Micrographs with fit CTF parameters worse than 6 Å were discarded, and further manual inspection was performed to remove obviously poor micrographs. Initial templates for particle picking were generated by performing blob picking followed by 2D classification in cryoSPARC, and several averages representing different views of BceAB were then used for template-based particle picking. Several rounds of 2D classification were performed to remove bad particles, and ab initio reconstruction in cryoSPARC was used to generate an initial 3D model for classification and refinement.
For the nucleotide-free conformation of wild-type BceAB, heterogeneous refinement in cryoSPARC was followed by classification with residual signal subtraction in Relion 3.0 (30) to isolate a particle population showing the highest resolution features in the TM and extracellular regions. Final nonuniform refinement and 3D variability analysis were performed in cryoSPARC. For the dataset containing the E169Q variant of BceA in BceAB bound to ATP, initial heterogeneous classification cryoSPARC revealed two distinct populations of particles corresponding to the nucleotide-free and ATP-bound conformations. Particles corresponding to the ATP-bound conformation were selected for final nonuniform refinement. Resolutions of all maps were calculated using the gold-standard Fourier shell correlation (FSC), and local resolutions were calculated in cryoSPARC.
Model Building and Refinement.
The quality of the cryo-EM map in the TM region was of sufficient quality to build all 10 TM helices of BceB de novo in COOT (31). All connecting loops between TM helices were also manually built, including the long linker between TM4 and TM5, which was disordered in the cryo-EM map of nucleotide-free BceAB but well-ordered in the ATP-bound conformation. An initial atomic model of BceA was generated using the SWISS-Model server (32) and the structure of the MacB nucleotide-binding domain (Protein Data Bank [PDB], accession No. 5LJ9) as a template. The BceA model was rigid body fit into the cryo-EM map and then manually adjusted in COOT. The C-terminal ? helix of BceA was not present in the homology model and was manually built in COOT.
To facilitate model building and refinement of the extracellular loop region of BceB (residues 325 to 513), we utilized the RoseTTAFold (33) algorithm available through the Robetta web server. The resulting model for the extracellular loop was docked into the cryo-EM map and manually adjusted in COOT. The ligand AUP was manually built using JLigand (34), and refinement restraints were generated with phenix.elbow (35). The final two prenyl units in the hydrophobic tail of AUP were disordered in the cryo-EM maps and were left unmodeled. The final complete atomic models of BceAB encompassing all residues of BceA and BceB were iteratively refined using phenix.real_space_refine (35) and manually adjusted in COOT. Model validation was performed with MolProbity (36).
Supplementary Material
Acknowledgments
We thank Dr. Sundharraman Subramanian and the Research Technology Support Facility (RTSF) Cryo-Electron Microscopy Facility at Michigan State University for assistance with cryo-EM screening and data collection. We are also grateful for the NIH Midwest Cryo-EM Consortium and Dr. Thomas Klose and Dr. Wen Jiang for providing microscope time and assistance with data collection at Purdue University. We thank members of the Michigan State University RTSF Mass Spectrometry and Metabolomics Core facility, in particular Dr. Dan Jones for assisting with MS data analysis. Finally, we are grateful to all members of the Orlando laboratory, Dr. Robert Hausinger, and Dr. Lee Kroos for careful reading of the manuscript and for providing constructive feedback.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123268119/-/DCSupplemental.
Data Availability
Atomic models and cryo-EM maps data have been deposited in PDB, accession Nos. 7TCG and 7TCH and Electron Microscopy Data Bank accession Nos. 25811 and 25812). The protein sequences and links to genetic databases describing the genes encoding BceA (accession No. O34697) and BceB (accession No. O34741) are available in Uniprot.
References
- 1.Egan A. J. F., Errington J., Vollmer W., Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 18, 446–460 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Malin J. J., de Leeuw E., Therapeutic compounds targeting lipid II for antibacterial purposes. Infect. Drug Resist. 12, 2613–2625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breukink E., de Kruijff B., Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–332 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Assoni L., et al. , Resistance mechanisms to antimicrobial peptides in gram-positive bacteria. Front. Microbiol. 11, 593215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohki R., et al. , The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49, 1135–1144 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Rietkötter E., Hoyer D., Mascher T., Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68, 768–785 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Kobras C. M., et al. , BceAB-type antibiotic resistance transporters appear to act by target protection of cell wall synthesis. Antimicrob. Agents Chemother. 64, e02241–e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dintner S., Heermann R., Fang C., Jung K., Gebhard S., A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J. Biol. Chem. 289, 27899–27910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dintner S., et al. , Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J. Bacteriol. 193, 3851–3862 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernard R., Guiseppi A., Chippaux M., Foglino M., Denizot F., Resistance to bacitracin in Bacillus subtilis: Unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189, 8636–8642 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritz G., et al. , A new way of sensing: Need-based activation of antibiotic resistance by a flux-sensing mechanism. mBio 6, e00975–e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemens R., Zaschke-Kriesche J., Khosa S., Smits S. H. J., Insight into two ABC transporter families involved in lantibiotic resistance. Front. Mol. Biosci. 4, 91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhard S., Mascher T., Antimicrobial peptide sensing and detoxification modules: Unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81, 581–587 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Falord M., Karimova G., Hiron A., Msadek T., GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 1047–1058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiron A., Falord M., Valle J., Débarbouillé M., Msadek T., Bacitracin and nisin resistance in Staphylococcus aureus: A novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81, 602–622 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Crow A., Greene N. P., Kaplan E., Koronakis V., Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. U.S.A. 114, 12572–12577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang X., et al. , Structural basis for bacterial lipoprotein relocation by the transporter LolCDE. Nat. Struct. Mol. Biol. 28, 347–355 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Sharma S., et al. , Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nat. Commun. 12, 4687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Thomas C., et al. , Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett. 594, 3767–3775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Economou N. J., Cocklin S., Loll P. J., High-resolution crystal structure reveals molecular details of target recognition by bacitracin. Proc. Natl. Acad. Sci. U.S.A. 110, 14207–14212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stone K. J., Strominger J. L., Mechanism of action of bacitracin: Complexation with metal ion and C 55 -isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U.S.A. 68, 3223–3227 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Ribeiro A. A., Guan Z., Raetz C. R. H., Identification of undecaprenyl phosphate-β-D-galactosamine in Francisella novicida and its function in lipid A modification. Biochemistry 48, 1162–1172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breazeale S. D., Ribeiro A. A., Raetz C. R. H., Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-L-arabinose. J. Biol. Chem. 277, 2886–2896 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Breazeale S. D., Ribeiro A. A., McClerren A. L., Raetz C. R. H., A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 280, 14154–14167 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Kallenberg F., Dintner S., Schmitz R., Gebhard S., Identification of regions important for resistance and signalling within the antimicrobial peptide transporter BceAB of Bacillus subtilis. J. Bacteriol. 195, 3287–3297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staroń A., Finkeisen D. E., Mascher T., Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55, 515–525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doerrler W. T., Raetz C. R. H., ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Suloway C., et al. , Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek M., et al. , Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebedev A. A., et al. , JLigand: A graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 68, 431–440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis I. W., et al. , MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375-83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic models and cryo-EM maps data have been deposited in PDB, accession Nos. 7TCG and 7TCH and Electron Microscopy Data Bank accession Nos. 25811 and 25812). The protein sequences and links to genetic databases describing the genes encoding BceA (accession No. O34697) and BceB (accession No. O34741) are available in Uniprot.